Abstract
Cervical cancer is still an important problem perplexing health management in developing countries. Previous studies have shown that cervical cancer cells show markers of aerobic glycolysis, suggesting that these tumors may secrete lactic acid. Through the biological characterization of lactate gene in tumor and its relationship with immune cells in tumor microenvironment, a lactate scoring system capable of evaluating cancer prognosis was constructed to explore the molecular mechanism of lactate metabolism disorder affecting prognosis. 29 hub genes in this study were differentially expressed in cervical cancer, including 24 genes related to lactate metabolism, LDHA in Lactate dehydrogenase (LDH) group, SLC16A3 in Monocarboxylate transporters (MCT) group and three Histone lactation modification related genes (EP300, ACAT1, ACACA). More importantly, we found that from an epigenetic point of view, histone lactation plays an important role in the pathogenesis and prognosis of cervical cancer. Mainly affect the prognosis of the disease through changes in the infiltration of plasmacytoid Dendritic Cell (pDC) and Central Memory T cell (Tcm) in the tumor immune microenvironment. Lactate inhibition may be a useful tool for anticancer therapy.
Keywords: Cervical cancer, Lactate, Tumor microenvironment, Post-translational modification, Immune responses
1. Introduction
The Tumor microenvironment (TME) is an active niche that shapes tumor pathogenesis and evolution [1]. The TME consists of malignant cells, immune cells, non-cancer cell stromas, fibroblasts, and the vasculature and lymphatics. Because MCTs are symporters, cotransporting lactate anions and protons, the intracellular pH (pHi) in cancer cells tends to be slightly alkaline compared to the extracellular space into which protons are excreted [2,3]. In normal cells, pHi approximates 7.2. However, in cancer cells pHi approximates 7.4 [4], or the pH of normal arterial blood. In contrast, an extracellular pH of 5.5–7.0 is common in cancers [4,5]. Lactate plays a central role in the bioenergetics and self-sufficiency of cancer cells.
Tumor-secreted lactate, a product of increased aerobic glycolysis of cancer cells, is associated with heightened metastatic incidence, increased angiogenesis, is responsible for metabolic reprogramming in adjacent tissues and can induce a pro-inflammatory state. In the study of lactic acid carcinogenesis, the role of oncogenes and mutated tumor suppressors is obviously to reprogram glycolysis for the production of lactic acid, thereby creating a concentration gradient for the exchange of lactic acid within, between cells and cells. The recent study has shown that the steps of the carcinogenic mechanism of lactic acid may be as follows: (1) increase glucose uptake; (2) increase the expression and activity of glycolytic enzymes; (3) decrease mitochondrial function; (4) Increase the production, accumulation and release of lactic acid; (5) Monocarboxylic acid transporters MCT1 and MCT4 are used for up-regulation of lactic acid exchange [6].
High glucose uptake and excessive lactate formation even in the presence of sufficient oxygen, which subsequently was referred to as the Warburg effect, also termed aerobic glycolysis, and remains a hallmark of cancer [7,8]. The high concentration of lactic acid in the tumor microenvironment has a strong immunosuppressive effect and mediates the immune escape of the tumor. Non-malignant lymphocytes or stromal cells, such as tumor-associated macrophages and cancer-associated fibroblasts, contribute to the accumulation of lactic acid in the tumor environment.
In breast cancer cells, the anaplerotic entry of pyruvate via Pyruvate Carboxylase (PC) into the TriCarboxylic Acid (TCA) cycle promoted an invasive phenotype by increasing their motility, although the underlying molecular mechanism remains elusive [[9], [10], [11]].
Lactate is an important carbon-containing metabolite in the glycolysis pathway of cells, and its biological function has been widely studied due to the existence of Warburg effect in tumor cells. As a chiral compound, lactic acid usually exists in the form of three optical isomers, namely D-body, L-body and racemic DL-body. Among them, l-lactate is the human body and the large. The main substances produced by glycolytic metabolism of most mammals increase significantly in pathological conditions such as tumors, sepsis, and autoimmune diseases. The latest research shows that, similar to other fatty acids, l-lactate can also form the corresponding fatty acyl-CoA-L-lactyl-CoA (L-lactyl-CoA). More importantly, It is reported for the first time that lactic acid is an important epigenetic regulatory molecule [12]. Under the action of the “writer” histone acetyltransferase p300 (EP300), histones are lactated and modified. l-lactate regulates the expression of macrophage polarization-related genes during immune activation.
Many processes are complicated and interpretation of data can be difficult. Our view is that dysregulated and chronic lactate exposure can be both cause and effect in cervix carcinogenesis.
2. Materials and methods
2.1. Data collection
The comprehensive analysis of somatic genome alterations were investigated from UCSC Xena (https://xenabrowser.net/datapages/) RNAseq data in Transcripts Per Million (TPM) format of The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and The Genotype-Tissue Expression (GTEx) (https://gtexportal.org/) uniformly processed by the toil process [13]. Through extraction of the CESC (Cervical squamous cell carcinoma and endocervical adenocarcinoma) data in TCGA and corresponding normal tissue data in GTEx, we classified the 306 samples of cervical tumor as malignant group and 3 samples of adjacent to cancer from TCGA together with ten normal cervix tissues from GTEx as no-malignant group. The RNAseq data in TPM format were log2 transformed for expression comparison between samples.
2.2. Select tumor lactate-based Metabolic Related Genes
The top 50 genes related to tumor lactate-based metabolic symbiosis were searched in the GeneCards database (https://www.genecards.org/). In our research, the inclusion criteria was identified with key word “lactate” association sorted by score in descending (Shown as Table 1). In this table, we list Gene symbol, Description of the genes, Functional Category, GeneCards Inferred Functionality Scores (GIFtS), GeneCards Identifiers (GC id) and association Score.
Table 1.
Top 50 tumor lactate-based metabolic related genes.
| Symbol | Description | Category | GIFtS | GC id | Score | |
|---|---|---|---|---|---|---|
| 1 | LDHB | Lactate Dehydrogenase B | Protein Coding | 46 | GC12M021635 | 64.51 |
| 2 | LDHA | Lactate Dehydrogenase A | Protein Coding | 51 | GC11P018394 | 55.6 |
| 3 | DARS2 | Aspartyl-TRNA Synthetase 2, Mitochondrial | Protein Coding | 42 | GC01P173824 | 45.49 |
| 4 | LDHD | Lactate Dehydrogenase D | Protein Coding | 40 | GC16M075111 | 40.12 |
| 5 | SLC16A1 | Solute Carrier Family 16 Member 1 | Protein Coding | 48 | GC01M112932 | 38.5 |
| 6 | LDHC | Lactate Dehydrogenase C | Protein Coding | 43 | GC11P018433 | 33.36 |
| 7 | LDHAL6B | Lactate Dehydrogenase A Like 6 B | Protein Coding | 40 | GC15P059206 | 25.27 |
| 8 | LDHAL6A | Lactate Dehydrogenase A Like 6 A | Protein Coding | 36 | GC11P018456 | 23.39 |
| 9 | EARS2 | Glutamyl-TRNA Synthetase 2, Mitochondrial | Protein Coding | 43 | GC16M023527 | 17.52 |
| 10 | SURF1 | SURF1 Cytochrome C Oxidase Assembly Factor | Protein Coding | 43 | GC09M133351 | 16.57 |
| 11 | MT-ND1 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 1 | Protein Coding | 33 | GCMTP003309 | 15.75 |
| 12 | UEVLD | UEV And Lactate/Malate Dehyrogenase Domains | Protein Coding | 36 | GC11M018554 | 15.62 |
| 13 | MT-ATP6 | Mitochondrially Encoded ATP Synthase Membrane Subunit 6 | Protein Coding | 33 | GCMTP008531 | 14.47 |
| 14 | MT-TL1 | Mitochondrially Encoded TRNA-Leu (UUA/G) 1 | RNA Gene | 16 | GCMTP003232 | 14.18 |
| 15 | POLG | DNA Polymerase Gamma, Catalytic Subunit | Protein Coding | 45 | GC15M089393 | 13.88 |
| 16 | MT-CO3 | Mitochondrially Encoded Cytochrome C Oxidase III | Protein Coding | 31 | GCMTP009209 | 13.67 |
| 17 | SCO2 | Synthesis Of Cytochrome C Oxidase 2 | Protein Coding | 44 | GC22M050523 | 13.66 |
| 18 | PC | Pyruvate Carboxylase | Protein Coding | 47 | GC11M066848 | 13.58 |
| 19 | NDUFS4 | NADH:Ubiquinone Oxidoreductase Subunit S4 | Protein Coding | 44 | GC05P053560 | 13.53 |
| 20 | PRL | Prolactin | Protein Coding | 43 | GC06M022287 | 13.25 |
| 21 | PDP1 | Pyruvate Dehydrogenase Phosphatase Catalytic Subunit 1 | Protein Coding | 46 | GC08P093857 | 12.99 |
| 22 | MT-ND4 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 4 | Protein Coding | 33 | GCMTP010762 | 12.93 |
| 23 | MT-CO2 | Mitochondrially Encoded Cytochrome C Oxidase II | Protein Coding | 34 | GCMTP007587 | 12.93 |
| 24 | MT-ND6 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 6 | Protein Coding | 33 | GCMTM014151 | 12.87 |
| 25 | INS | Insulin | Protein Coding | 47 | GC11M002159 | 12.69 |
| 26 | DGUOK | Deoxyguanosine Kinase | Protein Coding | 45 | GC02P073926 | 12.54 |
| 27 | SDHB | Succinate Dehydrogenase Complex Iron Sulfur Subunit B | Protein Coding | 48 | GC01M017338 | 12.51 |
| 28 | PET100 | PET100 Cytochrome C Oxidase Chaperone | Protein Coding | 30 | GC19P007632 | 12.49 |
| 29 | MT-ND5 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 5 | Protein Coding | 33 | GCMTP012339 | 12.15 |
| 30 | TACO1 | Translational Activator Of Cytochrome C Oxidase I | Protein Coding | 39 | GC17P063600 | 12.08 |
| 31 | LDHAP4 | Lactate Dehydrogenase A Pseudogene 4 | Pseudogene | 6 | GC09M014920 | 12 |
| 32 | FASTKD2 | FAST Kinase Domains 2 | Protein Coding | 39 | GC02P206766 | 11.88 |
| 33 | MT-CO1 | Mitochondrially Encoded Cytochrome C Oxidase I | Protein Coding | 34 | GCMTP005906 | 11.86 |
| 34 | LDHBP2 | Lactate Dehydrogenase B Pseudogene 2 | Pseudogene | 6 | GC0XM076334 | 11.83 |
| 35 | COX5A | Cytochrome C Oxidase Subunit 5 A | Protein Coding | 44 | GC15M074919 | 11.76 |
| 36 | MDH2 | Malate Dehydrogenase 2 | Protein Coding | 48 | GC07P076048 | 11.63 |
| 37 | SLC2A2 | Solute Carrier Family 2 Member 2 | Protein Coding | 49 | GC03M171014 | 11.58 |
| 38 | COX6B1 | Cytochrome C Oxidase Subunit 6B1 | Protein Coding | 43 | GC19P041420 | 11.56 |
| 39 | NDUFV1 | NADH:Ubiquinone Oxidoreductase Core Subunit V1 | Protein Coding | 46 | GC11P067680 | 11.52 |
| 40 | MT-TK | Mitochondrially Encoded TRNA-Lys (AAA/G) | RNA Gene | 15 | GCMTP008297 | 11.37 |
| 41 | TWNK | Twinkle MtDNA Helicase | Protein Coding | 33 | GC10P100993 | 11.35 |
| 42 | LDHAP1 | Lactate Dehydrogenase A Pseudogene 1 | Pseudogene | 5 | GC04M004896 | 11.29 |
| 43 | DLD | Dihydrolipoamide Dehydrogenase | Protein Coding | 50 | GC07P107890 | 11.27 |
| 44 | MT-TW | Mitochondrially Encoded TRNA-Trp (UGA/G) | RNA Gene | 13 | GCMTP005514 | 11.25 |
| 45 | LDHAP3 | Lactate Dehydrogenase A Pseudogene 3 | Pseudogene | 6 | GC02P041819 | 11.16 |
| 46 | LDHAP5 | Lactate Dehydrogenase A Pseudogene 5 | Pseudogene | 6 | GC10M118932 | 11.16 |
| 47 | LDHAL6CP | Lactate Dehydrogenase A Like 6C, Pseudogene | Pseudogene | 6 | GC12P063003 | 11.16 |
| 48 | NDUFA13 | NADH:Ubiquinone Oxidoreductase Subunit A13 | Protein Coding | 44 | GC19P019515 | 11.02 |
| 49 | COX4I1 | Cytochrome C Oxidase Subunit 4I1 | Protein Coding | 46 | GC16P085798 | 10.98 |
| 50 | SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A | Protein Coding | 48 | GC05P000208 | 10.86 |
2.3. Gene expression profiling interactive analysis
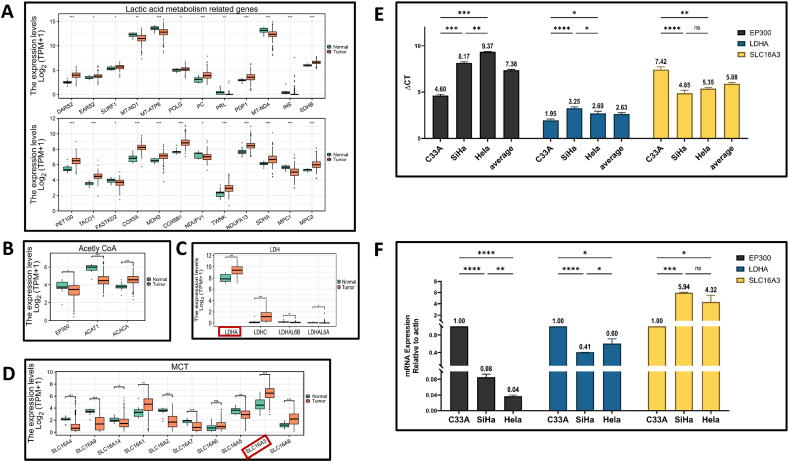
Gene expression in normal and cancerous tissues were compared in GEPIA2 (http://gepia2.cancer-pku.cn/#index). We show the expression according to gene classification by lactic acid metabolism related genes (Fig. 1A), Histone lactation modification related genes (Fig. 1B), LDH (Fig. 1C), MCT (Fig. 1D). Multidimensional cancer genomics informations were accessed from cBioPortal (www.cbioportal.org), select the two datasets of Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Firehose Legacy) and Cervical Squamous Cell Carcinoma (TCGA, PanCancer Atlas). Molecular Profiles include Mutations, Structural variants and Copy number alterations, through the “OncoPrint” module and “Cancer Types Summary” module.
