Abstract
PRIMPOL (primase-polymerase) is a recently discovered DNA primase-polymerase involved in DNA damage tolerance and replication stress response in eukaryotic cells. However, the detailed mechanism of the PRIMPOL response to replication stress remains elusive. Here, we demonstrate that replication-related factors, including replication protein A (RPA), regulate the accumulation of PRIMPOL in subnuclear foci in response to replication stress induced by replication inhibitors. Moreover, PRIMPOL works at G-quadruplexes (G4s) in human cells to resolve the replication stress induced by G4s. The formation of PRIMPOL foci persists throughout the cell cycle. We further demonstrate that PRIMPOL competes with RAD51 to resolve G4-induced replication stress. In conclusion, our results provide novel insight into the mechanism of PRIMPOL in G4s to resolve replication stress and competition between PRIMPOL (repriming)- and RAD51 (fork reversal)-mediated pathways, which indicates a new strategy to improve the tumor response to DNA-damaging chemotherapy by targeting the PRIMPOL pathway.
Keywords: G-quadruplex, PRIMPOL, RAD51, replication stress, RPA
Introduction
Accurate replication of genomic DNA is critical to maintaining chromosomal integrity. The replication machinery encounters numerous barriers, including DNA lesions, protein-DNA complexes, RNA-DNA hybrids, DNA secondary structures, and fragile sites, which can induce replication stress and stall replication progression [ 1, 2] . For instance, double-strand breaks induced by treatment with chemotherapeutic or DNA-damaging agents lead to genomic instability. Single-strand breaks caused by nucleotide depletion, ultraviolet (UV) irradiation or DNA secondary structures can block the replication fork and impede DNA replication [ 3, 4] . To ensure replication, these impediments must be overcome or bypassed. Studies on the effects of genotoxic agents on DNA replication are becoming increasingly important in cancer drug therapy.
Cells initiate replication stress responses, including protein-protein interactions and reorganization into subnuclear foci, to complete DNA replication and ultimately maintain genome stability. These foci represent the accumulation of proteins that respond to DNA damage repair. Numerous DNA damage response proteins relocalize into subnuclear foci upon genotoxic drug treatment [ 5, 6] . One example of this phenomenon is replication protein A (RPA), the most abundant single-stranded DNA-binding complex in human cells, which relocalizes to form foci when replication pressure is present [6]. Human RPA consists of three subunits, including RPA1 (70 kDa), RPA2 (32 kDa), and RPA3 (14 kDa), which are essential during DNA replication [7]. RPA recruits multiple binding partners for various roles, including initiating replication with MCM proteins [ 6, 8] , promoting replication fork reversal with RAD51, and resolving DNA secondary structures with BLM/WRN helicases [ 9, 10] . However, more investigation is required to explore whether RPA has other binding partners to facilitate the unfolding of G-quadruplexes (G4s).
DNA secondary structure G4s are four-stranded structures prone to form in G-rich sequences throughout the genome [11]. G4s are thought to play a role in regulating genomic DNA replication, such as inducing replication stress by blocking the replication fork and impeding DNA replication [12]. Moreover, G4s can be visualized in human cell lines by using the specific antibody BG4 and stabilized by G4 ligands, including pyridostatin (PDS) and tetra-(1-methylpyridine-4-yl)-porphyrin (TMPyP4), which can also be used to induce the formation of G4s [ 13, 14] . There is evidence that some DNA polymerases and helicases function in G4 replication, such that G4s can be resolved by BLM or WRN helicases and bypassed by REV1 to promote DNA synthesis [ 15, 16] .
PRIMPOL (primase-polymerase) was recently found to be a member of the archaeo-eukaryotic primase (AEP) superfamily enzyme containing both polymerase and primase activities [ 17, 18] . Several studies have demonstrated that PRIMPOL plays an essential role in the replication stress response in eukaryotic cells through its interaction with RPA [ 17‒ 21] . The accumulation of PRIMPOL on chromatin initiates DNA repriming downstream of the lesion or stalled replication forks following hydroxyurea (HU) or ultraviolet (UV) treatment [20]. Furthermore, the loss of PRIMPOL results in delayed recovery upon UV irradiation in human cells [22]. In addition, PRIMPOL has also been reported to play an indispensable role in bypassing G4s by repriming DNA synthesis downstream of G4s in vertebrate cells [ 23, 24] . However, further investigation is needed to reveal the detailed function of PRIMPOL in resolving G4s in human cells.
Recent studies have established that replication stress can be overcome by replication-coupled DNA repair pathways, including translesion synthesis (TLS), RAD51-mediated fork reversal, and repriming by DNA polymerases such as PRIMPOL [ 25, 26] . Replication fork reversal mediated by RAD51, SMARCAL1 or HLTF can handle replication stress by facilitating fork remodelling [ 25, 27] . When replication fork reversal is impaired due to deletion of related protein factors, stalled forks can be rescued by PRIMPOL repriming alternatively [ 2, 27, 28] . PRIMPOL-mediated repriming has been reported to antagonize transitional fork reversal in BRCA-deficient cells encountering replication stress [28].
