Abstract
Dihydrofolate reductase (DHFR) is the target of trimethoprim (TMP), which has been widely used in combination with sulfa drugs for treatment and prophylaxis of Pneumocystis carinii pneumonia. While the rat-derived P. carinii DHFR has been well characterized, kinetic studies of human-derived P. carinii DHFR, which differs from rat-derived P. carinii DHFR by 38% in amino acid sequence, have not been reported to date. Here we report on the expression and kinetic characterization of the recombinant human-derived P. carinii DHFR. The 618-bp coding sequence of the human-derived P. carinii DHFR gene was expressed in Escherichia coli. As determined by sodium dodecyl sulfate-polyacrylamide gel eletrophoresis, the purified enzyme had a molecular mass of 25 kDa, consistent with that predicted from the DNA sequence. Kinetic analysis showed that the Km values for dihydrofolate and NADPH were 2.7 ± 0.3 and 14.0 ± 4.3 μM, respectively, which are similar to those reported for rat-derived P. carinii DHFR. Inhibition studies revealed that both TMP and pyrimethamine were poor inhibitors of human-derived P. carinii DHFR, with Ki values of 0.28 ± 0.08 and 0.065 ± 0.005 μM, respectively, while trimetrexate and methotrexate were potent inhibitors, with Ki values of 0.23 ± 0.03 and 0.016 ± 0.004 nM, respectively. The availability of purified recombinant enzyme in large quantities should facilitate the identification of antifolate inhibitors with greater potency and higher selectivity for human-derived P. carinii DHFR.
Pneumocystis carinii pneumonia (PCP) remains a leading cause of morbidity and mortality in AIDS. Currently, one of the most widely used agents for treatment and prophylaxis of this infection is the combination of trimethoprim (TMP) and sulfamethoxazole (SMX). TMP inhibits dihydrofolate reductase (DHFR) (EC 1.5.1.3), which catalyzes the reduction of 7,8-dihydrofolate to 5,6,7,8-tetrahydrofolate in the presence of NADPH and is essential for biosynthesis of thymidylate, purine nucleotides, and several amino acids. Despite its obvious efficacy, this combination is complicated by frequent toxic and allergic side effects (19); moreover, there are increasing concerns about whether TMP truly contributes to the activity of this combination against P. carinii. It has been shown in vitro that TMP is a poor inhibitor of rat-derived P. carinii DHFR (2, 6, 7, 9, 22, 25) and that TMP alone is ineffective in the treatment of rat PCP (16, 26). Recently, mutations in the P. carinii dihydropteroate synthase gene, the target of sulfamides, have been reported in the United States (15, 21; Q. Mei, S. Gurunathan, H. Masur, and J. A. Kovacs, Letter, Lancet 351:1631, 1998) and Europe (11) and have been associated with prophylaxis and/or treatment failures of TMP-SMX, suggesting that P. carinii is developing resistance to sulfa drugs. In contrast, the DHFR gene did not show any mutations suggestive of drug resistance (21). This may reflect an absence of drug pressure on DHFR and supports the concept that TMP contributes little to the efficacy of the TMP-SMX combination against P. carinii.
While the rat-derived P. carinii DHFR has been well characterized in terms of its molecular and kinetic properties (2, 6, 7, 9, 17, 18, 22, 25), little is known about the human-derived P. carinii DHFR, which we have recently cloned and which differs from the rat-derived P. carinii DHFR by 38% in amino acid sequence (21). For designing potential antifolates for treatment of humans, the ideal target should be the DHFR of human P. carinii, since that is the pathogen for humans. However, the human-derived P. carinii is more difficult to study than the rat-derived P. carinii. Because the source of human-derived P. carinii organisms is very limited and because no reliable culture system for P. carinii is currently available, it is not feasible to isolate and purify native DHFR enzyme of human-derived P. carinii in a sufficient amount for detailed study. In fact, no enzyme from this organism has been purified. The primary goal of the present study was to produce catalytically active human-derived P. carinii DHFR enzyme in a bacterial system and thus to provide an abundant source of purified enzyme for detailed studies of the enzyme itself and, more importantly, for drug testing and design. We have also described a preliminary determination of the kinetic constants of the recombinant enzyme and its inhibitory properties against several commonly used antifolate drugs.
