Abstract
In most strains of Helicobacter pylori, mutational inactivation of the rdxA (HP0954) gene, which encodes a nitroreductase that converts metronidazole (MTZ) from a harmless prodrug to a mutagenic and bacteriocidal product, is sufficient to make this pathogen resistant to clinically significant levels of MTZ. Here we report that SS1, a strain with the special ability to colonize mice, is unusual in being susceptible to very low concentrations of MTZ (0.5 μg/ml) and in being especially difficult to mutate to MTZ resistance (Mtzr). These phenotypic traits were traced to expression in this strain of the normally quiescent H. pylori frxA gene (HP0642, an rdxA paralog) along with rdxA. Transformation tests using rdxA::cam and frxA::kan insertion mutant DNAs, with selection solely for the chloramphenicol and kanamycin resistance markers, and sequence analyses of frxA in spontaneous Mtzr derivatives of rdxA null mutant strains each showed that the development of Mtzr in SS1 required inactivation of both rdxA and frxA. Inactivation of either gene alone left SS1 susceptible to MTZ, although it was readily mutable from an MTZ-susceptible to an Mtzr phenotype. Reverse transcriptase PCR tests showed that frxA mRNA was at least 10-fold more abundant in SS1 than in reference strain 26695. It is proposed that these reductases play primarily nutritional roles during bacterial growth.
Helicobacter pylori is a genetically diverse gastric pathogen that chronically infects more than half of all people worldwide, often for years or decades. Although most infections are relatively benign, long-term H. pylori carriage is a major cause of peptic ulcer disease and is an early risk factor for gastric cancer, one of the most frequently lethal of malignancies in many societies (for reviews see references 22 and 29). The first culturing of H. pylori in the early 1980s led to a revolutionary merger of gastroenterology and infectious disease—the realization that ulcers could be cured and gastric cancer perhaps prevented by H. pylori eradication (5, 11, 20).
Metronidazole (MTZ), a synthetic nitroimidazole, is a key component of some of the most popular and affordable anti-H. pylori therapies worldwide, but its efficacy is reduced in many societies because large numbers of strains have become at least partially MTZ resistant (Mtzr) (7, 8, 10, 21). This resistance is attributable to (i) widespread use of MTZ against other infections (24), (ii) exposure of resident H. pylori strains to subtherapeutic levels of this drug, (iii) the mutagenic nature of products of MTZ activation (26), and (iv) induction of, as well as selection for, Mtzr mutants whenever this drug is used.
It has been shown that MTZ resistance in clinical isolates from diverse parts of the world is nearly always associated with loss-of-function mutations in rdxA (HP0954), the gene for a nitroreductase that normally activates MTZ and converts it from a harmless prodrug to a mutagenic and bacteriocidal agent (probably hydroxylamine) (6, 9, 15, 27). Mutational tests have indicated that rdxA inactivation is generally sufficient to confer resistance to moderate levels of MTZ (16 μg/ml, up from 1 or 1.5 μg/ml in most MTZ-susceptible [Mtzs] strains) (15). Higher-level resistance (e.g., to 32 or 64 μg/ml) is common among clinical isolates, however, and can be achieved by mutation in frxA (HP0642), a paralog of rdxA, and in additional chromosomal genes. In our study, inactivation of frxA in otherwise wild-type (rdxA+) strains did not significantly affect the instrinsic susceptibility of H. pylori cells to very low levels of MTZ (15). This suggested either (i) that frxA is expressed only weakly, if at all, relative to rdxA in wild-type H. pylori or (ii) that the reductase that it encodes does not act efficiently on MTZ. We note that another group (16a, 16b) has just argued that inactivation of either frxA or rdxA is sufficient to make typical H. pylori strains resistant to MTZ. Although results presented below suggest that their interpretation may be incorrect, our experiments and theirs were carried out using different protocols, and thus further analysis is needed.
