Abstract
Serratia marcescens Rio-5, one of 18 extended-spectrum β-lactamase (ESBL)-producing strains isolated in several hospitals in Rio de Janeiro (Brazil) in 1996 and 1997, exhibited a high level of resistance to aztreonam (MIC, 512 μg/ml) and a distinctly higher level of resistance to cefotaxime (MIC, 64 μg/ml) than to ceftazidime (MIC, 8 μg/ml). The strain produced a plasmid-encoded ESBL with a pI of 7.5 whose bla gene was not related to those of other plasmid-mediated Ambler class A ESBLs. Cloning and sequencing revealed a bla gene encoding a novel class A β-lactamase in functional group 2be, designated BES-1 (Brazil extended-spectrum β-lactamase). This enzyme had 51% identity with chromosomal class A penicillinase of Yersinia enterocolitica Y56, which was the most closely related enzyme and 47 to 48% identity with CTX-M-type β-lactamases, which were the most closely related ESBLs. In common with CTX-M enzymes, BES-1 exhibited high cefotaxime-hydrolyzing activity (kcat, 425 s−1). However, BES-1 differed from CTX-M enzymes by its significant ceftazidime-hydrolyzing activity (kcat, 25 s−1), high affinity for aztreonam (Ki, 1 μM), and lower susceptibility to tazobactam (50% inhibitory concentration [IC50], 0.820 μM) than to clavulanate (IC50, 0.045 μM). Likewise, certain characteristic structural features of CTX-M enzymes, such as Phe-160, Ser-237, and Arg-276, were observed for BES-1, which, in addition, harbored different residues (Ala-104, Ser-171, Arg-220, Gly-240) and six additional residues at the end of the sequence. BES-1, therefore, may be an interesting model for further investigations of the structure-function relationships of class A ESBLs.
Shortly after the introduction of extended-spectrum β-lactams such as cefotaxime, aztreonam, and ceftazidime, extended-spectrum β-lactamases (ESBLs) were characterized for members of the family Enterobacteriaceae, firstly in Europe (23, 47) and then worldwide. These enzymes hydrolyze extended-spectrum cephalosporins and aztreonam to varying extents but usually neither cephamycins (cefoxitin and moxalactam) nor carbapenems (imipenem and meropenem). A common feature of these enzymes is inhibition of their activity by clavulanic acid. According to the structural classification of Ambler et al. (1) and the latest functional scheme of Bush et al. (11), these ESBLs are generally class A enzymes of the 2be group, which arise as the result of a few amino acid substitutions from the common plasmid-mediated TEM and SHV-1 β-lactamases.
Since the first report of MEN-1 (CTX-M-1) at the beginning of the 1990s (3), non-TEM, non-SHV, class A ESBLs have been observed for strains of the family Enterobacteriaceae and in Pseudomonas aeruginosa. Except for the ESBL SFO-1, which is closely related to the chromosomal enzyme of Serratia fonticola (28), these ESBLs, especially the enzyme GES-1 (39), are distantly related not only to one another but also to chromosome-borne enzymes. However, two groups can be considered; on the one hand, the ESBLs of the growing family CTX-M (7), which cluster with the class A chromosomally encoded β-lactamases of Proteus vulgaris (36), S. fonticola (35), Citrobacter diversus (37), Klebsiella oxytoca (2), Burkholderia cepacia (52), and Yersinia enterocolitica (45), and on the other hand, the ESBLs of the PER type (5, 32), TLA-1 (46), and VEB-1 (40), which cluster with Bacteroides class A chromosomal β-lactamases (42, 48) and the β-lactamase CME-1 of Chryseobacterium (Flavobacterium) meningosepticum (43).
To estimate the diversity of ESBLs in Brazil, clinical strains that exhibited ESBL phenotypes in different species were collected in hospitals in Rio de Janeiro in 1996 and 1997. In this report, we describe a novel type of non-TEM, non-SHV, class A ESBL from a Serratia marcescens clinical isolate, designated BES-1 (Brazil extended spectrum).
MATERIALS AND METHODS
Strains and plasmids.
Table 1 shows the strains and plasmids used in this study. S. marcescens Rio-5 was isolated from the blood of a newborn hospitalized in the intensive care unit of a hospital in Rio de Janeiro, Brazil, in 1996. pRio-5 was the natural BES-1-encoding plasmid of S. marcescens Rio-5, and pClRio-5 was the recombinant plasmid, obtained by cloning the gene of BES-1 in phagemid vector pBK-CMV.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| S. marcescens Rio-5 | Clinical strain harboring the natural plasmid pRio-5 (Rio de Janeiro, Brazil, 1997) | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 44 |
| Plasmids | ||
| pRio-5 | Natural plasmid from S. marcescens Rio-5 containing blaBES-1 gene; resistance phenotype: ESBL | This study |
| pClRio-5 | Recombinant plasmid containing a 2.2-kb fragment with blaBES-1; resistance phenotype: ESBL, kanamycin | This study |
| pClRio-2 | Recombinant plasmid containing blaCTX-M-8; resistance phenotype: ESBL, kanamycin | 7 |
| pBK-CMV | Phagemid vector; resistance phenotype: kanamycin | Stratagene |
Plasmid study.
Conjugations of plasmids carrying the β-lactamase gene were performed by mating donor strains with in vitro-obtained rifampin- or nalidixic acid-resistant mutants of Escherichia coli HB101 (44) as recipient strains at 37 or 30°C in solid or liquid Mueller-Hinton medium. Transconjugants were selected on Mueller-Hinton agar containing rifampin (300 μg/ml) or nalidixic acid (150 μg/ml) and cefotaxime (2 μg/ml).
Electroporation of plasmid DNA into E. coli DH5α was performed according to the manufacturer's instructions (Bio-Rad, Richmond, Calif.). Transformants were selected on Mueller-Hinton agar containing cefotaxime (2 μg/ml).
Plasmid DNA was extracted and purified by alkaline lysis according to the Quiafilter protocol (Qiagen, Hilden, Germany). The plasmid size was determined after digestion with restriction endonucleases EcoRI and SalI (Boehringer Mannheim, Mannheim, Germany). Restriction fragments were visualized after electrophoresis in 0.8% agarose gels with a 1-kb DNA ladder (Eurogentec).
