Significance
Armadillo repeat-containing (ARM) proteins form an evolutionarily conserved protein family and participate in various cellular processes mainly by providing binding platforms for diverse proteins. However, it is unknown how a single ARM protein mediates its interactions with multiple binding partners so that one process does not hinder another. We used Vac8, the only ARM protein found in the budding yeast Saccharomyces cerevisiae, as a model to investigate the mode of action of ARM proteins. By performing X-ray crystallography of Vac8-containing protein complexes in combination with biochemical and microscopic analyses, this study provides a working model of how Vac8 differentially regulates three cellular processes (formation of an interorganellar contact site, an autophagy-like process, and organelle inheritance).
Keywords: armadillo repeats, organelle inheritance, Vac8, Vac17, Saccharomyces cerevisiae
Abstract
Vac8, a yeast vacuolar protein with armadillo repeats, mediates various cellular processes by changing its binding partners; however, the mechanism by which Vac8 differentially regulates these processes remains poorly understood. Vac8 interacts with Nvj1 to form the nuclear–vacuole junction (NVJ) and with Atg13 to mediate cytoplasm-to-vacuole targeting (Cvt), a selective autophagy-like pathway that delivers cytoplasmic aminopeptidase I directly to the vacuole. In addition, Vac8 associates with Myo2, a yeast class V myosin, through its interaction with Vac17 for vacuolar inheritance from the mother cell to the emerging daughter cell during cell divisions. Here, we determined the X-ray crystal structure of the Vac8–Vac17 complex and found that its interaction interfaces are bipartite, unlike those of the Vac8–Nvj1 and Vac8–Atg13 complexes. When the key amino acids present in the interface between Vac8 and Vac17 were mutated, vacuole inheritance was severely impaired in vivo. Furthermore, binding of Vac17 to Vac8 prevented dimerization of Vac8, which is required for its interactions with Nvj1 and Atg13, by clamping the H1 helix to the ARM1 domain of Vac8 and thereby preventing exposure of the binding interface for Vac8 dimerization. Consistently, the binding affinity of Vac17-bound Vac8 for Nvj1 or Atg13 was markedly lower than that of free Vac8. Likewise, free Vac17 had no affinity for the Vac8–Nvj1 and Vac8–Atg13 complexes. These results provide insights into how vacuole inheritance and other Vac8-mediated processes, such as NVJ formation and Cvt, occur independently of one another.
Armadillo repeat-containing (ARM) proteins are characterized by the presence of a repeating ~42 amino acid motif composed of three α-helices and are expressed from yeast to humans. They all share a closely related structure. Specifically, their tandem ARM repeat units fold together to form a versatile platform for interactions with various protein partners, which are involved in diverse cellular processes (1). β-catenin (Armadillo in Drosophila) is one of the best-characterized ARM proteins and mediates various essential functions, including cell adhesion and intracellular signaling (2). This single ARM protein mediates these diverse cellular functions by acting as an intracellular hub where many proteins are recruited and cooperate.
Vac8, a vacuolar membrane protein, is the only ARM protein found in the budding yeast Saccharomyces cerevisiae. Identical to its metazoan cousins, Vac8 mediates diverse cellular functions, such as interorganellar communication (3, 4), organelle inheritance (5, 6), and autophagy (7–9). Vac8 interacts with Nvj1, a nuclear envelope protein, to form an interorganellar membrane contact site termed the nuclear–vacuole junction (NVJ) (8, 10). The NVJ is thought to be a route through which ergosterol (yeast cholesterol) and phosphatidylinositol 4-monophosphate are transferred between the nuclear envelope and the vacuole (4). In addition, a unique type of nucleophagy, termed piecemeal microautophagy of the nucleus (PMN), is initiated at the NVJ under nutrient-deprived conditions (11–14). During PMN, part of the nucleus is directly invaginated into the lumen of the vacuole for degradation by vacuolar hydrolases. Vac8 is involved in homotypic vacuole membrane fusion (15–18), and Vac8 deletion causes mild vacuole fragmentation, an in vivo phenotype of vacuole fusion defects. Vacuoles isolated from vac8Δ yeast cells fuse poorly in vitro (17). Moreover, Vac8 enables a portion of vacuoles in mother cells to interact with actin cables for their delivery to the emerging daughter cells (buds) by binding to Vac17, which in turn interacts with Myo2, a motor protein that moves various cargoes along actin cables (5, 6, 19). Thus, Vac8 is essential for vacuole inheritance during mitosis, and vac8Δ yeast cells largely fail to share a portion of vacuoles with their daughter cells (5). Finally, Vac8 is involved in cytoplasm-to-vacuole targeting (Cvt) of aminopeptidase I (Ape1) (7). Ape1 is synthesized as an inactive form in the cytoplasm and transported to the lumen of vacuoles, where it is activated by vacuolar proteases. This Cvt pathway employs the core machinery and components used for autophagy, but it is a biosynthetic pathway that occurs constitutively even under nutrient-rich conditions (9). Although the single protein Vac8 mediates diverse cellular processes as described above, it is poorly characterized how these functions are coordinated and orchestrated. We previously reported that Vac8 adopts distinct quaternary structures depending on its binding partners and that these structures underlie the distinct functions of Vac8 (9). Two complexes of Vac8–Nvj1 adopt an arched shape for NVJ formation, whereas two complexes of Vac8–Atg13 exhibit a superhelical shape for Cvt (9).
In the current study, we determined the crystal structure of the Vac8–Vac17 complex, which connects vacuoles to Myo2 for vacuolar transport along actin cables during mitosis. One of the hallmarks of this structure, in comparison with the Vac8–Nvj1 and Vac8–Atg13 complexes, is that the Vac8–Vac17 complex exists as a heterodimer, instead of a heterotetramer, as we previously reported for the Vac8–Nvj1 and Vac8–Atg13 complexes (9). This structure suggests that binding of Vac17 to Vac8 blocks Vac8 dimerization by masking the critical region involved in Vac8 dimerization. By comparing the structure of the Vac8–Vac17 complex with those of the Vac8–Nvj1 and Vac8–Atg13 complexes, in combination with biochemical and cell biological analyses, we propose a working model of how the three distinct processes are differentially regulated by Vac8 in the cell so that one does not hinder another.
