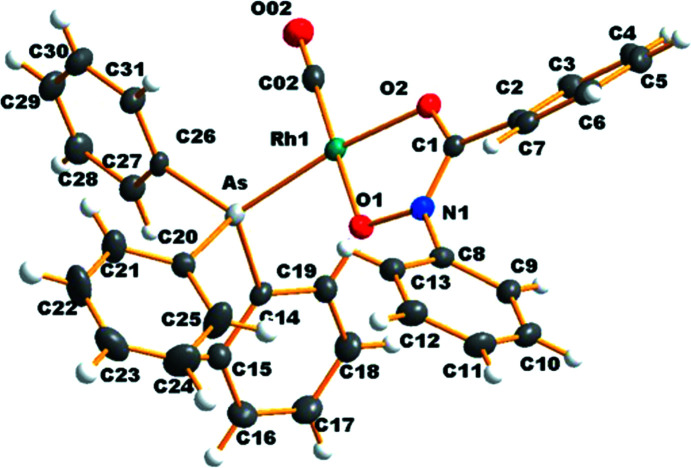
The molecular structure of the title compound, [Rh(BPHA)(CO)(AsPh3)] (where BPHA = N-benzoyl-N-phenylhydroxylaminate), has a distorted square-planar coordination environment around the central RhI atom, defined by a CO2As coordination set.
Keywords: crystal structure, rhodium, N-benzoyl-N-phenylhydroxamic acid, triphenylarsine
Abstract
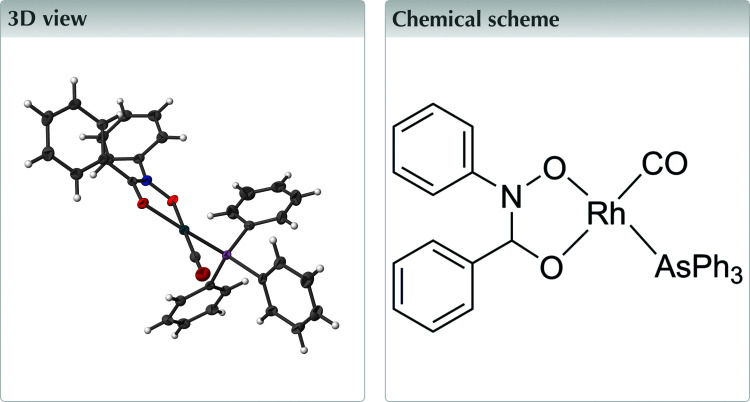
The molecule of the title compound, [Rh(C13H10NO2){As(C6H5)3}(CO)] or [Rh(BPHA)(AsPh3)(CO)] (BPHA is the N-benzoyl-N-phenylhydroxylaminate anion), comprises a bidentate N-benzoyl-N-phenylhydroxylaminate anion coordinating through the O atoms to the soft Lewis acid, rhodium(I), and two monodentate ligands, viz. triphenylarsine and carbonyl. The resulting CO2As coordination environment around the central RhI atom is distorted square planar.=
Structure description
The title complex, [Rh(BPHA)(AsPh3)(CO)], is composed of an O,O-bidentate N-benzoyl-N-phenylhydroxylaminate anion, a carbonyl ligand and a monodentate triphenylarsine ligand, all coordinating to the soft rhodium(I) metal atom (Fig. 1 ▸ and Table 1 ▸). The crystal structure is isotypic with that of [Rh(BPHA)(PPh3)(CO)] and shows similar Rh—O and Rh—C bond lengths (2.037/2.089 and 1.809 Å, respectively; Leipoldt & Grobler, 1982 ▸). The coordination environment in the molecule of [Rh(BPHA)(AsPh3)(CO)] is distorted square planar, as shown by the small O1—Rh—O2 bite angle of 79.53 (7)°, which is similar to the bite angles of related structures with O,O-binding five-membered chelate rings reported in the literature (Elmakki et al., 2017 ▸). The C02—Rh—O2 and C02—Rh—O1 angles involving the C02≡O02 carbonyl ligand were also found to deviate from ideal values, at 99.31 (9) and 178.39 (10)°, respectively, similar to those of related structures (Elmakki et al., 2016 ▸).
Figure 1.
The molecular structure of the title compound, showing atoms with displacement ellipsoids at the 50% probability level.