Only a few of the many different strains of H. pylori seem able to colonize mice (12, 16, 17, 18, 19, 25). One in particular, the SS1 or Sydney strain, has become particularly widely used in analyses of infection processes and host responses, in mutational tests of the importance of candidate bacterial genes, and in early assessments of drug and vaccine candidates. Of special relevance to the present study has been its use to model how MTZ resistance may develop during MTZ-based therapy that fails to fully eradicate H. pylori infection (14). Most (25 of 27) Mtzr mutants obtained from MTZ-treated mice infected with strain SS1 contained sequence changes in rdxA (13), as expected (9). One unanticipated result, however, was that the Mtzr mutants were rare, constituting only a small proportion of the H. pylori organisms recovered from the mice. Their rarity might be explained as a consequence of experimental design—of the researchers having allowed 1 month to elapse between the end of therapy and recovery of H. pylori. This explanation would assume that in the absence of MTZ, the Mtzr mutants were less vigorous than isogenic Mtzs parents, as has been proposed (4). A complementary explanation supposes that two or more genes need to be inactivated in SS1 in order for it to develop an Mtzr phenotype, rather than just one gene as in most strains.
Here we report that SS1 is especially susceptible to MTZ and is difficult to mutate to Mtzr. This is traced to expression of the normally quiescent frxA reductase gene, along with expression of its rdxA paralog, and the unusual need to inactivate both genes to achieve clinically significant resistance.
MATERIALS AND METHODS
H. pylori strain and culture conditions.
The H. pylori strain SS1 (18) used here was obtained from Adrian Lee via Kathryn Eaton and had been used previously by us to test whether the novel beta-beta prime RNA polymerase subunit fusion of H. pylori is important in vivo (23). Strain 26695 (1, 28) was originally from K. Eaton, and strain J99 (2) was provided by T. L. Cover and M. J. Blaser. These strains were grown on brain heart infusion agar (Difco) supplemented with 7% horse blood, 0.4% IsoVitaleX, and the antibiotics amphotericin B (8 μg/ml), trimethoprim (5 μg/ml), and vancomycin (6 μg/ml) and also with appropriate concentrations of MTZ when needed. Rifampin-resistant (Rifr) mutants were selected on medium with 5 μg of rifampin/ml. The plates were incubated at 37°C under microaerobic conditions (5% O2, 10% CO2, 85% N2). Rifr mutant frequencies were measured in five independent cultures, each derived recently from a different single colony and each growing exponentially.
The rdxA knockout mutant alleles rdxAΔ111 (a 111-bp in-frame deletion in the 630-bp rdxA gene), rdxA::cam (a null insertion mutant allele of rdxA containing a selectable chloramphenicol resistance [Camr] marker), and frxA::kan (a null mutant allele of frxA containing a selectable kanamycin resistance [Kanr] marker) have been described (15).
H. pylori transformation (electroporation) was carried out using standard methods. Replacement of the wild-type rdxA+ gene by the rdxAΔ111 allele was selected on media with 3 or 8 μg of MTZ per ml; transformants that had acquired the rdxA::cam and frxA::kan alleles were selected on media with 15 μg of chloramphenicol and 20 μg of kanamycin per ml, respectively.
Quantitative determination of MTZ sensitivity and resistance.
Young, exponentially growing cells were suspended from agar medium in phosphate-buffered saline buffer, a series of 10-fold dilutions of these suspensions was then prepared, and 10 μl of each dilution was spotted onto freshly prepared agar media containing appropriate concentrations of MTZ (0, 0.2, 0.5, 1.0, 1.5, 3, 8, 16, 32, or 64 μg/ml) as described previously (15). A strain was considered susceptible to a given concentration of MTZ if it decreased the efficiency of colony formation (i.e., efficiency of plating [EOP]) at least 10-fold or prolonged the time of incubation required for visible colonies to appear. This procedure, although somewhat labor-intensive, was far more sensitive for the research purposes described here than conventional determinations of MIC, which use fairly dense bacterial suspensions and typically score levels of drugs needed to completely block growth. When Mtzr mutants were rare (≤10−7), their frequency was estimated by spreading 50 μl of bacterial suspension (108 to 109 cells) on the surface of an entire petri plate of MTZ-containing agar. This is more accurate than our standard method of spotting 10-μl aliquots, when ≥107 cells are tested for mutation to resistance. Our protocols avoid complications that could stem from the mutagenicity of MTZ for H. pylori (26). We note that other investigators that had just argued that inactivation of frxA or of rdxA is sufficient to render SS1 or other Mtzs H. pylori strains Mtzr (16a, 16b) (a position with which we do not agree) had used a conventional MIC determination protocol.
DNA methods.