Fig. 1.
Hub genes of Tumor Lactate-based Metabolic Related Genes expression in CESC (RNA-seq TPM of tumor tissue and normal tissue). Difference of all the genes listed in this figure was statistically significant. (Wilcoxon rank sum test Significance mark. Ns: p ≥ 0.05, *: p < 0.05, **: p < 0.01; ***: p < 0.001). (A) Lactic acid metabolism related genes; (B) Histone lactation modification related genes; (C) Lactate dehydrogenase; (D) Monocarboxylic acid transporter; (E)–(F) EP300, LDHA and SLC16A expression in three different cell lines were examined using reverse transcription-quantitative PCR. All PCR data were calculated relative to β-actin and represent the average ± SD of triplicate samples.
2.4. Differentially expressed genes (DEGs)
In CESC, 309 samples are divided into high and low expression groups according to EP300 expression from low to high, take the first 50% as low expression group, the last 50% as high expression group. The total number of ID after removing null values is 38,095, of which 1740 ID meet the threshold of |log2FC|>1 and P < 0.05. Under this threshold, there are 902 high expression (logFC is positive) and 838 low expression (logFC is negative).
2.5. Translational expression signature
Transcriptional Regulatory Relationships Unraveled by Sentence-based Text Minig (TRRUST v2) (https://www.grnpedia.org/trrust/) was used to study the transcriptional regulation involved in CESC. Submit all the 29 significant genes in Fig. 1. Valid query group contains 26 genes: DARS2, EARS2, SURF1, POLG, PC, PRL, PDP1, INS, SDHB, PET100, TACO1, FASTKD2, COX5A, MDH2, COX6B1, NDUFV1, TWNK, NDUFA13, SDHA, MPC1, MPC2, EP300, ACAT1, ACACA, LDHA, SLC16A3; Invalid query group contains 3 genes: MT-ND1, MT-ATP6, MT-ND4; Query genes included in TRRUST contains 8 genes: EP300, LDHA, SURF1, SDHB, ACACA, PRL, PC, NDUFV1.
2.6. Tumor immune microenvironment characteristics
Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in CESC. In our study 24 kinds immune cells contain: activated DC (aDC), B cells, CD8 T cells, Cytotoxic cells, DC, Eosinophils, immature DC (iDC), Macrophages, Mast cells, Neutrophils, NK CD56bright cells, NK CD56dim cells, NK cells, plasmacytoid DC (pDC), T cells, T helper cells; T central memory (Tcm), T effector memory (Tem), T follicular helper (Tfh), T gamma delta (Tgd), Th1 cells, Th17 cells, Th2 cells, Treg. Research in ESTIMATE (https://bioinformatics.mdanderson.org/estimate/index.html) with scores for cervical cancer tumor purity, the level of stromal cells present, and the infiltration level of immune cells in tumor tissues based on expression data. This website is designed to view and download stromal, immune, and ESTIMATE scores for each sample across all TCGA tumor types and platforms.
2.7. Gene ontology (GO) term and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis and gene set enrichment analysis (GSEA)
29 hub genes GO and KEGG enrichment analysis, the functions of genes are divided into three categories: biological process (BP), cellular component (CC), and molecular function (MF). Using the GO database, we can get the relationship between the target gene at the three levels of CC, MF and BP. A total of 228 IDs were entered, of which, 225 Entre IDs were successfully converted, and the conversion ratio was 98.7%, under the conditions of P adj<0.05 and False Discovery Rate (FDR) < 0.2, there are 240 items for BP, 29 items for CC, 20 items for MF, and 10 items for KEGG.
EP300 GSEA using the curated hallmark gene set collection from the gene set (c2. cp.v7.2. symbols.gmt) from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). A total of 499 data sets satisfy with p adjust<0.05 and FDR (q value) < 0.25.
2.8. Survival analysis
CESC patient overall survival was downloaded from the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?). We aim to extend these findings through Kaplan–Meier and Multivariate Cox regression analysis of gene expression signatures with immune-oncology to find risk factors and nomograms for predicting disease progression.
2.9. Reverse transcription-quantitative PCR (RT-qPCR)
The MecDNA-HUtrC007Ce01 commercial chip (cat. No. 8*R100-M-20190104; Shanghai Outdo Biotech Co., Ltd.) contains cDNA reverse transcribed from RNA extracted from three cervical cancer cell lines: C33A, SiHa and HeLa. According to the manufacturer's instructions, qPCR was performed using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; RR820Q; Takara Bio, Inc.). Briefly, following the addition of 20 μl qPCR MasterMix into each well, the Axigen PlateMax Ultraclear Sealing Film (UC-500) was used to seal the chip, and it was placed on ice for 15 min to fully dissolve the freeze-dried cDNA. The chip was then centrifuged at 1750×g for 3 min at a temperature ramping rate of 2 °C/sec. qPCR was performed using a Roche LightCycler® 480II (Roche Diagnostics) with the following program: Initial denaturation (95 °C, 30 s); 40 cycles of denaturation (95 °C, 5 s), annealing (60 °C, 30 s) and elongation (95 °C, 5 s); final elongation (60 °C, 1 min) and a final hold (60 °C). The fold-change of gene expression was calculated using 2-(ΔCq experimental group-ΔCq control group). β-Actin was used as an internal control and primers are as follows: EP300 forward, 5′-CGAGGAGGAAGAGGTTGATGG-3'; EP300 reverse, 5′-AGTGGCTGGAGAGGGATGC-3'; LDHA forward, 5′-TCAGCCCGATTCCGTTACCT-3'; LDHA reverse, 5′-CCAGCAACATTCATTCCACTCC-3' (product length, 125 bp); SLC16A3 forward, 5′-CCTACTCCGTCTACCTCTTCA-3′, SLC16A3 reverse, 5′-CCACCGCCTCCATCAGCAG-3' (product length, 222 bp); human β-actin forward, 5′-GAAGAGCTACGAGCTGCCTGA-3'; human β-actin reverse, 5′-CAGACAGCACTGTGTTGGCG-3' (product length, 191 bp).
2.10. Statistical analysis
All analyses were performed in R version 3.6.3. Differentially expressed genes (DEGs) were identified using R package of mainly ggplot2 version 3.3.3. Chose the ssGSEA (GSVA package built-in algorithm) in immune infiltration algorithm [14] using R package of GSVA package version 1.34.0 [15]. ClusterProfiler package [16] was used for enrichment analysis and visualization. ID conversion were identified using R package of Org. Hs.eg.db package version 3.10.0. GSEA analysis use R package of mainly clusterProfiler package version 3.14.3 [16,17]. Survival data was acquisited by COX and survminer package version 0.4.9 and survival package version 3.2.10.
3. Results
3.1. Tumor lactate-based metabolic landscape
Lactate-producing cancer cells are characterized by increased aerobic glycolysis and excessive lactate formation, a phenomenon “Warburg effect” described by Otto Warburg 93 years ago [18]. The Warburg effect, describing heightened aerobic glycolysis in tumors, played a key role in launching the field of cancer metabolism. In Table 1, through the later GEPIA verification, the blank background includes six pseudogenes and 16 genes with no significant difference between tumor tissue and normal tissue in cervical cancer. Blue background represents lactate dehydrogenase family genes. Green background genes belong to Lactate transporters. Twenty-five pink background genes are lactate metabolism related genes. In the cytochrome c oxidase subunit group, due to the lowest correlation of COX4I1, only COX5A and COX6B1 are retained. Therefore, 24 lactate metabolism related genes with pink background are retained, as shown in Fig. 1A, among which 16 of them expressed up-regulated and 8 expressed down-regulated in cervical cancer tissues.
3.1.1. Divergent roles of mitochondrial function in cancer
Accelerated glycolysis caused by oncogenes and tumor suppressor mutations will deplete nicotinamide adenine dinucleotide (NAD+). The reduction of pyruvate to lactic acid supplements the level of NAD+ in the cytoplasm and regulates the balance of the cytoplasmic redox pair (NADH/NAD+) to continue glycolysis. Lactic acid enters the mitochondria and oxidized to pyruvate and then acetyl CoA (A-CoA) through the mitochondrial lactate oxidation complex (mLOC), which is composed of mitochondrial monocarboxylic acid transporter (mMCT), its stabilizer CD147, LDH and cytochrome oxidase (COx). Pyruvate can also enter the mitochondria through the mitochondrial pyruvate carrier (MPC) and be oxidized to A-CoA [6]. Based on the above reported literature, we studied Mitochondrial pyruvate carrier 1 (MPC1) and MPC2 (Fig. 1A) and the other nine monocarboxylate transporters were added (Fig. 1D). ALL these genes did not appear in Table 1. In MCT group, we chose SLC16A3 as hub gene because of its highest expression level in cervical cancer. Similarly, LDHA was selected as hub gene in LDH group (Fig. 1C). EP300 (E1A Binding Protein P300) functions as histone acetyltransferase that regulates transcription [[19], [20], [21]]. Acetylates all four core histones in nucleosomes. Histone acetylation gives an epigenetic tag for transcriptional activation [[22], [23], [24]]. In order to obtain the quantitative analysis results of the key lactic acid metabolism genes in three different cervical cancer cells, we used the cervical cancer cell line chip (C33A, SiHa, HeLa) for RT-PCR detection. According to the average value of ΔCT, the gene expression levels of the three cells are as follows: LDHA > SLC16A3>EP300, which is basically consistent with the results of bioinformatics analysis (Fig. 1E). The expression of EP300 and LDHA in SiHa and HeLa cells was lower than C33A, but the expression level of SLC16A3 was higher than C33A, and there was no statistically significant difference between HPV-positive SiHa and HeLa cells (Fig. 1F).