Table 1. Selected bond lengths (Å).
| Rh1—As | 2.3337 (4) | Rh1—O1 | 2.0338 (18) |
| Rh1—O2 | 2.0682 (17) | Rh1—C02 | 1.813 (3) |
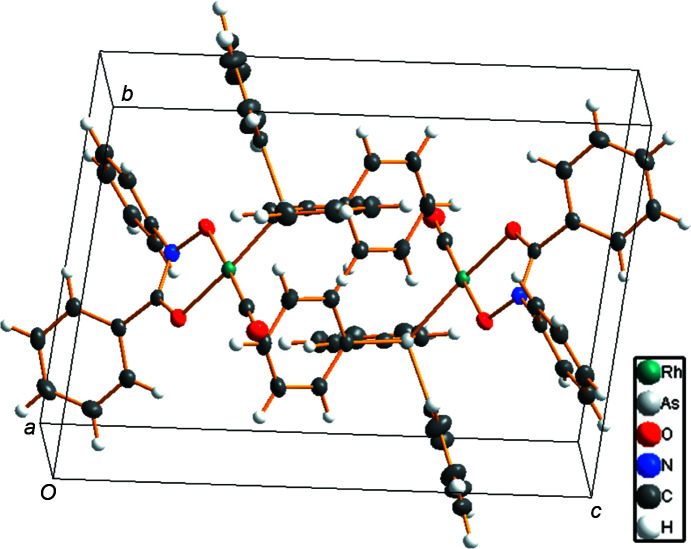
The crystal packing is dominated by van der Waals interactions (Fig. 2 ▸).
Figure 2.
An illustration of the molecular packing in the unit cell of [Rh(BPHA)(CO)(AsPh3)], viewed approximately along the a axis; atom labels have been omitted for clarity.
Synthesis and crystallization
A stepwise process was pursued in the complexation of the rhodium metal atom by the bidentate N-phenyl-N-benzoylhydroxylaminate anion. First, [RhCl(CO)2]2 was prepared in situ by heating RhCl3·3H2O in 5 ml of dimethylformamide under reflux for 30 min, followed by addition of the bidentate ligand to the reaction mixture, which then resulted in the formation of a dicarbonylrhodium species, [Rh(BPHA)(CO)2] (Leipoldt & Grobler, 1982 ▸). Rh(BPHA)(CO)2] (65 mg) was then dissolved in 5 ml of acetone. Triphenylarsine (AsPh3; 70 mg) was added to the reaction mixture under stirring, resulting in the immediate evolution of CO gas. The reaction mixture was then left to crystallize, resulting in the formation of yellow crystals suitable for X-ray analysis.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Rh(C13H10NO2)(C18H15As)(CO)] |
| M r | 649.36 |
| Crystal system, space group | Triclinic, P
|
| Temperature (K) | 100 |
| a, b, c (Å) | 9.5178 (17), 10.1995 (19), 14.589 (2) |
| α, β, γ (°) | 81.516 (6), 83.142 (6), 72.351 (7) |
| V (Å3) | 1330.7 (4) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.91 |
| Crystal size (mm) | 0.21 × 0.13 × 0.03 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.634, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 38485, 6425, 5747 |
| R int | 0.065 |
| (sin θ/λ)max (Å−1) | 0.660 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.030, 0.074, 1.07 |
| No. of reflections | 6425 |
| No. of parameters | 343 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 1.14, −0.61 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314623003553/wm4184sup1.cif
CCDC reference: 2257230
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The University of the Free State and the South African National Research Fund are acknowledged for funding.
