Significance
Recent hypotheses suggest that the last universal common ancestor (LUCA) was likely a diderm. Assuming that the primordial cell had only one membrane, it remains unknown how a second membrane could have evolved. Thermotoga is an early-branching phylum that is often placed near the root of the tree of life and as such can provide valuable information about the nature of LUCA. Characterization of the in situ structure of the cell envelope in Thermotoga maritima shows that the toga is composed of extensive protein arrays supporting small lipid patches. We propose a mechanism for OM biogenesis where a toga-like structure acted as an intermediate between monoderm and diderm cell envelope transitions.
Keywords: bacterial cell envelope, Thermotogae, cryo-ET, OM biogenesis, LUCA
Abstract
The presence of a cell membrane is one of the major structural components defining life. Recent phylogenomic analyses have supported the hypothesis that the last universal common ancestor (LUCA) was likely a diderm. Yet, the mechanisms that guided outer membrane (OM) biogenesis remain unknown. Thermotogae is an early-branching phylum with a unique OM, the toga. Here, we use cryo-electron tomography to characterize the in situ cell envelope architecture of Thermotoga maritima and show that the toga is made of extended sheaths of β-barrel trimers supporting small (~200 nm) membrane patches. Lipidomic analyses identified the same major lipid species in the inner membrane (IM) and toga, including the rare to bacteria membrane-spanning ether-bound diabolic acids (DAs). Proteomic analyses revealed that the toga was composed of multiple SLH-domain containing Ompα and novel β-barrel proteins, and homology searches detected variable conservations of these proteins across the phylum. These results highlight that, in contrast to the SlpA/OmpM superfamily of proteins, Thermotoga possess a highly diverse bipartite OM-tethering system. We discuss the implications of our findings with respect to other early-branching phyla and propose that a toga-like intermediate may have facilitated monoderm-to-diderm cell envelope transitions.
The bacterial cell envelope is a diverse and complex structure that provides a barrier between the cell and its environment and facilitates interactions between the two (1). The presence of one (monoderm) or two membranes (diderm) surrounding the cell affords a way to classify bacteria by their cell envelope architecture. In monoderm bacteria, exemplified by Bacillus subtilis, the cell is encapsulated by a single lipid bilayer, the cytoplasmic membrane, and a thick layer of peptidoglycan (PG). Diderm bacteria, such as Escherichia coli, have an inner membrane (IM), an outer membrane (OM), and a periplasm between them that contains a thin layer of PG (1). Many bacteria, however, deviate from the canonical monoderm and diderm architectures. For example, Cyanobacteria are diderm with a thick layer of PG (2), while Mollicutes lack an OM and PG altogether (3). Furthermore, members of the diderm phylum Deinococcus-Thermus have an IM, thick layer of PG, an OM lacking lipopolysaccharides (LPS), and a proteinaceous surface layer (S-layer) (4, 5). The bacterial cell envelope can further vary in lipid and protein composition, and the presence of a polysaccharide capsule. Studies into the diversity of the bacterial cell envelope structure and function can give valuable insight into its adaptive roles in various environment, as well as provide deeper understanding of the evolutionary paths of its major components.
The IM and OM in bacteria are structurally and functionally different. The IM contains α-helical proteins and sustains a proton gradient, while the OM is rich in β-barrel proteins and facilitates the free diffusion of small molecules into and out of the periplasm (6). In particular, the Omp85/BamA superfamily of OM proteins (OMPs) have distinct β-barrel tertiary structures and are essential for the proper incorporation of other β-barrel proteins in the OM. Broadly conserved membrane lipid components include glycolipids known as LPS, found in the outer leaflet of typical OMs. Another major feature of the cell envelope is the presence of a molecular tethering system used by diderm bacteria to anchor the OM to their PG. In the early-branching phyla (Terrabacteria), this is formed by the SlpA/OmpM superfamily of proteins. These proteins contain an N-terminal S-layer homology (SLH) domain that binds PG-associated secondary polymers, a long coiled-coil domain spanning the periplasm, and a C-terminal β-barrel component embedded in the OM (5, 7). The domain architecture of these proteins is conserved across most diderm Terrabacteria, and evolutionary scenarios involving SlpA/OmpM losses have been proposed as a mechanism for diderm-to-monoderm transition in Firmicutes (7). Overall, the presence of β-barrel proteins, particularly those of the BamA family, an OM-tethering system, and LPS glycolipids are considered hallmarks of archetypical diderm bacteria and are often used to predict the presence of a second membrane (8–10). Recent advances in metagenomics have expanded our appreciation of microbial diversity and provide an ample opportunity to study how and when major structural features of the bacterial envelope have evolved.
Thermotoga is a member of the clade Terrabacteria, which includes Cyanobacteria, Firmicutes, Actinobacteria, Deinococcus-Thermus, Chloroflexi, and the Candidate Phyla Radiation (CPR) (11, 12). Frequently placed near the base of the bacterial tree of life, Thermotogae are mostly anaerobic, hyperthermophilic bacteria that stain gram negative. Although their cell envelope consists of an IM and a thin layer of PG, due to their unique OM, they are considered atypical diderms (13). The OM, referred to as “toga,” is formed by an extended proteinaceous sheath that dissociates from the IM at the cell poles and as such is credited for their signature appearance. Genomic analyses revealed the absence of genes involved in LPS synthesis (lpx), indicating that Thermotoga maritima lacks LPS (8–10). Nevertheless, members of the phylum encode for an Omp85/BamA homolog that has been used as a proxy for the presence of an OM, calling into question the true nature of the toga (10). Another distinct feature of Thermotogae is the production of several unusual membrane lipids, including membrane-spanning lipids, widely present in archaea but rarely found in bacteria. The core of the membrane-spanning lipids is derived from C30 and C32 diabolic acids (DAs) and the head groups are formed by tetraether, tetraester, or mixed ether–ester linkages to phosphoglycerol head groups on one end and glycerol molecules on the other (14). Membrane-spanning lipids have been shown to organize into lipid monolayers with increased rigidity and decreased permeability compared to phospholipid bilayers (15), a possible adaptation to hyperthermal environments.
Insights into the structure of the cell envelope of Thermotogae mainly come from studies on T. maritima, the best characterized member of the phylum. It was proposed that Ompα and Ompβ (previously OmpA/B) were the major structural components of the toga (16–19). Ompα was shown to contain a 45-nm-long coiled-coil domain (17) and a conserved SLH domain (19, 20). A gene (ompβ) identified in an operon with ompα was predicted to form a β-barrel and proposed to form the extended arrays of the toga (19). More recently, Ompα and Ompβ were suggested to act analogously to the SlpA/OmpM tethering system (7, 21). Additional major structural components of the toga include the well-characterized hydrolases xylanase, amidase, and xylosidase, used by the bacteria to degrade complex carbohydrates present in the environment (22).
Here, we use a multidisciplinary approach including cryo-electron tomography (cryo-ET), MS-based proteomics, and LC-MS-based lipidomics to characterize the structure and composition of the cell envelope of T. maritima. Our tomograms reveal the in situ organization of the cell envelope in three dimensions to macromolecular resolution (~3 nm), fractionation and compositional analyses define the protein and lipid components of the IM and toga, and homology searches show the distribution of major toga and toga-associated proteins throughout the Thermotoga phylum. We discuss the evolutionary implications of our findings with respect to other early-branching phyla and propose a mechanism for OM biogenesis that implicates a toga-like structural intermediate.
