Significance
In adult mammals, blood cell production is sustained throughout life by hematopoietic stem and progenitor cells (HSPCs) that reside in the bone marrow in a specific microenvironment called the “niche.” Understanding how niche cells control HSPC homeostasis is a major challenge, since misregulation leads to severe pathologies such as leukemia. Many studies have highlighted the striking parallels that exist between Drosophila and mammalian hematopoiesis. In this study, we discover that the mechanosensing of blood flow by vascular niche cells regulates hematopoietic progenitor maintenance in the Drosophila hematopoietic organ. Furthermore, we identify molecular processes involved from mechanosensing to mechanotransduction.
Keywords: blood cell progenitors, blood flow, Piezo, cardiac tube, lymph gland
Abstract
Hematopoietic stem and progenitor cells maintain blood cell homeostasis by integrating various cues provided by specialized microenvironments or niches. Biomechanical forces are emerging as key regulators of hematopoiesis. Here, we report that mechanical stimuli provided by blood flow in the vascular niche control Drosophila hematopoiesis. In vascular niche cells, the mechanosensitive channel Piezo transduces mechanical forces through intracellular calcium upregulation, leading to Notch activation and repression of FGF ligand transcription, known to regulate hematopoietic progenitor maintenance. Our results provide insight into how the vascular niche integrates mechanical stimuli to regulate hematopoiesis.
In adult mammals, hematopoiesis takes place in bone marrow where hematopoietic stem and progenitor cells (HSPCs) are maintained by specialized microenvironments, or niches. HSPCs localize around arterioles and sinusoid blood vessels (1–9). An increasing number of studies investigate whether biomechanical cues impact HSPC functions. Biomechanical cues such as fluid mechanical forces, extracellular matrix stiffness, confined adhesiveness, and cell-intrinsic forces control HSPC properties through mechanotransduction (10, 11). Recent work revealed the importance of blood flow for HSPC emergence in vertebrate embryos. HSPCs emerge from the ventral floor of the dorsal aorta within the aorta–gonad–mesonephros (AGM) region, via a process termed endothelial-to-hematopoietic transition (12, 13). Two independent studies provided evidence that blood flow drives mouse HSPC emergence from the AGM (14, 15). Similar results are reported in zebrafish (16). Mouse and zebrafish embryos lacking blood circulation exhibit a reduced number of HSPCs and defects in their definitive hematopoiesis. Independent studies have identified that in endothelial cells exposed to biomechanical forces, multiple factors such as nitric oxide (NO), Ca2+, cAMP, and signaling pathways such as pProstaglandin E2, Wnt, Hedgehog, Notch (N), bBone mMorphogenetic protein, and vascular endothelial growth factor (VEGF) are activated (11). However, their potential interplay within the hemogenic endothelium and/or HSCs to achieve blood flow–dependent HSPC emergence in vertebrate embryos certainly deserves additional analyses. In adult vertebrate bone marrow, HSPCs are under the control of vascular niche cells which include endothelium cells that line the lumen of blood vessels, and perivascular cells (8, 9). Endothelial vascular niche cells are exposed to mechanical forces due to blood flow (10, 17). Whether the blood flow, by acting on endothelial cells, regulates HSPC properties remains poorly understood. Here, we used Drosophila as a model system to address this question.
The Drosophila larval hematopoietic organ, called the lymph gland, has emerged as an attractive in vivo model to study how hematopoietic progenitor maintenance and differentiation are regulated by their microenvironment (18–20). The lymph gland, aligned along the cardiac tube which corresponds to the vascular system, is composed of several pairs of lobes, with the anterior most lobe composed of an heterogeneous population of progenitors, differentiated blood cells, and the posterior signaling center (PSC) (21–23) (Fig. 1A). Two niches, the PSC and the cardiac tube, control lymph gland anterior-lobe homeostasis, maintaining the balance between hematopoietic progenitors and differentiated blood cells (20, 24–28). Cardiac cells produce the fibroblast growth factor (FGF) ligand Branchless (Bnl), activating its FGF receptor in a subpopulation of progenitors. Bnl signaling controls progenitor maintenance and prevents their differentiation into crystal cells and plasmatocytes, two mature blood cell types that differentiate under physiological conditions (29). The cardiac tube is composed of the aorta containing noncontractile cardiomyocytes and the posterior contractile heart chamber where cardiomyocytes, by contracting rhythmically, provide the propelling force for hemolymph/blood flow circulation within the cardiac lumen (30). The aorta is further subdivided into the anterior and posterior aortas, and the lymph gland is in contact with the anterior aorta (SI Appendix, Fig. S2A). In insects, hemolymph provides nutrients and conveys signals, but does not supply oxygen which is delivered via the tracheal system. Analyses of Drosophila cardiac physiology further established that, as in mammals, blood flow generates intracardiac mechanical forces (31). Here, we provide evidence that mechanosensing of blood flow by vascular niche cells modifies their secreted signals and subsequently impacts hematopoietic progenitor homeostasis.
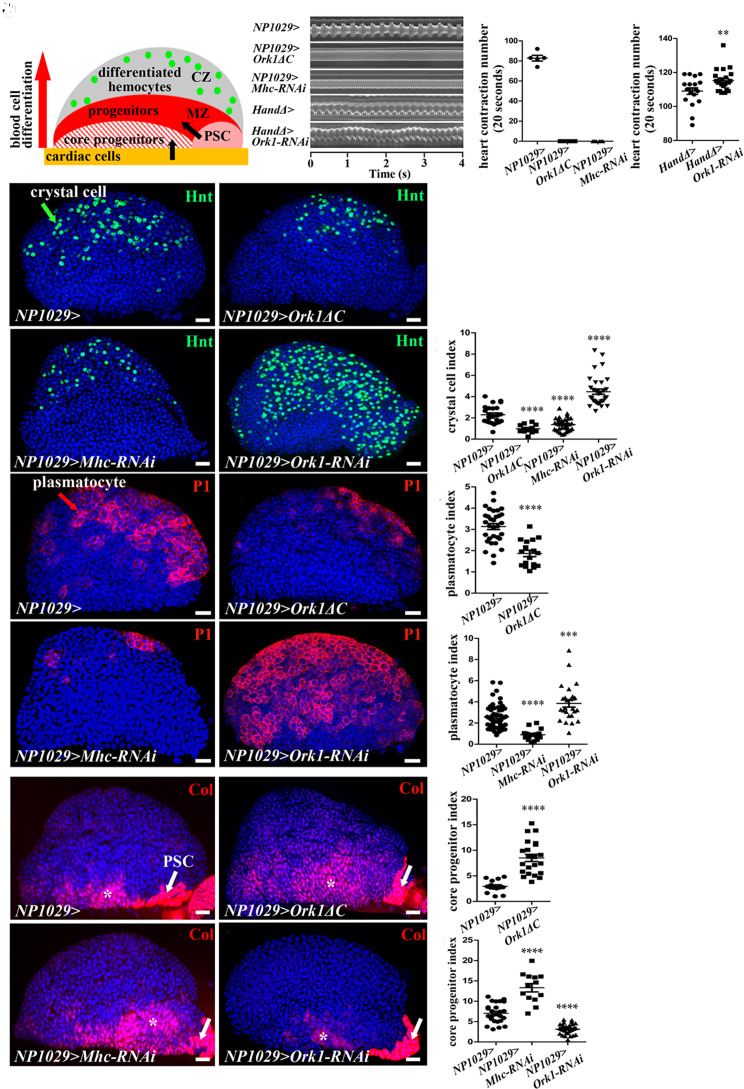
Fig. 1.
Heartbeat controls lymph gland homeostasis. (A) Schematic representation of a third-instar lymph gland anterior lobe. It is composed of progenitors (red) and core progenitors (hatched red) in a medullary zone (MZ), and a cortical zone, which contains differentiated hemocytes (CZ, green dots). The two niches, the PSC (pink) and the cardiac cells (orange), regulate different subsets of MZ progenitors (black arrows). (B) Kymograph of heartbeat in the control (NP1029> or HandΔ>) and when Ork1ΔC or Mhc-RNAi, or Ork1-RNAi are expressed with cardiac cell drivers. (C and D) Number of heart contractions per 20s. (E–H) Crystal cell differentiation (Hnt, green) when the heart is blocked (F and G) or accelerated (H). (I) Crystal cell index. (J, K, M, and N) Plasmatocyte differentiation (P1, red) when the heart is blocked (K and M) or accelerated (N). (L and O) Plasmatocyte index. (P, Q, S, and T) Col (red) labels core progenitors (*) and the PSC (arrow). (R and U) Coreprogenitor index. For all quantifications and figures, statistical analysis t test (Mann–Whitney nonparametric test) performed using GraphPad Prism 5 software. Error bars represent SEM and *P < 0.1; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns (not significant). Nuclei are labeled with Topro (blue). (Scale bars, 20 µm.)
Results
Heartbeat Regulates Lymph Gland Homeostasis.
