Abstract
Stenotrophomonas maltophilia is a nosocomial bacterial pathogen intrinsically resistant to several antibiotics. The mechanisms involved in this intrinsic multiresistance phenotype are poorly understood. A library of chromosomal DNA from a spontaneous multidrug-resistant S. maltophilia D457R mutant (A. Alonso and J. L. Martinez, Antimicrob. Agents Chemother. 41:1140–1142, 1997) was screened for complementation of erythromycin susceptibility on an antibiotic-hypersusceptible Escherichia coli ΔacrAB strain. Cloning and further analysis revealed that a 6-kbp region constituting a transcriptional unit was capable of complementing the antibiotic-susceptible phenotype of an E. coli ΔacrAB strain. We identified three open reading frames, smeD, smeE and smeF, which code for members of the membrane fusion protein, resistance nodulation division, and outer membrane factor families, respectively. Drug susceptibility assays indicated that the SmeDEF system cloned in E. coli mediates resistance to a wide range of antibiotics. Ethidium bromide and norfloxacin accumulation experiments in the presence and in the absence of carbonyl cyanide m-chlorophenylhydrazone showed that this system constitutes a drug efflux pump dependent on the membrane proton motive force. The presence of high levels of smeDEF mRNA in the multiresistant D457R mutant was consistent with the high levels of SmeF (formerly Omp54) observed in the same strain. In contrast, transcription levels of smeDEF in the D457 strain were tiny, which correlates with the low levels of SmeF observed for this strain. Also, for both the D457 and D457R strains, we observed growth phase-dependent regulation in which the highest level of transcription corresponded to early exponential phase, with transcription decreasing throughout the growth curve to undetectable levels at 24 h.
In the last decade, the gram-negative bacterium Stenotrophomonas maltophilia has emerged as a relevant nosocomial pathogen, usually associated with infections of immunocompromised patients. Although, the mechanisms involved in the virulence of S. maltophilia are poorly understood, this bacterium has been reported to be associated with bacteremia, endocarditis, infection of the respiratory and urinary tracts, meningitis, and ocular and gastrointestinal infections (for a review, see reference 10).
Infections caused by S. maltophilia are difficult to treat due to the intrinsic antibiotic resistance displayed by this bacterium (17, 40). Indeed, selective media developed to isolate S. maltophilia from clinical and environmental samples include antimicrobial agents (20). Two β-lactamases, L1 metallo- and L2 serine-β-lactamases, which allow many S. maltophilia isolates to be resistant both to β-lactams and combinations of β-lactams and β-lactamase inhibitors (46), have been characterized (47, 48). Quinolone resistance has been increasingly reported (46), and aminoglycoside-inactivating activity has been demonstrated for some isolates (22). The presence of a gene encoding the synthesis of a macrolide phosphotransferase with a gram-positive origin in the S. maltophilia D457R mutant has also been reported (3). Quite recently, one aminoglycoside, acetyl transferase, which is ubiquitously present in all S. maltophilia isolates and thus might contribute to the natural low susceptibility of S. maltophilia to aminoglycosides has been described (25). It should be noted that the low susceptibility showed by S. maltophilia strains can be considered an important virulence factor in patients under antibiotic treatment. In fact, prior exposure of the patient to antibiotics has been identified as an important risk factor associated with S. maltophilia infection or colonization (9, 10).
It has been elucidated that, although the low permeability of the outer membranes of gram-negative bacteria contributes to the intrinsic low susceptibilities of these microorganisms to some antibiotics, there should be other mechanisms that, synergically with this reduced permeability, produce significant levels of resistance (28, 35, 36). Indeed, multidrug resistance (MDR) efflux pumps together with the outer membrane barrier have been identified as the major mechanism of broad antibiotic resistance in Pseudomonas aeruginosa (26). MDR efflux pumps have been characterized for P. aeruginosa (24, 31, 38, 39) and Escherichia coli (23, 27), among others. Indeed, those determinants are probably found in most, if not all, bacterial species (42). With this point of view in mind, we speculated that S. maltophilia's typical MDR phenotype could be explained, at least in part, by the presence of such systems in the genome of this bacterial species. In fact, single-step spontaneous MDR mutants are easily selectable from clinical isolates of S. maltophilia upon incubation with antibiotics (2, 49). Furthermore, analysis of some of these mutants has demonstrated that they express outer membrane proteins (OMPs) immunologically related to OMPs involved in MDR in P. aeruginosa (49).