Susceptibilities to β-lactams.
MICs were determined by a dilution method on Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes la Coquette, France) with an inoculum of 104 CFU per spot. Antibiotics were provided as powders by SmithKline Beecham Pharmaceuticals (amoxicillin, ticarcillin, and clavulanate), Lederle Laboratories (piperacillin and tazobactam), Eli Lilly (Paris, France) (cephalothin), Roussel-Uclaf (cefotaxime and cefpirome), Glaxo Wellcome Research and Development (ceftazidime), and Bristol-Myers Squibb (cefepime).
Detection of ESBLs was performed with the standard double-disk synergy tests as described previously (21). Antibiotic disks for agar tests were obtained from Sanofi Diagnostics Pasteur.
Isoelectric focusing.
Isoelectric focusing was performed with 6% polyacrylamide gels containing ampholines (Pharmacia Biotech, Uppsala, Sweden) with a pH range of 3.5 to 10. Proteins were focused at a constant temperature (6°C) for 3 h at 1 W of constant power per cm with a Multiphor II flatbed apparatus (Pharmacia Biotech). After focusing, the β-lactamase activity was revealed with iodine agar by overlaying the polyacrylamide gel with an agar gel containing 0.6% (wt/vol) penicillin G, 6% (wt/vol) potassium iodide, and 0.6% (wt/vol) iodine. β-Lactamases with known pIs were used as standards: TEM-1 (pI 5.4), SHV-1 (pI 7.6), and MEN-1 (pI 8.6).
β-Lactamase preparation.
The ESBL-producing strain was grown in 6 liters of brain heart infusion broth containing cefotaxime at 2 μg/ml for 16 h at 37°C. The bacteria collected by centrifugation were suspended in morpholineethanesulfonic acid (MES)-NaOH (20 mM; pH 6.0; 20 ml/5 g of cells) and disrupted by ultrasonic treatment (four times for 30 s, each time at 20 W). After centrifugation (48,000 × g for 10 min at 4°C), the clarified supernatant was loaded onto an SP Sepharose column (10 ml; Amersham Pharmacia Biotech) equilibrated with MES-NaOH (50 mM; pH 6.0). After washing of the column with the same buffer, the bound proteins were eluted with a linear NaCl gradient (0 to 500 mM). After ultrafiltration-concentration (Centriprep YM-10; Amicon; Millipore Corporation, Bedford, Mass.), the β-lactamase-containing elution peak (spot test with nitrocefin as substrate) was loaded onto a Superose 12 (3.2 by 30 cm; Amersham Pharmacia Biotech) column that had been equilibrated and eluted with the buffer MES-NaOH (20 mM)–NaCl (100 mM; pH 6.0). The β-lactamase-containing elution peak was extensively dialyzed and concentrated by ultrafiltration against NaCl (100 mM) and stored at −20°C until use. The total protein concentration was estimated by the Bio-Rad protein assay (Bio-Rad) with bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) used as a standard. The purity of BES-1 extract was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). It was performed as described by Laemmli (26) with final acrylamide concentrations of 12 and 5% (wt/vol) for the separating and the stacking gels, respectively. Proteins were stained with Coomassie blue R-250 (Sigma Chemical Co.). After SDS-PAGE, renaturation of β-lactamase was performed as previously described (27), using the renaturation buffer Tris-HCl (100 mM)–Triton X-100 (2% [wt/vol]; pH 7.0). β-Lactamase activity was detected by overlaying the polyacrylamide gel with 0.5 mM nitrocefin (Oxoid, Paris, France) in 100 mM phosphate buffer (pH 7.0).
N-terminal protein sequence.
The N-terminal sequence of enzyme BES-1 was determined with a gas-phase sequencer as recommended by the manufacturer (Applied Biosystems, Foster City, Calif.) after loading the purified extract of BES-1 onto a polyvinylidene difluoride membrane (Millipore Corp.).
Determination of β-lactamase kinetic constants.
The Km and Kcat constants of the β-lactamase were obtained by a computerized microacidimetric method (25) with a purified extract. The enzyme BES-1 was purified from crude lysates of E. coli DH5α(pClRio-5). The concentrations of the inhibitors (clavulanate and tazobactam) required to inhibit enzyme activity by 50% (IC50s) were determined as described previously (7). IC50 and Ki were monitored with penicillin G (225 mM) as the reporter substrate.
β-Lactamase gene cloning.
Recombinant DNA manipulation and transformations were performed as described by Sambrook et al. (44). The T4 DNA ligase used was purchased from Boehringer Mannheim. The BES-1-encoding gene was cloned as follows. Natural plasmid pRio-5 was partially cleaved by Sau3A, and the resultant fragments were ligated into the BamHI site of pBK-CMV phagemid (Stratagene, La Jolla, Calif.). E. coli DH5α (44) was transformed by electroporation. The transformant harboring the recombinant BES-1-encoding plasmid pClRio-5 was selected on Mueller-Hinton agar supplemented with 2 μg of cefotaxime per ml.
DNA sequencing.
The insert of recombinant plasmid pClRio-5 was sequenced by the dideoxy chain termination procedure on both complementary strands using an Applied Biosystems sequencer (ABI 1377), as previously described (7).
Computer analysis.
The nucleotide sequence and the deduced protein sequence were analyzed with the software available on the Internet at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/, BLAST and ORF finder). A hydrophobic blot was obtained with the method of Nielsen et al. (31). The multiple sequence alignment and the phylogenic analysis, performed by the parsimony method, were carried out as previously described (7). BES-1 was compared with 23 class A β-lactamases: TEM-3 (49), SHV-2 (15), PSE-4 (6), GES-1 (39), VEB-1 (40), PER-1 (32), CEPA (42), CBLA (48), NMC-A (30), SME-1 (29), YENT (45), BCEP (52), PVUL (36), CDIV (37), SFO-1 (28), OXY-1 (2), OXY-2 (41), CTX-M-1 (MEN-1) (3, 4), CTX-M-2 (4), TOHO-1 (19), CTX-M-4 (16, 17), CTX-M-5 (10), and CTX-M-8 (7).
