Abstract
The upper respiratory tract is an obliged pathway for respiratory pathogens and a healthy microbiota may support the host's mucosal immunity preventing infection. We analyzed the nasopharyngeal microbiome in tuberculosis household contacts (HHCs) and its association with latent tuberculosis infection (TBI). A prospective cohort of HHCs was established and latent TBI status was assessed by serial interferon-γ release assay (IGRA). Nasopharyngeal swabs collected at baseline were processed for 16S rRNA gene sequencing. The 82 participants included in the analysis were classified as: (a) non-TBI [IGRA negative at baseline and follow-up, no active TB (n = 31)], (b) pre-TBI [IGRA negative at baseline but converted to IGRA positive or developed active TB at follow-up (n = 16)], and (c) TBI [IGRA positive at enrollment (n = 35)]. Predominant phyla were Actinobacteriota, Proteobacteria, Firmicutes and Bacteroidota. TBI group had a lower alpha diversity compared to non-TBI (padj = 0.04) and pre-TBI (padj = 0.04). Only TBI and non-TBI had beta diversity differences (padj = 0.035). Core microbiomes’ had unique genera, and genus showed differential abundance among groups. HHCs with established latent TBI showed reduced nasopharyngeal microbial diversity with distinctive taxonomical composition. Whether a pre-existing microbiome feature favors, are a consequence, or protects against Mycobacterium tuberculosis needs further investigation.
Subject terms: Microbiome, Tuberculosis
Introduction
Tuberculosis (TB) is an airborne infectious disease caused by the pathogenic bacteria Mycobacterium tuberculosis (Mtb). About a quarter of the global population is infected with Mtb1. The susceptibility to acquiring this infection depends on intensity2 and duration of exposure3. Host characteristics such as sex, age, genetics, immune status, nutrition, and environmental and social factors (crowding, poor ventilation, alcohol, smoking, and occupational risk) increase the risk of TB4,5; still, individuals with no known risk factors account for a large number of new cases.
The microbiota inhabiting our body provides a milieu that benefits the entire host-microbe system, influencing host homeostasis and immune response6,7, with each person having distinct microbe communities. A healthy microbiota supports the host’s immunity against infections by promoting mucosal barrier function, competing with pathogens, and priming immune responses. At the same time, its disruption can increase the risk of disease8,9. The upper respiratory tract is colonized by specialized resident bacterial assemblages, presumably preventing potential pathogens from overgrowing and disseminating toward the lungs, thereby functioning as gatekeepers to respiratory health10. The characteristics of the nasal and nasopharyngeal microbiome from infected and uninfected individuals have been shown to differ in upper respiratory tract infections such as sinusitis, otitis media, influenza, RSV, and SARS-CoV-211,12.
There is less evidence about the nasopharyngeal microbiota composition in lower respiratory tract bacterial infections. Two studies reported comparatively lower alpha diversity indices of microbial diversity in children with pneumonia13,14, while another study in adults did not. Nonetheless, the three studies reported that a more diverse microbiota is found in the nasopharynx of healthy individuals13–15. In addition, the experimental nasal inoculation of Streptococcus pneumoniae was shown to induce a shift in the nasopharyngeal microbiota of healthy adults correlating with the establishment of the bacterial carriage16.
In TB pathogenesis, the role of the respiratory tract microbiome remains largely unexplored. Thus, we assessed the nasopharyngeal microbiome in a prospective cohort of individuals exposed to Mtb to ascertain if there are differences in microbiome composition or abundance between individuals that acquired or not a latent TB infection, aiming to gain further insight into TB pathogenesis and susceptibility.
Results
Clinical and demographic data
From the enrolled cohort of 231 HHCs, 155 participants (67%) had complete follow-up and fulfilled inclusion/exclusion criteria for microbiome analysis (Supplementary Fig. S1). As per our classification, 44% were classified as TBI (n = 69), 41% as non-TBI (n = 63), and 15% as pre-TBI (n = 23).
From the 155 samples selected for 16S sequencing, 82 (53%) remained after the ASVs filtering process, with a considerable loss due to the initial low microbial biomass of the nasopharyngeal swabs collected samples. For analysis, 31 (38%) samples pertained to the Mtb uninfected (non-TBI), 16 (19%) from those acquiring a new Mtb infection on follow-up (pre-TBI), and 35 (43%) from HHCs displaying Mtb infection since the baseline evaluation (TBI). The demographic and clinical characteristics of HHCs included in the analysis are summarized in Table 1. Overall, 58.5% were female, the mean age was 34.5 (± 13.7) years, and all were residents in Chile of Hispanic/Latino descent, primarily migrants from Peru (58.5%). Individuals with TBI were slightly older than non-TBI and pre-TBI (p = 0.013). The comparison of the demographic and clinical characteristics of the entire cohort (composed of individuals whose samples were excluded, unsuccessfully sequenced, and successfully sequenced/analyzed) did not reveal substantial differences between groups and is summarized in Supplementary Table S1.
Table 1.
Clinical and epidemiological characterization of TB contacts included in the analysis (n = 82).
| Non-TBI (n = 31) |
Pre-TBI (n = 16*) |
TBI (n = 35†) |
p-value | |
|---|---|---|---|---|
| Female sex (n, %) | 16 (51.6%) | 10 (62.5%) | 22 (62.9%) | 0.611 |
| Mean age, years (SD) | 32.5 (11.8) | 27.9 (12.0) | 39.2 (14.7) | 0.013 |
| Countries, total (n, %) | 0.222 | |||
| Peru | 17 (54.8%) | 9 (56.3%) | 22 (62.9%) | |
| Chile | 9 (29.0%) | 1 (6.3%) | 8 (22.9%) | |
| Other Latin American countries | 5 (16.1%) | 6 (37.5%) | 5 (14.3%) | |
| Tobacco smoker (n, %) | 8 (25.8%) | 3 (18.8%) | 10 (28.6%) | 0.683 |
| Self-reported viral respiratory infections in last month (n, %) | 10 (32.3%) | 5 (31.3%) | 13 (37.1%) | 0.883 |
| Any antibiotic use—Last 6 months (n, %) | 7 (22.6%) | 1 (6.3%) | 5 (14.3%) | 0.386 |
| Probiotic or vitamin use—Last 3 months (n, %) | 7 (22.6%) | 3 (18.8%) | 2 (5.7%) | 0.125 |
| Proton pump inhibitor or antacids—Last 3 months (n, %) | 4 (12.9%) | 0 (0.0%) | 9 (25.7%) | 0.058 |
| Vegan or Vegetarian diet | 1 (3.2%) | 0 (0%) | 0 (0%) | 0.573 |
* n = 3 individuals (18.8%) developed active TB at follow-up. † n = 2 individuals (5.7%) developed active TB at follow-up. Statistical analysis were performed with analysis of variance (ANOVA) for continuous variables, and Fisher’s exact or Chi-squared test for categorical variables. Statistical significance was considered when p-values < 0.05 (two-tailed).