Results
The Toga Is Composed of Extended Arrays of β-barrel Trimers Supporting Small Lipid Patches.
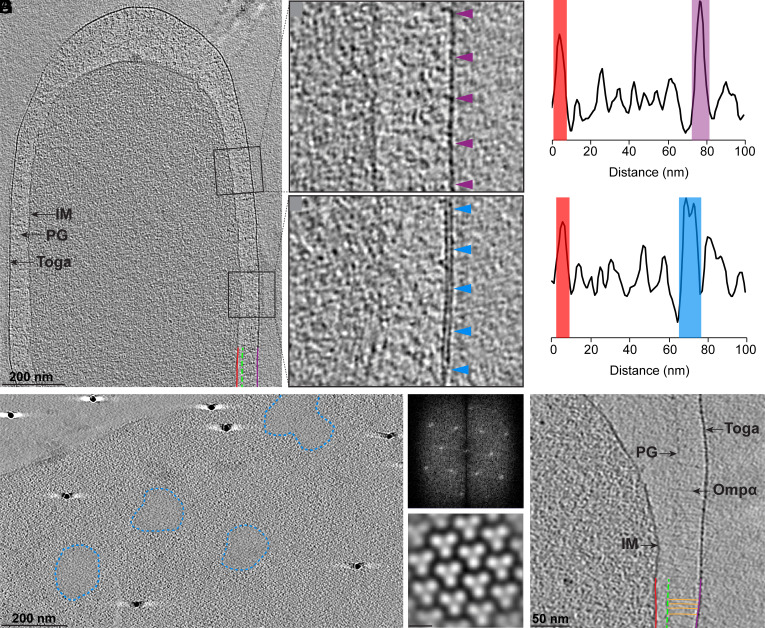
Cryo-electron tomography (cryo-ET) was used to visualize intact T. maritima cells (Fig. 1, Movie S1, n = 35). Tomograms revealed that the cell envelope consisted of an IM, a thin layer of PG, and a toga (Fig. 1A). Along the lateral sides of the cell, the periplasmic space was consistently ~70 ± 10 nm wide while it significantly varied at the poles (~450 ± 100 nm). The IM appeared as a 4-nm-thick monolayer, whereas the toga featured two distinct morphologies: 1) a 4-nm-thick monolayer (Fig. 1 B and C purple), and 2) a 7-nm-thick bilayer (Fig. 1 D and E light blue). Density profiles of the cell envelope at the monolayer and bilayer morphologies further supported the presence of a single and double layers, respectively (Fig. 1 C and E). Top (xz) views of the toga at the tips of cells revealed extended sheaths with a 2-dimensional pattern of β-barrel trimers (Fig. 1F, SI Appendix, Fig. S1 A and B, and Movie S2). Two-dimensional Fourier transforms and subtomogram averaging further confirmed that the β-barrel trimers formed a hexagonal array with 9.8-nm lattice spacing (Fig. 1F). In addition, top views revealed amorphous patches, ~100 to 200 nm in diameter that corresponded to the bilayer side views (Fig. 1F, blue outline). Based on these observations and the typical appearance of membranes as 7-nm bilayers, we concluded that the bilayers were composed of lipids and thus represent membrane patches. Notably, the presence of lipid patches smaller than the resolving power of cryo-ET (~3 nm) or individual lipids could also be excluded. Next, a continuous layer of ~50-nm fibrous densities was observed underneath the toga, connecting the toga to a thin layer of PG (Fig. 1G). Based on previous immunolabeling and freeze-etching studies localizing Ompα to the periplasm (17, 19), we concluded that the densities in our tomograms likely correspond to Ompα. Together with the β-barrel trimers of the toga, our tomograms revealed the molecular tethering system that connects the OM to the PG in T. maritima.
Fig. 1.
The toga of T. maritima is formed by extended protein arrays and small lipid patches. (A) A 20-nm thick tomographic slice showing the cell envelope architecture of T. maritima. IM, red, a thin layer of PG, green, and the toga (purple). (B and C) View of the toga protein monolayer (purple arrows) and corresponding density profile. (D and E) View of the toga lipid bilayer (light blue arrows) and corresponding density profile. (F) Top view of the toga showing extended protein arrays surrounding small lipid patches (blue dashed lines). 2D Fourier transform (Top Right) and subtomogram average (Bottom Right) reveal a hexagonal array of β-barrel trimers with 9.8 nm lattice spacing. (G) Numerous copies of Ompα (orange) span ~50 nm underneath the toga and connect the PG to the toga.
Additional Ultrastructural Features. Numerous tubular structures ~80-nm long and 10 nm in diameter were also observed spanning the periplasm and connecting the IM to the toga (SI Appendix, Fig. S1 A and C). Based on their similar appearance to type 4 pili (T4P)/type II secretion systems (T2SS) characterized in other bacteria and the detection of PilQ and PilA in the toga fraction using MS, we hypothesized that these represent a homologous system in T. maritima. Notably, the genome of T. maritima also encodes for homologs of pilQ (OM), pilA (pilin), pilC (IM), pilT (cytoplasm), and pilB (cytoplasm), further supporting the identity of the observed structures (23). Dividing cells revealed simultaneous invagination of the IM, PG-Ompα layer, and the toga. Filaments (~16 nm underneath the IM) were also observed at the division sites, likely corresponding to the conserved Z-ring protein, FtsZ (SI Appendix, Fig. S1D) (24–28).
Toga Lipid Patches and the IM Are Composed of the Same Major Species at Varying Proportions.
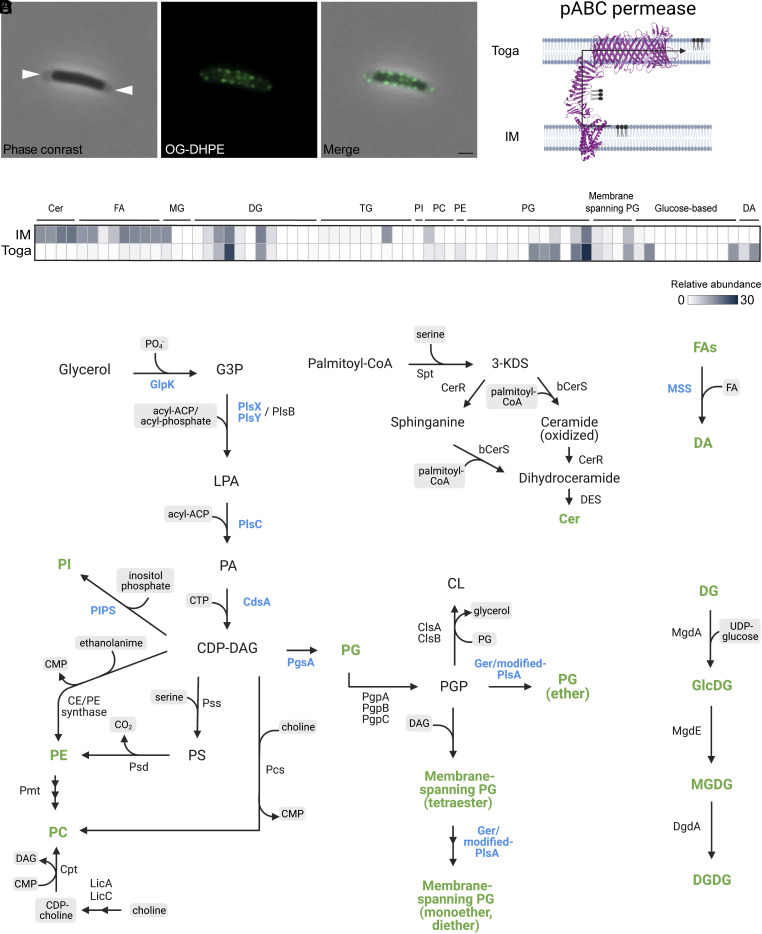
In order to visualize the distribution and abundance of lipid patches in the toga, we used the Oregon Green-1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine(OG-DHPE) lipid dye that incorporates nonspecifically into lipid bilayers. To distinguish the IM from the toga, we imaged cells in late-exponential phase. During that growth stage, the toga becomes significantly detached at the cell poles and can be clearly distinguished from the IM using phase contrast microscopy. Fluorescent light microscopy (fLM) of labeled cells revealed bright foci (~25 to 50 per cell) associated with the toga at the cell poles (Fig. 2A). The foci were ~200 nm in diameter (the resolution limit of fLM), consistent with the size of the lipid patches observed in our cryotomograms. Bright puncta were also observed along the lateral sides of cells likely due to staining of the toga. However, due to resolution limitations, the puncta in the toga could not be discerned from the IM. Weak uniform staining of the IM was observed when cells were stained for longer than 15 min, suggesting that the dye can diffuse through the toga and incorporate into the IM.