Nucleotide sequence accession number.
The blaBES gene nucleotide sequence data appear in the GenBank nucleotide sequence database under accession no. AF234999.
RESULTS
S. marcescens Rio-5 (Table 2) exhibited a resistance to broad-spectrum cephalosporins and a positive double-disk synergy test and produced three β-lactamases with pIs of 5.4, 7.5, and >8.6. PCR and DNA sequencing identified the β-lactamase with a pI of 5.4 as TEM-1. The β-lactamase with a pI of >8.6 is likely the chromosomal cephalosporinase of the S. marcescens isolate. No hybridizations were obtained with the probes of CTX-M and SHV types.
TABLE 2.
β-Lactam MICs for the BES-1-producing S. marcescens and E. coli DH5α(pClRio-5) in comparison with CTX-M-8-producing E. coli DH5α(pClRio-2) and E. coli DH5α
| β-Lactam | MIC (μg/ml) for strain:
|
|||
|---|---|---|---|---|
| S. marcescens Rio-5a | E. coli DH5α(pClRio-5)b | E. coli DH5α(pClRio-2)c | E. coli DH5α | |
| Amoxicillin | >2,048 | >2,048 | >2,048 | 2 |
| Amoxicillin + CLAd | 512 | 16 | 64 | 1 |
| Amoxicillin + TZBe | >2,048 | >2,048 | 64 | 1 |
| Ticarcillin | >2,048 | >2,048 | >2,048 | 2 |
| Ticarcillin + CLA | 512 | 32 | 256 | 2 |
| Ticarcillin + TZB | >2,048 | >2,048 | 128 | 2 |
| Piperacillin | 512 | 512 | 512 | 2 |
| Piperacillin + CLA | 64 | 4 | 4 | 2 |
| Piperacillin + TZB | 256 | 256 | 4 | 2 |
| Cephalothin | >1,024 | >1,024 | >1,024 | 4 |
| Cephalothin + CLA | >1,024 | 8 | 32 | 4 |
| Cephalothin + TZB | >1,024 | 512 | 16 | 4 |
| Cefoxitin | 32 | 4 | 4 | 2 |
| Cefotaxime | 64 | 64 | 32 | 0.06 |
| Cefotaxime + CLA | 4 | 0.06 | 0.06 | 0.06 |
| Cefotaxime + TZB | 16 | 16 | 0.06 | 0.06 |
| Aztreonam | 512 | 512 | 8 | 0.06 |
| Aztreonam + CLA | 4 | 0.25 | 0.12 | 0.06 |
| Aztreonam + TZB | 128 | 128 | 0.12 | 0.06 |
| Ceftazidime | 4 | 16 | 2 | 0.06 |
| Ceftazidime + CLA | 1 | 0.25 | 0.06 | 0.06 |
| Ceftazidime + TZB | 2 | 8 | 0.12 | 0.06 |
| Cefepime | 4 | 8 | 8 | 0.06 |
| Cefepime + CLA | 0.25 | 0.06 | 0.06 | 0.06 |
| Cefepime + TZB | 2 | 4 | 0.06 | 0.06 |
| Cefpirome | 4 | 16 | 16 | 0.06 |
| Cefpirome + CLA | 0.12 | 0.06 | 0.06 | 0.06 |
| Cefpirome + TZB | 2 | 8 | 0.06 | 0.06 |
| Imipenem | 0.5 | 0.25 | 0.25 | 0.06 |
S. marcescens Rio-5 producing β-lactamases BES-1 and TEM-1 and the chromosomally encoded cephalosporinase. pI, 7.5, 5.4, and >8.6.
E. coli DH5α harboring recombinant plasmid pClRio-5 which encodes β-lactamase BES-1. pI, 7.5.
E. coli DH5α harboring recombinant plasmid pClRio-2 which encodes β-lactamase CTX-M-8. pI, 7.7.
CLA, clavulanate at a fixed concentration of 2 μg/ml.
TZB, tazobactam at a fixed concentration of 4 μg/ml.
Transfer of β-lactam resistance.
No transconjugant was obtained by a mating-out assay in the conditions used. However, a transformant which exhibited an ESBL phenotype and produced two β-lactamases with pIs of 5.4 and 7.5 was obtained by electroporating the plasmid DNA content of S. marcescens Rio-5 into E. coli DH5α. The analysis of the plasmid content of this transformant revealed a 15.5-kb plasmid, pRio-5 (data not shown). No associated resistance markers were encoded by this plasmid.
Cloning and sequencing of the β-lactamase gene.
The ESBL gene encoded by pRio-5 was cloned in plasmid vector pBK-CMV. Different transformants which exhibited an ESBL phenotype and produced only the β-lactamases with a pI of 7.5 were obtained. One of these transformants contained a recombinant plasmid, pClRio-5, which harbored an insert of about 2 kb.
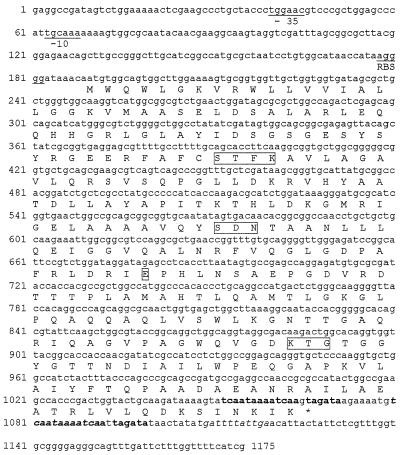
The insert of recombinant plasmid pClRio-5 was sequenced. It contained an open reading frame of 879 bp which had a G+C content of 60% (Fig. 1). This coding region did not have significant identity with previously described genes in the sequence databases. The initiation codon sequence was preceded by putative −35 and −10 consensus sequences and a putative ribosome-binding site. A terminator hairpin loop was detected 12 nucleotides from the stop codon (Fig. 1).
FIG. 1.
