Abstract
A hallmark of Huntington’s disease (HD) is a prolonged polyglutamine sequence in the huntingtin protein and, correspondingly, an expanded cytosine, adenine, and guanine (CAG) triplet repeat region in the mRNA. A majority of studies investigating disease pathology were concerned with toxic huntingtin protein, but the mRNA moved into focus due to its recruitment to RNA foci and emerging novel therapeutic approaches targeting the mRNA. A hallmark of CAG-RNA is that it forms a stable hairpin in vitro which seems to be crucial for specific protein interactions. Using in-cell folding experiments, we show that the CAG-RNA is largely destabilized in cells compared to dilute buffer solutions but remains folded in the cytoplasm and nucleus. Surprisingly, we found the same folding stability in the nucleoplasm and in nuclear speckles under physiological conditions suggesting that CAG-RNA does not undergo a conformational transition upon recruitment to the nuclear speckles. We found that the metabolite adenosine triphosphate (ATP) plays a crucial role in promoting unfolding, enabling its recruitment to nuclear speckles and preserving its mobility. Using in vitro experiments and molecular dynamics simulations, we found that the ATP effects can be attributed to a direct interaction of ATP with the nucleobases of the CAG-RNA rather than ATP acting as “a fuel” for helicase activity. ATP-driven changes in CAG-RNA homeostasis could be disease-relevant since mitochondrial function is affected in HD disease progression leading to a decline in cellular ATP levels.
Introduction
Huntington’s disease (HD) belongs to a group of nine neurodegenerative diseases, the so-called polyglutamine diseases, that are caused by the expansion of cytosine, adenine, and guanine (CAG) trinucleotide repeats located in the translated regions of functionally unrelated genes. Yet, they all have a number of shared molecular properties and symptoms, including their mRNAs being capable of forming ribonuclear foci that sequester proteins such as muscleblind protein 1 (MBNL1), leading to aberrant RNA splicing.1 In HD patients, one huntingtin (HTT) allele is expanded beyond the pathogenic threshold of 36 repeats,2 with the repeat length being inversely correlated with the age of onset of symptoms including involuntary movements and dementia.2 Both the mRNA transcript and the protein translated from the mutant allele exert pathogenic effects above this threshold.1 As such, the HTT mRNA appears to be a promising target for therapeutic intervention, e.g., by RNA interference.3,4 Alternatively, small molecules were reported that rescue disease-relevant features of myotonic dystrophies 15 and 26 and amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD)7 by either preventing RNA-binding proteins from interacting with their pathology-related target or degrading them by recruiting RNase L.
A key to such developments is a comprehensive understanding of the conformation and cellular distribution of HTT mRNA. In vitro experiments and simulations showed that CAG-repeat RNAs form a stable hairpin with increasing folding stability for higher repeat numbers due to the repetitive stable pairing of G and C nucleotides.8 In the context of HTT exon 1, base pairing with the CAG flanking sequences supports an ensemble of hairpin conformations not limited to the CAG-repeat region.9 Remarkably, extended CAG-RNA is retained in the nucleus within RNA foci inside nuclear speckles, leading to the sequestration of different RNA binding proteins.1,3,10 The binding affinity depends on the CAG-repeat length and, thus, the hairpin folding stability11 and is associated with disease pathology.1,10,11
Nuclear speckles contain high quantities of pre-mRNA splicing factors, transcription factors, and 3′-end RNA processing factors.12,13 They are classified as membraneless phase-separated organelles that exhibit liquid-like behavior and could play a functional role in the spatial organization of RNA splicing.14 Jain and Vale showed that CAG-repeat mRNA sequestered in nuclear speckles is mobile.15 In vitro experiments further revealed that CAG-repeat RNA itself forms clusters in condensates at concentrations as low as 25 nM.15 However, in these assemblies, the RNA does not retain dynamic behavior, and the condensates are thus classified as gels. Simulations suggest that this self-association behavior can be attributed to intermolecular base pairing of (partially) unfolded CAG hairpins.16 To explain the high mobility of CAG-RNA in nuclear speckles compared to its self-association in gels, Jain and Vale hypothesized that helicases in the nucleoplasm could remodel CAG-repeat RNA base pairing.15
The fact that nuclear speckles have a liquid-like character and the ability of CAG triplet repeat RNA to form gels suggest compelling research questions regarding CAG-RNA homeostasis: What is the molecular driving force of CAG-RNA-condensate association? In which conformation is the CAG-RNA sequestered? Do aberrant CAG entanglements/aggregates form preferentially in the nucleoplasm or in condensates? Is the sequestration of potentially toxic RNA a cytoprotective mechanism, e.g., by preventing non-native binding events in either of the environments? If this mechanism is exhausted, could nuclear speckles undergo pathogenic aberrant phase transitions?
In this study, we explore these questions by investigating the conformational equilibrium and mobility of CAG-RNA in different cellular environments and conditions.
Results
Sequestration of CAG-RNAs in Nuclear Speckles
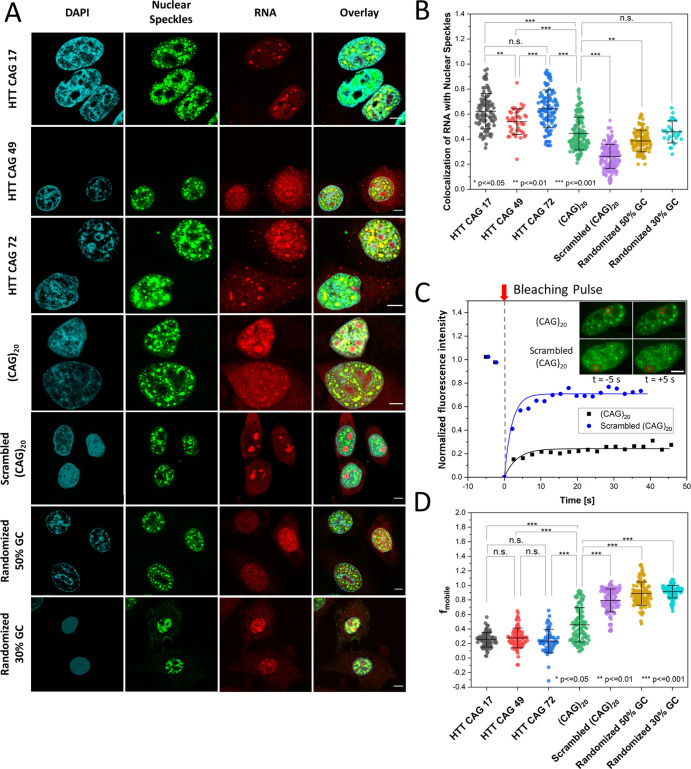
First, we investigated how the length, sequence, and flanking regions of the CAG repeats determine recruitment to nuclear speckles and how these properties determine mobility. Therefore, we microinjected fluorophore-labeled (CAG)20 RNA into HeLa cells (see the Supporting Information Materials and Methods section for details) at a stock solution concentration of 100 μM (if not stated otherwise), yielding a cellular concentration of approximately 1.4 μM that largely exceeds the endogenous CAG-repeat RNA concentration.3,17 Colocalization with nuclear speckles was determined using the SC35 nuclear speckle marker (Figure 1A). We compared (CAG)20 RNA to a scrambled sequence [scrambled (CAG)20] and to randomized sequences of the same length with a 30 and 50% GC content, respectively. We also included HTT exon 1 sequences with 17, 49, and 72 CAG repeats. These were employed at lower stock solution concentrations (100–200 ng/μL) as yielded after in vitro transcription and stochastic fluorescent labeling. While (CAG)20 RNA and all HTT exon 1 constructs were found to be predominantly localized in nuclear speckles, the randomized RNAs formed distinct small foci inside the nucleoplasm, and the scrambled RNA was mainly found in the nucleoli and the cytosol, with some small foci still being observed in the nucleoplasm [sample images: Figure 1A,C (inset) and evaluation of the focus size, number, and intensity: Figure S1].
Figure 1.