H. pylori genomic DNAs were isolated from confluent plate cultures using a Qiamp tissue kit (Qiagen Corporation, Chatsworth, Calif.) or by the cetyltrimethylammonium bromide-phenol method (3). PCR was carried out in 20-μl volumes containing 10 ng of genomic DNA, 10 pmol of each primer, 1 U of DNA polymerase (Promega) or high-fidelity Taq (Boehringer Mannheim), and 0.25 mmol of each deoxynucleoside triphosphate in standard PCR buffer. Reaction mixtures were incubated for 2 min at 94°C and then used for 30 cycles of 94°C for 40 s, 58°C for 40 s, and 72°C for 1 min per kilobase, with a final elongation step at 72°C for 10 min. PCR fragments were purified for sequencing with the Qiagen QIAquik PCR purification kit. Sequencing reactions were carried out using the Big Dye Terminator cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.), and products were run on an ABI automated sequencer in the Washington University Molecular Microbiology core facility. The primers used in these studies are listed in Table 1.
TABLE 1.
Primers used in these studies
| Primer | Sequence | PCR product size (bp) |
|---|---|---|
| rdxA-F | 5′-GCAGGAGCATCAGATAGTTCT-3′ | 886 |
| rdxA-R | 5′-GGGATTTTATTGTATGCTACAA-3′ | |
| rdxRT-F | 5′-GCATGCTGTGGTTGAATCTCAC-3′ | 347 |
| rdxRT-R | 5′-CGAGCGCCATTCTTGCAAGATGT-3′ | |
| rdxRT-F | 5′-GCATGCTGTGGTTGAATCTCAC-3′ | 238 |
| rdxRT-R2 | 5′-CCATGGCATTTTGTGATGGTTACT-3′ | |
| frxA-F | 5′-GGATATGGCAGCCGTTTATCATT-3′ | 780 |
| frxA-R | 5′-GAATAGGCATCATTTAAGAGATTA-3′ | |
| frxRT-F | 5′-GGACAGAGAACAAGTGGTTGCTT-3′ | 341 |
| frxRT-R | 5′-GCGAACCTAGAATTAGTGTCAT-3′ | |
| frxRT-F2 | 5′-CTTCAATCGGGCTTGAACCATGGA-3′ | 334 |
| frxRT-R2 | 5′-GCTGCCATCATCATGTTCGCCAT-3′ | |
| ureB-F | 5′-CGTCCGGCAATAGCTGCCATAGT-3′ | 463 |
| ureB-R | 5′-GTAGGTCCTGCTACTGAAGCCTTA-3′ |
RT-PCR analysis of mRNA levels.
Frozen bacterial cultures were streaked onto MTZ-free agar medium and incubated for 3 days, and the resulting bacterial growth was respread on fresh agar medium without or with MTZ (0 and 0.2 μg/ml for strain SS1; 0 and 1 μg/ml for strain 26695). Following 2 days of incubation, bacterial cells were collected and total RNA was prepared using a Qiagen RNeasy kit as recommended by the manufacturer. After elution from the RNeasy column, the RNA was treated with RNase-free DNase I, extracted twice with phenol-chloroform, and extracted once with chloroform-isoamyl alcohol. The RNA was precipitated with ammonium acetate (final concentration of 2.5 M) and 2.5 volumes of ice-cold ethanol, washed in 75% ethanol, and resuspended in RNase-free water. The integrity of the 16S and 23S rRNA was checked on a 1% agarose gel. Genomic DNA contamination was checked by PCR using Taq DNA polymerase without reverse transcriptase (RT). RT-PCR was carried out using the One-Step RT-PCR kit (Gibco-BRL) and primers frxRT-F and frxRT-R (for frxA mRNA), rdxRT-F and rdxRT-R (for rdxA mRNA), and ureB-F and ureB-R (for ureB mRNA). RT-PCR was carried out in a volume of 50 μl in a Perkin-Elmer GeneAmp PCR System 2400 thermal cycler under the following conditions: 50°C for 20 min, 94°C for 2 min, and 35 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 40 s, with a final incubation at 72°C for 10 min.
RESULTS
Extreme MTZ susceptibility of strain SS1.