Nucleotide sequence of the 1,175-bp fragment of pClRio-5 containing blaBES-1. The deduced amino acid sequence is designated in the single-letter code below the nucleotide sequence. The potential promoter sequences and ribosome-binding site are represented by −10 and −35 regions and RBS, respectively. Inverted repeat sequences possibly acting as terminators are in italics. Duplicated nucleotides are in boldface. Conserved residues in serine β-lactamases (SXXK, SDN, E, and KTG) are boxed.
The protein with 292 amino acid residues, which was deduced from the open reading frame, was designated BES-1. It contained the four structural elements characteristic of class A β-lactamases: S-X-X-K at positions 70 to 73, S-D-N at positions 130 to 132, E at position 166, and K-T-G at positions 234 to 236 (Fig. 1).
Homology with other β-lactamases.
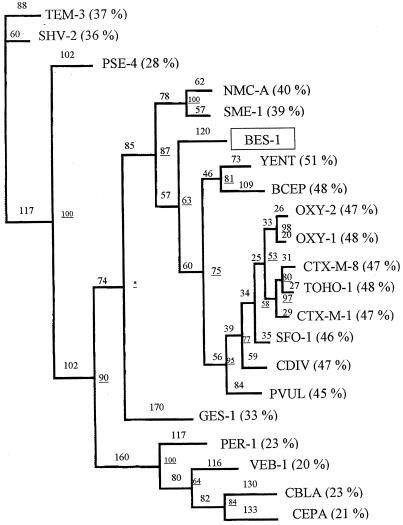
A phylogenic study was performed to relate BES-1 to the most closely related enzymes and to the representatives of major lineages of class A β-lactamases (Fig. 2). BES-1 exhibited the highest percentage of identity (51 to 45%) with the following group of gram-negative bacterial β-lactamases: the chromosomal class A β-lactamases of Y. enterocolitica Y56, B. cepacia 249, K. oxytoca, P. vulgaris R0104, and C. diversus CUV and the plasmid-borne β-lactamases of the CTX-M type and SFO-1. The phylogenic study located the enzyme BES-1 on a distinct branch between the cluster containing these class A β-lactamases and class A imipenemases NMC-A and SME-1 (Fig. 2).
FIG. 2.
Dendrogram for BES-1 and 20 other class A β-lactamases made according to the parsimony method. Branch lengths are to scale and are proportional to the number of amino acid changes. The percentages at the branch points (underlined numbers) refer to the numbers of times that a particular node was found in 100 bootstrap replications (the asterisk indicates uncertainty of nodes with bootstrap values of less than 50%). The distance along the vertical axis has no significance. Percent pairwise identities between BES-1 and the β-lactamases of the indicated types are indicated in parentheses.
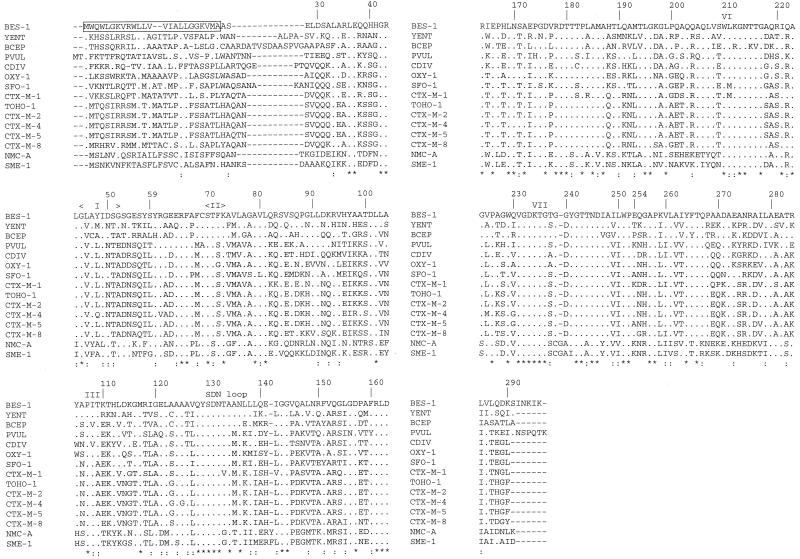
A multisequence alignment of the β-lactamase BES-1 and the most closely related enzymes is shown in Fig. 3. BES-1 possessed the highly conserved amino acid residues of class A β-lactamase, i.e., boxes I to VII (22), which interact with β-lactam compounds.
FIG. 3.
Alignments of the BES-1 amino acid sequence with its closest class A neighbors. Dots indicate amino acids identical to those of BES-1. Identical residues in all β-lactamases are indicated by asterisks; conservative substitutions are indicated by colons. The peptide signal of BES-1 is boxed. Roman numerals designate boxes described by Joris et al. (22). Amino acids are numbered according to the standard numbering scheme for the class A β-lactamases of Ambler et al. (1).
In addition, BES-1, like the most closely related enzymes, possessed only one cysteine at position 69. As in penicillinases TEM-1 and SHV-1, the Ω loop contained Arg-164 and Glu-179. Arg was observed at positions 220 and 276. Six more amino acid residues than in CTX-M-type ESBLs were observed for BES-1 at the end of the sequence. These residues were encoded by duplicated nucleotides (nucleotides 1052 to 1071) of the terminal hairpin loop (nucleotides 1080 to 1099) (Fig. 1).
β-Lactam susceptibility.
MICs of β-lactams for S. marcescens Rio-5 and for E. coli DH5α(pClRio-5) are listed in Table 2 and compared with those for CTX-M-8-producing E. coli DH5α(pClRio-2) and E. coli DH5α. The three β-lactamase-producing strains exhibited a high level of resistance to amoxicillin (MICs, >2,048 μg/ml), ticarcillin (MICs, >2,048 μg/ml), and cephalothin (MICs, ≥1,024 μg/ml).