Bacterial relative abundance in the nasopharynx of HHCs
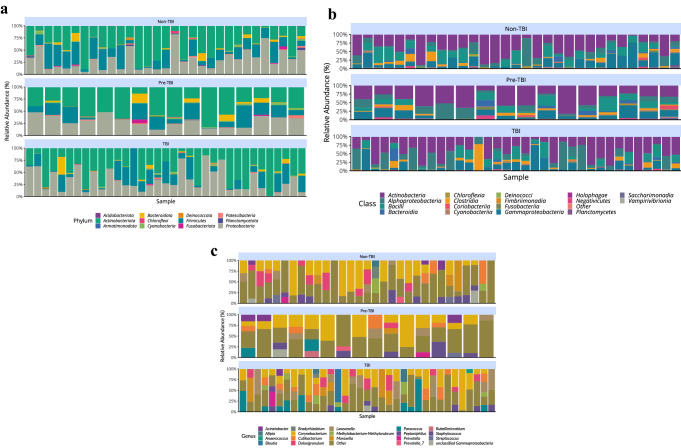
After dataset cleaning and filtering, 841 ASVs remained for evaluation. A descriptive analysis of relative abundance showed that for all HHCs, a total of 14 phyla were found, with the most predominant being: Actinobacteriota (43.8%), Proteobacteria (31.2%), Firmicutes (19.6%), and Bacteroidota (2.5%) (Table 2). There was variability in the predominance of less abundant phylum within each group (Supplementary Table S2) and a high degree of individual variability within groups (Fig. 1a). At the class level, for all HHCs, a total of 19 bacterial classes were found, dominating Actinobacteria (43.4%), Alphaproteobacteria (15.7%), Gammaproteobacteria (15.6%) and Bacilli (13.7%) (Table 3). Class domination varied for less abundant classes within each group (Supplementary Table S3) and a high individual variability within groups was also observed (Fig. 1b). At the genus level, the most frequently found were Corynebacterium (25%), Cutibacterium (7%), Lawsonella (6.9%), Dolosigranulum (6.1%), Paracoccus (5.7%) and Staphylococcus (5.5%). Groups exhibited a high degree of individual variability within each one (Fig. 1c). The Mycobacterium genus was not detected by 16S analysis in any of the analyzed participants, nor the bacteria itself by culture of the contralateral nasopharyngeal swabs.
Table 2.
Percentage distribution at phylum level for all nasopharyngeal samples analyzed after amplicon sequence variant filtering (n = 82).
| Phylum | Average (%) | Standard deviation (%) |
|---|---|---|
| Actinobacteriota | 43.820 | 23.256 |
| Proteobacteria | 31.242 | 20.099 |
| Firmicutes | 19.557 | 17.273 |
| Bacteroidota | 2.530 | 5.351 |
| Cyanobacteria | 1.014 | 1.402 |
| Patescibacteria | 0.753 | 1.092 |
| Fusobacteriota | 0.348 | 1.173 |
| Chloroflexi | 0.257 | 0.686 |
| Deinococcota | 0.206 | 0.557 |
| Planctomycetota | 0.168 | 0.664 |
| Acidobacteriota | 0.082 | 0.198 |
| Armatimonadota | 0.025 | 0.140 |
Figure 1.
Taxonomic classification of the nasopharynx microbiota in TB exposed household contacts (n = 82). Descriptive individual variability of relative abundance according to LTBI uninfected (non-TBI), new LTBI on follow-up (pre-TBI) and LTBI from baseline evaluation (TBI) groups at (a) Phylum, (b) Class and (c) Genus level.
Table 3.
Percentage distribution at class level for all nasopharyngeal samples analyzed after amplicon sequence variant filtering (n = 82).
| Class | Average (%) | Standard deviation (%) |
|---|---|---|
| Actinobacteria | 43.402 | 23.343 |
| Alphaproteobacteria | 15.651 | 19.288 |
| Gammaproteobacteria | 15.590 | 15.856 |
| Bacilli | 13.701 | 13.554 |
| Clostridia | 5.253 | 9.608 |
| Bacteroidia | 2.530 | 5.351 |
| Cyanobacteriia | 0.969 | 1.363 |
| Saccharimonadia | 0.753 | 1.092 |
| Negativicutes | 0.603 | 1.573 |
| Coriobacteriia | 0.394 | 1.471 |
| Fusobacteriia | 0.348 | 1.173 |
| Chloroflexia | 0.257 | 0.686 |
| Deinococci | 0.206 | 0.557 |
| Planctomycetes | 0.168 | 0.664 |
| Holophagae | 0.082 | 0.198 |
| Vampirivibrionia | 0.044 | 0.196 |
| Fimbriimonadia | 0.025 | 0.140 |
| Acidimicrobiia | 0.023 | 0.097 |
In addition, we also compared the relative abundance of the ASVs classified at the species level. Although the length of the analyzed region is too short to distinguish among all present microbial species, some unique matches based on the region analyzed can be distinguished (Supplementary Fig. S2). Among the taxa present, there are some taxa prevalent in a large fraction of the samples in all groups, such as Cutibacterium granulosum (6.1%). Some species were more prevalent (present in at least 50% of the samples) in some of the groups. For example, Dolosigranulum pigrum (7.5%) was present in more than 50% of the samples of the non-TBI group. In contrast, Kocuria rosea (1.1%) was found in more than 50% of the samples of the TBI group (Supplementary Fig. S3).