CAG-RNA colocalization with nuclear speckles and mobility in RNA foci. (A) Sample images showing the colocalization of CAG-RNAs (Alexa594) and nuclear speckles (SC35 nuclear speckle marker/Alexa488). Scale bars: 5 μm. (B) Statistical analysis of colocalization for the respective CAG-RNAs. (C) Normalized fluorescence intensity plotted against time for sample FRAP experiments with (CAG)20 and scrambled (CAG)20. Inset: pre- and post-bleach images of (CAG)20 (top row) and scrambled (CAG)20 (bottom row) foci in the nuclei of HeLa cells. The bleaching region is marked by a red circle. Scale bar: 5 μm. (D) Mobile fractions of CAG-RNAs in nuclear speckles (CAG-repeat RNAs) and other foci [scrambled (CAG)20, randomized 50% GC, and randomized 30% GC]. Statistical analyses shown in (B,D) were performed by a one-way ANOVA and post-hoc Tukey test (for sample sizes, see Table S1). Error bars show S.D. calculated by Gaussian error propagation (see the Supporting Information Materials and Methods section).
We further analyzed the extent of colocalization of the RNAs with nuclear speckles by an object-based approach measuring the overlap of RNA foci and nuclear speckles using CellProfiler18−21 (see the Supporting Information Materials and Methods section for details). (CAG)20 RNA showed a significantly higher colocalization share with nuclear speckles (45 ± 13%) compared to the scrambled and randomized sequences [scrambled (CAG)20: 26 ± 10%; randomized 50% GC: 39 ± 9%; and randomized 30% GC: 46 ± 9%] (Figure 1B). Colocalization of randomized 50% GC and randomized 30% GC was mainly caused by arbitrary overlaps between small RNA foci and nuclear speckles (Figure 1A). The highest colocalization was found for HTT exon 1 sequences including both the CAG tract and its physiological flanking sequences (HTT CAG 17: 62 ± 14%; HTT CAG 49: 54 ± 10%; and HTT CAG 72: 64 ± 15%). We did not find a significant difference between HTT exon 1 RNAs with physiological (HTT CAG 17) and pathologically expanded CAG repeats (HTT CAG 49 and HTT CAG 72).
To further corroborate these results, we conducted fluorescence recovery after photobleaching (FRAP) measurements in the different foci (Figure 1C). We found that the HTT exon 1 constructs exhibited the lowest mobility as shown by lowest mobile fraction (fmobile) values. fmobile is determined by the fraction of fluorescence that recovers throughout the course of a FRAP experiment (Figure 1C). This finding is in line with a strong association of the RNA with nuclear speckles. Again, no significant change in mobility between the different CAG repeat length extensions (HTT CAG 17: 25 ± 10%; CAG 49: 28 ± 14%; and CAG 72: 23 ± 16%) was found (Figure 1D). The (CAG)20 RNA showed slightly higher mobility compared to the HTT exon 1 RNAs [(CAG)20: 46 ± 24%], and the randomized and scrambled sequences of equal length showed significantly increased mobility [scrambled (CAG)20: 79 ± 16%; randomized 50% GC: 89 ± 16%; and randomized 30% GC: 91 ± 9%]. Microinjections at different (CAG)20 stock concentrations (10, 100, and 300 μM) showed that the mobility measurements were independent of the cellular concentration (Figure S2).
In comparison to earlier studies by Jain and Vale,15 who measured a mobile fraction of 83 ± 13% for a (CAG)49 sequence in nuclear speckles, the CAG-RNAs investigated here showed smaller mobile fractions [e.g., (CAG)20: 46 ± 24%]. In their earlier study, Jain and Vale conjugated the CAG-RNA with 12 MS2 hairpin loops binding to yellow fluorescent protein-labeled MS2-binding protein. In comparison to the Alexa Fluor dyes used here, their bulky detection system could potentially affect the phase behavior, e.g., by diminishing intermolecular CAG interactions in the condensate.
In summary, the colocalization and FRAP studies both show that an increased association of CAG-RNA sequences with nuclear speckles is accompanied by a decrease in mobility suggesting stronger association with the condensate. Association and interaction with nuclear speckles are specific to the CAG repetition motive and enhanced by the flanking sequences of HTT exon 1. For the here-studied HTT exon 1 constructs, these properties are independent of CAG-repeat length.
(CAG)20 RNA Hairpins Are Stable but Destabilized in Cells
Based on these results, we proceeded to study (CAG)20 RNA as a model system to analyze its conformational stability as a function of subcellular localization. We used fast relaxation imaging (FReI) that allows us to measure the folding kinetics and thermodynamics of biomolecules in cells with high spatio-temporal resolution by combining fluorescence resonance energy transfer (FRET) microscopy with fast temperature jumps induced by an infrared (IR) laser.22−24 The technique was previously used to study folding and phase behavior of superoxide dismutase 1,25,26 conformational dynamics and aggregation of Huntingtin exon 1,27,28 or the folding stability of 4U-RNA in the nucleus and cytoplasm of cells.29
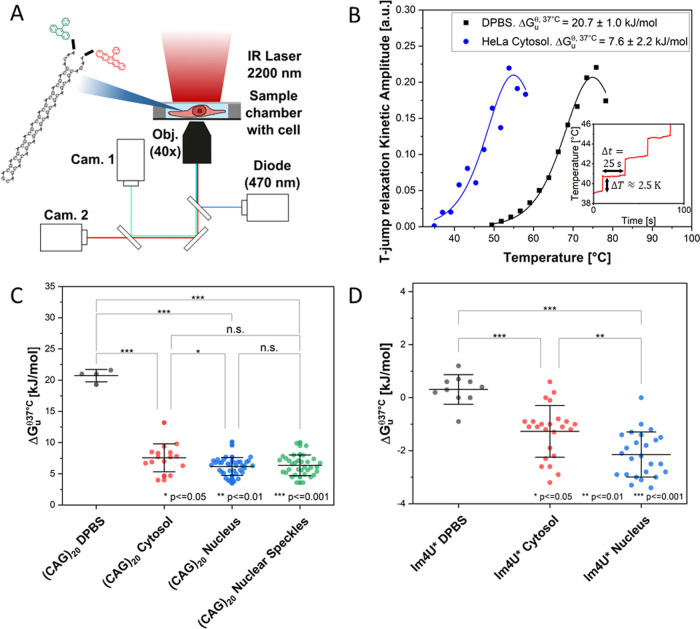
In preliminary work,30 we prepared (CAG)20 RNA for FReI experiments by terminal FRET labeling using Alexa Fluor 488 at the 5′-end and Alexa Fluor 594 at the 3′-end. Dyes were covalently connected to the RNA backbone via C6 linkers and by alkyne–azide click chemistry (5′-end) or amine coupling to a carbonic acid N-hydroxy succinimide ester (3′-end) (Figure 2A). We characterized the labeled RNA in vitro and established a tailored protocol to measure its folding stability by fast consecutive temperature jumps (Figure 2B, inset), recording the temperature-dependent unfolding kinetics within 8 min in a single living cell with every respective temperature jump lasting 25 s.30 We found that the (CAG)20 RNA forms stable hairpins in Dulbecco’s phosphate buffered saline (DPBS; pH 7.0–7.3, 2.7 mM KCl, 1.5 mM KH2PO4, 140 mM NaCl, 8 mM Na2HPO4) buffer solution (Tm = 349 ± 2 K, ΔGuθ,37°C = 20.7 ± 1.0 kJ/mol ),30 which was in agreement with previous studies.3,31 However, the folding stability was significantly decreased in the presence of cosolutes due to various factors such as transient chemical interactions30,32,33 or a decrease in water activity,30,33,34 leading to the question whether (CAG)20 RNA remains folded in the densely crowded cell or upon sequestration in nuclear speckles.
Figure 2.
Folding stability of (CAG)20 measured by FReI. (A) Schematic of the FReI microscope and (CAG)20 RNA (see Figure S4A for details). Thermal unfolding upon temperature jumps [induced by an IR laser (λ = 2200 nm)] is monitored by FRET [(Alexa Fluor 488/594); donor excitation: λ = 470 nm]. (B) Kinetic relaxation amplitudes measured at different temperatures and fitted by the Girdhar model.35 Inset: customized temperature jump protocol calibrated using the temperature-dependent fluorescence of Rhodamine B.38−40 (C,D) ΔGuθ,37°C of (CAG)20 (C) and lm4U* (D) RNA measured in DPBS and HeLa cell cytosol, nuclei, and nuclear speckles. Statistical analyses were performed by a one-way ANOVA and post-hoc Tukey test (for sample sizes, see Table S1). Error bars show S.D. calculated by Gaussian error propagation (see the Supporting Information Materials and Methods section).
