Significance
Human diseases frequently arise from defects in the mechanisms by which external cues are sensed and relayed to the interior of the cell. The proteins most widely targeted by existing therapeutic agents belong to a large family of cell surface receptors named G-protein-coupled receptors (GPCRs), which relay external cues by activating G-proteins in the interior of cells. Here, we report the surprising discovery of a synthetic small molecule that selectively targets G-proteins without compromising their ability to relay signals from GPCRs. Instead, this small molecule disrupts an atypical, GPCR-independent mechanism of G-protein signaling involved in cancer. This work reveals an alternative paradigm in targeting components of a signaling machinery with broad relevance in cellular communication in health and disease.
Keywords: GPCR, G protein, drug discovery, cancer
Abstract
Activation of heterotrimeric G-proteins (Gαβγ) by G-protein-coupled receptors (GPCRs) is a quintessential mechanism of cell signaling widely targeted by clinically approved drugs. However, it has become evident that heterotrimeric G-proteins can also be activated via GPCR-independent mechanisms that remain untapped as pharmacological targets. GIV/Girdin has emerged as a prototypical non-GPCR activator of G proteins that promotes cancer metastasis. Here, we introduce IGGi-11, a first-in-class small-molecule inhibitor of noncanonical activation of heterotrimeric G-protein signaling. IGGi-11 binding to G-protein α-subunits (Gαi) specifically disrupted their engagement with GIV/Girdin, thereby blocking noncanonical G-protein signaling in tumor cells and inhibiting proinvasive traits of metastatic cancer cells. In contrast, IGGi-11 did not interfere with canonical G-protein signaling mechanisms triggered by GPCRs. By revealing that small molecules can selectively disable noncanonical mechanisms of G-protein activation dysregulated in disease, these findings warrant the exploration of therapeutic modalities in G-protein signaling that go beyond targeting GPCRs.
G-protein-coupled receptors (GPCRs) mediate a large fraction of all transmembrane signaling in the human body, including responses triggered by every major neurotransmitter and by two-thirds of hormones (1). They are also the largest family of druggable proteins in the human genome, representing the target for over one-third of clinically approved drugs (2). To relay signals, GPCRs activate heterotrimeric G-proteins (Gαβγ) in the cytoplasm by promoting the exchange of GDP for GTP on Gα subunits, which results in a concomitant dissociation of Gβγ dimers (3). In turn, Gα-GTP and “free” Gβγ act on downstream effectors to propagate signaling. Signaling is turned off by the intrinsic GTPase activity of Gα, which leads to the reassociation of Gα with Gβγ. There is also a growing number of cytoplasmic proteins that modulate nucleotide handling by G-proteins, thereby exerting profound effects on the duration and amplitude of signaling (4–11).
In stark contrast to GPCRs, there are no clinically approved drugs for heterotrimeric G-proteins, despite their well-documented potential as pharmacological targets (12). Small-molecule inhibitors of Gβγ have been validated in some preclinical models (12, 13), but no drug-like small molecule that targets Gα subunits has been validated. There are, however, some natural cyclic depsipeptides that block α-subunits of the Gq/11 family with high specificity and potency (14). Unfortunately, because they inhibit G-protein activation en toto, these compounds could cause undesired side effects due to indiscriminate blockade of ubiquitous, physiologically relevant functions of their target G-proteins.
Perhaps, a more nuanced targeting approach that exploits disease-specific mechanisms of G-protein regulation could pave the way for new pharmacology. This idea is thwarted by the realization that the mechanisms of G-protein regulation beyond ubiquitous GPCR-mediated activation remain poorly understood in the absence of adequate tools to interrogate them. GIV (also known as Girdin) is a cytoplasmic protein that binds to Gαi subunits to promote G-protein signaling in a GPCR-independent manner (8, 15–17) and its expression in human primary solid tumors correlates with progression toward more invasive, metastatic stages in various types of cancer (18–20). Tumor cells depleted of GIV also fail to migrate in vitro or metastasize in mice (21). Here, we report the identification of a small molecule that binds to Gαi to selectively prevent GIV binding without disturbing other mechanisms by which the G-protein is regulated, including canonical GPCR-mediated signaling. We leverage this compound to establish that GIV-mediated activation of G-protein signaling favors proinvasive traits of cancer cells by operating downstream of receptor tyrosine kinases (RTKs) instead of downstream of GPCRs.
Results
High-Throughput Screen for Inhibitors of the GIV–Gαi Interaction.
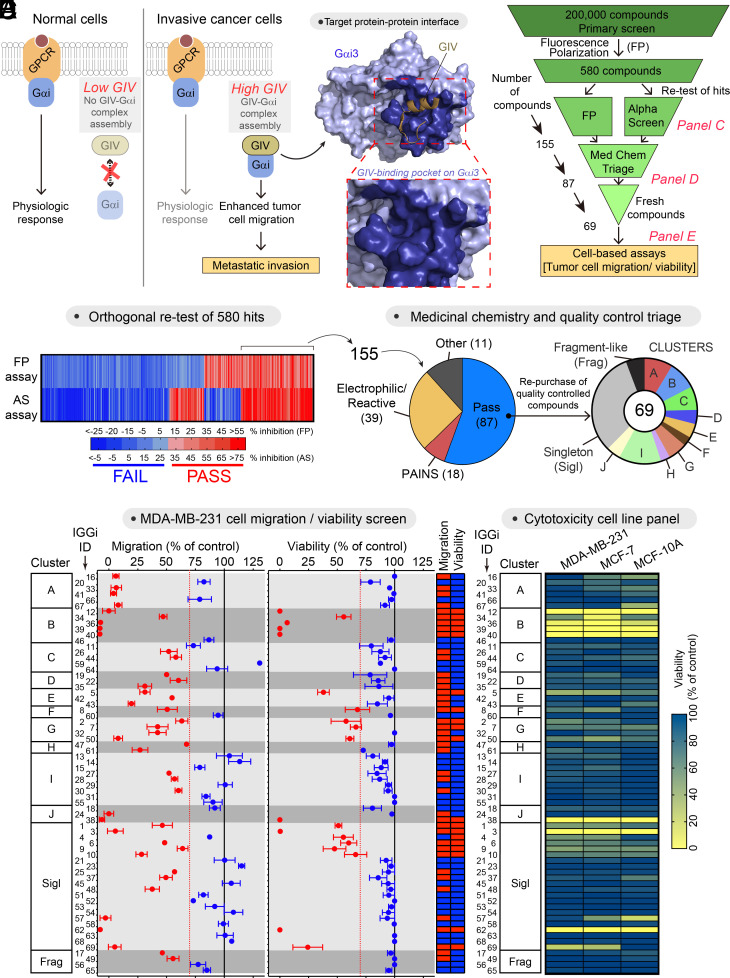