Bacterial diversity in the nasopharynx of HHCs
Alpha diversity
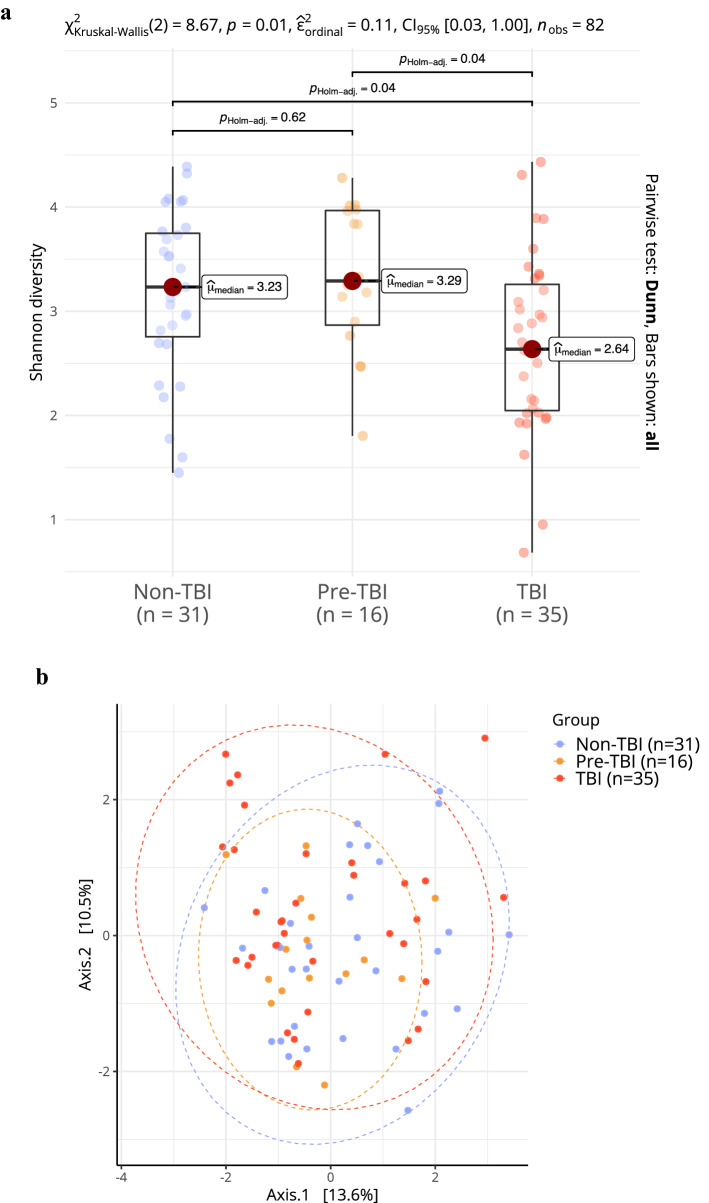
Community diversity composition between HHCs groups was compared using the Shannon diversity index. The comparison showed significant differences between the group medians (non-TBI: 3.23 [2.76–3.75]; pre-TBI: 3.29 [2.87–3.97]; TBI: 2.64 [2.05–3.26], p = 0.01). Posthoc analysis using the Wilcoxon test showed statistical differences between the non-TBI vs. TBI group (padj = 0.04) and the pre-TBI vs. TBI group (padj = 0.04) (Fig. 2a). These results indicate that the microbiome of the participants with a latent TB infection had lower alpha diversity than the non-infected, including those non-infected but subsequently acquiring Mtb on follow-up, suggesting that structural differences may arise after acquiring Mtb.
Figure 2.
Changes in microbial diversity in the nasopharynx microbiota in TB exposed household contacts (n = 82). (a) Alpha diversity in HHCs samples using Shannon index. (b) Principal Coordinate Analysis plot showing the Euclidian distance of the PhilR-transformed data for each of the samples. Differences between group composition were significant (p = 0.036), only the comparison between non-TBI versus TBI (p = 0.035). Statistical analysis for alpha diversity was performed with Kruskal–Wallis test followed by Wilcoxon test with Holm adjustment for multiple testing. For beta diversity, a permutational analysis of variance based on the Euclidean distance matrix was performed.
Beta diversity
The microbial community composition comparison showed significant differences between groups (p = 0.036) (Fig. 2b) that the dispersion of the centroids could not explain within the dataset (p = 0.233). Pairwise comparison indicated only the communities between the non-TBI and the TBI group were different (padj = 0.035), while those of pre-TBI and TBI (padj = 0.095) and non-TBI and pre-TBI (padj = 0.189) were not. Thus, infected and uninfected individuals showed differences in the beta diversity of microbial communities, but the smaller group of those who later acquired the infection did not.
Core microbiome
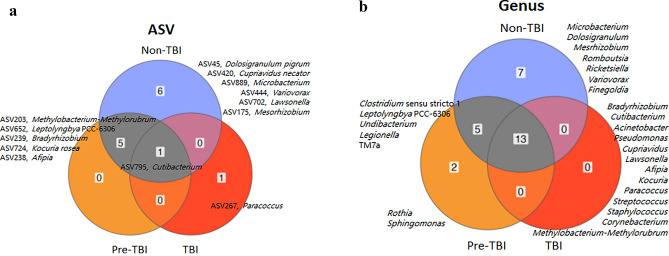
Evaluation of the core microbiome was performed for each HHCs group separately. At the ASV level, only a few ASVs were present in at least 50% of all samples from each group (non-TBI, 12 ASVs; pre-TBI, 6 ASVs; TBI, 2 ASVs). TBI and non-TBI, as well as TBI and pre-TBI, did not share any common ASVs. However, pre-TBI and non-TBI shared five ASVs (Methylobacterium-Methylorubrum, Afipia, Bradyrhizobium, Leptolyngbya PCC-6306, and Kocuria rosea), and TBI, pre-TBI and non-TBI shared only one (Cutibacterium granulosum) (Fig. 3a). Among unique ASVs in each group, six were associated with the non-TBI group (Dolosigranulum pigrum, Mesorhizobium, Cupriavidus necator, Variovorax, Lawsonella and Microbacterium), and 1 with the TBI group (Paracoccus).
Figure 3.
Core microbiome by TB infection status group, showing in each case ASVs or Genus that are present in at least 50% of the individuals within that group. (a) ASVs unique and shared among all groups. Names indicated the ASV number and its best taxonomic classification up to the species level. (b) Genus that are unique and shared between all groups. To identify taxonomic groups with statistically significant differences in abundance between groups was performed with the compositional approach implemented in ANCOMBC.