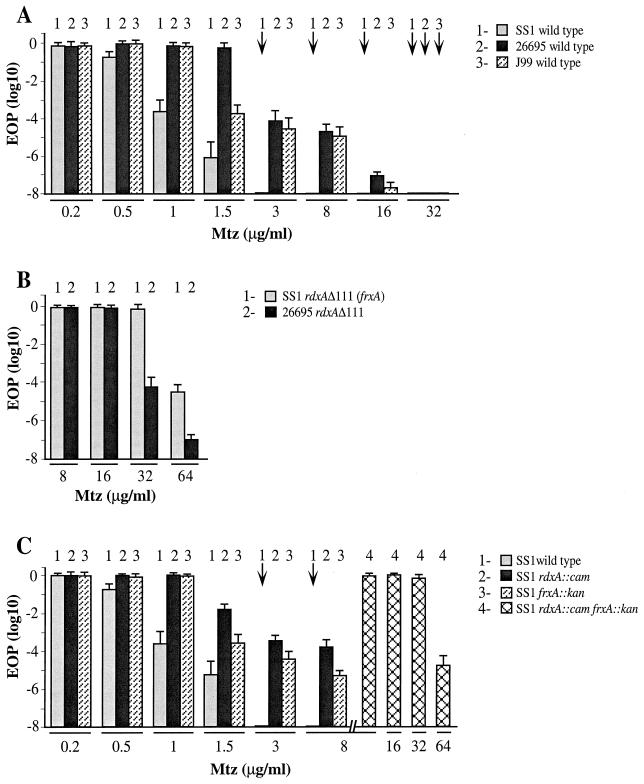
Initial tests indicated that H. pylori strain SS1 is susceptible to lower concentrations of MTZ than are most other Mtzs strains and is less likely than most to give rise to Mtzr mutant derivatives (Fig. 1A) (15). These tests involved the following steps: (i) spotting aliquots of young, exponentially growing cultures on media with appropriate low concentrations of MTZ and also on control medium with no MTZ, (ii) quantitating the EOP (i.e., efficiency of colony formation by single bacterial cells from appropriately diluted suspensions), and (iii) determining yields of new Mtzr mutants on media with MTZ at concentrations slightly higher than that required for lethality to the Mtzs parent. Figure 1A (column 1) shows that the EOP of SS1 on media with 1 and 1.5 μg of MTZ/ml was reduced ∼104- and ∼106-fold, respectively, relative to that on media with no MTZ or less MTZ (0.2 μg/ml). In addition, the colonies formed by SS1 on medium with 0.5 μg of MTZ per ml were unusually slow growing, requiring 6 days of incubation instead of 3 to 4 days to be visible to the naked eye. In contrast, most other clinical isolates from diverse parts of the world seemed to be fully resistant to at least 1 μg, and in most cases 1.5 μg, of MTZ per ml (15).
FIG. 1.
Profiles of intrinsic susceptibility and resistance to MTZ of H. pylori strains used in this study. Young, exponentially growing cultures were diluted, aliquots were spotted or spread on media with the indicated concentrations of MTZ, and colonies formed by aliquots of appropriate dilutions were counted, as detailed in Materials and Methods. Each mean and standard deviation survival value shown is based on two separate determinations from each of three young cultures, with each culture having been derived from a separate single colony isolate. (A) Strain SS1 and reference strains 26695 and J99 (references 18, 28, and 2, respectively). (B) rdxAΔ111 transformant derivatives of Mtzs strains. The designation SS1 rdxAΔ111 (frxA) refers to SS1 derivatives generated by transformation with rdxAΔ111 DNA and selection for resistance to 3 μg of MTZ per ml; these transformants were found to also contain point (frameshift) mutations in frxA. Strain 26695 rdxAΔ111 has been described previously (15) and does not contain a point mutation in frxA (J.-Y. Jeong and D. E. Berg, unpublished data). (C) Wild-type SS1 and isogenic derivatives with insertion mutations in rdxA, frxA, or both. These derivatives were generated by transformation and selection for the Camr or Kanr marker, not for Mtzr (reference 15; see also Materials and Methods). Downward arrows identify cases in which survival on MTZ-containing media was ≤10−8, relative to survival on media lacking MTZ.