Previous work indicates that expression of GIV at high levels in cancer cells might facilitate its association with Gαi, which in turn favors tumor cell migration and other prometastatic traits (8, 15–17, 22–25) (Fig. 1A). Moreover, characterization of the molecular basis for the GIV–Gαi interaction (Fig. 1A) revealed that this protein–protein interaction might be suitable for specific pharmacological disruption (26–28). These previous findings motivated us to pursue a small-molecule screen for inhibitors of the GIV–Gαi interaction. Using a fluorescence polarization (FP) assay that directly monitors GIV binding to Gαi3 (27), we obtained 580 hits from screening a collection of 200,000 compounds (Fig. 1 B and C). Of these, 155 tested positive for inhibition in both the primary FP assay and an orthogonal secondary assay (AlphaScreen®, AS) (27) (Fig. 1 C and D). After triage, 68 compounds were discarded based on unfavorable chemical properties, and only 69 of the remaining 87 compounds could be repurchased as fresh powder stocks (Fig. 1D and SI Appendix, Table S1). We named this set of compounds “IGGi,” for “Inhibitors of the GIV–Gαi interaction.” We next evaluated the performance of these 69 IGGi compounds in cell-based assays. In cancer cell lines that express high levels of GIV (e.g., the triple-negative metastatic breast cancer cell line, MDA-MB-231), loss of GIV or disruption of its ability to bind Gαi through mutagenesis impairs cell migration, but does not affect cell viability under standard in vitro culture conditions on plastic dishes (17, 21). We found that approximately one-third of the IGGi compounds impaired MDA-MB-231 cell migration without affecting viability (Fig. 1E), lending confidence on the ability of our biochemical screen to identify compounds with the desired biological activity. To further prioritize the 69 IGGi compounds, we excluded not only those with the undesired property of reducing MDA-MB-231 viability, but also those that reduced the viability of MCF-7 cells (a nonmetastatic breast cancer cell line that expresses low levels of GIV) or of MCF-10A (a nontransformed epithelial breast cell line) to eliminate molecules with nonspecific cytotoxicity (Fig. 1F). The remaining 44 compounds were tested in a tertiary GIV–Gαi binding assay based on GST-fusion pull-downs (PD) (Fig. 2A). As a positive control in this assay, we used NF023, a nonselective inhibitor of Gαi activity that also disrupts the GIV–Gαi interaction in cell-free systems (27, 29). Only one IGGi compound, IGGi-11, was found to inhibit Gαi3 binding to GIV in this assay. Despite the weak activity of this compound in MDA-MB-231 cell migration assays (Fig. 1E), we pursued its characterization further and experiments presented below indicated high specificity and suitability for cell-based systems upon analog development via relatively minor modification.
Fig. 1.
Small-molecule screening to identify inhibitors of the GIV–Gαi interaction. (A) Diagram depicting the rationale for targeting the GIV–Gαi interaction with small molecules. (B) Scheme of the full screening campaign. (C) Confirmation of hit compounds that inhibit the GIV–Gαi interaction in two orthogonal biochemical assays, FP and AS. (D) Triage of compounds based on unfavorable chemical properties and availability of quality controlled molecules. (E) Test of 69 IGGi compounds (100 μM) on MDA-MB-231 cell migration and viability. Red, <30% reduction; blue, >30% reduction. Mean ± SEM (N = 4). (F) Comparison of the effect of IGGi compounds (100 μM) on the viability of three breast cell lines, MDA-MB-231, MCF-7, and MCF-10A (mean of N = 3).
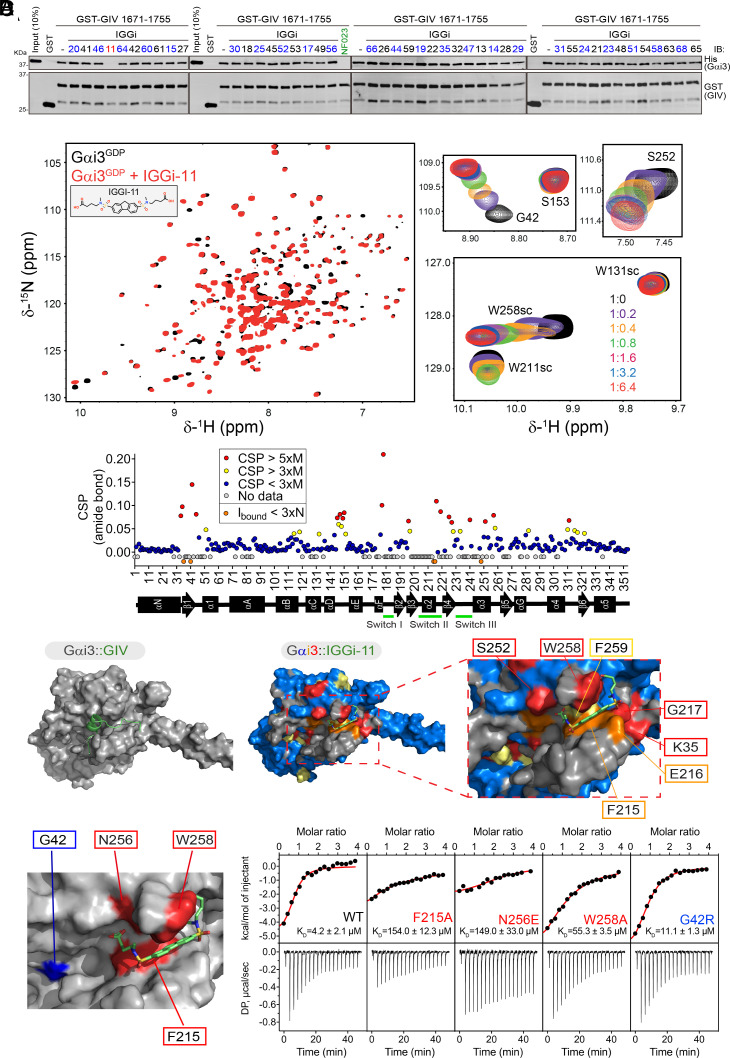
Fig. 2.
IGGi-11 binding to the GIV-interacting region of Gαi. (A) IGGi-11 disrupts GIV–Gαi binding in pull-down assays. His-Gαi3 was incubated with glutathione agarose-bound GST–GIV (aa 1671-1755) in the presence of the indicated compounds or the positive control NF023 at a concentration of 100 μM. After incubation and washes, bead-bound proteins were separated by SDS-PAGE and immunoblotted (IB) as indicated. Representative of 3 independent experiments. (B) Overlay of 1H–15N TROSY spectra of 2H,13C,15N–Gαi3–GDP in the absence or presence of IGGi-11. Selected regions from the overlaid spectra depicting representative perturbations in Gαi3 signals induced by increasing amounts of IGGI-11 are shown on the right. The scatter plot (bottom) corresponds to the quantification of IGGi-11-induced chemical shift perturbations (CSPs). Red, CSP > 5 times the median (M); yellow, CSP > 3xM; blue, CSP < 3xM; gray, no data. Reductions in signal intensity (Ibound) below three times the noise (N) are indicated in orange. (C) Comparison of models of IGGi-11 docked onto Gαi3 (Middle and Right, color coded according to NMR perturbations quantified in A) and GIV-bound Gαi3 (Left). (D) Quantification of IGGi-11 binding affinity (KD) for Gαi3 wild type (WT) or the indicated mutants using isothermal titration calorimetry (ITC). Data are representative of at least two independent experiments.