Fig. 2.
Lipid composition and distribution in the toga. (A) T. maritima cell in exponential growth labeled with OG-DHPE fluorescent dye. White arrows reveal areas of the toga distinctly separated from the IM. (Scale bar, 1 µm.) (B) Schematic demonstrating lipid transport from the IM to the toga via the cell envelope-spanning putative ABC permease. (C) Enrichment of major lipid classes in the IM and toga averaged over three biological replicates. (D) Map of known lipid synthetic pathways and their enzymes. Lipid species and enzymes detected in T. maritima are shown in green and blue, respectively. Abbreviations: G3P, glycerol-3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; CDP-DG, cytidine diphosphate-diacylglycerol; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine; PG, phosphatidylglycerol; PGP, phosphatidyl glycerophosphate; FA, fatty acid; CL, cardiolipin; DG, diacylglycerol; MG, monoacylglycerol; TG, triacylglycerol; Cer, ceramide; DA, diabolic acid.
T. maritima lacks genes associated with LPS synthesis; however, MS analysis of the toga detected the presence of a putative ABC permease (G4FGC1). Structure predictions and homology searches revealed that the protein had domains homologous to the Lpt lipid transport machinery in E. coli (29). In particular, the pABC permease had homologous domains to LptF/G, LptA, and LptD proteins associated with the IM, periplasm, and OM, respectively. Structure prediction revealed conserved tertiary folds within the pABC permease (Fig. 2B). Based on the characterization of these proteins in E. coli, we hypothesized that G4FGC1 in T. maritima encodes for a fusion pABC permease involved in lipid transport from the IM to the toga.
To identify the major lipid species and their abundancies, we performed lipidomic analysis on isolated fractions of the IM and toga. Overall, we found that the IM and toga shared the same types of polar lipids but in varying proportions (Fig. 2C). The IM was enriched in ceramides, free fatty acids, and phosphotidylcholine (PC), whereas the toga was enriched in phosphotidylglycerols and lipids containing unique glucose-based heads groups (Glu, diGlu, and deconyl-diGlu). Both fractions also contained mixed ether/ester membrane-spanning lipids with PG head groups. Notably, membrane-spanning DAs were detected only in the toga. All lipid species, including their relative abundances in the IM and toga, are listed in Dataset S1.
We used a combination of hidden Markov models (HMMs) and PSI BLAST searches to detect the presence of enzymes involved in lipid synthesis in the genome of T. maritima. We detected genes for PG and PI synthesis, as well as mixed ether/ester membrane-spanning lipids and DAs (Fig. 2D). Even though some lipids such as ceramides and DGs were abundant in the IM and toga, the known enzymes associated with their synthesis were absent from the genome of T. maritima. It is possible that these lipids are synthesized via novel or variant pathways or that these bacteria utilize retrograde transport and incorporate them from the environment.
The Toga Is Composed of Multiple Ompα and β-barrel Homologs.
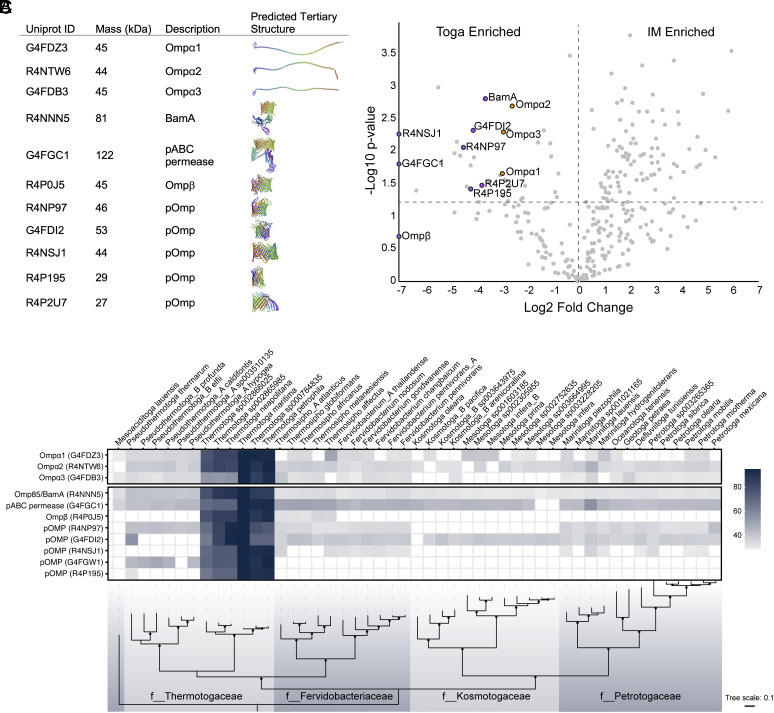
MS-based proteomics was used to characterize the protein content of the IM and the toga. Three of the most abundant proteins in the toga included Ompα homologs (Ompα1–3, ~45 kDa) (Fig. 3A). Tertiary structure prediction revealed that all the three homologs had a conserved SLH domain and formed extended coiled-coils, ~50 nm in length (Fig. 3A). Using AlphaFold2 Multimer (30), the three Ompα homologs were further predicted to be able to form trimers (SI Appendix, Fig. S3). An LysM-domain containing protein with a predicted α-helical structure (Q9WXR7) was also enriched and highly abundant in the toga. This protein could be functioning analogously to the Ompα homologs. Dataset S2 lists all detected proteins in the IM and toga with 95% confidence.
Fig. 3.
Major structural proteins of the toga. (A) Ompα1–3 and β-barrel proteins detected in the toga with their predicted tertiary structures. (B) Mass spectrometry-based proteomics reveal Ompα1–3 and β-barrel proteins are enriched in the toga. Ompα, orange dots; β-barrel proteins, purple dots. The significance cut off at P > 0.05 is indicated with a horizontal dashed line at 1.25 −log10 P-value. The zero-fold change is indicated with a vertical dashed line. (C) Homology conservation of toga-associated proteins across the Thermotogae phylum. PSI-BLAST hits are shaded by % amino acid similarity to T. maritima proteins. Species (Top) and family (f__) names (Bottom) are shown.