In this report, we describe the cloning and the characterization for the first time of an MDR efflux pump of S. maltophilia. Like other well-characterized gram-negative MDR determinants, the pump is composed of a membrane fusion protein, an energy-dependent transporter, and one OMP. Screening of the EMBL database yielded smeRSABC (accession no. AF173226) from S. maltophilia (in which “sme” stands for Stenotrophomonas multiple efflux), which is not associated with the multiresistance phenotype (L. Zhang, X. Li, and K. Poole, Pseudomonas '99: biotechnology and pathogenesis, abstr. 22, 1999). Indeed, sequence analysis revealed that the components of the efflux pumps of both systems differed substantially. To comply with current nomenclature, we have named the new system described in this article the smeDEF system.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids employed in this work are listed in Table 1. The S. maltophilia D457R strain is a spontaneous single-step multiresistance mutant (2) of the D457 clinical isolate that overexpresses the OMP SmeF (formerly Omp54). E. coli KZM120 contains an insertion in the efflux pump determinant acrAB (ΔacrAB::Tn903 Kanr) which renders the strain drug hypersusceptible and was a kind gift from Dzwokai Ma. E. coli AA68, a spontaneous rifampin-resistant clone, was obtained by plating E. coli KZM120 in Luria-Bertani (LB) medium containing 50 μg of rifampin per ml. E. coli AA81 and E. coli AA72 were obtained by P1 transduction (16) of a phage lysate prepared from KZM120. Cosmid pLAFR3 was a kind gift from Fernando Rojo. Strains were grown in LB medium (4) at 37°C, unless indicated otherwise.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. maltophilia | ||
| D457 strain | Bronchial-aspiration isolate | Laboratory collection 2 |
| D457R strain | D457 derivative strain with the MDR phenotype that overproduces Omp54 | Laboratory collection 2 |
| E. coli | ||
| KZM120 | ΔacrAB::Tn903 Kanr | Dzwokai Ma 29 |
| AA68 | KZM120 Rfr derivative, selected on 50 mg of rifampin/ml | This work |
| LE392 | supE44 supF58 hsdR514 galK2 galT22 metB1 trpR55 lacY1 | Laboratory collection 43 |
| AA81 | LE392 derivative, ΔacrAB::Tn903 Kanr | This work |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | Laboratory collection 43 |
| AA72 | TG1 derivative, ΔacrAB::Tn903 Kanr | This work |
| HB101 | supE44 hsdS20 (rB mB) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Laboratory collection 43 |
| Plasmids | ||
| pLAFR3 | Low-copy-number cloning cosmid, Tcr | Fernando Rojo 45 |
| pAS1 | pLAFR3 derivative that contains a 30-kbp BamHI DNA fragment from S. maltophilia D457R strain | This work |
| pUC19 | High-copy-number cloning vector, Apr | Laboratory collection 43 |
| pCK01 | Low-copy-number cloning vector, Cmr | Amparo Haro 13 |
| pAS2 | pCK01 derivative that contains smeDEF on the 9-kb digestion fragment E2 | This work |
| pRK2013 | KanroriV ColE1 RK2-mob+ RK2-tra+, helper plasmid in triparental matings | Laboratory collection 14 |
Kanr, kanamycin resistance; Rfr, rifampin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
Construction of a cosmid library, cloning procedures, and DNA sequencing.
Chromosomal DNA for library construction was obtained from the S. maltophilia D457R mutant as described previously (5) and partially digested with Bsp1431 (Fermentas). DNA fragments were separated on a 10 to 40% (wt/vol) sucrose gradient, and 0.5-ml aliquots were collected and analyzed electrophoretically on 0.5% agarose gel. Those samples that contained 20- to 25-kbp fragments were pooled, ligated to the alkaline-phosphatase-treated cosmid pLAFR3, linearized with BamHI (Fermentas), and introduced into phage particles by the lambda DNA in vitro packaging module (Amersham). A packaged reaction was used to infect E. coli AA81, and transconjugants complementing susceptibility were selected on media containing tetracycline (13 μg/ml), kanamycin (25 μg/ml), and erythromycin (9 μg/ml). To confirm the resistance phenotype displayed by transconjugants, cosmids were transferred conjugally into E. coli AA68 in the presence of the helper strain E. coli HB101(pRK2013) and plated on the appropriate medium to counterselect both donor and helper strains (8).
Restriction analysis of DNA and subcloning of the desired DNA fragments were performed by conventional methods (43). DNA sequencing was performed by the dideoxy chain termination method (43) with an ABI 373A automatic sequencer either by using the M13 universal primers or by primer walking.
DNA sequences were analyzed for open reading frames (ORFs) with the program CodonPreference from the University of Wisconsin Genetics Computer Group using a codon frequency table derived from highly expressed E. coli genes. Screening of the EMBL database was performed using the BLAST network service of the Swiss Institute of Bioinformatics.
Drug susceptibility measurements.
The MICs of antibiotics were determined with Mueller-Hinton (4) medium for S. maltophilia strains and LB medium for E. coli strains by the E-test (AB Biodisk, Olna, Sweden), according to the manufacturer's instructions. MICs of ethidium bromide were determined by the broth microdilution method (33) in Mueller-Hinton medium.
Protein analysis.
Whole-cell lysates and OMPs, obtained by differential solubilization in Triton X-100 as described previously (15), were analyzed on sodium dodecyl sulfate (SDS)–8% polyacrylamide gels using the Bio-Rad Protean minigel system and stained with GelCode Blue (Pierce). Protein concentration was determined by the bicinchoninic acid protein assay (Pierce), and molecular weight markers were from Bio-Rad.
For Western blot analysis, proteins were transferred to a polyvinylidene fluoride membrane (Millipore) and analyzed with polyclonal antibody raised against Omp54 at a final dilution of 1:2,000 (see below). Horseradish peroxidase-conjugated protein A (Sigma) was used at a final concentration of 0.25 μg/ml, and detection of immunoreactive bands was performed by chemiluminescence as described previously (41).
In gel digestion of proteins and sample preparation for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Digestion of proteins in excised gel plugs (in gel) was performed as described previously (44) with minor modifications. The excised gel plugs were washed in water and acetonitrile prior to reduction with 10 mM dithiothreitol and alkylation with 55 mM iodoacetamide and thereafter dried by vacuum centrifugation. Modified porcine trypsin (10 ng/μl, sequencing grade; Promega, Madison, Wis.) in digestion buffer (50 mM NH4HCO3, 300 ng of CaCl2 per μl) was added to the dry gel pieces, which were incubated on ice for 40 min for reswelling. After the supernatant was removed, 20 to 40 μl of digestion buffer was added and the digestion was continued at 37°C for 18 h.
A 0.5-μl aliquot of the digestion supernatant was deposited onto the stainless steel MALDI probe and allowed to dry at room temperature. Then, 0.5 μl of matrix solution (saturated a-cyano-4-hydroxycinnamic acid in 30% aqueous acetonitrile and 0.1% trifluoroacetic acid) was added and the supernatant was again allowed to dry at room temperature.
Samples were measured on a Reflex III MALDI-TOF mass spectrometer (Bruker-Franzen Analytic GmbH, Bremen, Germany) equipped with the SCOUT source in positive ion reflector mode. The ion acceleration voltage was 20 kV. The equipment was first externally calibrated employing protonated mass signals from a peptide mixture covering the 1,000- to 4,000-m/z range, and thereafter every spectrum was internally calibrated using signals arising from trypsin autoproteolysis.