At the genus level, we identified a core set of genera present in at least 50% of all samples within each group (non-TBI, 25; pre-TBI, 20; TBI, 13). There were 13 genera shared between the three of them (Acinetobacter, Afipia, Bradyrhizobium, Corynebacterium, Cupriavidus, Cutibacterium, Kocuria, Lawsonella, Methylobacterium-Methylorubrum, Paracoccus, Pseudomonas, Staphylococcus and Streptococcus), five between non-TBI and pre-TBI (Clostridium sensu stricto 1, Legionella, Undibacterium, Leptolyngbya PCC-6306, and TM7a), and none between pre-TBI and TBI, and non-TBI and TBI. Additionally, the non-TBI group had seven unique genera (Finegoldia, Romboutsia, Dolosigranulum, Mesrhizobium, Ricketsiella, Variovorax and Microbacterium). In contrast, the pre-TBI had only two (Sphingomonas and Rothia), and the TBI had none (Fig. 3b).
Differentially abundant genera between HHC groups
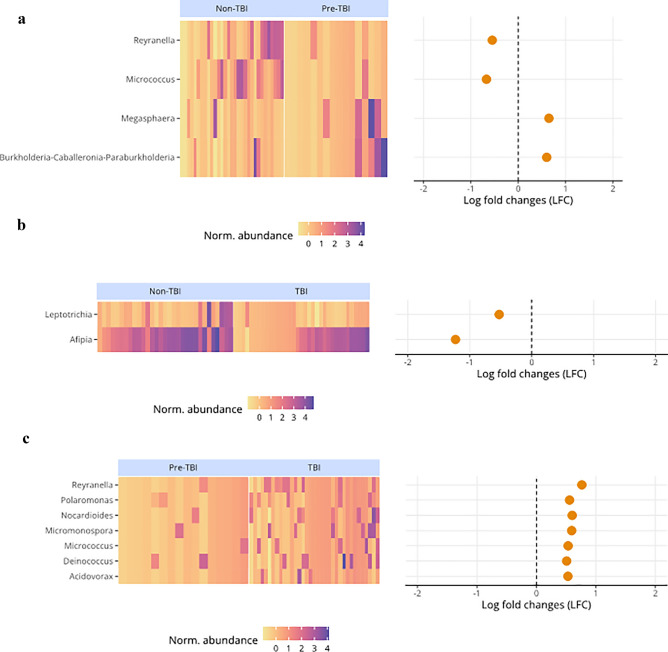
We used compositional-based analysis to evaluate differentially abundant genera between the three HHCs groups. Statistically significant differences (log fold changes) were found for non-TBI compared to pre-TBI, with Reyranella and Micrococcus being more abundant in the non-TBI group. At the same time, Megasphaera and Burkholderia-Caballeronia-Paraburkholderia were more abundant in the pre-TBI group (Fig. 4a). In addition, for non-TBI vs. TBI, Leptotrichia and Afipia were more abundant in the non-TBI group (Fig. 4b). For pre-TBI vs. TBI, seven genera, Reyranella, Polaromonas, Nocardiodes, Micromonospora, Micrococcus, Deinococcus, and Acidivorax, were more abundant in the TBI group (Fig. 4c). Finally, Reyranella was consistently lower in the pre-TBI dataset.
Figure 4.
Genera with statistically significant differential abundance comparing household contacts (n = 82) TB infection status. (a) non-TBI vs. pre-TBI, (b) non-TBI vs. TBI, and (c) pre-TBI vs. TBI.
Discussion
In the present study, we assessed the nasopharyngeal microbiome of a population of HHCs of patients with active TB. We found that individuals with an acquired latent TBI had a lower alpha diversity than uninfected individuals. Our results are consistent with reports of both nasal and oropharynx samples of patients with active PTB disease, who had a less diverse microbiota compared to healthy controls17, as well as the analysis of gut microbiota of children with PTB that reported a decreased microbial diversity18. Nonetheless, the findings observed in microbiota diversity in contacts who were about to acquire the infection remain to be confirmed in a larger cohort. Previous work by our group and others has reported that transient Mtb localization in the upper respiratory tract mucosa can rarely occur after exposure to a PTB case19,20. However, the persistence, replication and direct effect of Mtb in nasopharyngeal mucosa is unlikely, as we did not detect Mtb in the nasopharyngeal samples.
The interaction between intestinal microbial communities and the resident immune cells in the mucosa can lead to modulation of local and systemic immune response; an increased susceptibility to specific respiratory diseases (the “gut–lung axis”) has been proposed to develop when microbial dysbiosis occurs21. In murine models, broad-spectrum antibiotics-driven changes in the gut microbial composition affect the survival, burden, and dissemination of Mtb in the lungs, liver, and spleen compared to the mice with intact microbiota. Furthermore, fecal transplantation succeeds in restraining Mtb growth in the lungs preventing its dissemination22. Of interest, both macaques challenged with Mtb and household contacts having a latent TBI are less likely to progress to active TB when infected with Helicobacter pylori23. Although less explored, the upper and lower respiratory tract may also display immune system/microbiota interactions. In mice models, nasal instillation of Staphylococcus aureus attenuates influenza-mediated lethal inflammation in the lungs by inducing M2 alveolar macrophages24, and Lactobacillus plantarum administration protects against lethal pneumovirus infection in the lungs by TLR2 and NOD2 receptor signaling25.
The present study’s overall analysis of nasopharyngeal microbiota showed that for all HHCs, the predominant phyla were Actinobacteriota, Proteobacteria, Firmicutes and Bacteroidota. These results are consistent with other reports of the nasopharyngeal microbiota, with variations in their relative abundances17,26–31, and with microbiome studies of nasal and oropharynx samples from patients with active TB compared to uninfected individuals17. Interestingly, HHCs not acquiring latent TBI had a higher relative abundance mean (above 5%) of Corynebacterium, Dolosigranulum, Cutibacterium and Staphylococcus, similar to what is found in healthy non-exposed individuals32, while HHCs with an establish LTBI or those about to acquire a latent TBI had a higher relative abundance of Corynebacterium, Lawsonella, Paracoccus, Cutibacterium, Dolosigranulum, Moraxella, and Corynebacterium, Staphylococcus, Cutibacterium, Lawsonella and Paracoccus, respectively.