The BES-1-producing strains were characterized by their level of resistance to aztreonam (MICs, 512 μg/ml), which was 8- to 64-fold higher than that to the other aminothiazol β-lactams (MICs, 4 to 64 μg/ml). For BES-1- and CTX-M-8-producing strains, MICs of cefotaxime (MICs, 32 to 64 μg/ml) were 2- to 16-fold higher than those of ceftazidime (MICs, 2 to 16 μg/ml), cefpirome (MICs, 4 to 16 μg/ml), and cefepime (MICs, 4 to 8 μg/ml).
Clavulanate partially or totally restored the activity of the β-lactams against BES-1- and CTX-M-8-producing strains (Table 2). In contrast to clavulanate, tazobactam was much less effective in decreasing the MICs of extended-spectrum β-lactams for BES-1-producing strains: the MICs of cefotaxime, ceftazidime, and aztreonam associated with tazobactam were 16, 2 to 8, and 128 μg/ml, respectively.
Biochemical properties of β-lactamase BES-1.
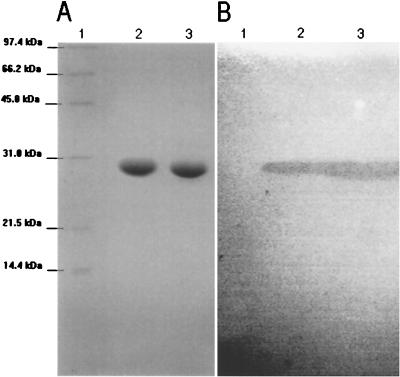
The purified proteins appeared on SDS-polyacrylamide gels as a band of 28.5 kDa for BES-1 (28.2 kDa for CTX-M-8), which exhibited β-lactamase activity (Fig. 4). The overall recovery of enzyme from clarified extract was around 65%. The specific activity of purified β-lactamase (≥97% pure) BES-1 was 85 μmol · min−1 · mg of protein−1, determined with 225 μM benzylpenicillin as the substrate.
FIG. 4.
Electrophoresis analysis of BES-1 and CTX-M-8 purified extracts. (A) SDS-polyacrylamide gel stained with Coomassie brilliant blue R-250. (B) Zymogram detection of β-lactamase activity with nitrocefin after renaturation treatment of SDS-polyacrylamide gel. Lanes: 1, protein molecular mass reference; 2, purified extract of β-lactamase CTX-M-8; 3, purified extract of β-lactamase BES-1.
The amino-terminal sequence of the purified protein was determined to be ASELDSALAR. The signal peptide comprised therefore the first 24 amino acids, as found by hydrophobicity analysis. Thus, the putative mature enzyme consisted of 268 amino acid residues with a calculated molecular mass of 28,619 Da.
The substrate and inhibition profiles of BES-1 were determined with purified extract (Tables 3 and 4). BES-1, like CTX-M-8, had high catalytic efficiency against penicillins, cephalothin, and cefuroxime. BES-1 had lower catalytic activity against penicillins (kcat, 6 to 28 s−1) and cephalothin (kcat, 173 s−1) than did CTX-M-8 (kcat, 17 to 150 s−1 and 1,600 s−1, respectively) but had slightly better affinity (Km, 3 to 18 μM) for these substrates than did CTX-M-8 (Km, 11 to 87 μM).
TABLE 3.
Substrate profile of β-lactamases BES-1 and CTX-M-8
| Substrate | Value for enzyme:
|
|||||
|---|---|---|---|---|---|---|
| BES-1
|
CTX-M-8
|
|||||
| kcat (s−1) | Km (μM) | Kcat/Km (μM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) | |
| Penicillin G | 28 | 5 | 5.6 | 150 | 11 | 13.6 |
| Amoxicillin | 22 | 5 | 4.4 | 55 | 12 | 4.6 |
| Ticarcillin | 6 | 3 | 2.0 | 17 | 14 | 1.2 |
| Piperacillin | 7 | 3 | 2.3 | 74 | 19 | 3.9 |
| Cephalothin | 173 | 18 | 9.6 | 1,600 | 87 | 18.4 |
| Cefuroxime | 116 | 19 | 6.1 | 35 | 12 | 2.9 |
| Cefotaxime | 425 | 95 | 4.5 | 72 | 74 | 1.0 |
| Cefpirome | 708 | 950 | 0.745 | 455 | 1,200 | 0.379 |
| Cefepime | 240 | 1,100 | 0.218 | 144 | 990 | 0.145 |
| Aztreonam | 1 | 1a | 1.0 | 13 | 800 | 0.061 |
| Ceftazidime | 25 | 1,000 | 0.025 | 2 | >500a | <0.002 |
| Cefoxitin | NDb | 22a | NDb | 5a | ||
| Imipenem | NDb | <1a | NDb | 1.5a | ||
Km values were determined as Ki by substrate competition with penicillin G.
No catalytic activity detected.
TABLE 4.
Inhibitor profile of β-lactamase BES-1 in comparison with those of enzymes CTX-M-8 and TEM-1
| Enzyme | IC50 (μM) of drug:
|
||
|---|---|---|---|
| Clavulanate | Tazobactam | Sulbactam | |
| BES-1 | 0.045 | 0.820 | 4.5 |
| CTX-M-8 | 0.036 | 0.010 | 4.0 |
| TEM-1 | 0.085 | 0.090 | 8.0 |
BES-1 and CTX-M-8 had better catalytic activity against methoximinocephems (cefuroxime, cefotaxime, cefpirome, and cefepime) than against carboxypropyloximino β-lactams (ceftazidime and aztreonam) and hence are cefotaximase-type ESBLs.
For BES-1, ceftazidime was a better substrate (kcat, 25 s−1) than was aztreonam (kcat, 1 s−1). Surprisingly, however, BES-1 had high affinity to this monobactam (aztreonam Ki, 1 μM) and thus higher catalytic efficiency against aztreonam (kcat/Km, 1 μM−1 s−1) than against ceftazidime (kcat/Km, 0.025 μM−1 s−1). BES-1 exhibited at least 10-fold-higher catalytic activity than did CTX-M-8 against ceftazidime.
