Abstract
Lung cancer is a complex disease influenced by a variety of genetic and environmental factors. The cytokine interleukin 1 encoded by IL1B is an important mediator of the inflammatory response, and is involved in a variety of cellular activities. The effect of single nucleotide polymorphisms (SNP) at IL1B has been investigated in relation to cancer with inconsistent results. This Northeastern-Chinese case–control study involving 627 cases and 633 controls evaluated the role of three haplotype-tagging single nucleotide polymorphisms (htSNP) (rs1143633, rs3136558 and rs1143630) representing 95% of the common haplotype diversity across the IL1B gene and assessed interactions with IL1B, PPP1R13L, POLR1G and smoking duration in relation to lung cancer risk. The analyses of five genetic models showed associations with lung cancer risk for rs1143633 in the dominant model [adjusted-OR (95% CI) = 0.67 (0.52–0.85), P = 0.0012] and rs3136558 in the recessive model [adjusted-OR (95% CI) = 1.44 (1.05–1.98), P = 0.025]. Haplotype4 was associated with increased lung cancer risk [adjusted-OR (95% CI) = 1.55 (1.07–2.24), P = 0.021]. The variant G-allele of rs1143633 was protective in smoking sub-group of > 20 years. Using multifactor dimensionality reduction (MDR) analyses, we identified the three best candidate models of interactions and smoking-duration or IL1B rs1143633 as main effect. In conclusion, our findings suggest that IL1B SNP rs1143633 may associate with lower risk of lung cancer, confirming previously identified marker; IL1B SNP rs3136558 and haplotype4 consisting of IL1B htSNPs may associate with increasing risk of lung cancer; interactions of IL1B with POLR1G or PPP1R13L or smoking-duration, which is independent or combined, may involve in risk of lung cancer and lung squamous cell carcinoma.
Subject terms: Cancer, Biomarkers, Molecular medicine
Introduction
Lung cancer is an important and prevalent cause of cancer-related death worldwide and constitutes a serious public health problem. Lung cancer is a complex disease influenced by a variety of genetic and environmental factors. Susceptibility gene/single nucleotide polymorphisms (SNP) have been linked to lung cancer risks. Tobacco remains the leading risk factor for lung cancer1. Chronic lung diseases that entail chronic inflammation have been suspected to play a role in the pathogenesis of lung cancer1. Another possible mechanism may involve gene–gene or gene–environment interactions in relation to lung cancer2.
IL1B (interleukin 1 beta) (HGNC ID: 5992, Gene ID: 3553) is located on chromosome 2q14.1. The protein encoded by IL1B is a member of the interleukin 1 cytokine family (https://www.ncbi.nlm.nih.gov/gene/3553). The IL1 cluster consists of three related genes: IL1A, IL1B, and IL1RA, which encode the signal proteins IL1A, IL1B, and their receptor, IL1RA, respectively3. IL1A and IL1B are pro-inflammatory cytokines, whereas IL1RA is an anti-inflammatory cytokine and competes with IL1A and IL1B for binding to the IL1 receptors4. The interleukin 1 cytokine is an important mediator of the inflammatory response, and is involved in a variety of cellular activities, including cell proliferation, differentiation, apoptosis (https://www.ncbi.nlm.nih.gov/gene/3553) and innate immunity5. Cytokines are known as important regulators in cancer and are involved in inflammatory and immunological responses6.
The IL1B gene has been extensively investigated in relation to cancers and inflammatory and infectious diseases. The effect of SNPs at IL1B has been investigated in relation to cancer with inconsistent results in predominantly European and Asian populations6–13. Polymorphisms in cytokine genes may be functional14, modify the inflammatory and immune responses and therefore modulate risk of lung cancer. Non-functional polymorphisms may be genetically linked to functional polymorphisms. There are only few studies involving IL1B SNPs and lung cancer risk, retrieving only one study of Caucasia-Danes7. The molecular mechanism underlying carcinogenic IL1B inflammatory-induced lung cancer is still unclear completely.
Two genes PPP1R13L [protein phosphatase 1 regulatory subunit 13 like] (HGNC ID: 18838, Gene ID: 10848) and POLR1G [RNA polymerase I subunit G, Previous symbols CD3EAP: CD3e molecule, epsilon-associated protein] (HGNC ID: 24219, Gene ID: 10849) located on chromosome 19q13.32 relate to DNA repair and cell survival and cell proliferation, respectively. We previously reported that PPP1R13L rs1970764, POLR1G rs967591 and rs735482 were associated with lung cancer or interacted in relation to lung cancer risk among both Caucasian Danes and Chinese15–18. IL1B, PPP1R13L and POLR1G had the effect on apoptosis and DNA repair pathways.
IL1B belongs to pathways of cytokines and inflammatory response, immune system, and lung fibrosis. PPP1R13L and POLR1G share pathway of gene expression (transcription) [https://www.ncbi.nlm.nih.gov/gene/3553, /10848, /10849]. Significant advances in statistical approaches make it possible considering the main pathways to which the genes belongs and possible covariates, as required in the analysis of complex traits19. A deeper understanding of common pathways of inflammation and cancer may increase our understanding of the role of inflammation and cancer20.
In this case–control study of Northeastern-Chinese, we explored the role of haplotype-tagging single nucleotide polymorphisms (htSNP) tagging 95% of common haplotypes across the IL1B gene on lung cancer risk. And we assessed gene–gene or gene–gene–environment interactions between genes of pathway of cytokines and inflammatory response and gene expression (transcription) pathway related to lung cancer risk, including the interaction between IL1B htSNPs, PPP1R13L and POLR1G risk SNPs and smoking-duration.
Results
Population characteristics
The baseline characteristics of the lung cancer patients and healthy controls are shown in Table 1. The IL1B three htSNPs were evaluated in a Northeastern-Chinese hospital-based case–control study involving 627 lung cancer cases and 633 controls. There were no statistically significant differences for the distribution of age and sex between cases and controls. However, there were more cases with family history of cancer and longer smoking history (> 20 years) (both P < 0.0001).
Table 1.
Distribution of selected characteristics in Chinese case–control study population.
| Characteristics | Cases n (%) | Controls n (%) | P value |
|---|---|---|---|
| Over all | 627 | 633 | |
| Age (years) | |||
| Mean (± SD) | 58 (± 10.4) | 58 (± 10.5) | 0.846a |
| ≤ 40 | 29 (4.6%) | 29 (4.6%) | |
| 41–50 | 109 (17.4%) | 125 (19.7%) | |
| 51–60 | 222 (35.4%) | 214 (33.8%) | 0.748b |
| > 60 | 267 (42.6%) | 265 (41.9%) | |
| Sex | |||
| Female | 185 (29.5%) | 184 (29.1%) | |
| Male | 442 (70.5%) | 449 (70.9%) | 0.86b |
| Family history of cancer | |||
| No | 536 (85.5%) | 628 (99.2%) | |
| Yes | 91 (14.5%) | 5 (0.8%) | < 0.0001b |
| Smoking duration | |||
| Never | 241 (38.4%) | 333 (52.6%) | |
| ≤ 20 (years) | 104 (16.6%) | 98 (15.5%) | < 0.0001b |
| > 20 (years) | 282 (45.0%) | 202 (31.9%) | |
| Histopathology | |||
| Lung squamous cell carcinoma | 266 (42.4%) | ||
| Lung adenocarcinoma | 251(40.0%) | ||
| Other | 110 (17.6%) | ||
aFor Mann–Whitney U test (P < 0.0001 for Shapiro–Wilk test).
bFor χ2 test (two-sided), boldface indicates statistical significance.