IGGi-11 Binds to the GIV-Interacting Region of Gαi.
We reasoned that inhibitors of the GIV–Gαi interaction should bind to the G-protein because our primary screening assay used a small peptide fragment of GIV unlikely to harbor enough structural features to accommodate a small molecule. Using NMR spectroscopy, we found that IGGi-11 caused dose-dependent CSPs in the amide bond signals of discrete amino acids of isotopically labeled (2H–13C–15N) Gαi3 (Fig. 2B and SI Appendix, Fig. S1), indicating compound binding. In contrast, another IGGi compound, IGGi-41, that was a potent inhibitor of MDA-MB-231 cell migration (Fig. 1E) but did not disrupt GIV–Gαi binding (Fig. 2A), did not cause NMR signal perturbations (SI Appendix, Fig. S2). These results suggested that IGGi-11 binds specifically to Gαi3. When IGGi-11-induced NMR perturbations were overlaid on a structural model of IGGi-11-bound Gαi3 and compared to a structural model of the GIV–Gαi3 complex, several of the amino acids with the largest perturbations (S252, W258, F259, F215, E216, G217, and K35) clustered around the predicted docking site for IGGi-11 and overlapped with the binding area for GIV (Fig. 2C). To directly test whether IGGi-11 binds on this predicted site located in the groove between the α3 helix and the conformationally dynamic Switch II (SwII) region, we carried out ITC experiments with WT Gαi3 or mutants. We found that three different mutations in the predicted binding site for IGGi-11 on Gαi3 (F215A, N256E, and W258A) lead to large decreases in compound binding affinity (>10 to 30-fold), whereas another mutation in an amino acid adjacent to the predicted binding site (G42R) did not (Fig. 2D). All mutant proteins fold properly and remain functional based on multiple assays (26). The estimated equilibrium dissociation constant (KD) for the Gαi3/IGGi-11 interaction based on ITC was ~4 μM (Fig. 2D), which was in good agreement with estimates based on curve fits of CSPs observed in NMR experiments (0.9 to 4.6 μM, SI Appendix, Fig. S1). IGGi-11 also blocked GIV binding to Gαi3 in FP assays with an inhibition constant (Ki) of ~14 μM (SI Appendix, Fig. S3A). Consistently, IGGi-11 also inhibited the ability of GIV to promote the steady-state GTPase activity of Gαi3, which reports increased nucleotide exchange in vitro (25) (SI Appendix, Fig. S3B). Although GIV does not bind to Gαo subunits, which belong to the same Gi/o family as Gαi3, it does bind to the other Gαi isoforms: Gαi1 and Gαi2 (15, 25). Unsurprisingly, IGGi-11 blocked GIV binding to Gαi1 or Gαi2 in FP assays with a potency similar to that observed for Gαi3 (SI Appendix, Fig. S3A). Together, these results indicate that IGGi-11 binds to the GIV-interacting site of Gαi proteins with low micromolar affinity, thereby precluding the formation of the GIV–Gαi complex in vitro.
IGGi-11 Does Not Affect GIV-Independent Aspects of G-Protein Regulation and Function.
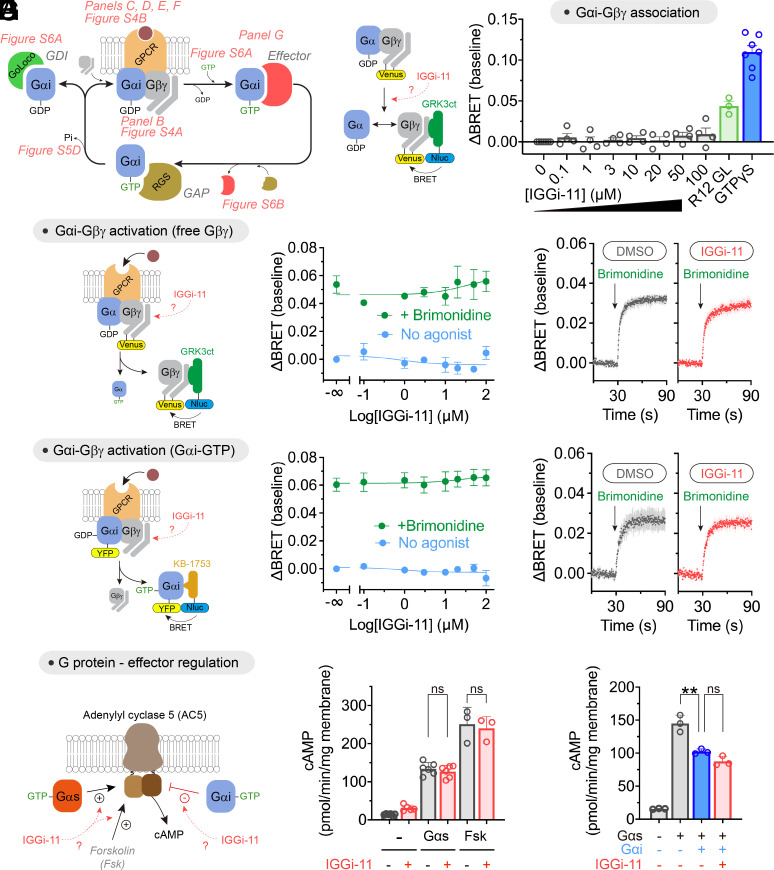
A concern with targeting Gαi is the potential on-target but nonetheless undesired effects that may result due to the many functions of G-proteins. The activity of Gα subunits depends on the ability to handle nucleotides (GDP/GTP exchange, GTP hydrolysis), on proteins that regulate their activity [Gβγ, GPCRs, guanine nucleotide dissociation inhibitors (GDIs), and GTPase accelerating proteins (GAPs)], or on how they regulate other proteins that propagate signaling (effectors) (Fig. 3A). With this in mind, we set out to thoroughly address the potential effect of IGGi-11 on G-protein functions other than those mediated via GIV binding by using isolated cell membranes or purified proteins. First, we tested the effect of IGGi-11 on the association of Gβγ with Gα using a bioluminescence resonance energy transfer (BRET) assay (30, 31). We found that concentrations of IGGi-11 up to 100 μM did not cause the dissociation of Gβγ from Gαi3 (Fig. 3B), whereas incubation with a positive control peptide (R12 GL, 25 μM) or a nonhydrolyzable GTP analog (GTPγS, 300 μM) did. Similar observations were made with three other Gα subunits that belong to the same family as Gαi (i.e., Gαo), or to different ones (i.e., Gαq and Gα13) (SI Appendix, Fig. S4A), indicating that IGGi-11 does not disrupt Gαβγ heterotrimers. Using the same assay, we assessed the effect of IGGi-11 on GPCR-mediated activation of G-proteins, which results in the dissociation of Gβγ from Gα. We found that concentrations of IGGi-11 up to 100 μM did not interfere with the ability of agonist-stimulated GPCRs to activate Gi3, Go, Gq, or G13 heterotrimers (Fig. 3C and SI Appendix, Fig. S4B). Rapid kinetic assays further confirmed that IGGi-11 did not alter the rate of Gβγ dissociation upon GPCR activation (Fig. 3D). Moreover, the rate and extent of Gβγ–Gαi3 reassociation upon GPCR signal termination was unaffected by IGGi-11 (SI Appendix, Fig. S4C). Similar observations were made for the reassociation of Gβγ with other Gα subunits, like Gαo or Gαq (SI Appendix, Fig. S4 D and E). These observations indicate that IGGi-11 not only fails to disrupt preformed Gαβγ (as in Fig. 3B), but also that it does not interfere with the association of trimers. As an alternative to assess GPCR-mediated activation of G-proteins, we used another BRET-based biosensor (32) that directly monitors the formation of GTP-bound Gαi3 (Fig. 3E). We found that neither amplitude nor kinetics of Gαi3–GTP formation upon GPCR stimulation were affected by IGGi-11 (Fig. 3 E and F). We also found that IGGi-11 did not interfere with the spontaneous exchange of GDP for GTP on Gαi3 using three independent assays: BRET-based GTPγS binding to Gαi in isolated membranes (SI Appendix, Fig. S5A), binding of fluorescently labeled GTPγS to purified Gαi (SI Appendix, Fig. S5B), or steady-state GTPase activity of purified Gαi with radiolabeled GTP (SI Appendix, Fig. S5C). We also found that IGGi-11 did not affect the hydrolysis of GTP to GDP by purified Gαi (SI Appendix, Fig. S5D).