BamA and Ompβ were readily detected in the toga and predicted to form β-barrels (Fig. 3A). In addition, we detected six previously uncharacterized proteins with predicted β-barrels folds and Sec-dependent signal sequences. These putative OMPs (pOMPs) included proteins with UniprotID G4FGC1 (pABC permease), R4NP97, G4GDI2, R4NSJ1, R4P195, and R4P195. Except for Ompβ, all pOMPs, BamA, pABC permease, and Ompα1–3 homologs were significantly enriched in the toga (Fig. 3B). All the eight putative β-barrel proteins had extensive hydrophobic surfaces along the central portion and charged belts at the top and bottom (SI Appendix, Fig. S4). Using AlphaFold2 Multimer (31), all pOMPs (except BamA and pABC permease) could assemble into trimers with dimensions consistent with the subtomogram average of the β-barrel trimers in the toga (SI Appendix, Fig. S4). The models had high per-residue confidence (predicted local-distance difference test) values, an indicator for high accuracy of the model (SI Appendix, Fig. S5).
Homologs of the toga-associated proteins were conserved among multiple members of the family Thermotogaceae; however, only Ompα1, Ompα2, BamA, and pABC permease were found conserved in all members of the phylum (Fig. 3C). Using a two-step approach, SLH domains were identified in 124 proteins across the Thermotogae, 117 of which also had a coiled-coil domain (Dataset S3). This approach was used to identify Ompα homologs with shared overall domain architecture, despite significant (up to 70%) sequence variation in the coil-coiled region. Notably, Ompβ was poorly conserved, with homologs detected only in the genera Thermotoga and Thermosipho. Contrary to previous studies (16, 32), Ompβ was found in an operon with Ompα2 solely in the genus Thermotoga, and in an operon with either Ompα1 or Ompα2 in the genus Thermosipho. Of the pOMPs with predicted β-barrel tertiary structure, G4FDI2 and R4NP97 were best conserved, with some exceptions in Pseudothermotoga and Kosmotogaceae families, respectively. G4FGW1 and R4P195 were conserved only in the Thermotogaceae, whereas R4NSJ1 was only consistently found in the genera Thermotoga and Fervidobacterium. Together with the structure predictions, our results suggested that Ompα1, Ompα2, G4FDI2, and R4NP97 were conserved among all members of the phylum and likely the main constituents of the toga.
Other Proteins Detected in High Abundance in the Toga. The most abundant protein detected in the toga fraction was xylanase A, an enzyme that hydrolyses xylan into xylose consistent with the use of xylose as a carbon source in our growth media (Dataset S2). Maltodextrin glucosidase and β-xylosidase, enzymes involved in polysaccharide hydrolysis and transport, respectively (33), along with β-grasp domain containing protein, were also enriched in the toga.
Discussion
In this study, we characterized the unique cell envelope architecture of T. maritima and determined the lipid and protein composition of the toga. Previous studies have shown that the toga is composed of extended arrays of β-barrel trimers (16) and have proposed Ompβ as the major component (19, 32). Ompα was recognized as the periplasmic protein likely connecting the OM to PG, and since it was encoded in an operon with Ompβ, the two were proposed as the major structural components of the toga. Our tomograms confirm that the toga is composed of extended proteinaceous sheaths of β-barrel trimers (Fig. 1F). In addition to Ompβ, we detect five other highly abundant pOMPs with predicted β-barrel tertiary structures and three Ompα homologs in the toga (Fig. 3). We further show that Ompβ is not enriched in the toga, the operon structure of Ompα–Ompβ is poorly conserved, and homologs are found only in members of the Thermotoga and Thermosipho genera. In contrast, two novel pOMPs (G4FDI2 and R4NP97) and Ompα1 and Ompα2 were best conserved among all phylum members, leading us to propose them as the major structural components of the toga.
Recent bioinformatic analyses have shown that all diderm early-branching phyla have a conserved molecular tethering system that connects the OM to the PG (7). Encoded by the SlpA/OmpM superfamily of proteins, it is characterized by consisting of an N-terminal SLH domain, a coiled-coil domain, and a β-barrel C-terminal domain. Since Thermotogae lacked a homolog to the SlpA/OmpM protein, Ompα and Ompβ were proposed to act analogously. Based on our current study, we show that multiple Ompα and β-barrel proteins are present in the toga, and all members of the phylum have at least one homolog of each conserved in their genomes. Since the existence of a toga has been documented in all Thermotogae classes (34–43) and Thermotogae are known for their highly dynamic genomes (44), we conclude that members of the phylum have developed a bipartite OM-tethering system with high degree of plasticity. With the exception of BamA and pABC permease, none of the β-barrel proteins in Thermotogae have homologs in other phyla, which further suggests a unique evolutionary pathway of their OM-tethering system. Notably, the β-barrel fold is highly conserved among bacteria as it is likely related to function rather than protein sequence. The need for different OMPs in the toga may have evolved as an adaptation to extreme environments since different expression patterns could provide alternative properties. Since all Ompα and pOMPs were predicted to form trimers, we hypothesize that multiple proteins and combination of proteins could be assembling the proteinaceous sheath of the toga.
In addition to the extended arrays of β-barrel trimers, we detect the presence of lipid patches in the toga. Though the same lipid species were found in the IM and the toga, certain lipids were differentially enriched. Intriguingly, the membrane-spanning PG-ester/ether lipids, suggested to stabilize membranes at high temperatures (45), were found equally in the IM and the toga, whereas DAs were enriched in the toga (Fig. 2). The detection of a pABC permease with homologous domains to the LptF, LptG, and LptA proteins of the E. coli Lpt machinery provides a mechanism for lipid transport from the IM to the toga, analogous to other systems (46). We speculate that the presence of lipid patches in the toga is essential for proper function of membrane-embedded complexes such as BamA, the pABC permease, and T4P/T2SS system.
Evolutionary Implications.
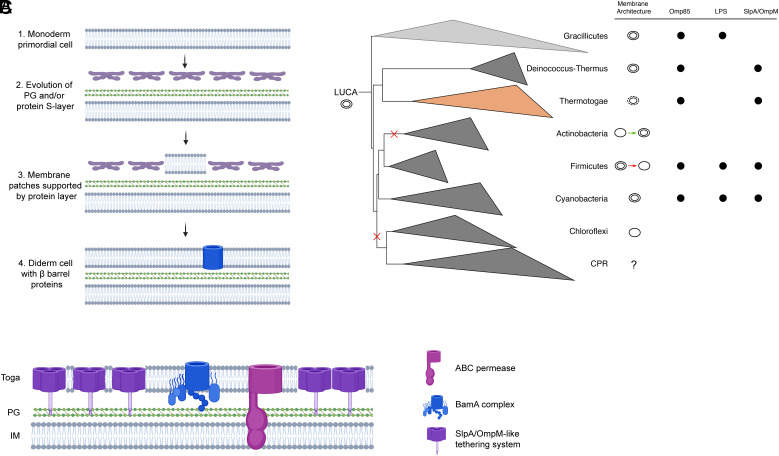
Major questions about the biogenesis and evolution of the cell envelope, particularly the OM of diderm bacteria, remain unresolved. Current phylogenomic evidence suggests that the last universal common ancestor (LUCA) was a diderm (10, 12). Yet, only three mechanistic models have been put forward describing how a diderm cell envelope could have evolved de novo: 1) Cavalier–Smith proposed that two half-cells fused to make a protocell with a double-membrane envelope (47), 2) Blobel theorized that protein–protein interactions on the surface of lipid vesicles could have mediated the formation of a diderm ancestor (48), and 3) Tocheva et al proposed that an ancient sporulation-like event could have transitioned a monoderm primordial cell into a diderm LUCA (49).