Production of polyclonal anti-Omp54 antibody.
Electrophoretic bands from an outer membrane preparation of the S. maltophilia D457R mutant corresponding to Omp54 were excised from a preparative SDS-polyacrylamide gel, crushed in the presence of liquid N2 until a fine dust was obtained, and resuspended in 50% phosphate-buffered saline. This solution was mixed in a 1:1 ratio with Freund's adjuvant (complete for the first injection only) (Sigma Chemical Co., St. Louis, Mo.) immediately before injection. One New Zealand White male rabbit weighing 2.5 kg was injected intramuscularly and subcutaneously with a total of 100 μg of Omp54 protein, and the rabbit was boosted by subcutaneous injection four times at 2-week intervals. Preimmune serum was obtained from the central ear artery prior to the first injection, and total serum was obtained after euthanasia of the rabbit. Western blot analysis to determine the specificity of total serum indicated that, although Omp54 was immunoreactive, the lipopolysaccharides of S. maltophilia protein preparations were detected by the serum, giving a high background. To improve the specificity, serum was incubated with OMPs of the S. maltophilia D457 strain for 48 h at 4°C; the outer membrane fraction was then pelleted by centrifugation at 40,000 × g for 1 h at 10°C, and the supernatant containing the antibody was recovered. After verification of loss of specificity to lipopolysaccharide and retention of immunodetection of Omp54, the supernatant was used in further Western blot analysis.
Drug accumulation assays.
The intracellular accumulation of norfloxacin (6) and ethidium bromide (34) in E. coli strains was analyzed by fluorometric methods as described previously. Briefly, mid-logarithmic cells were recovered after 10 min of centrifugation at 4,000 × g at 4°C, washed, and concentrated sixfold in 50 mM NaPO4 (pH 7.0)–1 mM MgSO4–0.2% glucose. The suspension was incubated for 10 min at 37°C before we proceeded with the accumulation assays. Ethidium bromide was added to the suspension to a final concentration of 10 μg/ml, and accumulation was recorded continuously by change of fluorescence (λexcite, 530 nm; λemit, 600 nm) on a Hitachi F-2500 spectrofluorometer. After 5 min, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to a final concentration of 100 μM and fluorescence was recorded for another 5 min. Norfloxacin accumulation experiments were performed by adding to the cellular suspensions quinolone to a final concentration of 10 μg/ml. After 10 min of incubation at 37°C, the suspension was divided in halves and CCCP was added to one of them to reach a final concentration of 100 μM. After another 10 min of incubation, 0.5-ml triplicate aliquots were recovered from each bacterial suspension and treated as described previously (6) and fluorescence (λexcite, 281 nm; λemit, 600 and 440 nm) was measured. The amount of quinolone accumulated was determined by comparison with the fluorescence shown by known concentrations of norfloxacin standards. Accumulation was compared with protein concentration in each sample.
RNA analysis.
Total RNA from the S. maltophilia D457 and D457R strains was obtained using guanidine thiocyanate-based Tri Reagent-LS (Molecular Research Center Inc.) according to the manufacturer's instructions. Residual DNA was removed by treatment with RNase-free DNase I (Boehringer Mannheim) at 37°C for 15 min. The reaction mixture was extracted twice with acid phenol, and RNA was precipitated with ethanol and dissolved in water. The RNA concentration and purity were estimated by measuring UV absorption at A260 and A280 (43).
For Northern blot analysis, 25 μg of total RNA was electrophoresed on 1% agarose under denaturing conditions (formaldehyde-formamide procedure [43]) and transferred to Hybond-N (Amersham) according to the manufacturer's instructions. RNA molecular weight markers were from Boehringer Mannheim. The membrane was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) to verify that RNA levels in each lane were comparable (data not shown). Membranes were subjected to overnight hybridization and subsequent washings under stringent conditions at 55°C with an smeD probe. The smeD (150-bp product) probe was prepared by PCR amplification of pAS1 using primer 1 (5′-CCAAGAGCCTTTCCGTCAT-3′) and primer 2 (5′-TCTCGGACTTCAGCGTGAC-3′). The reaction mixture (50 μl) contained 0.2 mM (each) dCTP, dTTP, and dGTP, 0.32 mM [32P]dATP (50 μCi), 0.5 μM each primer, 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 100 ng of pAS1, and 1.0 U of Taq DNA polymerase. The mixture was heated for 90 s at 94°C, followed by 35 cycles of 30 s at 94°C, 60 s at 58°C, a 90-s extension step at 72°C, and, finally, one 10-min extension cycle at 72°C before the end of the reaction. The obtained PCR product was purified with Micro Bio-Spin chromatography columns (Bio-Rad), according to the manufacturer's instructions and added to the hybridization buffer at a final concentration of 106 cpm/ml.
Nucleotide sequence accession number.
The nucleotide sequence of smeDEF was submitted to the EMBL database under accession number AJ252200.
RESULTS
Cloning of the smeDEF operon.
In a previous work (2), we analyzed a spontaneous MDR mutant (D457R mutant) obtained from the D457 S. maltophilia clinical isolate. Resistance to erythromycin in the MDR D457R strain is increased compared to that of the parental D457 strain (Table 2); therefore, selection with this antibiotic should allow growth of clones harboring a gene(s) that complements the susceptible background of E. coli AA81. This strain was obtained by P1 transduction of the acrAB deletion from E. coli KZM120 as described in Materials and Methods, and it is hypersusceptible to several antibiotics (data not shown) because it lacks acrAB, the major MDR determinant from E. coli. A pLAFR3-based cosmid library of the S. maltophilia D457R mutant was thus constructed in E. coli AA81, and colonies able to grow on erythromycin were selected. Although the number of transformants plated was high (approximately 104 colonies), only one colony grew up on the selective medium. To confirm that resistance was due to the cosmid (pAS1), this extrachromosomal element was conjugally transferred to the hypersusceptible strain E. coli AA68 (which also lacks acrAB [Table 1]). Introduction of pAS1 in E. coli AA68 conferred resistance not only to erythromycin but also to other nonrelated antibiotics (Table 2), indicating that an MDR determinant is encoded by the S. maltophilia DNA fragment present in this cosmid. Increases in MICs for E. coli AA68(pAS1) compared with those for the control strain E. coli AA68(pLAFR3) reached ratios of more than 256 times for erythromycin, 16 times for chloramphenicol, and 6 times for members of the quinolone family. No significant changes were observed in the MICs of amikacin and β-lactams.