In the present study, we demonstrated that RPA regulates PRIMPOL accumulation in subnuclear foci to respond to DNA damage repair. Additionally, PRIMPOL plays an essential role in responding to G4-induced replication stress. Furthermore, the formation of PRIMPOL foci persists throughout the cell cycle. Afterwards, we observed that PRIMPOL might compete with RAD51 to localize at DNA damage sites to resolve G4s-induced replication stress. Altogether, our study provides novel insights into the mechanism of PRIMPOL in replication stress responses.
Materials and Methods
Cell culture
U2OS and HEK 293T cells purchased from the American Type Culture Collection (ATCC; Manassas, USA) were grown in DMEM (Corning, New York, USA). HeLa cells purchased from ATCC were grown in 1640 medium (Corning). All media were supplemented with 10% fetal bovine serum (Lonsera, Canelones, Uruguay) and 1% penicillin-streptomycin (HyClone, Logan, USA, and cells were cultured at 37°C with 5% CO 2. Cells were treated with 2 mM hydroxyurea (HU; Sigma-Aldrich, Darmstadt, Germany), 10 μM pyridostatin (PDS; Sigma-Aldrich), or 50 μM TMPyP4 (Sigma-Aldrich) for 24 h.
Plasmids and transfection
The shRNA constructs against PRIMPOL were generated using the pLKO.1 vector. PRIMPOL cDNA was cloned into the pLenti-3FLAG vector. Transfections of HEK 293T cells were performed using polyethyleneimine (PEI; Polysciences Madison, Wisconsin, USA) in OPTI-MEM medium (Gibco, Carlsbad, USA). Viral particles were produced by HEK 293T cells in 10-cm dishes transfected with 7.5 μg of pSPAX2 and 2.5 μg of pMD2.G packaging plasmids, together with 10 μg pLKO.1-shPRIMPOL or pLenti-3FLAG-PRIMPOL vectors. Supernatants carrying viral particles were harvested for viral infection, and PRIMPOL-knockdown and PRIMPOL-overexpressing cell lines were constructed. Cells were transfected with siRNAs (GenePharma, Shanghai, China) using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, USA). The sequences of shRNA and siRNA used are as follows: shScramble: 5′-CCTAAGGTTAAGTCGCCCTCG-3′; shPRIMPOL-1: 5′-GTCAGGTTCTCAGATACTTTA-3′; shPRIMPOL-2: 5′-CCCATAAGAGTAATAATAT-3′; siNC (negative control): 5′-UUCUCCGAACGUGUCACGUTT-3′; siRPA1: 5′-AACACUCUAUCCUCUUUCAUG-3′; and siRAD51: 5′-GAGCUUGACAAACUACUUCdTdT-3′.
Antibodies
The antibodies used are as follows: anti-FLAG-tag (Cell Signaling Technology, Danvers, USA), anti-RPA2 (Abcam, Cambridge, UK), anti-β-tubulin (Immunoway, Plano, USA), anti-PRIMPOL (Abcam), anti-RAD51 (Abcam), goat-anti-mouse Alexa Fluor 488 (Thermo Scientific, Waltham, USA), goat-anti-mouse Alexa Fluor 555 (Thermo Scientific), goat-anti-rabbit Alexa Fluor 488 (Thermo Scientific), and goat-anti-rabbit Alexa Fluor 555 (Thermo Scientific).
Western blot analysis
Cell extracts were harvested and separated by SDS-PAGE, and then transferred to PVDF membranes (Millipore, Billerica, USA). The membranes were blocked with 5% non-fat milk for 1 h at room temperature, washed, and incubated with specific antibodies overnight at 4°C. The membranes were washed again and incubated with the corresponding HRP-conjugated secondary antibodies (Immunoway). Finally, protein bands were detected using a chemiluminescence system (Millipore). The primary antibodies used are as follows: anti-FLAG-tag (Cell Signaling Technology), anti-RPA2 (Abcam), anti-β-tubulin (Immunoway), anti-PRIMPOL (Abcam), and anti-RAD51 (Abcam).
Co-immunopercipitation (Co-IP) and mass spectrometry
The pLenti-3FLAG or pLenti-3FLAG-PRIMPOL vector was transfected into HEK 293T cells using polyethyleneimine (PEI). Cells were lysed with NP-40 buffer containing 50 mM Tris, 150 mM NaCl, 1% NP-40, and protease inhibitor. After being bound by anti-FLAG beads (Sigma-Aldrich) at 4°C overnight, proteins were eluted by FLAG peptide (0.2 mg/mL; Sigma-Aldrich) and resolved by SDS-PAGE. Protein bands were detected by western blot analysis. For silver staining and mass spectrometry, eluted proteins were resolved by SDS-PAGE and subsequently analyzed using the Pierce Silver Stain kit (Thermo Scientific) according to manufacturer’s protocol. Specific bands were excised for mass spectrometry sequencing and data analysis [29].