A primary function of the Drosophila heart is to pump hemolymph throughout the body in an open circulatory system (30). We asked whether mechanical constraints exerted by blood flow, resulting from cardioblast contraction, could modify lymph gland hematopoiesis. To stop blood flow, we either expressed a constitutively active form of Ork1 (Ork1-deltaC, Ork1ΔC), a potassium channel subunit required to set heartbeat frequency (32), or knocked down the Myosin heavy chain (Mhc, necessary for muscle contraction), specifically in cardiac cells using two independent Gal4 drivers NP1029 and HandΔ (29, 33). Ork1ΔC expression in cardiac cells starting from late L1 larval stage or Mhc knockdown starting from L2 larval stage both resulted in heartbeat arrest (Fig. 1 B and C, Movies S1–S3, and SI Appendix, Fig. S1C) without affecting the formation of the heart lumen (SI Appendix, Fig. S1 A and B). In all these contexts, heart failure resulted in decreased differentiation of both crystal cells (labeled by Hindsight, Hnt antibody) (Fig. 1 E–G and I and SI Appendix, Fig. S1 E, F, H and I) and plasmatocytes (P1 antibody labeling) (Fig. 1 J–M and O). In the following experiments, we used without distinction any of these four combinations to block heartbeat. Conversely and in agreement with published data (32), an increased heart rate was observed when Ork1-RNAi was expressed with the HandΔ cardiac driver (Fig. 1 B and D and Movies S4 and S5). This resulted in increased crystal cell and plasmatocyte differentiation compared to control (Fig. 1 H, I, N, and O). We then analyzed the effect of heart arrest on hematopoietic progenitors. The transcription factor Collier (Col) (also known as Knot) is strongly expressed in PSC niche cells and at lower levels in core progenitors, a subset of hematopoietic progenitors maintained by cardiac cell signals (26, 29, 34) (Fig. 1A). Compared to wild type, higher numbers of Col+ progenitors were recorded when heartbeat was blocked, indicating that heartbeat controls core progenitor maintenance (NP1029>Ork1Δc and NP1029>Mhc-RNAi, Fig. 1 P–S and U and HandΔ>Ork1ΔC in SI Appendix, Fig. S1 J–L). Conversely, when heartbeat was increased, a lower number of Col+ progenitors was recorded (NP1029>Ork1-RNAi, Fig. 1 T and U). Since the PSC is also known to control lymph gland homeostasis (25, 34–39), we looked at PSC cell numbers with Antennapedia (Antp) immunostaining, upon heartbeat arrest. No difference in PSC cell numbers was found (NP1029>Ork1ΔC, SI Appendix, Fig. S1 M–O). Altogether, these data establish that the heartbeat, most likely through blood flow regulation, triggers progenitor differentiation into mature blood cells. These results suggest that mechanical stimuli resulting from blood flow are critical regulators of hematopoiesis.
Mechanosensitive Ion Channel Piezo Controls Lymph Gland Homeostasis.
Heartbeat imposes mechanical constraints on cardiac cells (31). Piezo encodes a mechanosensitive cation channel that directly senses mechanical tension in lipid bilayers (40) and is responsible for mechanoreception in many cell types (41, 42). A single piezo gene is present in the Drosophila genome (42–46). Piezo is expressed in larval cardiomyocytes, but no function in these cells has been reported so far (42, 47). To document its expression, we used both piezo-Gal4, where Gal4 is under the control of a piezo enhancer, (42) and the Piezo knock-in Gal4 allele piezo-Gal4 (KI) (43) to express UAS-mcd8GFP. In both cases, GFP was expressed in cardiac cells in L2 and L3 larvae and at low levels in a subset of crystal cells in L3 larvae (Fig. 2 A and B and SI Appendix, Fig. S2 A, B, D, and E). We investigated whether Piezo in cardiac cells regulates lymph gland hematopoiesis. In two null mutant contexts, piezo-KO (Fig. 2 C, D, and G) and piezo-Gal4(KI) (43) (SI Appendix, Fig. S2 F–H), crystal cell indexes were lower than those in control. Similar results were obtained by knocking down piezo in cardiac cells with RNAi (NP1029>piezo-RNAi, Fig. 2 E, F, and H and HandΔ>piezo-RNAi in SI Appendix, Fig. S2 I–L), or by overexpressing mPiezo1-2336-Myc where the Myc tag insertion in the last extracellular loop impairs channel conductivity without affecting gene expression or trafficking of the protein to the membrane (44, 48) (HandΔ>mPiezo1-2336-Myc, SI Appendix, Fig. S2 M–O). When piezo was knocked down in cardiac cells (NP1029>piezo-RNAi), plasmatocyte (Fig. 2 N, O, and R) and Col+ progenitor indexes (Fig. 2 P, Q, and S) were lower and higher than the control, respectively. Altogether, these data establish that piezo activity in cardiac cells regulates lymph gland homeostasis. To determine whether this function is specifically required in the anterior aorta cells which are in direct contact with the lymph gland, we knocked down piezo using the 76E11-Gal4 driver expressed only in the anterior aorta cells of late L2/early L3 larvae (SI Appendix, Fig. S2P). piezo knockdown starting from the L2 stage, with the Gal80ts system (49), led to a reduced crystal cell index (SI Appendix, Fig. S2 Q–S), which means that piezo is required in anterior aorta cells in late L2-L3 larval stages to control blood cell differentiation. Finally, we showed that piezo knockdown in cardiac cells does not affect heartbeat (HandΔ>piezo-RNAi, SI Appendix, Fig. S2 T and U and Movie S6). In summary, our data indicate that the function of piezo is required noncell-autonomously in L3 larval anterior aorta cells to control lymph gland homeostasis. While Piezo reduction in MZ progenitors does not affect crystal cell differentiation (SI Appendix, Fig. S3 A–F), its reduction in crystal cells using the lz-gal4 driver led to increased crystal cell numbers (lz>piezo-RNAi, SI Appendix, Fig. S2 B and C). This indicates that piezo also ensures a cell-autonomous function in the control of crystal cell numbers.
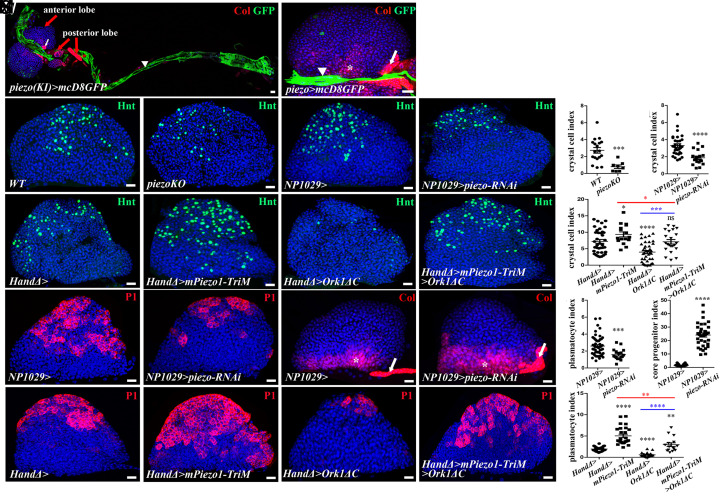
Fig. 2.
Piezo expressed in cardiac cells controls lymph gland homeostasis. (A) piezo-gal4(KI)>mcD8-GFP. Col (red) labels PSC (white arrow) and subsets of lymph gland cells in the anterior and posterior lobes (red arrows). In L3 larvae, piezo (green) is expressed in cardiac cells (white arrowhead). (B) Higher magnification of lymph gland anterior lobe where mcD8-GFP (green) is expressed under piezo-Gal4 in cardiac cells (arrowhead). (C–F) Fewer crystal cells (Hnt, green) are observed in a piezo null mutant (D), or when piezo is knocked down in cardiac cells (F). (G and H) Crystal cell index. (I–L) Active form of piezo (mPiezo1-TriM) with arrested heart (Ork1ΔC) restores wild-type crystal cell numbers (L). (M) Crystal cell index. (N and O) Compared to the control, (N) fewer plasmatocytes (P1, red) are seen when piezo is knocked down in cardiac cells (O). (R) Plasmatocyte index. (P and Q) Col (red) labels core progenitors (*) and the PSC (arrow). (S) Core–progenitor index. (T and U) Active form of piezo (mPiezo1-TriM) leads to increased plasmatocyte numbers (U). (V and W) Active form of piezo (mPiezo1-TriM) with arrested heart (Ork1ΔC) restores wild-type plasmatocyte numbers (W). (X) Plasmatocyte index.