TABLE 2.
Antibiotic susceptibilities of S. maltophilia and E. coli strains expressing or not expressing the MDR determinant SmeDEF
| Strain | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TC | CM | AK | IMP | TICAR-CLV | ERY | NDX | NFX | OFX | EtBr | |
| S. maltophilia | ||||||||||
| D457 strain | 6 | 1.5 | 24 | >32 | 4 | 48–64 | 8 | 6 | 3 | 500 |
| D457R strain | 16 | 12 | 16 | >32 | 6 | >256 | 128 | 64 | >32 | 500 |
| E. coli | ||||||||||
| AA68(pLAFR3) | ND | 0.38 | 0.50 | 0.125 | 0.50 | 1.0 | 1.0 | <0.016 | 0.008 | 4 |
| AA68(pAS1) | ND | 6 | 0.50 | 0.125 | 0.38 | >256 | 6 | 0.032 | 0.047 | 125 |
| AA72(pCK01) | 0.38 | ND | 0.19 | 0.094 | 1.0 | 1.5–2 | 1.0 | <0.016 | 0.012 | 4 |
| AA72(pAS2) | 1.0 | ND | 0.19 | 0.19 | 0.75 | 32–48 | 3.0 | 0.032 | 0.064 | 125 |
TC, tetracycline; CM, chloramphenicol; AK, amikacin; IMP, imipenem; TICAR-CLV, ticarcillin-clavulanic acid; ERY, erythromycin; NDX, nalidixic acid; NFX, norfloxacin; OFX, ofloxacin; EtBr, ethidium bromide; ND, not done.
Sequence analysis of smeDEF.
Digestion of pAS1 with EcoRI and HindIII revealed that the cosmid pAS1 contains an insert of approximately 30 kbp. Several different restriction fragments from this insert were subcloned into pUC19 and partially sequenced using the forward and reverse M13 universal primers. DNA sequence from one of the ends of fragment E2 (approximately 9 kb in size) showed homology to several members of the membrane fusion protein family. A 6-kbp region was sequenced by primer walking from this DNA fragment. Three ORFs were identified. The first ORF (smeD) spanned nucleotides 82 to 1,266 of E2 and encodes a protein of 394 amino acids with a predicted molecular mass of 40,918 Da. The second ORF (smeE, nucleotides 1,279 to 4,401) encodes a protein of 1,040 amino acids with a predicted molecular mass of 111,311 Da. The third ORF (smeF, nucleotides 4,495 to 5,895) encodes a protein of 466 amino acids with a predicted molecular mass of 50,028 Da. Pairwise analysis of amino acidic alignments of each of the three ORFs to proteins in the EMBL database revealed homology to several components of efflux pumps from gram-negative bacteria. SmeD showed homology to members of the membrane fusion protein family (37), in which highest similarity was to the E. coli MDR determinants AcrA (29) and AcrE (23). Both proteins were 48% identical to SmeD. Interestingly, the level of similarity to SmeA from S. maltophilia (accession number AF173226) was lower and the identity to SmeD was 41%. A putative lipoprotein modification site (SLAIAATUAAC) was identified beginning from amino acid 12 at the N terminus of SmeD.
The second ORF, SmeE, showed homology to several proteins of the root nodulation and division (RND) family. AcrB (29) and AcrF (23) from E. coli had the highest similarities, being 61 and 58% identical to SmeE, respectively. SmeB (accession number AF173226) from S. maltophilia was 51% identical to SmeE. Analysis of transmembrane-spanning (TMS) regions revealed that SmeE has 12 predicted TMS regions, and as with other RND members (37), two conserved periplasmic loops were identified between TMS regions 1 and 2 and between TMS regions 7 and 8.
The third ORF, SmeF, showed homology to several OMPs. Highest similarity was found with SmeC (accession number AF173226) from S. maltophilia, which was 42% identical to SmeF. A putative lipoprotein modification site (SIAATLALAGC) beginning at amino acid 14 at the N terminus of SmeF was identified. It has been recently described that in vitro-obtained S. maltophilia MDR mutants might overexpress an OMP (SmeM) putatively involved in MDR and immunologically similar to OprM from P. aeruginosa (49). Comparison of the available sequence of an internal peptide of SmeM with the deduced sequences of SmeF and other OMPs involved in MDR systems indicated that SmeM and SmeF are not the same protein (Fig. 1), although they share several conserved amino acids in this region.
FIG. 1.
Alignment of a conserved region of SmeF with the same region in the S. maltophilia OMP SmeM and other efflux OMPs (OprM and OprJ from P. aeruginosa and SprC from Pseudomonas putida). Residues conserved in all proteins are indicated with an asterisk, and residues conserved in four proteins are indicated by a dot. The numbers at the right indicate the positions of the amino acid sequences in the proteins.
Omp54 and SmeF expression.