Co-immunofluorescence (Co-IF) assay
Cells were grown on coverslips, fixed in 2% paraformaldehyde for 20 min, permeabilized with 0.5% Triton X-100 for 20 min, and blocked in PBS buffer containing 0.1% gelatin and 0.1% bovine serum albumin for 1 h. Anti-PRIMPOL, RAD51 and RPA antibodies were diluted at 1:200 and incubated with the slides for 1 h. After being washed, the slides were incubated with Alexa Fluor secondary antibodies (1:2000; Thermo Scientific) for 30 min, dehydrated using an ethanol series, and stained with DAPI (Life Technologies, New York, USA). A fluorescence microscope (Nikon, Tokyo, Japan) was used to detect fluorescence. For G4s detection, cells on coverslips were treated with extraction buffer (1 M HEPES, 1 M NaCl, 1 M MgCl 2, 1.5 M sucrose, and 0.5% NP-40) for 15 min at room temperature before being blocked. For G4s staining, the anti-BG4 antibody (plasmid) was obtained from Dr Shankar Balasubramanian’s lab (University of Cambridge, Cambridge, UK) and diluted at 1:1000 and incubated with the slides, followed by incubation with an anti-FLAG-tag antibody diluted at 1:500. Then, coverslips were prepared as above.
Cell cycle synchronization and analysis
Cells were synchronized to G1 phase using a double-thymidine blocking assay. Cells were first treated with 2 mM thymidine (Sigma-Aldrich) for 21 h, washed and released for 11 h, and then treated again with 2 mM thymidine for 20 h. Cells were synchronized to S phase using a double-thymidine blocking method. Cells were first treated with 2 mM thymidine for 21 h, washed and released for 8 h, treated again with 2 mM thymidine for 16 h and then released for 4 h. Cells were synchronized to G2 phase using thymidine and CDK1i (RO3306). Cells were first treated with 2 mM thymidine for 21 h, washed and released for 8 h, and then treated with 10 μM RO3306 (MCE, Princeton, USA) for 21 h. For EdU staining, cells were pulsed with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) for 2 h, harvested and washed with PBS. Then, the cells were fixed with ice-cold 70% ethanol and stained with 100 μg/mL propidium iodide (PI; Beyotime Biotechnology, Shanghai, China). Cell cycle distribution was determined with a FACS Calibur flow cytometer (BD, Monmouth Junction, USA).
EdU and PRIMPOL costaining
Cells were pulsed with 10 μM EdU for 30 min and stained using the Click-it Alexa Fluor 555 EdU Imaging kit (Thermo Scientific). After EdU staining, slides were blocked and incubated with anti-PRIMPOL antibody as described in the co-immunofluorescence assay.
Cell counting kit-8 assay
Cell viability was assessed using a cell counting kit-8 (Biosharp, Hefei, China). Cells were seeded in 96-well plates (1000 HeLa cells/well or 500 U2OS cells/well) and treated with PDS (10 μM) at 37°C for 1, 2, 3, 4 and 5 days. Fresh culture media containing 10% CCK-8 was added to each well and incubated at 37°C for 1 h. The absorbance was measured at 450 nm with a microplate reader (Bio-Rad, Hercules, USA).
Statistical analysis
Data were obtained from at least three independent experiments and shown as the mean±SEM. P values were determined using a two-tailed test. Data analysis was performed using GraphPad. Statistical significance was set at P<0.05.
Results
PRIMPOL interacts with replication-related factors in response to replication stress
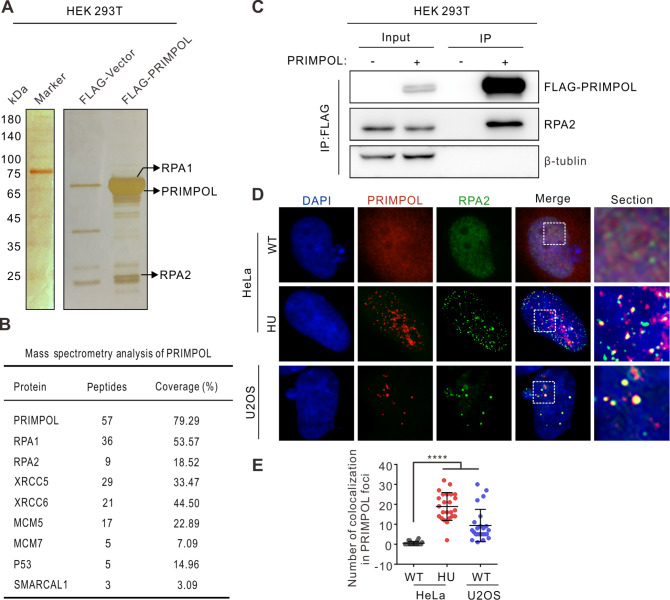
To investigate the mechanism of the PRIMPOL response to replication stress, proteins interacting with PRIMPOL were detected by mass spectrometry in HEK 293T cells overexpressing FLAG-PRIMPOL. The results revealed that numerous proteins were associated with PRIMPOL in vivo, including the single-stranded DNA binding protein RPA, DNA repair proteins XRCC5 and XRCC6, chromosome maintenance proteins MCM5 and MCM7, DNA damage response protein P53, and SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 SMARCAL1 ( Figure 1A,B). Previous studies reported that PRIMPOL DNA synthesis could be stimulated by the mtDNA replicative helicase Twinkle in vitro [30], reminding us that MCM and SMARCAL1 may play a similar role in facilitating the process of PRIMPOL.