Due to its unique cell envelope and ancient origin, the Thermotoga phylum provides valuable insight into OM biogenesis. In particular, our study reveals that the toga of T. maritima is composed of extended protein arrays supporting small membrane patches. Based on these observations, we propose a model for OM biogenesis where a toga-like structure could have served as a structural intermediate between monoderm-to-diderm transitions (Fig. 4A). Briefly, due to the seemingly simpler architecture, we hypothesize that the primordial cell was monoderm (Fig. 4A, step 1). Additional surface layers (protein and/or PG) evolved over time to support the primordial cell (step 2). Lipid flux exchange likely facilitated accumulation of lipids in the surface layer, forming a structure analogous to the toga (step 3). Notably, a key property of the surface layer would be the ability to stabilize lipid bilayers. Over time, continuous buildup of the lipid molecules would have given rise to a second membrane (step 4). Since BamA is membrane embedded and essential for proper folding of other β-barrel proteins, our model suggests that the β-barrel fold is ancient and likely coevolved with the diderm cell envelope.
Fig. 4.
The toga as a model for OM biogenesis. (A) Major evolutionary steps outlining a proposed mechanism for OM biogenesis: 1) the primordial cell was likely monoderm, 2) PG and/or a protein surface layer evolved to support the bilayer of the primordial cell, 3) the abundance of surface proteins was able to support small membrane patches (analogous to the toga), 4) lipid accumulation extended to form a second membrane. Since the β-barrel fold (blue) is ancient, it likely evolved concurrently with the OM. (B) A phylogenetic tree of Terrabacteria depicting the major early-branching phyla Witwinowski et al. 7. Diversity in the cell envelopes could be explained with losses and gains of the OM as shown as red and green arrows for Firmicutes and Actinobacteria, respectively. Membrane architecture annotations are based on cryo-ET studies (except CPR, uncultured). Presence of OM signature features such as Omp85/BamA superfamily, LPS synthetic pathway, SlpA/OmpM tethering system, is indicated with black circles. (C) Cell envelope model for T. maritima based on current study showing major structural components of the toga. The SlpA/OmpM-like tethering system is likely composed of multiple Ompα and β-barrel proteins.
A diderm LUCA has significant implications for the cell envelope diversity in Terrabacteria, including multiple OM-losses and de novo biogenesis (Fig. 4B). For example, it is now clear that the last common ancestor of Firmicutes was diderm and that the monoderm members lost their OM (50, 51), possibly through a perturbation of their OM-tethering system (52). Monoderm and diderm members are found in Actinobacteria as well; however due to the unique nature of the MOM in Corynebacteriales, the phylum likely started off as monoderm and evolved a novel OM (53). Deinococcus-Thermus and Cyanobacteria retained their OMs, whereas Chloroflexi and CPR lost theirs (52). Intriguingly, a diderm LUCA would suggest that Thermotogae reverted their OM into a toga intermediate, a notion that remains to be explored further.
Materials and Methods
Strains and Growth Conditions.
Thermotoga maritima MSB8 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) German Collection of Microorganisms and Cell Cultures and grown in DSMZ medium 1,232 or TMB medium with 0.5% xylose (54). T. maritima is an anaerobic and hyperthermophilic organism. As such, great care was taken to provide cells with an optimal growth environment. Liquid media were sterilized by passing through a 0.22-μ filter, then flushed with N2 gas to remove oxygen. Uninoculated media were left to equilibrate in the anaerobic chamber overnight before inoculation. Resazurin, an oxygen sensor, was added to the growth medium to indicate oxygen presence (bright pink). The anaerobic growth media were inoculated in a BactronEZ anaerobic growth chamber (Sheldon Manufacturing Inc.) maintained at an atmosphere of 5% H2, 10% CO2, and 85% N2. Hungate tubes were sealed before removing them from the anaerobic chamber and incubated at 80 °C for 16 h to reach mid-exponential growth phase. Cultures were manipulated in the anaerobic chamber and maintained in anaerobic conditions for all experiments unless otherwise stated.
Cryo-ET, Data Reconstruction, and Subtomogram Averaging.
To prepare grids for cryo-ET, a small volume of culture was mixed anaerobically with 15 nm or 20 nm gold fiducial markers. Cells were loaded onto glow-discharged Cu grids (R2/2, Quantifoil), plunge-frozen in liquid ethane–propane, and cooled to liquid nitrogen temperatures within 1 min of oxygen exposure using the Vitrobot Mark IV (ThermoFisher Scientific). Grids were stored in liquid nitrogen until cryo-ET data collection. Tilt series were collected using SerialEM (55) on a Titan Krios 300 kV transmission electron microscope (ThermoFisher Scientific) equipped with a Falcon III direct electron detector. Data collection conditions were −6 µm defocus, 120 e/Å total dose, +/− 60° oscillations, with 1° tilt increments. Tilt series were aligned using IMOD, contrast transfer function (CTF) was corrected using CTFFind4 and novaCTF, and 3D reconstructions were calculated using IMOD with the back-weighted projection method (56–59). Density profiles were calculated using ImageJ (60). For subtomogram averaging of the toga, 1,584 subvolumes were extracted and averaged with Dynamo (61). Alignment was refined eight times without applying symmetry.
Membrane Labeling and Fluorescence Light Microscopy (fLM).
In order to detect lipids in the toga, we used the OG 488 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine (OG-DHPE, ThermoFisher Scientific). OG-DHPE is a synthetic PE lipid analog coupled with a fluorophore that embeds nonspecifically into lipid bilayers. Prior to labeling, cells were grown for 16 h at 80 °C, then stained with 10 µg/mL OG-DHPE directly in the culture medium for 15 min under anoxic conditions. The cells were washed once with PBS pH 7.5 and slides for fLM were prepared in the anaerobic chamber. The cells were imaged immediately with phase contrast and fLM using Zeiss Axio Examiner equipped with a 100×/1.46 oil immersion lens. Total imaging time was less than 5 min. Images were collected and processed using Zen Blue 2.6 software. The cells remained viable with consistent fLM staining patterns for at least 30 min after oxygen exposure.
IM and Toga Isolation.