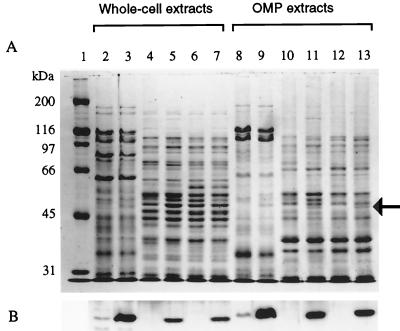
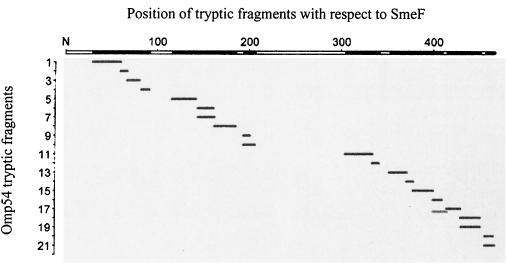
The S. maltophilia D457R mutant overexpresses an OMP (Omp54) which is diagnostic for MDR and is present in clinical isolates of S. maltophilia showing an antibiotic MDR phenotype (2). To know whether the SmeDEF determinant was related to Omp54, a polyclonal antibody was raised as described in Materials and Methods against this OMP. Whole-cell lysates and OMPs of the S. maltophilia D457 and D457R strains and of E. coli AA81, AA81(pAS1), AA68, and AA68(pAS1) were obtained and analyzed on SDS-8% polyacrylamide gels and blotted for immunodetection with the polyclonal antibody. The outer membrane fraction of both E. coli strains harboring pAS1 contained a new protein, which was not detected in the control strains (Fig. 2A). This protein was of the same size as Omp54, and Western blot analysis revealed that it was immunoreactive with the anti-Omp54 antibody (Fig. 2B). The immunoreactive band was also detectable by Western blot analysis of the whole-cell lysates of both E. coli strains harboring pAS1 but not of the parental strains. Together, these data strongly suggest that the Omp54 overexpressed in the D457R mutant and that SmeF are the same protein. Further confirmation of the identity of the protein was obtained by mass spectrometry analysis of the fragments generated with trypsin of the Omp54 band excised from an SDS-polyacrylamide gel. The experimental masses of 21 obtained fragments, spanning all along the protein, fit exactly (differences of less than 0.08 Da) with those predicted from the analysis of the smeF sequence (Fig. 3), indicating that SmeF and Omp54 are indeed the same protein. Three predicted tryptic fragments were absent in the mass spectrometry analysis. One, spanning amino acids 243 to 305, has a mass of 6,365.45 Da and probably was not efficiently transferred due to its size. The other two fragments were located at the N terminus of the SmeF protein, one from amino acids 1 to 7 (size, 846.43 Da) and another from amino acids 8 to 30 (size, 2263.21 Da). In this case, the lack of these low-molecular-size peptides strongly suggests that SmeF is a processed protein, a feature common to other OMPs.
FIG. 2.
Analysis of SmeF expression by bacterial strains containing the smeDEF operon. (A) Protein profiles obtained by SDS-8% polyacrylamide gel electrophoresis of protein extracts from S. maltophilia and E. coli strains either expressing or not expressing or not expressing smeDEF. (B) Results of Western blot analysis of the same samples using an anti-SmeF antibody. In all cases, samples contained 5 μg of protein. Lanes 2 to 7, whole-cell protein extracts; lanes 8 to 13, OMP fractions. Slight differences in protein mobilities were observed between whole-cell extracts and OMP fractions. Lane 1, molecular mass standards; lanes 2 and 8, S. maltophilia D457 strain; lanes 3 and 9, S. maltophilia D457R strain; lanes 4 and 10, E. coli AA81; lanes 5 and 11, E. coli AA81(pAS1); lanes 6 and 12, E. coli AA68; lanes 7 and 13, E. coli AA68(pAS1). The arrow shows the position of SmeF. Note that the protein is expressed only in E. coli strains containing smeDEF genes. Also, SmeF is overexpressed in the MDR S. maltophilia D457R mutant and is detectable, although at a low level, in the wild-type D457 strain.
FIG. 3.
Mass spectrometry analysis of tryptic fragments obtained from Omp54. The masses of the fragments obtained after in gel tryptic digestion of Omp54 were compared with those deduced from the smeF sequence. Black rectangles on the x axis indicate that the experimentally determined masses and the deduced masses were identical within an absolute error of less than 0.08 Da.
SmeDEF is an efflux pump upon expression in E. coli.
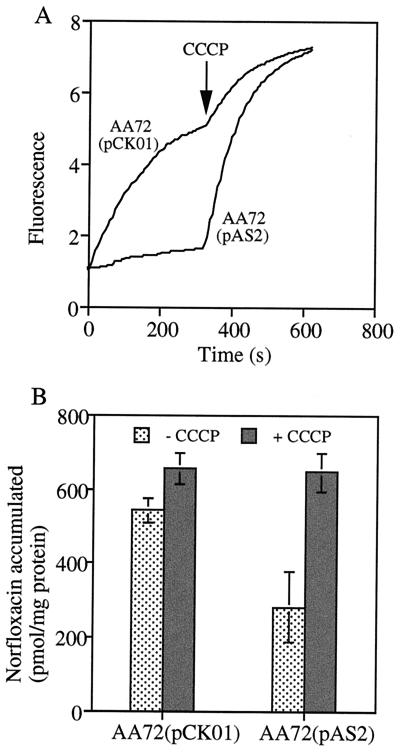
To determine whether the smeDEF operon indeed encodes an efflux pump, the E2 fragment from pAS1, which contains the whole smeDEF operon, was subcloned downstream from the lac promoter into the low-copy-number vector pCK01. The obtained recombinant plasmid, hereafter named pAS2, was introduced in the ΔacrAB E. coli strain AA72. Western blot analysis of total cell extract with polyclonal anti-SmeF antibody confirmed that SmeF is expressed in E. coli AA72 (data not shown) even without IPTG (isopropyl-β-d-thiogalactopyranoside) induction. Analysis of MICs of several unrelated antibiotics (Table 1) demonstrated that AA72 harboring pAS2 rendered a multiple-antibiotic resistance phenotype. To determine if reduced susceptibility could be explained by impaired uptake of such drugs, intracellular accumulation of ethidium bromide and norfloxacin were performed in the absence and in the presence of the proton uncoupler CCCP. The intracellular accumulations of ethidium bromide (Fig. 4A) and norfloxacin (Fig. 4B) were reduced, respectively, 3.1- and 1.9-fold in the smeDEF-expressing strain E. coli AA72(pAS2) compared with levels in the control strain E. coli AA72(pCK01). Treatment with CCCP increased the accumulation of norfloxacin and ethidium bromide, reaching the same level in the smeDEF-expressing strain E. coli AA72(pAS2) as in the controls after 5 min of incubation with the uncoupler agent. These results indicate that smeDEF is an efflux pump determinant whose activity is linked to the membrane potential. The changes in the MICs for E. coli AA72(pAS2), which expresses smeDEF, upon comparison with those for the parental strain E. coli AA72(pCK01) indicate that the range of antibiotics for which this pump is active includes tetracycline, erythromycin, and the quinolone family of antibiotics, but it seems that it is not effective in extruding amikacin or β-lactams.