Figure 1 .
PRIMPOL interacts with replication-related factors in response to replication stress
(A) Silver staining of HEK 293T cells transiently transfected with FLAG-vector or FLAG-PRIMPOL. (B) The proteins associated with PRIMPOL were identified by mass spectrometry, as indicated. (C) Co-IP assay indicated the interaction between PRIMPOL and RPA2. (D) Immunofluorescence (IF) assay was carried out using anti-RPA2 (green) and anti-PRIMPOL (red) antibodies in HeLa and U2OS cells. HeLa cells were treated with HU (2 mM) for 24 h. DAPI (blue). The colocalized signals are shown in enlarged images. HU, hydroxyurea. (E) Quantification of the number of colocalizations in PRIMPOL foci. **** P<0.0001.
Nuclear reorganization induces protein relocalization into discrete subnuclear foci, which is a hallmark of the cellular response to genotoxic drug-induced replication stress [ 5, 6] . Here, we blocked genomic DNA synthesis in three different cell lines, HeLa, HEK 293T and U2OS, with hydroxyurea (HU) and ultraviolet light (UV-C), and then determined the subcellular localization of PRIMPOL. A significant increase was observed in PRIMPOL foci after replication stress in all three cell types ( Supplementary Figure S1A,B), suggesting that PRIMPOL plays an essential role in the replication stress response. In addition, RPA was reported to be a critical factor in DNA replication and replication stress responses. Upon replication stress, RPA gains negative charge through phosphorylation and is focalized to form subnuclear foci [6].
Here, our results showed that PRIMPOL could interact with RPA, as previously reported ( Figure 1C) [ 20, 21, 31] . Additionally, our data revealed that PRIMPOL and RPA could focalize together after induction of replication stress in HeLa cells ( Figure 1D,E). Conversely, PRIMPOL formed distinct nuclear foci with RPA in U2OS cells ( Figure 1D,E) without genotoxic agent treatment, indicating that U2OS cells may have suffered from more severe replication stress. Collectively, our results indicated that PRIMPOL might interact with replication-related factors, including RPA, MCM proteins and SMARCAL1, in response to replication stress induced by replication inhibitors.
PRIMPOL is required to resolve G4 structure-induced replication stress
G4s can aggravate replication stress by inducing replication fork stalling. Since previous studies have reported that PRIMPOL could bind G4s in vitro and restart DNA synthesis downstream of G4s in vertebrate cells [ 23, 24] , we wondered if PRIMPOL could contribute to resolving G4-induced replication fork stalling. First, the G4 stabilizers TMPyP4 and PDS were used to cause more G4-induced replication fork stalling [13]. We verified that the formation of G4s was significantly increased in HeLa, HEK 293T, and U2OS cells treated with TMPyP4 or PDS compared to control cells ( Supplementary Figure S2A and Figure 2A). To address whether PRIMPOL binds to G4s in cells, we performed coimmunofluorescence assay to detect the colocalization between PRIMPOL and G4s using the PRIMPOL and G4 structure-specific antibody BG4. As shown in Figure 2B, colocalization was observed between PRIMPOL and G4 in human cells. Although only 20% of PRIMPOL was localized to G4 in U2OS cells ( Supplementary Figure S2B), the colocalization between PRIMPOL and G4 was significantly increased in HeLa and U2OS cells upon G4 formation by treatment with the G4 stabilizer PDS, compared to control cells ( Figure 2C). The percentage of nuclei with PRIMPOL foci and the number of PRIMPOL foci per cell were significantly increased in HeLa, HEK 293T, and U2OS cells upon G4 formation by TMPyP4 or PDS treatment compared to control cells ( Figure 2D‒F and Supplementary Figure S2C,D). Our results suggested that PRIMPOL plays roles in G4-dependent and G4-independent replication stress.
Figure 2 .
PRIMPOL is required to resolve G4 structure-induced replication stress
(A) Quantification of the number of G4 foci in HeLa, HEK 293T, and U2OS cells in Supplementary Figure S2A. The cells were treated with TMPyP4 (50 μM) or PDS (10 μM) for 24 h. (B) Representative images of PRIMPOL (red) and G4 (green) colocalization in HeLa and U2OS cells. White arrows indicate colocalizations. The cells were treated with PDS (10 μM) for 24 h. (C) Quantification of the colocalization of PRIMPOL and G4 in control and PDS-treated cells. (D) Representative IF showing the nuclear foci formation of PRIMPOL in HeLa, HEK 293T, and U2OS cells. The cells were treated with TMPyP4 (50 μM) or PDS (10 μM) for 24 h. DAPI (blue); PRIMPOL (green). (E,F) Quantification of the percentage of nuclei with PRIMPOL foci and the number of PRIMPOL foci per cell. Approximately 200 cells were analysed. (G) The expression of PRIMPOL is shown in the PRIMPOL-knockdown clones of HeLa and U2OS cells. (H) Representative images of G4 foci in control (scramble) and PRIMPOL-knockdown HeLa and U2OS cells. DAPI (blue); G4 (red). (I) Quantification of the number of G4 foci per cell. (J,K) Approximately 1000 HeLa cells and 500 U2OS cells were seeded. Cell viability was assessed for 5 days by CCK8 assays. * P<0.05, ** P<0.001, *** P<0.001, **** P<0.0001.