We therefore microinjected (CAG)20 RNA into HeLa cells and measured its folding stability by FReI (Figure 2A). Analysis of the temperature-dependent unfolding kinetics (Figure 2B) allowed us to calculate the melting temperature Tm, first-order cooperativity parameter g(1) (the slope at the inflection point of a thermal melting curve), and the standard free energy of unfolding at 37 °C: ΔGuθ,37°C = (310 K – Tm)·g(1).35 ΔGu was used to calculate the equilibrium constant K and folded and unfolded fractions of the RNA at 37 °C (K37°C, ffolded37°C, = funfolded) (see the Supporting Information Materials and Methods section for details) with up to 1.5–2 μm of spatial resolution in live cells.
The model is based on the two-state assumption which is supported by single-molecule FRET measurements of (CAG)20 RNA in DPBS under native and destabilizing conditions (Figure S3). Instead of the commonly used sigmoidal fitting procedure which requires fully resolved folded and unfolded state baselines, we used a previously established a kinetic method to analyze in-cell melting curves with a limited accessible temperature range.35 This method allows for a thermodynamic analysis with only a few data points recorded beyond Tm (Figure 2B).
The results are summarized in Tables S2 and S3, and ΔGuθ,37°C values are compared in Figure 2C,D. The analysis showed that (CAG)20 RNA was largely destabilized in the cellular environment compared to dilute DPBS buffer solution [ΔGu(DPBS) = 20.7 ± 1.0 kJ/mol30 and ΔGuθ,37°C(cytosol) = 6.8 ± 2.1 kJ/mol]. Despite the major destabilization, it is important to note that the RNA was still mostly folded under physiological conditions (ΔGu(cytosol) = 6.8 ± 2.1 kJ/mol, meaning that only 6.7% of the RNA was unfolded at 37 °C). The decrease in ΔGuθ,37°C was also large compared to that under any in vitro condition measured previously (e.g., ΔGu(DPBS + 300 g/L Ficoll 70) = 18.0 ± 1.0 kJ/mol; ΔGuθ,37°C(DPBS + 300 g/L ethylene glycol) = 12.0 ± 3.0 kJ/mol; ΔGu(HeLa cell lysate) = 13.3 ± 0.5 kJ/mol).30 Thus, crowding effects such as the decrease in water activity or non-specific interactions of the RNA and cosolutes could not solely account for the observed decrease in folding stability.
To further interpret these results, a low-melting but stable RNA hairpin, the Salmonella fourU RNA thermometer [lm4U*; *denoting the destabilizing mutation (C23U)], was studied for comparison to CAG hairpins. The secondary structure in comparison to that of a CAG hairpin is shown in Figure S4A,B. The RNA is located in the 5′-untranslated region (5′-UTR) of the aggregation–suppression protein and functions as a temperature sensor in free-living microorganisms.36,37 lm4U* was shown to fold in a two-state manner in HeLa cells,29 and since it is a prokaryotic RNA functioning as a temperature-sensitive control element in free-living organisms, it is not engaged in specific cellular functions in HeLa cells. lm4U* is marginally stable in DPBS at 37 °C (ΔGuθ,37°C(DPBS) = 0.3 ± 0.6 kJ/mol) and slightly destabilized in cells ΔGu(cytosol) = −1.3 ± 1.0 kJ/mol, ΔGuθ,37°C(nucleus) = −2.1 ± 0.8 kJ/mol) (Figure 2D). Compared to (CAG)20 RNA, the destabilization in cells is small and is explained by compensating crowding effects. While the loss in water activity and transient chemical interactions destabilize lm4U*, excluded volume effects lead to a counteracting stabilizing effect.29
We then analyzed the folding stability in nuclear speckles for (CAG)20 RNA [lm4U* does not associate with nuclear speckles under any condition (Figure S5)]. We found a similar ΔGuθ,37°C in nuclear speckles and the bulk nucleoplasm (ΔGu(nucleus) = 6.0 ±1.4 kJ/mol; ΔGuθ,37°C(nuclear speckles) = 6.4 ± 1.6 kJ/mol (Figure 2C). This shows that (CAG)20 RNA associates with nuclear speckles in the folded rather than unfolded hairpin conformation (92.3% of the RNA is folded in nuclear speckles) and that condensate association is not associated with a conformational transition under physiological conditions.
However, we observed that the (CAG)20 RNA is enriched in nuclear speckles upon stepwise heating (Figure S6A–C). In particular, at temperatures beyond the melting point where the RNA is mostly unfolded, strong migration to the foci was observed (Figure S6A–C). We found that the association of the RNA with nuclear speckles is partially irreversible as the partitioning coefficient (PC) between the nuclear speckles and surrounding nucleoplasm does not recover its initial value at 37 °C (Figure S6A,B). Since the RNA unfolds reversibly in a two-state manner in the cytoplasm and nucleoplasm and also under various crowded conditions in vitro, it is not expected that irreversibly misfolded CAG entanglements cause this effect. We analyzed single temperature jumps of FReI experiments performed in nuclear speckles at high temperatures (e.g., 55 °C). Despite this high temperature, (CAG)20 RNA unfolding showed no indication of self-association due to intermolecular FRET (Figure S7B), which would be expected in this case due to the high ratio of labeled to endogenous CAG-RNA. In fact, FReI is very sensitive to such self-association events upon unfolding.22,27,28,41 Measurements of (CAG)20 RNA in concentrated adenosine triphosphate (ATP) solutions (15 and 20 mM) led to microscopically visible aggregates, showing that self-association could be indeed detected for the (CAG)20 RNA construct (Figure S8). Thus, the data suggest that CAG-RNA entanglements do not form in speckles (or the nucleoplasm), even at high temperatures and at concentrations much higher than those of endogenous CAG-RNA levels.
ATP Destabilizes (CAG)20 RNA and Maintains Its Mobility in Cells
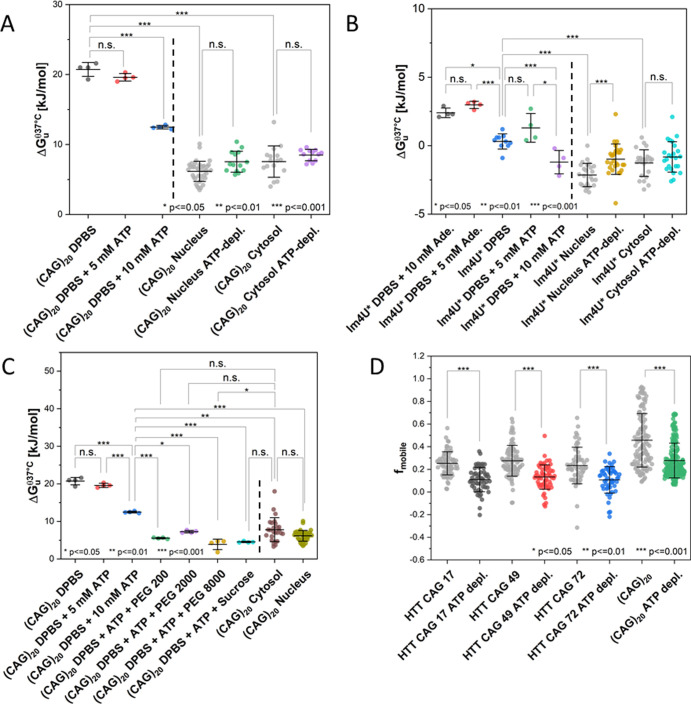
Inspired by the previous work of Jain and Vale, who found a profound role of ATP in maintaining the liquid-like properties of CAG repeats in nuclear speckles (e.g., by enabling helicase activity),15 we investigated the role of ATP in determining (CAG)20 RNA folding stability and mobility in speckles and the nucleoplasm. We first conducted in vitro experiments in the absence of ATP-dependent helicases and other cofactors or binding partners. We found that (CAG)20 RNA is increasingly destabilized with rising ATP concentrations (ΔGuθ,37°C(DPBS) = 20.7 ± 1.0 kJ/mol; ΔGu(DPBS + 5 mM ATP) = 19.6 ± 0.5 kJ/mol; ΔGuθ,37°C(DPBS + 7.5 mM ATP) = 17.1 ± 4.9 kJ/mol; and ΔGu(DPBS + 10 mM ATP) = 12.5 ± 0.3 kJ/mol) (Figures 3A and S4A). In comparison, lm4U* is slightly stabilized at 5 mM and also largely destabilized at 10 mM (ΔGuθ,37°C(DPBS) = 0.3 ± 0.6 kJ/mol; ΔGu(DPBS + 5 mM ATP) = 1.3 ± 1.0 kJ/mol; and ΔGuθ,37°C(DPBS + 10 mM ATP) = −1.2 ± 0.9 kJ/mol) (Figure 3B). All experiments were performed in magnesium-free DPBS buffer. Thus, an indirect effect of ATP on ΔGu by chelating magnesium ions and removing them from the RNA can be ruled out.