MATERIALS AND METHODS
Construction of recombinant plasmid and expression of recombinant DHFR.
Cloning of the human-derived P. carinii DHFR gene has been previously described (21). To eliminate the single intron in the gene, we employed the thermal cycled fusion PCR method described by Kahn et al. (14), in which four primers were involved. Primer FR331 (5′-GGATCCATGGATTGGCAAAAGTCATTGAC-3′) and primer FR1018 (5′-AAGCTTGCTTCAAACCTTGTGTAACGCG-3′) were complementary to the sequence at the 5′ and the 3′ ends of human-derived P. carinii DHFR-coding region (21) and contained BamHI and HindIII restriction sites added to the 5′ ends (underlined), respectively. Primer FR577 (5′-CAAGAGGTTCTTGATTTAGGAGGTGGAGCGTACCATGCAAG-3′) and primer FR659 (5′-CTTGCATGGTACGCTCCACCTCCTAAATCAAGAACCTCTTG-3′) were complementary to each other, and each contained 5′- and 3′-flanking sequences of the DHFR intron. Two fragments flanking the intron were first amplified in two separate PCRs using human-derived P. carinii genomic DNA and primers FR331-FR577 and FR659-FR1018, respectively. Aliquots of the two initial PCR products were then diluted and mixed together, along with primers FR331 and FR1018, to amplify the entire DHFR-coding region without an intron. The PCRs were carried out with a touchdown protocol as described previously (21).
The final PCR product was gel purified, subcloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.), and sequenced as described previously (21). The coding sequence was cloned into the BamHI and HindIII sites of the pET28a(+) expression vector (Novagen, Inc., Madison, Wis.), which expresses the recombinant protein with an N-terminal extension of 34 residues including a track of six histidines, which allows purification of the recombinant product using a nickel affinity column. The fidelity of the expression construct was confirmed by DNA sequencing, and the recombinant plasmid pET-DHFR was transformed into Escherichia coli strain BL21(DE3).
A single colony containing pET-DHFR was cultured at 37°C overnight in 5 ml of Luria broth supplemented with 30 μg of kanamycin per ml. One milliliter of the overnight culture was grown at 37°C in 400 ml of Luria broth supplemented with 30 μg of kanamycin per ml until the optical density at 600 nm reached 0.6, and then the culture was incubated at 30°C for an additional 2.5 h in the presence of 0.4 mM isopropyl-β-d-thiogalactoside (IPTG) to induce the expression of the recombinant protein.
Purification of the recombinant DHFR enzyme.
Cell paste harvested from the induced culture was resuspended in binding buffer (5 mM imidazole, 500 mM NaCl, and 20 mM Tris-HCl, pH 7.9) and lysed by sonication. After centrifugation at 18,000 × g and 4°C for 20 min, the supernatant was collected and loaded onto a precharged His · Bind column (Novagen). The column was washed once with binding buffer and once with 60 mM imidazole–500 mM NaCl–20 mM Tris-HCl (pH 7.9). The recombinant protein was eluted with 1 M imidazole–500 mM NaCl–20 mM Tris-HCl (pH 7.9) and then dialyzed against 50 mM Tris-HCl (pH 7.2). Each step of the isolation and purification procedure was monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) using precast 4 to 20% gradient Tris-glycine gels (Novex, San Diego, Calif.). Protein concentrations were determined by the method of Bradford (5) using a protein assay kit from Bio-Rad Laboratories (Hercules, Calif.). Glycerol was added to the purified DHFR preparation to a final concentration of 40% (vol/vol) to stabilize the enzyme activity.
Enzyme activity assay.