Results
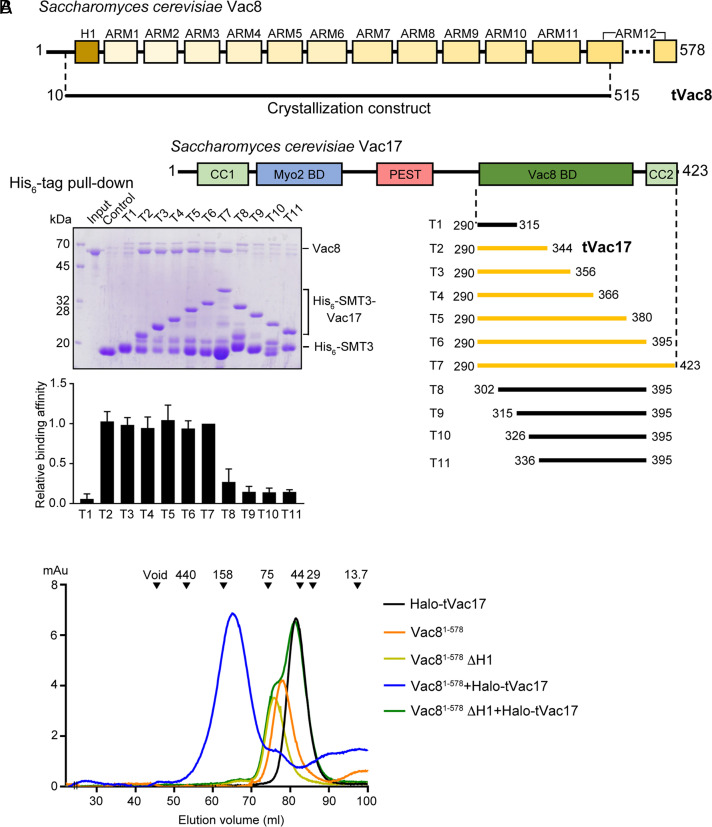
Functional Dissection of the Vac8-Binding Region of Vac17.
S. cerevisiae Vac17 functions as an adapter between the vacuolar membrane protein Vac8 and Myo2, a class V myosin motor protein that transports a subset of fragmented vacuoles via actin cables from mother cells to daughter cells during cell divisions (20, 21), and is composed of 423 amino acid residues (Fig. 1A). The Vac8-binding domain of Vac17 was previously mapped to residues 290 to 380 (19, 20). In vitro binding experiments further narrowed down the Vac8-binding domain to residues 290 to 344 (T2; tVac17), which bound Vac8 comparably as the entire domain (T5; 290 to 380) (Fig. 1A). These results were confirmed by size-exclusion chromatography (SEC) (Fig. 1B and SI Appendix, Fig. S1). Vac8 lacking the H1 helix did not associate with tVac17 (green line), indicating that the H1 helix is critical for the Vac8–tVac17 interaction. In addition, no oligomerization of the Vac8–tVac17 complex was observed (blue line). This is in contrast with the Vac8–Nvj1 and Vac8–Atg13 complexes, in which Vac8 forms a homodimer and thereby causes the complex to be a heterotetramer (8, 9).
Fig. 1.
Mapping the interaction between Vac8 and Vac17. (A) Schematic diagrams show the domain structures of S. cerevisiae Vac8 and Vac17. SDS-PAGE (Left below) shows the result of His6-tag pull-down assays to characterize the Vac8-binding region of Vac17. His-SMT3-fused Vac17 was mixed with full-length Vac8 proteins (see Materials and Methods for details). The relative quantification data (n = 3) of the pull-down assays using the T7 fragment as a reference are shown below. The series of Vac17 constructs (T1 to T11) used in the experiments are shown with bars (Right below). Constructs colored yellow bind to Vac8, while constructs colored black do not, according to pull-down experiments. Based on the results, we crystallized tVac8 comprising residues 10 to 515 in complex with tVac7 (residues 290 to 344). (B) Analysis of the direct interactions between Vac8 and Vac17 by SEC. The chromatograms show the behavior of Vac8 (full length 1 to 578 and lacking the H1 helix), Halo-tVac17, and a mixture of the two. The standard molecular masses are shown above the chromatograms to indicate the relative molecular masses of samples (ferritin, 440 kDa; aldolase, 158 kDa; conalbumin, 75 kDa; ovalbumin, 44 kDa; carbonic anhydrase, 29 kDa; and ribonuclease A, 13.7 kDa).
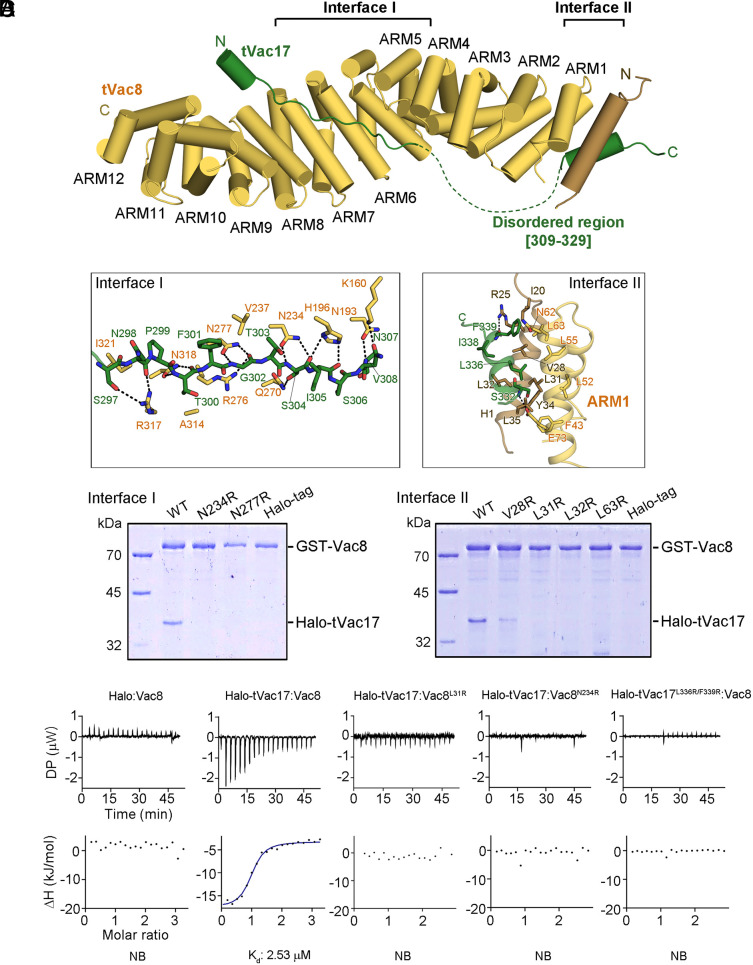
Structure of tVac8 Complexed with tVac17.