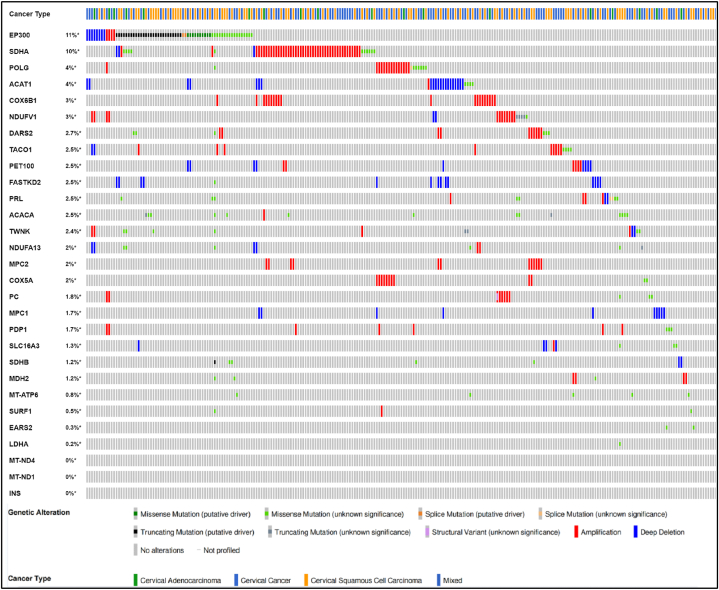
ACAT1 (Acetyl-CoA Acetyltransferase 1) is one of the enzymes that catalyzes the last step of the mitochondrial beta-oxidation pathway, an aerobic process breaking down fatty acids into acetyl-CoA [[25], [26], [27]]. ACACA (Acetyl-CoA Carboxylase Alpha) catalyzes the carboxylation of acetyl-CoA, the first and rate-limiting step of de novo fatty acid biosynthesis [[28], [29], [30]]. By converting pyruvate to acetyl-CoA (AcCoA), the pyruvate dehydrogenase (PDH) complex (PDC) links glycolysis and the TCA cycle [31]. Therefore, we classified the key enzymes ACAT1 and ACACA in the metabolic pathway of acetyl CoA together with EP300 as histone lactation modification related genes. Finally we selected 29 genes as hub genes in our research, as shown in Fig. 2, with the type and frequency of genomic changes in CESC.
Fig. 2.
The genomic alteration types and alteration frequency of association genes in CESC were analyzed through the “OncoPrint” module and “Cancer Types Summary” module. We chose two cervical squamous cell carcinoma datasets with 607 cases for further analysis by using cBioPortal (http://www.cbioportal.org/). (Ranked by genetic alteration percentage).
3.1.2. Lactate dehydrogenase
LDHA is responsible for the conversion of pyruvate into lactate and NAD+, whereas LDHB converts lactate into pyruvate, fueling oxidative metabolism. LDHA express higher in CESC tumor tissue, while LDHB expression has no statistically significant in cervical cancer (Fig. 1C and Table 1). Genetic alteration of LDHA is low in CESC, which is account about only 0.2% (Fig. 2).
3.1.3. Lactate transporters in cancer
Lactate is transported into and out of cells by 14 members of the of monocarboxylate transporters (MCTs) family with different isoforms (MCT1–4). MCT1 (also known as SLC16A1), MCT2 (also known as SLC16A7), MCT3 (also known as SLC16A8), and MCT4 (also known as SLC16A3), and two sodium-coupled lactate cotransporters (SLC5A12, SLC5A8) have been described [32]. RNA-seq expression increased in tumor tissue are SLC16A1 (P < 0.01), SLC16A3 (P < 0.001), SLCA16A8 (P < 0.001), among which the highest expression in tumor tissues is SLC16A3. Decreased expression in tumor are SLC16A4 (P < 0.001), SLC16A9 (P < 0.001), SLC16A14 (P < 0.05), SLC16A2 (P < 0.001), SLC16A7 (P < 0.001), SLC16A5 (P < 0.01) (Fig. 1D). The second highest expression in cervical cancer is MCT1 (SLC16A1) (P < 0.01). SLC16A3 genetic alteration in CESC is 1.3%, including Missense Mutation (unknown significance), Amplification, and Deep Deletion (Fig. 2). MCT4 is associated with glycolytic metabolism and poor-prognosis cancer [33].
Query genes included in TRRUST contains eight, seven of them are statistically significant (Table 2, P < 0.01): EP300, LDHA, SURF1, SDHB, ACACA, PRL and PC. Key factors (P < 0.01) are MYC, HIF1A, CREB1, AR. MYC regulates three of these genes, ranking first, indicating that MYC is a key transcription factor regulating lactic acid metabolism. MYC family has been shown to increase cancer cell glycolysis and lead to lactate accumulation within the tumor microenvironment [34]. The second important transcription factor is HIF1A. Cancers with activating mTOR mutations accumulate high HIF1 levels due to increased HIF1A transcription and translation [35]. MYC cooperates with HIF-1 in activating lactate dehydrogenase-A (LDHA) that encode glycolytic proteins.
Table 2.
Key regulated factor of Lactic acid metabolism biomarker in CESC.
| Key Factor | Description | Regulated gene | P value | FDR | |
|---|---|---|---|---|---|
| 1 | MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | EP300/LDHA↑/SURF1↑ | 0.000305 | 0.00153 |
| 2 | HIF1A | hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | LDHA↑/SDHB↓ | 0.00537 | 0.00837 |
| 3 | CREB1 | cAMP responsive element binding protein 1 | ACACA↑/PRL | 0.00628 | 0.00837 |
| 4 | AR | androgen receptor | EP300↑/PC↑ | 0.0067 | 0.00837 |
| 5 | SP1 | Sp1 transcription factor | LDHA↑/NDUFV1↑ | 0.129 | 0.129 |
3.2. Lactate shuttle for carcinogenesis
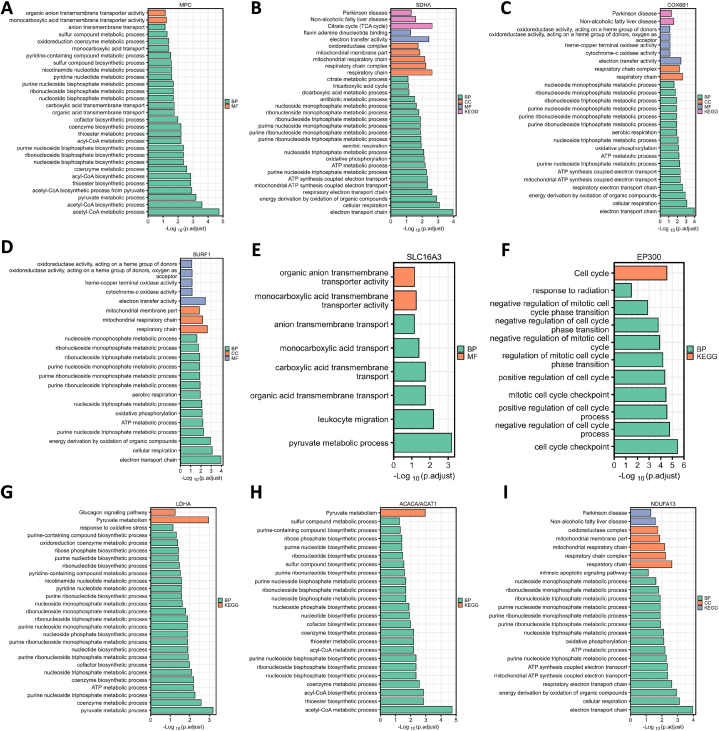
GO-KEGG enrichment of 29 hub genes in Fig. 3, we only retention the nine significant genes (P < 0.05): MPC BP-acetly-CoA metabolic process, MF-monocarboxylic acid transmembrane transporter activity (Fig. 3A); SDHA-KEGG-TCA cycle, MF-electron transfer activity, CC-respiratory chain, BP-electron transport chain (Fig. 3B); COX6B1 KEGG-non-alcoholic fatty liver disease, MF-electron transfer activity, CC-respiratory chain, BP-electron transport chain (Fig. 3C); SURF1 MF-electron transfer activity, CC-respiratory chain, BP-electron transport chain (Fig. 3D); SLC16A3 MF-monocarboxylic acid transmembrane transporter activity, BP-pyruvate metabolic process (Fig. 3E); EP300 KEGG-cell cycle, BP-cell cycle checkpoint (Fig. 3F); LDHA KEGG-pyruvate metabolism, BP-pyruvate metabolic process (Fig. 3G); ACAT1/ACACA KEGG-pyruvate metabolism, BP-acetyl-CoA metabolic process (Fig. 3H); NDUFA13 KEGG-no-alcoholic fatty liver disease, CC-respiratory chain, BP-electron transport chain (Fig. 3I). Among the nine significant genes, only EP300 and ACAT1 decreased in tumor tissues. However, the expression of ACACA is indeed increased in cervical cancer tissues. Therefore, we speculate whether there is an effect of protein post-translational modification on carcinogenic mechanism in the central law of gene transcription. Increased lactic acid content, also defined as lactagenesis, caused by genetic mutations is the cause and purpose of the Warburg effect, and lactic acid metabolism and signal transduction disorders are the key factors for carcinogenesis [18]. Lactate stimulates release of vascular endothelial growth factor (VEGF) for wound healing and repair [36,37]. In cancer, lactic acid stimulates the expression of VEGF protein in endothelial cells in a concentration-dependent manner [[38], [39], [40], [41]]. When LDHA inhibitor oxamate is used to block lactic acid production, angiogenesis is greatly reduced [42]. LDH knockout can inhibit cancer cell proliferation [43,44], and targeted inhibition of MCT can block the transport of lactic acid across the cancer cell membrane and reduce angiogenesis and cell migration [[45], [46], [47]].
Fig. 3.
GO-KEGG cluster analysis. The X axis represents “- log (adj. P)”, the greater the value, the stronger the significance. The Y axis represents GO term. Each color represents an enrichment, including (BP, CC, MF, KEGG). Here list (A) MPC; (B) SDHA; (C) COX6B1; (D) SURF1; (E) SLC16A3; (F) EP300; (G) LDHA; (H) ACACA/ACAT1; (I) NDUFA13. (R package: mainly ggplot2 package).