The minor-allele frequencies were determined among the controls (G = 0.46, C = 0.38 and A = 0.16 for rs1143633, rs3136558 and rs1143630, respectively). The minor-allele frequency of rs1143633 was similar to one in HapMap-HCB (Han Chinese in Beijing) reported by NCBI dbSNP database (https://www.ncbi.nlm.nih.gov/snp) (P = 0.745) whereas rs3136558 (P = 0.002) and rs1143630 (P = 0.027) were not (Table 2). The genotype distributions in the control group were in Hardy–Weinberg equilibrium for rs1143633 (P = 0.29), rs3136558 (P = 0.26), and rs1143630 (P = 0.77).
Table 2.
Characteristics of IL1B three htSNPs selected and SNPs in PPP1R13L and POLR1G.
| Gene/chromosome/rsa | Position | Functional consequence | Base change | Allele frequency in HapMap HCBb | Control MAFc in this study |
|---|---|---|---|---|---|
| IL1B Chr2q14.1 | |||||
| rs1143633 | 112832890 | Intron | A/G | A0.5465/G0.4535 | G0.46 |
| rs3136558 | 112833698 | Intron | T/C | T0.7442/C0.2558 | C0.38 |
| rs1143630 | 112834078 | Intron | C/A | C0.9024/A0.0976 | A0.16 |
| PPP1R13L Chr19q13.32 | |||||
| rs1970764 | 45387615 | Intron | A/G | No | G0.48d |
| POLR1G Chr19q13.32 | |||||
| rs967591 | 45406676 | 5′ UTR | G/A | G0.5250/A0.4750e | A0.42d |
| rs735482 | 45408744 | Exon, missense | A/C | A0.5581/C0.4419 | C0.45d |
aInformation from NCBI SNP database (GRCh38.p12) and HapMap database. From 3′–5′.
bHan Chinese in Beijing.
cMinor allele frequency.
dFrom previous result (Yin et al.18), here this is employed for interaction analysis.
eCHB + JPT (Han Chinese in Beijing + Japanese from 1000GENOMES).
Selected IL1B htSNPs and lung cancer risk
Genotype distributions and lung cancer risk for the three IL1B htSNPs in co-dominant, dominant, recessive, over-dominant and log-additive models after adjustment smoking-duration were analyzed (Table 3). For whole study group, rs1143633 in the dominant model [Odd Ratio (95% confidence interval): adjusted-OR (95% CI) = 0.67 (0.52–0.85), P = 0.0012] (and also including co-dominant model and log-additive model) and rs3136558 in the recessive model [adjusted-OR (95% CI) = 1.44 (1.05–1.98), P = 0.025] (and also including co-dominant model) showed in association with lung cancer risk. No significant models with association were found for rs1143630. For subgroup stratified by smoking duration, G variant-allele of rs1143633 showed the protective effect in > 20 years subgroup (Table 4). No significant associations were found for two other htSNPs (data not shown). For subgroup stratified by histopathology, rs1143633 in the log-additive model [adjusted-OR (95% CI) = 0.59 (0.47–0.75), P < 0.0001] and rs3136558 in the log-additive model [adjusted-OR (95% CI) = 1.35 (1.08–1.69), P = 0.0086] were associated with the disease risk in the subgroup of lung squamous cell carcinoma (Table 3). No significant association was found for subgroups of lung adenocarcinoma and other histoathology (data not shown).
Table 3.
Associations of single htSNP in IL1B with lung cancer risk.
| Type/modela/rs | Genotype | Controls 633 n (%) |
Cases 627 n (%) |
OR (95 CI)b,c | P valuec | AICd |
|---|---|---|---|---|---|---|
| Whole group: lung cancer | ||||||
| rs1143633 (A>G) | ||||||
| Co-dominant | A/A | 169 (28.2%) | 224 (36.4%) | 1.00 | ||
| G/A | 311 (51.9%) | 289 (47%) | 0.68 (0.53–0.89) | 0.0044 | 1651.4 | |
| G/G | 119 (19.9%) | 102 (16.6%) | 0.62 (0.44–0.87) | |||
| Dominant | A/A | 169 (28.2%) | 224 (36.4%) | 1.00 | ||
| G/A-G/G | 430 (71.8%) | 391 (63.6%) | 0.67 (0.52–0.85) | 0.0012 | 1649.7 | |
| Recessive | A/A-G/A | 480 (80.1%) | 513 (83.4%) | 1.00 | ||
| G/G | 119 (19.9%) | 102 (16.6%) | 0.78 (0.58–1.05) | 0.1 | 1657.6 | |
| Over-dominant | A/A-G/G | 288 (48.1%) | 326 (53%) | 1.00 | ||
| G/A | 311 (51.9%) | 289 (47%) | 0.81 (0.65–1.02) | 0.077 | 1657.1 | |
| Log-additive | – | – | – | 0.77 (0.65–0.91) | 0.0021 | 1650.8 |
| rs3136558 (T>C) | ||||||
| Co-dominant | T/T | 223 (37.2%) | 214 (34.8%) | 1.00 | ||
| C/T | 296 (49.4%) | 293 (47.6%) | 1.02 (0.79–1.31) | 0.079 | 1657.2 | |
| C/C | 80 (13.4%) | 108 (17.6%) | 1.46 (1.03–2.06) | |||
| Dominant | T/T | 223 (37.2%) | 214 (34.8%) | 1.00 | ||
| C/T-C/C | 376 (62.8%) | 401 (65.2%) | 1.11 (0.88–1.41) | 0.38 | 1659.5 | |
| Recessive | T/T-C/T | 519 (86.6%) | 507 (82.4%) | 1.00 | ||
| C/C | 80 (13.4%) | 108 (17.6%) | 1.44 (1.05–1.98) | 0.025 | 1655.2 | |
| Over-dominant | T/T-C/C | 303 (50.6%) | 322 (52.4%) | 1.00 | ||
| C/T | 296 (49.4%) | 293 (47.6%) | 0.91 (0.73–1.15) | 0.44 | 1659.6 | |
| Log-additive | – | – | – | 1.16 (0.99–1.37) | 0.073 | 1657 |
| rs1143630 (C>A) | ||||||
| Co-dominant | C/C | 422 (70%) | 407 (66.9%) | 1.00 | ||
| A/C | 164 (27.2%) | 190 (31.2%) | 1.19 (0.92–1.53) | 0.21 | 1655.7 | |
| A/A | 17 (2.8%) | 11 (1.8%) | 0.67 (0.31–1.46) | |||
| Dominant | C/C | 422 (70%) | 407 (66.9%) | 1.00 | ||
| A/C-A/A | 181 (30%) | 201 (33.1%) | 1.14 (0.89–1.46) | 0.29 | 1655.7 | |
| Recessive | C/C-A/C | 586 (97.2%) | 597 (98.2%) | 1.00 | ||
| A/A | 17 (2.8%) | 11 (1.8%) | 0.64 (0.29–1.38) | 0.25 | 1655.5 | |
| Over-dominant | C/C-A/A | 439 (72.8%) | 418 (68.8%) | 1.00 | ||
| A/C | 164 (27.2%) | 190 (31.2%) | 1.21 (0.94–1.55) | 0.15 | 1654.7 | |
| Log-additive | – | – | – | 1.07 (0.86–1.33) | 0.54 | 1656.5 |
| Histopahtology subgroup: lung squamous cell carcinomae | ||||||
| rs1143633 (A>G) | ||||||
| Log-additive | – | – | – | 0.59 (0.47–0.75) | < 0.0001 | 980.8 |
| rs3136558 (T>C) | ||||||
| Log-additive | – | – | – | 1.35 (1.08–1.69) | 0.0086 | 994.5 |
aDominant model: AB (Heterozygote) + BB (Homozygous variant-type) versus AA (Homozygous wild-type), Recessive model: BB versus AA + AB, Co-dominant model: AB versus AA and BB versus AA, Over-dominant model: AB versus AA + BB, Log-additive model: Analysis of trend where AA is ‘0’, AB is ‘1’ and BB is ‘2’.