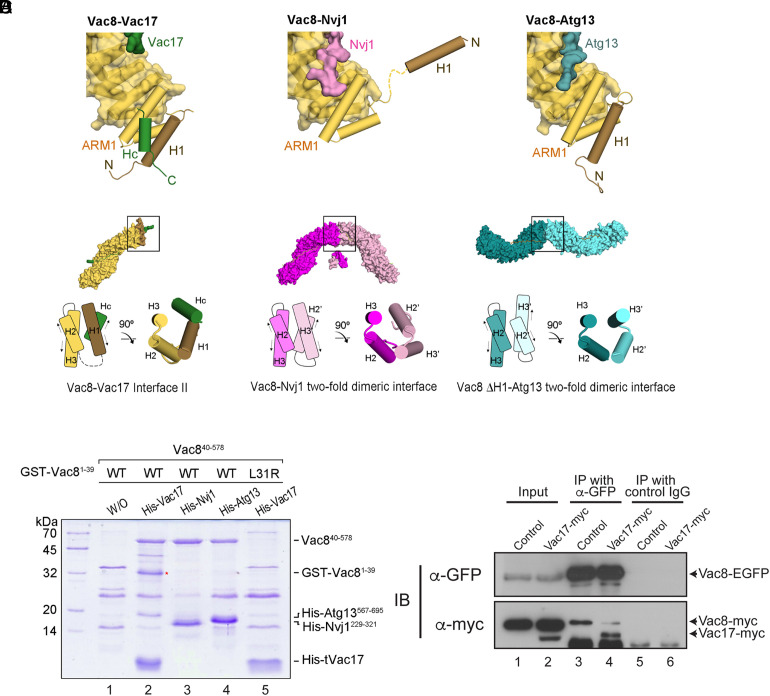
To investigate the interaction between Vac8 and Vac17, we attempted to solve high-resolution structures of the tVac8–tVac17 complex. Truncated Vac8 (tVac8; residues 10 to 515) was used as described in our previous studies (8, 9). The crystals of tVac8–tVac17 were grown in a reservoir buffer containing SOKALAN CP 42 and sarcosine as the primary precipitant. The structure was solved by the molecular replacement method using the Vac8 coordinates (PDB code: 5XJG) as a search model and refined to 2.1 Å resolution (see SI Appendix, Table S1 for crystallographic details). Fig. 2A shows the overall structure of tVac8 complexed with tVac17 in a ribbon diagram. The conformation of tVac8 in the tVac8–tVac17 complex largely resembles that in the tVac8–Nvj1 and tVac8–Atg13 complexes with a root mean square deviation (rmsd) value of 1.665 and 1.463 Å, respectively (8, 9). However, tVac17 binds to tVac8 in a notably different manner from Nvj1 and Atg13. Specifically, tVac17 makes contacts with tVac8 in an antiparallel, bipartite manner at two binding interfaces (Interfaces I and II) (Fig. 2 A and B). In Interface I, the N-terminal region of tVac17 forms a short helix (referred to as HN) consisting of residues 290 to 294 followed by an extended loop (residues 295 to 308). This extended loop binds across the central ARM4–ARM7 domains of Vac8. Intriguingly, the structure of this extended loop resembles that of the equivalent region of Nvj1 (residues 301 to 314) bound to Vac8 (SI Appendix, Fig. S2). However, the C-terminal region of tVac17 (residues 332 to 340, referred to as HC) forms an α-helix and associates with both the ARM1 domain and the H1 helix of tVac8. This structural feature is consistent with the SEC finding that the absence of the H1 helix abolished the interaction of tVac8 with tVac17 (Fig. 1B). The two interfaces are connected by a ~20 amino acid linker, which is disordered in the electron density map (Fig. 2A, green dotted line).
Fig. 2.
Crystal structure of the tVac8–tVac17 complex. (A) Crystal structure of yeast tVac8 (yellow) in complex with tVac17 (green). The structure was determined by molecular replacement using tVac8 as a search model (PDB ID: 5XJG) and refined to 2.1 Å resolution. See Materials and Methods for details of the crystallographic analysis. (B) Close-up view of the interfaces of the tVac8–tVac17 complex. In Interface I, the tVac17 peptide is drawn as a ball-and-stick representation in green, with oxygen and nitrogen atoms colored red and blue, respectively. Vac8 residues involved in the interaction with Vac17 are shown in yellow. In Interface II, the Hc helix of Vac17 (green) mainly makes hydrophobic contact with the H1 helix (brown) and ARM1 helices (yellow). Black dashed lines indicate intermolecular hydrogen bonds. (C) The role of Vac8 residues in the interaction with tVac17 was assessed through GST pull-down experiments using point mutants. (D) ITC titration curves for binding of Halo-tVac17 to wild-type or mutant Vac8. The Upper panel shows primary data. The Lower panel shows data fitted to binding isotherms in order to calculate affinities.
Interfaces of the tVac8–tVac17 Complex.
In Interface I, the HN helix of tVac17 does not directly contact tVac8, but the following extended stretch tightly associates with the ARM domains of tVac8 mainly through a hydrogen-bond network between the side-chain atoms of tVac17 and tVac8. In particular, the carbonyl oxygen atoms of N298, T300, G302, S304, and I305 from tVac17 are recognized by the side chains of R317, N318, N277, N234, and H196 from tVac8, respectively. By contrast, hydrophobic interactions mainly contribute to the association between tVac17 and tVac8 in Interface II. Specifically, the side chain of F339 from tVac17 is involved in hydrophobic interactions with the side chains of I20 and V28 from the H1 helix as well as the side chains of N62 and L63 from ARM1. Furthermore, L336 of tVac17 forms a hydrophobic core with the side chains of V28, L31, L32, Y34, and L35 of the H1 helix as well as the side chains of L52 and L55 of ARM1. To examine whether the hydrogen-bond network (Interface I) and hydrophobic core (Interface II) are critical for the tight association between tVac8 and tVac17, as expected based on the crystal structure, we constructed a series of full-length Vac8 mutants carrying a single amino acid substitution and analyzed their abilities to associate with Halo-tag-conjugated tVac17 (Halo-tVac17) (Fig. 2C). Mutation of N234 or N277 in Interface I of Vac8 completely abolished the Vac8–tVac17 interaction. Similarly, mutation of V28, L31, L32 (H1 helix), or L63 (ARM1) in Interface II of Vac8 markedly or completely disrupted the Vac8–tVac17 interaction. These results were further corroborated by isothermal titration calorimetry (ITC) analysis (Fig. 2D and SI Appendix, Table S2). Halo-tVac17 bound to Vac8 with a Kd of 2.53 μM, but no meaningful interaction was observed when an amino acid residue (L31 or N234) critical for the Vac8–Vac17 association was mutated. A similar result was obtained when both L336 and F339 in Interface II of Vac17 were mutated.
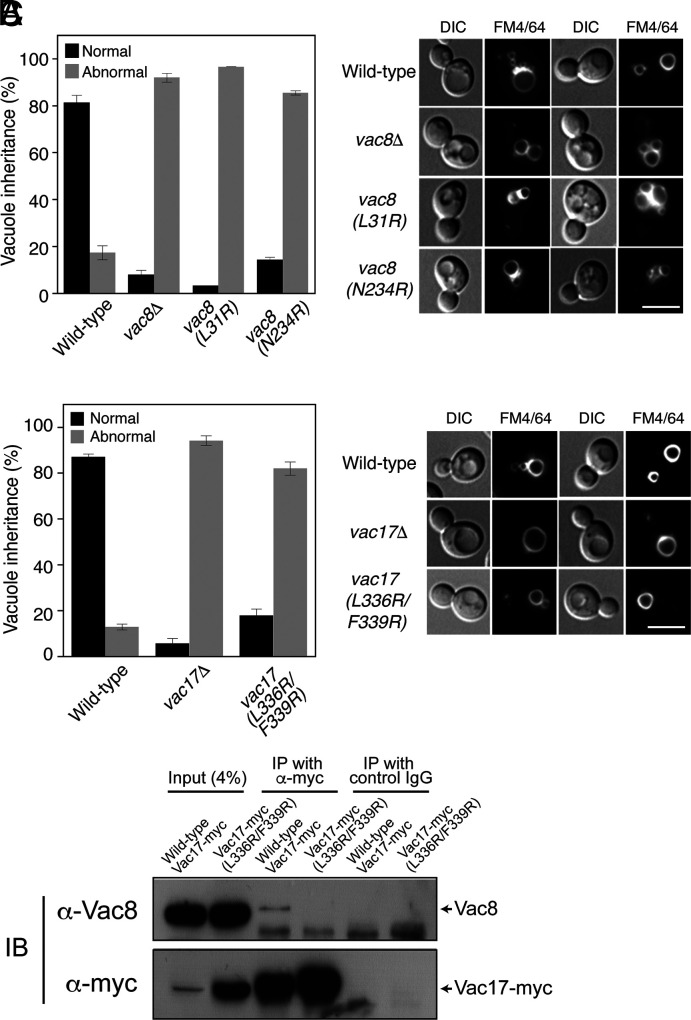
The Interaction Interfaces of Vac8–Vac17 Are Crucial for Vacuole Inheritance In Vivo.