full crystallographic data
Crystal data
| [Rh(C13H10NO2)(C18H15As)(CO)] | Z = 2 |
| Mr = 649.36 | F(000) = 652 |
| Triclinic, P1 | Dx = 1.621 Mg m−3 |
| a = 9.5178 (17) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.1995 (19) Å | Cell parameters from 9339 reflections |
| c = 14.589 (2) Å | θ = 2.3–28.3° |
| α = 81.516 (6)° | µ = 1.91 mm−1 |
| β = 83.142 (6)° | T = 100 K |
| γ = 72.351 (7)° | Plate, yellow |
| V = 1330.7 (4) Å3 | 0.21 × 0.13 × 0.03 mm |
Data collection
| Bruker APEXII CCD diffractometer | 5747 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.065 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | θmax = 28.0°, θmin = 2.1° |
| Tmin = 0.634, Tmax = 0.746 | h = −12→12 |
| 38485 measured reflections | k = −13→13 |
| 6425 independent reflections | l = −19→19 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.030 | H-atom parameters constrained |
| wR(F2) = 0.074 | w = 1/[σ2(Fo2) + (0.0141P)2 + 1.8613P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max = 0.002 |
| 6425 reflections | Δρmax = 1.14 e Å−3 |
| 343 parameters | Δρmin = −0.61 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Rh1 | 0.25715 (2) | 0.53736 (2) | 0.27266 (2) | 0.01779 (6) | |
| As | 0.12957 (3) | 0.72691 (2) | 0.35181 (2) | 0.01699 (6) | |
| O2 | 0.39711 (19) | 0.38301 (18) | 0.19949 (12) | 0.0210 (4) | |
| O1 | 0.42339 (19) | 0.62308 (18) | 0.22612 (13) | 0.0217 (4) | |
| O02 | 0.0140 (2) | 0.4121 (2) | 0.33366 (15) | 0.0313 (4) | |
| N1 | 0.5303 (2) | 0.5365 (2) | 0.17371 (15) | 0.0207 (4) | |
| C1 | 0.5119 (3) | 0.4183 (2) | 0.15914 (16) | 0.0169 (4) | |
| C26 | −0.0798 (3) | 0.7588 (2) | 0.38511 (18) | 0.0197 (5) | |
| C8 | 0.6478 (3) | 0.5925 (3) | 0.13234 (17) | 0.0198 (5) | |
| C14 | 0.2129 (3) | 0.7244 (3) | 0.46672 (17) | 0.0189 (5) | |
| C20 | 0.1411 (3) | 0.9043 (3) | 0.28753 (17) | 0.0207 (5) | |
| C15 | 0.2025 (3) | 0.8463 (3) | 0.50321 (19) | 0.0241 (5) | |
| H15 | 0.1470 | 0.9331 | 0.4741 | 0.029* | |
| C7 | 0.6819 (3) | 0.3597 (3) | 0.01447 (18) | 0.0212 (5) | |
| H7 | 0.6631 | 0.4551 | −0.0082 | 0.025* | |
| C27 | −0.1418 (3) | 0.7798 (3) | 0.47479 (19) | 0.0237 (5) | |
| H27 | −0.0816 | 0.7815 | 0.5218 | 0.028* | |
| C13 | 0.6118 (3) | 0.7151 (3) | 0.07297 (19) | 0.0241 (5) | |
| H13 | 0.5113 | 0.7637 | 0.0637 | 0.029* | |
| C2 | 0.6182 (3) | 0.3193 (3) | 0.10122 (17) | 0.0190 (5) | |
| C19 | 0.2907 (3) | 0.5971 (3) | 0.51116 (18) | 0.0237 (5) | |
| H19 | 0.2958 | 0.5138 | 0.4873 | 0.028* | |
| C02 | 0.1092 (3) | 0.4594 (3) | 0.31095 (18) | 0.0224 (5) | |