3.3. Lactate modulation of immune responses in tumor microenvironments
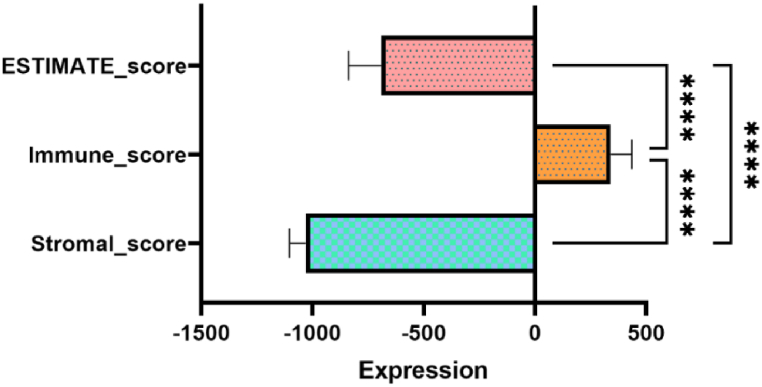
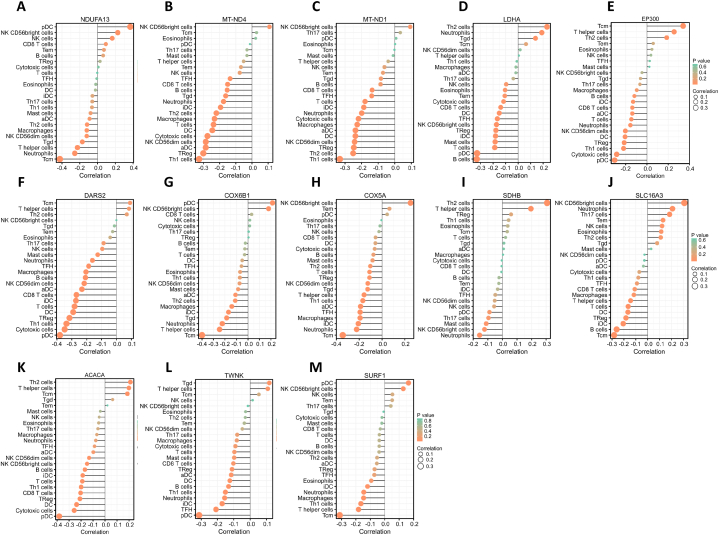
We found that the expression of immune-related genes and stromal related genes increased as the immune score and stromal score (Fig. 4) increased, respectively. Tumor purity is related to the level of stromal cells, which is opposite to the infiltration level of immune cells in tumor tissues. These results suggest that the immune and stromal scores can be used to evaluate the expression of genes related to immune activity and stromal activity, respectively. The correlation between 29 hub genes and 24 immune cells infiltration in cervical cancer samples was analyzed in our research. Sixteen genes with Spearman degree less than 0.3 were excluded, and the infiltration of the remaining 13 genes in cervical cancer samples with immune cells are shown in Table 3 and Fig. 5. The microenvironment of tumor tissue has the characteristics of immunosuppression and tumorigenicity. Through the previous GO-KEGG analysis and discussion, the lactic acid produced by cancer cells can be further secreted into the extracellular space, and play a key role in promoting cancer progression by creating an active niche to shape the pathogenesis and evolution of tumors. This study found that pDC and Tcm were related to most lactate metabolism related genes in the microenvironment of cervical cancer. Lactic acid is a bridge between metabolism, immunity and cancer.
Fig. 4.
Expression of Tumor purity, Immune activity and Stromal-related genes in cervical cancer (****, P < 0.001).
Table 3.
Immune Infiltration of Lactic acid metabolism related genes in CESC.
| EP300 | SLC16A3 | ACACA | LDHA | COX5A | COX6B1 | DARS2 | MT-ND1 | MT-ND4 | NDUFA13 | SDHB | SURF1 | TWNK | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aDC | −0.138* | −0.036 | −0.098 | −0.019 | −0.188*** | −0.109 | −0.232*** | −0.237*** | −0.286*** | −0.089 | 0.008 | −0.055 | −0.115* |
| B cells | −0.117* | −0.261*** | −0.168** | −0.330*** | −0.075 | −0.024 | −0.208*** | −0.085 | −0.153** | 0.063 | −0.023 | −0.038 | −0.131* |
| CD8 T cells | −0.137* | −0.109 | −0.204*** | −0.138* | −0.057 | 0.031 | −0.269*** | −0.136* | −0.150** | 0.094 | −0.005 | −0.031 | −0.102 |
| Cytotoxic cells | −0.284*** | −0.071 | −0.257*** | −0.116* | −0.065 | 0.015 | −0.348*** | −0.219*** | −0.275*** | 0.009 | −0.004 | −0.019 | −0.090 |
| DC | −0.205*** | −0.166** | −0.238*** | −0.151** | −0.059 | −0.051 | −0.291*** | −0.237*** | −0.245*** | −0.024 | −0.006 | −0.038 | −0.124* |
| Eosinophils | 0.047 | 0.114* | −0.053 | −0.104 | −0.005 | −0.069 | −0.041 | 0.002 | 0.022 | −0.019 | 0.036 | −0.093 | −0.028 |
| iDC | −0.130* | −0.205*** | −0.186 | −0.180** | −0.208*** | −0.168** | −0.273*** | −0.186** | −0.194*** | −0.057 | −0.033 | −0.119* | −0.171** |
| Macrophages | −0.095 | −0.116* | −0.069 | −0.018 | −0.192*** | −0.140* | −0.205*** | −0.227*** | −0.231*** | −0.121* | 0.004 | −0.147* | −0.081 |
| Mast cells | 0.024 | 0.029 | −0.040 | −0.184** | −0.083 | −0.087 | −0.125* | −0.009 | −0.031 | −0.078 | −0.111 | −0.020 | −0.098 |
| Neutrophils | −0.155** | 0.208*** | −0.077 | 0.189*** | −0.222*** | −0.224*** | −0.161** | −0.200*** | −0.175 | −0.256*** | −0.156** | −0.145* | −0.156** |
| NK CD56bright cells | −0.046 | 0.308*** | −0.150** | −0.172** | 0.249*** | 0.179** | −0.000 | 0.093 | 0.107 | 0.228*** | −0.123* | 0.128* | −0.014 |
| NK CD56dim cells | −0.202*** | −0.027 | −0.133* | 0.017 | −0.125* | −0.074 | −0.217*** | −0.242*** | −0.285*** | −0.122* | −0.053 | −0.039 | −0.048 |
| NK cells | 0.038 | 0.121* | −0.048 | −0.095 | −0.027 | 0.016 | −0.094 | −0.062 | −0.076 | 0.165** | −0.057 | 0.054 | 0.015 |
| pDC | −0.303*** | −0.028 | −0.382*** | −0.328*** | 0.045 | 0.213*** | −0.379*** | 0.008 | −0.013 | 0.367*** | −0.089 | 0.165** | −0.313*** |
| T cells | −0.146* | −0.163** | −0.189*** | −0.187** | −0.109 | −0.030 | −0.282*** | −0.179** | −0.238*** | −0.003 | 0.022 | −0.037 | −0.096 |
| T helper cells | 0.255*** | −0.144* | 0.199*** | 0.006 | −0.157** | −0.246*** | 0.084 | −0.040 | −0.052 | −0.226*** | 0.195*** | −0.181** | 0.108 |
| Tcm | 0.340*** | −0.282*** | 0.186** | 0.063 | −0.344*** | −0.400*** | 0.091 | −0.002 | 0.028 | −0.424*** | 0.033 | −0.312*** | 0.054 |
| Tem | 0.059 | 0.129* | 0.020 | −0.106 | 0.065 | −0.028 | −0.025 | −0.071 | −0.068 | 0.072 | −0.025 | 0.052 | −0.035 |
| TFH | 0.026 | −0.090 | −0.087 | −0.165 | −0.191*** | −0.057 | −0.187** | −0.139* | −0.135* | −0.009 | −0.049 | −0.072 | −0.209*** |
| Tgd | −0.060 | 0.079 | 0.063 | 0.139* | −0.126* | −0.185** | −0.013 | −0.083 | −0.163** | −0.171** | 0.010 | −0.008 | 0.117* |
| Th1 cells | −0.220*** | −0.079 | −0.199*** | −0.010 | −0.169** | −0.071 | −0.336*** | −0.332*** | −0.327*** | −0.063 | 0.042 | −0.166** | −0.151** |
| Th17 cells | −0.066 | 0.184** | −0.059 | −0.038 | −0.018 | 0.005 | −0.086 | 0.034 | −0.031 | −0.059 | −0.095 | 0.042 | −0.079 |
| Th2 cells | 0.187** | 0.109 | 0.211*** | 0.231*** | −0.103 | −0.112 | 0.070 | −0.251*** | −0.220*** | −0.118* | 0.307*** | −0.054 | −0.029 |
| TReg | −0.214*** | −0.183** | −0.210*** | −0.177** | −0.110 | 0.004 | −0.315*** | −0.247*** | −0.301*** | 0.028 | 0.059 | −0.070 | −0.110 |
Fig. 5.
Visualization of the correlation between 13 genes and 23 immune cells infiltration in cervical cancer tissue samples. (A) NDUFA13; (B) MT-ND4; (C) MT-ND1; (D) LDHA; (E) EP300; (F) DARS2; (G) COX6B1; (H) COX5A; (I) SDHB; (J) SLC16A3; (K) ACACA; (L) TWNK; (M) SURF1. (aDC: activated DC; iDC: immature DC; pDC: Plasmacytoid DC; Tcm: T central memory; Tem: T effector memory; Tfh: T follicular helper; Tgd: T gamma delta). The Y axis represents the immune cells, while the X axis represents the correlation of molecules and the corresponding cells (negative number: negatively correlated). The size of the circle represents the degree of correlation, and the larger the circle, the higher the degree of correlation. The greater the height of the rod (distance from 0), the higher the correlation (positive number represents positive correlation, negative number represents negative correlation). The depth of the circle represents the p value.
We suspect that the increased lactic acid content in the cervical cancer tumor microenvironment is related to the inhibition of DC and T regulatory cells. Tumor cells can secrete large amounts of lactic acid in the extracellular microenvironment, leading to acidosis, angiogenesis and immunosuppression. Lactic acid can change the polarization of macrophages (Activation of the ERK–STAT3 pathway; HIF1A stabilization; Activation of GPR132 and Notch; Histone lactylation), produce M2 inflammatory characteristic phenotype (↑IL-6, ↑VEGF, ↑ARG1, ↑CCL5) and inhibit the activation and proliferation of CD8+ T cells (Inhibition of lactate efflux; Acidic pH environment; Reduced NAD availability), natural killer (NK) cells and dendritic cells. Lactic acid has a positive effect on the metabolic characteristics of CD4+CD25+ regulatory T (Treg) cells, enabling them to maintain and enhance their immunosuppressive function in acidic TME. Lactic acid can regulate the metabolism of innate and adaptive immune cells, participate in angiogenesis and tissue remodeling, and promote tumor growth and invasion [[48], [49], [50], [51]].
3.4. Prognostic value of lactated metabolism related genes and risk score constructed associated with tumor-infiltrating immune cells
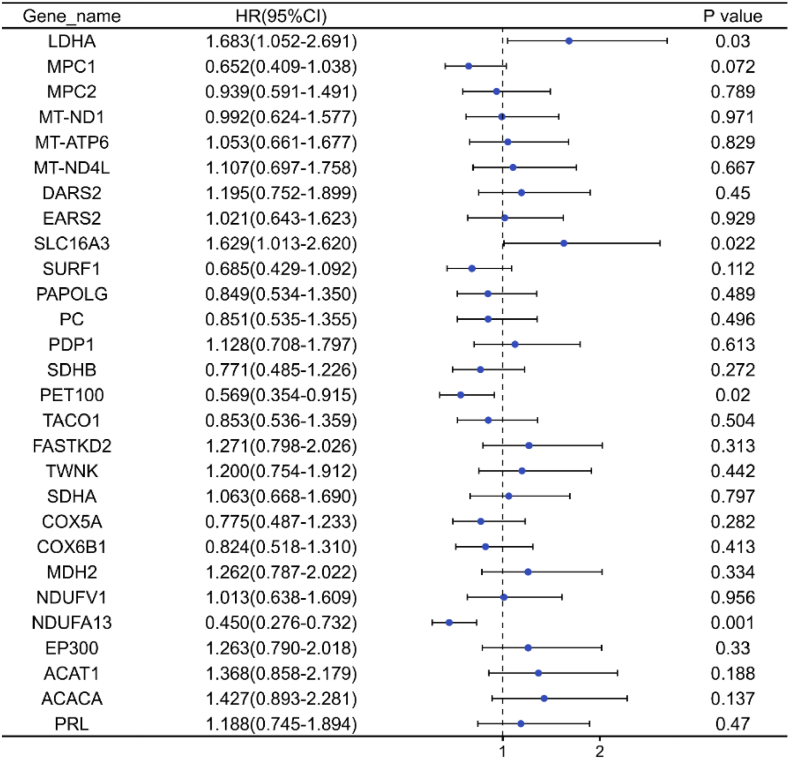
According to the forest map of Fig. 6, four genes with statistical significance were screened: LDHA (HR: 1.683), SLC16A3 (HR: 1.629), PET100 (HR: 0.569), and NDUFA13 (HR: 0.450). On the basis of multi-factor regression analysis, the four genes and immune factors were analyzed by multi-factor analysis.