bOR (odd ratio), 95% CI (95% confidence interval), adjusted for smoking duration.
cBoldface indicates statistical significance.
dAkaike’s Information Criterion.
eOnly list the best significant models.
Table 4.
rs1143633's genotypes within smoking-duration.
| Smoking-duration | Genotype | Controls | Cases | OR (95% CI) | P value |
|---|---|---|---|---|---|
| Never | AA | 96 | 87 | 1.0 | |
| GA | 163 | 110 | 0.75 (0.51–1.09) | 0.126 | |
| GG | 60 | 38 | 0.70 (0.42–1.15) | 0.159 | |
| GA + GG | 223 | 148 | 0.73 (0.51–1.05) | 0.087 | |
| ≤ 20 (years) | AA | 30 | 39 | 1.00 | |
| GA | 44 | 49 | 0.86 (0.46–1.60) | 0.628 | |
| GG | 17 | 13 | 0.59 (0.25–1.40) | 0.227 | |
| GA + GG | 61 | 62 | 0.78 (0.43–1.41) | 0.415 | |
| > 20 (years) | AA | 43 | 98 | 1.00 | |
| GA | 104 | 130 | 0.55 (0.35–0.85)a | 0.007a | |
| GG | 42 | 51 | 0.53 (0.31–0.92)a | 0.022a | |
| GA + GG | 146 | 181 | 0.54 (0.36–0.83)a | 0.004a |
aBoldface indicates statistical significance.
Analysis of linkage disequilibrium (LD) and haplotype
Linkage disequilibrium analysis was examined. The analyses showed that LD was moderate between rs1143633 and rs3136558 (D′ value = 0.5814) and between rs3136558 and rs1143630 (D′ value = 0.462), and very low between rs1143633 and rs1143630 (D′ value = 0.0701) in present population (Table 5). Haplotype association analysis of IL1B htSNPs with lung cancer risk showed that haplotype4 (rs1143633A-rs3136558C-rs1143630A) was associated with increased risk of lung cancer after adjustment for smoking duration [adjusted-OR (95% CI) = 1.55 (1.07–2.24), P = 0.021] and that haplotype2 was marginally associated with increased risk of lung cancer (Table 6).
Table 5.
Pair-wise linkage disequilibrium analysis for IL1B three htSNPs.
| Statistic/rs number | |||
|---|---|---|---|
| D′ statistic | rs1143633 | rs3136558 | rs1143630 |
| rs1143633 | – | 0.5814 | 0.0701 |
| rs3136558 | – | – | 0.462 |
| rs1143630 | – | – | – |
| r statistic | rs1143633 | rs3136558 | rs1143630 |
|---|---|---|---|
| rs1143633 | – | − 0.4095 | 0.0365 |
| rs3136558 | – | – | 0.2568 |
| rs1143630 | – | – | – |
| P value | rs1143633 | rs3136558 | rs1143630 |
|---|---|---|---|
| rs1143633 | – | 0 | 0.0742 |
| rs3136558 | – | – | 0 |
| rs1143630 | – | – | – |
Table 6.
Haplotype association of IL1B htSNPs with lung cancer risk.
| Haplotypea,b | Constructionc | Control frequency | Case frequency | OR (95% CI) | P value |
|---|---|---|---|---|---|
| 1 | GTC | 0.3274 | 0.295 | 1.00 | – |
| 2 | ACC | 0.2323 | 0.2531 | 1.26 (1.00–1.59) | 0.054 |
| 3 | ATC | 0.2328 | 0.24 | 1.17 (0.91–1.49) | 0.22 |
| 4 | ACA | 0.0693 | 0.0947 | 1.55 (1.07–2.24)d | 0.021d |
| 5 | GTA | 0.0523 | 0.04 | 0.86 (0.51–1.46) | 0.58 |
| 6 | GCC | 0.0432 | 0.0374 | 0.93 (0.55–1.59) | 0.8 |
| 7 | GCA | 0.0353 | 0.0284 | 0.91 (0.49–1.69) | 0.77 |
| Rare | - | - | - | 1.67 (0.49–5.71) | 0.42 |
aAdjusted for smoking duration, Global haplotype association P-value = 0.1
bThree-locus order: rs1143633- rs3136558- rs1143630.
cUnderlined indicates minor allele.
dBoldface indicates statistical significance.