Fig. 3.
Lack of effect of IGGi-11 on G-protein coupling to GPCRs and effectors. (A) Diagram of key steps and protein interactions involved in Gαi-subunit functions. (B) IGGi-11 does not dissociate Gβγ from Gαi3 in membranes isolated from HEK293T cells expressing a BRET-based biosensor for free Gβγ, whereas two positive controls do (a GoLoco peptide derived from RGS12, R12 GL, 25 μM; and GTPγS 300 μM). (C–F) IGGi-11 does not affect GPCR-mediated activation of Gi3 as determined by the dissociation of Gαi3–Gβγ heterotrimers (C and D) or the formation of Gαi3–GTP (E and F) using BRET-based biosensors. In C and E, membranes isolated from HEK293T cells expressing the α2A adrenergic receptor were treated with the indicated concentrations of IGGi-11 with (green) or without (blue) stimulation with a receptor agonist (brimonidine, 1 μM) for 2 min before BRET measurements. In D and F, BRET was continuously measured in real time in the presence of 100 μM IGGi-11 or vehicle (1% dimethyl sulfoxide (DMSO), v:v). (G) IGGi-11 does not interfere with G-protein-mediated regulation of adenylyl cyclase. Membranes isolated from HEK293T cells expressing adenylyl cyclase 5 were treated with IGGi-11 (100 μM), purified Gαs (0.5 μM), purified myristoylated Gαi1 (Gαi, 1 μM), and forskolin (Fsk, 10 μM) in the combinations indicated in the graphs. Mean ± SEM (N ≥ 3). **P < 0.01, ANOVA.
Next, we evaluated the potential impact of IGGi-11 on the ability of active, GTP-bound Gαi proteins to engage and modulate effectors. First, we observed that IGGi-11 did not cause NMR signal perturbations in the α3/SwII region of GTPγS-loaded Gαi3 (SI Appendix, Fig. S6A), which contrasts with the observations obtained for GDP-loaded Gαi3 (Fig. 2B and SI Appendix, Fig. S1) and suggests lack of compound binding to active G-proteins. Consistent with this, we also found that IGGi-11 did not inhibit the interaction between purified Gαi3 and KB-1753, an effector-like peptide that binds to the α3/SwII region of Gαi-GTP (33) (SI Appendix, Fig. S6B). We then tested whether IGGi-11 affected the regulation of a bona fide effector of Gαi, i.e., adenylyl cyclase (Fig. 3G). In membranes from cells expressing adenylyl cyclase 5, IGGi-11 did not affect either activation mediated by purified Gαs or inhibition mediated by purified Gαi (Fig. 3G). The compound did not affect adenylyl cyclase activity either under basal conditions or upon direct, G-protein-independent activation with forskolin (Fig. 3G).
Finally, we assessed whether IGGi-11 would preclude the binding of Gαi to other G-protein regulators like GDIs that contain a GoLoco motif (4, 5), or GTPase-accelerating proteins (GAPs) of the regulators of G-protein signaling (RGS) family (6, 7). We found that IGGi-11 did not inhibit the interaction of Gαi3 with the GoLoco motif responsible for the GDI activity of RGS12 (R12 GL, SI Appendix, Fig. S6B) or with the GAP RGS4 (SI Appendix, Fig. S6C).
Taken together, our results indicate that IGGi-11 specifically inhibits GIV binding to Gαi without interfering with any other major function of Gαi, including nucleotide binding and hydrolysis, association with Gβγ subunits and other cytoplasmic regulators, activation by GPCRs, or modulation of effectors.
Validation of an IGGi-11 Analog with Increased Activity in Cells.
After establishing the specificity of IGGi-11 for the target GIV–Gαi complex in vitro, we sought to determine its biological activity in cells. We found that preincubation of MDA-MB-231 cells with IGGi-11 inhibited their ability to migrate only marginally (SI Appendix, Fig. S7A). We reasoned that this could be due to low membrane permeability because IGGi-11 contains two negatively charged carboxylate groups (SI Appendix, Fig. S7A). To overcome this, we generated IGGi-11me, an analog in which the carboxylates were esterified with methyl groups. We hypothesized that esterification would increase membrane permeability by eliminating the charges of the carboxylates, and that cytoplasmic esterases would revert the modification to produce IGGi-11, thereby enabling enhanced inhibitory activity in cells (SI Appendix, Fig. S7A). Indeed, preincubation of MDA-MD-231 cells with IGGi-11me efficiently reduced their ability to migrate when fetal bovine serum (FBS) was used as the chemoattractant (SI Appendix, Fig. S7A). In contrast, IGGi-11me did not affect MDA-MB-231 cell migration when the chemoattractant was SDF-1α, an agonist specific for the Gi-coupled GPCR CXCR4 (SI Appendix, Fig. S7A). This observation is significant for two reasons. First, the effect of IGGi-11me does not blunt MDA-MB-231 cell migration nonspecifically even at a relatively high concentration (up to 100 μM). Second, IGGi-11me does not interfere with mechanisms of migration shared between canonical GPCR-Gi signaling and signaling mediated by GIV-Gi, the intended target of IGGi-11me. Also as desired, IGGi-11me (or IGGi-11) did not affect the viability of MDA-MB-231 or MCF-10A cells (SI Appendix, Fig. S7B), consistent with previous observations that depletion of GIV does not affect MDA-MB-231 cell growth under similar conditions (17). We confirmed that IGGi-11me had higher permeability than IGGi-11 by using a parallel artificial membrane permeability assay (SI Appendix, Fig. S7C). We also confirmed that IGGi-11me was converted to IGGi-11 by esterases present in the cytosol of MDA-MB-231 cells (SI Appendix, Fig. S7D), which is a critical step because IGGi-11me would be a poor inhibitor of GIV–Gαi3 binding at the concentrations tested in cells due to its lower potency compared to IGGi-11 (SI Appendix, Fig. S7E). These results indicate that IGGi-11me serves as a prodrug that allows the action of the active GIV–Gαi inhibitor compound, IGGi-11, in cells.