| C6 | 0.7729 (3) | 0.2591 (3) | −0.03845 (19) | 0.0250 (5) | |
| H6 | 0.8153 | 0.2862 | −0.0978 | 0.030* | |
| C28 | −0.2931 (3) | 0.7983 (3) | 0.4956 (2) | 0.0304 (6) | |
| H28 | −0.3356 | 0.8119 | 0.5570 | 0.036* | |
| C9 | 0.7931 (3) | 0.5246 (3) | 0.15114 (19) | 0.0232 (5) | |
| H9 | 0.8160 | 0.4439 | 0.1950 | 0.028* | |
| C3 | 0.6475 (3) | 0.1796 (3) | 0.13401 (19) | 0.0247 (5) | |
| H3 | 0.6039 | 0.1518 | 0.1928 | 0.030* | |
| C18 | 0.3607 (3) | 0.5920 (3) | 0.59044 (19) | 0.0280 (6) | |
| H18 | 0.4137 | 0.5054 | 0.6211 | 0.034* | |
| C11 | 0.8703 (3) | 0.6955 (3) | 0.0429 (2) | 0.0296 (6) | |
| H11 | 0.9472 | 0.7295 | 0.0106 | 0.036* | |
| C31 | −0.1692 (3) | 0.7556 (3) | 0.3171 (2) | 0.0268 (6) | |
| H31 | −0.1268 | 0.7393 | 0.2559 | 0.032* | |
| C12 | 0.7241 (3) | 0.7658 (3) | 0.0274 (2) | 0.0290 (6) | |
| H12 | 0.7011 | 0.8487 | −0.0144 | 0.035* | |
| C16 | 0.2733 (3) | 0.8406 (3) | 0.58217 (19) | 0.0284 (6) | |
| H16 | 0.2669 | 0.9234 | 0.6069 | 0.034* | |
| C4 | 0.7403 (3) | 0.0805 (3) | 0.0812 (2) | 0.0298 (6) | |
| H4 | 0.7616 | −0.0149 | 0.1044 | 0.036* | |
| C10 | 0.9049 (3) | 0.5768 (3) | 0.1046 (2) | 0.0275 (6) | |
| H10 | 1.0053 | 0.5304 | 0.1155 | 0.033* | |
| C17 | 0.3531 (3) | 0.7141 (3) | 0.6246 (2) | 0.0301 (6) | |
| H17 | 0.4033 | 0.7106 | 0.6779 | 0.036* | |
| C5 | 0.8022 (3) | 0.1204 (3) | −0.0057 (2) | 0.0300 (6) | |
| H5 | 0.8644 | 0.0523 | −0.0423 | 0.036* | |
| C29 | −0.3810 (3) | 0.7971 (3) | 0.4275 (2) | 0.0332 (6) | |
| H29 | −0.4841 | 0.8107 | 0.4420 | 0.040* | |
| C25 | 0.2759 (3) | 0.9299 (3) | 0.2743 (2) | 0.0390 (7) | |
| H25 | 0.3614 | 0.8592 | 0.2938 | 0.047* | |
| C24 | 0.2892 (4) | 1.0577 (4) | 0.2327 (2) | 0.0417 (8) | |
| H24 | 0.3829 | 1.0747 | 0.2256 | 0.050* | |
| C21 | 0.0196 (3) | 1.0048 (3) | 0.2526 (2) | 0.0324 (6) | |
| H21 | −0.0736 | 0.9871 | 0.2583 | 0.039* | |
| C23 | 0.1672 (4) | 1.1593 (3) | 0.2019 (2) | 0.0368 (7) | |
| H23 | 0.1750 | 1.2480 | 0.1759 | 0.044* | |
| C30 | −0.3194 (3) | 0.7760 (3) | 0.3380 (2) | 0.0340 (7) | |
| H30 | −0.3802 | 0.7755 | 0.2911 | 0.041* | |
| C22 | 0.0335 (4) | 1.1316 (3) | 0.2091 (3) | 0.0442 (8) | |
| H22 | −0.0499 | 1.1994 | 0.1841 | 0.053* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Rh1 | 0.01456 (9) | 0.02186 (10) | 0.01630 (10) | −0.00485 (7) | 0.00038 (7) | −0.00246 (7) |
| As | 0.01309 (12) | 0.02045 (12) | 0.01611 (13) | −0.00335 (9) | −0.00019 (9) | −0.00199 (9) |
| O2 | 0.0163 (8) | 0.0257 (9) | 0.0210 (9) | −0.0066 (7) | 0.0018 (7) | −0.0040 (7) |