Since piezo knockdown in cardiac cells and heart failure led to similar lymph gland defects, we asked whether both were functionally linked. Expression of a constitutively active form of Piezo, mPiezo1-TriM (44), in cardiac cells (HandΔ>) led to a significant increase in both crystal cell and plasmatocyte differentiation (Fig. 2 I, J, M, T, U, and X). In addition, activation of Piezo function in larvae with reduced heart contractility (HandΔ>mPiezo1-TriM>Ork1ΔC) rescued lymph gland defects as it restored a wild-type number of crystal cells (Fig. 2 L and M) and partially rescued both plasmatocytes (Fig. 2 W and X) and Col+ progenitors (SI Appendix, Fig. S3 G–K). Overall, these data indicate that heartbeat and the mechanoreceptor Piezo are functionally linked and support the proposition that heartbeat—at least in part—acts via Piezo to control lymph gland homeostasis.
Piezo and Heartbeat Control Expression of the FGF Ligand Bnl.
Drosophila cardiac cells correspond to the vascular niche. The FGF ligand Bnl secreted by cardiac cells activates in L3 larvae the FGF pathway in hematopoietic progenitors, which in turn controls lymph gland homeostasis (29). To investigate whether Bnl levels in the cardiac tube are dependent on piezo and/or on heartbeat, we analyzed endogenous Bnl expression using the bnl:GFP endo knock-in allele (50). When the heart was arrested (NP1029>Mhc-RNAi) or piezo knockdown (HandΔ>piezo-RNAi) in cardiac cells, higher bnl:GFP endo expression was observed in the aorta (Fig. 3 A–G), indicating that piezo and heartbeat control Bnl protein levels in the aorta. We then established that Piezo and heartbeat repress bnl transcription, since bnl transcripts, as quantified by in situ hybridization, were higher when either piezo was knocked down (HandΔ>piezo-RNAi) or the heart arrested (NP1029>Ork1ΔC) (Fig. 3 H–L).
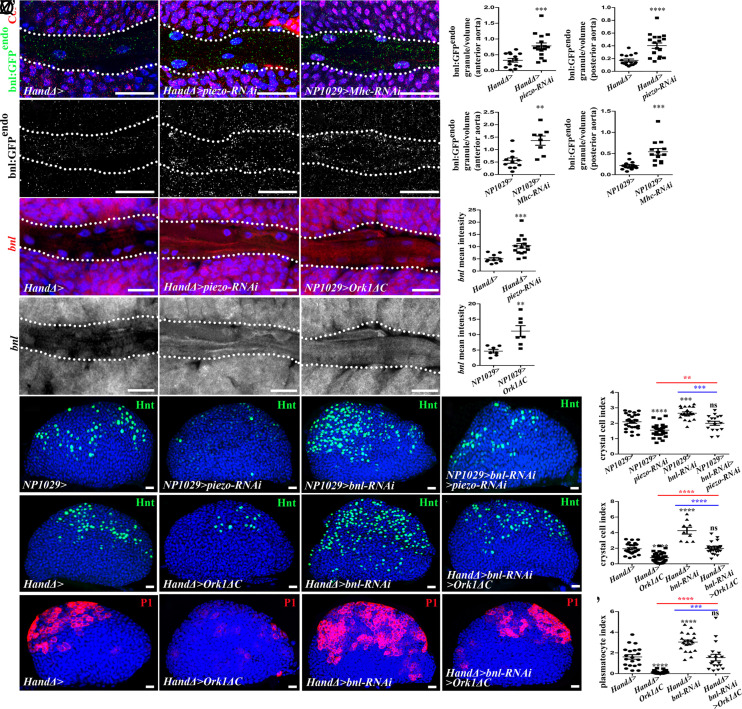
Fig. 3.
Heartbeat and Piezo control expression of FGF ligand Branchless (Bnl) in cardiac cells and thus regulate lymph gland hematopoiesis. (A–C’) Close-up view of larval anterior aorta expressing bnl:GFPendo (green in A–C, white in A’–C’) in the aorta and Col (red) in core progenitors (A–C). Dotted lines indicate cardiac tube outline. bnl:GFPendo expression is higher when piezo is knocked down or the heart is blocked (B-–C’). (D–G) Quantification of bnl:GFP endo in anterior or posterior aorta. (H–J’) Close-up view of cardiac tube when piezo-RNAi (I–I’) or Ork1ΔC (J–J’) is expressed in cardiac cells; bnl transcripts (red in H–J, white in H’–J’) in anterior aorta cells increase in both conditions. (K–L) Quantification of bnl expression. (M–P) Crystal cell (Hnt, green) differentiation increases when bnl is knocked down (O) and restored to wild type when piezo and bnl are simultaneously knocked down (P) in cardiac cells. (R–U) Crystal cell differentiation rescued when bnl is knocked down in cardiac cells simultaneously with heart arrest (U). (Q and V) Crystal cell index. (W–Z) Plasmatocyte (P1, red) differentiation increases when bnl is knocked down in cardiac cells (Y) and restored to wild type when bnl is knocked down in cardiac cells simultaneously with heart arrest (Z). (Z’) Plasmatocyte index.
To investigate the functional link between bnl and piezo in cardiac cells and lymph gland hematopoiesis, we monitored crystal cell and plasmatocyte differentiation and asked whether we could rescue the reduced blood cell differentiation due to piezo knockdown or heart arrest, by knocking down bnl in cardiac cells. While the expression of bnl-RNAi in cardiac cells (NP1029>bnl-RNAi) increased the number of crystal cells (Fig. 3 O and Q) and plasmatocytes (Fig. 3 W, Y, and Z’), which is in agreement with the previous report (29), piezo-RNAi reduced the number of crystal cells (Fig. 2 F and H and Fig. 3 N and Q) and plasmatocytes (Figs. 2 O and R and 3 W, Y, and Z’). Simultaneous expression of bnl-RNAi and piezo-RNAi (NP1029>bnl-RNAi>piezo-RNAi) restored wild-type crystal cell numbers (Fig. 3 P and Q). Altogether, these data indicate that Piezo and Bnl are functionally linked and Piezo functions at least in part by repressing bnl expression. Likewise, crystal cell (Fig. 3 U and V) and plasmatocyte (Fig. 3 Z–Z’) differentiation were partially rescued when bnl-RNAi was expressed in a context where the heart was blocked (HandΔ>bnl-RNAi>Ork1ΔC). In conclusion, heartbeat, Piezo, and Bnl are functionally linked and heartbeat and Piezo control lymph gland hematopoiesis by—at least in part—repressing bnl expression in cardiac cells. All these data lead us to propose that the blood flow driven by heart beat activates the mechanosensitive channel Piezo in aorta cells, which in turn represses bnl transcription and therefore regulates lymph gland homeostasis.
Notch Acts Downstream of Piezo to Repress bnl Expression.
To further decipher the Piezo-dependent mechanotransduction mechanism in cardiac cells, we first analyzed Ca2+ levels, since Piezo triggers upregulation of cytosolic Ca2+ (51–54). Using the Ca2+ sensor GCaMP3 (55), we observed that knocking down piezo in cardiac cells (NP1029>piezo-RNAi, Fig. 4 A–D) or blocking heartbeat (NP1029>Mhc-RNAi, SI Appendix, Fig. S4 A–D) led to decreased fluorescence in the aorta compared to the control, reflecting reduced Ca2+ levels. Thus heartbeat, via blood flow, probably activates Piezo, which in turn regulates cytosolic Ca2+ levels in aorta cells.
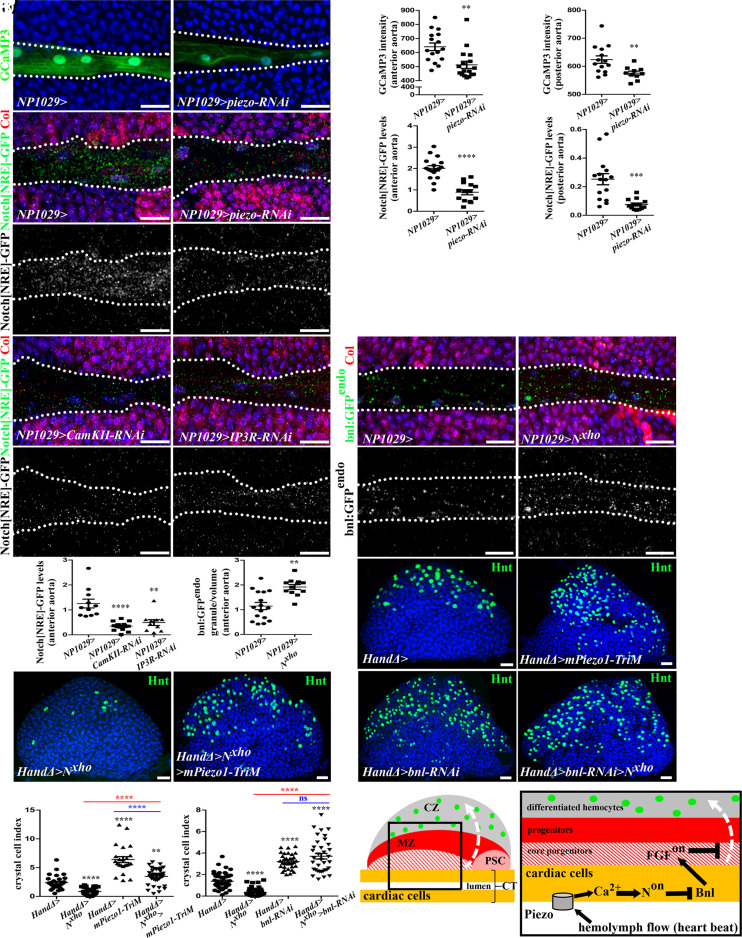
Fig. 4.