Fig. 6.
Forest plot of risk assessment of genes related to lactate metabolism.
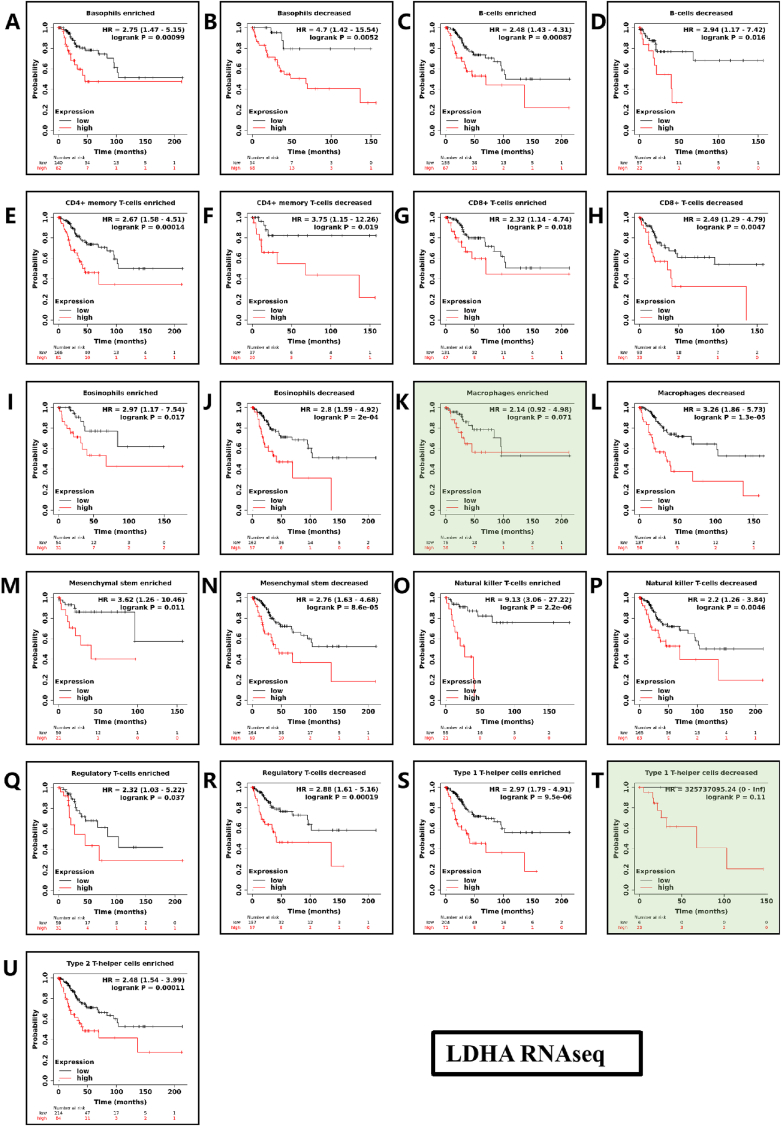
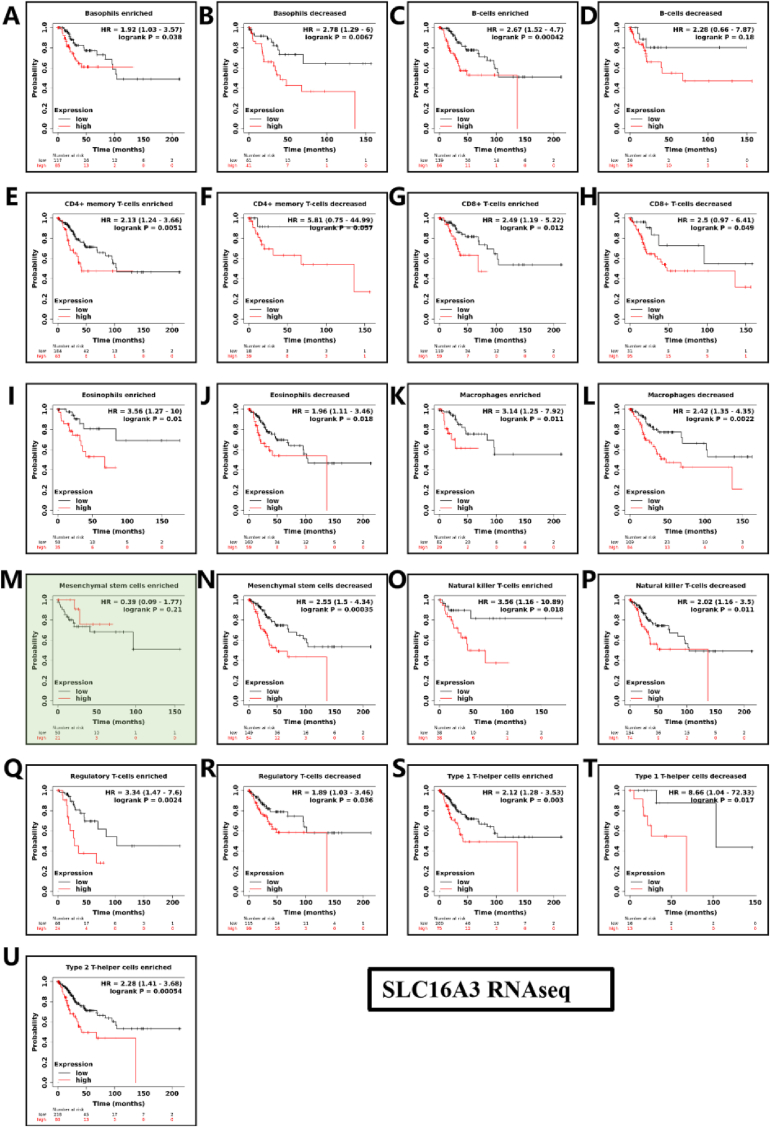
Kaplan-Meier survival curves according to high and low expression of LDHA, SLC16A3 in immune cell subgroups in CESC. In the case of Macrophages enriched and Type1 T-helper cells decreased immune cell infiltration in TME, the expression of LDHA is not statistically significant for the prognostic risk, showned as light green background in Fig. 7. In the case of Mesenchymal stem cells enriched immune cell infiltration in TME, the expression of SLC16A3 is not statistically significant for the prognostic risk showned as light green background in Fig. 8. According to Fig. 7, Fig. 8, the HR of expression of LDHA and SLC16A3 with the high and low expression of immune cells were both greater than 1 (No statistically significant immune cells were excluded, P > 0.05). LDHA and SLC16A3 low expression in immune cell infiltration can reduce the immunosuppression in the tumor microenvironment and have a good prognosis for cervical cancer. In the two types of helper T cells enriched state, the high expression of SLC16A3 truncated the survival curve, indicating that the prognosis of SLC16A3 in the helper T cells enriched state is worse than LDHA. We speculate that SLC16A3 has a poorer on the prognosis of cervical cancer. Therefore, in the multivariate analysis in Fig. 9, we optimized SLC16A3.
Fig. 7.
Kaplan-Meier survival curves according to high and low expression of LDHA in immune cell subgroups in CESC. Correlations between LDHA expression and OS in different immune cell subgroups in CESC patients were estimated by Kaplan-Meier plotter. (A) Basophils enriched; (B) Basophils decreased; (C) B-cells enriched; (D) B-cells decreased; (E) CD4+ memory T-cells enriched; (F) CD4+ memory T-cells decreased; (G) CD8+ memory T-cells enriched; (H) CD8+ memory T-cells decreased; (I) Eosinophils enriched; (J) Eosinophils decreased; (K) Macrophages enriched; (L) Macrophages decreased; (M) Mesenchymal stem enriched; (N) Mesenchymal stem decreased; (O) Natural killer T-cells enriched; (P) Natural killer T-cells decreased; (Q) Regulatory T-cells enriched; (R) Regulatory T-cells decreased; (S) Type 1 T-helper cells enriched; (T) Type 1 T-helper cells decreased; (U) Type 2 T-helper cells enriched.
Fig. 8.
Kaplan-Meier survival curves according to high and low expression of SLC16A3 in immune cell subgroups in CESC. Correlations between SLC16A3 expression and OS in different immune cell subgroups in CESC patients were estimated by Kaplan-Meier plotter. (A) Basophils enriched; (B) Basophils decreased; (C) B-cells enriched; (D) B-cells decreased; (E) CD4+ memory T-cells enriched; (F) CD4+ memory T-cells decreased; (G) CD8+ memory T-cells enriched; (H) CD8+ memory T-cells decreased; (I) Eosinophils enriched; (J) Eosinophils decreased; (K) Macrophages enriched; (L) Macrophages decreased; (M) Mesenchymal stem enriched; (N) Mesenchymal stem decreased; (O) Natural killer T-cells enriched; (P) Natural killer T-cells decreased; (Q) Regulatory T-cells enriched; (R) Regulatory T-cells decreased; (S) Type 1 T-helper cells enriched; (T) Type 1 T-helper cells decreased; (U) Type 2 T-helper cells enriched.
Fig. 9.
Multivariable model (Nomogram chart of Cox regression) of CESC to PFI, DFS, OS.
CTLA4 (Cytotoxic T-Lymphocyte Associated Protein 4) gene is a member of the immunoglobulin superfamily and encodes a protein which transmits an inhibitory signal to T cells. The affinity of CTLA4 for its natural B7 family ligands, CD80 and CD86, is considerably stronger than the affinity of their cognate stimulatory coreceptor CD28. We performed multi-factor COX regression on HUB genes and immune cells and found that SLC16A3 and CTLA4 were statistically significant after univariate and multivariate prognostic analysis. Nomogram will filter out any sample with missing information. We chooses the better Concordance as shown in Fig. 9. In this kind model the Concordance (C)-index in OS COX analysis is 0.650 (P < 0.01), C-index in Disease Free Survival (DSS) COX analysis is 0.663 (P < 0.01), and C-index in PFI COX analysis is 0.663 (P < 0.01).
3.5. Post-translational protein modification as epigenetic modification by histone lactylation
The latest research founds a chemical modification called lactylation — the addition of a lactyl (La) group to the lysine amino-acid residues in the tails of histone proteins [12]. Histone lactylation mediates gene expression to promote an M2-like Phenotype. Under the hypoxia condition of M1 macrophages, glucose is incompletely oxidized to lactate metabolite. Lactic acid can produce lactyl coenzyme A, which contributes a lactyl group to the lysine tail of histone through acetyltransferase P300 (EP300), resulting in a modification called lactyl lysine. In transcription assays, histone lactylation promotes gene transcription similar to histone acetylation. Histone lactation modifies genes that activate the wound healing pathway, resulting in a Macrophage M2 like phenotype. ACAT1 as a mitochondrial protein acetyltransferase and most importantly, stable knockdown of ACAT1 attenuates tumor growth. EP300 (P < 0.05) and ACAT1 (P < 0.001) express decrease in CESC tumor samples (Fig. 1B). ACAT1 is hijacked to contribute to the Warburg effect in human cancer [52]. EP300 are highly conserved, ubiquitously expressed enzymes that belong to the KAT3 family of acetyltransferases [53]. EP300 gene encodes a histone acetyltransferase (HAT), which influences transcription through chromatin remodeling and has tumor suppressor activity [54].