IGGi-11me Inhibits GIV-Dependent Cancer Cell Signaling.
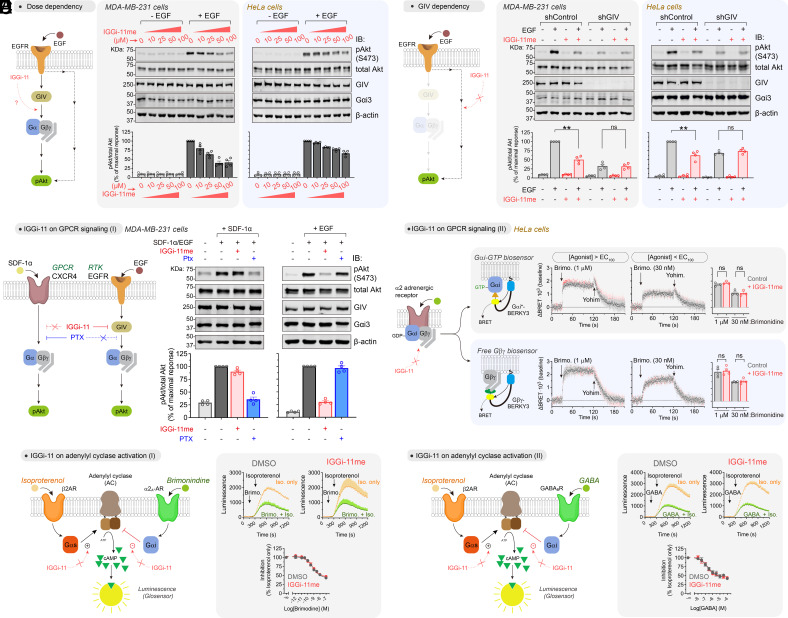
Previous work has shown that GIV mediates the activation of Akt downstream of various receptor tyrosine kinases (RTKs), including the epidermal growth factor receptor (EGFR), and other surface receptors via G-protein (i.e., Gβγ)-dependent activation of PI3K (8, 15, 17, 23, 25, 34–36). We found that IGGi-11me reduced, in a dose-dependent manner, the phosphorylation of Akt at S473 (pAkt) upon epidermal growth factor (EGF) stimulation in two cell lines, MDA-MB-231 and HeLa, indicating reduced Akt activity (Fig. 4A). The lack of complete Akt inhibition is consistent with the known existence of GIV-independent mechanisms utilized by EGFR to activate PI3K–Akt signaling (37). In fact, the extent of IGGi-11me-mediated inhibition of Akt was similar to that observed upon depletion of GIV in these cell lines (Fig. 4B). Moreover, IGGi-11me failed to further reduce Akt activation in GIV-depleted cells even at the maximal concentration of compound tested (100 μM), indicating that it does not affect GIV-independent mechanisms of Akt activation downstream of EGFR (Fig. 4B). These results suggest that, even at relatively high concentrations, IGGi-11me does not have nonspecific effects on Akt signaling because it lacks an inhibitory effect in the absence of the intended target. Also, IGGi-11me did not change the total amount of GIV or Gαi (Fig. 4 A and B), supporting that its mechanism of action is the disruption of the interaction of the two proteins, rather than indirectly altering their abundance. These results are consistent with the idea that IGGi-11me specifically inhibits GIV-dependent G-protein signaling in cancer cells.
Fig. 4.
IGGi-11me specifically inhibits GIV-dependent G-protein cell signaling. (A) IGGi-11me inhibits EGF-stimulated Akt activation (phospho-serine 473, pAkt S473) in MDA-MB-231 and HeLa cells. Cells were preincubated with the indicated concentrations of IGGi-11me and stimulated with EGF (1.6 nM for MDA-MB-231 or 50 nM for HeLa) for 5 min before lysis and immunoblotting. (B) IGGi-11me (100 μM) does not inhibit EGF-stimulated Akt activation in GIV-depleted cells. GIV-depleted cells (shGIV) or control cells (shControl) were treated as in A. (C) IGGi-11me does not block Akt activation upon stimulation of the GPCR CXCR4. MDA-MB-231 cells were preincubated with IGGi-11me (100 μM) or pertussis toxin (PTX, 100 ng/mL) and stimulated with SDF-1α (100 ng/mL for 10 min) or EGF (1.6 nM for 5 min) before processing as in A. (D) IGGi-11me does not affect GPCR-mediated modulation of G-protein activity. HeLa cells expressing BRET biosensors for Gαi-GTP (Gαi*-BERKY3) or free Gβγ (Gβγ-BERKY3) were preincubated with IGGi-11me (100 μM) and sequentially treated with the α2 adrenergic agonist brimonidine and the antagonist yohimbine (25 μM) during real-time BRET measurements as indicated in the figure. (E and F) IGGi-11me does not affect GPCR-mediated modulation of cAMP responses. HEK293T cells expressing Glosensor, a luminescence-based cAMP sensor, and either α2A-AR (E) or GABABR (F) were pretreated with IGGi-11me (100 μM) or DMSO before measuring luminescence. Cells were treated with isoproterenol (100 nM) with or without prestimulation with brimonidine (E) or GABA (F) as indicated. The concentration of brimonidine and GABA in the kinetic traces shown was 1 μM and 100 μM, respectively. All results are mean ± SEM (N ≥ 3). **P < 0.01; ns, P > 0.05, ANOVA.
IGGi-11me Does Not Affect GIV-Independent G-Protein Cell Signaling.