| O1 | 0.0176 (8) | 0.0245 (9) | 0.0235 (10) | −0.0066 (7) | 0.0048 (7) | −0.0087 (7) |
| O02 | 0.0258 (10) | 0.0340 (10) | 0.0364 (12) | −0.0152 (8) | 0.0031 (8) | −0.0022 (9) |
| N1 | 0.0172 (10) | 0.0252 (10) | 0.0200 (11) | −0.0067 (8) | 0.0012 (8) | −0.0046 (8) |
| C1 | 0.0154 (11) | 0.0209 (11) | 0.0138 (11) | −0.0045 (9) | −0.0014 (8) | −0.0012 (8) |
| C26 | 0.0126 (10) | 0.0187 (11) | 0.0253 (13) | −0.0023 (8) | 0.0003 (9) | −0.0014 (9) |
| C8 | 0.0196 (11) | 0.0245 (12) | 0.0178 (12) | −0.0091 (9) | 0.0011 (9) | −0.0066 (9) |
| C14 | 0.0139 (11) | 0.0261 (12) | 0.0159 (12) | −0.0045 (9) | −0.0007 (9) | −0.0031 (9) |
| C20 | 0.0212 (12) | 0.0222 (11) | 0.0182 (12) | −0.0065 (9) | 0.0007 (9) | −0.0019 (9) |
| C15 | 0.0234 (13) | 0.0254 (12) | 0.0220 (13) | −0.0053 (10) | −0.0005 (10) | −0.0033 (10) |
| C7 | 0.0176 (11) | 0.0281 (12) | 0.0190 (12) | −0.0074 (10) | −0.0039 (9) | −0.0023 (9) |
| C27 | 0.0212 (12) | 0.0280 (13) | 0.0215 (13) | −0.0086 (10) | 0.0014 (10) | −0.0014 (10) |
| C13 | 0.0223 (12) | 0.0258 (12) | 0.0260 (14) | −0.0085 (10) | −0.0019 (10) | −0.0056 (10) |
| C2 | 0.0144 (11) | 0.0244 (12) | 0.0191 (12) | −0.0053 (9) | −0.0025 (9) | −0.0056 (9) |
| C19 | 0.0224 (12) | 0.0245 (12) | 0.0218 (13) | −0.0039 (10) | −0.0005 (10) | −0.0024 (10) |
| C02 | 0.0218 (12) | 0.0228 (12) | 0.0191 (13) | −0.0018 (10) | −0.0015 (10) | −0.0019 (9) |
| C6 | 0.0183 (12) | 0.0381 (14) | 0.0191 (13) | −0.0076 (11) | −0.0006 (10) | −0.0074 (10) |
| C28 | 0.0236 (13) | 0.0376 (15) | 0.0277 (15) | −0.0089 (11) | 0.0093 (11) | −0.0058 (12) |
| C9 | 0.0202 (12) | 0.0259 (12) | 0.0235 (13) | −0.0059 (10) | −0.0029 (10) | −0.0042 (10) |
| C3 | 0.0257 (13) | 0.0275 (13) | 0.0218 (13) | −0.0086 (10) | −0.0020 (10) | −0.0038 (10) |
| C18 | 0.0286 (14) | 0.0314 (13) | 0.0204 (14) | −0.0045 (11) | −0.0044 (11) | 0.0010 (10) |
| C11 | 0.0248 (13) | 0.0402 (15) | 0.0294 (15) | −0.0187 (12) | 0.0039 (11) | −0.0069 (12) |
| C31 | 0.0197 (12) | 0.0335 (14) | 0.0267 (14) | −0.0051 (10) | −0.0008 (10) | −0.0087 (11) |
| C12 | 0.0314 (14) | 0.0333 (14) | 0.0258 (15) | −0.0154 (12) | −0.0009 (11) | −0.0023 (11) |
| C16 | 0.0336 (15) | 0.0320 (14) | 0.0210 (14) | −0.0097 (12) | −0.0012 (11) | −0.0083 (11) |
| C4 | 0.0307 (14) | 0.0233 (12) | 0.0339 (16) | −0.0036 (11) | −0.0030 (12) | −0.0074 (11) |
| C10 | 0.0185 (12) | 0.0381 (15) | 0.0286 (15) | −0.0095 (11) | 0.0004 (10) | −0.0115 (11) |
| C17 | 0.0311 (15) | 0.0407 (16) | 0.0190 (14) | −0.0101 (12) | −0.0046 (11) | −0.0040 (11) |