The IM and toga were isolated from a 2-g wet weight cell pellet from 1.5 L of culture following established protocols (62). Briefly, cells were incubated for 16 h at 80 °C and spun down at 4000 g for 5 min under anaerobic conditions. The cells were washed once in anaerobic PBS to remove medium contaminates and frozen at −80 ˚C. Cell pellets were resuspended in 10 mL lysis buffer (25 mM Tris pH 7.4, 5 mM ETDA, 500 mM NaClO4, 1 mM PMSF, 40 µg/mL lysozyme, 20 U/mL DNase) and passed two times through a homogenizer at 50 psi. Unlysed cells and large debris were removed by centrifugation at 4,000 g for 10 min. The supernatant, containing the whole membrane fraction, was isolated by ultracentrifugation at 100,000 g for 2 h. The membrane pellet was washed three times with 10 mL wash buffer (25 mM Tris pH 7.4, 5 mM ETDA, 500 mM NaClO4) and pelleted each time at 100,000 g for 1 h. To separate the IM from the toga, the whole membrane fraction was resuspended in 200 µL wash buffer and layered on a sucrose gradient containing 1.2 mL of 70% (w/w), 2 mL of 50% (w/w), and 1.6 mL of 20% (w/w) sucrose in wash buffer and spun at 100,000 g for 2 h. Bands were removed from the top and were washed once with 20 mL wash buffer to remove excess sucrose. The bands were resuspended in wash buffer and run separately on a second sucrose gradients. Individual bands were removed from the top and washed once in wash buffer to remove excess sucrose. An NADH oxidase assay (Sigma) was used to assess the quality of the fractionation. As expected, the toga fractions had no NADH oxidase activity, whereas the IM fractions had NADH oxidase indicative of successful separation of the IM from the toga.
Lipid Extraction and Analysis.
Biological triplicates of the IM and toga fractions were analyzed in technical triplicates of their lipid composition using liquid chromatography-mass spectrometry (LC-MS) analysis. Lipids were extracted in methanol:acetonitrile: water (2:2:1, vol:vol:vol) and methyl-tert-butyl ether. Solvent was evaporated from the lipid containing fraction using vacuum concentration. The resulting dried residue was reconstituted in isopropanol:acetonitrile (1:1, vol:vol), and lipid profiling of cellular membranes was carried out using a reverse-phase Acquity UPLC BEH C18 column (1.0 × 100 mm, 1.7 µm, Waters) on an Agilent 1,290 Infinity II ultra-high-performance liquid chromatography system (Agilent Technologies) coupled with Bruker Impact II electrospray-ionization quadrupole time-of-flight mass spectrometer (Bruker Daltonics). Lipid identification was completed on MS-DIAL 4.7 tool based on mass accuracy, isotope ratio, and retention time along with MS/MS similarity against publicly available libraries. MS-DIAL 4.7 can be downloaded at the PRIMe website (http://prime.psc.riken.jp/) (63). Raw lipidomics data were collected in positive and negative ion modes, after intensity correction and normalization (post quality control (QC) calibration). QC calibration was done to obtain more accurate fold changes: The relative standard deviation (rsd) of the QC injections (at the beginning, middle, and end of the injection sequence) was below the threshold of 0.25. Relative abundance was calculated as an average percentage of all mass spectrometry intensities in a given sample from three technical replicates +/− 1 SD. Enrichment was calculated using a Student’s t test (P < 0.05) to determine whether the average of technical triplicates for IM and toga were significantly different for each biological replicate. Lipid species were denoted as enriched if they were significantly different in two out of three biological replicates. It should be noted that intact polar lipid species have diverse degrees of ionization efficiencies and hence the relative intensities of different components do not necessarily reflect their actual relative abundance. Additional details of the lipidomics analysis can be found in SI Appendix.
Protein Identification and Structure Prediction.
Triplicates of IM and toga fractions from the sucrose gradients were visualized on an SDS-PAGE gel and excised for subsequent digestion with trypsin for MS analysis in triplicate. Tryptic peptides were analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (ThermoFisher Scientific) operated with Xcalibur (version 4.0.21.10) and coupled to a Scientific Easy-nLC 1200 system. Monoisotopic precursors were searched against the T. maritima MSB8 proteome (Uniprot ID 243274) using RawConverter (v1.1.0.18; The Scripps Research Institute). Peptides identified with a score having a confidence higher than 95% were kept for further analysis (Datasets S4 and S5). Scaffold 4 (Proteome Software Inc.) was used for statistical analysis and to generate the volcano plot. Additional details of the proteomics analysis can be found in SI Appendix. Protein localization was predicted using pSORTb (64), β-barrel prediction was done using BOMP and PRED-TMBB2 (65, 66), and tertiary structures were predicted using RoseTTAFold (67). Trimer structure predictions were performed using AlphaFold2 Multimer v2.3.0 (31), and the surface hydrophobicity and electrostatic charge for the highest ranked model were computed in ChimeraX (68). Per-residue confidence plots for all structures were generated using Alphapickle and are available in SI Appendix, Figs. S3 and S5.
Phylogeny and Sequence-Based Homology Searches.
To generate a phylogenetic tree of the phylum Thermotogae, the maximum-likelihood algorithm was used to extract an alignment of 120 conserved bacterial single-copy marker genes (69) from publicly available Thermotogae members using the Genome Taxonomy Database GTDB-Tk v1.4.0 software (70). The tree was constructed using IQ-TREE v2.1.4 with 1,000 ultrafast bootstraps and the substitution model LG+F+R6, as determined by ModelFinder (71, 72). Representative Coprothermobacterota genomes served as an outgroup. Publicly available phylogenetic tree data, created from the alignment of concatenated RNA polymerase subunits β, β′, and elongation factor IF-2 proteins rooted between Gracilicutes and Terrabacteria, were modified to represent early-branching phyla (7). Both trees were visualized using iTOL (v4) (73).
To assess the genomes of Thermotoga members for the presence of homologs of toga-associated proteins detected with MS-based proteomics in T. maritima, we performed gene prediction using Prodigal on publicly available Thermotoga genomes from the GTDB to generate proteomes with locus tag information (69, 74). PSI-BLAST searches were performed on these proteomes using four iterations and an E value of 1e−10. To detect Ompα proteins, we used a two-step approach. First, we searched all Thermotoga genomes for proteins with an SLH domain using the Pfam profile PF00395. Next, we inspected all sequences with N-terminal SLH domains for the presence of a central coiled-coil segment using PCOILS (75). Since none of the identified proteins had a C-terminal β-barrel component, the presence of a β-barrel in an operon with the identified Ompα homologs was assessed manually.
To identify lipid biosynthetic enzymes, we searched the proteomes of T. maritima (Uniprot IDs UP000013901 and 3AUP00000818) using a curated list of HMMs and reference protein sequences. HMM searches were performed using HMMER with default hmmsearch settings (http://hmmer.org/), and PSI-BLAST searches were conducted using four iterations and an E value of 1e−10.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Tomogram of a T. maritima cell. 3-dimensional volume shows the cell envelope of T. maritima (corresponds to Fig. 1A). The toga is dissociating at the tip of the cell. Side views of monolayers and bilayers of the OM are clearly visible.
Tomogram of the toga of a T. maritima cell. 3-dimensional volume of the tip of a T. maritima cell shows the extended toga. The monolayer and bilayer side views are highlighted with purple and blue arrows, respectively (Fig. S1). The extended protein arrays and lipid patches are outlined with purple dashed lines.
Acknowledgments
This project was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to E.I.T. (RGPIN 04345). D.L.S. was supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (546024) and A.H. was supported by a Natural Sciences and Engineering Research Council of Canada Post Graduate Scholarship (552674). We thank Drs. Claire Atkinson and Florian Rossman and the High Resolution Macromolecular Cryo-Electron Microscopy facility at the University of British Columbia for assistance with microscope operation and tilt series acquisition. We thank Dr. Laurent Brechenmacher at the Southern Alberta Mass Spectrometry Center for proteomics analysis. We also thank Dr. Nicole Bale and Dr. Diana Sahonero-Canaves for helpful discussions.