Piezo regulates Ca2+ levels and activates N signaling, which controls blood cell differentiation by repressing bnl transcription in cardiac cells. (A and B) GCaMP3 Ca2+ sensor (green) decreases when piezo is knocked down in cardiac cells. (C and D) Quantification of Ca2+ sensor signal. (E, F, I, and J) Enlarged view of larval cardiac tube expressing Notch[NRE]-GFP (green) and Col (red) in core progenitors (E, F, I, and J) and Notch[NRE]-GFP (white) in (E', F', I', and J’). Dotted lines indicate cardiac tube outline. Notch[NRE]-GFP expression decreases when piezo is knocked down (F–F') and when Ca2+ levels are decreased (I - J’) in cardiac cells. (G, H, and M) Quantification of Notch[NRE]-GFP. (K - L’) bnl:GFPendo expression increased when N signaling inhibited (L). (N) Quantification of bnl:GFPendo in the anterior aorta. (O–T) Crystal cell (Hnt, green) differentiation is reduced when N signaling is inhibited (Q) and rescued when Piezo is simultaneously constitutively activated (R). Crystal cell differentiation defect observed when N is inhibited (Nxho) is rescued by simultaneous bnl knockdown (T). (U and V) Crystal cell index. (W) Representation of third instar larval lymph gland anterior lobe. Blood flow resulting from heartbeat activates Piezo in cardiac/vascular cells leading to Ca2+ increase, which activates N signaling and in turn, represses bnl expression in vascular cells. Bnl normally produced by vascular cells activates FGF pathway in progenitors, where it is required for their maintenance at the expense of their differentiation. The white dashed arrow indicates blood cell differentiation.
We next focused on Notch (N) signaling since Piezo, by modulating Ca2+ levels, regulates N signaling in several cell types such as mouse endothelial cells (56), zebrafish cardiac valve cells (57), and Drosophila gut cells (43). Moreover, in Drosophila, bnl transcription is repressed by N signaling in the embryonic tracheal system (58). We therefore hypothesized that in cardiac cells Piezo could activate N signaling, which in turn would repress bnl expression. We therefore investigated the role of N signaling in cardiac cells.
We analyzed the expression of three reporters of N activation: Notch-responsive element (Notch[NRE]-GFP) corresponding to a N enhancer (59), NRE-GFP, a synthetic reporter for N pathway activation, and E(spl)mbeta-GFP, which reports on canonical N pathway activation (60). With all the three reporters, GFP expression was detected in cardiac cells, confirming that the N pathway is activated there (Fig. 4E and SI Appendix, Fig. S4 K and L). Notch[NRE]-GFP expression was lower than control when a dominant negative form of N (Nxho) was expressed in cardiac cells (NP1029>N xho, SI Appendix, Fig. S4 E, F', and H). These results demonstrate that Notch[NRE]-GFP is a bona fide reporter of N activation in cardiac cells. Upon either piezo knockdown in cardiac cells (NP1029>piezo-RNAi, Fig. 4 F–H) or heartbeat block (NP1029>Mhc-RNAi, SI Appendix, Fig. S4 G–G', I, and J), Notch[NRE]-GFP expression was reduced, thus establishing that heartbeat and piezo are required for N activation in cardiac cells. Furthermore, we showed that N signaling activation is dependent on Ca2+ levels, since Notch[NRE]-GFP expression was reduced when Ca2+ levels were decreased in cardiac cells, either through CamKII knockdown (CamKII-RNAi) or through the inhibition of ER-mediated Ca2+ release in the cytosol (IP3R-RNAi) (61) (Fig. 4 I, J', and M). Collectively, these data indicate that Piezo activates N signaling through the control of intracellular Ca2+ levels.
Next, we investigated the relationship between N signaling and bnl and determined whether N signaling activation in cardiac cells is required for lymph gland blood cell differentiation. Bnl levels, as visualized with bnl:GFP endo, were higher in the cardiac tube when the N pathway was inhibited (NP1029>N Xho, Fig. 4 K, L, and N, and HandΔ>Notch-RNAi, SI Appendix, Fig. S4 M–O). Analyzing blood cell differentiation indicated that when N signaling was inhibited, both crystal cells (HandΔ>N xho, Fig. 4 Q and U; NP1029>N xho, SI Appendix, Fig. S5 A–C and HandΔ>N-RNAi, SI Appendix, Fig. S5 D–F) and plasmatocyte (HandΔ>N-RNAi and HandΔ>N xho SI Appendix, Fig. S5 G–J) numbers were lower than those of the control. Finally, analysis of Col+ progenitors indicated an increased core progenitor index (HandΔ>N xho, SI Appendix, Fig. S5 K–M). Altogether, these data indicate that activation of N signaling in cardiac cell represses bnl expression in these cells and controls lymph gland homeostasis. To further characterize the molecular mechanism of N signaling in cardiac cells, we analyzed the function of several members of this pathway. The E(spl) complex contains several genes which are direct targets of N signaling and encode transcriptional repressors (60). kuzbanian (kuz) encodes an ADAM metallopeptidase that regulates N activation by cleavage, and Mastermind (Mam) is the coactivator of NICD that regulates N target gene expression (60). To address whether N activation in cardiac cells acts via the E(spl) complex, we expressed in cardiac cells, E(spl)mbeta-Act which contains the VP16 activation domain and converts the E(spl)mbeta from a repressor to an activator. E(spl)mbeta-Act therefore turns on genes which E(spl)mbeta normally represses. We observed increased bnl:GFP-endo levels in the aorta (HandΔ>E(spl)mbeta-Act, SI Appendix, Fig. S4 P–R). This supports that N signaling represses Bnl levels in aorta cells through the E(spl) complex. Then, we looked at blood cell differentiation. A reduction in crystal cell numbers was observed when expressing in cardiac cells the active form of E(spl)mbeta (HandΔ>E(spl)mbeta-Act, SI Appendix, Fig. S6 E and F), or a dominant negative form of mastermind (MamDN) (NP1029>mamDN, SI Appendix, Fig. S6 A–C), or when kuz function was inhibited by expressing a dominant-negative form (KuzDN) or kuz-RNAi (HandΔ>kuzDN, SI Appendix, Fig. S6 I and K, and NP1029>kuz-RNAi, SI Appendix, Fig. S6 M and N). Reduced plasmatocyte differentiation was also observed (NP1029>kuz-RNAi, SI Appendix, Fig. S7 A–C and HandΔ>kuzDN, SI Appendix, Fig. S7 F and H). Collectively, these data establish that the canonical N signaling pathway is activated in aorta cells and noncell autonomously regulates lymph gland homeostasis.
Since Piezo activation and N loss of function have opposite effects on blood cell differentiation, we asked whether both were functionally linked. Expression of a constitutively active form of Piezo (mpiezo1-TriM) in a context of N inhibition by expressing Nxho or KuzDN led to increased crystal cell numbers compared to inactivation of N signaling alone (HandΔ>NXho>mPiezo1-TriM, Fig. 4 R and U, and HandΔ>kuzDN>mPiezo1-TriM, SI Appendix, Fig. S6 J–K). Similar results were obtained for plasmatocyte differentiation (HandΔ>kuzDN>mPiezo1-TriM, SI Appendix, Fig. S7 G and H). Altogether, these data show that Piezo and N are involved in the same process and that Piezo—at least in part—functions through N signaling. We then investigated functional links between N signaling activation and Bnl. Knockdown of bnl in cardiac cells compensated for the crystal cell differentiation defect due to N signaling inhibition (HandΔ>bnl-RNAi>N xho, Fig. 4 T and V, and HandΔ>bnl-RNAi>kuzDN, SI Appendix, Fig. S7I). Similar results were obtained for plasmatocytes (HandΔ>bnl-RNAi>kuzDN, SI Appendix, Fig. S7J). Col+ progenitor index is similar to bnl-RNAi when bnl-RNAi was coexpressed with N xho (HandΔ>bnl-RNAi>N xho, SI Appendix, Fig. S7K), indicating that bnl-RNAi is epistatic to N inhibition. Collectively, our data are consistent with the model that in cardiac cells Piezo activates canonical N signaling, which subsequently represses bnl expression, which in turn regulates lymph gland hematopoiesis (Fig. 4W).
Temporal Control of Lymph Gland Hematopoiesis by Blood Flow.