| C5 | 0.0222 (13) | 0.0328 (14) | 0.0337 (16) | −0.0007 (11) | −0.0025 (11) | −0.0153 (12) |
| C29 | 0.0153 (12) | 0.0405 (16) | 0.0431 (18) | −0.0075 (11) | 0.0029 (12) | −0.0081 (13) |
| C25 | 0.0228 (14) | 0.0416 (17) | 0.048 (2) | −0.0094 (12) | −0.0029 (13) | 0.0112 (14) |
| C24 | 0.0386 (17) | 0.0452 (18) | 0.046 (2) | −0.0252 (15) | 0.0003 (15) | 0.0052 (15) |
| C21 | 0.0230 (13) | 0.0269 (13) | 0.0418 (18) | −0.0032 (11) | 0.0029 (12) | 0.0005 (12) |
| C23 | 0.0518 (19) | 0.0258 (13) | 0.0319 (17) | −0.0152 (13) | 0.0118 (14) | −0.0041 (12) |
| C30 | 0.0199 (13) | 0.0419 (16) | 0.0415 (18) | −0.0075 (12) | −0.0050 (12) | −0.0101 (13) |
| C22 | 0.0405 (18) | 0.0228 (14) | 0.058 (2) | 0.0006 (13) | 0.0035 (16) | 0.0053 (14) |
Geometric parameters (Å, º)
| Rh1—As | 2.3337 (4) | C6—C5 | 1.379 (4) |
| Rh1—O2 | 2.0682 (17) | C28—H28 | 0.9500 |
| Rh1—O1 | 2.0338 (18) | C28—C29 | 1.376 (4) |
| Rh1—C02 | 1.813 (3) | C9—H9 | 0.9500 |
| As—C26 | 1.933 (2) | C9—C10 | 1.395 (4) |
| As—C14 | 1.932 (2) | C3—H3 | 0.9500 |
| As—C20 | 1.939 (2) | C3—C4 | 1.386 (4) |
| O2—C1 | 1.301 (3) | C18—H18 | 0.9500 |
| O1—N1 | 1.367 (3) | C18—C17 | 1.387 (4) |
| O02—C02 | 1.144 (3) | C11—H11 | 0.9500 |
| N1—C1 | 1.319 (3) | C11—C12 | 1.387 (4) |
| N1—C8 | 1.437 (3) | C11—C10 | 1.375 (4) |
| C1—C2 | 1.479 (3) | C31—H31 | 0.9500 |
| C26—C27 | 1.388 (4) | C31—C30 | 1.384 (4) |
| C26—C31 | 1.392 (4) | C12—H12 | 0.9500 |
| C8—C13 | 1.387 (4) | C16—H16 | 0.9500 |
| C8—C9 | 1.385 (4) | C16—C17 | 1.381 (4) |
| C14—C15 | 1.395 (3) | C4—H4 | 0.9500 |
| C14—C19 | 1.393 (3) | C4—C5 | 1.391 (4) |
| C20—C25 | 1.371 (4) | C10—H10 | 0.9500 |
| C20—C21 | 1.384 (4) | C17—H17 | 0.9500 |
| C15—H15 | 0.9500 | C5—H5 | 0.9500 |
| C15—C16 | 1.388 (4) | C29—H29 | 0.9500 |
| C7—H7 | 0.9500 | C29—C30 | 1.385 (4) |
| C7—C2 | 1.397 (4) | C25—H25 | 0.9500 |
| C7—C6 | 1.391 (4) | C25—C24 | 1.390 (4) |
| C27—H27 | 0.9500 | C24—H24 | 0.9500 |
| C27—C28 | 1.395 (4) | C24—C23 | 1.371 (5) |
| C13—H13 | 0.9500 | C21—H21 | 0.9500 |
| C13—C12 | 1.385 (4) | C21—C22 | 1.391 (4) |
| C2—C3 | 1.389 (4) | C23—H23 | 0.9500 |
| C19—H19 | 0.9500 | C23—C22 | 1.374 (5) |
| C19—C18 | 1.390 (4) | C30—H30 | 0.9500 |
| C6—H6 | 0.9500 | C22—H22 | 0.9500 |
| O2—Rh1—As | 171.06 (5) | C29—C28—H28 | 119.8 |
| O1—Rh1—As | 91.66 (5) | C8—C9—H9 | 120.6 |
| O1—Rh1—O2 | 79.53 (7) | C8—C9—C10 | 118.8 (2) |
| C02—Rh1—As | 89.53 (8) | C10—C9—H9 | 120.6 |
| C02—Rh1—O2 | 99.31 (9) | C2—C3—H3 | 119.9 |