Moreover, to further investigate whether PRIMPOL functions at G4s in cells, two stable PRIMPOL-knockdown clones were made by infecting HeLa and U2OS cells with shRNA-encoding lentivirus to PRIMPOL. The level of depletion was measured by western blot analysis ( Figure 2G). Our results revealed that the number of G4s was significantly increased in PRIMPOL-knockdown cells compared to that in the nontargeting control HeLa and U2OS cells ( Figure 2H,I), suggesting that PRIMPOL responds to G4-induced replication stress. Next, cell growth was detected by the CCK-8 assay to test whether PRIMPOL depletion affects cell sensitivity to G4s-inducing reagent. We observed that the cell viability was decreased after HeLa and U2OS cells were treated with PDS ( Figure 2J,K). Moreover, cell growth was further inhibited when PRIMPOL was depleted in HeLa and U2OS cells treated with PDS ( Figure 2J,K), indicating the essential role of PRIMPOL in resolving G4-induced replication stress. Our results showed an approximately 2- fold increase in the number of G4 foci formed in U2OS cells compared to that in HeLa cells ( Figure 2A,H,I). Consistent with the data in Figure 1D and Supplementary Figure S1, we observed that the percentage of nuclei with PRIMPOL foci in U2OS WT cells was much higher than that in untreated HeLa or HEK 293T WT cells ( Figure 2D,E). We suppose that the increased percentage of U2OS cells with PRIMPOL foci may contribute to resolving G4s. Overall, the current data supported the hypothesis that PRIMPOL works at G4s in human cells in response to G4-induced replication stress.
The formation of PRIMPOL foci persists throughout the cell cycle
To investigate the PRIMPOL functional foci formation during different cell cycles, U2OS cells were synchronized to the G1, S, and G2 phases with thymidine block and CDK1 inhibitor treatment as described ( Supplementary Figure S3). The cell cycle profiles are presented in Figure 3A. EdU assay was used to assess the replicative progression and identify cells in the S phase [32]. Immunostaining was used to elucidate the formation of PRIMPOL foci in different cell cycle phases ( Figure 3B). The results showed no statistically significant difference in the number of PRIMPOL foci per cell among G1, G2, and S phases ( Figure 3C), suggesting that PRIMPOL persisted throughout the cell cycle. However, the number of PRIMPOL-EdU colocalized foci was greater in S-phase cells than in G2-phase cells ( Figure 3D). Moreover, approximately 30% of the S-phase cells containing PRIMPOL foci exhibited ≥5 PRIMPOL-EdU colocalized foci, which is much higher than G2-phase cells (10%) ( Figure 3E). Together, the above data indicated that although PRIMPOL foci existd throughout the cell cycle, they mainly function in S-phase replication and G2-phase homology-directed DNA synthesis, which may be used to respond to G4-induced replication stress [ 33‒ 36] .
Figure 3 .
The formation of PRIMPOL foci persists throughout the cell cycle
(A) U2OS cells were synchronized to G1, S and G2 phases respectively. Representative FACS profile by PI staining. (B) PRIMPOL and EdU were detected by IF in synchronized cells. White arrows indicate colocalizations. (C) Quantification of the number of PRIMPOL foci in the synchronized cells containing PRIMPOL foci. (D) Quantification of the colocalization of PRIMPOL and EdU per cell. (E) Quantification of the percentage of nuclei with ≥5 PRIMPOL-EdU colocalized foci.** P<0.001, **** P<0.0001.
PRIMPOL competes with RAD51 to resolve G4-induced replication stress
Since PRIMPOL interacts with RPA in response to replication stress ( Figure 1), we sought to investigate whether RPA is essential for forming PRIMPOL foci. Hence, a small interfering RNA (siRNA) was used to knockdown RPA in U2OS cells. Western blot analysis was used to determine the level of depletion ( Figure 4A). Depleting RPA drastically reduced the formation of PRIMPOL foci compared with control cells ( Figure 4B,C), suggesting that PRIMPOL resolves replication stress by interacting with RPA.
Figure 4 .
PRIMPOL competes with RAD51 to resolve G4-induced replication stress
(A) Cells were transiently transfected with control, RPA siRNA, or RAD51 siRNA. Western blot analysis showing the knockdown of RPA and RAD51 in HeLa, HEK 293T, and U2OS cells. (B) Representative images showing the formation of PRIMPOL foci in RPA-knockdown, RAD51-knockdown, and control cells. DAPI (blue); PRIMPOL (red). (C) Quantification of the number of PRIMPOL foci per cell. More than 100 cells were analysed. (D) Representative images of G4 foci in the control (NC) and RPA- or RAD51-knockdown HeLa, HEK 293T, and U2OS cells. DAPI (blue); G4 (red). (E) Quantification of the number of G4 foci per cell. (F) Representative images of RAD51 (green) and G4 (red) colocalizations in control (scramble) and PRIMPOL-knockdown HeLa and U2OS cells. White arrows indicate colocalizations. (G) Quantification of the colocalization of PRIMPOL and G4 per cell. PRIMPOL and RAD51 were detected by IF in U2OS cells. DAPI (blue); PRIMPOL (red); RAD51 (green). * P<0.05, ** P<0.001, *** P<0.001, **** P<0.0001.