BES-1 and CTX-M-8, like TEM-1, were susceptible to the β-lactam inhibitors clavulanate and sulbactam (Table 4). Sulbactam was approximately 2 orders of magnitude less effective than was clavulanate. However, tazobactam, which is as efficient as clavulanate against TEM-1 (IC50, 0.090 versus 0.085 μM) and slightly better than clavulanate against CTX-M-8 (IC50, 0.010 versus 0.036 μM), was 20-fold less efficient than clavulanate against BES-1 (IC50, 0.820 versus 0.045 μM).
DISCUSSION
The starting point of this work was the observation of the clinical strain S. marcescens Rio-5, which exhibited resistance to aztreonam and cefotaxime and, to a lesser extent, to ceftazidime and had a positive double-disk synergy test. In the course of cloning, a novel bla gene was characterized, which induced this ESBL phenotype. The deduced protein was a novel class A β-lactamase, designated BES-1, which had only 51% identity with the most closely related β-lactamase (chromosomal penicillinase of Y. enterocolitica). BES-1 is distantly related to either the CTX-M or GES-1 enzyme that has been isolated from South American isolates (7, 39). The origin of BES-1 thus remains unknown.
The SHV- and TEM-type ESBLs, first observed in 1985 (23, 47) shortly after the introduction of broad-spectrum cephalosporins such as cefotaxime, aztreonam, and ceftazidime, resulted from point mutations of widespread class A penicillinases. Non-SHV- and non-TEM-type ESBLs, MEN-1, PER-1, VEB-1, TLA-1, GES-1, and CTX-M enzymes, have been characterized since 1992 (3, 4, 5, 7, 10, 16, 32, 39, 40, 46). Some of these ESBL-encoding genes were mobile, since blaVEB-1 and blaGES-1 are located in mobile cassettes of class 1 integrons (39, 40), blaPER genes are chromosomal and plasmidic (5, 13, 32), and blaCTX-M-3 and blaCTX-M-8 genes are observed in different plasmids (7, 33). The type 1 integrase gene and associated resistance markers were not detected in the natural BES-1-encoding plasmid. In the same way, amplification performed with primers specific for 5′ and 3′ conserved regions of type 1 integrons did not reveal their presence (data not shown).
In view of its susceptibility to clavulanate and its enzymatic properties against extended-spectrum cephalosporins, BES-1 could be classified in the 2be group of the functional classification (11). Like CTX-M enzymes, BES-1 is an ESBL with a strong cefotaxime-hydrolyzing activity inducing a distinctly higher level of resistance to cefotaxime than to ceftazidime. However, BES-1 exhibited a catalytic activity against ceftazidime and an affinity for aztreonam 10- and 1,000-fold higher, respectively, than those of CTX-M enzymes (3, 10). The activity of BES-1 against ceftazidime and aztreonam has been observed to a lesser extent with SHV-4 (34).
The residue Thr-237, observed for some TEM-type β-lactamases and implicated in cefotaxime-hydrolyzing activities (24), is observed for BES-1. This residue, Thr-237, which has the same hydrogen-bonding capacity as Ser, could be involved, in association with Arg-276, in the cefotaximase activity of BES-1, as previously suggested for residues Ser-237 and Arg-276 of CTX-M enzymes (3, 16, 17, 18, 19). Four glycine residues, including Gly-232, which is frequently observed for ESBL enzymes (18), were present in BES-1, as they were in CTX-M enzymes. They could increase the flexibility of strand β3, one wall of the active-site cavity. BES-1, like CTX-M enzymes, contains residue Phe-160. This residue, conserved in non-ESBL β-lactamases, is responsible for the lack of a hydrogen bond between the N and C termini of the Ω loop (18). In addition, Ser-171 was observed for BES-1, as in PER-1 and in the Streptomyces albus G β-lactamase (9, 14). It eliminates the hydrogen bond between residues 171 and 164 (14). Thus, Ser-171 and Phe-160 of BES-1 could increase movement of the Ω loop (positions 162 to 179), thereby facilitating the hydrolysis of bulky β-lactams. The glycine residues of strand β3 and Phe-160 and Ser-171 of BES-1 could facilitate binding to and/or hydrolysis of expanded-spectrum cephems.
BES-1, which exhibited ceftazidime-hydrolyzing activity, harbored neither the residues in positions 164 and 179 of the Ω loop (positions 162 to 179) nor residues Lys-104 and Lys-240 of SHV- and TEM-type ESBLs (24). Residue 104 could contribute to the precise positioning of the SDN loop (residues 130 to 132), which is involved in substrate binding and catalysis (38). In addition, Lys-104, frequently observed for TEM-type ESBLs, is thought to interact with the carboxylic acid group in the alkyloximino substituent of ceftazidime and aztreonam (24). The polar residue Asn-104 of CTX-M-type ESBLs is replaced in BES-1 by the small nonpolar residue Ala-104, which may contribute to the particular properties of BES-1.
The mutation Glu-240→Lys plays a major role in the extended-spectrum activity of TEM- and SHV-type ESBLs against ceftazidime and aztreonam. CTX-M enzymes contain the acid residue Asp in position 240, which could repel the propyloximino carboxylic acid group of ceftazidime and aztreonam and impair their binding. Residue Gly-240, observed for both BES-1 and PER-1 (8), may enhance the binding of aztreonam but not correct positioning. This could explain the poor catalytic activity and the high affinity of BES-1 against this substrate.
Arg-220 in the S. albus G β-lactamase, Arg-244 in some class A enzymes, and Arg-276 in CTX-M enzymes play an important role in enzyme catalysis (18–20). BES-1 is the first enzyme to harbor two of these residues: residue Arg-220, located at the end of helix H10, and residue Arg-276, located at the beginning of helix H11. Given the structure of Toho-1 (18), it is likely that the side chain of Arg-220 plays a role in the catalytic process, whereas Arg-276 is more probably involved in the extension of substrate specificity. The association of the two residues Arg-220 and Arg-276, which extend their side chains close to one another, and the six additional amino acids observed in helix H11 of BES-1 could lead to changes in positioning of Arg-276 and could be responsible for the enzymatic properties of BES-1.