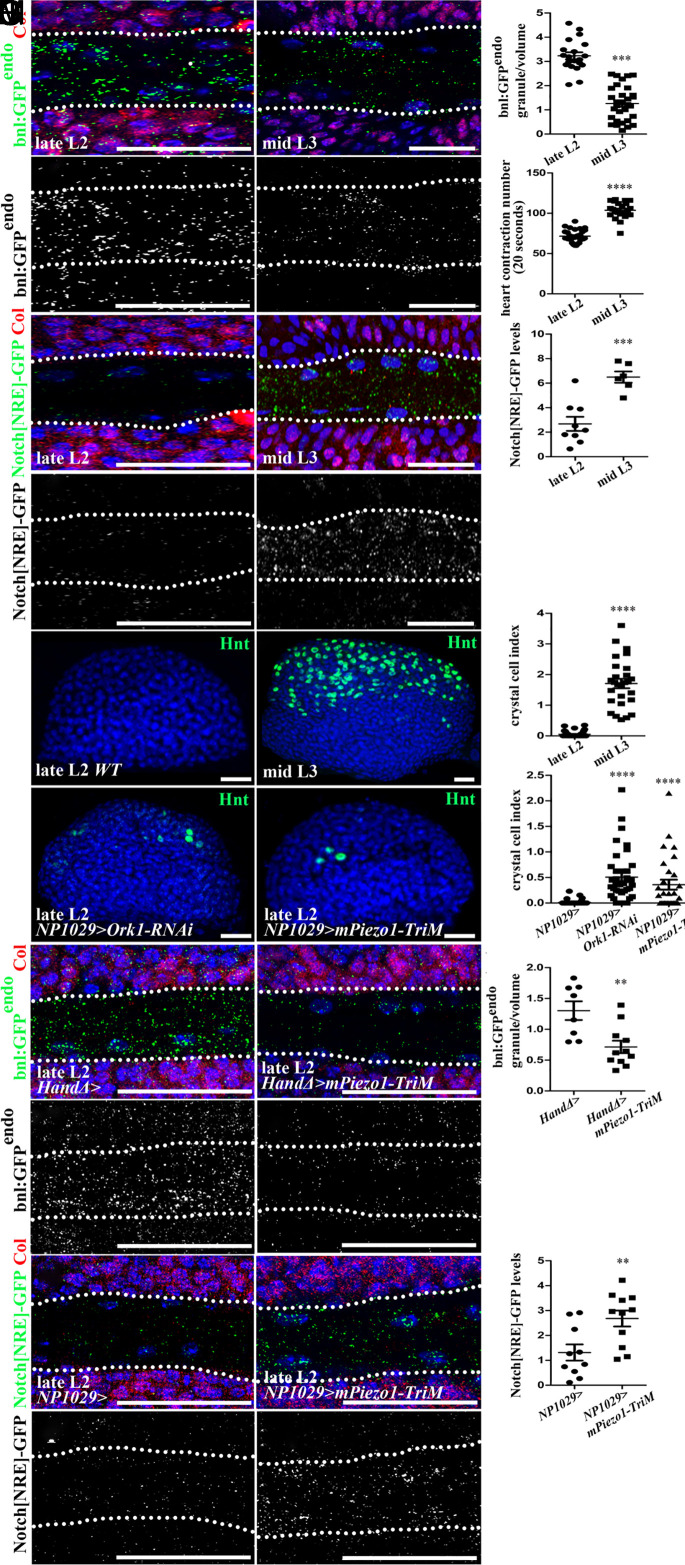
We finally wondered what the physiological relevance of lymph gland hematopoiesis control by heartbeat could be. While there were neither crystal cells (Fig. 5 H–J) nor plasmatocytes (SI Appendix, Fig. S8 A and B) in late L2 lymph glands, many were found in mid L3 larvae, consistent with published data that blood cell differentiation initiates in L3 larvae, mature blood cells being seldom found in L2 larvae (20). Strikingly, we discovered that heartbeat rate increases slightly between L2 and L3 larval stages (Fig. 5D and Movies S7 and S8). We then looked at Bnl expression and N activation. We observed a decrease in bnl:GFP endo (Fig. 5 A–C) and an increase in Notch[NRE]-GFP (Fig. 5 E–G) levels in the aorta of mid L3 compared to late L2 larvae. Next, we tested whether temporal differentiation of lymph gland blood cells is dependent on heartbeat and/or Piezo activation. We found that when heartbeat rate was increased (Ork1-RNAi) or when Piezo was constitutively activated (mPiezo1-TriM) from L1 to L2 larval stages, premature crystal cell (Fig. 5 K–M) and plasmatocyte (SI Appendix, Fig. S8 A and C–F) differentiation occurred in late L2 larvae.
Fig. 5.
Physiological modulation of heartbeat rate during larval development regulates hematopoiesis. (A–B’) Close-up view of cardiac tube in larvae expressing bnl:GFPendo (green in A and B and white in A' and B’) and Col (red) in core progenitors (A and B) in late L2 (A and A’) and mid L3 larvae (B and B’). Dotted lines indicate cardiac tube outline. bnl:GFPendo decreases in mid L3 compared to late L2 larvae. (C) Quantification of bnl:GFPendo in the anterior aorta. (D) Heart contraction number per 20 s in late L2 and mid L3 larvae. (E–F’) Notch[NRE]-GFP (green in E and F and white in E’–F’) and Col (red) in core progenitors (E and F). Dotted lines indicate cardiac tube outline. Notch[NRE]-GFP increases in mid L3 compared to late L2 larvae. (G) Quantification of Notch[NRE]-GFP in the anterior aorta. (H, I, K, and L) While crystal cells (Hnt, green) are seldom found in late L2 (H), they massively differentiate in mid L3 larvae (I). Premature crystal cell differentiation is observed in late L2 larvae when heartbeat is accelerated (K) or when an active form of Piezo is expressed in cardiac cells (L). (J and M) Crystal cell index. (N–O’) bnl:GFPendo (green in N and O, white in N’–O’) and Col (red) in core progenitors (N and O). Dotted lines indicate cardiac tube outline. bnl:GFPendo expression is decreased in late L2 larvae when an active form of Piezo (mPiezo1-TriM) is expressed in cardiac cells (O–O’). (P) Quantification of bnl:GFPendo in the anterior aorta. (Q–R’) Notch[NRE]-GFP (green in Q and R, white in Q’–R’) and Col (red) in core progenitors (Q and R). Notch[NRE]-GFP expression is increased in late L2 larvae when an active form of Piezo is expressed in cardiac cells (R–R’). (S) Quantification of Notch[NRE]-GFP in the anterior aorta.
Finally, we investigated Bnl levels and N activity when Piezo was constitutively activated (mPiezo1-TriM) in cardiac cells from L1 to L2 stages on. In late L2 anterior aorta, a decrease in bnl:GFP endo (HandΔ>mPiezo1-TriM, Fig. 5 N–P) and an increase in Notch[NRE]-GFP (NP1029>mPiezo1-TriM, Fig. 5 Q–S) compared to control were observed. These results indicate that the premature activation of Piezo in L2 larval cardiac cells induces Notch activation, which in turn represses bnl expression. Collectively, these data suggest that the temporal control, between L2 and L3 larval stages, of lymph gland blood cell differentiation is achieved through physiological modulation of the heartbeat rate.
Discussion
In Drosophila larvae, cardiac cells act as vascular niche cells to regulate lymph gland homeostasis. Here, we provide evidence that the underlying mechanism of this regulation involves Piezo-mediated mechanosensing of blood flow. During larval development, this mechanism adapts lymph gland hematopoiesis to the heartbeat. We provide the demonstration of a mechanotransduction pathway, in hematopoietic vascular niche cells, to promote blood cell homeostasis in the lymph gland. Our findings show that Piezo, activated by blood flow, promotes blood progenitor maintenance via activation of the N signaling pathway in vascular niche cells, which in turn represses expression of the FGF ligand Bnl. N signaling represses bnl transcription in cardiac cells via E(spl) genes. Bnl secreted by vascular niche cells noncell-autonomously activates the FGF pathway in hematopoietic progenitors for their maintenance. The regulatory cascade identified in this study where Piezo activates N signaling which in turn represses Bnl expression, appears specific to cardiac cells. Indeed, although N, Bnl, and Piezo have a cell-autonomous function for crystal cell formation, N or Bnl loss-of-function in crystal cells results in similar defects, which is the opposite of the phenotype that would be expected if N inhibited Bnl expression (62, 63). Besides demonstrating a key role of Piezo in vascular niche cells, we identify that the canonical N signaling pathway is also required in these cells. While the activation of Ca2+ by Piezo is well established, it remains largely unclear how Ca2+activates N signaling. Caolo et al. (56) provided an explanation for the activation of N signaling by shear stress in mouse adult endothelial cells. Activation of the Piezo1 channel by mechanical force (e.g., fluid flow) causes increased intracellular Ca2+ levels, which stimulate the enzymatic activity of the metalloendopeptidase ADAM 10, to cause S2 and S3 proteolytic cleavage of N1 and in turn the release of the N1 intracellular domain. The result is activation of N target gene expression. Kuz is the Drosophila ortholog of ADAM10, and we establish that Kuz function is required in cardiac cells to regulate lymph gland homoeostasis. Whether in the lymph gland Ca2+ levels control Kuz function is an interesting possibility which remains to be investigated.