Our study is among the few that have analyzed the human nasopharyngeal microbiome in the latent TBI33 and, to our knowledge, the first to examine highly exposed HHCs from a prospective cohort acquiring or not Mtb infection at follow-up. However, there are several reports about gut34,35 and lungs’ microbiome characteristics of patients with established active PTB17,36–39, recurrent TB, and TB treatment failure40, with most studies including patients undergoing antimicrobial therapy. Regarding latent TBI, our findings differ from those of a study involving a comparatively small cohort size, in which the nasopharyngeal community diversity and microbial community composition of healthy controls (n = 18) were similar to that of latent TBI individuals (n = 19)33. These dissimilarities could lie in our control group that despite not having acquired the infection had being highly exposed to Mtb. Also, the differences could arise in the smaller number of patients included in that report and in the analytic approach, as the authors sequenced the V3-V4 region of the 16S rRNA gene and evaluated microbial diversity by looking at operational taxonomic units (OTUs) with a 97% or higher level of sequence similarity. In the current study, we sequenced the V1-V3 region which allows better resolution (even at the species level) for samples from human oral and respiratory tracts41, and also used ASVs, which allows for improved discrimination of ecological patterns42. Although not the scope of our study, the same authors found differences at the phylum and class levels between active TB and latent TBI, but not in alpha nor in beta diversity.
The lung microbiome resembles the healthy upper respiratory tract microbiome10, and whether a healthy microbiota in the nasopharynx can prevent Mtb establishment in the lower respiratory tract epithelium is unknown. Our study does not support this, as we did not find significant differences in microbiome composition (beta diversity) and relative abundance between HHCs acquiring a new Mtb infection at follow-up from those who remained uninfected. Notwithstanding, the core microbiome showed that the genus Dolosigranulum was one among which detection was only in the uninfected individuals. Dolosigranulum and its combination with Corynebacterium are a potential keystone species in the upper respiratory tract microbiota. Reports have shown an association with nasal health in chronic rhinosinusitis43, as well as a protective role in in vitro experiments against Staphylococcus aureus43 and S. pneumoniae colonization and otitis media in children44,45. This protective effect might be mediated by Dolosigranulum’s antimicrobial properties (anti-inflammatory and barrier-enhancing) by exploiting TLR2/6 interaction mechanisms within the host cells and L-lactic acid production43 or by its capacity to produce antibiotics or bioactive molecules that may play key roles in interspecies interactions with its microbial neighbors, e.g., for niche competition46. Both species are part of the healthy nasopharyngeal microbiota, even though Dolosigranulum has been rarely isolated from patients with bacteremia and queratitis47. Nonetheless, carriage surveys focused on detection of S. pneumoniae indicates the nasopharynx as the natural habitat of this bacterium47. Additionally, Variovax, also present only in non-infected HHCs, is a member of the human oral flora, has been previously identified as exclusive in nasal/oropharyngeal swabs of healthy participants compared to COVID-19 patients48, and it is also known for its strong biofilm formation49.
In our study, uninfected individuals who later acquired Mtb infection had a core microbiome that included the genus Rothia. Evidence shows that enrichment of lung microbiota with oral taxa such as Rothia induces host cellular mucosal immunity of the Th17/neutrophilic phenotype while suppressing innate immunity. Moreover, dysbiosis can predispose to or exacerbate conditions locally and at distal body sites50. We also found that individuals who later acquired Mtb showed an enrichment of the opportunistic pathogen Megasphaera, which has also been reported to be more abundant in the oropharyngeal microbiota of individuals with viral pulmonary infections as in COVID-1951. Taken together, we may speculate that the lack of Dolosigranulum, together with the presence Rothia and Megasphaera reflect a dysbiosis that might have rendered individuals more susceptible to Mtb infection.
The core’s microbiome present in at least 50% of Mtb infected individuals was represented only by Paracoccus, a gram-negative non-motile which genus, previously associated with infections, including leprosy and other skin infections where the skin microbiota had a relatively higher abundance of the bacteria compared to healthy controls52,53.
Among the present study limitations were the loss of samples due to low biomass recovery in the nasopharyngeal sampling process, leading to low DNA sequencing quality. Additionally, we found mild differences in participant's age that may have partially contributed to the observed differences between groups. Although, this does not seem to be clinically relevant as our participants were young (median age 34.5) and alterations in nasal microbiota communities start in middle-aged adults (40–65 years)54, similar to what has been found for gut microbiome that is relative stable in adults55. A further limitation of the study is that despite being embedded in a prospective cohort, the microbiome analysis was done only at the baseline visit (cross-sectional sampling), so spurious ecological associations cannot be ruled out. The study of samples at follow-up visits would have allowed assessment of changes in microbiome composition before and after Mtb acquisition as well as assessing whether it is the LTBI that induces changes in the microbiota composition or conversely, whether the upper respiratory tract microbiota structure confers susceptibility to acquiring this infection.
In conclusion, our study shows that HHCs with established latent TBI have differences in the nasopharyngeal microbiome with reduced alpha diversity and different bacterial communities compared to uninfected individuals. Whether an immune modulation of all respiratory microbiota ensuing Mtb infection occurs, as it has been described for active TB disease, remains to be established, as well as the protective role it might grant.
Methods
Patient selection
A prospective cohort study of household contacts (HHCs) of active pulmonary TB (PTB) cases (only acid-fast smear-positive index cases), aged 15 years or older (n = 231), was conducted between September 2017 and February 2020 in Santiago, Chile56. Contacts found to have active TB at baseline, reporting prior TB, autoimmune diseases, pregnancy, HIV infection, use of immunosuppressants, or inhalation drugs were not invited to participate. All HHCs were recruited within the first month after the TB diagnosis of the index case and screened for symptoms. Two nasopharyngeal swabs (one for 16S rRNA gene-targeted sequencing and another for Mtb culture) were taken at the baseline evaluation. Additionally, a chest X-ray to exclude secondary TB cases and an interferon-γ release assay (IGRA) test (QuantiFERON®-TB Gold Plus (QFT), QIAGEN, Hilden, Germany) were done at enrollment to evaluate TB infection (TBI) status. IGRA testing was repeated at a 12-weeks follow-up visit for all HHCs with negative IGRA results at the baseline evaluation, and participants were telephonically followed-up for 1 year to screen for symptoms of new active TB cases developing.