FIG. 4.
Intracellular accumulation of drugs by E. coli strains containing or not containing smeDEF. (A) Accumulation of ethidium bromide; (B) accumulation of norfloxacin. In both cases, the proton uncoupler CCCP was added at a final concentration of 100 μM. Note that the accumulation of both drugs is much lower for the E. coli strain AA72(pAS2) encoding smeDEF than for the control strain AA72(pCK01). Accumulation is restored to reach the same level in both strains after treatment with CCCP. The increased accumulation of the control strain AA72(pCK01) in the presence of CCCP is probably the consequence of the activities of endogenous E. coli pumps other than acrAB and that mediating ethidium bromide efflux.
Growth-phase regulation of SmeDEF expression in wild-type and MDR S. maltophilia mutants.
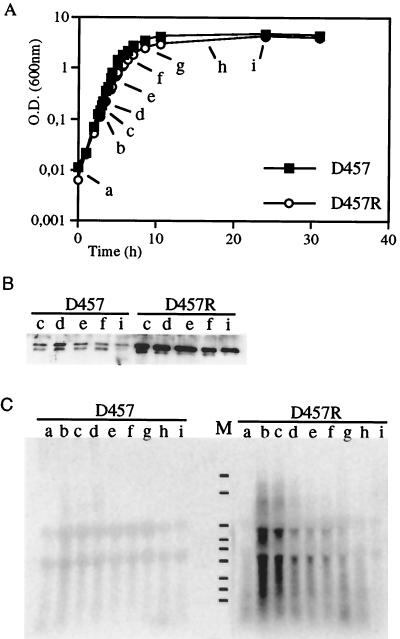
In order to know if the expression of smeDEF is increased in the S. maltophilia D457R mutant compared to that in the parental D457 strain, both strains were grown in liquid LB medium at 37°C and samples for protein and RNA analysis were withdrawn at several points throughout the growth curve (Fig. 5A). Western blot analysis revealed that expression of SmeF by the wild-type D457 strain was low but that this protein was heavily expressed in the D457R strain (Fig. 5B). Levels of SmeF seemed to be constant throughout growth, although a small reduction in the amount of the protein was detectable at 24 h. These data indicate either that the expression of this OMP is constant throughout the cell cycle or that it is very stable.
FIG. 5.
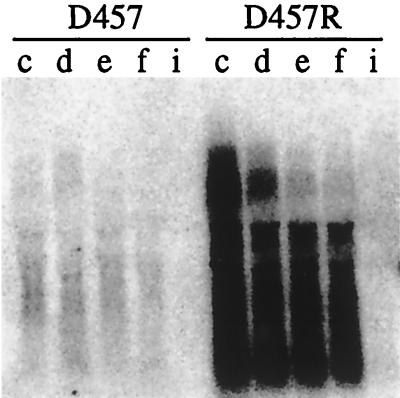
Growth-dependent analysis of smeDEF expression in S. maltophilia D457 and D457R strains. (A) Samples were withdrawn throughout the growth curve. O.D., optical density. (B) Results of Western blot analysis using an anti-SmeF antibody of protein extracts obtained along the growth cycle either from the wild-type S. maltophilia D457 strain or from the D457R mutant. Note that SmeF protein is much more highly expressed in the D457R mutant than in the D457 strain and that expression of this protein is nearly constant throughout the cell cycle, with a small reduction at the stationary phase of growth. (C) Results of a Northern blot analysis, using an smeD probe, of RNAs obtained at different points along the growth curve of the S. maltophilia D457 and S. maltophilia D457R strains. Lane M, RNA molecular size markers (from top to bottom: 6.9, 4.7, 2.7, 1.8, 1.5, 1.0, 0.6, 0.4, and 0.3 kb). Notice the strong induction of smeDEF in the D457R mutant strain at early exponential phase. Also, the low levels of smeDEF transcripts observed for the wild-type D457 strain are remarkable.
In order to analyze smeDEF mRNA transcripts, Northern blot analysis was performed using an smeD probe as described in Materials and Methods. A transcript whose size (5.8 kb) is consistent with the dimension of the entire smeDEF operon was detected (Fig. 5C) in samples obtained from the S. maltophilia D457R mutant. Some other major transcripts with smaller molecular sizes were also detected, indicating either the processing of smeDEF mRNA or the degradation of larger mRNA species. Interestingly, the levels of transcripts in D457R samples revealed a growth-dependent regulation in such a way that highest transcription corresponded to early exponential phase and decreased gradually throughout the growth curve, reaching undetectable levels at 24 h. In contrast, transcription levels of smeDEF mRNA in D457 samples were very low throughout the growth curve. This low basal level of transcription is consistent with the low levels of the SmeF protein in D457 samples (Fig. 5B). The increased expression of both smeDEF RNA and SmeF protein in the S. maltophilia D457R mutant compared with levels in the D457 parental strain strongly suggests that SmeDEF is the MDR determinant, the expression of which is increased in the previously analyzed D457R mutant (2).