Dihydrofolate (dihydrofolic acid) and NADPH were purchased from Sigma Chemical Co. (St. Louis, Mo.). The catalytic activity of DHFR was measured essentially as described previously for rat-derived P. carinii DHFR (18). The standard assay mixture (0.25 ml) contained 0.1 mM NADPH, 0.1 mM dihydrofolate, 160 mM Tris-HCl (pH 7.2), 160 mM KCl, and the enzyme preparation. Assays were performed at 37°C in 96-well flat-bottom microplates (Costar Corp., Corning, N.Y.) using a SpectraMAX Plus spectrophotometer interfaced with a Macintosh computer running Softmax Pro 2.2.1 software (Molecular Devices Corp., Sunnyvale, Calif.). The absorbance was read at 340 nm using the kinetic mode with a reading interval of 2 or 4 s for a duration of 10 min. One unit was defined as the amount of enzyme required to reduce 1 μmol of dihydrofolate per min, based on a molar extinction coefficient of 12,300 M−1 cm−1 at 340 nm (13). For kinetic studies, enzyme concentrations were chosen such that the initial reaction velocity was linear over the 10-min assay.
Determination of Km values for enzyme substrates.
To determine the Michaelis constant (Km) values for NADPH and dihydrofolate, initial reaction velocity measurements were performed in a five-by-five or four-by-four matrix of NADPH (2.5 to 40 μM) and dihydrofolate (0.5 to 20 μM) concentrations. Reaction velocity is expressed as micromoles of dihydrofolate reduced per minute per milliliter of sample. Reactions were initiated by the addition of enzyme. Km values were calculated by primary and secondary Hanes plots (10) as described previously (24).
Determination of IC50s and Ki values for enzyme inhibitors.
TMP, methotrexate, and pyrimethamine were purchased from Sigma Chemical Co., and trimetrexate was a generous gift from Carmen J. Allegra (National Cancer Institute, National Institutes of Health, Bethesda, Md.). The concentrations of inhibitors required to achieve 50% inhibition of the enzyme reaction (IC50s) were determined at 25 μM dihydrofolate and 75 μM NADPH. Assays were started by the addition of dihydrofolate after preincubation of the enzyme with the inhibitor. IC50s were estimated by interpolation of plots of the percentage of inhibition against the concentration of inhibitor. The inhibition constant (Ki) values for TMP and pyrimethamine were determined by primary Hanes plots and secondary Dixon plots as described previously (24). The Ki values for trimetrexate and methotrexate were estimated by Henderson analysis (12) as described previously (3). Graphs were plotted by using Cricket Graph (version 1.3.2; Cricket Software, Malvern, Pa.) or Microsoft Excel software (98 Macintosh edition; Microsoft Corp., Redmond, Wash.). The values of the correlation coefficient, slope, and intercept were calculated by linear regression analysis.
RESULTS
PCR amplification of the DHFR-coding region.
To engineer a plasmid containing the human-derived P. carinii DHFR coding sequence with no intron, the 5′-end (287-bp) and 3′-end (421-bp) fragments flanking the P. carinii DHFR intron were first amplified separately by two PCRs from human-derived P. carinii genomic DNA. These two fragments were then fused by another round of PCR, resulting in a 667-bp fragment. Subsequent sequencing confirmed that this fused fragment contained the correct P. carinii DHFR-coding sequence (618 bp) without intron and the 3′-end noncoding sequence (49 bp).
Expression and purification of the recombinant enzyme.