To investigate whether Interfaces I and II of the Vac8–Vac17 complex are critical for vacuole inheritance in vivo, wild-type or mutant yeast cells were grown in the presence of FM 4-64, a lipophilic, fluorescent dye that selectively stains vacuoles, and vacuole segregation into daughter cells during cell divisions was analyzed. While most wild-type daughter cells successfully inherited vacuoles from their mother cells during cell divisions, daughter cells lacking Vac8 failed to receive vacuoles, as previously reported (5) (Fig. 3A and SI Appendix, Fig. S3A). Consistent with the results of in vitro binding experiments and ITC analysis (Fig. 2 C and D), severe defects in vacuole inheritance were observed in yeast cells bearing the Vac8(L31R) or Vac8(N234R) mutant (Fig. 3A). Similarly, mutant daughter cells expressing Vac17(L336R/F339R), which had no detectable affinity for Vac8 (Fig. 2D), exhibited strong defects in vacuole inheritance comparable with vac17Δ cells (Fig. 3B). Consistent with the previous finding that a Vac17 mutant defective for Vac8 binding is more resistant than wild-type Vac17 to Dma1-mediated ubiquitinylation followed by proteasomal degradation (20, 22), the expression level of Vac17(L336R/F339R) was markedly higher than that of wild-type Vac17 (SI Appendix, Fig. S3 B and C). These results clearly show that the binding interfaces between Vac8 and Vac17 predicted by the tVac8–tVac17 crystal structure (Fig. 2 A and B) and confirmed by in vitro binding assays (Fig. 2 C and D) are critical for vacuole inheritance in vivo. Consistently, our coimmunoprecipitation experiments revealed that the mutant Vac17 (L336R/F339R) did not interact with Vac8, although its expression was markedly higher than that of wild-type Vac17 (Fig. 3C). Likewise, nearly no interaction was observed between wild-type Vac17 and mutant Vac8 (L31R) or Vac8 (N234R) (SI Appendix, Fig. S3D).
Fig. 3.
Disruption of the interaction between Vac8 and Vac17 markedly impairs vacuole inheritance. (A) Interaction sites of Vac8 in Interfaces I and II are important for vacuole inheritance. Vacuoles of yeast strains expressing wild-type or mutant Vac8 were cultured with FM 4-64 in YPD medium at 30 °C for 30 min. After removing free dye, cells were resuspended in fresh YPD medium and further grown at 30 °C for 3 h. FM 4-64 fluorescence on vacuoles was observed by fluorescence microscopy. Representative images from each cell type are shown (Right), and the bar graph shows the quantification of vacuole inheritance (Left). More than 100 cells per strain were examined in each experiment. Data represent the means ± SEM (error bar; n = 3). (Scale bar: 5 μm). (B) The interaction site of Vac17 in Interface II is important for vacuole inheritance. Experiments were performed as described in Fig. 3A. Yeast cells expressing wild-type or mutant Vac17 were used. Representative images from each cell type are shown (Right) and the graph shows quantification of vacuole inheritance (Left). More than 100 cells per strain were examined in each experiment. Data represent the means ± SEM (error bar; n = 3). (Scale bar: 5 μm). (C) The L336R/F339R mutation largely abolishes the interaction between Vac8 and Vac17. Yeast spheroplasts were detergent-solubilized, and detergent-insoluble material was removed by centrifugation. The resulting postcentrifugation supernatants were precleared by incubation with protein A Sepharose and treated with anti-myc antibodies or control mouse IgG. Protein A Sepharose was then added, and bound proteins were eluted with SDS sample buffer for Sodium Dodecyl Sulfate (SDS)-PolyAcrylamide Gel Electrophoresis (PAGE) analysis followed by immunoblotting using anti-myc and anti-Vac8 antibodies.
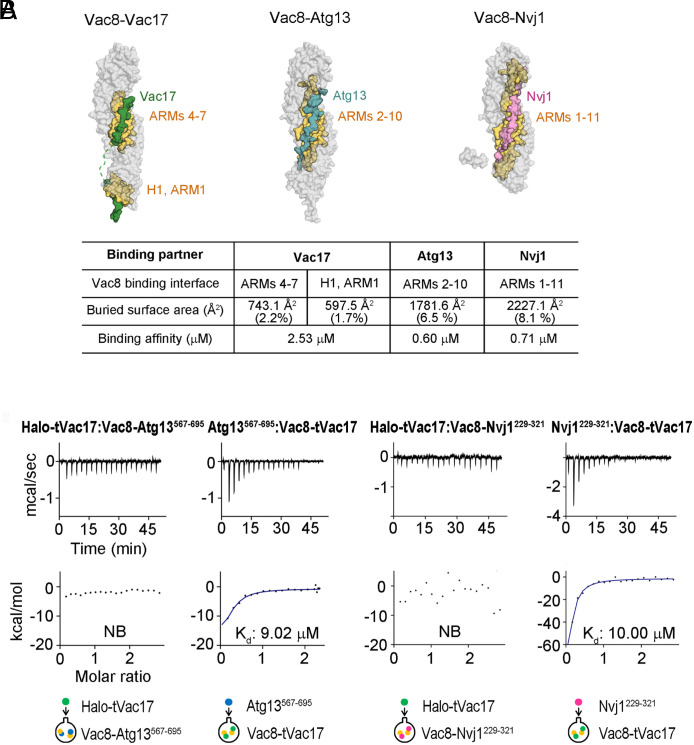
Structural Comparison of the Vac8–Vac17 Complex and the Vac8–Nvj1 and Vac8–Atg13 Complexes.