Figure 3.
(CAG)20 and lm4U* RNA folding stability and mobility as a function of the ATP concentration. A dotted line is used to separate in vitro and in cell data. (A,B) ΔGuθ,37°C measured for (CAG)20 and lm4U* in vitro (DPBS) and in cells with and without ATP depletion. (C) ΔGu for (CAG)20 in different cosolutes mimicking the cellular environment. If not stated otherwise, the ATP concentration is 10 mM, and cosolute concentrations are 300 g/L. Values from the HeLa cytosol and nucleus are taken from A and shown for comparison. (D) Mobile fraction fmobile of (CAG)20 and HTT exon 1 RNA in nuclear speckles under normal and ATP-depleted conditions. Data acquired under physiological conditions (Figures 1D and 2C,D) are shown in gray for comparison. Statistical Analysis was performed by a one-way ANOVA and post-hoc Tukey test (for sample sizes, see Table S1). Error bars show S.D. calculated by Gaussian error propagation (see the Supporting Information Materials and Methods section).
Experiments using adenosine alone showed an increase in folding stability (ΔGuθ,37°C(lm4U*; DPBS + 5 mM adenosine) = 3.0 ± 0.3 kJ/mol; ΔGu(lm4U*; DPBS + 10 mM adenosine) = 2.4 ± 0.3 kJ/mol; see Figure S4), suggesting that the triphosphate group is required to mediate the destabilization. In accordance with similar observations regarding proteins,42 the triphosphate could increase the solubility of ATP, thus facilitating its interaction as a cosolute with the RNA. Furthermore, the extent of destabilization of (CAG)20 RNA by ATP is remarkable since similar ΔGuθ,37°C values were only found for small cosolutes like ethylene glycol, PEG 200, or sucrose.30 However, this destabilization occurred at significantly lowered water activity and cosolute concentrations that are about 50-fold higher. Neither crowding nor ATP destabilization alone could mimic in-cell folding stability, but remarkably, we found that solutions containing crowding agents [PEG 200, PEG 2000, PEG 8000, and sucrose (300 g/L)] in addition to 10 mM ATP (Figure 3C) could do so. This shows that crowders and ATP both play a role in determining (CAG)20 RNA folding stability under cellular conditions. However, further studies are required to define the contribution of ATP in cell-mimicking environments, accounting also for changes, e.g., in pH or ionic strength.
To probe whether changes in the ATP concentration also affect the folding stability of (CAG)20 and lm4U* in cells, we conducted ATP depletion experiments, adding final concentrations of 1 mM KCN and 10 mM 2-deoxyglucose to the medium.43 ATP depletion was monitored by the ATeam ATP concentration sensor (see Figure S9 and Supporting Information Materials and Methods for further details),43,44 and (CAG)20 RNA folding stability was assessed at the minimum ATP level. The normalized FRET ratio decreased from 1.0 to 0.6, corresponding to a decrease in the cellular ATP concentration by 2–3 mM.45 As ATP levels are estimated to be 2–3 mM in human cells,46 it is assumed that ATP is completely depleted from the cells. Indeed, we found that ATP depletion caused a minor stabilization of (CAG)20 RNA by ΔΔGuθ,37°C ≈ 1.5 kJ/mol in both the cytosol and nucleus (ΔGu(cytosol, ATP – depl.) = 8.5 ± 0.8 kJ/mol, ΔGuθ,37°C(nucleus, ATP – depl.) = 7.5 ± 1.5 kJ/mol) of HeLa cells (Figure 3A). For lm4U*, an even smaller stabilizing effect was observed (ΔΔGu(cytosol) ≈ 0.5 kJ/mol, ΔΔGuθ,37°C(nucleus) ≈ 1.1 kJ/mol) (Figure 3B).
Moreover, ATP depletion led to a significant decrease in mobility (10–20%) of (CAG)20 and HTT exon 1 RNA recruited to nuclear speckles [HTT CAG 17: 11 ± 11%; HTT CAG 49: 13 ± 11%; HTT CAG 72: 11 ± 12%; and (CAG)20: 28 ± 15%] (Figure 3D). Remarkably, ATP depletion fully prevented further heat-induced recruitment of (CAG)20 from the nucleoplasm to the nuclear speckles and completely immobilized RNA that had already been recruited to the nuclear speckles prior to the beginning of the experiment. PCs were independent of temperature (see Figure S6D,E).15
These results are in line with the observations by Jain and Vale who found mobile fractions of 83 ± 13% under normal and 23 ± 7% under ATP-depleted conditions. In their study, Jain and Vale suggested that ATP depletion led to a decrease in mobility due to a decline in activity of RNA chaperones with unwinding helicase activity (e.g., from DEAD box family chaperones15). From this hypothesis, it can be deduced that unfolding of RNA hairpins, e.g., by RNA helicases should lead to decreased hairpin folding stability. However, since our in vitro results showed that ATP largely decreases CAG hairpin folding stability as a cosolute in the absence of RNA-unwinding helicases, we propose that the measured changes in both folding stability and mobility may be caused by direct interactions of ATP with the RNA.
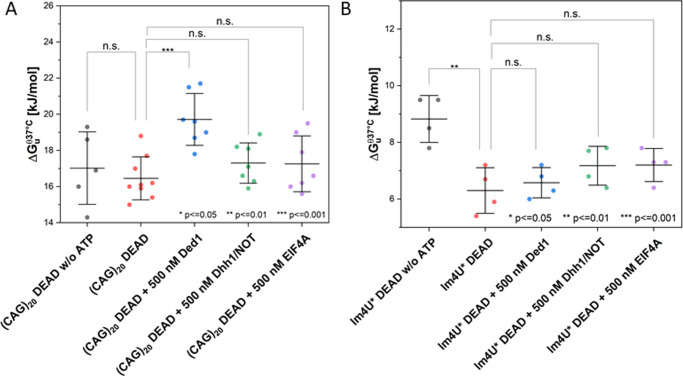
To further test this hypothesis, we performed in vitro experiments employing three different DEAD box helicases (Ded1, Dhh1, and EIF4A). Indeed, we found that the helicases did not change RNA folding stability compared to the respective buffer solution (Figure 4). This could be rationalized by the known limited RNA helicase capacity of DEAD box chaperones. Their helicase activity decreases with the increasing number and folding stability of base pairs within an RNA duplex as the binding of the protein’s RecA domain to the RNA has to outcompete binding interactions within the RNA. This process becomes ineffective at more than 15 base pairs or even less if the duplex mostly consists of C–G pairs.47,48 (CAG)20 and lm4U* comprise 18 and 14 base pairs, respectively. In the case of (CAG)20, all of these are highly stable C–G pairs. Thus, it can be assumed that DEAD box chaperones do not modulate the folding stability and mobility of CAG repeat hairpins, especially not for the more elongated and stable hairpins linked to the disease. Mobility and folding stability of these RNAs are rather governed by ATP as a cosolute.
Figure 4.