Strain SS1 was also unusual in its very low Mtzr mutant frequency: ≤10−8, in contrast to ∼10−4 with most other strains (Fig. 1A) (15); the normal (∼10−4) frequency reflects both induction and selection for loss-of-function mutations in the rdxA gene (15, 26). In control experiments, new Rifr mutant derivatives of SS1 were found at frequencies of ∼2 × 10−8 (Table 2). This is about 10-fold lower than that observed using the 26695 reference strain under the same conditions (26), and it illustrates that H. pylori strains may differ in their intrinsic mutability. More important, however, this Rifr mutant frequency was increased nearly 100-fold by growth on medium with 0.5 μg of MTZ/ml, a partially inhibitory concentration. Equivalent frequencies of spontaneous and MTZ-induced mutation to Rifr were observed with an Mtzr derivative of SS1 (Table 2), as expected (26). We infer that SS1 is not immutable, despite the rarity of Mtzr mutants in young cultures. One explanation, which is supported by analyses described below, involves two different genes, each contributing independently to the special MTZ susceptibility of SS1, and a need to inactivate each of them to achieve an Mtzr phenotype.
TABLE 2.
Strain SS1 susceptibility to spontaneous and Mtz-induced mutation
| Strain | Growth conditiona (μg of MTZ/ml) | Mean no. of Rifr colonies/108 cellsb |
|---|---|---|
| Wild-type SS1 | 0 | 2 ± 1 |
| 0.5 | 144 ± 35 | |
| Mtzr SS1c | 0 | 2 ± 1 |
| 32 | 44 ± 14 | |
| 64 | 322 ± 181 |
Bacteria were grown on media without MTZ or with the indicated concentrations of MTZ for 48 h before being spread on medium with rifampin, essentially as described in reference 26.
Data are the mean values and standard deviations of five independent populations.
This strain contains the rdxAΔ111 allele that was used in DNA transformation and also contains a frameshift mutation in frxA (loss of one AT pair in a run of seven AT pairs at position 48). This additional point mutation was selected (and may have been induced as well) when transformed cells were spread on medium with 3 μg of MTZ/ml (see reference 26).
rdxA and frxA null mutant transformants of SS1.
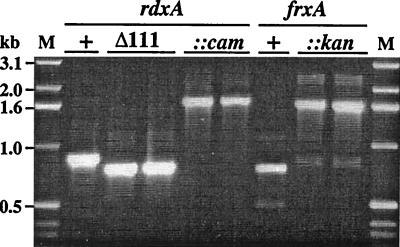
Four tests indicated that inactivation of both frxA and rdxA was needed to generate an Mtzr phenotype in SS1. First, transformation of SS1 with rdxAΔ111 DNA yielded a few Mtzr transformants on media with 3 or 8 μg of MTZ per ml (estimated frequency, ∼10−7), which were shown by PCR (Fig. 2) to contain the rdxAΔ111 allele in place of the wild-type rdxA+ allele. However, these transformants were resistant to 32 μg of MTZ per ml, a level that is attained in other strains only by inactivation of both frxA and rdxA and that is twice the level attained in typical strains by knocking out rdxA alone (Fig. 1B) (15). Second, SS1 transformants were generated with rdxA::cam and frxA::kan insertion mutant DNAs, with selection for Camr and Kanr phenotypes, respectively, rather than for an Mtzr phenotype. These transformants, which were verified by PCR (Fig. 2), were still susceptible to MTZ (albeit 1 μg/ml instead of 0.5 μg/ml). However, the rdxA mutant derivatives and the frxA mutant derivatives of SS1 were each much more easily mutated to Mtzr (frequency, ∼10−4) than was their parent (∼10−8) (Fig. 1C). The new Mtzr mutant derivatives of rdxA::cam transformants were also resistant to 32 μg of MTZ per ml even though they had been selected only on media with 3 or 8 μg of MTZ per ml. Third, an SS1 derivative with null rdxA and frxA alleles was then generated by transformation of SS1 frxA::kan with rdxA::cam DNA and selection for Camr. The resultant rdxA::cam frxA::kan strain was resistant to 32 μg of MTZ per ml. Fourth, Mtzr transformants of SS1 generated with the rdxAΔ111 allele with selection for an Mtzr phenotype were found by DNA sequencing of PCR-amplified frxA DNA to contain 1-bp deletion (frameshift) mutations in poly(A) tracts at positions 48 (three cases) and 310 (one case) in this HP0642 gene (for nucleotide sequence positions, see http://www.tigr.org/tdb/CMR/ghp/htmls/SplashPage.html). Based on these four results, we concluded that the unusual MTZ susceptibility of SS1 and its relative inability to mutate to Mtzr stems from the unusual activity of its FrxA reductase on low concentrations of MTZ in addition to the apparently quite normal activity of its RdxA reductase.