Structure comparison analysis revealed that the overall structure of the tVac8–tVac17 complex closely resembles that of the tVac8–Nvj1 and tVac8–Atg13 complexes, but the binding mode of Vac17 to tVac8 is remarkably different from that of Nvj1 and Atg13. The sequences of Nvj1 and Atg13 important for Vac8 binding do not form any secondary structure, such as α-helices (SI Appendix, Fig. S2A). In the Vac8–Atg13 complex, Atg13 forms a long extended loop comprising residues 660 to 685 and this loop tightly binds across the inner groove generated by the central ARM repeats (ARM2–ARM10) of Vac8, thereby generating a buried surface area (BSA) of 1781 Å2 (Fig. 4A). Likewise, residues 292 to 321 of Nvj1 adopt an elongated loop structure and extensively contact the apposed groove generated by ARM1–ARM11 of Vac8, producing a BSA of 2227.1 Å2. However, tVac17 (residues 290 to 344) forms two helices (HN and HC) in distant regions from each other (Interfaces I and II) (SI Appendix, Figs. S2A and S4). The extended loop structure of tVac17 binds exclusively to ARM4–ARM7 of Vac8, and the disordered region following the loop seems too flexible to interact with Vac8 (Figs. 2A and 4A). The interactions mediated by ARM4–ARM7 of Vac8 are similar among Vac17, Nvj1, and Atg13, despite their low sequence similarity (SI Appendix, Fig. S2A). In these contact regions, the main-chain atoms of Vac17, Nvj1, and Atg13 are mostly recognized by the conserved side chains of Vac8 (SI Appendix, Fig. S2B). Although ARM2–ARM3 of Vac8 also contribute to its interactions with Nvj1 and Atg13, the region of Vac17 close to ARM2–ARM3 seems to be disordered (SI Appendix, Figs. S2B and S5A). Thus, structural flexibility may explain why no interaction with Vac8 was observed in this region of Vac17. However, this flexibility may allow the HC helix to properly associate with ARM1 in Interface II (Figs. 2 A and B and 4A and SI Appendix, Fig. S5A). The reduced area of Vac17 that contacts the central ARMs of Vac8, compared with those of Nvj1 and Atg13, also explains the weaker binding affinity of Vac17 (2.53 μM) for Vac8 compared with that of Nvj1 (0.71 μM) and Atg13 (0.60 μM) (Fig. 4A).
Fig. 4.
Structural comparison of the Vac8–Vac17, Vac–Atg13, and Vac8–Nvj1 complexes. (A) Surface representations of the Vac8–Vac17, Vac8–Atg13 (PDB ID: 6KBM), and Vac8–Nvj1 (PDB ID: 5XJG) complexes are displayed in the same orientation. ARM domains of Vac8 involved in the interactions with its binding partners are highlighted in yellow and the BSAs generated by the associations were calculated. Vac17, Atg13, and Nvj1 are colored green, cyan, and pink, respectively. (B) Competitive binding assays using ITC. Halo-tVac17 was titrated into Vac8–Atg13567–695 or Vac8–Nvj1229–321 complexes, and Atg13567–695 and Nvj1229–321 were titrated into Vac8–tVac17 complexes. The schematic diagrams below show the proteins used in the sample cell and syringe for experiments.
As shown in SI Appendix, Fig. S6 and a recent study (23), vacuole segregation is followed by nuclear inheritance during cell divisions in budding yeast. Interestingly, vacuole segregation occurred independently of preformed NVJs in mother cells. In the vast majority of mother cells, NVJs were maintained throughout cell division. Thus, the Vac8–Vac17 interaction for vacuole inheritance during cell division did not seem to affect preformed NVJs. To provide a molecular premise for this phenomenon, we examined whether Vac17 could replace Nvj1 in preformed Vac8–Nvj1 complexes to form the Vac8–Vac17 complex by ITC (Fig. 4B). The relatively low affinity of Vac17 for Vac8 (Kd = 2.53 μM, Fig. 4A) may explain, at least in part, why an increase in Vac17 expression in early mitosis (6, 20) does not markedly affect the integrity of preformed NVJs. In accordance with this idea, no detectable binding was observed when Halo-tagged tVac17 was added to preformed Vac8–Nvj1229-321 complexes (Fig. 4B). Consistently, Nvj1 had a much lower affinity for Vac8–tVac17 complexes (Kd = 10.0 μM) than for free Vac8 (Kd = 0.71 μM) (Fig. 4B), suggesting that Vac8 bound to Vac17 during vacuole inheritance can participate in NVJ formation by interacting with Nvj1 only after it dissociates from Vac17 although it is unlikely that Nvj1 and Vac17 compete for Vac8 in vivo because the endogenous expression of Vac8 was reported to be approximately an order of magnitude higher than that of its main binding partners (24).
Vac17 Clamps the H1 Helix and ARM1 of Vac8 and Thereby Blocks Vac8 Dimerization.
Our previous studies (8, 9) reported that the H1 helix of Vac8 plays a critical role in Vac8 self-association. The structures of the Vac8–Nvj1 and Vac8–Atg13 complexes also revealed that the H1 helix is flexibly connected to ARM1 via nine amino acid residues (residues 37 to 45). Thus, dynamic organization of the H1 helix may regulate Vac8 self-association through ARM1, which provides the interface for Vac8 dimerization. Indeed, the crystal structure of the Vac8–Nvj1 complex showed that the H1 helix is released from ARM1 and exposed to solvent. By contrast, the H1 helix directly contacts the ARM1 helices in the structure of the Vac8–Atg13 complex (Fig. 5A and SI Appendix, Fig. S5). The HC helix of Vac17 in Interface II associates with the ARM1 helices and H1 helix simultaneously (Figs. 2A and 5A). This tight interaction seems to prevent the H1 helix from being separated from ARM1, which blocks exposure of the interface for Vac8 dimerization (Fig. 5 A and B).
Fig. 5.
Vac17 is required for the interaction between the H1 helix and ARM1 of Vac8. (A) Structural comparison highlighting the H1 helix and ARM1 of Vac8 in the Vac8–Vac17 (Left), Vac8–Nvj1 (Middle), and Vac8–Atg13 (Right) complexes. The color scheme is the same as in Fig. 4A. (B) Surface representations show the crystal structures of the Vac8–Vac17, Vac8–Nvj1 (PDB ID: 5XJG), and Vac8 (ΔH1)–Atg13 (PDB ID: 6KBN) complexes in the crystallographic asymmetric unit. Previous studies (8, 9) revealed that Vac8 bound to Nvj1 or Atg13 self-associates through ARM1 (highlighted by black boxes). The diagram compares the organization of ARM1 helices at the twofold dimeric interface of Vac8. Arrows indicate the direction of the helix axis. In the Vac8–Vac17 complex, the geometry of the HC helix of Vac17 and the H1 helix of Vac8 highly resembles that of the H2′ and H3′ helices of ARM1 from the Vac8 counter molecule in the structure of the Vac8–Nvj1 complex. (C) Pull-down experiments show that Vac17 is required for the interaction between the H1 helix and ARM1 of Vac8. The supernatants of E. coli cells coexpressing GST-Vac8 (residues 1 to 39) and Vac8 (residues 40 to 578) with his6-tagged tVac17, Nvj1229–321, or Atg13567–695 were incubated with 10 μL Ni–NTA agarose beads. Proteins were analyzed by 12% SDS-PAGE and Coomassie blue staining (see Materials and Methods for experimental details). The red asterisk indicates GST-Vac81–39 (H1 helix) bound to the Vac8 armadillo repeat domain in the presence of tVac17. (D) Vac8–Vac17 interaction blocks Vac8 dimerization. Yeast spheroplasts were prepared from vac8Δ VAC17 yeast cells expressing both Vac8-myc and Vac8-EGFP with or without Vac17-myc overexpression from the GPD1 promoter (see SI Appendix, Table S3 for details of the yeast strains used). After detergent solubilization of the spheroplasts, detergent-insoluble material was removed by centrifugation. The resulting postcentrifugation supernatants were precleared by incubation with protein A Sepharose and treated with anti-myc antibodies or control mouse IgG. Protein A Sepharose was then added, and bound proteins were eluted with SDS sample buffer for SDS-PAGE analysis followed by immunoblotting using anti-myc and anti-GFP antibodies.