Analysis of multifactor dimensionality reduction (MDR) approach
Table 7 summarizes the best candidate models of interactions of selected attributes related to lung cancer risk using MDR approach. Three best models were set for whole study group: in the combined interaction analysis of IL1B, PPP1R13L, POLR1G and smoking-duration, both the two-factor model and the three-factor model were statistically significant, but the three-factor model (IL1B rs3136558, POLR1G rs967591 and smoking duration) had a relatively higher values of balanced accuracy overall of 0.6062 and cross-validation consistency of 8/10 that was significant at the P-value 0.0100–0.0110. In the conjoined interaction analysis of IL1B and smoking-duration, two-factor model, the three-factor model and the fourth-factor model were all statistically significant, but the fourth-factor model (IL1B rs1143633, rs3136558 and rs1143630 and smoking duration) had a relatively higher values of balanced accuracy overall of 0.6112 and cross-validation consistency of 10/10 that was significant at the P-value 0.0040–0.0050. In the joint interaction analysis of IL1B, PPP1R13L, POLR1G, both the two-factor model and the three-factor model were statistically significant, but the three-factor model (IL1B rs1143633, PPP1R13L rs1970764, POLR1G rs735482) had a relatively higher values of balanced accuracy overall of 0.5973 and cross-validation consistency of 10/10 that was significant at the P-value 0.0130–0.0140. Smoking-duration presented interaction main-effect in model consisting of IL1B htSNPs-PPP1R13L and POLR1G SNPs-smoking duration or IL1B htSNPs-smoking duration. IL1B rs1143633 presented interaction main-effect in model consisting of IL1B htSNPs. For histopathology study subgroup: only conjoined interaction analysis of IL1B and smoking-duration was performed. In the subgroup of squamous cell carcinoma, both the two-factor model (IL1B rs1143633 and smoking duration) and the four-factor model (IL1B rs1143633, rs3136558 and rs1143630 and smoking duration) had relatively higher values of balanced accuracy overall of and cross-validation consistency of 10/10 that were statistically significant at the P-values, and smoking duration showed obvious main effects. In the subgroup of other histopathology, the two-factor model had statistical significance. In the subgroup of lung adenocarcinoma, no interaction was identified.
Table 7.
The best candidate models of interactions of selected attributes related to lung cancer by MDR approach.
| Model factora,b | Attribute | Bal. Acc.c overall | Bal. Acc. CVd training | Bal. Acc. CV testing | CV consistency | P valuee |
|---|---|---|---|---|---|---|
| Whole group | ||||||
| Lung cancer | ||||||
| IL1B + PPP1R13L + POLR1G + smoking-duration | ||||||
| One-factor | Smoking | 0.5708 | 0.5708 | 0.5708 | 10/10 | 0.0040–0.0050 |
| Two-factor | Smoking | |||||
| rs1143633 | 0.5849 | 0.5866 | 0.5649 | 5/10 | 0.0090–0.0100 | |
| Three-factor | Smoking | |||||
| rs3136558 | ||||||
| rs967591 | 0.6062 | 0.6084 | 0.5602 | 8/10 | 0.0100–0.0110 | |
| IL1B + smoking-duration | ||||||
| One-factor | Smoking | 0.5708 | 0.5708 | 0.5708 | 10/10 | < 0.0010 |
| Two-factor | Smoking | |||||
| rs1143633 | 0.5849 | 0.5862 | 0.5721 | 6/10 | < 0.0010 | |
| Three-factor | Smoking | |||||
| rs1143633 | ||||||
| rs1143630 | 0.5944 | 0.5964 | 0.5619 | 8/10 | < 0.0010 | |
| Fourth-factor | Smoking | |||||
| rs1143633 | ||||||
| rs3136558 | ||||||
| rs1143630 | 0.6112 | 0.6139 | 0.5588 | 10/10 | 0.0040–0.0050 | |
| IL1B + PPP1R13L + POLR1G | ||||||
| Two-factor | rs1143633 | |||||
| rs1970764 | 0.5682 | 0.5689 | 0.5507 | 9/10 | 0.0440–0.0450 | |
| Three-factor | rs1143633 | |||||
| rs1970764 | ||||||
| rs735482 | 0.5973 | 0.5987 | 0.5576 | 10/10 | 0.0130–0.0140 | |
| IL1B | ||||||
| One-factor | rs1143633 | 0.5451 | 0.5451 | 0.5451 | 10/10 | 0.0290–0.0300 |
| Histopahtology subgroup | ||||||
| Lung squamous cell carcinoma | ||||||
| IL1B + smoking-duration | ||||||
| One-factor | Smoking | 0.6315 | 0.6318 | 0.6227 | 10/10 | < 0.0010 |
| Two-factor | Smoking | |||||
| rs1143633 | 0.6532 | 0.6533 | 0.6472 | 10/10 | < 0.0010 | |
| Three-factor | Smoking | |||||
| rs1143633 | ||||||
| rs1143630 | 0.6619 | 0.6645 | 0.6123 | 5/10 | < 0.0010 | |
| Fourth-factor | Smoking | |||||
| rs1143633 | ||||||
| rs3136558 | ||||||
| rs1143630 | 0.6785 | 0.6827 | 0.5871 | 10/10 | 0.0030–0.0040 | |
| Other | ||||||
| IL1B + smoking-duration | ||||||
| Two-factor | Smoking | |||||
| rs3136558 | 0.6178 | 0.6181 | 0.6073 | 10/10 | 0.0090–0.0100 | |
aAnalyzed by MDR 3.0.3. dev. Jar, data of PPP1R13L + CD3EAP from previous study (Yin et al.18).
bOnly list statistical significant models.
cBalanced accuracy.
dCross-validation.
eP value based on 1000 permutation test. Boldface indicates statistical significance.
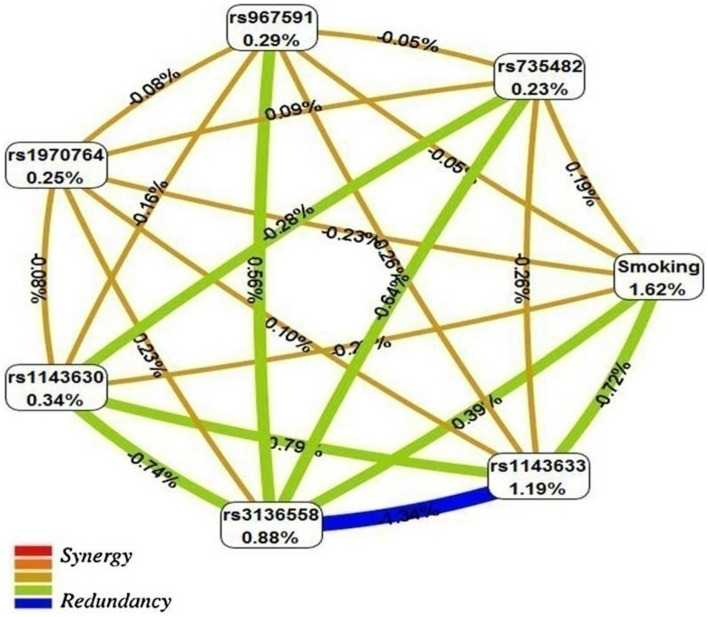
Figure 1 shows the interaction entropy model from the interaction analysis of IL1B htSNPs, PPP1R13L and POLR1G SNPs and smoking-duration built using the MDR software. The entropy-based model indicated that some values between 7 attribute interaction presented medium-level interaction or independence whereas degree of synergy interaction was not apparent in the current analysis between 7 attributes.
Figure 1.
Interaction entropy model. This graphical model, describes the percent entropy that is explained by each selected attributes or pair-wise combination in our study population. Positive percent entropy indicates information gain (IG) or synergy and negative percent indicates lack of information gain (IG) or redundancy. Schematic coloration used in the visualization tools represents a continuum from synergy (i.e. non-additive) to redundancy. Red represents a high degree of synergy interaction, orange a lesser degree (both colors are not apparent in the current analysis), brown represents medium-level interaction or independence; and green and blue represent redundancy between the markers. This image was created by MDR software (3.0.3. dev. Jar) (https://sourceforge.net/projects/mdr/)36.