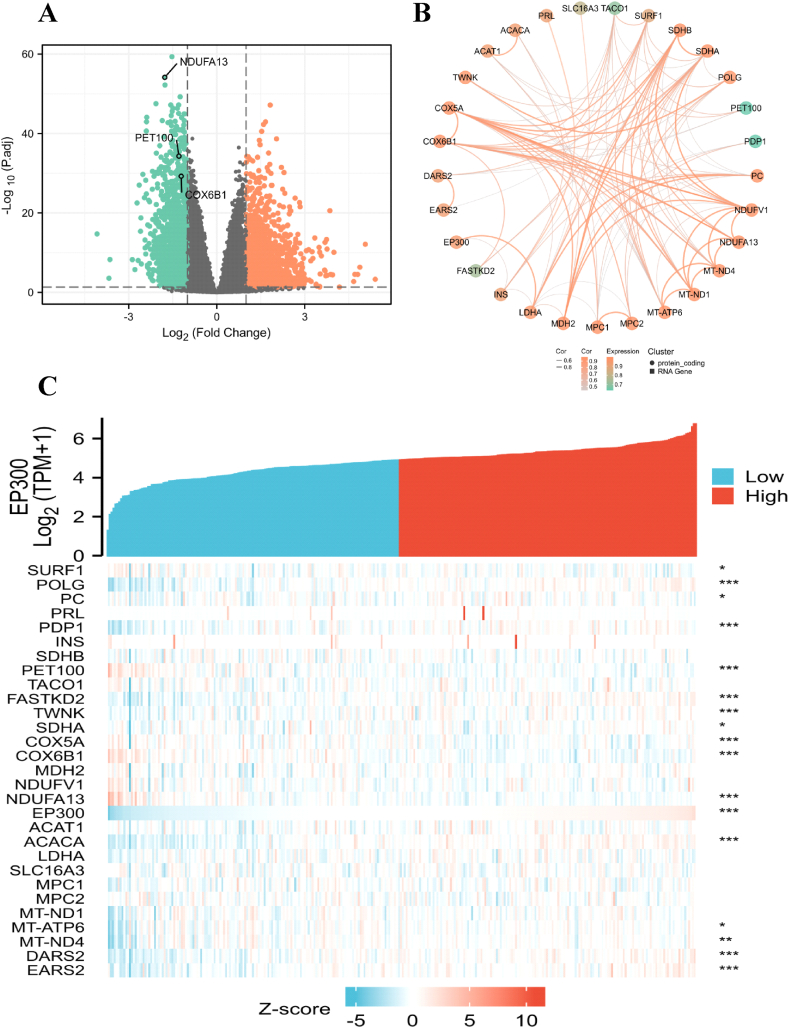
As shown in Fig. 2, EP300 genetic alteration is highest about 11%, including Missense Mutation, Amplification, Deep Deletion, and truncating mutation. Our study found that EP300 played a bridge role in tumor immunosuppression in lactate metabolism and carcinogenesis. The total number of Gene IDs in CESC is 56,493. The expression value of EP300 is averagely divided into high and low expression groups, simulating the effect of similar over-expression or knockdown of this gene. The different molecules in the two groups are analyzed to infer the molecules that may be related to EP300. Among them, there are 1740 IDs that meet the |log2 (FC)|>1 and P < 0.05 threshold. Under this threshold, the number of high expression (logFC is positive) in the High group is 902, in the low group, the number of low expression (logFC is negative) is 838 (Fig. 10A). Fig. 10B visualize a heat map showing the correlation of 29 lactic acid metabolism-related genes. EP300 has greater significance of POLG (P < 0.001), PDP1 (P < 0.001), PET100 (P < 0.001), FASTKD2 (P < 0.001), TWNK (P < 0.001), COX5A (P < 0.001), COX6B1 (P < 0.001), NDUFA13 (P < 0.001), ACACA (P < 0.001), DARS2 (P < 0.001), EARS2 (P < 0.001) (Fig. 10C).
Fig. 10.
Differential Analysis of EP300 Single Gene Expression in CESC. (A) Volcano map of EP300 DEGs in CESC, where red represents upregulated genes and light green represents downregulated genes. (B) The (circle-curve) correlation diagram of the 29 statistically significant genes related to lactate metabolism genes in CESC. (C) Correlation between EP300 and Other genes related to lactate metabolism, Significance mark: ns: p ≥ 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001).
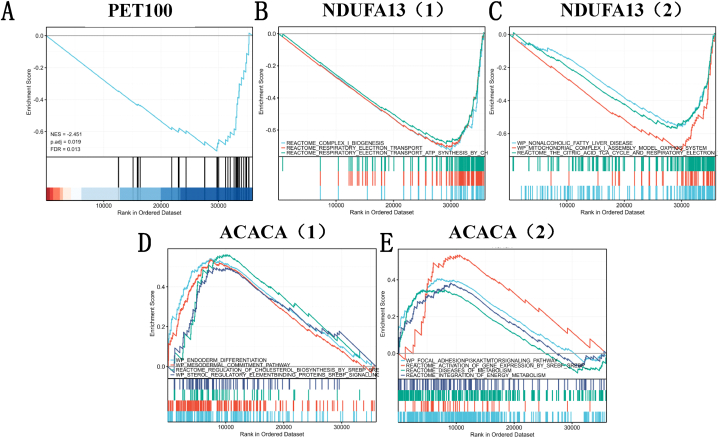
Fig. 11 visualized the EP300 single GSEA result, using the workflow described as follow: First, conduct EP300 single gene difference analysis. After obtaining the results, extract the molecular column and the corresponding logFC for GSEA analysis, that is, single gene GSEA analysis meeting the threshold (P < 0.05 and FDR<0.25). The molecular sieves of NES top meeting the threshold were selected to select six of 29 hub genes:.LDHA, PET100, SLC16A3, MPC, NDUFA13 and ACACA. However, it was found in Fig. 10C that there was no significant difference in LDHA、MPC and SLC16A3 with EP300 Single gene differential expression. We enriched all the pathway functions involved in the three genes: PET100, NDUFA13 and ACACA. Chosing the maximum value of the pathway NES of each gene corresponds to the reaction, it was found that NDUFA13 enriched in “REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS”, ACACA in “WP_ENDODERM_DIFFERENTIATION”, PET100 in “WP_MITOCHONDRIAL_CIV_ASSEMBLY”. NES score of PET and NDUFA13 is negative while NES score of ACACA is positive. All the pathways involved above are related to lactic acid metabolism.
Fig. 11.
EP300 GSEA is used to annotate the malignant hallmarks of these genes with different expression levels. (A) PET100; (B)–(C) NDUFA13; (D)–(E) ACACA. The abscissa is the sorted gene list, and the ordinate is the value of signal2 noise. The area ratio of gray shadow can reflect the size of signal2 noise between groups as a whole, that is, the difference between. (Here, it is considered that the threshold of significant enrichment is: false discovery rate (FDR) < 0.25 and p. adjust <0.05).
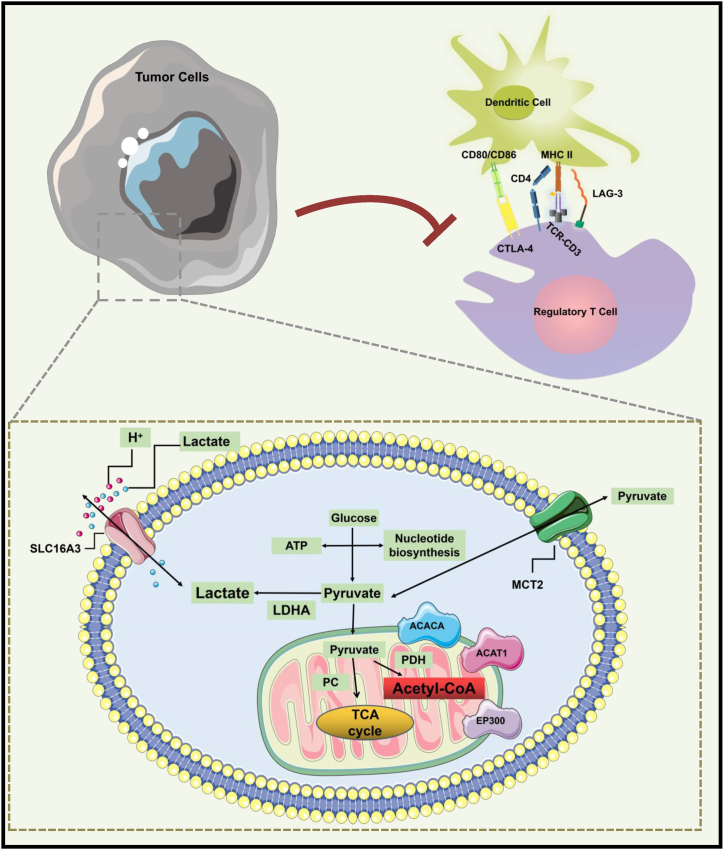
Lactate production is determined by the balance between glycolysis and mitochondrial metabolism. Figure 12 shows the Schematic of lactate metabolism in tumor-immune microenvironment. Lactate contributes to the immune escape in different ways. The role of lactate in shaping the tumor microenvironment due to its key role in carcinogenesis, tumor-associated microenvironment. When LDHA increased, excessive lactic acid in the cytoplasm affected the post-translational modification of histones through the lactylation of histones (EP300 protein was consumed), high concentrations of lactate lead to its conversion into pyruvate by LDH, resulting in the production of NADH and consequent inhibitory feedback on glycolysis. Decreased EP300 leads to inhibition of glucose metabolism demonstrating that EP300 maybe a promoter of glycolysis in tumor. Different immune cells in tumor microenvironment depend on different metabolic pathways. Recent studies [55,56] have shown that aerobic glycolysis plays an important role in immune cell activation. EP300 decreased and inhibited glycolysis in the tumor microenvironment. This study found that the function of Tcm cells closely related to ep300 was inhibited by this glycolysis-low cervical cancer tumor, which promoted the immune escape. At the same time, the residual lactate was transported outside the cell membrane through Monocarboxylic acid transporter-SLC16A3 in the TME also inhibition the function of dendritic cells and T cells, which proof that Lactate contributes to the immune escape.
Fig. 12.
Tumor cell lactate metabolism pathway and immune microenvironment model diagram.
4. Discussion
Tumor-secreted lactate, a product of increased aerobic glycolysis of cancer cells, is associated with heightened metastatic incidence, increased angiogenesis, is responsible for metabolic reprogramming in adjacent tissues and can induce a pro-inflammatory state. In glycolytic tumors, the lactic acid level of cancer cells is significantly increased to 40 times [57,58], and is highly correlated with the aggressiveness of cancer and low survival rate. Our integrated multi-omics approach revealed the role of lactate in TME and immune infiltration of cervical cancer. Assess genes related to lactate metabolism at RNA and protein levels. There are differences between cervical cancer and normal tissues (Fig. 1). Mutations of oncogenes and tumor suppressor genes directly increase the expression of MCTs, this phenomenon indicates. In cancer, MCTs allow lactic acid to continue its journey outside the cell, realizing its mediating role as a carcinogenic mediator. Inhibition of MCT1 can inhibit growth and canceration. The highly coordinated metabolic reprogramming in cancer cells to glycolysis and lactic acid production does not just stop at the end of the Warburg effect. A recent study found that blocking the lactate transporter MCT1 reduces the proliferation of breast cancer cells coexpressing MCT1 and MCT4 [59] and reduces HIF1A (Table 2) induced angiogenesis in cervix squamous carcinoma [47,60]. MYC (Table 2) turnover is required for histone acetylation to chromatin and transcriptional elongation. Activation of MYC target genes occurs in response to histone acetylation, while repression occurs when histones are deacetylated [61]. This suggests that lactic acid metabolism may be involved in reprogramming genes for lactose production and carcinogenesis in the transcriptional network.
