Abstract
Antibiotic resistance is nowadays a major public health issue. Rapid antimicrobial susceptibility tests (AST) are one of the options to fight this deadly threat. Performing AST with single-cell sensitivity that is rapid, cheap, and widely accessible, is challenging. Recent studies demonstrated that monitoring bacterial nanomotion by using atomic force microscopy (AFM) upon exposure to antibiotics constitutes a rapid and highly efficient AST. Here, we present a nanomotion detection method based on optical microscopy for testing bacterial viability. This novel technique only requires a very basic microfluidic analysis chamber, and an optical microscope equipped with a camera or a mobile phone. No attachment of the microorganisms is needed, nor are specific bacterial stains or markers. This single-cell technique was successfully tested to obtain AST for motile, nonmotile, gram-positive, and gram-negative bacteria. The simplicity and efficiency of the method make it a game-changer in the field of rapid AST.
Keywords: nanomotion, bacteria, antibiotic, optical microscopy
The recent proliferation of bacterial resistance to antibiotics is among the most serious public health concerns (1). An efficient way to limit the proliferation of resistant bacteria consists in starting the therapeutic process with the most efficient drug (2), which is only possible if rapid antibiotic susceptibility testing (AST) methods are available. Unfortunately, traditional replication-based AST methods typically last 24 h, and even up to 1 month for slow-growing bacteria. Recently, some rapid AST techniques have been developed, such as whole genome sequencing, matrix-assisted laser desorption/ionization time-of-flight spectrometry, Fourier transform infrared spectroscopy, and various microfluidics-based technologies (3). However, these methods are complex and need sophisticated and costly equipment. A couple of years ago, we demonstrated that virtually all living organisms on Earth oscillate at a nanometric scale. These oscillations can be used to monitor their life/death state (4). We used this “signature of life” to develop nanomotion detectors that conduct AST in a timeframe of 1 to 2 h. The technique consists of attaching the organism of interest onto a cantilever and monitoring its oscillations as a function of time before and during the treatment with antibiotics (5). To simplify the technique, we recently demonstrated that traditional optical microscopes equipped with a camera can also detect nanomotions of cells such as yeast cells (Saccharomyces cerevisiae, Candida albicans, Candida lusitaniae, and Candida glabrata) (6). Their sensitivity or resistance to antifungals could be assessed in a simple and rapid way by monitoring cellular oscillations. Since individual cells are resolved, this technique, referred to as optical nanomotion detection (ONMD), is also single-cell sensitive.
In the present contribution, we modified and extended this detection method to bacterial AST. Clinical and research-relevant strains, such as Escherichia coli (gram-negative, motile), Staphylococcus aureus (gram-positive nonmotile), Lactobacillus rhamnosus (gram-positive, nonmotile), and Mycobacterium smegmatis (nonmotile and nonpathogenic bacterial model for tuberculosis) were analyzed. The experiments successfully demonstrated that ONMD monitors not only life–death transitions upon exposure to different chemicals but also variations in the bacterial metabolic level induced by different nutrient concentrations. We demonstrate that the technique only requires a basic traditional optical microscope, a camera/mobile phone, and dedicated software to achieve rapid, single-cell sensitive, attachment-, and label-free AST.
Results and Discussion
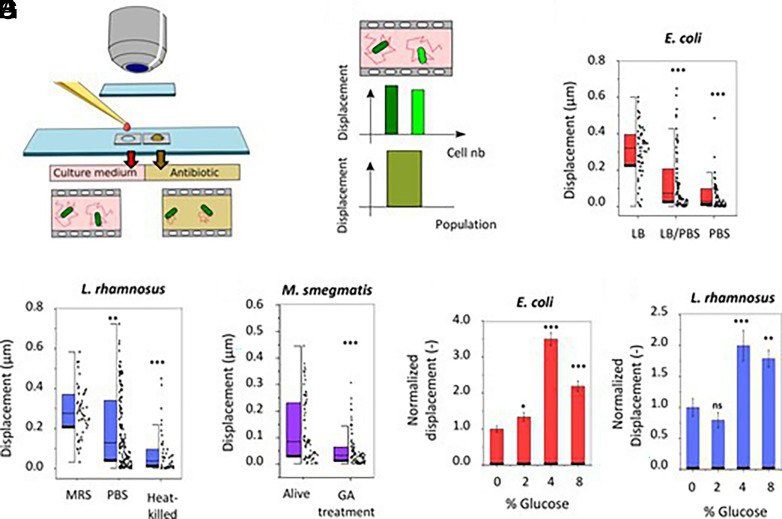
The organisms of interest were placed in a homemade analysis chamber and their displacements were recorded with a traditional optical microscope and a video camera/mobile phone (Fig. 1A). Cellular displacements were measured by tracking every single cell with a subpixel resolution (7) (Fig. 1B) or by highlighting in false colors the regions of the field of view that changed the most between consecutive frames. In the first set of experiments, we investigated the effect of the dilution or the replacement of the bacterial growth medium with phosphate-buffered saline. The lack of nutrients induced a dramatic decrease in the displacements of motile E. coli, and nonmotile L. rhamnosus (Fig. 1 C and D). In these experiments, we also explored the ability of ONMD to monitor life-dead states in these bacterial species. Differences in nanomotion between living and glutaraldehyde (GA)-killed M. smegmatis were highlighted by this technique (Fig. 1E). M. smegmatis is used as a model organism for Mycobacterium tuberculosis whose traditional (replication rate-based) antibiotic sensitivity tests last several weeks.