Author contributions
D.L.S. and E.I.T. designed research; D.L.S., A.H., P.B., L.S., and E.I.T. performed research; D.L.S., A.H., P.B., L.S., T.H., and E.I.T. analyzed data; and D.L.S., A.H., and E.I.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010), 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoiczyk E., Hansel A., Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J. Bacteriol. 182, 1191–1199 (2000), 10.1128/jb.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balish M. F., Mycoplasma pneumoniae, an underutilized model for bacterial cell biology. J. Bacteriol. 196, 3675–3682 (2014), 10.1128/JB.01865-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexton D. L., Burgold S., Schertel A., Tocheva E. I., Super-resolution confocal cryo-CLEM with cryo-FIB milling for in situ imaging of deinococcus radiodurans. Curr. Res. Struct. Biol. 4, 1–9 (2022), 10.1016/j.crstbi.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Kügelgen A., van Dorst S., Alva V., Bharat T. A. M., A multidomain connector links the outer membrane and cell wall in phylogenetically deep-branching bacteria. Proc. Natl. Acad. Sci. U.S.A. 119, e2203156119 (2022), 10.1073/pnas.2203156119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz G. E., The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565, 308–317 (2002), 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 7.Witwinowski J., et al. , An ancient divide in outer membrane tethering systems in bacteria suggests a mechanism for the diderm-to-monoderm transition. Nat. Microbiol. 7, 411–422 (2022), 10.1038/s41564-022-01066-3. [DOI] [PubMed] [Google Scholar]
- 8.Sutcliffe I. C., A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470 (2010), 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Taib, et al. , Genome-wide analysis of the firmicutes illuminates the diderm/monoderm transition. Nat. Ecol. Evol. 4, 1661–1672 (2020), 10.1038/s41559-020-01299-7. [DOI] [PubMed] [Google Scholar]
- 10.Megrian D., Taib N., Witwinowski J., Beloin C., Gribaldo S., One or two membranes? Diderm firmicutes challenge the gram-positive/gram-negative divide. Mol. Microbiol. 113, 659–671 (2020), 10.1111/mmi.14469. [DOI] [PubMed] [Google Scholar]
- 11.Gupta R. S., Bhandari V., Phylogeny and molecular signatures for the phylum thermotogae and its subgroups. Antonie Van Leeuwenhoek 100, 1–34 (2011), 10.1007/s10482-011-9576-z. [DOI] [PubMed] [Google Scholar]
- 12.Coleman G. A., et al. , A rooted phylogeny resolves early bacterial evolution. Science 372, eabe0511 (2021), 10.1126/science.abe0511. [DOI] [PubMed] [Google Scholar]
- 13.Huber R., et al. , Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic euhacteria growing up to 90°. Arch. Microbiol. 144, 324–333 (1986). [Google Scholar]
- 14.Sahonero-Canavesi D. X., et al. , Changes in the distribution of membrane lipids during growth of thermotoga maritima at different temperatures: Indications for the potential mechanism of biosynthesis of ether-bound diabolic acid (membrane-spanning) lipids. Appl. Environ. Microbiol. 88, e0176321 (2021), 10.1128/AEM.01763-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong P. L., Archaebacterial bipolar tetraether lipids: Physico-chemical and membrane properties. Chem. Phys. Lipids 163, 253–265 (2010), 10.1016/j.chemphyslip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Rachel R., Engel A. M., Huber R., Stetter K. O., Baumeister W., A porin-type protein is the main constituent of the cell envelope of the ancestral eubacterium thermotoga maritima. FEBS Lett. 262, 64–68 (1990). [Google Scholar]
- 17.Engel A., Cejka Z., Lupas A., Lottspeich F., Baumeister W., Isolation and cloning of OmpA, a coiled-coil protein spanning the periplasmic space of the ancestral eubacterium thermotoga maritima. EMBO J. 11, 4369–4378 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupas A., et al. , Model structure of the Omp alpha rod, a parallel four-stranded coiled coil from the hyperthermophilic eubacterium thermotoga maritima. J. Mol. Biol. 248, 180–189 (1995), 10.1006/jmbi.1995.0210. [DOI] [PubMed] [Google Scholar]
- 19.Petrus A. K., et al. , Genes for the major structural components of thermotogales species’ togas revealed by proteomic and evolutionary analyses of OmpA and OmpB homologs. PLoS One 7, e40236 (2012), 10.1371/journal.pone.0040236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesnage S., et al. , Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. Embo J. 19, 4473–4484 (2000), 10.1093/emboj/19.17.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Kugelgen A., van Dorst S., Alva V., Bharat T. A. M., A multidomain connector links the outer membrane and cell wall in phylogenetically deep-branching bacteria. Proc. Natl. Acad. Sci. U.S.A. 119, e2203156119 (2022), 10.1073/pnas.2203156119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conners S. B., et al. , Microbial biochemistry, physiology, and biotechnology of hyperthermophilic thermotoga species. FEMS Microbiol. Rev. 30, 872–905 (2006), 10.1111/j.1574-6976.2006.00039.x. [DOI] [PubMed] [Google Scholar]
- 23.Nelson K. E., et al. , Evidence for lateral gene transfer between archaea and bacteria from genome sequence of thermotoga maritima. Nature 399, 323–329 (1999), 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 24.Megrian D., Taib N., Jaffe A. L., Banfield J. F., Gribaldo S., Ancient origin and constrained evolution of the division and cell wall gene cluster in bacteria. Nat. Microbiol. 7, 2114–2127 (2022), 10.1038/s41564-022-01257-y. [DOI] [PubMed] [Google Scholar]
- 25.Khanna K., Lopez-Garrido J., Sugie J., Pogliano K., Villa E., Asymmetric localization of the cell division machinery during bacillus subtilis sporulation. Elife 10, e62204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z., Trimble M. J., Brun Y. V., Jensen G. J., The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708 (2007), 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szwedziak P., Wang Q., Bharat T. A., Tsim M., Löwe J., Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3, e04601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Q., et al. , Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis. EMBO J. 36, 10 (2017), 10.15252/embj.201696235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., Kahne D., Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016), 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans R., et al. , Protein complex prediction with AlphaFold-multimer. bioRxiv [Preprint] (2021). 10.1101/2021.10.04.463034 (Accessed 5 August 2022). [DOI]
- 31.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2022). 10.1101/2021.10.04.463034 (Accessed 6 February 2023). [DOI]
- 32.Engel A. M., Brunen M., Baumeister W., The functional properties of OmpB, the regularly arrayed porin of the hyperthermophilic bacteriium Thermotoga maritima. FEMS Microbiol. Lett. 109, 231–236 (1993). [Google Scholar]
- 33.Meissner H., Liebl W., Thermotoga maritima maltosyltransferase, a novel type of maltodextrin glycosyltransferase acting on starch and malto-oligosaccharides. Eur. J. Biochem. 258, 1050–1058 (1998), 10.1046/j.1432-1327.1998.2581050.x. [DOI] [PubMed] [Google Scholar]
- 34.Urios L., et al. , Thermosipho atlanticus sp. nov., a novel member of the thermotogales isolated from a mid-atlantic ridge hydrothermal vent. Int. J. Syst. Evol. Microbiol. 54, 1953–1957 (2004), 10.1099/ijs.0.63069-0. [DOI] [PubMed] [Google Scholar]
- 35.Podosokorskaya O. A., et al. , Fervidobacterium riparium sp. nov., a thermophilic anaerobic cellulolytic bacterium isolated from a hot spring. Int. J. Systemat. Evol. Microbiol. 61, 2697–2701 (2011), 10.1099/ijs.0.026070-0. [DOI] [PubMed] [Google Scholar]