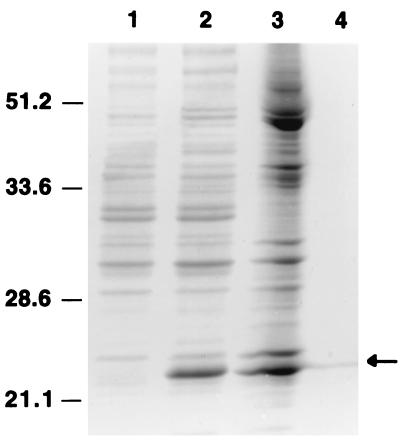
The coding region obtained by thermal cycled fusion PCR was placed in the expression vector pET28a(+) and introduced into E. coli strain BL21(DE3). Upon induction with IPTG, lysates of cells containing the pET-DHFR construct showed a 25-kDa protein, which was not present in cells containing vector alone (Fig. 1). Our initial expression at 37°C resulted in large amounts of enzyme in inclusion bodies and only trace amounts of enzyme in the soluble fraction. We attempted to solubilize the inclusion bodies using either guanidine or urea and then to refold the enzyme, but subsequent enzyme assays did not show any DHFR activity. Therefore, we expressed the recombinant enzyme at a lower temperature (30°C) and found that the amounts of soluble enzyme increased adequately to permit purification without solubilization, even though most of the recombinant protein mass remained in insoluble aggregates (data not shown). The recombinant enzyme in soluble extracts was readily purified by affinity chromatography using His · Bind columns. By SDS-PAGE (Fig. 1), the purified enzyme migrated as a single band of about 25 kDa, consistent with the predicted molecular mass of 23,407 Da calculated from the DNA sequence (21). From 1 liter of culture we could obtain approximately 1.9 mg of purified enzyme, which had an initial specific activity of 10.8 U/mg of protein. The presence of the N-terminal extension with a His tag did not appear to interfere with the enzyme activity, and thus we did not remove it from the recombinant enzyme. Previous studies of other recombinant enzymes expressed in pET vectors have shown that the presence of an N-terminal His tag does not affect the enzyme kinetics (20, 27).
FIG. 1.
Coomassie blue-stained SDS-polyacrylamide gel of recombinant human-derived P. carinii DHFR. Lane 1, crude extract from cells containing vector pET28(+) alone; lane 2, crude extract from cells containing pET-DHFR; lane 3, soluble extract from cells containing pET-DHFR; lane 4, purified DHFR preparation (arrow). The migrations of protein size markers (kilodaltons) are indicated at the left.
We examined the storage conditions for purified enzyme and found that the enzyme stored in 40% glycerol at −20°C was relatively stable over a period of 5 to 6 months, with gradual loss of enzyme activity. In the absence of glycerol, an approximately 60% loss of activity was observed after 1 week of storage at −20°C.
Kinetic constants of the recombinant enzyme.
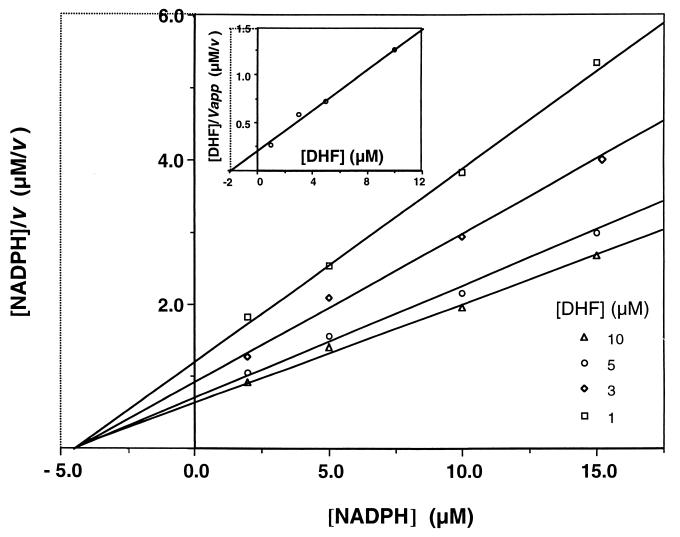
The Km values for dihydrofolate (Fig. 2) and NADPH were determined to be 2.7 ± 0.3 and 14.0 ± 4.3 μM, respectively. These values are similar (within severalfold) to those of rat-derived P. carinii, for which the Km values were reported to be 1.77 to 17.7 and 1.38 to 40.2 μM for dihydrofolate and NADPH, respectively (7, 18, 22, 25).
FIG. 2.