In addition, BES-1 exhibited resistance to the inhibitor tazobactam, whereas CTX-M enzymes are susceptible to all β-lactam inhibitors. Surprisingly, the respective IC50s of sulbactam and clavulanate for BES-1 are close to those observed for CTX-M-8 and TEM-1. Thus, this is the first report of a β-lactamase that exhibits selective resistance to tazobactam. The residues in positions 69, 244, 275, and 276 responsible for the resistance of TEM-type enzymes to the β-lactam inhibitors were not observed for BES-1. For TEM-1 β-lactamase, molecular modeling suggests the existence of one hydrogen bond between the triazole ring of tazobactam and the side chain of Arg-244 via a water molecule (12). In BES-1, the absence of Arg-244 associated with the environment created by Arg-220 and Arg-276 could suppress interaction or produce unfavorable interactions between the triazole ring of tazobactam and BES-1.
In conclusion, the functional activity of BES-1 against cefotaxime resembles that of CTX-M-type ESBLs. However, BES-1 exhibits particular enzymatic properties against alkyloximinocephems, monobactams, and β-lactam inhibitors. Thus, BES-1 could be an interesting model for further investigations of the structure-function relationships of class A ESBLs. S. marcescens Rio-5 was the sole BES-1-producing strain from among 18 ESBL-producing Enterobacteriaceae strains, which produced mainly SHV-type ESBLs (10 strains out of 18) and CTX-M-type enzymes (5 strains out of 18). The recent emergence of non-SHV- and non-TEM-type ESBLs could be the result of mobilization of genes from environmental strains to clinical strains as observed with the blaSFO-1 gene (28). The G+C content of blaBES-1 (60%), not typical of Enterobacteriaceae genes (around 50%), is compatible with this hypothesis. It will be interesting to investigate the spread of BES-1 or to detect BES-1-like enzymes.
ACKNOWLEDGMENTS
We thank Angeline Serre, Rolande Perroux, Marlène Jan, and Dominique Rubio for technical assistance. We are also grateful to Ekkehard Collatz, Laboratoire de Recherche Moleculaire sur les Antibiotiques (LRMA), INSERM E0004, Universite Paris VI, for his help in N-terminal protein sequencing.
This work was supported in part by a grant from M.E.N.R.T. (Ministère de l'Education Nationale, de la Recherche et de la Technologie).
REFERENCES
- 1.Ambler R P, Coulson A F W, Frère J-M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Ohta M, Kido N, Mori M, Ito H, Komatsu T, Fujii Y, Kato N. Chromosomal β-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum β-lactam antibiotics. Antimicrob Agents Chemother. 1989;33:63–70. doi: 10.1128/aac.33.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Mangold P, Amann S, Akalin E, Ang O, Bal C, Casellas J M. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother. 1996;40:616–620. doi: 10.1128/aac.40.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boissinot M, Levesque R C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC1 beta-lactamase crystal structure. J Biol Chem. 1990;265:1225–1230. [PubMed] [Google Scholar]
- 7.Bonnet R, Sampaio J L M, Labia R, de Champs C, Sirot D, Chanal C, Sirot J. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob Agents Chemother. 2000;44:1936–1942. doi: 10.1128/aac.44.7.1936-1942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouthors A T, Dagoneau-Blanchard N, Naas T, Nordmann P, Jarlier V, Sougakoff W. Role of residues 104, 164, 166, 238 and 240 in the substrate profile of PER-1 beta-lactamase hydrolysing third-generation cephalosporins. Biochem J. 1998;15:1443–1449. doi: 10.1042/bj3301443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouthors A T, Delettré J, Mugnier P, Jarlier V, Sougakoff W. Site-directed mutagenesis of residues 164, 170, 171, 179, 220, 237 and 242 in PER-1 beta-lactamase hydrolysing expanded-spectrum cephalosporins. Protein Eng. 1999;12:313–318. doi: 10.1093/protein/12.4.313. [DOI] [PubMed] [Google Scholar]
- 10.Bradford P A, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1213. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaibi E B, Péduzzi J, Farzaneh S, Barthélémy M, Sirot D, Labia R. Clinical inhibitor-resistant mutants of the beta-lactamase TEM-1 at amino-acid position 69: kinetic analysis and molecular modeling. Biochim Biophys Acta. 1998;1382:38–46. doi: 10.1016/s0167-4838(97)00127-1. [DOI] [PubMed] [Google Scholar]
- 13.Danel F, Hall L M, Gur D, Akalin H E, Livermore D M. Transferable production of PER-1 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:281–294. doi: 10.1093/jac/35.2.281. [DOI] [PubMed] [Google Scholar]
- 14.Dideberg O, Charlier P, Wery J-P, Dehottay P, Dusart J, Erpicum T, Frère J-M, Ghuysen J-M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987;245:911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbarg-Chenon A, Godard V, Labia R, Nicolas J C. Nucleotide sequence of SHV-2 beta-lactamase gene. Antimicrob Agents Chemother. 1990;34:1444–1446. doi: 10.1128/aac.34.7.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazouli M, Legakis N J, Tzouvelekis L S. Effect of substitution of Asn for Arg-276 in the cefotaxime-hydrolyzing class A β-lactamase CTX-M-4. FEMS Microbiol Lett. 1998;169:289–293. doi: 10.1111/j.1574-6968.1998.tb13331.x. [DOI] [PubMed] [Google Scholar]
- 17.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibuka A, Taguchi A, Ishiguro M, Fushinobu S, Ishii Y, Kamitori S, Okuyama K, Yamaguchi K, Konno M, Matsuzawa H. Crystal structure of the E166A mutant of extended-spectrum β-lactamase Toho-1 at 1.8 Å resolution. J Mol Biol. 1999;285:2079–2087. doi: 10.1006/jmbi.1998.2432. [DOI] [PubMed] [Google Scholar]
- 19.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob-Dubuisson F, Lamotte-Brasseur J, Dideberg O, Joris B, Frère J-M. Arginine 220 is a critical residue for the catalytic mechanism of the Streptomyces albus G beta-lactamase. Protein Eng. 1991;4:811–819. doi: 10.1093/protein/4.7.811. [DOI] [PubMed] [Google Scholar]