Lactic acid is used by cancer cells to promote cancer growth and mitochondrial metabolism [62]. Lactic acid is also the substrate of alanine. Lactic acid shuttle enables aerobic cancer cells to use lactic acid produced by hypoxic cells in tumors [34,63,64]. Therefore, thousands of genes up-regulated (DARS2, PC, PDP1, SDHB, PET100, COX5A, MDH2, COX6B1, TWNK, SDHA, MPC2) (Fig. 1A, P < 0.001) by lactate may collectively represent a transcriptional network that participates in the reprogramming of cells for lactate generation and carcinogenesis. We know that pyruvic acid is transported into mitochondria through MPC, which is composed of MPC1 and MPC2 subunits. Human MPC1 and MPC2 exist in the form of homodimer or heterodimer. Recent literature data show that [[65], [66], [67]], the down-regulation or deletion of MPC1 is related to various types of cancer, while MPC2 plays an important role in the control of hepatic gluconeogenesis. A heteroligomeric complex of MPC1 and MPC2 is necessary and sufficient for this transport of pyruvate. The expression of MPC1 and MPC2 should be decreased in cancer cells. In this study, we found that the expression of MPC1 was down-regulated in cervical cancer, but the expression of MPC2 was up-regulated, which may be similar to the conclusion in the literature, most cancers exhibit decreased expression of MPC1 and MPC2, with MPC1 being the most strongly and consistently affected [68]. The down regulation of MPC1 has been shown to accelerate progression of cervical cancer. EP300 genetic alteration is highest (Fig. 2A) in our reasearch, with a decrease expression in CESC tumor samples (Fig. 1B). It plays an important role in regulating cell growth and blocking the promotion of cancerous tumors [69]. Next, we have researched the mechanism of EP300 in cervical cancer. We consider histone lactylation as post-protein modification plays an important role in cervical carcinogenesis, especially the clinical prognosis of cervical cancer. The main enrichment pathway of lactic acid metabolism-related hub genes show molecular function is MCT transporter activity (Fig. 3A, E), biological process are pyruvate metabolism (Fig. 3E, G), acetyl-CoA metabolic process (Fig. 3A, H). Lactate as a substrate for the generation of lactyl-CoA for lysine lactylation on histones to promote gene expression. Of note, hypoxia also increases the production of lactate by glycolysis (Fig. 3B, C, D, I). Hypoxia is characterized by decreased flux through the tricarboxylic acid (TCA) cycle in mitochondria, leading to the reduction of metabolites, such as acetyl-CoA. Specifically, the tumor hypoxia environment induces the increase of LDHA, and then negatively regulates the entry of pyruvate into the cell TCA cycle by promoting the production of lactate and inhibiting its conversion to acetyl-CoA, respectively. Hypoxia also affects the activity of electron transport chain (ETC).
In the previous result analysis, the immune cells most closely related to the genes related to lactate metabolism are pDC and Tcm, who have the number of immune cells with correlation degree greater than 0.3 ranked first and second, respectively, 6 and 5. EP300, ACACA, LDHA, DARS2, NDUFA13 and TWNK are highly correlated with pDC, of which only NDUFA13 is positively correlated. EP300, COX5A, COX6B1, NDUFA13 and SURF1 were highly correlated with Tcm, and only EP300 was positively correlated with Tcm. The immune effects of the two genes are completely opposite: EP300 is negatively correlated with pDC while positively correlated with Tcm. On the other hand, NDUFA13 is oppositely correlated (Table 3, Fig. 5). The gene protein NDUFA13 (also known as GRIM-19) is found with a functional component of mitochondrial complex I. High lactic acid in TME may hinder the formation and accumulation of dendritic cells. Gene prognosis analysis found four genes (LDHA, SLC16A3, PET100, and NDUFA13) have statistical significance with the prognosis of cervical cancer (Fig. 6). KM curve research proved that the mechanism of LDHA and SLC16A3 affecting the prognosis may be related to the infiltration of immune cells in the tumor microenvironment (Fig. 7, Fig. 8). We selected lactic acid metabolism hub genes and immune marker factors for multi-factor model analysis of cervical cancer prognosis. Verify that the effective model is shown in Fig. 9. The identification of more potent and selective LDHA inhibitors and SLC16A3 inhibitors could open up new perspectives in anticancer therapies in the near future.
Fig. 1 shows that among the six genes of EP300, ACAT1, ACACA, LDHA, PC and SLC16A3 in Fig. 12, LDHA, ACACA and SLC16A3 are elevated in cervical cancer, while the expression of EP300 in cervical cancer is reduced. Combined with Fig. 10, there is no significant correlation between EP300 and LDHA, SLC16A3, only ACACA has signficant relationship with EP300 expression. After analyzing the EP300 single GSEA, pathways mentioned in Fig. 11, ACACA was concentrated in the EP300 high-expression group (NES<0). PET and NDUFA13 was concentrated in the EP300 low-expression group (NES<0). A preliminary study was discussed earlier in this study that cervical cancer is EP300 downregulated tumors. Therefore, although ACACA was upregulated in cervical cancer, its effect on EP300 function is faintly.
The expression of EP300 is decreased in cervical cancer, which reduces the histone lactation modification and transcriptional mutation of cervical cancer cells, so as to stabilize the proliferation of cancer cells. On the other hand, the decrease of ACAT1 can affect the metabolism of acetyl-CoA in mitochondria, further increase the release of lactic acid to the tumor microenvironment and inhibit the imuune and antitumor properties of Tcm. The decrease of EP300 is related to the antitumor inhibition of Tcm. Due to the characteristics of resident memory T cell subsets (Trm), they are located in tissues that are difficult to reach by circulating T cells, especially in cervical tissue of reproductive tract. This leads us to speculate that this change in lactate metabolism may be more closely related to Trm. Here is no doubt that macrophages are the most abundant in the tumor microenvironment. We speculate that the change of immune microenvironment in cervical cancer is mainly related to macrophages.
CD8+ or CD4+ T cells are activated by DC cells expressing MHC class I or II molecules on their surfaces, respectively. There is increasing evidence that different DC types support their functions by establishing unique and strictly regulated metabolic processes [70]. Tumor-associated macrophages (TAM) are a heterogeneous cell population. TAMs are usually the most abundant infiltrating leukocytes in most tumors and are mainly considered to have tumor-promoting effects. In addition to promoting angiogenesis and metastasis, these also include immunosuppressive effects. TAMs may also suppress T cell functions through the direct engagement of T cell inhibitory and apoptotic receptors. Treg Cell-Mediated Suppression through Modulation of APCs mechanism (Fig. 12): (1) CTLA-4 interacts with CD80 and CD86 on DC to block the ability to activate immature T cells. (2) The combination of LAG-3 and MHC II induces TAM mediated inhibitory signaling pathway and blocks the maturation and antigen presentation ability of DC. Pyruvate oxidation requires PDH activity. The resulting acetyl CoA production and further accumulation of acetyl CoA inhibit its consumption enzyme acetyl CoA carboxylase 1 through phosphorylation induction. This study shows that in cervical tumor tissue, lactic acid promotes tumor growth and invasion, generates macrophages with anti-inflammatory (M2 like) phenotype through activated macrophage polarization, and inhibits the activation and proliferation of T cells and dendritic cells. At the same time, lactic acid has a positive effect on the metabolic characteristics of CD4 + CD25 + Treg cells, maintaining its persistence and enhancing immunosuppressive function in acidic TME.
The clarification of the metabolic characteristics of tumors and inflammatory diseases brings new ideas for therapeutic intervention, aiming to adjust the bioavailability of metabolites, correct the imbalanced immune state, and trigger a beneficial tumor immunosuppressive response.
5. Conclusions
Tumor microenvironment (TME) is characterized by the presence of various cell types, including tumor, stromal and immune cells, as well as blood vessels. In this poor metabolic microenvironment, tumor cells use most nutrients, which affects the function of invasive immune cells. In addition, due to the Warburg effect, tumor cells secrete a large amount of lactic acid to the extracellular microenvironment, resulting in acidosis, angiogenesis and immunosuppression. In our research, lactate is recognized as a molecule capable of suppressing immune responses, through inhibition of T cells and dendritic cells in cervical cancer. In glycolytic tumors, the increased lactic acid level of cervical cancer cells increases the invasion and poor survival of tumors. Exciting, this study illustrates from a new perspective of epigenetics that histone lactation at the level of protein post-translational modification mediates gene expression and promotes the cachexia of tumor after immunosuppression. We defend EP300 as an inhibitor of anti-tumor immune response in cervical cancer via metabolic modulation. Inhibition of lactate synthesis or transport can reduce DC and T cell-mediated immunosuppression in cervical cancer. Therefore, lactate inhibition may be a useful tool for anticancer therapy. At the same time, there is a significant difference in the expression of HPV positive and negative cervical cancer, so whether the effect of post-translational modification is related to HPV virus still needs further study in the future.
Author contributions
Conceptualization: Xiaoyue Yang. And Weipei Zhu.; methodology: Weipei Zhu.; software, investigation: Wenjing Zhang; data curation: Xiaoyue Yang.; writing—original draft preparation: Xiaoyue Yang.; writing—review and editing: Xiaoyue Yang.; visualization: Xiaoyue Yang.; supervisions: Xiaoyue Yang.; funding acquisition: Xiaoyue Yang and Weipei Zhu. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Research on early diagnosis of cervical cancer based on terahertz technology” in Zhenjiang social development project, Project number: SH2020051; “Observation and study on the clinical efficacy of a vaginal gel for the treatment of female genital HPV infection and related diseases” 2019 Key Laboratory of Pharmacodynamics and Safety Evaluation of Traditional Chinese Medicine in Jiangsu Province, China Project number: JKLPSE201906; “Study on the Innovation of Distance Teaching Mode of Gynecological Laparoscopic Surgery under the Background of New Medicine” Project of Cooperative Education between Industry and Education in 2020, Project number: 202,002,144,004. Open Project of State Key Laboratory of Radiology and Radiation Protection“LINC00460/miR-361–3p/Gli1 Pathway and Radiosensitivity of Cervical Cancer Cells”, Project number: GZK1202106; “Effect of Sun's Abdominal Acupuncture on Brain Iron Content and Rehabilitation in Patients with Post-stroke Depression Based on Magnetic Sensitive Imaging” in Zhenjiang social development project, Project number: SH2019039.
Data availability statement
All methods in the study were carried out in accordance with relevant guidelines and regulations. The raw data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Professor Zhang Hong's theoretical guidance of the Second Affiliated Hospital of Suzhou University, and Professor Su Zhaoliang's theoretical guidance of Jiangsu University Medical College.