Determination of the Km for dihydrofolate (DHF). Initial velocity measurements (micromoles per minute per milliliter) were performed at various concentrations of NADPH in the presence of different fixed concentrations of DHF, and the results were fitted to the Hanes equation (10) a/V = a/Vm + Km/Vm, where a is the substrate concentration, V is the reaction velocity, and Vm is the maximal velocity. In the primary plot of [NADPH]/V versus [NADPH], each DHF concentration gives a straight line whose slope represents the reciprocal of the apparent maximal velocity (Vapp). In the secondary plot (inset), [DHF]/Vapp is plotted against [DHF], and the intercept on the [DHF] axis is the value for −Km. This experiment was repeated five times, and representative results are shown.
Inhibitory properties of antifolate drugs.
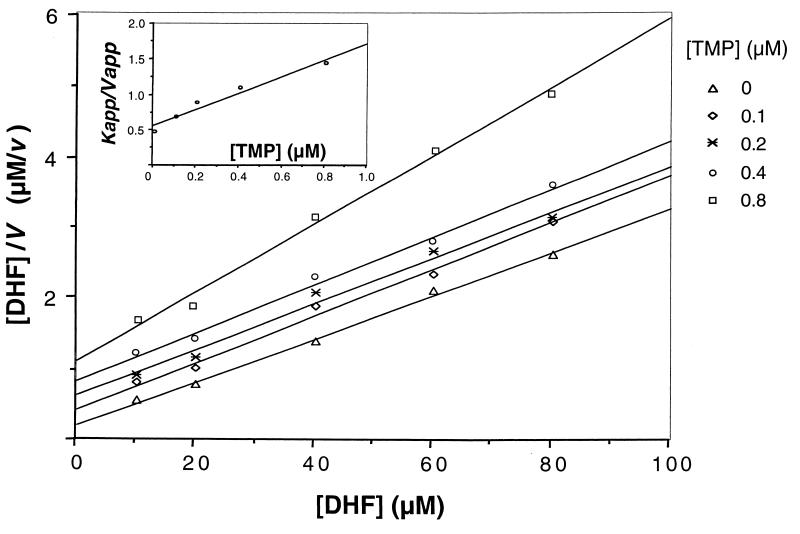
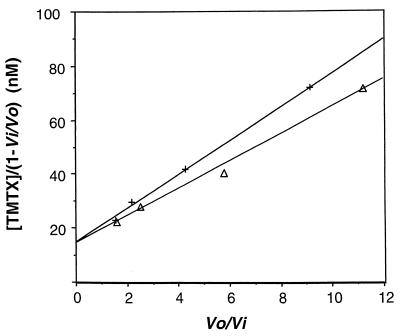
Preliminary determination of the IC50s of four antifolates against human-derived P. carinii DHFR showed that TMP and pyrimethamine appeared to be weak inhibitors, with IC50s in the micromolar range, while trimetrexate and methotrexate were much stronger inhibitors, with IC50s in the nanomolar range (Table 1). When the reaction velocities with TMP or pyrimethamine were analyzed using a Hanes plot, the results indicated that both were competitive inhibitors of DHFR. A secondary Dixon plot revealed that the Ki values for TMP and pyrimethamine were 0.28 ± 0.08 μM (five assays) (Fig. 3) and 0.065 ± 0.005 μM (two assays), respectively (Table 1). Both methotrexate and trimetrexate are slow, tight-binding inhibitors, and the steady-state reaction velocities were analyzed with Henderson plots (Fig. 4). The Ki values for methotrexate and trimetrexate were determined from three assays to be 0.016 ± 0.004 and 0.23 ± 0.03 nM, respectively (Table 1).
TABLE 1.
Inhibition of recombinant human-derived P. carinii DHFR by antifolates
| Inhibitor | IC50 (μM) (n)a | Ki (μM) (n)a |
|---|---|---|
| TMP | 4.85 ± 0.21 (2) | 0.28 ± 0.08 (5) |
| Pyrimethamine | 0.24 ± 0.03 (3) | 0.065 ± 0.005 (2) |
| Trimetrexate | (7.5 ± 2.7) × 10−3 (3) | (2.3 ± 0.3) × 10−4 (3) |
| Methotrexate | (6.2 ± 1.3) × 10−4 (2) | (1.6 ± 0.4) × 10−5 (3) |
n is the number of replicate experiments that were performed. Values are means and standard deviations for the replicate experiments.