Participants were classified as: (a) “non-TBI” if the IGRA was negative both at baseline and at the 12-weeks evaluation, and they did not develop active TB on 1-year follow-up; (b) “pre-TBI” if the IGRA was negative at baseline evaluation but converted to positive IGRA at 12-weeks, or if the participant developed active TB on 1-year follow-up, and (c) “TBI” if the IGRA test was already positive at the baseline visit. Considering the known variability of the IGRA test around the positivity cut-off57, we included in the TBI category only HHCs with a positive IGRA result at baseline with CD4 or CD4-CD8 tubes IFN-γ values well above the positivity cut-off (≥ 1.0 UI/ml). Individuals who did not complete follow-up or had lower positive IGRA results (≤ 0.35 – 0.99 UI/ml) were excluded from the analysis (Supplementary Fig. S1).
Ethical approval
The study has ethical approval from the Institutional Review Board of the Pontificia Universidad Católica de Chile. All eligible participants provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations.
Nasopharyngeal sample collection
Nasopharyngeal samples were collected with Flocked swabs (Copan®, California, United States). For 16S sequencing, samples were stored in 2 ml of DNA preservative solution tubes (Norgen Biotek Corp., Ontario, Canada), kept on ice and then stored within 1 to 4 h of collection at − 80 °C until processing. Only one nasopharyngeal swab sample per participant—collected at the baseline visit—was analyzed. For Mtb culture, samples were stored in 2 ml PBS tubes and kept at room temperature until processing.
16S rRNA gene sequencing
Frozen swabs were thawed at room temperature and vortexed. Total DNA extraction of 500 μl was performed using the Saliva DNA Isolation Kit (Norgen Biotek Corp., Ontario, Canada) according to the manufacturer's supplementary protocol instructions at Instituto de Ciencias Biomédicas, Universidad de Chile. Frozen samples were shipped and sequenced at the Genomics Center of the University of Minnesota (Minneapolis, United States). Briefly, the genomic DNA (gDNA) content was quantified via qPCR of the 16S rRNA gene and used to normalize the gDNA concentration samples before library preparation. Libraries were prepared by amplifying the V1-V3 region of the 16S rRNA gene with 25 PCR cycles using the Nextera sequencing kit (Illumina, California, United States). Sequencing was performed with MiSeq 600 cycle v3 kit.
Data processing and analysis
Amplicon sequencing variant generation
Sequencing data were processed with the DADA2 package58 in R version 4.2.059 to generate amplicon sequencing variants (ASVs). Sequence quality was manually inspected to define trimming parameters. For sequence filtering and trimming, DADA2 function filterAndTrim was used with the following parameters: trimLeft = c(20,18), truncLen = c(290,275), maxEE = c(5,7), maxN = 0, truncq = 0. Error rates were estimated from the total dataset with the function learnErrors and the following parameters: max_consist = 20, nbases = 1e8. ASVs were generated with the dada function in pseudo-pooling mode. Read pairs were merged using the mergePairs function with minOverlap = 10 and maxMismatch = 1. Finally, the removal of chimeric sequences was done with the removeBimeraDenovo function using a consensus approach. The resulting ASVs were annotated using the assignTaxonomy function of DADA2 against the SILVA 138 database60. A phylogenetic tree was constructed for all ASVs by aligning them using MAAFT version 7.47561 and FastTree version 2.1.1062 using a General Time Reversible (GTR) model.
ASV analysis
ASV filtering
ASVs were removed if present in two different set control (preservation buffer and two water controls) used for library construction before sequencing. After ASVs generation, samples with less than 1,000 reads were also removed in a second filtering stage, as well as any ASV within the dataset classified as either contaminant (Chloroplast, Mitochondria or Eukaryota) or taxa with low prevalence (1 ASV in 1 sample at the Phylum level – Abditibacteriota, Campylobacterota, Cloacimonadota, Entotheonellaeota, Elusimicrobiota, Halanaerobiaeota, Hydrogenedentes, Methylomirabilota, Nitrospinota, Nitrospirota, SAR324 clade, Sumerlaeota, Synergistota, WOR-1, WS2, and WPS-2), keeping only ASVs present in at least four samples (5% of the dataset).
Microbiome analysis, alpha and beta diversity
Composition analysis of the cleaned dataset (841 ASVs) was done using phyloseq63 and its visualization with the ggplot2 package64. Core microbiome analysis was performed using the microbiome package65. A core microbiome for each group (non-TBI, pre-TBI and TBI) was generated using a detection threshold of 0.001 and a prevalence of 50% of the samples within the group at the ASV and genus classification level.
For alpha diversity analysis, Shannon values were calculated using phyloseq63 for each sample. A Kruskal–Wallis test followed by a Wilcoxon test with Holm adjustment for multiple testing was applied to assess pairwise differences. Analysis and visualization were performed using the ggstatsplot package66.
Beta diversity was estimated using the compositional data (raw counts) and processed with the PhILR package67 to normalize counts with an isometric log-ratio transformation using the phylogenetic tree information (distance of the ASVs present in the samples). Sample distance was calculated on the transformed dataset with Euclidean distance using the phyloseq package63. To evaluate statistical differences between group clustering, a permutational analysis of variance was performed on the Euclidean distance matrix using the adonis function of the vegan package68. The variance homogeneity assumption was evaluated using the functions betadisperser and permutest of the vegan package.
Differentially abundant taxa
The compositional approach implemented in ANCOMBC69 that allows for compositional analysis of the microbiome data with bias correction was used to identify taxonomic groups with statistically significant differences in abundance between HHCs groups. This method acknowledges sources of variation in microbiome datasets, including unequal sampling fractions and differences in sequencing efficiency, to attempt to limit the effects of group unevenness due to the microbiome variability among individuals in our study. The ASVs were summarized at the genus level, and all three pairwise comparisons of the groups (non-TBI vs. TBI, non-TBI vs. pre-TBI, and pre-TBI vs. TBI) were performed using default parameters. Multiple hypothesis testing was corrected with the Benjamini–Hochberg method with a false discovery rate of 0.05; the resulting differentially abundant ASVs were filtered, keeping only those with a log fold change of at least 0.5.