The stalled replication forks could be resolved by RAD51-mediated fork reversal. There is a balance between fork repriming and reversal in eukaryotes [28]. Therefore, we explored the competition between PRIMPOL and RAD51 at the stalled replication forks. PRIMPOL foci were determined when RAD51 was depleted, and the results showed that RAD51 depletion with siRNA increased PRIMPOL foci formation ( Figure 4A‒C). Moreover, the formation of G4s was examined in HeLa, HEK 293T, and U2OS cells when RPA or RAD51 was depleted ( Figure 4A,D). The number of G4 foci per cell was significantly increased in HeLa, HEK 293T, and U2OS cells compared with that in control cells when RPA or RAD51 was depleted ( Figure 4E), suggesting that RPA and RAD51 responded to G4-induced replication stress. To determine whether PRIMPOL competes with RAD51 to resolve G4-induced replication stress, the colocalization between RAD51 and G4s was examined ( Figure 4F). Our results revealed that the colocalization between RAD51 and G4s was significantly increased in HeLa and U2OS cells when PRIMPOL was depleted ( Figure 4F,G), suggesting that PRIMPOL competes with RAD51 to resolve G4-induced replication stress. We also detected the formation of RPA foci in PRIMPOL-knockdown cells ( Supplementary Figure S4A). The results showed that RPA foci were decreased when PRIMPOL was depleted ( Supplementary Figure S4B). A previous study reported that PRIMPOL could increase the affinity of RPA for ssDNA [37], which is consistent with our results. RPA foci were decreased when PRIMPOL was depleted possibly because PRIMPOL weakened the affinity of RPA for ssDNA. Next, the PRIMPOL and RAD51 foci were determined in PRIMPOL-overexpressing cells. Our results showed that RAD51 foci could not be detected in 80% of PRIMPOL-overexpressing cells possibly because RAD51 was suppressed when PRIMPOL was overexpressed, suggesting that PRIMPOL competes with RAD51 to cope with replication stress ( Supplementary Figure S4C,D). Overall, these results indicated that PRIMPOL (repriming)-mediated pathways compete with RAD51 (fork reversal)-mediated pathways in response to G4-induced replication stress.
Discussion
PRIMPOL is a recently discovered DNA polymerase possessing dual enzymatic activities, including polymerase and primase, which play a role in DNA damage tolerance and replication stress response in eukaryotic cells [ 17‒ 19] . However, the detailed mechanism by which PRIMPOL responds to replication stress remains unclear. In this study, we demonstrated that PRIMPOL competes with RAD51 to resolve G4-induced replication stress in human cells via physical interaction with RPA.
Accurate DNA replication is essential for genome stability. Replication stress may arise from extracellular and intracellular factors, leading to replication fork stalling. The DNA damage response signaling pathway may be activated in response to replication stress, and associated nuclear proteins may be reorganized to form subnuclear foci [ 5, 6] . Previous studies reported that RPA relocalizes to subnuclear foci to respond to replication stress [7]. Indeed, our results indicated that RPA forms subnuclear foci with PRIMPOL throughout the cell cycle ( Figure 1D and Figure 3), suggesting that PRIMPOL is essential for responding to replication stress.
RPA can interact with multiple binding partners to facilitate various functions, including initiating replication with MCM proteins [ 6, 8] and promoting replication fork reversal with RAD51. Here, we confirmed the interaction between PRIMPOL and RPA ( Figure 1). In addition, the loss of RPA abolished the formation of PRIMPOL foci, suggesting that the formation of PRIMPOL foci relies on the presence of RPA ( Figure 4A‒C). In other words, RPA is required for the recruitment or regulation of PRIMPOL to the stalled replication forks or DNA damage sites.
DNA secondary structure G4s can also induce replication stress by blocking the replication fork and impeding DNA replication [4]. G4s must be overcome or bypassed to ensure that replication continues. RPA has been reported to resolve DNA secondary structures by interacting with BLM/WRN helicases [ 9, 10] . Given that PRIMPOL has primase activity, PRIMPOL was speculated to be recruited to the stalled replication forks by RPA to facilitate G4-induced stalled forks repriming at the downstream site [ 23, 24] . Here, our results showed that PRIMPOL could localize at G4s in vivo and that PRIMPOL foci significantly increased after treatment with G4 stabilizers ( Figure 2B‒F). PRIMPOL depletion affected cell sensitivity to G4-inducing reagent and increased G4s ( Figure 2G‒K). These data further supported that PRIMPOL plays an essential role in the response to replication stress induced by G4s. Further studies will be valuable to explore a more detailed mechanism by which PRIMPOL reprimes downstream of G4s.