Next, we set out to further assess the specificity of IGGi-11me in cell signaling. Although IGGi-11 does not interfere with GIV-independent mechanisms of G-protein regulation in vitro (Fig. 3 and SI Appendix, Figs. S4–S6), confirmation that the same holds for IGGi-11me in cells was warranted given the relatively high concentrations of compounds needed to block GIV-dependent signaling. First, we compared side by side the effect of a maximal concentration of IGGi-11me (100 μM) on GIV-dependent and GIV-independent G-protein signaling in the same cell line (MDA-MB-231) with the same readout (Akt activation). For GIV-dependent G-protein signaling, we stimulated cells with EGF as in Fig. 4 A and B, whereas for GIV-independent G-protein signaling, we stimulated cells with SDF-1α, an agonist for the endogenously expressed Gi-coupled GPCR CXCR4 (Fig. 4C). We found that IGGi-11me inhibited Akt activation in response to EGF but not in response to SDF-1α (Fig. 4C), indicating that it does not interfere with GPCR-mediated G-protein signaling. In contrast, PTX, which precludes Gαi activation by GPCRs but not by GIV (38), efficiently blocked activation of Akt in response to SDF-1α but not to EGF (Fig. 4C). These results indicate that IGGi-11me specifically targets GIV-dependent G-protein signaling mechanisms in cells without interfering with canonical GPCR-mediated G-protein signaling. This result is in good agreement with the lack of effect of IGGi-11me on SDF-1α-stimulated migration of MDA-MB-231 cell (SI Appendix, Fig. S7A), further supporting the notion that the compound does not affect signaling mechanisms shared between GPCR-Gi and GIV-Gi pathways. To substantiate this point, we assessed the effect of IGGi-11me on GPCR signaling by using BRET-based biosensors that directly monitor the activation of endogenous G-proteins (32). More specifically, HeLa cells expressing biosensors for either Gαi-GTP or free Gβγ were treated with a maximal concentration of IGGi-11me (100 μM) exactly under the same conditions that led to decreased GIV-dependent Akt activation after EGF stimulation in this cell line (Fig. 4 A and B). We found that G-protein responses elicited by stimulation of endogenous α2 adrenergic receptors with maximal (>EC100) or submaximal (<EC100) concentrations of a cognate agonist were unaltered by IGGi-11me (Fig. 4D). Not only were the amplitudes and rates of the activation responses unchanged, but the rates of deactivation upon GPCR blockade with an antagonist also remained the same (Fig. 4D), indicating that the compound does not have effects on nucleotide exchange or hydrolysis rates, or the dissociation or reassociation of G-protein heterotrimers upon GPCR-mediated modulation. We went on to determine whether, in addition to not having an effect at the level of G-protein regulation, IGGi-11me also lacked an effect on a well-established downstream signaling readout like the second messenger cAMP. For this, we measured cAMP regulation by GPCRs in HEK293T cells in the presence and absence of a maximal concentration of IGGi-11me (100 μM). Endogenous β-adrenergic receptors were stimulated with isoproterenol to elevate cAMP levels via Gαs, and this response was modulated by the stimulation of exogenously expressed α2A adrenergic receptors (Fig. 4E) or GABAB receptors (Fig. 4F), which suppress the isoproterenol-stimulated cAMP response via Gαi. We found that IGGi-11me had no effect on 1) the cAMP response to isoproterenol in the absence of Gi-mediated regulation, or 2) the efficacy or potency of Gi-mediated inhibition of the isoproterenol response by any of the two GPCRs tested Fig. 4 E and F). Taken together, these results show that IGGi-11me does not interfere with GIV-independent G-protein signaling, including that elicited by canonical GPCR/G-protein signaling pathways.
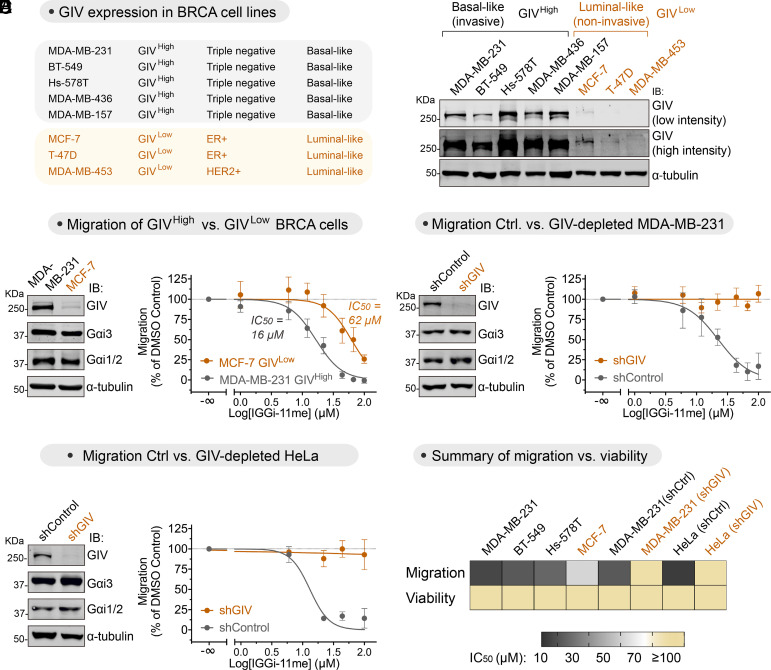
IGGi-11me Specifically Inhibits GIV-Dependent Tumor Cell Migration.
Previous evidence indicates that GIV is expressed at high levels in metastatic cancers, and that formation of the GIV–Gαi complex favors cell migration (15, 18–20). Consistent with some of these observations, we found that invasive breast cancer (BRCA) cell lines prone to metastasis expressed higher levels of GIV (GIVHigh) than noninvasive breast cancer cell lines (GIVLow) (Fig. 5A). IGGi-11me was approximately four times more potent inhibiting the migration of MDA-MB-231 cells (GIVHigh) than that of MCF-7 cells (GIVLow) (Fig. 5B). This difference in IGGi-11me sensitivity could not be attributed to differences in Gαi protein abundance because they were present in similar amounts in both cell lines (Fig. 5B). While we could not test the effect of IGGi-11me on the GIVLow cell lines T47D and MDA-MB-453 because they lacked measurable migration, we found that IGGi-11me inhibited cell migration in the GIVHigh cell lines BT-549 and Hs578T with a potency similar to that seen for MDA-MB-231 cells (SI Appendix, Fig. S8A). Thus, despite the different genetic background of these cell lines bearing different drivers of cancer traits, the common denominator is that GIV expression (and presumably the formation of a GIV–Gαi complex) correlates with sensitivity to IGGi-11me. To further assess the specificity of IGGi-11me in inhibiting GIV-dependent tumor cell migration, we tested its effect on GIV-depleted MDA-MB-231 cells. We found that, compared to control cells, IGGi-11me had no effect on MDA-MB-231 cell migration upon GIV depletion even when tested at maximal concentration (100 μM) (Fig. 5C). Similar observations were made with GIV-depleted HeLa cells, which are of different origin and genetic background than that of MDA-MB-231 (Fig. 5D). Moreover, GIV-depleted MDA-MB-231 cells or Hela cells contained amounts of Gαi proteins similar to those in their corresponding control cells (Fig. 5 C and D), further supporting that the inhibition of cell migration exerted by IGGi-11me is GIV-dependent. These results are also consistent with the findings shown in SI Appendix, Fig. S7A indicating that IGGi-11me does not have an effect on CXCR4-promoted cell migration, which rule out that IGGi-11me affects mechanisms of migration shared between canonical GPCR-Gi signaling and signaling mediated by GIV-Gi. Furthermore, the inhibition of cell migration by IGGi-11me was not a consequence of reduced cell viability, as the latter was not affected by the compound in any of the cell lines investigated (Fig. 5E and SI Appendix, Fig. S8B). These findings indicate that IGGi-11me specifically blocks GIV-dependent tumor cell migration, implying that the disruption of the GIV–Gαi complex hinders the proinvasive features of GIVHigh cancer cells.