ΔGuθ,37°C for (CAG)20 (A) and lm4U* (B) in the presence of DEAD box helicases in DEAD buffer (containing 5 mM ATP if not specified otherwise). Statistical analysis was performed by a one-way ANOVA and post-hoc Tukey test (for sample sizes, see Table S1). Error bars show S.D. calculated by Gaussian error propagation (see the Supporting Information Materials and Methods section).
Molecular Dynamics Simulations Reveal ATP Interactions with the Unwound State of (CAG)20
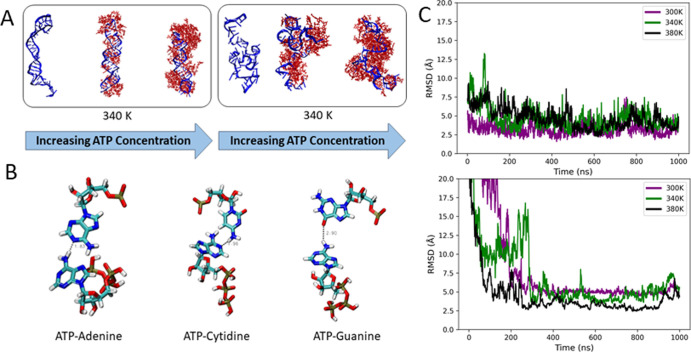
To investigate the mechanism of RNA destabilization by ATP, we performed large-scale molecular dynamics (MD) simulations. We used ATP and salt concentrations (see Table S4) and different temperatures (300, 340, and 380 K) to reproduce the experimental conditions. As even small RNAs of 50–80 nucleic acids tend to fold on the millisecond regime,49 it is currently impossible to simulate reversible (un)folding via conventional atomistic MD simulations, even though coarse-grained simulations are possible.50 Instead, we performed independent equilibrium MD simulations with two different starting conformations—the native, folded hairpin (named “native state” from here on) and a partially unfolded, unwound hairpin-like state (named “unwound state” hereon; see Figure S10). Final structures from all simulations with ATP molecules surrounding the RNA are shown in Figures 5A and S11. In the absence of ATP, native hairpin RNA was stable at 300 K but destabilized at 340 and 380 K. Specifically, with increasing temperature, there was greater variation in the chi torsion angles which characterizes the relative nucleobase/ribose orientation (Figure S12). In the unwound RNA, the presence of ATP led to fewer fluctuations in the root-mean-square-deviation (RMSD) with respect to the average nucleic backbone structure at 300 K (Figure S13). The time required for the RMSD to plateau is reduced in the presence of ATP at 300 and 380 K. This indicates that the backbone of the unwound RNA stabilized faster under these conditions. At 340 K and the 5 mM ATP concentration, the unwound RNA backbone also stabilized faster. However, with a 10 mM ATP concentration, the backbone RMSD stabilized after ∼450 ns, similar to that observed in the absence of ATP (Figure 5).
Figure 5.
(A) Final structures from MD simulations of native (left) and unwound (CAG)20 RNA (right) at increasing ATP concentrations (left to right) from simulations at 340 K (see Figures S11 for other temperatures). ATP concentrations were 0, 5, and 10 mM. (B) Hydrogen bond formation between the ATP’s adenine residue and the RNA strand’s nucleobases adenine, cytidine, and guanine. (C) RMSD of native (top) and unwound states (bottom) of (CAG)20 RNA between each momentaneous configuration and the average structure over the course of the simulation with the 5 mM ATP concentration (see Figure S13 for other concentrations).
We also observed an opening of the hairpin and exposition of the nucleobases for intermolecular binding interactions with ATP (see Figure 5B). Previous findings by Lambert and Draper also observed that urea destabilized RNA hairpin structures by preferential formation of H-bonds with the nucleobases, and similar results were reported for the interaction of ATP with proteins.32,42 This supports our notion that ATP replaces Watson–Crick base-pairing interactions, which subsequently leads to the enthalpic destabilization of the RNA backbone (note that increased temperature causes an entropic destabilization). To investigate and quantify this formation of hydrogen bonding interactions between ATP and RNA moieties, we used a donor–acceptor distance cutoff of 0.4 nm and a minimum donor–hydrogen–acceptor angle of 120° as the criteria (Figure 6, Table S5, and Figures S14 and S15). In all cases, ATP formed, on average, more hydrogen bonds with the atoms involved in Watson–Crick base pairing rather than with the RNA backbone’s phosphate or ribose moiety. Preferential formation of ATP–nucleobase hydrogen bonding interactions in the unwound state relative to the native state suggests that ATP destabilizes the RNA hairpin. An exception is at 5 mM ATP and 380 K, where the average number of ATP–nucleobase hydrogen bonds for native and unwound states was roughly the same. However, the average number of ATP–ribose hydrogen bonds in the unwound RNA was larger, and so the overall number of ATP-RNA interactions increased. The high local concentration of ATP close to the backbone also affects the surrounding hydration shell and biomolecular stability,51−53 as the phosphate groups of the backbone offer direct hydration sides and interactions sites for the counter ions and ATP.52,53 Quantifying these hydration shell changes would be exciting future work by complementing, e.g., small-angle X-ray scattering of time-averaged counterion distributions with MD simulations.54
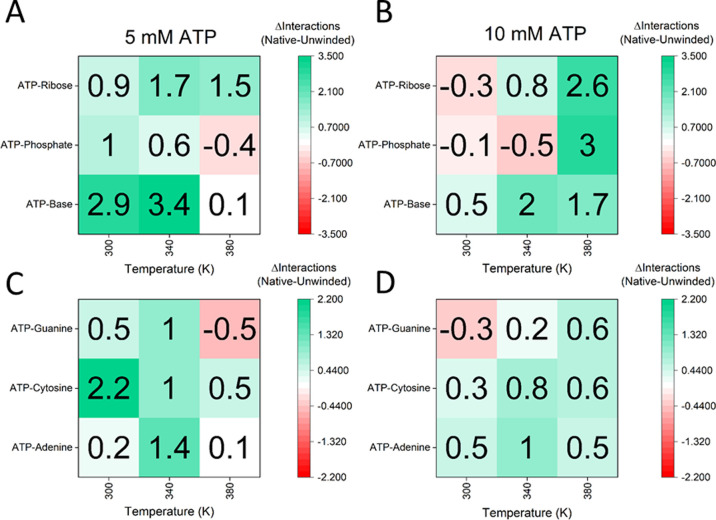
Figure 6.
Changes in interaction of ATP with the (CAG)20 RNA’s ribose, phosphate, and base moieties between native and unwound states at 5 and 10 mM ATP and different temperatures. Average numbers of interactions between ATP and the different RNA moieties were counted, and differences between folded and unwound states were calculated [thresholds: distance <0.4 nm, angle (donor–hydrogen–acceptor) > 120°. Absolute values: see Table S5, Figure S14, and Figure S15]. (A,B) The main shift is observed in interactions between ATP and the nucleobases. Interactions with phosphate or ribose increase much less or even decrease. (C,D) ATP–base interactions from (A) and (B) broken down to the different nucleobases. ATP mainly interacts with adenine and cytosine.
To summarize, the MD simulations show an enthalpic destabilization of (CAG)20 RNA caused by preferential interactions of ATP with the nucleobases in the unwound state, which is different from an entropically stabilized unwound state caused by elevated temperatures. This finding is in line with results reported in the literature and further corroborates our experimental observations.