References
- 1.Chen W., Zheng R., Baade P.D., et al. Cancer statistics in China, 2015. CA A Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Gillies R.J., Raghunand N., Karczmar G.S., et al. MRI of the tumor microenvironment. J. Magn. Reson. Imag.: An Official J. Int. Soc. Magn. Res. Med. 2002;16(4):430–450. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- 3.Stubbs M., McSheehy P.M.J., Griffiths J.R., et al. Causes and consequences of tumour acidity and implications for treatment. Mol. Med. Today. 2000;6(1):15–19. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 4.Webb B.A., Chimenti M., Jacobson M.P., et al. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer. 2011;11(9):671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 5.Justus C.R., Dong L., Yang L.V. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front. Physiol. 2013;4:354. doi: 10.3389/fphys.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San-Millán I., Brooks G.A. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38(2):119–133. doi: 10.1093/carcin/bgw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Racker E. Bioenergetics and the problem of tumor growth: an understanding of the mechanism of the generation and control of biological energy may shed light on the problem of tumor growth. Am. Sci. 1972;60(1):56–63. [PubMed] [Google Scholar]
- 8.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Bergers G., Fendt S.M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer. 2021;21(3):162–180. doi: 10.1038/s41568-020-00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phannasil P., Israr-ul H.A., El Azzouny M., et al. Mass spectrometry analysis shows the biosynthetic pathways supported by pyruvate carboxylase in highly invasive breast cancer cells. Biochim. Biophys. Acta, Mol. Basis Dis. 2017;1863(2):537–551. doi: 10.1016/j.bbadis.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phannasil P., Thuwajit C., Warnnissorn M., et al. Pyruvate carboxylase is up-regulated in breast cancer and essential to support growth and invasion of MDA-MB-231 cells. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0129848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D., Tang Z., Huang H., et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vivian J., Rao A.A., Nothaft F.A., Ketchum C., Armstrong J., Novak A., Pfeil J., Narkizian J., Deran A.D., Musselman-Brown A., Schmidt H., Amstutz P., Craft B., Goldman M., Rosenbloom K., Cline M., O'Connor B., Hanna M., Birger C., Kent W.J., Patterson D.A., Joseph A.D., Zhu J., Zaranek S., Getz G., Haussler D., Paten B. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017 Apr 11;35(4):314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G., Mlecnik B., Tosolini M., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14(1):1–15. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 19.Tropberger P., Pott S., Keller C., Kamieniarz-Gdula K., Caron M., Richter F., Li G., Mittler G., Liu E.T., Bühler M., Margueron R., Schneider R. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013 Feb 14;152(4):859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 20.Delvecchio M., Gaucher J., Aguilar-Gurrieri C., Ortega E., Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 2013 Sep;20(9):1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 21.Ogryzko V.V., Schiltz R.L., Russanova V., Howard B.H., Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996 Nov 29;87(5):953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 22.Galarneau L., Nourani A., Boudreault A.A., et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 2000;5(6):927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 23.Selleck W., Fortin I., Sermwittayawong D., et al. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell Biol. 2005;25(13):5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki H., Schiltz L., Chiu R., et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature. 2000;405(6783):195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 25.Fukao T., Yamaguchi S., Tomatsu S., Orii T., Frauendienst-Egger G., Schrod L., Osumi T., Hashimoto T. Evidence for a structural mutation (347Ala to Thr) in a German family with 3-ketothiolase deficiency. Biochem. Biophys. Res. Commun. 1991 Aug 30;179(1):124–129. doi: 10.1016/0006-291x(91)91343-b. [DOI] [PubMed] [Google Scholar]
- 26.Wakazono A., Fukao T., Yamaguchi S., Hori T., Orii T., Lambert M., Mitchell G.A., Lee G.W., Hashimoto T. Molecular, biochemical, and clinical characterization of mitochondrial acetoacetyl-coenzyme A thiolase deficiency in two further patients. Hum. Mutat. 1995;5(1):34–42. doi: 10.1002/humu.1380050105. [DOI] [PubMed] [Google Scholar]
- 27.Fukao T., Nakamura H., Song X.Q., Nakamura K., Orii K.E., Kohno Y., Kano M., Yamaguchi S., Hashimoto T., Orii T., Kondo N. Characterization of N93S, I312T, and A333P missense mutations in two Japanese families with mitochondrial acetoacetyl-CoA thiolase deficiency. Hum. Mutat. 1998;12(4):245–254. doi: 10.1002/(SICI)1098-1004(1998)12:4<245::AID-HUMU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Colbert C.L., Kim C.W., Moon Y.A., Henry L., Palnitkar M., McKean W.B., Fitzgerald K., Deisenhofer J., Horton J.D., Kwon H.J. Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 2010 Nov 2;107(44):18820–18825. doi: 10.1073/pnas.1012736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim C.W., Moon Y.A., Park S.W., Cheng D., Kwon H.J., Horton J.D. Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 2010 May 25;107(21):9626–9631. doi: 10.1073/pnas.1001292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunkeler M., Hagmann A., Stuttfeld E., Chami M., Guri Y., Stahlberg H., Maier T. Structural basis for regulation of human acetyl-CoA carboxylase. Nature. 2018 Jun;558(7710):470–474. doi: 10.1038/s41586-018-0201-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Lu J., Kulkarni S., Zhang W., Gorka J.E., Mandel J.A., Goetzman E.S., Prochownik E.V. Metabolic and oncogenic adaptations to pyruvate dehydrogenase inactivation in fibroblasts. J. Biol. Chem. 2019 Apr 5;294(14):5466–5486. doi: 10.1074/jbc.RA118.005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halestrap A.P. The monocarboxylate transporter family—structure and functional characterization. IUBMB Life. 2012;64(1):1–9. doi: 10.1002/iub.573. [DOI] [PubMed] [Google Scholar]
- 33.Baek G.H., Yan F.T., Hu Z., et al. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014;9(6):2233–2249. doi: 10.1016/j.celrep.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Cascone T., McKenzie J.A., Mbofung R.M., et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metabol. 2018;27(5):977–987. e4. doi: 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briggs K.J., Koivunen P., Cao S., et al. Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell. 2016;166(1):126–139. doi: 10.1016/j.cell.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar N.P., Joseph R., Kamaraj T., et al. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J. Gen. Virol. 2008;89(8):1945–1948. doi: 10.1099/vir.0.83628-0. [DOI] [PubMed] [Google Scholar]
- 37.Trabold O., Wagner S., Wicke C., et al. Lactate and oxygen constitute a fundamental regulatory mechanism in wound healing. Wound Repair Regen. 2003;11(6):504–509. doi: 10.1046/j.1524-475x.2003.11621.x. [DOI] [PubMed] [Google Scholar]
- 38.Dhup S., Kumar Dadhich R., Ettore Porporato P., et al. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr. Pharmaceut. Des. 2012;18(10):1319–1330. doi: 10.2174/138161212799504902. [DOI] [PubMed] [Google Scholar]
- 39.Polet F., Feron O. Endothelial cell metabolism and tumour angiogenesis: glucose and glutamine as essential fuels and lactate as the driving force. J. Intern. Med. 2013;273(2):156–165. doi: 10.1111/joim.12016. [DOI] [PubMed] [Google Scholar]
- 40.Sonveaux P., Copetti T., De Saedeleer C.J., et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckert S., Farrahi F., Aslam R.S., et al. Lactate stimulates endothelial cell migration. Wound Repair Regen. 2006;14(3):321–324. doi: 10.1111/j.1743-6109.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y.H., Hunt M.C., Mancini R.A., et al. Mechanism for lactate-color stabilization in injection-enhanced beef. J. Agric. Food Chem. 2006;54(20):7856–7862. doi: 10.1021/jf061225h. [DOI] [PubMed] [Google Scholar]
- 43.Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9(6):425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 44.Shim H., Dolde C., Lewis B.C., et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doherty B., Haugh H., Lyon F. Social enterprises as hybrid organizations: a review and research agenda. Int. J. Manag. Rev. 2014;16(4):417–436. [Google Scholar]
- 46.Draoui N., Feron O. Lactate shuttles at a glance: from physiological paradigms to anti-cancer treatments. Disease models & mechanisms. 2011;4(6):727–732. doi: 10.1242/dmm.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonveaux P., Copetti T., De Saedeleer C.J., et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Certo M., Tsai C.H., Pucino V., et al. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2020:1–11. doi: 10.1038/s41577-020-0406-2. [DOI] [PubMed] [Google Scholar]
- 49.Batista-Gonzalez A., Vidal R., Criollo A., et al. New insights on the role of lipid metabolism in the metabolic reprogramming of macrophages. Front. Immunol. 2020;10:2993. doi: 10.3389/fimmu.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Certo M., Elkafrawy H., Pucino V., et al. Endothelial cell and T‐cell crosstalk: targeting metabolism as a therapeutic approach in chronic inflammation. Br. J. Pharmacol. 2021;178(10):2041–2059. doi: 10.1111/bph.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braaten T.J., Brahmer J.R., Forde P.M., et al. Immune checkpoint inhibitor-induced inflammatory arthritis persists after immunotherapy cessation. Ann. Rheum. Dis. 2020;79(3):332–338. doi: 10.1136/annrheumdis-2019-216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan J., Lin R., Xia S., et al. Tetrameric acetyl-CoA acetyltransferase 1 is important for tumor growth. Mol. Cell. 2016;64(5):859–874. doi: 10.1016/j.molcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia D., Augert A., Kim D.W., et al. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov. 2018;8(11):1422–1437. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClure B.J., Heatley S.L., Kok C.H., et al. Pre-B acute lymphoblastic leukaemia recurrent fusion, EP300-ZNF384, is associated with a distinct gene expression. Br. J. Cancer. 2018;118(7):1000–1004. doi: 10.1038/s41416-018-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nolt B., Tu F., Wang X., et al. Lactate and immunosuppression in sepsis. Shock. 2018;49(2):120. doi: 10.1097/SHK.0000000000000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinfeld B.I., Rathmell W.K., Kim T.K., Rathmell J.C. The therapeutic implications of immunosuppressive tumor aerobic glycolysis. Cell. Mol. Immunol. 2022 Jan;19(1):46–58. doi: 10.1038/s41423-021-00727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holm E., Hagmüller E., Staedt U., et al. Substrate balances across colonic carcinomas in humans. Cancer Res. 1995;55(6):1373–1378. [PubMed] [Google Scholar]
- 58.Brizel D.M., Scher R.L., Walenta S., et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 2000;3(48):177. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 59.Hong C.S., Graham N.A., Gu W., et al. MCT1 modulates cancer cell pyruvate export and growth of tumors that Co-express MCT1 and MCT4. Cell Rep. 2016 Feb 23;14(7):1590–1601. doi: 10.1016/j.celrep.2016.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonveaux P., Végran F., Schroeder T., et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 2008;118(12):3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapinas K., Grandy R., Ghule P., et al. The abbreviated pluripotent cell cycle. J. Cell. Physiol. 2013;228(1):9–20. doi: 10.1002/jcp.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez-Outschoorn U.E., Sotgia F., Lisanti M.P. Power surge: supporting cells “fuel” cancer cell mitochondria. Cell Metabol. 2012;15(1):4–5. doi: 10.1016/j.cmet.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 63.Kennedy K.M., Scarbrough P.M., Ribeiro A., et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0075154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rey S., Semenza G.L. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc. Res. 2010;86(2):236–242. doi: 10.1093/cvr/cvq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCommis K.S., Chen Z., Fu X., McDonald W.G., Colca J.R., Kletzien R.F., Burgess S.C., Finck B.N. Loss of mitochondrial pyruvate carrier 2 in the liver leads to defects in gluconeogenesis and compensation via pyruvate-alanine cycling. Cell Metabol. 2015;22:682–694. doi: 10.1016/j.cmet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takaoka Y., Konno M., Koseki J., Colvin H., Asai A., Tamari K., Satoh T., Mori M., Doki Y., Ogawa K., et al. Mitochondrial pyruvate carrier 1 expression controls cancer epithelial-mesenchymal transition and radioresistance. Cancer Sci. 2019;110:1331. doi: 10.1111/cas.13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang X.P., Chen Q., Li Y., Wang Y., Zou H.B., Fu W.J., Niu Q., Pan Q.G., Jiang P., Xu X.S., et al. Mitochondrial pyruvate carrier 1 functions as a tumor suppressor and predicts the prognosis of human renal cell carcinoma. Lab. Invest. 2019;99:191. doi: 10.1038/s41374-018-0138-0. [DOI] [PubMed] [Google Scholar]
- 68.Bensard C.L., Wisidagama D.R., Olson K.A., et al. Regulation of tumor initiation by the mitochondrial pyruvate carrier. Cell Metabol. 2020;31(2):284–300. doi: 10.1016/j.cmet.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma S., Jiang T., Jiang R. Constructing tissue-specific transcriptional regulatory networks via a Markov random field. BMC Genom. 2018;19(10):65–77. doi: 10.1186/s12864-018-5277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du X., Chapman N.M., Chi H. Emerging roles of cellular metabolism in regulating dendritic cell subsets and function. Front. Cell Dev. Biol. 2018;6:152. doi: 10.3389/fcell.2018.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All methods in the study were carried out in accordance with relevant guidelines and regulations. The raw data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.