FIG. 2.
PCR tests that show replacement of wild-type alleles by deletion or insertion mutant alleles of the rdxA and frxA genes. The rdxA gene segment was PCR amplified from SS1 wild-type (lanes +), rdxAΔ111 (lanes Δ111), and rdxA::cam (lanes ::cam) strains using primers rdxA-F and rdxA-R. The frxA gene segment was PCR amplified from SS1 wild-type and frxA::kan (lanes ::kan) strains using primers frxA-F and frxA-R. Lane M indicates a 1-kb DNA ladder (size marker). Each of the lanes is from a separate transformant.
frxA mRNA is abundant in strain SS1.
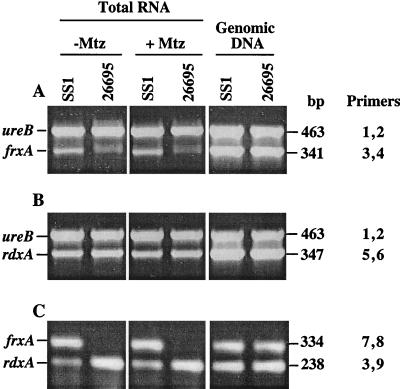
RT-PCR assays were carried out to estimate the levels of frxA and rdxA mRNAs relative to an internal ureB mRNA standard (Fig. 3A and B) or relative to each other (Fig. 3C). The data showed that frxA mRNA is of very low abundance in reference strain 26695 and at least 10-fold more abundant in SS1, whereas rdxA mRNA was less abundant than frxA mRNA in strain SS1. These levels were not affected by 2 days of growth with low (near-inhibitory) levels of MTZ. Thus, the high FrxA activity in strain SS1 probably stems from an unusually high level of frxA mRNA, not from a hyperactive form of the FrxA reductase enzyme.
FIG. 3.
RT-PCR amplification of the rdxA, frxA, and ureB gene segments from total RNA of strains 26695 and SS1. Ten nanograms of total RNA was used in each RT-PCR. Plus and minus signs indicate products from cells grown with and without MTZ, respectively, as detailed in Materials and Methods. The primers used are as follows: 1, ureB-F; 2, ureB-R; 3, frxRT-F; 4, frxRT-R; 5, rdxRT-F; 6, rdxRT-R; 7, frxRT-F2; 8, frxRT-R2; and 9, rdxRT-R2 (for sequences, see Table 1). The two pairs of primers indicated were used together in each RT-PCR or PCR. Lanes showing products made in control PCRs, using genomic DNAs as a template and Taq polymerase, are marked “Genomic DNA.”
DISCUSSION
We report here that the mouse-colonizing H. pylori strain SS1 is unusual in its susceptibility to very low levels of MTZ and in the very low frequency of resistant mutants found when cultures are spread on MTZ-containing media. These characteristics were traced to expression of the normally quiescent frxA reductase gene, along with apparently normal expression of its paralog rdxA, and a special requirement that both genes be inactivated if SS1 is to become Mtzr. The frxA gene seems not to be well expressed in most other Mtzs H. pylori strains, since they can be rendered Mtzr by inactivation of rdxA alone but not by inactivation of frxA alone (15). In principle, the high level of frxA mRNA in SS1 can be attributed to unusual production of a transcription activator or to escape from negative regulation, which might entail loss of a transcription repressor, gain of a constitutive promoter, or even stabilization of frxA mRNA.
Independent of the mechanism of abnormal frxA expression in SS1, the results of this study may have a special significance because SS1 and mice are widely used to study events during acute and chronic infection and to test possible drug and vaccine targets. SS1-infected mice also seemed promising as a model for how MTZ resistance emerges during human infection (14, 15). In a first test, 25 of 27 Mtzr SS1 isolates recovered from MTZ-treated mice (therapy chosen to suppress but not fully eradicate infection) contained mutations in rdxA (13) equivalent to those found in Mtzr clinical isolates from natural human infections (9, 15, 27). No need for mutations in other genes had been anticipated in those mouse studies, and none were sought.