Discussion
Studies addressing IL1B SNPs in cancer
Previous epidemiology studies have identified association of IL1B SNPs with cancers risk but with inconsistent results6–13. A Caucasian-Danish prospective study showed that variant allele carriers of IL1B SNP rs1143627 (− 31T>C) were at increased risk of lung cancer [Dominant model: IRR (incidence rate ratio) (95% CI) = 1.51 (1.08–2.12)]7. An Asian-Chinese case–control study reported IL1B SNPs rs16944 (− 511G>A) and rs1143623 (C>G) was associated with decreased breast cancer risk [co-dominant model: OR (95% CI) = 0.60 (0.41–0.90), P = 0.034; co-dominant model: OR (95% CI) = 0.65 (0.45–0.94), P = 0.023]6. A Russian case–control study revealed a significant association between IL1B SNP rs1143623 (− 1473G>C) and risk of rectal cancer [co-dominant model: OR (95% CI) = 1.67 (1.06–2.63), P = 0.048]8. An Asian-Chinese case–control study reported association between IL1B SNP rs1143634 (+ 3954C>T) and gastric cancer risk [co-dominant model: OR [95% CI) = 6.93 (3.13–15.36)]4. A Asian-Korean study suggested that IL1B SNPs rs1143633 (A>G) and rs1143627 (T>C) protected against hepatocellular carcinoma [dominant model: OR (95% CI) = 0.59 (0.37–0.94), P = 0.027; dominant model: OR (95% CI) = 0.56 (0.34–0.91), P = 0.019] and IL1B SNP rs3917356 (G>A) increased the risk of hepatocellular carcinoma [recessive model: OR (95% CI) = 2.58 (1.53–4.33), P < 0.001]9. A Caucasian-Danish prospective case-cohort study showed that variant genotypes of IL1B SNPs rs4848306 (− 3737C>T) and rs1143623 (− 1464G>C) were associated with colorectal cancer risk [dominant model: IRR (95% CI) = 0.81 (0.68–0.97), P = 0.02; dominant model: IRR (95% CI) = 1.22 (1.04–1.44), P = 0.02]10.
A Caucasian-American community-based case–control study (The Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening) reported the common rs16944 and rs1143634 SNPs of IL1B did not seem to play a role in prostate cancer risk11. A Chinese case–control study reported that IL1B SNP rs1143634 was not associated with gastric cancer risk12. A Asian-Chinese case–control study of inflammation-related genes involved in wound healing reported that studied IL1B SNPs were not associated with oesophageal squamous cell carcinoma13.
Main findings, implications and strengths of study
In this Northeastern-Chinese case–control study, we examined three htSNPs tagging 95% of the haplotyping diversity of IL1B known to be involved in the inflammatory response and associated with cancer risks in previously studies. To the best of our knowledge, this is the first study to evaluate three htSNPs tagging 95% of the haplotyping diversity of IL1B and to assess specific interactions between IL1B htSNPs, PPP1R13L, POLR1G risk SNPs and smoking-duration in relation to a lung cancer risk.
From a classical case–control approach in whole study, our main finding is that variant G-allele of IL1B SNP rs1143633 (A>G) associated with lower risk of lung cancer under dominant model and that variant C-allele of IL1B SNP rs3136558 (T>C) was at increased risk of lung cancer. When stratified by smoking-duration, IL1B SNP rs1143633 was specifically associated with lung cancer risk among long-term smokers (> 20 years). When stratified by histopathology, it should be noted that the studied IL1B htSNPs were only associated with risks among patients with lung squamous cell carcinoma and not among patients with lung adenocarcinoma. These results again suggest that the pathogenesis of the two subtypes may be different in genetic factors and gene changes21. The haplotype analysis of IL1B three htSNPs revealed positive association with lung cancer risk for the haplotype4 encompassing the variant alleles of rs3136558 and rs1143630. Haplotype2, which also encompasses the wild-type allele of rs1143633 and the variant allele of rs3136558, was also marginally associated with increased lung cancer risk. The other haplotypes encompassing the variant allele of rs1143630 were not associated with lung cancer risk. This suggests that haplotypes2 and 4 are in linkage with the functional IL1B polymorphism. The present result for IL1B SNP rs1143633 replicates for the finding from hepatocellular carcinoma study in Asians-Korean9. The present result of IL1B SNP rs3136558 (T>C) agrees with the finding that variant-allele carriers of IL1B SNP were at increased lung cancer risk among Caucasus-Danes7.
We did not correct for multiple testing. In this study, we included 3 SNPs, and thus, it could be argued that the threshold for statistical significance should be 0.05/3 = 0.0167. However, the current study is hypothesis driven, and the SNPs were selected to be in linkage disequilibrium with 95% of the genetic variation in IL1B.
The integration of genetic variants in risk prediction models beyond the traditional epidemiological covariates has been considered as the way forward in lung cancer risk prediction modeling22. Using a MDR approach: for whole study group, smoking-duration or IL1B rs1143633 was observed respectively as single main effect in one-factor model. The values of balanced accuracy overall and cross-validation consistency raised along with the increasing number of the factors. This phenomenon indicates the presence of interaction, meaning that the effect change of smoking-duration or rs1143633 at different levels depends on the level of another or several factors. Its existence shows that the effects of several factors studied simultaneously are not independent of each other. Special interactions between smoking duration, IL1B rs3136558 and POLR1G rs967591; smoking-duration, IL1B rs1143633, rs3136558 and rs1143630; and IL1B rs1143633, PPP1R13L rs1970764 and POLR1G rs735482 were observed in relation to lung cancer risk. The medium-level interactions were found between most markers. For histopathology study subgroup: smoking-duration was as main effect and positive interactions were only seen in subgroup of lung squamous cell carcinoma. These results add new evidence to our previous study17. The results again suggests that lung squmacarcinoma cell carcinoma displays the strongest relation with tobacco-smoking than lung adenocarcinoma17. Overall MDR results show that smoking duration as the main effect and the interactions between IL1B htSNP and PPP1R13L SNP and POLR1G SNP and smoking duration play critical roles in the occurrence of lung cancer and lung squamous cell carcinoma.