- 21.Jarlier V, Nicolas M H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 22.Joris B, Ledent P, Dideberg O, Fonze E, Lamotte-Brasseur J, Kelly J A, Ghuysen J M, Frère J M. Comparison of the sequences of class A β-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991;35:2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliebe C, Nies B A, Meyer S F, Tolxdorff-Neutzling R M, Wiedeman B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labia R, Andrillon J, Le Goffic F. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 1973;33:42–44. doi: 10.1016/0014-5793(73)80154-1. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Massidda O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto Y, Inoue M. Characterization of SFO-1, a plasmid-mediated inducible class A beta-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother. 1999;43:307–313. doi: 10.1128/aac.43.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas T, Livermore D M, Nordmann P. Characterization of an LysR family protein, SmeR from Serratia marcescens S6, its effect on expression of the carbapenem-hydrolyzing β-lactamase Sme-1, and comparison of this regulator with other β-lactamase regulators. Antimicrob Agents Chemother. 1995;39:629–637. doi: 10.1128/AAC.39.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naas T, Nordmann P. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc Natl Acad Sci USA. 1994;91:7693–7697. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palucha A, Mikiewicz B, Hryniewicz W, Gniadkowski M. Concurrent outbreaks of extended-spectrum β-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J Antimicrob Chemother. 1999;44:489–499. doi: 10.1093/jac/44.4.489. [DOI] [PubMed] [Google Scholar]
- 34.Péduzzi J, Barthélémy M, Tiwari K, Mattioni D, Labia R. Structural features related to hydrolytic activity against ceftazidime of plasmid-mediated SHV-type CAZ-5 β-lactamase. Antimicrob Agents Chemother. 1989;33:2160–2163. doi: 10.1128/aac.33.12.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Péduzzi J, Farzaneh S, Reynaud A, Barthélémy M, Labia R. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolysing class A beta-lactamase from Serratia fonticola CUV. Biochim Biophys Acta. 1997;1341:58–70. doi: 10.1016/s0167-4838(97)00020-4. [DOI] [PubMed] [Google Scholar]
- 36.Péduzzi J, Reynaud A, Baron P, Barthélémy M, Labia R. Chromosomally encoded cephalosporin-hydrolysing β-lactamase of Proteus vulgaris R0104 belongs to Ambler's class A. Biochim Biophys Acta. 1994;1207:31–39. doi: 10.1016/0167-4838(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 37.Perilli M G, Franceschini N, Segatore B, Amicosante G, Oratore A, Duez C, Joris B, Frère J M. Cloning and nucleotide sequencing of the gene encoding the beta-lactamase from Citrobacter diversus. FEMS Microbiol Lett. 1991;67:79–84. doi: 10.1016/0378-1097(91)90448-j. [DOI] [PubMed] [Google Scholar]
- 38.Petit A, Maveyraud L, Lenfant F, Samama J P, Labia R, Masson J M. Multiple substitutions at position 104 of β-lactamase TEM-1: assessing the role of this residue in substrate specificity. Biochem J. 1995;305:33–40. doi: 10.1042/bj3050033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynaud A, Péduzzi J, Barthélémy M, Labia R. Cefotaxime-hydrolysing activity of the β-lactamase of Klebsiella oxytoca D488 could be related to a threonine residue at position 140. FEMS Microbiol Lett. 1991;81:185–192. doi: 10.1016/0378-1097(91)90301-p. [DOI] [PubMed] [Google Scholar]
- 42.Rogers M B, Parker A C, Smith C J. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob Agents Chemother. 1993;37:2391–2400. doi: 10.1128/aac.37.11.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossolini G M, Franceschini N, Lauretti L, Caravelli B, Riccio M L, Galleni M, Frère J-M, Amicosante G. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaCME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob Agents Chemother. 1999;43:2193–2199. doi: 10.1128/aac.43.9.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Seoane A, Francia M V, Garcia Lobo J-M. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother. 1992;36:1049–1052. doi: 10.1128/aac.36.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva J, Aguilard C, Ayala G, Estrada M A, Garza-Ramos U, Lara-Lemus R, Ledezma L. TLA-1: a new plasmid-mediated extended-spectrum β-lactamase from Escherichia coli. Antimicrob Agents Chemother. 2000;44:997–1003. doi: 10.1128/aac.44.4.997-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirot D, Sirot J, Labia R, Morand A, Courvalin P, Darfeuille-Michaud A, Perroux R, Cluzel R. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel β-lactamase. J Antimicrob Chemother. 1987;20:323–334. doi: 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- 48.Smith C J, Bennett T K, Parker A C. Molecular and genetic analysis of the Bacteroides uniformis cephalosporinase gene, cblA, encoding the species-specific β-lactamase. Antimicrob Agents Chemother. 1994;38:1711–1715. doi: 10.1128/aac.38.8.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sougakoff W, Goussard S, Courvalin P. The TEM-3 β-lactamase, which hydrolyzes broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino acid substitutions. FEMS Microbiol Lett. 1988;56:343–348. [Google Scholar]
- 50.Swaren P, Maveyraud L, Raquet X, Cabantous S, Duez C, Pedelacq J D, Mariotte-Boyer S, Mourey L, Labia R, Nicolas-Chanoine M H, Nordmann P, Frère J-M, Samama J P. X-ray analysis of the NMC-A β-lactamase at 1.64-Å resolution, a class A carbapenemase with broad substrate specificity. J Biol Chem. 1998;273:26714–26721. doi: 10.1074/jbc.273.41.26714. [DOI] [PubMed] [Google Scholar]
- 51.Tassios P T, Gazouli M, Tzelepi E, Milch H, Kozlova N, Sidorenko S, Legakis N J, Tzouvelekis L S. Spread of a Salmonella typhimurium clone resistant to expanded-spectrum cephalosporins in three European countries. J Clin Microbiol. 1999;37:3774–3777. doi: 10.1128/jcm.37.11.3774-3777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trepanier S, Prince A, Huletsky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997;41:2399–2405. doi: 10.1128/aac.41.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]