Discussion
Sequestration and retention of RNA in nuclear speckles are a hallmark of several triplet repeat expansion diseases.12 Here, we investigated CAG-RNA recruitment to nuclear speckles under different cellular conditions as a function of RNA conformation and mobility. First, the comparison of (CAG)20 RNA with randomized and scrambled CAG sequences corroborated the CAG-repeat sequence specificity found in earlier studies.15,55 The comparison of (CAG)20 with HTT exon 1 RNAs showed that the flanking sequences slightly increased the colocalization with nuclear speckles and decreased the mobility. This could be explained by previous studies that suggested a profound role of the flanking sequences in HTT exon 1 folding, in particular suppressing CAG hairpin formation at low CAG repeat lengths.9 The authors suggested that a GCUGC moiety located in the 5′ UTR and the (CCG)7-10 and GCUGCUGC motifs in the flanking regions could provide additional binding valency to form an intermolecular RNA network. A high binding valency is commonly associated with biomolecules that engage with biomolecular condensates.56 Given the high RNA concentration inside the nuclear speckles,12,57 this would explain the decreased mobility and the increased association with nuclear speckles. Alternatively, the same binding valency could also be used to form extended hairpin structures which could then be bound by hairpin-binding proteins such as MBNL1 or MID1.1,11
An unexpected result was that both the recruitment and mobility of HTT exon 1 RNA were independent of CAG repeat length, with a similar behavior of constructs with repeat numbers below (n = 17) and above (n = 49, 72) the pathogenic threshold. This is, however, in agreement with fluorescence in situ hybridization studies reporting mostly small differences in the partitioning of CAG-RNA between the nucleoplasm and nuclear speckles across the pathogenic threshold.10,15,55 Significant differences were only observed for very high repeat expansions (>120) that are only rarely found in patients.58,59 Our results thus suggest that CAG repeat RNA recruitment to nuclear speckles itself may not be directly related to disease.55 However, pathogenic processes may still be induced by elongated repeats in speckles. Furthermore, the repeat threshold for such processes may be increased to higher repeat numbers due to somatic expansion and thus not detected in our experiments.60
Next, we investigated the conformations of CAG-repeat RNA recruited to nuclear speckles. It was expected that RNA may unfold in nuclear speckles due to its increased binding valency in the unfolded state. As such, previous studies found that single-stranded RNA56 and destabilized proteins26 are preferentially recruited by stress granules. Recruitment of the RNA in its unfolded state would lead to a significant decrease in folding stability in the nuclear speckles (e.g., ΔGuθ,37°C(nuclear speckles) < 0) in comparison to the nucleoplasm (can be shown in a thermodynamic cycle). However, at physiological temperature, we observed that folding stabilities were equal in both compartments with the RNAs being mostly folded (>90%), which suggests that no conformational transitions occur upon RNA migration. Heating led to further recruitment of the (CAG)20 RNA to nuclear speckles, increasing the PC, in particular near Tm with strongly increasing fractions of unfolded RNA. Importantly, we could not observe any homotypic interactions (entanglement, self-association) of unfolded (CAG)20 RNA. Thus, in contrast to the hypothesis by Jain and Vale,15 we think that aggregation and gelation of the CAG-RNA (although observed in vitro even with low CAG-repeat expansion) do not occur in cells and nuclear speckles. We suggest that CAG-RNA binding proteins like MBNL11 or MID111 could rather sequester the RNA in nuclear speckles. However, due to the high temperatures applied to the cells, this process could also be caused by non-physiological interactions such as the binding of aberrant temperature-unfolded proteins. Thus, experiments under different cell stresses are needed to further investigate this hypothesis.
The metabolite ATP emerged as a crucial factor to promote unfolding of RNAs [both for (CAG)20 and lm4U*] in the cell, with a CAG-specific function in enabling recruitment of the RNA to and its mobility in nuclear speckles. For the RNAs investigated here, MD simulations and in vitro experiments showed that the effect can be attributed to a direct interaction of the nucleobases with ATP molecules, rather than ATP acting as ‘a fuel’ for RNA-unwinding helicase activity.15 However, on the cellular level, it is important to note that the results do not exclude synergistic unfolding by helicases or unfolding by ATP-dependent RNA-binding proteins.
The function of ATP acting as a cosolute to maintain biomolecular processes in cells could explain why the cellular ATP concentration (5–10 mM)46 exceeds the level required to sustain energy-consuming processes by a factor of up to 1000.25,42,43,61 In line with this hypothesis, previous studies showed that elevated ATP levels could increase protein solubility and maintain the liquid-like properties of nucleoli,62−64 stress granules,64,65 and, in general, the cytoplasm of eukaryotes and prokaryotes.42,61,64,66,67
In this study, similar trends for folding stability in the presence of ATP were observed for both (CAG)20 and lm4U* RNA. However, quantitatively, the effects on (CAG)20 were 5-fold larger, despite the two constructs having a similar number of base pairs. This could be explained by the profound mismatches in the (CAG)20 hairpin which provide more space for ATP to interact with the RNA’s nucleobases. lm4U* does not contain such mismatches as it consists of longer stretches of dsRNA. Especially at low concentrations, ATP mainly interacts with the cytosine and adenine nucleobases, further corroborating this assumption. A remarkable fact is that in the case of lm4U*, the in-cell folding stability can be matched by cell-mimicking crowding agents, whereas for (CAG)20, ATP is additionally required. These findings suggest that ATP-induced destabilization occurs also in other hairpins. However, further studies are required to test this hypothesis and to investigate possible biological implications. Due to their comparable molecular composition,42 other nucleoside phosphates such as adenosine diphosphate, guanosine triphosphate, or cytidine 5′-triphosphate are expected to show similar effects, but their impact is assumed to be smaller due to their lower concentration in cells.
Conclusions
The recruitment of RNA to nuclear speckles is specific to CAG-repeat RNA. Although the CAG hairpin is largely destabilized in cells, it remains folded in the cytoplasm, the nucleoplasm, and in nuclear speckles under physiological conditions. The metabolite ATP is crucial to destabilize the hairpin and maintain its mobility and nuclear speckle association by direct nucleobase interactions between ATP and RNA. Since mitochondrial function is affected in HD disease progression, leading to a decline in cellular ATP levels,68,69 the changes in CAG-RNA homeostasis could be linked to disease pathology. We hypothesize that the stabilized folded states of CAG-repeat RNA under ATP depletion could preferentially bind to transcription and translation factors such as MBNL1 and MID1, resulting in an upregulation of the expression of mutant HTT protein. In a self-amplifying mechanism, mutant HTT could enhance mitochondrial dysfunction, leading to a further decline in ATP levels. In fact, this mechanism would be most detrimental to striatal neurons (involved in HD disease pathology) with a high ATP content as these are most affected by decreasing ATP levels.46,68,69
Acknowledgments
We thank Oliver Brylski, Paolo Carloni, David Gnutt, Sibylle Krauß, and Giulia Rossetti for helpful discussions. We furthermore acknowledge Sibylle Krauß for providing us the plasmids used for transcribing the HTT exon 1 RNA sequences. S.E. acknowledges support from the German Research Foundation DFG-SPP 2191 “Molecular Mechanisms of Functional Phase Separation” (project numbers 402723784 and 419071406). K.W. was supported by grants from the Swiss National Science Foundation (project numbers SNF 31003A_179275 and CRSII5_193740). A.S. acknowledges support from the German Research Foundation under GRK2450 and from the Helmholtz Association Initiative and Networking Fund (project no. ZT-I-0003). A.S. also gratefully acknowledges the Gauss Centre for Supercomputing e.V. (www.gauss-centre.eu) for funding this project by providing computing time through the John von Neumann Institute for Computing (NIC) on the GCS Supercomputer JUWELS at the Jülich Supercomputing Centre (JSC).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c13653.