Fig. 5.
IGGi-11me blocks GIV-dependent tumor cell migration. (A) Basal-like invasive breast cancer (BRCA) cell lines express higher amounts of GIV (GIVHigh) than luminal-like noninvasive BRCA cell lines (GIVLow) as determined by immunoblotting. (B) IGGi-11me inhibits cell migration more potently in MDA-MB-231 cells (GIVHigh) than in MCF-7 cells (GIVLow). Chemotactic cell migration toward FBS was determined in the presence of the indicated concentrations of IGGi-11me using a modified Boyden-chamber assay. (C and D) IGGi-11me-mediated inhibition of tumor cell migration is lost upon depletion of GIV from MDA-MB-231 (C) or HeLa (D) cells. GIV-depleted cells (shGIV) or control cells (shControl) were processed as described in B. (E) IGGi-11me impairs tumor cell migration without affecting cell viability. Heatmap comparing the half-maximal inhibitory concentration (IC50) of IGGi-11me on cell migration or viability of the indicated cell lines. IC50 values were determined from results shown in this figure or in SI Appendix, Fig. S8. Cell viability was determined upon incubation with IGGi-11me for 24 h, which is longer than the times cells were exposed to the compound in cell migration assays. All results are mean ± SEM (N ≥ 3).
IGGi-11me Inhibits Cancer Cell Growth in Tumor-Like Contexts.
GIV-depleted MDA-MB-231 cells fail to metastasize in mouse xenograft models, which correlates well with the effects of GIV depletion on tumor cell migration and invasion (21). Unfortunately, we could not test the effect of IGGi-11me on cancer mouse models to assess metastasis because preliminary results showed that IGGi-11me is rapidly degraded in plasma. As an alternative, we set out to investigate the impact of IGGi-11me on the behavior of MDA-MB-231 cells in a tumor-like context. This was motivated by previous observations that loss of GIV does not affect the growth of tumor cells, including MDA-MB-231, on plastic dishes (17, 21), but hinders growth in three-dimensional Matrigel cultures (17), which account for tumor cell interactions with the extracellular matrix and recapitulate many of the behavioral features of cancer cells in tumors in situ (39). We found that IGGi-11me mimicked previous observations (17) upon loss of GIV in Matrigel cultures—i.e., MDA-MB-231 became smaller and more organized acinar structures than control cells, resulting in an overall reduction of cell growth (SI Appendix, Fig. S9 A and B). In contrast, IGGi-11me did not affect the growth of nontransformed MCF-10A breast cells in Matrigel cultures even when tested at a maximal concentration (100 μM) (SI Appendix, Fig. S9B), suggesting that the effect on MDA-MB-231 was not due to nonspecific toxicity. As a second model to assess the effect of IGGi-11me on cancer cell growth in a tumor-like environment, we pretreated MDA-MB-231 cells with IGGi-11me and assessed their ability to form tumors when implanted subcutaneously as xenografts in mice (SI Appendix, Fig. S9C). We found that IGGi-11me-treated cells formed tumors less efficiently than controls (SI Appendix, Fig. S9C). In contrast, when cells treated and prepared in the same way as above were seeded on plastic dishes and grown under standard cell culture conditions, IGGi-11me had no effect (SI Appendix, Fig. S9C), suggesting that the effect on xenograft growth is specific to the tumor-like context for MDA-MB-231 cells recapitulated in mice. Together, these results indicate that disruption of the GIV–Gαi interaction by IGGi-11me prevents cancer cell growth in tumor-like contexts.
Discussion
In this work, we identify and characterize a chemical probe of broad utility for dissecting atypical mechanisms of cellular communication mediated by G-proteins with important biomedical implications not only for cancer, but also for fibrosis, and male fertility, among other maladies (8, 9, 17, 18, 20, 40, 41). From a broader perspective, this work provides the proof of principle for a modality of pharmacological targeting in heterotrimeric G-protein signaling that deviates from the widespread focus on GPCRs or the direct ablation of G-protein activity en toto. This modality consists of targeting G-proteins to selectively disrupt specific mechanisms by which they are regulated. IGGi-11 disrupts Gαi binding to GIV but not to many of its other binding partners, despite them physically engaging the same region of Gαi as GIV. This region includes the SwII, which is dynamic and adopts different conformations depending on the protein partner bound to Gαi. Although it is tempting to speculate that the selectivity of IGGi-11 may arise from its relative ability to interact with these different conformations, the structural basis for the action of IGGi-11 remains to be fully elucidated. The targeting modality described here follows the path opened by recent advances on small-molecule inhibitors for another GTPase, KRas, in reshaping the traditional definition of what constitutes a druggable target (42, 43). The main limitations of IGGi-11(me) relate to its modest affinity and poor stability in plasma. Because its chemical scaffold is synthetically tractable, IGGi-11 may further serve as a lead compound to develop analogs with improved potency and pharmacokinetic properties that could have therapeutic value.
Materials and Methods
Chemical compounds of interest were purchased from reliable vendors or synthesized in-house, and tested in in vitro assays, including NMR, BRET assays, or different protein–protein binding experiments following previously established procedures that are described in detail in SI Appendix. Cell-based experiments to assess the efficacy and specificity of compounds were also carried out using previously established procedures and/or cell lines, including cell migration assays using modified Boyden chambers, immunoblotting, and signaling assays, all of which are described in detail in SI Appendix along with the animal experiments measuring xenograft tumor growth by luminescence bioimaging.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by NIH grant R01GM130120 and the Karin Grunebaum Cancer Research Foundation (to M.G.-M). J.Z. was supported by a Dahod International Scholar Award, and A.L. was supported by a F31 Ruth L. Kirschstein NRSA Predoctoral Fellowship (F31NS115318). F.J.B. was supported by Spanish Government grant PID2020-113225GB-I00. M.F.-G was supported by Spanish Government fellowship PRE-2018-085788. We thank the ICCB-Longwood Screening Facility at Harvard Medical School, S. Whelan (Boston University), F. Seta (Boston University), N. Ganem (Boston University), and J. B. Blanco-Canosa (Institute for Advanced Chemistry of Catalonia) for access to instrumentation and reagents. We thank N. Merino (CIC bioGUNE, Spain) for help with the purification of Gαi3 protein used in NMR studies, M. Rico (CIC bioGUNE, Spain) for access to NMR spectrometers, and A. González-Magaña (CIC bioGUNE, Spain) for preliminary ITC experiments. We thank the following investigators for providing DNA plasmids: K. Martemyanov (The Scripps Research Institute, Jupiter, FL), N. Lambert (Augusta University, Augusta, GA), P. Wedegaertner (Thomas Jefferson University, Philadelphia, PA), J. Blumer (Medical University of South Carolina, Charleston, SC), J. Sondek (University of North Carolina, Chapel Hill, NC), N. Artemyev (University of Iowa) C. Dressauer (University of Texas Health Science Center at Houston, TX), and M. Linder (Cornell University).