Structure comparison analysis (Fig. 5B) also revealed that the organization of the four helices (the H2 and H3 helices from ARM1 of Vac8, the H1 helix of Vac8, and the HC helix of Vac17) in the Vac8–Vac17 complex highly resembles that of the H2 and H3 helices from ARM1 of one Vac8 molecule and the H2′ and H3′ helices from ARM1 of the other Vac8 molecule in a Vac8 homodimer in the structure of the Vac8–Nvj1 complex. The organization of the four helices in the Vac8–Vac17 complex is as if the H2′ and H3′ helices of one Vac8 molecule in the Vac8–Nvj1 tetramer are replaced by the H1 helix of Vac8 and the HC helix of Vac17 (Fig. 5B, compare Interface II of the Vac8–Vac17 complex with the dimeric interface of the Vac8–Nvj1 complex). This explains how the four-helix bundle structure forms and stabilizes in Interface II of the Vac8–Vac17 complex. Thus, the Vac8–Vac17 complex cannot form a heterotetramer, unlike the Vac8–Nvj1 and Vac8–Atg13 complexes. This conclusion is well supported by our SEC experiment (Fig. 1B and SI Appendix, Fig. S1).
To test whether the HC helix is indeed required for interaction between the H1 helix and ARM1 of Vac8, we performed in vitro binding assays (Fig. 5C). In the absence of any his6-tagged protein, only nonspecific binding was observed (Fig. 5C, lane 1). In the presence of his6-tagged tVac17, the H1 helix (GST-Vac81-39) interacted with Vac840-578 (lane 2). By contrast, neither his6-tagged Nvj1 nor his6-tagged Atg13 induced binding of the H1 helix to Vac840-578 (lanes 3 and 4). The his6-tagged tVac17-mediated interaction between the H1 helix and Vac840-578 seems to depend on the Hc helix because the H1 helix bearing a mutation that disrupts binding of the HC helix (9) did not associate with Vac8 even in the presence of his6-tagged tVac17 (lane 5). These data strongly suggest that the HC helix of Vac17 is crucial for the stable association of the H1 helix with ARM1 of Vac8. In addition, because Vac8 dimerization is critical for its interactions with Nvj1 and Atg13 (8, 9), these results correlate well with the observation that the affinity of Vac8–tVac17 for Atg13 or Nvj1 was about 14 to 15 times lower than that of free Vac8 for Atg13 or Nvj1, respectively (Fig. 4). Finally, to examine whether the Vac17–Vac8 interaction affects Vac8 dimerization in vivo, we performed coimmunoprecipitation experiments using yeast cells expressing both GFP-conjugated Vac8 and myc-tagged Vac8. Vac8-myc coprecipitated with Vac8-EGFP, indicative of Vac8 dimerization (Fig. 5D). Overexpression of myc-tagged Vac17 in these cells markedly reduced the amount of Vac8-myc that coprecipitated with Vac8-EGFP, suggesting that the Vac17–Vac8 interaction blocks Vac8 dimerization. Consistently, overexpression of Vac17-myc lowered the level of the Vac8–Nvj1 complex (SI Appendix, Fig. S7).
Discussion
ARM proteins are evolutionarily conserved from yeast to humans, although their cellular roles have become different and diverse during evolution. Even though the cellular processes in which ARM proteins participate markedly differ between yeast and humans (e.g., yeast Vac8 vs. human β-catenin), their ability to interact with diverse proteins has not changed during evolution. It remains a long-standing mystery how ARM proteins spatiotemporally interact with their binding partners to perform diverse cellular functions, some of which are completely unrelated. Vac8, a vacuolar protein present in the budding yeast S. cerevisiae, one of the simplest eukaryotic organisms, provides a facile tool to investigate how ARM proteins participate in diverse cellular events by changing their binding partners.
How does Vac8 participate in NVJ formation and vacuole inheritance simultaneously during mitosis in vivo? Nvj1-GFP largely localized to NVJs in wild-type cells but dispersed along the nuclear envelope in vac8Δ cells (SI Appendix, Fig. S8), consistent with previous studies (8, 10), suggesting that the vast majority of Nvj1 is involved in NVJ formation. By contrast, Vac8-GFP was distributed all over the vacuolar membrane, although GFP signals were stronger at NVJs (10), implying that a considerable amount of free Vac8 molecules is present on the vacuolar membrane and thus available for Vac17 binding. Indeed, Vac8 was reported to be present in one order of magnitude in excess of its main binding partners in vivo (24). Therefore, preformed Vac8–Nvj1 complexes can be preserved while vacuole inheritance is under way mediated by Vac8–Vac17–Myo2 complexes during mitosis. Consistently, live imaging of yeast cells undergoing mitosis (SI Appendix, Fig. S6) revealed that the portions of the vacuole that did not participate in NVJs in a mother cell started to segregate into an emerging bud as small vesicles. Upon arrival of fragmented vacuoles at the daughter cell, Vac17 is rapidly degraded by a well-known mechanism (20), which may further accumulate free Vac8 in the vacuole of the daughter cell. In the meantime, the nuclear membrane was elongated and segregated into the daughter cell, and free Vac8 in vacuoles of the daughter cell became available again for binding of Nvj1 from the segregated nuclear membrane to form a new NVJ in the daughter cell.
Cvt is a biosynthetic pathway that constitutively delivers cytoplasmic Ape1 to the lumen of vacuoles for its activation by vacuolar proteases; however, nutrient starvation further stimulates the transport of cytoplasmic Ape1 to vacuoles by rapidly increasing the availability of the machinery that Cvt and autophagy share. As shown in SI Appendix, Fig. S9, vacuole inheritance was not affected by nitrogen starvation, which markedly enhanced vacuolar transport of Ape1–EGFP, suggesting that Vac8 and Vac17 can interact normally even when Atg13 actively associates with Vac8 for elevated Cvt. These results are consistent with our ITC data (Fig. 4): Atg13 had a much lower affinity for Vac8–tVac17 complexes (Kd = 9.02 μM) than that for free Vac8 (Kd = 0.60 μM). In addition, Vac17 had nearly no affinity for Vac8–Atg13 complexes. Furthermore, Vac17 is degraded immediately after vacuole inheritance is completed because it contains a PEST sequence, a signal for rapid protein degradation (20), whereas Vac8 exists in excess of Vac17 (24). Thus, the steady-state level of Vac17 is unlikely to be high enough to hinder Vac8-mediated Cvt.
A Working Model of How Vac8 Coordinates Three Distinct Cellular Processes.