FIG. 3.
Determination of the Ki for the inhibitor TMP. Initial velocities (micromoles per minute per milliliter) were determined at various concentrations of dihydrofolate (DHF) in the presence of different fixed concentrations of TMP, and the results were fitted to the equation a/V = a/Vm + Km(1 + i/Ki)/Vm, which is derived from the Dixon equation (8), where V is the reaction velocity, a is the dihydrofolate concentration, Vm is the maximal velocity, and i is the inhibitor concentration. Plots of [DHF]/V versus [DHF] at different concentrations of TMP give straight lines with y intercepts of Km(1 + i/Ki)/Vm, which refer to Kapp/Vapp. These y intercepts were replotted against [TMP] (inset), and the intercept on the [TMP] axis is the value for −Ki. This experiment was repeated five times, and representative results are shown.
FIG. 4.
Determination of the Ki for the tight-binding inhibitor trimetrexate (TMTX). The enzyme was preincubated with 75 μM NADPH and various concentrations of TMTX (between 4.0 and 64 nM), and then the reaction was started by adding 45 μM (Δ) or 90 μM (+) dihydrofolate (DHF). Steady-state velocities were measured and fitted to the Henderson equation (3, 12) It/(1 − Vi/V0) = Ki(1 + At/Ka) V0/Vi + Et, where It is the total concentration of inhibitor, Vi is the velocity in the presence of inhibitor, V0 is the velocity without inhibitor, Ki is the inhibition constant, At is the concentration of competing substrate DHF, Ka is the Michaelis constant for the DHF, and Et is the enzyme concentration. Plots of It/(1 − Vi/V0) against V0/Vi give straight lines with slopes of Ki(1 + At/Ka); then Ki = slope/(1 + At/Ka). The crossing lines on the ordinate suggested competitive inhibition. This experiment was repeated three times, and representative results are shown.
DISCUSSION
In the present study, we have successfully expressed the human-derived P. carinii DHFR gene to high levels in E. coli and have characterized the kinetics of the purified enzyme. Our data show that the kinetic properties of human- and rat-derived P. carinii DHFRs are similar. We examined the inhibitory properties of several commonly used antifolates against human-derived P. carinii DHFR (Table 1). Based on the Ki values, TMP was the weakest inhibitor, pyrimethamine was 4-fold stronger than TMP, and trimetrexate and methotrexate were 1,200- and 18,000-fold stronger than TMP, respectively. Compared to the reported Ki values of these inhibitors for human DHFR (4), only TMP showed a relatively favorable selectivity for human-derived P. carinii DHFR (the Ki ratio for human versus human-derived P. carinii DHFR is 3.0), while the other three inhibitors appeared to be selective for human DHFR (the Ki ratios for human versus human-derived P. carinii DHFR varied from 0.008 for trimetrexate to 0.5 for methotrexate). This is consistent with the inhibition profiles of rat-derived P. carinii DHFR described previously (2, 6, 7, 9, 22, 25).
The combination of TMP and SMX is utilized for treatment and prophylaxis of human PCP because it was shown to be effective in animal and human trials, but in fact, this particular fixed combination was originally chosen for trials because it had been formulated for bacterial indications and was readily available. TMP binds much more tightly to bacterial DHFRs than to eukaryotic DHFRs. The Ki for TMP with human DHFR is around 1 μM, whereas it is only 80 pM for E. coli DHFR (4). The Ki values for TMP with both rat-derived P. carinii DHFR (0.15 μM) (22) and human-derived P. carinii DHFR (0.28 μM in this study) are intermediate but more closely resemble that of human than that of E. coli DHFR. This is consistent with the fact that P. carini is a fungus. Very potent DHFR inhibitors such as trimetrexate and piritrexim are active as single agents against both murine and human PCP (1, 23), but TMP alone appears to be ineffective in the treatment of rat PCP (16, 26). While there is increasing evidence showing that the P. carinii dihydropteroate synthase is developing mutations that confer resistance to sulfa drugs (11, 15, 21; Mei et al., Letter, Lancet 351:1631, 1998), no mutations likely to confer drug resistance have been identified in the DHFR gene to date (21). This may reflect an absence of drug pressure on DHFR, suggesting that TMP is not an effective inhibitor of P. carinii DHFR. Alternatively, there may be other mechanisms that can also lead to resistance, such as increased production of native DHFR, that would not be identified by screening for mutations.