Author contributions
J.Z., V.D., A.B., F.J.B., and M.G.-M. designed research; J.Z., V.D., M.F.-G., S.D., A.I.d.O., J.-C.P., A.L., and Q.C. performed research; J.Z., V.D., M.F.-G., S.D., A.I.d.O., F.J.B., and M.G.-M analyzed data; and J.Z., V.D., and M.G.-M. wrote the paper.
Competing interests
Boston University has filed a provisional patent application related to the content of this manuscript in which M.G.-M. is listed as an inventor.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Vincent DiGiacomo, Email: Vincent.DiGiacomo@gmail.com.
Mikel Garcia-Marcos, Email: mgm1@bu.edu.
Data, Materials, and Software Availability
All study data and protocols are included and can be accessed directly in the article and/or SI Appendix. No code or software was generated for this study, and data were not deposited in a public database.
Supporting Information
References
- 1.Weis W. I., Kobilka B. K., The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser A. S., Attwood M. M., Rask-Andersen M., Schioth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilman A. G., G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Sato M., Blumer J. B., Simon V., Lanier S. M., Accessory proteins for G proteins: Partners in signaling. Annu. Rev. Pharmacol. Toxicol. 46, 151–187 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Siderovski D. P., Willard F. S., The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 1, 51–66 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross E. M., Wilkie T. M., GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Dohlman H. G., Thorner J., RGS proteins and signaling by heterotrimeric G proteins. J. Biol. Chem. 272, 3871–3874 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Marcos M., Ghosh P., Farquhar M. G., GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J. Biol. Chem. 290, 6697–6704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiacomo V., Marivin A., Garcia-Marcos M., When heterotrimeric G proteins are not activated by G protein-coupled receptors: Structural insights and evolutionary conservation. Biochemistry 57, 255–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cismowski M. J., et al. , Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J. Boil. Chem. 275, 23421–23424 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Tall G. G., Ric-8 regulation of heterotrimeric G proteins. J. Recept. Signal Transduct. Res. 33, 139–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell A. P., Smrcka A. V., Targeting G protein-coupled receptor signalling by blocking G proteins. Nat. Rev. Drug Discov. 17, 789–803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonacci T. M., et al. , Differential targeting of Gbetagamma-subunit signaling with small molecules. Science 312, 443–446 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kostenis E., Pfeil E. M., Annala S., Heterotrimeric Gq proteins as therapeutic targets? J. Biol. Chem. 295, 5206–5215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Marcos M., Ghosh P., Farquhar M. G., GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 3178–3183 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leyme A., et al. , Specific inhibition of GPCR-independent G protein signaling by a rationally engineered protein. Proc. Natl. Acad. Sci. U.S.A. 114, E10319–E10328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leyme A., Marivin A., Perez-Gutierrez L., Nguyen L. T., Garcia-Marcos M., Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. J. Cell Biol. 210, 1165–1184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh P., Garcia-Marcos M., Farquhar M. G., GIV/Girdin is a rheostat that fine-tunes growth factor signals during tumor progression. Cell Adh. Migr. 5, 237–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Marcos M., et al. , Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 25, 590–599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh P., Heterotrimeric G proteins as emerging targets for network based therapy in cancer: End of a long futile campaign striking heads of a Hydra. Aging 7, 469–474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P., et al. , An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 68, 1310–1318 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Leyme A., Marivin A., Garcia-Marcos M., GIV/Girdin (Galpha-interacting, vesicle-associated Protein/Girdin) creates a positive feedback loop that potentiates outside-in integrin signaling in cancer cells. J. Boil. Chem. 291, 8269–8282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Marcos M., et al. , Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 1961–1966 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma G. S., et al. , Therapeutic effects of cell-permeant peptides that activate G proteins downstream of growth factors. Proc. Natl. Acad. Sci. U.S.A. 112, E2602–E2610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Marcos M., Ghosh P., Ear J., Farquhar M. G., A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J. Biol. Chem. 285, 12765–12777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Opakua A. I., et al. , Molecular mechanism of Galphai activation by non-GPCR proteins with a Galpha-Binding and Activating motif. Nat. Commun. 8, 15163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiGiacomo V., et al. , The Galphai-GIV binding interface is a druggable protein-protein interaction. Sci. Rep. 7, 8575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalogriopoulos N. A., et al. , Structural basis for GPCR-independent activation of heterotrimeric Gi proteins. Proc. Natl. Acad. Sci. U.S.A. 116, 16394–16403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollins B., Kuravi S., Digby G. J., Lambert N. A., The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell. Signal. 21, 1015–1021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuho I., et al. , Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal. 8, ra123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maziarz M., et al. , Revealing the activity of trimeric G-proteins in live cells with a versatile biosensor design. Cell 182, 770–785 e716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston C. A., et al. , Minimal determinants for binding activated G alpha from the structure of a G alpha(i1)-peptide dimer. Biochemistry 45, 11390–11400 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh P., et al. , A G{alpha}i-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol. Boil. Cell 21, 2338–2354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C., et al. , Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol. Biol. Cell 25, 3654–3671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Marcos M., Ear J., Farquhar M. G., Ghosh P., A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol. Biol. Cell 22, 673–686 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemmon M. A., Schlessinger J., Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Marcos M., Complementary biosensors reveal different G-protein signaling modes triggered by GPCRs and non-receptor activators. ELife 10, e65620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debnath J., Brugge J. S., Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5, 675–688 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Minn A. J., et al. , Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynoso S., et al. , GIV/Girdin, a non-receptor modulator for Galphai/s, regulates spatiotemporal signaling during sperm capacitation and is required for male fertility. ELife 10, e69160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Sanchez I., et al. , GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat. Commun. 5, 4451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skoulidis F., et al. , Sotorasib for lung cancers with KRAS p. G12C mutation. N. Engl. J. Med. 384, 2371–2381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data and protocols are included and can be accessed directly in the article and/or SI Appendix. No code or software was generated for this study, and data were not deposited in a public database.