Statistical analysis
Clinical and demographic data analyses were performed in RStudio59,70. Comparisons between groups were performed with analysis of variance (ANOVA) for continuous variables, and Fisher’s exact or Chi-squared test for categorical variables. Statistical significance was considered when p-values < 0.05 (two-tailed).
Supplementary Information
Acknowledgements
This work was supported by ANID/CONICYT FONDECYT [1171570 and 1211225 to M.E.B, 1191874 to R.N, 11140666 and 1221209 to J.A.U.].
Author contributions
M.E.B. and R.N. worked in the conception and design of the work, C.R.-T. and M.E.B. wrote the main manuscript text, J.A.U. made the microbiome statistical analyses and prepared the figures, C.R.-T. made clinical and demographic statistical analyses and prepared the tables. All authors reviewed the manuscript.
Data availability
The datasets generated and/or analyzed during the current study are available in the European Nucleotide Archive (ENA), Project Accession number PRJEB61078.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-34052-8.
References
- 1.Global tuberculosis report 2020. (Geneva: World Health Organization; 2020. Licence: CC BY-NC-SA 3.0 IGO).
- 2.Acuña-Villaorduna C, et al. Intensity of exposure to pulmonary tuberculosis determines risk of tuberculosis infection and disease. Eur. Respir. J. 2018;51:1. doi: 10.1183/13993003.01578-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reichler MR, et al. Duration of exposure among close contacts of patients with infectious tuberculosis and risk of latent tuberculosis infection. Clin. Infect. Dis. 2020;71:1627–1634. doi: 10.1093/cid/ciz1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narasimhan P, Wood J, Macintyre CR, Mathai D. Risk factors for tuberculosis. Pulm Med. 2013;2013:828939. doi: 10.1155/2013/828939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salgame P, Geadas C, Collins L, Jones-Lopez E, Ellner JJ. Latent tuberculosis infection–Revisiting and revising concepts. Tuberculosis (Edinb.) 2015;95:373–384. doi: 10.1016/j.tube.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall A, Pirofski LA. What is a host? Incorporating the microbiota into the damage-response framework. Infect. Immun. 2015;83:2–7. doi: 10.1128/IAI.02627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young VB. The role of the microbiome in human health and disease: An introduction for clinicians. BMJ. 2017;356:831. doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Lun CY, Tsui SK. Metagenomics: A New Way to Illustrate the Crosstalk between Infectious Diseases and Host Microbiome. Int. J. Mol. Sci. 2015;16:26263–26279. doi: 10.3390/ijms161125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldman AJ, Balskus EP. The human microbiota, infectious disease, and global health: Challenges and opportunities. ACS Infect. Dis. 2018;4:14–26. doi: 10.1021/acsinfecdis.7b00232. [DOI] [PubMed] [Google Scholar]
- 10.Man WH, de SteenhuijsenPiters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventero MP, et al. Nasopharyngeal Microbial Communities of Patients Infected With SARS-CoV-2 That Developed COVID-19. Front. Microbiol. 2021;12:637430. doi: 10.3389/fmicb.2021.637430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HM, Kang JH. Effects of nasopharyngeal microbiota in respiratory infections and allergies. Clin. Exp. Pediatr. 2021;64:543–551. doi: 10.3345/cep.2020.01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai W, et al. The concordance between upper and lower respiratory microbiota in children with Mycoplasma pneumoniae pneumonia. Emerg. Microbes Infect. 2018;7:92. doi: 10.1038/s41426-018-0097-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakwinska O, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J. Clin. Microbiol. 2014;52:1590–1594. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haak BW, et al. Bacterial and viral respiratory tract microbiota and host characteristics in adults with lower respiratory tract infections: A case-control study. Clin. Infect. Dis. 2022;74:776–784. doi: 10.1093/cid/ciab568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremers AJ, et al. The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome. 2014;2:44. doi: 10.1186/2049-2618-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botero LE, et al. Respiratory tract clinical sample selection for microbiota analysis in patients with pulmonary tuberculosis. Microbiome. 2014;2:29. doi: 10.1186/2049-2618-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Zhu Y, Liao Q, Wang Z, Wan C. Characterization of gut microbiota in children with pulmonary tuberculosis. BMC Pediatr. 2019;19:445. doi: 10.1186/s12887-019-1782-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warndorff DK, et al. Polymerase chain reaction of nasal swabs from tuberculosis patients and their contacts. Int. J. Leprosy. 1996;64:404–408. [PubMed] [Google Scholar]
- 20.Balcells ME, et al. M. tuberculosis DNA detection in nasopharyngeal mucosa can precede tuberculosis development in contacts. Int. J. Tuberc. Lung Dis. 2016;20:848–852. doi: 10.5588/ijtld.15.0872. [DOI] [PubMed] [Google Scholar]
- 21.Budden KF, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 22.Khan N, et al. Alteration in the Gut Microbiota Provokes Susceptibility to Tuberculosis. Front. Immunol. 2016;7:529. doi: 10.3389/fimmu.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry, S. et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One5, e8804. 10.1371/journal.pone.0008804 (2010). [DOI] [PMC free article] [PubMed]
- 24.Wang J, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice TA, et al. Signaling via pattern recognition receptors NOD2 and TLR2 contributes to immunomodulatory control of lethal pneumovirus infection. Antiviral Res. 2016;132:131–140. doi: 10.1016/j.antiviral.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin T, et al. Super-dominant pathobiontic bacteria in the nasopharyngeal microbiota as causative agents of secondary bacterial infection in influenza patients. Emerg. Microbes Infect. 2020;9:605–615. doi: 10.1080/22221751.2020.1737578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling Z, et al. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genom. 2013;14:390. doi: 10.1186/1471-2164-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nardelli C, et al. Nasopharyngeal microbiome signature in COVID-19 positive patients: Can we definitively get a role to fusobacterium periodonticum? Front Cell Infect Microbiol. 2021;11:625581. doi: 10.3389/fcimb.2021.625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edouard S, et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis. 2018;37:1725–1733. doi: 10.1007/s10096-018-3305-8. [DOI] [PubMed] [Google Scholar]