The finding that inactivation of two genes, frxA as well as rdxA, is needed to make strain SS1 Mtzr helps explain why Mtzr derivatives of strain SS1 were so uncommon among the H. pylori organisms recovered from MTZ-treated mice (only 1 to 4% of the total). Indeed, given this need for two different mutations, were it not for the mutagenicity of products of MTZ activation (26); also this study), it would have been quite astonishing that any Mtzr derivatives of SS1 were ever obtained. However, our finding that Mtzr derivatives of SS1 could be obtained by transformation with rdxA mutant DNA and selection on MTZ agar and that such derivatives contain additional point mutations in frxA illustrates the potency of mutagenesis and selection in the emergence of phenotypes with complex (multigenic) bases. MTZ-induced mutation may also help explain why another group (16a, 16b) observed Mtzr H. pylori after transformation of Mtzs cells with frxA mutant DNAs. More generally, the mutagenicity of products of MTZ activation highlights a significant public health concern: that frequent MTZ use against diverse infections may contribute to the emergence of resistance in H. pylori to other useful anti-H. pylori drugs, such as clarithromycin, and, more generally, perhaps also speed host-specific adaptation and the evolution of virulence (26).
The possibility that the low frequency of Mtzr mutants among H. pylori isolates from MTZ-treated mice may reflect loss of fitness relative to their Mtzs parents also merits consideration. Indeed, we have recently found culture conditions in which Mtzr H. pylori strains grow less well than their Mtzs parents, and accordingly we have begun testing for fitness effects of rdxA and frxA inactivation in vivo. More generally, we are using the emerging understanding of genes involved in MTZ susceptibility and resistance to investigate how quantitative differences in activities of nonessential metabolic enzymes might affect the capacity of a given strain to infect different individual human hosts and the nature and severity of disease that such infections can cause.
ACKNOWLEDGMENTS
We are grateful to Adrian Lee for freely distributing SS1 to the Helicobacter community prior to publication. We thank Robert H. Gilman, Paul S. Hoffman, Asish K. Mukhopadhyay, and Alan J. Parkinson for many stimulating discussions.
This work was supported by NIH grants AI38166 and DK53727 to D.E.B. and Research Core Center Grant P30 DK52574 to the Division of Gastroenterology, Washington University Medical Center.
REFERENCES
- 1.Akopyants N S, Eaton K A, Berg D E. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect Immun. 1995;63:116–121. doi: 10.1128/iai.63.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1998. p. 1.1.1. [Google Scholar]
- 4.Berg D E, Gilman R H, Lelwala-Guruge J, Srivastava K, Valdez Y, Watanabe J, Miyagi J, Akopyants N S, Ramirez-Ramos A, Yoshiwara T H, Recavarren S, Leon-Barua R. Helicobacter pylori populations in Peruvian patients. Clin Infect Dis. 1997;25:996–1002. doi: 10.1086/516081. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M J. Helicobacter pylori and gastric diseases. Br Med J. 1998;316:1507–1510. doi: 10.1136/bmj.316.7143.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debets-Ossenkopp Y J, Pot R G J, van Westerloo D J, Goodwin A, Vandenbroucke-Grauls C M J E, Berg D E, Hoffman P S, Kusters J G. Insertion of mini-IS605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob Agents Chemother. 1999;43:2657–2662. doi: 10.1128/aac.43.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards D I. Nitroimidazole drugs—action and resistance mechanisms. I. Mechanisms of action. J Antimicrob Chemother. 1993;31:9–20. doi: 10.1093/jac/31.1.9. [DOI] [PubMed] [Google Scholar]
- 8.Glupczynski Y. Antimicrobial resistance in Helicobacter pylori: a global overview. Acta Gastro-enterol Belg. 1998;61:357–366. [PubMed] [Google Scholar]
- 9.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J O, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 10.Graham D Y. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 11.Graham D Y, Go M F. Antibacterial treatment of gastric ulcers. N Engl J Med. 1995;333:190. doi: 10.1056/NEJM199507203330312. [DOI] [PubMed] [Google Scholar]