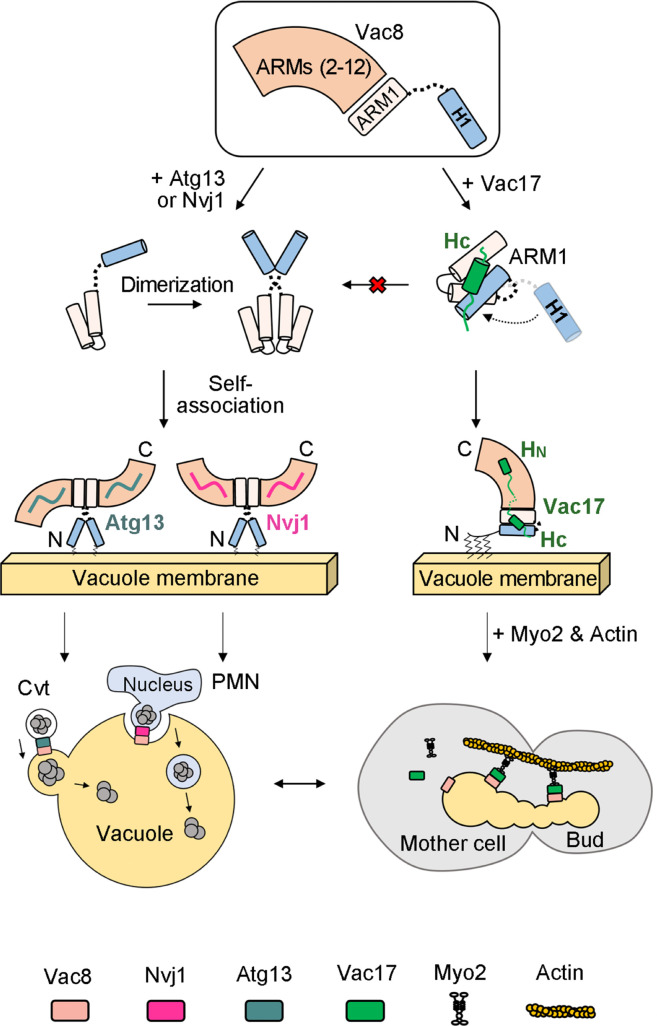
Based on the data presented in this study, together with those reported in our previous studies regarding the structures of the Vac8–Nvj1 and Vac8–Atg13 complexes (8, 9), we propose a working model in which Vac8 mediates three distinct cellular processes by changing its binding partners (Fig. 6). Upon binding of Nvj1 or Atg13 to Vac8, the H1 helix is released from ARM1 and the interaction interface for Vac8 dimerization is exposed. Then, the Vac8–Nvj1 or Vac8–Atg13 complex self-associates to form a heterotetramer, which in turn mediates NVJ formation or initiates Cvt, respectively. When Vac8 binds to Vac17, the HC helix of Vac17 simultaneously interacts with both the H1 helix and part of ARM1, bringing them into close proximity and thereby preventing exposure of the interaction interface for Vac8 dimerization. This enables Vac8–Vac17 complexes to form and stably perform their functions even though the affinity of Vac17 for Vac8 is much lower than those of Nvj1 and Atg13. The resulting heterodimeric Vac8–Vac17 complexes are recruited to actin cables through the interaction between Myo2 and Vac17, and vacuoles are transported to emerging daughter cells during mitosis. Once the Vac8–Vac17 complex has formed, Nvj1 and Atg13 are unlikely to disrupt it because their affinities for preformed Vac8–Vac17 complexes are 14 to 15-fold lower than those for free Vac8 (Fig. 4). Finally, as previously reported (24), Vac8 exists in excess of to its main binding partners, which makes it unlikely that Vac17, Nvj1, and Atg13 compete for binding to Vac8 in vivo.
Fig. 6.
A working model of how Vac8 mediates vacuole inheritance in comparison with the Cvt and PMN pathways. This study, together with our previous studies (8, 9), offers a working model of how the single ARM protein Vac8 regulates three distinct cellular processes, namely, formation of NVJs (which is a prerequisite for PMN), Cvt, and vacuole inheritance. In the presence of Nvj1 or Atg13, Vac8 dimerizes to form a serpent column-like or arch-shaped heterotetramer, respectively. Using different quaternary structures, Vac8 may perform two distinct functions (formation of NVJs and Cvt). However, in the presence of Vac17, Vac8 fails to dimerize because binding of Vac17 clamps the H1 helix and ARM1, masking the interaction interface for Vac8 dimerization. The Vac8–Vac17 heterodimer interacts with Myo2, which in turn moves along actin cables for vacuole transport during mitosis.
Vac8 belongs to a family of ARM proteins that contain several 42-amino acid repeat domains and are homologous to Drosophila armadillo protein. Although ARM proteins of higher eukaryotic organisms mediate cellular processes that completely differ from those mediated by Vac8 in budding yeast, they have retained their mode of action during evolution. Specifically, they provide a binding platform for diverse proteins. In higher organisms, ARM proteins play critical roles in cell adhesion, signal transduction, regulation of mitochondrial functions, and even tumorigenesis. Several human ARM proteins, such as ARMC1–10, ARMC12, and ARMCX1–6, have recently received increasing attention because they are highly conserved in mammals and implicated to function in various human diseases (25). Despite this increasing attention, it remains poorly understood how these proteins can change their binding partners to mediate various cellular functions at the atomic level, mainly due to a lack of high-resolution structures of protein complexes containing these proteins and their binding partners. The current study and our previous studies (8, 9) revealed three distinct structures of Vac8 depending on its binding partner (Vac17, Nvj1, or Atg13). Vac8 forms a heterotetramer when its binds to Nvj1 or Atg13, and binding of Nvj1 or Atg13 to Vac8 seems to induce Vac8 dimerization (8, 9). Strikingly, the overall structure of the Vac8–Nvj1 heterotetramer differs from that of the Vac8–Atg13 heterotetramer. Specifically, the Vac8–Nvj1 heterotetramer forms an arch-shaped structure, whereas the Vac8–Atg13 heterotetramer has an extended helical structure. Even more surprisingly, this study found that Vac8–Vac17 exists as a heterodimer. One of the modes of action by which Vac8 regulates various cellular processes is via formation of distinct quaternary structures depending on its binding partner. It would be very intriguing if future studies reveal that ARM proteins also adopt this strategy in higher eukaryotic cells.
Vacuole inheritance seems to be well coordinated with cell cycle progression. Thus, it is reasonable to assume that the interactions of Vac8 with various binding partners are also governed by cell cycle regulators. These include cyclin-dependent kinases and their corresponding phosphatases, which mediate phosphorylation and dephosphorylation in a cell cycle–dependent manner, respectively. Ptc1, a type 2C protein phosphatase, is reportedly required to maintain the steady-state level of vacuole-specific receptors, such as Vac8, as well as that of Vac17. Vac8 contains two potential phosphorylation sites in the N-terminal region; therefore, we investigated whether mutations of these residues affect vacuole inheritance. No defect in vacuole inheritance was observed when the phosphorylation site was replaced by alanine (phosphorylation defective) or aspartate (phosphorylation mimetic) (SI Appendix, Fig. S10), although these mutations significantly affect NVJ formation (SI Appendix, Fig. S11) and Cvt (8, 9). These results suggest that another phosphorylation site in Vac8 is regulated by cell cycle–dependent signaling or that Vac17 is the major target for cell cycle–dependent regulation of vacuole inheritance as suggested previously (22).
Materials and Methods
Cloning, Protein Expression, and Purification.