- 36.Roumagnac M., et al. , Responses to the hydrostatic pressure of surface and subsurface strains of pseudothermotoga elfii revealing the piezophilic nature of the strain originating from an oil-producing well. Front. Microbiol. 11, 588771 (2020), 10.3389/fmicb.2020.588771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiPippo J. L., et al. , Kosmotoga olearia gen. nov., sp. nov., a thermophilic, anaerobic heterotroph isolated from an oil production fluid. Int. J. Systemat. Evol. Microbiol. 59, 2991–3000 (2009), 10.1099/ijs.0.008045-0. [DOI] [PubMed] [Google Scholar]
- 38.Miranda-Tello E., et al. , Petrotoga mexicana sp. nov., a novel thermophilic, anaerobic and xylanolytic bacterium isolated from an oil-producing well in the gulf of mexico. Int. J. Systemat. Evol. Microbiol. 54, 169–174 (2004), 10.1099/ijs.0.02702-0. [DOI] [PubMed] [Google Scholar]
- 39.Reysenbach A.-L., et al. , Mesoaciditoga lauensis gen. nov., sp. nov., a moderately thermoacidophilic member of the order thermotogales from a deep-sea hydrothermal vent. Int. J. Systemat. Evol. Microbiol. 63, 4724–4729 (2013), 10.1099/ijs.0.050518-0. [DOI] [PubMed] [Google Scholar]
- 40.Jayasinghearachchi H. S., Lal B., Oceanotoga teriensis gen. nov., sp. nov., a thermophilic bacterium isolated from offshore oil-producing wells. Int. J. Systemat. Evol. Microbiol. 61, 554–560 (2011), 10.1099/ijs.0.018036-0. [DOI] [PubMed] [Google Scholar]
- 41.Semenova E. M., Grouzdev D. S., Tourova T. P., Nazina T. N., Physiology and genomic characteristics of geotoga petraea, a bacterium isolated from a low-temperature petroleum reservoir (Russia). Microbiology 88, 662–670 (2019). 10.1134/S0026261719060171. [DOI] [Google Scholar]
- 42.Nesbø C. L., et al. , Mesotoga prima gen. nov., sp. nov., the first described mesophilic species of the thermotogales. Extremophiles 16, 387–393 (2012), 10.1007/s00792-012-0437-0. [DOI] [PubMed] [Google Scholar]
- 43.Alain K., et al. , Marinitoga piezophila sp. nov., a rod-shaped, thermo-piezophilic bacterium isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int. J. Systemat. Evol. Microbiol. 52, 1331–1339 (2002), 10.1099/00207713-52-4-1331. [DOI] [PubMed] [Google Scholar]
- 44.Nesbø C. L., et al. , Evidence for extensive gene flow and thermotoga subpopulations in subsurface and marine environments. ISME J. 9, 1532–1542 (2015), 10.1038/ismej.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siliakus M. F., van der Oost J., Kengen S. W. M., Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21, 651–670 (2017), 10.1007/s00792-017-0939-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutcliffe I. C., A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 18, 464–470 (2010), 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Cavalier-Smith T., Obcells as proto-organisms: Membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J. Mol. Evol. 53, 555–595 (2001), 10.1007/s002390010245. [DOI] [PubMed] [Google Scholar]
- 48.Blobel G., Intracellular protein topogenesis. Proc. Natl. Acad. Sci. U.S.A. 77, 1496–1500 (1980), 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tocheva E. I., Ortega D. R., Jensen G. J., Sporulation, bacterial cell envelopes and the origin of life. Nat. Rev. Microbiol. 14, 535–542 (2016), 10.1038/nrmicro.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tocheva E. I., et al. , Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146, 799–812 (2011), 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taib N., et al. , Genome-wide analysis of the firmicutes illuminates the diderm/monoderm transition. Nat. Ecol. Evol. 4, 1661–1672 (2020), 10.1038/s41559-020-01299-7. [DOI] [PubMed] [Google Scholar]
- 52.Witwinowski J., et al. , An ancient divide in outer membrane tethering systems in bacteria suggests a mechanism for the diderm-to-monoderm transition. Nat. Microbiol. 7, 411–422 (2022), 10.1038/s41564-022-01066-3. [DOI] [PubMed] [Google Scholar]
- 53.Vincent A. T., et al. , The mycobacterial cell envelope: A relict from the past or the result of recent evolution? Front. Microbiol. 9, 2341 (2018), 10.3389/fmicb.2018.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y., Zhou Q., Wu K., Li X.-Q., Shao W.-L., A highly efficient method for liquid and solid cultivation of the anaerobic hyperthermophilic eubacterium thermotoga maritima. FEMS Microbiol. Lett. 259, 254–259 (2006), 10.1111/j.1574-6968.2006.00273.x. [DOI] [PubMed] [Google Scholar]
- 55.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005), 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996), 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 57.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017), 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turoňová B., Schur F. K. M., Wan W., Briggs J. A. G., Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4 Å. J. Struct. Biol. 199, 187–195 (2017), 10.1016/j.jsb.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015), 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012), 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castaño-Díez D., Kudryashev M., Stahlberg H., Dynamo catalogue: Geometrical tools and data management for particle picking in subtomogram averaging of cryo-electron tomograms. J. Struct. Biol. 197, 135–144 (2017), 10.1016/j.jsb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Rachel R., Engel A. M., Huber R., Stetter K. O., Baumeister W., A porin-type protein is the main constituent of the cell-envelope of the ancestral eubacterium thermotoga maritima. FEBS Lett. 262, 64–68 (1990), 10.1016/0014-5793(90)80155-C. [DOI] [Google Scholar]
- 63.Lai Z., et al. , Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53–56 (2018), 10.1038/nmeth.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu N. Y., et al. , PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010), 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berven F. S., Flikka K., Jensen H. B., Eidhammer I., BOMP: A program to predict integral beta-barrel outer membrane proteins encoded within genomes of gram-negative bacteria. Nucleic Acids Res. 32, W394–W399 (2004), 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsirigos K. D., Elofsson A., Bagos P. G., PRED-TMBB2: Improved topology prediction and detection of beta-barrel outer membrane proteins. Bioinformatics 32, i665–i671 (2016), 10.1093/bioinformatics/btw444. [DOI] [PubMed] [Google Scholar]
- 67.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021), 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen F. P., et al. , UCSF chimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021), 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parks D. H., et al. , A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018), 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 70.Parks D. H., et al. , GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794 (2022), 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017), 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minh B. Q., et al. , IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020), 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019), 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010), 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gabler F., et al. , Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinformat. 72, e108 (2020), 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Tomogram of a T. maritima cell. 3-dimensional volume shows the cell envelope of T. maritima (corresponds to Fig. 1A). The toga is dissociating at the tip of the cell. Side views of monolayers and bilayers of the OM are clearly visible.
Tomogram of the toga of a T. maritima cell. 3-dimensional volume of the tip of a T. maritima cell shows the extended toga. The monolayer and bilayer side views are highlighted with purple and blue arrows, respectively (Fig. S1). The extended protein arrays and lipid patches are outlined with purple dashed lines.
Data Availability Statement
All study data are included in the article and/or SI Appendix.