There is evidence of causal relationships between chronic infection, inflammation, and cancer23. An inflammatory microenvironment is an essential component of the tumor microenvironment (TME)20. The lung presents a unique milieu in which tumors progress in collusion with the TME. Inflammation plays an important role in the pathogenesis of lung cancer, and pulmonary disorders in lung cancer patients such as chronic obstructive pulmonary disease (COPD) and emphysema, constitute co-morbid conditions and are independent risk factors for lung cancer24. Chronic inflammation is a key feature of COPD and could be a potential driver of lung cancer development25. The chronic inflammatory microenvironment is associated with the release of various pro-inflammatory and oncogenic mediators including cytokines IL1B20. Excessive and uncontrolled releases of pro-inflammatory cytokines such as IL1B were increased in severe corona-virus disease 2019 (COVID-19) patients26. Environmental and occupational toxicants may induce pulmonary inflammation27–29. Chronic inflammation has been linked to several human diseases and also to initiation and promotion of cancer. High-expression of the promoter of IL1B SNP rs1143627 (− 31T>C) was induced in the human lung epithelial NCI-H2009 cells (Human lung adenocarcinoma cell line) treated with cigarette-smoke condensate30. Release of inflammasome products, such as IL1B and cytokine storms are hallmarks of COVID-19 infection and smoking may critically exacerbate COVID-19-related inflammation31. Present interaction study have added evidence that related to inflammation and immunity IL1B, which is independent or combined with other factors such as smoking, is involved in lung cancer risk.
Potential functional roles of selected IL1B htSNPs
SNP Function Prediction (FuncPred)32 indicated that IL1B rs1143630 has significant conservation score = 0.004 in three htSNP analyses about nsSNP (non-synonymous coding SNPs), splicing regulation, stop Codon, polyphen prediction, SNPs3D prediction, TFBS (transcription factor-binding site) prediction, miRNA binding site prediction, regulatory potential score, and conservation score by present data of SNPinfo Web Server. However we observed rs1143633 was the most important htSNP in this study, and we identified haplotype4 as a candidate to be in linkage with the functional SNP. Several functional SNPs have been identified in IL1B14.
Limitations
We have several study limitations. Power-test analyses for current study showed that for rs1143633, we had 91% chance of detecting OR = 0.67 at the 0.05 significant level using two-sided tests under the dominant model, showing that the sample size is reasonable and can meet the reasonable confidence level conditions. We had 80% or 81% chance for rs3136558 or rs1143630 respectively, detecting OR = 1.4 at the 0.05 significant level using two-sided tests under the dominant model, indicating that further larger population-based studies are warranted to confirm present findings. In addition, the haplotype analysis suggests that the studied SNPs are in linkage with the functional polymorphism. Thus functional studies of the polymorphisms under study would reveal whether the polymorphisms are functional or whether the observed associations are due to linkage with the functional polymorphism. Although the current research improves the efficiency of controlling confounding factors by matching age, sex and ethnic between cases and controls, which cannot directly control other confounding factors such as smoking-duration.
Conclusions
Our findings suggest that IL1B SNP rs1143633 may associate with lower risk of lung cancer, confirming previously identified marker; IL1B SNP rs3136558 and haplotype4 consisting IL1B htSNPs (rs1143633A-rs3136558C-rs1143630A) may associate with increased risk of lung cancer; interactions of IL1B with POLR1G or PPP1R13L or smoking-duration, which is independent or combined, may involve in risk of lung cancer and lung squamous cell carcinoma. These interesting findings should be sought in further validation with larger prospective cohorts. These could be used as a clinical biomarkers in lung cancer.
Materials and methods
Study population
1260 individuals were enrolled in this hospital-based case–control study including 627 cases and 633 controls as previously report18. The patients with lung cancer were diagnosed based on standard clinical and histological criteria. Eligible cases were previously untreated (no chemotherapy or radiotherapy for cancer prior to recruitment). Cancer-free controls (matched on: sex same, age ± 3 years and ethnic same) were selected from the orthopedics wards in the same area. Demographic and covariate data were obtained from medical records and questionnaires by personal interview with professional physicians. All participants were unrelated ethnic Han Chinese from Northeast China. Stratification criteria were defined as follows: age (10 years intervals), sex, family history of cancer, smoking duration (20 years intervals) and histopathology (3 subgroups).
htSNPs in IL1B gene determined
htSNPs of the IL1B gene were achieved from region of chromosome 12 of the International HapMap Project [http://www.hapmap.org, HapMap Data Rel 27 PhaseII + III, Feb09, on NCBI B36 assembly, dbSNP [the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) b26], by applying the TagSNPs software online and approaches of the algorithm-Tagger-pairwiseTagging. Qualified criteria: r2-cut off at 0.8 and minor allele frequency (MAF)-cut off at 0.05 in Han Chinese in Beijing (HCB) samples. Three htSNPs (rs1143633, rs3136558 and rs1143630) (the dbSNP database: https://www.ncbi.nlm.nih.gov/snp/?term=rs1143633, https://www.ncbi.nlm.nih.gov/snp/?term=rs3136558 and https://www.ncbi.nlm.nih.gov/snp/?term=rs1143630) were selected representing 95% of the common haplotype diversity across the IL1B gene. Table 2 displays the information of IL1B three htSNPs and three risk SNPs in Chr19q13.3 sub-region. Three risk SNPs of Chr19q13.3 were previously reported16,18,33. The data of three risk SNPs in PPP1R13L and POLR1G were used for analyses of gene–gene and gene–gene–environment interaction in current study.
DNA isolation and genotyping
A volume of 5 mL of whole blood with ethylenediamine tetraacetic acid (EDTA) anticoagulation was taken from each volunteer. Genomic DNA of whole blood samples was drawn with the Puregene DNA Isolation Kit or FlexiGene DNA kit 250 (Gentra Systems, Minneapolis, MN, USA or Qiagen, Germany) following the product' s instructions. Genotyping of rs1143633 (A>G), rs3136558 (T>C) and rs1143630 (C>A) of the IL1B gene was executed with the genotyping assay of ligase detection reaction coupled with polymerase chain reaction (LDR-PCR) as previously published34 in Shanghai Generay Biotechnology Co. Ltd. (P. R. China). Genotypes of PPP1R13L rs1970764 (A>G) and POLR1G rs967591 (G>A) and rs735482 (A>C) have been previously reported16. The software of Primer Premier 5.0 was used for design primers. The sequences (5′–3′) of primers and probes of IL1B three htSNPs are displayed in Table 8. Each group of LDR probes consisted of 1 common probe and 2 discriminating probes for the 2 alleles. For the PCR reactions, the DNA concentration was 50 ng–100 ng/μL and DNA purity was OD260/OD280 = 1.8–2.0. The genotyping procedure was in summary: performed PCR reactions, completed LDR reactions and sequenced LDR products. The genotyping call-rate was 96.35% for the IL1B three htSNPs. As quality control: pure water was used as negative control and 20% samples including cases and controls were genotyped twice, yielding 100% identical results.
Table 8.