Our data support the hypothesis that mechanical forces exerted by blood flow activate Piezo in cardiac cells to regulate lymph gland homeostasis. However, it cannot be completely ruled out that a biochemical interaction between Ork1 and Myosin with Piezo in anterior aorta cells may induce a conformational change of Piezo leading to its opening, in a process independent of mechanics. Applying mechanical constraints on cardiac cells to determine whether there is an impact on lymph gland hematopoiesis would certainly complete our data. Unfortunately, such an experiment to test the consequences on lymph gland hematopoiesis is technically impossible. Indeed, the lymph gland is very sensitive to many kinds of global stresses, as for example a needle puncture (64) or forceps-squeezing (65) of whole larvae induces lamellocyte differentiation and abnormal lymph gland hematopoiesis. Finally, whether rhythmic cardioblast contractions transduce mechanical stimuli not only to cardiac cells but also to lymph gland cells, and in particular to the core progenitors which are close to cardiac cells, remains a possibility.
Since heartbeat rate is different in L2 and L3 larvae, and in turn regulates lymph gland hematopoiesis during larval development, one point to consider is how heartbeat is controlled during larval development. While the Drosophila adult heart receives neuronal input to regulate cardiac activity (66), the larval heart lacks innervation. The current dogma is that the larval cardiac impulse is only myogenic (67). However, the larval heartbeat rate can be modulated by neurotransmitters and neuromodulators provided by hemolymph, such as serotonin, GABA, octopamine, acetylcholine, and dopamine (68–70). Recent work established that larval cardiac cells express neurotransmitter receptors, such as those for acetylcholin (Ach) and serotonin (5HT); both Ach and 5HT act directly in heart cells to modulate heartbeat (67, 69, 71). Whether under physiological conditions neurotransmitters circulating in the hemolymph modulate heart rate between L2 and L3 larval stages remains unknown. More work is required to identify which factors are involved and to decipher, at the molecular level, how this regulation is achieved.
That blood flow promotes the emergence of definitive HSPCs from the AGM in vertebrate developing embryos is well established (11, 14–16). Signals generated by hemodynamic forces and the molecular mechanisms involved are beginning to be deciphered. Whether blood flow regulates adult hematopoiesis in bone marrow remains poorly understood. The microvasculature in the bone marrow forms a complex and irregular network of interconnecting sinusoidal and arterioles. It has been proposed that blood flow velocity and wall shear stress are important factors in HSPC homing (72). Yet, no role for blood flow on endothelial vascular niche cells has been established. However, in the bone marrow, niche cells can sense mechanical forces and regulate hematopoiesis. A recent study established that physical exercise, by acting on hematopoietic niche cells, can stimulate the immune system (17). Shen et al. identified a population of mouse periarteriolar niche cells in the bone marrow and physical exercise, which activates Piezo1 on their surface, leading to expression and secretion of stem cell factor required to maintain nearby common lymphoid progenitors. These progenitors are able to differentiate into lymphocytes and fight bacterial infections (17, 73). This finding raises the exciting possibility that in bone marrow, mechanosensing of blood flow by endothelial niche cells might regulate HSPC properties. Given the significant parallels between Drosophila lymph gland and mammalian bone marrow hematopoiesis, there is good possibility that our regulation involving blood flow, Piezo, and N signaling could be conserved in mammals. Interestingly, mPiezo1 is expressed in mouse endothelial cells of developing blood vessels and is required for the development of the vascular system during embryogenesis (74). While N signaling plays many roles on the main cellular components of bone marrow, including HSPCs, the pathway is also activated in endothelial cells and stimulates long-term self-renewal of HSPCs, through the angiocrine release of N ligands (75, 76). Collectively, these data allow us to propose that as in Drosophila, mammalian bone marrow endothelial cells may sense blood flow mechanical stimulation and accordingly modify their transcriptional program and production of signals, which in turn may control HSPC properties.
Materials and Methods
Fly Strains.
w1118 (wild type, WT), UAS-mcD8GFP (26), handΔ-gal4 (29), NP1029-gal4 (77), tep4-gal4 and dome-gal4 (26), bnl:GFPendo (50), HandC-GFP (78), UAS-Ork1ΔC and UAS-Ork1-RNAi (32), UAS-Piezo-RNAi1 (line 1), UAS-mPiezo1-2336-Myc and UAS-mPiezo1-TriM (44), UAS-Mhc-RNAi1 (line 1) (F.Schnörrer), Notch[NRE]-GFP and UAS-Notch-RNAi (59), NRE-GFP and UAS-MamDN (60), UAS-Nxho and UAS-E(spl)mbeta-Act (S.Bray). UAS-E(spl)mbeta-Act contains the VP16 activation domain that replaces the C-terminal WRPW motif of E(spl)mbeta and converts the protein from a repressor to an activator (79). Other strains were provided by Bloomington (BL) and Vienna Drosophila Resource Center (VDRC): UAS-GCaMP3 (BL32116), UAS-Dicer (BL24650), 76E11-Gal4 (BL39933), tub-Gal80ts (BL7019), UAS-piezo-RNAi2 (line 2) (VDRC v25780), UAS-Mhc-RNAi2 (line 2) (BL26299), piezo-Gal4 (KI) (BL78335), piezo-Gal4 (enhancer) (BL59266), piezo (KO) (BL58770), UAS-bnl-RNAi (VDRC GD5730), UAS-CaMKII-RNAi (BL29401), UAS-IP3R-RNAi (BL25937), E(spl)mbeta-GFP (BL65294), UAS-kuz-RNAi (BL66958), UAS-KuzDN (BL6578), UAS-MamDN, and UAS-E(spl)mbeta-Act (79). Crosses and subsequent raising of larvae until late L1/early L2 stage were performed at 22 °C, before shifting larvae to 25 °C until their dissection at L3 stage. For RNAi treatments, the same procedure was followed except that larva were shifted to 29 °C until their dissection. UAS-Dicer was introduced and at least two independent RNAi lines per gene were tested. Controls correspond to Gal4 drivers with UAS-Dicer crossed with w1118. For Gal80ts temperature shift experiments with Mhc-RNAi or 76E11-Gal4, crosses were initially maintained at 18 °C (permissive temperature) for 3 d after egg laying, and then shifted to 29 °C until dissection.
Antibodies.
Primary antibodies were mouse anti-Hnt (1/100, Hybridoma Bank), mouse anti-Col (1/40) (26), mouse anti-P1 (1/30, I. Ando, Institute of Genetics, Biological Research Center of the Hungarian Academy of Science, Szeged, Hungary), mouse anti-Antp (1/100, Hybridoma Bank), chicken anti-GFP (1/500, Abcam). Secondary antibodies were Alexa Fluor-488 and -555 conjugated antibodies (1/1,000, Molecular Probes) and goat anti-Chicken Alexa Fluor-488 (1/800, Molecular Probes). Nuclei were labeled with TOPRO3 (ThermoFisher Scientific). For detecting bnl:GFPendo and Notch[NRE]-GFP, GFP immunostainings were performed.
Heart Rate Measurements and Movies.
Larvae expressed HandC-GFP with GFP labeling of cardioblasts and pericardial cells. Larvae were anaesthetized with FlyNap (Carolina Biological). Five microliters of FlyNap was put on a piece of cotton in a closed chamber and larvae were put in the chamber for 160 s. Anesthetized larvae were deposited on a glass slide for recording with a binocular macroscope (SMZ18, Nikon). The anesthetized larvae were kept at 25 °C for 10 min before recording. For each individual, 20 s-recordings were performed, and this was repeated twice with 10 min of rest in the dark. To measure heart contraction, movies were converted into a kymograph using Fiji software. For each larva, the number of heartbeat contractions during 20 s corresponds to the average of three measurements. At least 10 larvae per genotype were scored, and experiments were reproduced at least three times. All recorded larvae survived and gave rise to an adult. Statistical analyses employing t test (Mann–Whitney nonparametric test) were performed using GraphPad Prism 5 software.
Quantifications.
In all experiments, optimized confocal sections were performed on Leica SP8 microscope for three-dimensional (3D) reconstructions, and since the number of fluctuating lymph gland differentiated blood cells depends on the larval stage, and to limit discrepancies, all genotypes were always analyzed in parallel. For all quantifications, statistical analyses employing t-test (Mann–Whitney nonparametric test) were performed using GraphPad Prism 5 software.
Blood Cell and Progenitor Quantification.