Experimental procedures; sample sizes for post-hoc Tukey tests; fitting results; variation of the ATP concentration and position; summary of the average number of hydrogen bonds; sequences of RNA constructs; sequences of primers; number, size, and cumulative relative intensity of the foci; concentration dependency, results from single-molecule FRET experiments; secondary structures; acceptor fluorescence; local PCs and the PC ratio; background-corrected acceptor fluorescence intensity curves and D/A intensities; aggregation; relative change in the ATP concentration; final structures; torsion wheels; and RMSD (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Wojciechowska M.; Krzyzosiak W. J. CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol. 2011, 8, 565–571. 10.4161/rna.8.4.15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos R. A. Huntington’s disease: a clinical review. Orphanet J. Rare Dis. 2010, 5, 40–47. 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mezer M.; Wojciechowska M.; Napierala M.; Sobczak K.; Krzyzosiak W. J. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011, 39, 3852–3863. 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar S.; van der Gaag B.; Cortese F. A. B. RNAi mechanisms in Huntington’s disease therapy: siRNA versus shRNA. Transl. Neurodegener. 2017, 6, 30. 10.1186/s40035-017-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata A.; Nakamori M.; Nakatani K. Modulating RNA secondary and tertiary structures by mismatch binding ligands. Methods 2019, 167, 78–91. 10.1016/j.ymeth.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Wagner-Griffin S.; Abe M.; Benhamou R. I.; Angelbello A. J.; Vishnu K.; Chen J. L.; Childs-Disney J. L.; Disney M. D. A Druglike Small Molecule that Targets r(CCUG) Repeats in Myotonic Dystrophy Type 2 Facilitates Degradation by RNA Quality Control Pathways. J. Med. Chem. 2021, 64, 8474–8485. 10.1021/acs.jmedchem.1c00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J. A.; Aikawa H.; Fuerst R.; Li Y.; Ursu A.; Meyer S. M.; Benhamou R. I.; Chen J. L.; Khan T.; Wagner-Griffin S.; van Meter M. J.; Tong Y.; Olafson H.; McKee K. K.; Childs-Disney J. L.; Gendron T. F.; Zhang Y.; Coyne A. N.; Wang E. T.; Yildirim I.; Wang K. W.; Petrucelli L.; Rothstein J. D.; Disney M. D. Ribonuclease recruitment using a small molecule reduced c9ALS/FTD r(G4C2) repeat expansion in vitro and in vivo ALS models. Sci. Transl. Med. 2021, 13, eabd5991 10.1126/scitranslmed.abd5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesiolka A.; Jazurek M.; Drazkowska K.; Krzyzosiak W. J. Structural Characteristics of Simple RNA Repeats Associated with Disease and their Deleterious Protein Interactions. Front. Cell. Neurosci. 2017, 11, 97. 10.3389/fncel.2017.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busan S.; Weeks K. M. Role of context in RNA structure: flanking sequences reconfigure CAG motif folding in huntingtin exon 1 transcripts. Biochemistry 2013, 52, 8219–8225. 10.1021/bi401129r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M.; Krzyzosiak W. J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011, 20, 3811–3821. 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauß S.; Griesche N.; Jastrzebska E.; Chen C.; Rutschow D.; Achmüller C.; Dorn S.; Boesch S. M.; Lalowski M.; Wanker E.; Schneider R.; Schweiger S. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat. Commun. 2013, 4, 1511. 10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- Galganski L.; Urbanek M. O.; Krzyzosiak W. J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L.; Lamond A. I.. Nuclear Speckles Cold Spring Harbor Perspectives in Biology; Cold Spring Harbor Lab, 2011; Vol. 3 ( (2), ), p a00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. E.; Regev O. Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res. 2021, 49, 636–645. 10.1093/nar/gkaa1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.; Vale R. D. RNA phase transitions in repeat expansion disorders. Nature 2017, 546, 243–247. 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T.; Hori N.; Thirumalai D. Condensates in RNA repeat sequences are heterogeneously organized and exhibit reptation dynamics. Nat. Chem. 2022, 14, 775–785. 10.1038/s41557-022-00934-z. [DOI] [PubMed] [Google Scholar]
- Fiszer A.; Wroblewska J. P.; Nowak B. M.; Krzyzosiak W. J. Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells. Genes 2016, 7, 132. 10.3390/genes7120132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E.; Jones T. R.; Lamprecht M. R.; Clarke C.; Kang I. H.; Friman O.; Guertin D. A.; Chang J. H.; Lindquist R. A.; Moffat J.; Golland P.; Sabatini D. M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L.; Jones T. R.; Fraser A.; Bray M.-A.; Logan D. J.; Madden K. L.; Ljosa V.; Rueden C.; Eliceiri K. W.; Carpenter A. E. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 2011, 27, 1179–1180. 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. R.; Swain-Bowden M. J.; Lucas A. M.; Carpenter A. E.; Cimini B. A.; Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinf. 2021, 22, 433. 10.1186/s12859-021-04344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C.; Goodman A.; Chernyshev V.; Kamentsky L.; Cimini B. A.; Karhohs K. W.; Doan M.; Ding L.; Rafelski S. M.; Thirstrup D.; Wiegraebe W.; Singh S.; Becker T.; Caicedo J. C.; Carpenter A. E. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbinghaus S.; Dhar A.; McDonald J. D.; Gruebele M. Protein folding stability and dynamics imaged in a living cell. Nat. Methods 2010, 7, 319–323. 10.1038/nmeth.1435. [DOI] [PubMed] [Google Scholar]
- Dhar A.; Gruebele M. Fast Relaxation Imaging in Living Cells. Curr. Protoc. Protein Sci. 2011, 65, 28.1.1–28.1.19. 10.1002/0471140864.ps2801s65. [DOI] [PubMed] [Google Scholar]
- Guo M.; Xu Y.; Gruebele M. Temperature dependence of protein folding kinetics in living cells. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 17863–17867. 10.1073/pnas.1201797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnutt D.; Timr S.; Ahlers J.; König B.; Manderfeld E.; Heyden M.; Sterpone F.; Ebbinghaus S. Stability Effect of Quinary Interactions Reversed by Single Point Mutations. J. Am. Chem. Soc. 2019, 141, 4660–4669. 10.1021/jacs.8b13025. [DOI] [PubMed] [Google Scholar]
- Samanta N.; Ribeiro S. S.; Becker M.; Laborie E.; Pollak R.; Timr S.; Sterpone F.; Ebbinghaus S. Sequestration of Proteins in Stress Granules Relies on the In-Cell but Not the In Vitro Folding Stability. J. Am. Chem. Soc. 2021, 143, 19909–19918. 10.1021/jacs.1c09589. [DOI] [PubMed] [Google Scholar]
- Büning S.; Sharma A.; Vachharajani S.; Newcombe E.; Ormsby A.; Gao M.; Gnutt D.; Vöpel T.; Hatters D. M.; Ebbinghaus S. Conformational dynamics and self-association of intrinsically disordered Huntingtin exon 1 in cells. Phys. Chem. Chem. Phys. 2017, 19, 10738–10747. 10.1039/c6cp08167c. [DOI] [PubMed] [Google Scholar]
- Vöpel T.; Bravo-Rodriguez K.; Mittal S.; Vachharajani S.; Gnutt D.; Sharma A.; Steinhof A.; Fatoba O.; Ellrichmann G.; Nshanian M.; Heid C.; Loo J. A.; Klärner F.-G.; Schrader T.; Bitan G.; Wanker E. E.; Ebbinghaus S.; Sanchez-Garcia E. Inhibition of Huntingtin Exon-1 Aggregation by the Molecular Tweezer CLR01. J. Am. Chem. Soc. 2017, 139, 5640–5643. 10.1021/jacs.6b11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M.; Gnutt D.; Orban A.; Appel B.; Righetti F.; Winter R.; Narberhaus F.; Müller S.; Ebbinghaus S. RNA Hairpin Folding in the Crowded Cell. Angew. Chem., Int. Ed. 2016, 55, 3224–3228. 10.1002/anie.201510847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautke A. C.; Ebbinghaus S. Folding Stability and Self-Association of a Triplet-Repeat (CAG) 20 RNA Hairpin in Cytomimetic Media. ChemSystemsChem 2020, 3, 28. 10.1002/syst.202000052. [DOI] [Google Scholar]