Clearly, there is a need to discover inhibitors with greater potency and higher selectivity for human-derived P. carinii DHFR. The availability of purified recombinant enzyme in large quantities should facilitate such studies. One approach is to screen available antifolate libraries (6). Another approach is to determine the tertiary structure of the purified enzyme with the aim of designing antifolates that can optimally target the active sites of the enzyme. Such studies are under way.
ACKNOWLEDGMENTS
We thank Mark Cowan for helpful suggestions about the PCR methodology and Carmen J. Allegra for reviewing the manuscript.
REFERENCES
- 1.Allegra C J, Chabner B A, Tuazon C U, Ogata-Arakaki D, Baird B, Drake J C, Simmons J T, Lack E E, Shelhamer J H, Balis F, et al. Trimetrexate for the treatment of Pneumocystis carinii pneumonia in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1987;317:978–985. doi: 10.1056/NEJM198710153171602. [DOI] [PubMed] [Google Scholar]
- 2.Allegra C J, Kovacs J A, Drake J C, Swan J C, Chabner B A, Masur H. Activity of antifolates against Pneumocystis carinii dihydrofolate reductase and identification of a potent new agent. J Exp Med. 1987;165:926–931. doi: 10.1084/jem.165.3.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baccanari D P, Joyner S S. Dihydrofolate reductase hysteresis and its effect of inhibitor binding analyses. Biochemistry. 1981;20:1710–1716. doi: 10.1021/bi00510a002. [DOI] [PubMed] [Google Scholar]
- 4.Blakley R L. Eukaryotic dihydrofolate reductase. Adv Enzymol Relat Areas Mol Biol. 1995;70:23–102. doi: 10.1002/9780470123164.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Broughton M C, Queener S F. Pneumocystis carinii dihydrofolate reductase used to screen potential antipneumocystis drugs. Antimicrob Agents Chemother. 1991;35:1348–1355. doi: 10.1128/aac.35.7.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delves C J, Ballantine S P, Tansik R L, Baccanari D P, Stammers D K. Refolding of recombinant Pneumocystis carinii dihydrofolate reductase and characterization of the enzyme. Protein Expr Purif. 1993;4:16–23. doi: 10.1006/prep.1993.1003. [DOI] [PubMed] [Google Scholar]
- 8.Dixon M, Webb E C. Enzymes. 2nd ed. New York, N.Y: Academic Press; 1964. [Google Scholar]
- 9.Edman J C, Edman U, Cao M, Lundgren B, Kovacs J A, Santi D V. Isolation and expression of the Pneumocystis carinii dihydrofolate reductase gene. Proc Natl Acad Sci USA. 1989;86:8625–8629. doi: 10.1073/pnas.86.22.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanes C S. Studies on plant amylases. I. The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem J. 1932;26:1406–1421. doi: 10.1042/bj0261406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helweg-Larsen J, Benfield T L, Eugen-Olsen J, Lundgren J D, Lundgren B. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet. 1999;354:1347–1351. doi: 10.1016/S0140-6736(99)03320-6. [DOI] [PubMed] [Google Scholar]
- 12.Henderson P J. Steady-state enzyme kinetics with high-affinity substrates or inhibitors. A statistical treatment of dose-response curves. Biochem J. 1973;135:101–107. doi: 10.1042/bj1350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillcoat B L, Nixon P F, Blakley R L. Effect of substrate decomposition on the spectrophotometric assay of dihydrofolate reductase. Anal Biochem. 1967;21:178–189. doi: 10.1016/0003-2697(67)90179-0. [DOI] [PubMed] [Google Scholar]