Fig. 1.
Optical nanomotion detection of bacteria. (A) Sample preparation method. (B) The data analysis software calculates the displacements of single cells (two green columns) as well as the mean displacement of a full cellular population (one large green column). Effect of the nutrient concentration on (C) E. coli (n > 50) and (D) L. rhamnosus (n > 41), heating on (D) L. rhamnosus (n > 41), 5% GA incubation on (E) M. smegmatis (n > 75), and the glucose concentration on (F) E. coli (n > 286) and on (G) L. rhamnosus (n > 144). Student's t test: ***P < 0.001; **P < 0.01; *P < 0.05. ♦: single-cell displacement.
Next, we assessed the effect of the carbon source, glucose, on bacterial nanomotion. We incubated E. coli and L. rhamnosus cells in their respective culture medium with different glucose concentrations. By analyzing the cellular nanomotions, we noticed that increasing the glucose concentration up to 4% (m/v) increased the E. coli nanomotion by 350% whereas a further increase in glucose concentration induced a drop in the bacterial movements (Fig. 1F). This phenomenon can be explained by the inhibitory effect of high glucose concentrations on E. coli flagella (8). The results are fully consistent with previously published data obtained by AFM-based nanomotion detection (9). The effect of the glucose on L. rhamnosus also induced a significant increase in its nanomotion (Fig. 1G).
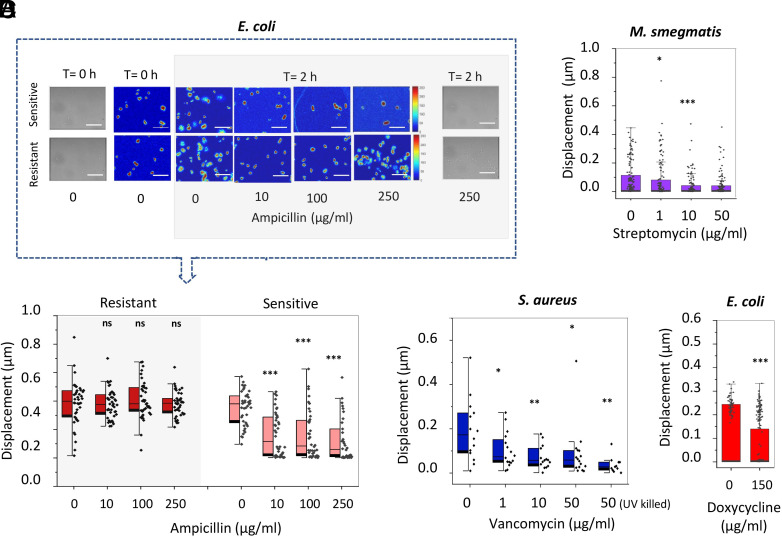
Finally, we explored the capacity of the ONMD technique to monitor bacterial sensitivity to antibiotics in their exponential phase. For this purpose, we incubated various bacterial species with different concentrations of chemicals and monitored their nanomotion after exposure to the drugs. The sensitive E. coli strain showed a statistically significant decrease in its average nanomotion after exposure for 2 h to ampicillin (Fig. 2A) whereas the resistant strain was unaffected. Similarly, a reduction in the average nanomotion was revealed for M. smegmatis after 5 h incubation with streptomycin (Fig. 2B). The ONMD analysis of a bacterial populations (i.e., displacement analysis of the full field of view) highlighted the response of S. aureus to different concentrations of vancomycin (Fig. 2C). In this case too, the total displacement of the whole cellular population was significantly reduced. Finally, E. coli exposed to doxycycline for 2 h, decreased their nanomotion by 40% demonstrating that the technique is sensitive to bactericidal as well bacteriostatic antibiotics (Fig. 2D). In all previously described experiments, we considered dead bacteria’s displacements close to purely Brownian motion to which living cells’ nanomotion was compared.
Fig. 2.