- 30.Rouchka EC, et al. Induction of interferon response by high viral loads at early stage infection may protect against severe outcomes in COVID-19 patients. Sci. Rep. 2021;11:15715. doi: 10.1038/s41598-021-95197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen EK, et al. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2:22. doi: 10.1186/2049-2618-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Boeck I, et al. Comparing the healthy nose and nasopharynx microbiota reveals continuity as well as niche-specificity. Front. Microbiol. 2017;8:2372. doi: 10.3389/fmicb.2017.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, et al. Alterations in the nasopharyngeal microbiota associated with active and latent tuberculosis. Tuberculosis. 2022 doi: 10.1016/j.tube.2022.102231. [DOI] [PubMed] [Google Scholar]
- 34.Eribo OA, et al. The gut microbiome in tuberculosis susceptibility and treatment response: guilty or not guilty? Cell Mol. Life Sci. 2020;77:1497–1509. doi: 10.1007/s00018-019-03370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naidoo CC, et al. The microbiome and tuberculosis: state of the art, potential applications, and defining the clinical research agenda. Lancet Respir. Med. 2019;7:892–906. doi: 10.1016/s2213-2600(18)30501-0. [DOI] [PubMed] [Google Scholar]
- 36.Cheung, M. K. et al. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One8, e54574, doi:10.1371/journal.pone.0054574 (2013). [DOI] [PMC free article] [PubMed]
- 37.Cui Z, et al. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol. 2012;12:276. doi: 10.1186/1471-2180-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong BY, Paulson JN, Stine OC, Weinstock GM, Cervantes JL. Meta-analysis of the lung microbiota in pulmonary tuberculosis. Tuberculosis (Edinb.) 2018;109:102–108. doi: 10.1016/j.tube.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, et al. Metagenomic analysis of the lung microbiome in pulmonary tuberculosis—A pilot study. Emerg. Microbes Infect. 2020;9:1444–1452. doi: 10.1080/22221751.2020.1783188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, J. et al. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS One8, e83445, doi:10.1371/journal.pone.0083445 (2013). [DOI] [PMC free article] [PubMed]
- 41.Escapa IF, et al. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. mSystems. 2018;3:1. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Boeck I, et al. The nasal mutualist Dolosigranulum pigrum AMBR11 supports homeostasis via multiple mechanisms. iScience. 2021;24:1078. doi: 10.1016/j.isci.2021.102978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laufer, A. S. et al. Microbial communities of the upper respiratory tract and otitis media in children. mBio2, e00245–00210. 10.1128/mBio.00245-10 (2011). [DOI] [PMC free article] [PubMed]
- 45.Pettigrew MM, et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugger SD, et al. Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere. 2020;5:1. doi: 10.1128/mSphere.00852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laclaire L, Facklam R. Antimicrobial susceptibility and clinical sources of Dolosigranulum pigrum cultures. Antimicrob Agents Chemother. 2000;44:2001–2003. doi: 10.1128/AAC.44.7.2001-2003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rueca M, et al. Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int. J. Environ. Res. Public Health. 2021;18:1. doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamieson WD, Pehl MJ, Gregory GA, Orwin PM. Coordinated surface activities in Variovorax paradoxus EPS. BMC Microbiol. 2009;9:124. doi: 10.1186/1471-2180-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mindt, B. C. & DiGiandomenico, A. Microbiome Modulation as a Novel Strategy to Treat and Prevent Respiratory Infections. Antibiotics (Basel)11, 1. 10.3390/antibiotics11040474 (2022). [DOI] [PMC free article] [PubMed]
- 51.Ma S, et al. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target Ther. 2021;6:191. doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bayal N, et al. Structural aspects of lesional and non-lesional skin microbiota reveal key community changes in leprosy patients from India. Sci. Rep. 2021;11:3294. doi: 10.1038/s41598-020-80533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Rensburg, J. J. et al. The Human Skin Microbiome Associates with the Outcome of and Is Influenced by Bacterial Infection. mBio6, e01315–01315, doi:10.1128/mBio.01315-15 (2015). [DOI] [PMC free article] [PubMed]
- 54.Kumpitsch C, Koskinen K, Schopf V, Moissl-Eichinger C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019;17:87. doi: 10.1186/s12915-019-0703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilmanski T, et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021;3:274–286. doi: 10.1038/s42255-021-00348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balcells ME, et al. Systematic screening of adult household contacts of patients with active pulmonary tuberculosis reveals high rates of new infections during follow-up. Rev. Med. Chil. 2020;148:151–159. doi: 10.4067/s0034-98872020000200151. [DOI] [PubMed] [Google Scholar]
- 57.Wikell, A. et al. The Impact of Borderline Quantiferon-TB Gold Plus Results for Latent Tuberculosis Screening under Routine Conditions in a Low-Endemicity Setting. J. Clin. Microbiol.59, e0137021. 10.1128/JCM.01370-21 (2021). [DOI] [PMC free article] [PubMed]
- 58.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R: A language and environment for statistical computing. https://www.R-project.org/ (2021).
- 60.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One5, e9490. 10.1371/journal.pone.0009490 (2010). [DOI] [PMC free article] [PubMed]
- 63.McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One8, e61217. 10.1371/journal.pone.0061217 (2013). [DOI] [PMC free article] [PubMed]
- 64.Wickham, H. ggplot2. (Springer Science+Business Media, 2009).
- 65.Lahti, L. & Shetty, S. Tools for microbiome analysis in R. http://microbiome.github.com/microbiome (2017).
- 66.Patil, I. Visualizations with statistical details: The 'ggstatsplot' approach. J. Open Source Softw.6, 10.21105/joss.03167 (2021).
- 67.Silverman, J. D., Washburne, A. D., Mukherjee, S. & David, L. A. A phylogenetic transform enhances analysis of compositional microbiota data. Elife6, doi:10.7554/eLife.21887 (2017). [DOI] [PMC free article] [PubMed]
- 68.Oksanen, J. et al. (2020). vegan: Community Ecology Package. R package version 2.5–7. https://CRAN.R-project.org/package=vegan.
- 69.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.RStudio: Integrated Development for R. RStudio, PBC.http://www.rstudio.com/. (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the European Nucleotide Archive (ENA), Project Accession number PRJEB61078.