Vac8, Atg13, and Nvj1 proteins were prepared as described previously (8, 9). For Vac17 proteins, DNA fragments encoding Vac17 were amplified by PCR using S. cerevisiae genomic DNA and cloned into the pET28b-SMT3 vector. For coexpression of tVac8 and tVac17, the recombinant plasmids pGEX-6P-1-tVac8 and pET28b-SMT3-tVac17 were both transformed into BL21 (DE3) Escherichia coli cells. Cells were grown in LB medium at 37 °C until optical density at 600 nm (OD600) reached 0.6 and protein expression was induced by addition of 0.35 mM isopropyl β-D-1-thiogalactopyranoside at 18 °C for 18 h. The proteins were purified by Ni2+-chelated HiTrap column chromatography (GE Healthcare, Chicago, IL, USA). The His6-SUMO and GST tags were cleaved by addition of Ulp1 and PreScission proteases at ratios of 1:500 and 1:200 (w/v), respectively. The proteins were further purified by SEC (Superdex 200 column, GE Healthcare) in buffer A (25 mM Tris-HCl pH 7.5, 150 mM NaCl, and 5 mM DTT). All mutants were generated by PCR-based mutagenesis, confirmed by DNA sequencing, and purified as described above.
Crystallization and Structure Determination.
The tVac8–tVac17 complex was concentrated to 10 mg/mL using Amicon centrifugal filter units (MERCK, USA). This complex was crystallized using the hanging-drop vapor diffusion method at 4 °C by mixing 1 μL protein solution with 1 μL reservoir buffer comprising 25% (w/v) SOKALAN cp42, 100 mM Tris-HCl pH 8.5, and 100 mM sarcosine. The crystals were harvested in a solution comprising well solution plus 30% (v/v) glycerol and flash frozen in liquid nitrogen. Diffraction data were collected on the 5C beamline of the Pohang Accelerator Laboratory at an X-ray wavelength of 0.9919 Å. Data were processed with the HKL2000 program (26). The crystal structure was determined by molecular replacement with this program, using the coordinates of Vac8 (PDB ID: 5XJG) as a search model (8). Following rigid-body and positional refinement of the model with the phaser (PHENIX), the complete sequence of tVac17 (residues 290 to 308 and residues 330 to 344) could be positioned into residual electron density. Structures were built using Coot (27) and refined with PHENIX (28). The final model was refined to R/Rfree values of 0.1875/0.2233. Crystallographic data are summarized in SI Appendix, Table S1.
Accession Numbers.
The coordinates and crystallographic structure factors for the Vac8–Vac17 complex have been deposited in the Protein Data Bank (PDB) under the accession code 7YCJ.
Yeast Strains and Vacuole Staining with FM 4-64.
The yeast strains used in this study are listed in SI Appendix, Table S3. Vacuoles of yeast cells were labeled with FM 4-64 as previously described (29) with minor modifications. Cells were inoculated in 5 mL YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) and grown overnight at 30 °C with mild shaking. Thereafter, cells were diluted to an OD600 of 0.3 in fresh YPD medium, mixed with FM 4-64 (Molecular Probes) at a final concentration of 1 μM, and grown at 30 °C with mild shaking. After 30 min, cells were harvested by centrifugation (3,000 × g) at room temperature for 1 min, resuspended in fresh medium, and further grown for 3 h at 30 °C. Finally, cells were harvested by centrifugation (3,000 × g) at room temperature for 1 min, resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.47 mM KH2PO4, pH 7.4), and analyzed using a fluorescence microscope (Nikon Eclipse Ti-U) equipped with a Nikon Plan Apo 100×, 1.45/NA oil immersion objective. Following expression of GFP-conjugated Nvj1, NVJs were analyzed under a fluorescence microscope as described above.
Additional methods are described in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the staff of the 5C beamline at the Pohang Accelerator Laboratory for assistance with synchrotron facilities. This research was supported by a grant from the 2022 Joint Research Project (1.220065.01) of Institutes of Science and Technology, the Cell Logistics Research Center (2016R1A5A1007318), and grants from the National Research Foundation of Korea (NRF-2021M3A9G8022417, NRF-2022R1A2C1007314, and NRF-2021R1A2C2009550) funded by the Korean government.
Author contributions
C.L. and Y.J. designed research; Hyejin Kim, Jihyeon Park, Hyunwoo Kim, N.K., Jumi Park, E.J., S.Y.Y., J.A.R.D., C.L., and Y.J. performed research; Hyejin Kim, Jihyeon Park, Hyunwoo Kim, N.K., Jumi Park, C.L., and Y.J. analyzed data; and Jihyeon Park, C.L., and Y.J. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. C.U. is a guest editor invited by the Editorial Board.
Contributor Information
Changwook Lee, Email: changwook@unist.ac.kr.
Youngsoo Jun, Email: junys@gist.ac.kr.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Tewari R., Bailes E., Bunting K. A., Coates J. C., Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 20, 470–481 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Valenta T., Hausmann G., Basler K., The many faces and functions of beta-catenin. EMBO J. 31, 2714–2736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egea P. F., Mechanisms of non-vesicular exchange of lipids at membrane contact sites: Of shuttles, tunnels and funnels. Front. Cell Dev. Biol. 9, 784367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manik M. K., Yang H., Tong J., Im Y. J., Structure of yeast OSBP-related protein Osh1 reveals key determinants for lipid transport and protein targeting at the nucleus-vacuole junction. Structure 25, 617–629.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Wang Y. X., Catlett N. L., Weisman L. S., Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 140, 1063–1074 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisman L. S., Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37, 435–460 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Scott S. V., et al. , Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 275, 25840–25849 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Jeong H., et al. , Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure. Proc. Natl. Acad. Sci. U.S.A. 114, E4539–E4548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J., et al. , Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways. Autophagy 16, 991–1006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X., et al. , Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell 11, 2445–2457 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts P., et al. , Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kvam E., Goldfarb D. S., Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 3, 85–92 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Krick R., et al. , Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 19, 4492–4505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millen J. I., Krick R., Prick T., Thumm M., Goldfarb D. S., Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy 5, 75–81 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Wang Y. X., Kauffman E. J., Duex J. E., Weisman L. S., Fusion of docked membranes requires the armadillo repeat protein Vac8p. J. Biol. Chem. 276, 35133–35140 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Seeley E. S., Wickner W., Merz A. J., Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357–369 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Veit M., Laage R., Dietrich L., Wang L., Ungermann C., Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 20, 3145–3155 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohde J., Dietrich L., Langosch D., Ungermann C., The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J. Biol. Chem. 278, 1656–1662 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Weisman L. S., Organelles on the move: Insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7, 243–252 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Tang F., et al. , Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature 422, 87–92 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa K., et al. , Identification of an organelle-specific myosin V receptor. J. Cell Biol. 160, 887–897 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yau R. G., et al. , Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev. Cell 28, 520–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K. W., Lu M. S., Iwamoto Y., Drubin D. G., Pedersen R. T. A., A preferred sequence for organelle inheritance during polarized cell growth. J. Cell Sci. 134, jcs258856 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho B., Baryshnikova A., Brown G. W., Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Huang Y., et al. , Structures, functions, evolutions, interactions, and diseases. Front. Mol. Biosci. 8, 791597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vida T. A., Emr S. D., A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.