The sequences (5′–3′) of primers and probes for IL1B three htSNPs examined.
| rs Number | Primers | Probes |
|---|---|---|
| rs1143633 | ||
| Forward primer | CTACTGGTGTTGTCATCAGAC | |
| Reverse primer | AGCTTTTTTGCTGTGAGTCCC | |
| Common probe | -P-CCTCGCCTCACGAGGCCTGCCCTTC-HEX- | |
| Discriminating probe G | TTTTCCTCCAAGAAATCAAATTTTGCCG | |
| Discriminating probe A | TCCTCCAAGAAATCAAATTTTGCCA | |
| rs3136558 | ||
| Forward primer | TCGCCATGTTGGCCAGGCTGG | |
| Reverse primer | GTGGTCCTGCCAGGAACCATG | |
| Common probe | -P-CTTTAGAAGCTCGGGATTCTTTCAATTT-HEX- | |
| Discriminating probe C | TTTTCACGCCTGGCCCAGAGAGGGATGAC | |
| Discriminating probe T | TTTTTTTCACGCCTGGCCCAGAGAGGGATGAT | |
| rs1143630 | ||
| Forward primer | GAATAGCCTGTAAGGTGTCAG | |
| Reverse primer | GGATGCAGTAAGCCAAGATTG | |
| Common probe | -P-CTTAACCTCCTTGAGCTTCAGAGAGTTTTTTTTT-FAM- | |
| Discriminating probe A | TTTTTTTTTTTTCCTGCTGTGTGCCCTTGAGTACACA | |
| Discriminating probe C | TTTTTTTTTTTTTTTCCTGCTGTGTGCCCTTGAGTACACC | |
Statistical analysis
Selected characteristics of cases and controls, allele frequencies, genotype frequencies, Hardy–Weinberg equilibrium, co-dominant model; dominant model; recessive model; over-dominant model; and log-additive model for case–control association of each single-locus, haplotype associations, and pair-wise LD, unconditional logistic regression for measurement of OR (95% CI) after adjusting smoking-duration, The Shapiro–Wilk test and Mann–Whitney U test were explored employing SPSS© v16.0 (SPSS Inc, Chicago, IL, USA) or SNPStats program35. Akaike’s Information Criterion (AIC) is a standard to measure the goodness of fit for statistical model. AIC criterion was used: give priority to model with the lowest AIC value35. Haplotypes with frequency < 0.01 among both cases and controls were excluded from the analysis. The interaction analyses of gene–gene and gene–gene-smoking duration in relation to lung cancer risk were conducted employing platform of MDR. This software (3.0.3. dev. Jar)36 is an updated version where permutation testing has been added into the main MDR program. The MDR method is nonparametric and free model. MDR has rational power for identifying interactions between two or more loci in relatively small samples. MDR has excellent power for identifying high-order gene–gene interactions. MDR can be directly used to case–control and discordant-sib-pair studies36. MDR conducts selection and evaluation of model by cross-validation and permutation-test. Balanced accuracy cross-validation training and balanced accuracy cross-validation testing indicate the accuracy rate in the training set and the testing set, respectively. Whose range is 0–1. The larger the number, the higher the accuracy rate. Cross-validation consistency indicates the consistency rate of cross-validation. Permutation-test = 1000 was set according to the instructions36. The P value was less than 0.05 was considered statistically significant. The possible functionality of IL1B three htSNPs was assessed using the web tool: SNPinfo31 in silico analysis. Power test was examined employing online statistical software: Unmatched Case/Control Studies (https://www.stat.ubc.ca/~rollin/stats/ssize/caco.html).
Ethics approval
The study protocol was approved by Human Genetic Resource Administration of China (HGRAC) (no. [2001] 015) and was conducted in accordance with the Helsinki Declaration. Every study participant agreed to participate in the study.
Author contributions
J.Y. contributed to conceptualization, study design, funding acquisition, project administration, data analysis, data interpretation, writing-original draft preparation and writing-review and editing; U.V. contributed to data interpretation and writing-review and editing; Y.M. and C.W. and Z.S. and S.D. contributed to data acquisition; H.W. and Y.Z. contributed to data analysis. All authors approved the final manuscript.
Funding
This work was supported by grant from the National Natural Science Foundation of China (Grant nos. 30571016 and 81072384).
Data availability
The information of selected htSNPs across the IL1B gene was from the dbSNP database: https://www.ncbi.nlm.nih.gov/snp/?term=rs1143633, rs3136558 and rs1143630. All data generated during this study are included in this published article. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bade BC, Dela Cruz CS. Lung cancer 2020: Epidemiology, etiology, and prevention. Clin. Chest Med. 2020;41:1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Simonds NI, Ghazarian AA, Pimentel CB, Schully SD, Ellison GL, Gillanders EM, Mechanic LE. Review of the gene–environment interaction literature in cancer: What do we know? Genet. Epidemiol. 2016;40:356–365. doi: 10.1002/gepi.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuzhalin A. The role of interleukin DNA polymorphisms in gastric cancer. Hum. Immunol. 2011;72:1128–1136. doi: 10.1016/j.humimm.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhang WH, Wang XL, Zhou J, An LZ, Xie XD. Association of interleukin-1B (IL-1B) gene polymorphisms with risk of gastric cancer in Chinese population. Cytokine. 2005;30:378–381. doi: 10.1016/j.cyto.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Wooff Y, Man SM, Aggio-Bruce R, Natoli R, Fernando N. IL-1 family members mediate cell death, inflammation and angiogenesis in retinal degenerative diseases. Immunology. 2019;10:1618. doi: 10.3389/fimmu.2019.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo X, Li M, Yang Y, Liang T, Yang H, Zhao X, Yang D. Interleukin gene polymorphisms in Chinese Han population with breast cancer, a case–control study. Oncotarget. 2017;9:17994–18001. doi: 10.18632/oncotarget.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel U, Christensen J, Wallin H, Friis S, Nexø BA, Raaschou-Nielsen O, Overvad K, Tjønneland A. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat. Res. 2008;639:89–100. doi: 10.1016/j.mrfmmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Kutikhin AG, Yuzhalin AE, Volkov AN, Zhivotovskiy AS, Brusina EB. Correlation between genetic polymorphisms within IL-1B and TLR4 genes and cancer risk in a Russian population: A case–control study. Tumour. Biol. 2014;35:4821–4830. doi: 10.1007/s13277-014-1633-6. [DOI] [PubMed] [Google Scholar]
- 9.Tak KH, Yu GI, Lee MY, Shin DH. Association between polymorphisms of interleukin 1 family genes and hepatocellular carcinoma. Med. Sci. Monit. 2018;24:3488–3495. doi: 10.12659/MSM.907524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen V, Holst R, Kopp TK, Tjønneland A, Vogel U. Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX-2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case-cohort study. PLoS ONE. 2013;8:e78366. doi: 10.1371/journal.pone.0078366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaud DS, Daugherty SE, Berndt SI, Platz EA, Yeager M, Crawford ED, Hsing A, Huang WY, Hayes RB. Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL-8, and IL-10 and risk of prostate cancer. Cancer Res. 2006;66:4525–4530. doi: 10.1158/0008-5472.CAN-05-3987. [DOI] [PubMed] [Google Scholar]