- 14.Kahn S M, Jiang W, Borner C, O'Driscoll K, Weinstein I B. Construction of defined deletion mutants by thermal cycled fusion: applications to protein kinase C. Tech J Methods Cell Mol Biol. 1990;2:27–30. [Google Scholar]
- 15.Kazanjian P, Locke A B, Hossler P A, Lane B R, Bartlett M S, Smith J W, Cannon M, Meshnick S R. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in AIDS patients. AIDS. 1998;12:873–878. doi: 10.1097/00002030-199808000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Kluge R M, Spaulding D M, Spain A J. Combination of pentamidine and trimethoprim-sulfamethoxazole in therapy of Pneumocystis carinii pneumonia in rats. Antimicrob Agents Chemother. 1978;13:975–978. doi: 10.1128/aac.13.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacs J A, Allegra C J, Beaver J, Boarman D, Lewis M, Parrillo J E, Chabner B, Masur H. Characterization of de novo folate synthesis in Pneumocystis carinii and Toxoplasma gondii: potential for screening therapeutic agents. J Infect Dis. 1989;160:312–320. doi: 10.1093/infdis/160.2.312. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs J A, Allegra C J, Masur H. Characterization of dihydrofolate reductase of Pneumocystis carinii and Toxoplasma gondii. Exp Parasitol. 1990;71:60–68. doi: 10.1016/0014-4894(90)90008-z. [DOI] [PubMed] [Google Scholar]
- 19.Kovacs J A, Masur H. Pneumocystis carinii pneumonia: therapy and prophylaxis. J Infect Dis. 1988;158:254–259. doi: 10.1093/infdis/158.1.254. [DOI] [PubMed] [Google Scholar]
- 20.Kozaki A, Kamada K, Nagano Y, Iguchi H, Sasaki Y. Recombinant carboxyltransferase responsive to redox of pea plastidic acetyl-CoA carboxylase. J Biol Chem. 2000;275:10702–10708. doi: 10.1074/jbc.275.14.10702. [DOI] [PubMed] [Google Scholar]
- 21.Ma L, Borio L, Masur H, Kovacs J A. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J Infect Dis. 1999;180:1969–1978. doi: 10.1086/315148. [DOI] [PubMed] [Google Scholar]
- 22.Margosiak S A, Appleman J R, Santi D V, Blakley R L. Dihydrofolate reductase from the pathogenic fungus Pneumocystis carinii: catalytic properties and interaction with antifolates. Arch Biochem Biophys. 1993;305:499–508. doi: 10.1006/abbi.1993.1453. [DOI] [PubMed] [Google Scholar]
- 23.Queener S F, Bartlett M S, Jay M A, Durkin M M, Smith J W. Activity of lipid-soluble inhibitors of dihydrofolate reductase against Pneumocystis carinii in culture and in a rat model of infection. Antimicrob Agents Chemother. 1987;31:1323–1327. doi: 10.1128/aac.31.9.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rancourt S L, Walker V K. Kinetic characterization of dihydrofolate reductase from Drosophila melanogaster. Biochem Cell Biol. 1990;68:1075–1082. doi: 10.1139/o90-161. [DOI] [PubMed] [Google Scholar]
- 25.Sirawaraporn W, Edman J C, Santi D V. Purification and properties of recombinant Pneumocystis carinii dihydrofolate reductase. Protein Expr Purif. 1991;2:313–316. doi: 10.1016/1046-5928(91)90088-z. [DOI] [PubMed] [Google Scholar]
- 26.Walzer P D, Kim C K, Foy J M, Linke M J, Cushion M T. Inhibitors of folic acid synthesis in the treatment of experimental Pneumocystis carinii pneumonia. Antimicrob Agents Chemother. 1988;32:96–103. doi: 10.1128/aac.32.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zomer A W, Michels P A, Opperdoes F R. Molecular characterisation of Trypanosoma brucei alkyl dihydroxyacetone-phosphate synthase. Mol Biochem Parasitol. 1999;104:55–66. doi: 10.1016/s0166-6851(99)00141-3. [DOI] [PubMed] [Google Scholar]