- Sobczak K.; Michlewski G.; de Mezer M.; Kierzek E.; Krol J.; Olejniczak M.; Kierzek R.; Krzyzosiak W. J. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010, 285, 12755–12764. 10.1074/jbc.m109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D.; Draper D. E. Denaturation of RNA secondary and tertiary structure by urea: simple unfolded state models and free energy parameters account for measured m-values. Biochemistry 2012, 51, 9014–9026. 10.1021/bi301103j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S.; Karimata H.; Ohmichi T.; Kawakami J.; Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004, 126, 14330–14331. 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- Takahashi S.; Sugimoto N. Stability prediction of canonical and non-canonical structures of nucleic acids in various molecular environments and cells. Chem. Soc. Rev. 2020, 49, 8439–8468. 10.1039/d0cs00594k. [DOI] [PubMed] [Google Scholar]
- Girdhar K.; Scott G.; Chemla Y. R.; Gruebele M. Better biomolecule thermodynamics from kinetics. J. Chem. Phys. 2011, 135, 015102. 10.1063/1.3607605. [DOI] [PubMed] [Google Scholar]
- Rinnenthal J.; Klinkert B.; Narberhaus F.; Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010, 38, 3834–3847. 10.1093/nar/gkq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T.; Heidrich N.; Brantl S.; Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007, 65, 413–424. 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- Fu R.; Xu B.; Li D. Study of the temperature field in microchannels of a PDMS chip with embedded local heater using temperature-dependent fluorescent dye. Int. J. Therm. Sci. 2006, 45, 841–847. 10.1016/j.ijthermalsci.2005.11.009. [DOI] [Google Scholar]
- Ross D.; Gaitan M.; Locascio L. E. Temperature measurement in microfluidic systems using a temperature-dependent fluorescent dye. Anal. Chem. 2001, 73, 4117–4123. 10.1021/ac010370l. [DOI] [PubMed] [Google Scholar]
- Samy R.; Glawdel T.; Ren C. L. Method for microfluidic whole-chip temperature measurement using thin-film poly(dimethylsiloxane)/rhodamine B. Anal. Chem. 2008, 80, 369–375. 10.1021/ac071268c. [DOI] [PubMed] [Google Scholar]
- Brylski O.; Shrestha P.; House P. J.; Gnutt P.; Mueller J. W.; Ebbinghaus S. Disease-related protein variants of the highly conserved enzyme PAPSS2 show marginal stability and aggregation in cells. Front. Mol. Biosci. 2022, 9, 860387. 10.3389/fmolb.2022.860387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehringer J.; Do T.-M.; Touraud D.; Hohenschutz M.; Khoshsima A.; Horinek D.; Kunz W. Hofmeister versus Neuberg: is ATP really a biological hydrotrope?. Cell Rep. Phys. Sci. 2021, 2, 100343. 10.1016/j.xcrp.2021.100343. [DOI] [Google Scholar]
- Brylski O.; Shrestha P.; Gnutt P.; Gnutt D.; Mueller J. W.; Ebbinghaus S. Cellular ATP Levels Determine the Stability of a Nucleotide Kinase. Front. Mol. Biosci. 2021, 8, 790304. 10.3389/fmolb.2021.790304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera I.; Iwasaki T.; Imamura H.; Noji H.; Nagai T. Reversible Dimerization of Aequorea victoria Fluorescent Proteins Increases the Dynamic Range of FRET-Based Indicators. ACS Chem. Biol. 2010, 5, 215–222. 10.1021/cb900263z. [DOI] [PubMed] [Google Scholar]
- Lerchundi R.; Huang N.; Rose C. R. Quantitative Imaging of Changes in Astrocytic and Neuronal Adenosine Triphosphate Using Two Different Variants of ATeam. Front. Cell. Neurosci. 2020, 14, 80. 10.3389/fncel.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut T. W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. 10.1007/bf00928361. [DOI] [PubMed] [Google Scholar]
- Weis K.; Hondele M. The Role of DEAD-Box ATPases in Gene Expression and the Regulation of RNA-Protein Condensates. Annu. Rev. Biochem. 2022, 91, 197–219. 10.1146/annurev-biochem-032620-105429. [DOI] [PubMed] [Google Scholar]
- Rogers G. W.; Richter N. J.; Merrick W. C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999, 274, 12236–12244. 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- Hyeon C.; Thirumalai D. Chain length determines the folding rates of RNA. Biophys. J. 2012, 102, L11–L13. 10.1016/j.bpj.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz B.; Faber M.; Verma A.; Klumpp S.; Schug A. Differences between cotranscriptional and free riboswitch folding. Nucleic Acids Res. 2014, 42, 2687–2696. 10.1093/nar/gkt1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbinghaus S.; Kim S. J.; Heyden M.; Yu X.; Heugen U.; Gruebele M.; Leitner D. M.; Havenith M. An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 20749–20752. 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laage D.; Elsaesser T.; Hynes J. T. Water Dynamics in the Hydration Shells of Biomolecules. Chem. Rev. 2017, 117, 10694–10725. 10.1021/acs.chemrev.6b00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerhut B. P. The mutual interactions of RNA, counterions and water - quantifying the electrostatics at the phosphate-water interface. Chem. Commun. 2021, 57, 12880–12897. 10.1039/d1cc05367a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiel M.; Götz M.; Klein A.; Coquelin D.; Floca R.; Schug A. Dynamic particle swarm optimization of biomolecular simulation parameters with flexible objective functions. Nat. Mach. Intell. 2021, 3, 727–734. 10.1038/s42256-021-00366-3. [DOI] [Google Scholar]
- Urbanek M. O.; Jazurek M.; Switonski P. M.; Figura G.; Krzyzosiak W. J. Nuclear speckles are detention centers for transcripts containing expanded CAG repeats. Biochim. Biophys. Acta 2016, 1862, 1513–1520. 10.1016/j.bbadis.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Guillén-Boixet J.; Kopach A.; Holehouse A. S.; Wittmann S.; Jahnel M.; Schlüßler R.; Kim K.; Trussina I. R. E. A.; Wang J.; Mateju D.; Poser I.; Maharana S.; Ruer-Gruß M.; Richter D.; Zhang X.; Chang Y.-T.; Guck J.; Honigmann A.; Mahamid J.; Hyman A. A.; Pappu R. V.; Alberti S.; Franzmann T. M. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilık İ. A.; Aktaş T. Nuclear speckles: dynamic hubs of gene expression regulation. FEBS J. 2022, 289, 7234–7245. 10.1111/febs.16117. [DOI] [PubMed] [Google Scholar]
- Costa M. d. C.; Magalhães P.; Guimarães L.; Maciel P.; Sequeiros J.; Sousa A. The CAG repeat at the Huntington disease gene in the Portuguese population: insights into its dynamics and to the origin of the mutation. J. Hum. Genet. 2006, 51, 189–195. 10.1007/s10038-005-0343-8. [DOI] [PubMed] [Google Scholar]
- Hećimović S.; Klepac N.; Vlasić J.; Vojta A.; Janko D.; Skarpa-Prpić I.; Canki-Klain N.; Marković D.; Bozikov J.; Relja M.; Pavelić K. Genetic background of Huntington disease in Croatia: Molecular analysis of CAG, CCG, and Delta2642 (E2642del) polymorphisms. Hum. Mutat. 2002, 20, 233. 10.1002/humu.9055. [DOI] [PubMed] [Google Scholar]
- Monckton D. G. The Contribution of Somatic Expansion of the CAG Repeat to Symptomatic Development in Huntington’s Disease: A Historical Perspective. J. Huntingt. Dis. 2021, 10, 7–33. 10.3233/jhd-200429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.; Malinovska L.; Saha S.; Wang J.; Alberti S.; Krishnan Y.; Hyman A. A. ATP as a biological hydrotrope. Science 2017, 356, 753–756. 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- Feric M.; Vaidya N.; Harmon T. S.; Mitrea D. M.; Zhu L.; Richardson T. M.; Kriwacki R. W.; Pappu R. V.; Brangwynne C. P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P.; Mitchison T. J.; Hyman A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 4334–4339. 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y.; Brangwynne C. P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Jain S.; Wheeler J. R.; Walters R. W.; Agrawal A.; Barsic A.; Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry B. R.; Surovtsev I. V.; Cabeen M. T.; O’Hern C. S.; Dufresne E. R.; Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 2014, 156, 183–194. 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder M. C.; Midtvedt D.; Franzmann T.; Nüske E.; Otto O.; Herbig M.; Ulbricht E.; Müller P.; Taubenberger A.; Maharana S.; Malinovska L.; Richter D.; Guck J.; Zaburdaev V.; Alberti S. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 2016, 5, e09347 10.7554/elife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano M.; Galvan L.; Déglon N.; Brouillet E. Mitochondria in Huntington’s disease. Biochim. Biophys. Acta 2010, 1802, 52–61. 10.1016/j.bbadis.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Jodeiri Farshbaf M.; Ghaedi K. Huntington’s Disease Mitochondria. Neurotox. Res. 2017, 32, 518–529. 10.1007/s12640-017-9766-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.