To our knowledge, PRIMPOL-mediated repriming plays an essential role in the replication stress response [ 25, 27] . In addition, replication fork reversal is an alternative pathway to cope with replication stress. Replication fork reversal mediated by RAD51, SMARCAL1 or HLTF can deal with replication stress by facilitating fork remodelling [2]. Furthermore, there is a balance between fork repriming and reversal in eukaryotes [28]. Hence, we supposed that there is an equilibrium between PRIMPOL- and RAD51-mediated pathways in response to replication stress. Indeed, depletion of RAD51 with siRNA increased PRIMPOL foci formation ( Figure 4A‒C). RAD51 foci decreased in PRIMPOL-overexpressing cells ( Supplementary Figure S4C,D). Consistent with our hypothesis, when replication fork reversal is impaired due to deletion of RAD51, HLTF or SMARCAL1, stalled forks can be rescued by PRIMPOL repriming [ 25, 27, 28] . In a study by Quinet et al. [28], PRIMPOL-mediated repriming was found to suppress replication fork reversal in BRCA-deficient cells. In addition, the accumulation of RAD51 on chromatin was increased due to loss of PRIMPOL in UV-C-treated cells [38], while loss of RAD51 induced PRIMPOL-dependent excessive fork elongation after the cells were treated with UV-C [39]. In another study by Junyeop Lee et al. [40], BRCA2 was found to promote RAD51 recruitment to G4. Here, we verified the colocalization between RAD51 and G4s and revealed that the colocalization between RAD51 and G4s was significantly increased in PRIMPOL-knockdown cells, suggesting that PRIMPOL competes with RAD51 to resolve G4-induced replication stress. However, further investigation is required to examine how PRIMPOL- or RAD51-mediated pathways are selected to respond to G4-induced replication stress.
In summary, our current work provides novel insights into the mechanism by which PRIMPOL in G4s resolves replication stress via interaction with RPA. This study provides evidence that PRIMPOL could work at G4s in vivo to resolve replication stress in human cells. In addition, the PRIMPOL-mediated pathway alone or in combination with the RAD51-mediated pathway could serve as novel molecular targets to improve the tumor response to DNA-damaging chemotherapy.
Acknowledgments
We are grateful for the excellent technical support from the Basic Research Center of Tianjin Medical University.
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinica online.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (No. 32170762), the Tianjin Health Research Project (No. 19YFZCSY00600), and the Natural Science Foundation of Tianjin City (No. 19JCJQJC63500).
References
- 1.Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, Zou L. ATR protects the genome against R loops through a MUS81-triggered feedback loop. Mol Cell. . 2020;77:514–527. doi: 10.1016/j.molcel.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinet A, Lemaçon D, Vindigni A. Replication fork reversal: players and guardians. Mol Cell. . 2017;68:830–833. doi: 10.1016/j.molcel.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista LF, Kaina B, Meneghini R, Menck CF. How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat Res Rev Mutat Res. . 2009;681:197–208. doi: 10.1016/j.mrrev.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Frasson I, Pirota V, Richter SN, Doria F. Multimeric G-quadruplexes: a review on their biological roles and targeting. Int J Biol Macromolecules. . 2022;204:89–102. doi: 10.1016/j.ijbiomac.2022.01.197. [DOI] [PubMed] [Google Scholar]
- 5.Panichnantakul P, Patel A, Tse EY, Wyatt HD. An open-source platform to quantify subnuclear foci and protein colocalization in response to replication stress. DNA Repair. . 2021;105:103156. doi: 10.1016/j.dnarep.2021.103156. [DOI] [PubMed] [Google Scholar]
- 6.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response. Cell. . 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen DD, Kim EY, Sang PB, Chai W. Roles of OB-fold proteins in replication stress. Front Cell Dev Biol. . 2020;8:574466. doi: 10.3389/fcell.2020.574466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakaya R, Takaya J, Onuki T, Moritani M, Nozaki N, Ishimi Y. Identification of proteins that may directly interact with human RPA. J Biochem. . 2010;148:539–547. doi: 10.1093/jb/mvq085. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol Cell. . 2017;65:832–847.e4. doi: 10.1016/j.molcel.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W, Rokutanda N, Takeuchi J, Lai Y, Maruyama R, Togashi Y, Nishikawa H, et al. HERC2 facilitates BLM and WRN helicase complex interaction with RPA to suppress G-quadruplex DNA. Cancer Res. . 2018;78:6371–6385. doi: 10.1158/0008-5472.CAN-18-1877. [DOI] [PubMed] [Google Scholar]