Nanomotion of bacteria upon exposure to antibiotics. (A) Optical, changing pixels images and cell displacement of sensitive (n > 34) and resistant (n = 42) E. coli after 2 h ampicillin incubation; Scale bar: 10 µm. (B) Nanomotion of M. smegmatis exposed to increasing concentration of streptomycin during 5 h (n > 100). (C) Whole population nanomotion of S. aureus after 2 h vancomycin exposure and UV-killed cells as dead population control (n > 11). (D) Nanomotion of E. coli exposed to doxycycline during 2 h (n > 50). Student’s t test: ***P < 0.001; **P < 0.01; *P < 0.05; ns: not significant. ♦: single-cell (A, B, and D) or single population (C) displacement.
Conclusion
Rapid identification of the most appropriate antibiotic is key in limiting the spread of multi-resistant bacteria. The present technique is rapid, extremely simple to set up, possesses a single-cell sensitivity, is attachment- and label-free, and significantly faster and cheaper than other existing fast AST methods (10). Importantly, it is independent of the bacterial replication rate, the structure of their cell wall, or motility. This technique allowed us to study bacteria viability and assess nanomotion under different nutrient and antibiotics conditions. Its simplicity opens novel avenues to fast AST, specifically in developing countries. Additional application domains could include the search for living organisms in extreme environments in a chemistry-independent manner, such as on some planets in the Universe.
Materials and Methods
A 0.6 to 1 µL droplet of the bacteria solution is used to fill a microfluidic analysis chamber that is eventually placed under a traditional optical microscope. Alternatively, to limit bacteria adhesion to the analysis chamber, the bacteria suspensions are mixed before the measurement in oil. Typically, the organisms are observed with an optical microscope and 10-s movies are recorded (30 fps). A homemade MATLAB (https://www.mathworks.com) software eventually detects every single-cell displacement during 200 frames and analyses the distance traveled by the cells. Detailed descriptions are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This research was funded by the Belgian Federal Science Policy Office (Belspo) and the European Space Agency, grant number PRODEX project Flumias Nanomotion; The Research Foundation – Flanders (FWO), grant numbers AUGE/13/19 and I002620; FWO-SNSF, grant number 310030L_197946; FWO-SB-FDW PhD grant of V.R., M.I.V., and S.K. were supported by Swiss National Science Foundation (SNSF) grants CRSII5_173863. We thank G. Longo, S. Dinarelli, K. Brown, and R. A. Floto for providing the M. smegmatis; D. Sanglard (CHUV or Centre Hospitalier Universitaire Vaudois) for providing P2 lab facilities.
Author contributions
R.G.W. and S.K. designed research; M.I.V., E.R., and A.B. performed research; R.G.W. and S.K. contributed new reagents/analytic tools; M.I.V., E.R., A.B., C.Y., V.R., R.G.W., and S.K. analyzed data; and M.I.V., E.R., A.B., C.Y., V.R., R.G.W., and S.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Data, Materials, and Software Availability
Data have been deposited in the BioStudies database (EMBL-EBI), accession: S-BSST1065 (11). The code is available in GitHub at: https://github.com/BacteriaNanomotion/Bacteria-Nanomotion.git (12).
Supporting Information
References
- 1.van Belkum A., et al. , Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 18, 299–311 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Milani R. V., et al. , Reducing inappropriate outpatient antibiotic prescribing: Normative comparison using unblinded provider reports. BMJ Open Qual. 8, e000351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaprou G. D., Bergšpica I., Alexa E. A., Alvarez-Ordóñez A., Prieto M., Rapid methods for antimicrobial resistance diagnostics. Antibiotics 10, 209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasas S., et al. , Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. U.S.A. 112, 378–381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mustazzolu A., et al. , A rapid unraveling of the activity and antibiotic susceptibility of mycobacteria. Antimicrob. Agents Chemother. 63, e02194–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willaert R. G., et al. , Single yeast cell nanomotions correlate with cellular activity. Sci. Adv. 6, eaba3139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guizar-Sicairos M., Thurman S. T., Fienup J. R., Efficient subpixel image registration algorithms. Opt. Lett. OL 33, 156–158 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Notley-McRobb L., Death A., Ferenci T., The relationship between external glucose concentration and cAMP levels inside Escherichia coli: Implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology (Reading) 143, 1909–1918 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Longo G., et al. , Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotech. 8, 522–526 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Vasala A., Hytönen V. P., Laitinen O. H., Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell Infect. Microbiol. 10, 308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villalba M. I., et al. , Simple optical nanomotion method for single-bacterium viability and antibiotic response testing. BioStudies. https://www.ebi.ac.uk/biostudies/studies/S-BSST1065?query=S-BSST1065. Deposited 12 April 2023. [DOI] [PMC free article] [PubMed]
- 12.BacteriaNanomotion, Bacteria-Nanomotion (folder: Nanomotion software A). Github. https://github.com/BacteriaNanomotion/Bacteria-Nanomotion. Deposited 12 April 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Data have been deposited in the BioStudies database (EMBL-EBI), accession: S-BSST1065 (11). The code is available in GitHub at: https://github.com/BacteriaNanomotion/Bacteria-Nanomotion.git (12).