The growth-dependent regulation of smeDEF transcripts, observed in D457R samples, was not clearly detectable in D457 samples as a consequence of some unspecific hybridization with rRNAs. We thus decided to carry out another Northern blot analysis, in which extensive washes to remove the smeD probe in order to avoid the background and extended exposure times might allow the analysis of smeDEF expression in the wild-type D457 strain. Figure 6 shows the results of this analysis. Although the levels of the transcripts cannot be quantified for the D457R mutant because the autoradiogram is overexposed, it is clear that the same growth-dependent regulation of smeDEF was detectable for both the D457R and D457 samples. These data strongly suggest that expression of the smeDEF system is regulated at at least two levels. One level is skipped in the D457R mutant, and its loss allows the increased expression of smeDEF in this mutant strain compared with its expression in the D457 wild-type strain. The other level is the growth-dependent regulation of smeDEF expression, which is detectable in both wild-type and MDR mutant strains.
FIG. 6.
Growth-dependent analysis of smeDEF expression in the S. maltophilia D457 strain. Due to the low level of smeDEF transcripts observed for the S. maltophilia D457 strain in previous experiments (Fig. 3), a new Northern blot analysis of samples obtained throughout the growth curves of both the D457 and D457R strains (Fig. 3A) was performed as described in the text. The growth-phase regulation of the amount of smeDEF RNA in the wild type was similar to that in the MDR mutant.
DISCUSSION
To gain some insight into the mechanisms that allow the reduced susceptibility of S. maltophilia, we decided to study the presence of MDR determinants in this bacterial species. MDR efflux pumps from gram-negative bacteria are composed of three proteins located in the inner membrane, the periplasmic space, and the outer membrane. These proteins form a channel capable of extruding a broad range of substances from inside the bacterial cell through a proton motive force-dependent mechanism (36, 37). Synthesis of MDR determinants is usually down-regulated under standard laboratory conditions (36), so that we decided to clone S. maltophilia MDR determinants from a spontaneous nonrepressed MDR mutant previously obtained in our laboratory (2). To make that, a cosmid-based library was made and expressed in the ΔacrAB E. coli strain AA81. Functional selection for antibiotic-resistant clones allowed the isolation and further sequencing of the first MDR efflux pump determinant (smeDEF) so far characterized for S. maltophilia. The results of Northern and Western blot analyses, together with mass spectrometry data, presented in this work support the identity of Omp54 and SmeF. Another determinant sharing the characteristics of an efflux determinant has also been recently sequenced from S. maltophilia; however, those authors indicate that it is not involved in antibiotic resistance (L. Zhang et al., Pseudomonas '99, abstr. 22).
SmeDEF overexpression increased the MICs of several antibiotics both for S. maltophilia and for the heterologous host E. coli, indicating that it is a broad-range MDR determinant. The fact that its expression in E. coli reduces the accumulation of structurally different compounds by a mechanism that is dependent on bacterial membrane potential indicates that smeDEF encodes all the elements needed for the synthesis of a functionally active MDR efflux pump similar to others so far described. Indeed, the gene organization of the smeDEF operon is similar to that of operons of other efflux systems of gram-negative bacteria (37, 42). Highest homology was found between SmeE and components of the RND family. The membrane fusion proteins of these systems also showed high similarities, although the similarities were lower than for the members of the RND family. The lowest similarities were found between the outer membrane components of efflux pumps; interestingly, SmeF showed the highest homology to SmeC, an OMP from another efflux determinant recently described for S. maltophilia (accession no. AF173226). Protein sequence analysis strongly suggests that SmeD and SmeF display lipid attachment sites at the N terminus; indeed, early attempts at sequencing the N terminus of SmeF failed. Predictions of TMS regions in SmeE were also consistent with structural characteristics of the members of the RND family (37); 12 TMS regions and two external loops situated between TMS regions 1 and 2 and TMS regions 7 and 8 were identified.
Together, these data indicate that SmeDEF is an antibiotic efflux determinant similar to others so far described for gram-negative bacteria (37), which thus might contribute to the intrinsic susceptibility of S. maltophilia to different drugs. A recent work has shown that this bacterial species might have several different MDR determinants, some of which are immunologically related with those previously characterized for P. aeruginosa (49). It is noteworthy that expression of SmeDEF strongly increases the MIC of erythromycin. Erythromycin is commonly used for the treatment of infections by gram-positive bacteria; however, gram-negative organisms are barely susceptible to this antibiotic. It has been speculated that this reduced susceptibility may be due to a reduced permeability of cellular envelopes to erythromycin. However, some MDR determinants from gram-negative bacteria are capable of extruding this antibiotic (1, 11, 32), as occurs with SmeDEF. Searching for inhibitors of those MDR determinants involved in erythromycin extrusion might allow us to increase the susceptibilities of gram-negative bacteria and thus allow us to introduce this antibiotic into the armamentarium for the treatment of gram-negative infections.
Expression of SmeDEF has been analyzed by Northern and Western blotting both for the wild-type D457 strain and for the derepressed D457R mutant. In both cases, a maximum amount of smeDEF was observed at the beginning of the exponential phase and decreased to undetectable levels after 24 h of growth. Western blot analysis demonstrated, however, that the amount of SmeF is nearly constant throughout cell cycle, with a small reduction at 24 h. These data might be explained either by the presence of another internal promoter which drives smeF expression and is not regulated by the cell cycle or because this protein is very stable and thus its amount is maintained, although smeDEF RNA levels decrease.