- 12.Guruge J L, Falk P G, Lorenz R G, Dans M, Wirth H P, Blaser M J, Berg D E, Gordon J I. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc Natl Acad Sci USA. 1998;95:3925–3930. doi: 10.1073/pnas.95.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenks P J, Ferrero R L, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 14.Jenks P J, Labigne A, Ferrero R L. Exposure to metronidazole in vivo readily induces resistance in Helicobacter pylori and reduces the efficacy of eradication therapy in mice. Antimicrob Agents Chemother. 1999;43:777–781. doi: 10.1128/aac.43.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong J-Y, Mukhopadhyay A K, Dailidiene D, Wang Y, Velapatiño B, Gilman R H, Parkinson A J, Nair G B, Wong B C Y, Lam S K, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea M L, Ito Y, Kersulyte D, Lee H-K, Gong Y, Goodwin A, Hoffman P S, Berg D E. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J Bacteriol. 2000;182:5082–5090. doi: 10.1128/jb.182.18.5082-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleanthous H, Myers G A, Georgakopoulos K M, Tibbitts T J, Ingrassia J W, Gray H L, Ding R, Zhang Z-Z, Lei W, Nichols R, Lee C K, Ermak T H, Monath T P. Rectal and intranasal immunizations with recombinant urease induce distinct local and serum immune responses in mice and protect against Helicobacter pylori infection. Infect Immun. 1998;66:2879–2886. doi: 10.1128/iai.66.6.2879-2886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Kwon D H, Kato M, El-Zaatari F A, Osato M S, Graham D Y. Frame-shift mutations in NAD(P)H flavin oxidoreductase encoding gene (frxA) from metronidazole resistant Helicobacter pylori ATCC43504 and its involvement in metronidazole resistance. FEMS Microbiol Lett. 2000;188:197–202. doi: 10.1111/j.1574-6968.2000.tb09193.x. [DOI] [PubMed] [Google Scholar]
- 16b.Kwon D-H, El-Zaatari F A K, Kato M, Osato M S, Reddy R, Yamaoka Y, Graham D Y. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:2133–2142. doi: 10.1128/aac.44.8.2133-2142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee A. Animal models of gastroduodenal ulcer disease. Bailliere's Best Pract Res Clin Gastroenterol. 2000;14:75–96. doi: 10.1053/bega.2000.0060. [DOI] [PubMed] [Google Scholar]
- 18.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 19.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 20.Marshall B J. Helicobacter pylori. Am J Gastroenterol. 1994;89(Suppl. 8):S116–S128. [PubMed] [Google Scholar]
- 21.Megraud F. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology. 1998;115:1278–1282. doi: 10.1016/s0016-5085(98)70101-5. [DOI] [PubMed] [Google Scholar]
- 22.Parsonnet J. Helicobacter and gastric adenocarcinoma. In: Parsonnet J, editor. Microbes and malignancy: infection as a cause of human cancers. New York, N.Y: Oxford University Press; 1999. pp. 372–408. [Google Scholar]
- 23.Raudonikiene A, Zakharova N, Su W W, Jeong J Y, Bryden L, Hoffman P S, Berg D E, Severinov K. Helicobacter pylori with separate beta- and beta′-subunits of RNA polymerase is viable and can colonize conventional mice. Mol Microbiol. 1999;32:131–138. doi: 10.1046/j.1365-2958.1999.01336.x. [DOI] [PubMed] [Google Scholar]
- 24.Samuelson J. Why metronidazole is active against both bacteria and parasites. Antimicrob Agents Chemother. 1999;43:1533–1541. doi: 10.1128/aac.43.7.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirai M, Arichi T, Nakazawa T, Berzofsky J A. Persistent infection by Helicobacter pylori down-modulates virus-specific CD8+ cytotoxic T cell response and prolongs viral infection. J Infect Dis. 1998;177:72–80. doi: 10.1086/513827. [DOI] [PubMed] [Google Scholar]
- 26.Sisson G, Jeong J-Y, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg D E, Hoffman P S. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tankovic J, Lamarque D, Delchier J-C, Soussy C-J, Labigne A, Jenks P J. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:608–613. doi: 10.1128/aac.44.3.608-613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K, Klenk H, Gill S, Dougherty B, Nelson K, Quackenbush J, Zhou L, Kirkness E, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H, Glodek A, McKenney K, Fitzegerald L, Lee N, Adams M, Hickey E, Berg D, Gocayne J, Utterback T, Peterson J, Kelley J, Cotton M, Weidman J, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W, Borodovsky M, Karp P, Smith H, Fraser C, Venter J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 29.Westblom T U, Czinn S J, Nedrud J G, editors. Current topics in microbiology and immunology. 241. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Berlin, Germany: Springer Press; 1999. [Google Scholar]