- 12.Yuan LJ, Jin TB, Yin JK, Du XL, Wang Q, Dong R, Wang SZ, Cui Y, Chen C, Lu JG. Polymorphisms of tumor-related genes IL-10, PSCA, MTRR and NOC3L are associated with the risk of gastric cancer in the Chinese Han population. Cancer Epidemiol. 2012;36:e366–372. doi: 10.1016/j.canep.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Golozar A, Beaty TH, Gravitt PE, Ruczinski I, Qiao YL, Fan JH, Ding T, Tang ZZ, Etemadi A, Hu N, Hyland PL, Wang L, Wang C, Dawsey SM, Freedman ND, Abnet CC, Goldstein AM, Taylor PR. Oesophageal squamous cell carcinoma in high-risk Chinese populations: Possible role for vascular epithelial growth factor A. Eur. J. Cancer. 2014;50:2855–2865. doi: 10.1016/j.ejca.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Wilkins LM, Aziz N, Cannings C, Wyllie CD, Bingle C, Rogus J, Beck JD, Offenbacher S, Cork MJ, Rafie-Kolpin M, Hsieh CM, Kornman KS, Duff GW. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum. Mol. Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- 15.Vogel U, Laros I, Jacobsen NR, Thomsen BL, Bak H, Olsen A, Bukowy Z, Wallin H, Overvad K, Tjønneland A, Nexø BA, Raaschou-Nielsen O. Two regions in chromosome 19q13.2–3 are associated with risk of lung cancer. Mutat. Res. 2004;546:65–74. doi: 10.1016/j.mrfmmm.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Yin J, Wang H, Vogel U, Wang C, Ma Y, Hou W, Zhang Y, Guo L, Li X. Fine-mapping markers of lung cancer susceptibility in a sub-region of chromosome19q13.3 among Chinese. Oncotarget. 2016;7:60929–60939. doi: 10.18632/oncotarget.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin J, Hou W, Vogel U, Li X, Ma Y, Wang C, Wang H, Sun Z. TP53 common variants and interaction with PPP1R13L and CD3EAP SNPs and lung cancer risk and smoking behavior in a Chinese population. Biomed. J. 2022;45:169–178. doi: 10.1016/j.bj.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin J, Hou W, Vogel U, Ma Y, Wang C, Wang H, Sun Z. Interaction between common variants of MDM2 and PPP1R13Land CD3EAP and TP53 SNPs in relation to lung cancer risk among Chinese. Ann. Transl. Med. 2020;8:934. doi: 10.21037/atm-19-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curk T, Rot G, Zupan B. SNPsyn: Detection and exploration of SNP-SNP interactions. Nucleic Acids Res. 2011;39:W444–449. doi: 10.1093/nar/gkr321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khandia R, Munjal A. Interplay between inflammation and cancer. Adv. Protein Chem. Struct. Biol. 2020;119:199–245. doi: 10.1016/bs.apcsb.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Chen JW, Dhahbi J. Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci. Rep. 2021;11:13323. doi: 10.1038/s41598-021-92725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youn RP, Hopkins RJ. Incorporating genomic data into multivariate risk models for lung cancer. Genet. Med. 2013;15:667–668. doi: 10.1038/gim.2013.89. [DOI] [PubMed] [Google Scholar]
- 23.Gomes M, Teixeira AL, Coelho A, Araújo A, Medeiros R. The role of inflammation in lung cancer. Adv. Exp. Med. Biol. 2014;816:1–23. doi: 10.1007/978-3-0348-0837-8_1. [DOI] [PubMed] [Google Scholar]
- 24.Mittal V, El Rayes T, Narula N, McGraw TE, Altorki NK, Barcellos-Hoff MH. The microenvironment of lung cancer and therapeutic implications. Adv. Exp. Med. Biol. 2016;890:75–110. doi: 10.1007/978-3-319-24932-2_5. [DOI] [PubMed] [Google Scholar]
- 25.Parris BA, O'Farrell HE, Fong KM, Yang IA. Chronic obstructive pulmonary disease (COPD) and lung cancer: Common pathways. J. Thorac. Dis. 2019;11(Suppl 17):S2155–S2172. doi: 10.21037/jtd.2019.10.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crisci CD, Ardusso LRF, Mossuz A, Müller L. A precision medicine approach to SARS-CoV-2 pandemic management. Curr. Treat. Options Allergy. 2020;8:1–19. doi: 10.1007/s40521-020-00258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danielsen PH, Knudsen KB, Štrancar J, Umek P, Koklič T, Garvas M, Vanhala E, Savukoski S, Ding Y, Madsen AM, Jacobsen NR, Weydahl IK, Berthin GT, Poulsen SS, Schmid O, Wolff H, Vogel U. Effects of physicochemical properties of TiO(2) nanomaterials for pulmonary inflammation, acute phase response and alveolar proteinosis in intratracheally exposed mice. Toxicol. Appl. Pharmacol. 2020;386:114830. doi: 10.1016/j.taap.2019.114830. [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen KM, Gren L, Malmborg VB, Shukla PC, Tunér M, Essig YJ, Krais AM, Clausen PA, Berthing T, Loeschner K, Jacobsen NR, Wolff H, Pagels J, Vogel UB. Particle characterization and toxicity in C57BL/6 mice following instillation of five different diesel exhaust particles designed to differ in physicochemical properties. Part Fibre. Toxicol. 2020;17:38. doi: 10.1186/s12989-020-00369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendtsen KM, Brostrøm A, Koivisto AJ, Koponen I, Berthing T, Bertram N, Kling KI, Maso MD, Kangasniemi O, Poikkimäki M, Loeschner K, Clausen PA, Wolff H, Jensen KA, Saber AT, Vogel U. Airport emission particles: exposure characterization and toxicity following intratracheal instillation in mice. Part Fibre. Toxicol. 2019;16:23. doi: 10.1186/s12989-019-0305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart K, Haugen A, Zienolddiny S. Allele-specific induction of IL1B -31T/C promoter polymorphism by lung carcinogens. Mutat Res. 2008;656:14–18. doi: 10.1016/j.mrgentox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Lee AC, Chakladar J, Li WT, Chen C, Chang EY, Wang-Rodriguez J, Ongkeko WM. Tobacco, but not nicotine and flavor-less electronic cigarettes, induces ACE2 and immune dysregulation. Int. J. Mol. Sci. 2020;21:5513. doi: 10.3390/ijms21155513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, Taylor JA. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37(Web Server issue):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou W, Yin J, Vogel U, Sun Z, Liang D. 19p13.3-GADD45B common variants and 19q13.3-PPP1R13L and 19q13.3-CD3EAP in lung cancer risk among Chinese. Chem. Biol. Interact. 2017;277:74–78. doi: 10.1016/j.cbi.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Z, Xiao J, Jiang Y, Zhang S, Yu M, Zhao J, Wei D, Cao H. A novel method based on ligase detection reaction for low abundant YIDD mutants detection in hepatitis B virus. Hepatol. Res. 2006;34:150–155. doi: 10.1016/j.hepres.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The information of selected htSNPs across the IL1B gene was from the dbSNP database: https://www.ncbi.nlm.nih.gov/snp/?term=rs1143633, rs3136558 and rs1143630. All data generated during this study are included in this published article. The data that support the findings of this study are available from the corresponding author upon reasonable request.