Crystal cells were visualized by immunostaining with antibodies against Hnt. Plasmatocytes and core progenitors were labeled by P1 and Col immunostaining, respectively. Nuclei were labeled by TOPRO3. The number of crystal cells and volume (in µm3) of each anterior lymph gland lobe were measured using Fiji software and 3DSuite plugin (80). Crystal cell index corresponds to [(number of crystal cells)/(primary lobe volume)] × 100. Plasmatocyte or progenitor indexes correspond to (plasmatocyte or progenitor volume/anterior lobe volume) × 10. At least 15 anterior lobes per genotype were scored, and experiments were reproduced at least three times.
Quantification of UAS-GCaMP3 Intensity.
The volume of the portion of the cardiac tube localized in between the two lymph gland anterior lobes was measured. For GCaMP3, mean intensity of cytoplasmic GFP was quantified using Fiji software and the 3DSuite plugin (80). The mean intensity of GCaMP3 corresponds to the sum of GCaMP3 intensity per cardiac tube volume. Quantifications for anterior and posterior aorta were performed independently using different lymph gland sets.
The anterior aorta corresponds to the cardiac tube which lies in between the two LG anterior lobes. At least 20 cardiac tubes per genotype were scored, and experiments were reproduced at least three times.
Quantification of bnl::GFP and Notch[NRE]-GFP Dots, and bnl Expression.
A region of interest (ROI) including the cardiac tube portion located in between the anterior lymph gland lobes was selected. To normalize with respect to background in tissue preparations, a ROI in the lymph gland cortical zone was selected and used to quantify background level. The volume of ROI and number of Bnl::GFP or Notch[NRE]-GFP granules per ROI were quantified using Fiji software and DiAna plugin (81). The number of GFP granules per volume was calculated. Notch[NRE]-GFP and bnl::GFP levels correspond to GFP granules measured in the cardiac tube minus GFP granules measured in the cortical zone. Quantification for anterior and posterior aortas was performed independently using different lymph gland sets. At least 10 cardiac tubes per genotype were scored. For bnl expression levels, volume of cardiac tube and bnl fluorescence were quantified using Fiji software and 3DSuite plugin (80). bnl mean intensity corresponds to bnl fluorescence/volume. At least 6 cardiac tubes per genotype were scored. All experiments were reproduced at least three times.
Supplementary Material
Appendix 01 (PDF)
heartbeat in control NP1029, UAS-dicer; mcD8-GFP>w1118
heart is arrested in NP1029, UAS-Dicer, mcD8-GFP>Ork1ΔC
heart is arrested in NP1029, UAS-Dicer, mcD8-GFP>Mhc-RNAi
heartbeat in control HandΔ, UAS-Dicer; HandC-GFP> w1118
heartbeat is increased in HandΔ, UAS-Dicer; HandC-GFP>Ork1-RNAI
heartbeat in HandΔ, UAS-Dicer; HandC-GFP>piezo- RNAi
heartbeat in L2 larvae in HandΔ UAS-Dicer; HandC-GFP> w1118
heartbeat in L3 larvae in HandΔ UAS-Dicer; HandC-GFP> w1118
Acknowledgments
We thank H. Bouhkatmi, S. Bray, F. Schnorrer, Y. N. Jan, S. Roy, BL and Vienna Stock Center and the Transgenic RNAi Project (TRiP) at Harvard Medical School for fly strains; I. Ando, A. Moore, and T. Trenczek for antibodies; and V. Gobert, M. Haenlin, G. Lebreton, M. Meister, B. Monier, C. Monod, A. Vincent, and X. Wang for critical reading of the manuscript. We are grateful to B. Ronsin and S. Bosch for assistance with confocal microscopy (Platform TRI); J. Favier, V. Nicolas, and A. Destenable for fly culture; and B. Monier and X. Wang for their advice during the course of Y.T.’s thesis. Research in the authors’ laboratory is supported by the CNRS, University Toulouse III, FRM (Fondation pour la Recherche Médicale) DEQ20180339171 (M.C.) and FDT202106012829 (Y.T.), La Ligue Contre le Cancer 31 (M.C.), La Société Française d’Hématologie (SFH), the China Scholarship Council (Y.T.), and the CNRS « Groupement de recherche 3740 ». Portions of this work were developed from the doctoral dissertation of Mr. Y.T.
Author contributions
Y.T. and M.C. designed research; Y.T., I.M.-P., and X.L. performed research; Y.T., I.M.-P., N.V., and M.C. analyzed data; and Y.T. and M.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Torres L. S., et al. , Recent advances in “sickle and niche” research–Tribute to Dr. Paul S Frenette [in English]. Stem. Cell Rep. 17, 1509–1535 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acar M., et al. , Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal [in English]. Nature 526, 126–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baryawno N., et al. , A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia [in English]. Cell 177, 1915–1932.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crane G. M., Jeffery E., Morrison S. J., Adult haematopoietic stem cell niches [in English]. Nat. Rev. Immunol. 17, 573–590 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Pinho S., Frenette P. S., Haematopoietic stem cell activity and interactions with the niche [in English]. Nat. Rev. Mol. Cell Biol. 20, 303–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramasamy S. K., et al. , Regulation of hematopoiesis and osteogenesis by blood vessel-derived signals [in English]. Annu. Rev. Cell Dev. Biol. 32, 649–675 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Tikhonova A. N., et al. , The bone marrow microenvironment at single-cell resolution [in English]. Nature 569, 222–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comazzetto S., Shen B., Morrison S. J., Niches that regulate stem cells and hematopoiesis in adult bone marrow [in English]. Dev. Cell 56, 1848–1860 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunisaki Y., et al. , Arteriolar niches maintain haematopoietic stem cell quiescence [in English]. Nature 502, 637–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton P. D., Dumbali S., Wenzel P. L., Mechanoregulation in hematopoiesis and hematologic disorders [in English]. Curr. Stem. Cell Rep. 6, 86–95 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., et al. , Biomechanical cues as master regulators of hematopoietic stem cell fate [in English]. Cell Mol. Life Sci. 78, 5881–5902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kissa K., Herbomel P., Blood stem cells emerge from aortic endothelium by a novel type of cell transition [in English]. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Boisset J. C., et al. , In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium [in English]. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Adamo L., et al. , Biomechanical forces promote embryonic haematopoiesis [in English]. Nature 459, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.North T. E., et al. , Hematopoietic stem cell development is dependent on blood flow [in English]. Cell 137, 736–748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., et al. , A blood flow-dependent klf2a-NO signaling cascade is required for stabilization of hematopoietic stem cell programming in zebrafish embryos [in English]. Blood 118, 4102–4110 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Shen B., et al. , A mechanosensitive peri-arteriolar niche for osteogenesis and lymphopoiesis [in English]. Nature 591, 438–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letourneau M., et al. , Drosophila hematopoiesis under normal conditions and in response to immune stress [in English]. FEBS Lett. 590, 4034–4051 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Morin-Poulard I., Tian Y., Vanzo N., Crozatier M., Drosophila as a model to study cellular communication between the hematopoietic niche and blood progenitors under homeostatic conditions and in response to an immune stress [in English]. Front. Immunol. 12, 719349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee U., Girard J. R., Goins L. M., Spratford C. M., Drosophila as a genetic model for hematopoiesis [in English]. Genetics 211, 367–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung S. H., Evans C. J., Uemura C., Banerjee U., The Drosophila lymph gland as a developmental model of hematopoiesis [in English]. Development 132, 2521–2533 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Cho B., et al. , Single-cell transcriptome maps of myeloid blood cell lineages in Drosophila [in English]. Nat. Commun. 11, 4483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girard J. R., et al. , Paths and pathways that generate cell-type heterogeneity and developmental progression in hematopoiesis [in English]. Elife 10, e67516 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebestky T., Jung S. H., Banerjee U., A serrate-expressing signaling center controls Drosophila hematopoiesis. Genes. Dev. 17, 348–353 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal L., Martinez-Agosto J. A., Evans C. J., Hartenstein V., Banerjee U., A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors [in English]. Nature 446, 320–324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krzemien J., et al. , Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre [in English]. Nature 446, 325–328 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Mondal B. C., et al. , Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance [in English]. Cell 147, 1589–1600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morin-Poulard I., et al. , Identification of bipotential blood cell/nephrocyte progenitors in Drosophila: Another route for generating blood progenitors [in English]. Front. Cell. Dev. Biol. 10, 834720 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Destalminil-Letourneau M., Morin-Poulard I., Tian Y., Vanzo N., Crozatier M., The vascular niche controls Drosophila hematopoiesis via fibroblast growth factor signaling [in English]. Elife 10, e64672 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotstein B., Paululat A., On the morphology of the Drosophila heart [in English]. J. Cardiovasc. Dev. Dis. 3, 15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choma M. A., Suter M. J., Vakoc B. J., Bouma B. E., Tearney G. J., Physiological homology between Drosophila melanogaster and vertebrate cardiovascular systems [in English]. Dis. Model Mech. 4, 411–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalevee N., Monier B., Senatore S., Perrin L., Semeriva M., Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel [in English]. Curr. Biol. 16, 1502–1508 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Perrin L., Monier B., Ponzielli R., Astier M., Semeriva M., Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev. Biol. 272, 419–431 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Oyallon J., et al. , Two independent functions of collier/early B cell factor in the control of Drosophila blood cell homeostasis [in Enlgish]. PLoS One 11, e0148978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennetier D., et al. , Size control of the Drosophila hematopoietic niche by bone morphogenetic protein signaling reveals parallels with mammals [in Enlgish]. Proc. Natl. Acad. Sci. U.S.A. 109, 3389–3394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin-Poulard I., et al. , Vascular control of the Drosophila haematopoietic microenvironment by Slit/Robo signalling [in English]. Nat. Commun. 