- 11.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. . 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 12.Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. . 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 13.Marsico G, Chambers VS, Sahakyan AB, McCauley P, Boutell JM, Antonio MD, Balasubramanian S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. . 2019;47:3862–3874. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SY, Chang EYC, Lim J, Kwan HH, Monchaud D, Yip S, Stirling PC, et al. G-quadruplexes mark alternative lengthening of telomeres. NAR Cancer. . 2021;3:zcab031. doi: 10.1093/narcan/zcab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. . 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiavone D, Guilbaud G, Murat P, Papadopoulou C, Sarkies P, Prioleau MN, Balasubramanian S, et al. Determinants of G quadruplex‐induced epigenetic instability in REV 1‐deficient cells. EMBO J. . 2014;33:2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Im JS, Lee KY, Dillon LW, Dutta A. Human Primpol1: a novel guardian of stalled replication forks. EMBO Rep. . 2013;14:1032–1033. doi: 10.1038/embor.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. . 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mourón S, Rodriguez-Acebes S, Martínez-Jiménez MI, García-Gómez S, Chocrón S, Blanco L, Méndez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol. . 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 20.Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, Sun Y, et al. hPrimpol1/CCDC111 is a human DNA primase‐polymerase required for the maintenance of genome integrity. EMBO Rep. . 2013;14:1104–1112. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilliam TA, Brissett NC, Ehlinger A, Keen BA, Kolesar P, Taylor EM, Bailey LJ, et al. Molecular basis for PrimPol recruitment to replication forks by RPA. Nat Commun. . 2017;8:15222. doi: 10.1038/ncomms15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey LJ, Teague R, Kolesar P, Bainbridge LJ, Lindsay HD, Doherty AJ. PLK1 regulates the PrimPol damage tolerance pathway during the cell cycle. Sci Adv. . 2021;7:eabh1004. doi: 10.1126/sciadv.abh1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estep KN, Butler TJ, Ding J, Brosh RM. G4-Interacting DNA helicases and polymerases: potential therapeutic targets. Curr Med Chem. . 2019;26:2881–2897. doi: 10.2174/0929867324666171116123345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiavone D, Jozwiakowski SK, Romanello M, Guilbaud G, Guilliam TA, Bailey LJ, Sale JE, et al. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol Cell. . 2016;61:161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirman S, Quinet A, Wood M, Meroni A, Cybulla E, Jackson J, Pegoraro S, et al. Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells. Mol Cell. . 2021;81:4026–4040.. doi: 10.1016/j.molcel.2021.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Tang TS, Guo C. Regulation of translesion DNA synthesis in mammalian cells. Environ Mol Mutagen. . 2020;61:680–692. doi: 10.1002/em.22359. [DOI] [PubMed] [Google Scholar]
- 27.Bai G, Kermi C, Stoy H, Schiltz CJ, Bacal J, Zaino AM, Hadden MK, et al. HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol Cell. . 2020;78:1237–1251.e7. doi: 10.1016/j.molcel.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinet A, Tirman S, Jackson J, Šviković S, Lemaçon D, Carvajal-Maldonado D, González-Acosta D, et al. PRIMPOL-Mediated adaptive response suppresses replication fork reversal in BRCA-Deficient Cells. Mol Cell. . 2020;77:461–474.e9. doi: 10.1016/j.molcel.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai X, Lu C, Jin J, Tian S, Guo Z, Chen P, Zhai G, et al. Development of a DNA-Templated peptide probe for photoaffinity labeling and enrichment of the histone modification reader proteins. Angew Chem Int Ed. . 2016;55:7993–7997. doi: 10.1002/anie.201602558. [DOI] [PubMed] [Google Scholar]
- 30.Stojkovič G, Makarova AV, Wanrooij PH, Forslund J, Burgers PM, Wanrooij S. Oxidative DNA damage stalls the human mitochondrial replisome. Sci Rep. . 2016;6:28942. doi: 10.1038/srep28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilliam TA, Jozwiakowski SK, Ehlinger A, Barnes RP, Rudd SG, Bailey LJ, Skehel JM, et al. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. . 2015;43:1056–1068. doi: 10.1093/nar/gku1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chippalkatti R, Suter B. 5-Ethynyl-2′-deoxyuridine/phospho-histone H3 dual-labeling protocol for cell cycle progression analysis in Drosophila neural stem cells . J Vis Exp. . 2021, doi: 10.3791/62642 doi: 10.3791/62642. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JM, Yadav T, Ouyang J, Lan L, Zou L. Alternative lengthening of telomeres through two distinct break-induced replication pathways. Cell Rep. . 2019;26:955–968. doi: 10.1016/j.celrep.2018.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma P, Dilley RL, Zhang T, Gyparaki MT, Li Y, Greenberg RA. RAD52 and SLX4 act nonepistatically to ensure telomere stability during alternative telomere lengthening. Genes Dev. . 2019;33:221–235. doi: 10.1101/gad.319723.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. . 2016;539:54–58. doi: 10.1038/nature20099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Özer Ö, Bhowmick R, Liu Y, Hickson ID. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget. . 2018;9:15836–15846. doi: 10.18632/oncotarget.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Jiménez MI, Lahera A, Blanco L. Human PrimPol activity is enhanced by RPA. Sci Rep. . 2017;7:783. doi: 10.1038/s41598-017-00958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, et al. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell. . 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallerga MB, Mansilla SF, Federico MB, Bertolin AP, Gottifredi V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc Natl Acad Sci USA. . 2015;112:E6624–6633. doi: 10.1073/pnas.1508543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Sung K, Joo SY, Jeong JH, Kim SK, Lee H. Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis. Nat Commun. . 2022;13:3396. doi: 10.1038/s41467-022-31156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]