The regulation of the expression of smeDEF occurs then at two independent levels. First, the system is repressed in the wild-type D457 strain, a repression that is retrieved in the MDR D457R strain. Second, the expression of the system is regulated by growth phase, a regulation that is maintained in both strains. The signals which allow expression of the usually down-regulated MDR systems are poorly understood. Some of these systems are activatable by natural signal molecules like salicylate (7, 30). Induction of an MDR phenotype by toxic substances such as solvents (19, 21) and heavy metals (18) has also been described. Gene fusion experiments have demonstrated that the expression of acrAB from E. coli (29) and mexABOprM from P. aeruginosa (12) is increased by stress conditions and in stationary growth phase. Unlike with this growth-phase regulation, increased expression of smeDEF is observed at early exponential phase whereas expression of the system in stationary phase is nearly null. It is for the first time that this type of regulation is observed for MDR systems. Based on the expression of other determinants (see above), it was speculated that MDR systems, might have a physiological role during stationary-phase stress (29). However, our data do not support such a role for smeDEF. A search of published sequences of bacterial genomes has demonstrated the presence of multiple MDR determinants in bacterial chromosomes (42). This high redundancy probably indicates that they do not share the same physiological function. Hence, it is not strange that their expression responds in different ways to environmental and physiological signals.
ACKNOWLEDGMENTS
Thanks are given to Dzwokai Ma for the gift of E. coli KZM120 and to E. Campanario and A. Varas for technical assistance. E. Camafeita and the Proteomics Facility of the CNB are also acknowledged by their help with the mass spectrometry analysis.
This research was aided in part by grant 08.2/022/98 from CAM. A. Alonso is a recipient of a fellowship from Gobierno Vasco.
REFERENCES
- 1.Aires J R, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Martínez J L. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1140–1142. doi: 10.1128/aac.41.5.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso A, Martínez J L. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob Agents Chemother. 2000;44:1778–1782. doi: 10.1128/aac.44.7.1778-1782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlas R M. Handbook of microbiological media. London, United Kingdom: CRC Press, Inc.; 1993. [Google Scholar]
- 5.Bagdasarian M, Bagdasarian M M. Gene cloning and expression. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 406–417. [Google Scholar]
- 6.Chapman J S, Georgopapadakou N H. Fluorometric assay for fleroxacin uptake by bacterial cells. Antimicrob Agents Chemother. 1989;33:27–29. doi: 10.1128/aac.33.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 9.Denton M, Todd N J, Littlewood J M. Role of anti-pseudomonal antibiotics in the emergence of Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1996;15:402–405. doi: 10.1007/BF01690098. [DOI] [PubMed] [Google Scholar]
- 10.Denton M, Kerr K G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans K, Poole K. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol Lett. 1999;173:35–39. doi: 10.1111/j.1574-6968.1999.tb13481.x. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez S, de Lorenzo V, Perez-Martin J. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 14.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuoka T, Masuda N, Takenouchi T, Sekine N, Iijima M, Ohya S. Increase in susceptibility of Pseudomonas aeruginosa to carbapenem antibiotics in low-amino-acid media. Antimicrob Agents Chemother. 1991;35:529–532. doi: 10.1128/aac.35.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg R B, Benber R A, Steicher S L. Direct selection for P1-sensitive mutants of enteric bacteria. J Bacteriol. 1974;118:810–814. doi: 10.1128/jb.118.3.810-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock R E. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin Infect Dis. 1998;27(Suppl. 1):S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez A, Mellado R P, Martinez J L. Metal accumulation and vanadium-induced multidrug resistance by environmental isolates of Escherichia hermannii and Enterobacter cloacae. Appl Environ Microbiol. 1998;64:4317–4320. doi: 10.1128/aem.64.11.4317-4320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isken S, Santos P M A C, deBont J A M. Effect of solvent adaptation on the antibiotic resistance in Pseudomonas putida S12. Appl Microbiol Biotechnol. 1997;48:642–647. [Google Scholar]
- 20.Juhnke M E, des Jardin E. Selective medium for isolation of Xanthomonas maltophilia from soil and rhizosphere environments. Appl Environ Microbiol. 1989;55:747–750. doi: 10.1128/aem.55.3.747-750.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieboom J, Dennis J J, Zylstra G J, de Bont J A. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J Bacteriol. 1998;180:6769–6772. doi: 10.1128/jb.180.24.6769-6772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King B A, Shannon K P, Phillips I. Aminoglycoside 6′-N acetyltransferase production by an isolate of Pseudomonas maltophilia. J Antimicrob Chemother. 1978;4:467–468. doi: 10.1093/jac/4.5.467-a. [DOI] [PubMed] [Google Scholar]
- 23.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 24.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 25.Lambert T, Ploy M C, Denis F, Courvalin P. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1999;43:2366–2371. doi: 10.1128/aac.43.10.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X Z, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 29.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 30.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 34.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 36.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 39.Poole K, Gotoh N, Tsujimoto H, Zhao Q X, Wada A, Yamasaki T, Neshat S, Yamagishi J I, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 40.Quinn J P. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin Infect Dis. 1998;27:S117–S124. doi: 10.1086/514912. [DOI] [PubMed] [Google Scholar]
- 41.Renart J, Behrens M M, Fernández-Renart M, Martinez J L. Immunoblotting techniques. In: Diamandis E P, Christopoulos T K, editors. Immunoassay. San Diego, Calif: Academic Press, Inc.; 1996. pp. 537–554. [Google Scholar]
- 42.Saier M H, Paulsen I T, Sliwinski M K, Pao S S, Skurray R A, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 45.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vartivarian S, Anaissie E, Bodey G, Sprigg H, Rolston K. A changing pattern of susceptibility of Xanthomonas maltophilia to antimicrobial agents: implications for therapy. Antimicrob Agents Chemother. 1994;38:624–627. doi: 10.1128/aac.38.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh T R, MacGowan A P, Bennett P M. Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1460–1464. doi: 10.1128/aac.41.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, MacGowan A P, Bennett P M. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Li X Z, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother. 2000;44:287–293. doi: 10.1128/aac.44.2.287-293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]