7, 11634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokusumi Y., Tokusumi T., Stoller-Conrad J., Schulz R. A., Serpent, suppressor of hairless and U-shaped are crucial regulators of hedgehog niche expression and prohemocyte maintenance during Drosophila larval hematopoiesis [in English]. Development 137, 3561–3568 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldeosingh R., Gao H., Wu X., Fossett N., Hedgehog signaling from the Posterior Signaling Center maintains U-shaped expression and a prohemocyte population in Drosophila [in English]. Dev. Biol. 441, 132–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benmimoun B., Polesello C., Haenlin M., Waltzer L., The EBF transcription factor Collier directly promotes Drosophila blood cell progenitor maintenance independently of the niche [in English]. Proc. Natl. Acad. Sci. U.S.A. 112, 9052–9057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coste B., et al. , Piezo proteins are pore-forming subunits of mechanically activated channels [in English]. Nature 483, 176–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkers L., Mechioukhi Y., Coste B., Piezo channels: From structure to function. Pflugers Arch. 467, 95–99 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Kim S. E., Coste B., Chadha A., Cook B., Patapoutian A., The role of Drosophila Piezo in mechanical nociception [in English]. Nature 483, 209–212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He L., Si G., Huang J., Samuel A. D. T., Perrimon N., Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel [in English]. Nature 555, 103–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y., et al. , The mechanosensitive ion channel piezo inhibits axon regeneration [in English]. Neuron 102, 373–389.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokusumi Y., Tokusumi T., Schulz R. A., The nociception genes painless and Piezo are required for the cellular immune response of Drosophila larvae to wasp parasitization [in English]. Biochem. Biophys. Res. Commun. 486, 893–897 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Suslak T. J., et al. , Piezo is essential for amiloride-sensitive stretch-activated mechanotransduction in larval Drosophila dorsal bipolar dendritic sensory neurons [in English]. PLoS One 10, e0130969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zechini L., et al. , Piezo buffers mechanical stress via modulation of intracellular Ca(2+) handling in the Drosophila heart [in English]. Front. Physiol. 13, 1003999 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coste B., et al. , Piezo1 ion channel pore properties are dictated by C-terminal region [in English]. Nat. Commun. 6, 7223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGuire S. E., Mao Z., Davis R. L., Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila [in English]. Sci. STKE 2004, pl6 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Du L., Sohr A., Yan G., Roy S., Feedback regulation of cytoneme-mediated transport shapes a tissue-specific FGF morphogen gradient [in English]. Elife 7, e38137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cinar E., et al. , Piezo1 regulates mechanotransductive release of ATP from human RBCs [in English]. Proc. Natl. Acad. Sci. U.S.A. 112, 11783–11788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J., et al. , Piezo1 integration of vascular architecture with physiological force [in English]. Nature 515, 279–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gudipaty S. A., et al. , Mechanical stretch triggers rapid epithelial cell division through Piezo1 [in English]. Nature 543, 118–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pathak M. M., et al. , Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells [in English]. Proc. Natl. Acad. Sci. U.S.A. 111, 16148–16153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakai J., Ohkura M., Imoto K., A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein [in English]. Nat. Biotechnol. 19, 137–141 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Caolo V., et al. , Shear stress activates ADAM10 sheddase to regulate Notch1 via the Piezo1 force sensor in endothelial cells [in English]. Elife 9, e50684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duchemin A. L., Vignes H., Vermot J., Mechanically activated piezo channels modulate outflow tract valve development through the Yap1 and Klf2-Notch signaling axis [in English]. Elife 8, e44706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikeya T., Hayashi S., Interplay of Notch and FGF signaling restricts cell fate and MAPK activation in the Drosophila trachea [in English]. Development 126, 4455–4463 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Simon R., Aparicio R., Housden B. E., Bray S., Busturia A., Drosophila p53 controls Notch expression and balances apoptosis and proliferation [in English]. Apoptosis 19, 1430–1443 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Zacharioudaki E., Bray S. J., Tools and methods for studying Notch signaling in Drosophila melanogaster [in English]. Methods 68, 173–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shim J., et al. , Olfactory control of blood progenitor maintenance [in English]. Cell 155, 1141–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherjee T., Kim W. S., Mandal L., Banerjee U., Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells [in English]. Science 332, 1210–1213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tattikota S. G., et al. , A single-cell survey of Drosophila blood [in English]. Elife 9, e54818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans C. J., Liu T., Girard J. R., Banerjee U., Injury-induced inflammatory signaling and hematopoiesis in Drosophila [in English]. Proc. Natl. Acad. Sci. U.S.A. 119, e2119109119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tokusumi Y., Tokusumi T., Schulz R. A., Mechanical stress to Drosophila larvae stimulates a cellular immune response through the JAK/STAT signaling pathway [in English]. Biochem. Biophys. Res. Commun. 502, 415–421 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Dulcis D., Levine R. B., Innervation of the heart of the adult fruit fly, Drosophila melanogaster [in English]. J. Comp. Neurol. 465, 560–578 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Dasari S., Cooper R. L., Direct influence of serotonin on the larval heart of Drosophila melanogaster [in English]. J. Comp. Physiol. B. 176, 349–357 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Zornik E., Paisley K., Nichols R., Neural transmitters and a peptide modulate Drosophila heart rate [in English]. Peptides 20, 45–51 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Majeed Z. R., Stacy A., Cooper R. L., Pharmacological and genetic identification of serotonin receptor subtypes on Drosophila larval heart and aorta [in English]. J. Comp. Physiol. B. 184, 205–219 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Titlow J. S., Rufer J. M., King K. E., Cooper R. L., Pharmacological analysis of dopamine modulation in the Drosophila melanogaster larval heart [in English]. Physiol. Rep. 1, e00020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malloy C. A., Ritter K., Robinson J., English C., Cooper R. L., Pharmacological identification of cholinergic receptor subtypes on Drosophila melanogaster larval heart [in English]. J. Comp. Physiol. B. 186, 45–57 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Bixel M. G., et al. , Flow dynamics and HSPC homing in bone marrow microvessels [in English]. Cell Rep. 18, 1804–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sacma M., Geiger H., Exercise generates immune cells in bone [in English]. Nature 591, 371–372 (2021). [DOI] [PubMed] [Google Scholar]
- 74.Ranade S. S., et al. , Piezo1, a mechanically activated ion channel, is required for vascular development in mice [in English]. Proc. Natl. Acad. Sci. U.S.A. 111, 10347–10352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Azizidoost S., et al. , The role of notch signaling in bone marrow niche [in English]. Hematology 20, 93–103 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Butler J. M., et al. , Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells [in English]. Cell Stem Cell 6, 251–264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monier B., Astier M., Semeriva M., Perrin L., Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart [in English]. Development 132, 5283–5293 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Sellin J., Albrecht S., Kolsch V., Paululat A., Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand [in English]. Gene. Expr. Patterns 6, 360–375 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Jennings B. H., Tyler D. M., Bray S. J., Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins [in English]. Mol. Cell Biol. 19, 4600–4610 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ollion J., Cochennec J., Loll F., Escude C., Boudier T., TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization [in English]. Bioinformatics 29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gilles J. F., Dos Santos M., Boudier T., Bolte S., Heck N., DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis [in English]. Methods 115, 55–64 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
heartbeat in control NP1029, UAS-dicer; mcD8-GFP>w1118
heart is arrested in NP1029, UAS-Dicer, mcD8-GFP>Ork1ΔC
heart is arrested in NP1029, UAS-Dicer, mcD8-GFP>Mhc-RNAi
heartbeat in control HandΔ, UAS-Dicer; HandC-GFP> w1118
heartbeat is increased in HandΔ, UAS-Dicer; HandC-GFP>Ork1-RNAI
heartbeat in HandΔ, UAS-Dicer; HandC-GFP>piezo- RNAi
heartbeat in L2 larvae in HandΔ UAS-Dicer; HandC-GFP> w1118
heartbeat in L3 larvae in HandΔ UAS-Dicer; HandC-GFP> w1118
Data Availability Statement
All study data are included in the article and/or SI Appendix.