| C02—Rh1—O1 | 178.39 (10) | C4—C3—C2 | 120.2 (3) |
| C26—As—Rh1 | 118.78 (7) | C4—C3—H3 | 119.9 |
| C26—As—C20 | 103.15 (10) | C19—C18—H18 | 120.1 |
| C14—As—Rh1 | 112.68 (7) | C17—C18—C19 | 119.7 (3) |
| C14—As—C26 | 104.81 (11) | C17—C18—H18 | 120.1 |
| C14—As—C20 | 100.97 (11) | C12—C11—H11 | 119.7 |
| C20—As—Rh1 | 114.44 (8) | C10—C11—H11 | 119.7 |
| C1—O2—Rh1 | 111.54 (15) | C10—C11—C12 | 120.6 (3) |
| N1—O1—Rh1 | 110.33 (13) | C26—C31—H31 | 119.9 |
| O1—N1—C8 | 114.38 (19) | C30—C31—C26 | 120.3 (3) |
| C1—N1—O1 | 119.1 (2) | C30—C31—H31 | 119.9 |
| C1—N1—C8 | 126.2 (2) | C13—C12—C11 | 119.9 (3) |
| O2—C1—N1 | 119.3 (2) | C13—C12—H12 | 120.0 |
| O2—C1—C2 | 117.1 (2) | C11—C12—H12 | 120.0 |
| N1—C1—C2 | 123.5 (2) | C15—C16—H16 | 120.1 |
| C27—C26—As | 122.12 (19) | C17—C16—C15 | 119.8 (3) |
| C27—C26—C31 | 119.6 (2) | C17—C16—H16 | 120.1 |
| C31—C26—As | 118.27 (19) | C3—C4—H4 | 119.9 |
| C13—C8—N1 | 118.2 (2) | C3—C4—C5 | 120.2 (3) |
| C9—C8—N1 | 120.5 (2) | C5—C4—H4 | 119.9 |
| C9—C8—C13 | 121.3 (2) | C9—C10—H10 | 119.9 |
| C15—C14—As | 121.74 (19) | C11—C10—C9 | 120.1 (3) |
| C19—C14—As | 118.28 (19) | C11—C10—H10 | 119.9 |
| C19—C14—C15 | 119.9 (2) | C18—C17—H17 | 119.6 |
| C25—C20—As | 118.6 (2) | C16—C17—C18 | 120.7 (3) |
| C25—C20—C21 | 118.7 (3) | C16—C17—H17 | 119.6 |
| C21—C20—As | 122.7 (2) | C6—C5—C4 | 119.7 (2) |
| C14—C15—H15 | 120.1 | C6—C5—H5 | 120.1 |
| C16—C15—C14 | 119.9 (2) | C4—C5—H5 | 120.1 |
| C16—C15—H15 | 120.1 | C28—C29—H29 | 120.0 |
| C2—C7—H7 | 120.2 | C28—C29—C30 | 120.1 (3) |
| C6—C7—H7 | 120.2 | C30—C29—H29 | 120.0 |
| C6—C7—C2 | 119.5 (2) | C20—C25—H25 | 119.5 |
| C26—C27—H27 | 120.2 | C20—C25—C24 | 121.0 (3) |
| C26—C27—C28 | 119.7 (3) | C24—C25—H25 | 119.5 |
| C28—C27—H27 | 120.2 | C25—C24—H24 | 120.0 |
| C8—C13—H13 | 120.4 | C23—C24—C25 | 120.0 (3) |
| C12—C13—C8 | 119.1 (3) | C23—C24—H24 | 120.0 |
| C12—C13—H13 | 120.4 | C20—C21—H21 | 119.9 |
| C7—C2—C1 | 123.2 (2) | C20—C21—C22 | 120.3 (3) |
| C3—C2—C1 | 117.0 (2) | C22—C21—H21 | 119.9 |
| C3—C2—C7 | 119.7 (2) | C24—C23—H23 | 120.3 |
| C14—C19—H19 | 120.1 | C24—C23—C22 | 119.5 (3) |
| C18—C19—C14 | 119.8 (2) | C22—C23—H23 | 120.3 |
| C18—C19—H19 | 120.1 | C31—C30—C29 | 120.0 (3) |
| O02—C02—Rh1 | 178.5 (2) | C31—C30—H30 | 120.0 |
| C7—C6—H6 | 119.7 | C29—C30—H30 | 120.0 |
| C5—C6—C7 | 120.7 (3) | C21—C22—H22 | 119.8 |
| C5—C6—H6 | 119.7 | C23—C22—C21 | 120.3 (3) |
| C27—C28—H28 | 119.8 | C23—C22—H22 | 119.8 |
| C29—C28—C27 | 120.4 (3) | ||
| Rh1—O2—C1—N1 | 4.4 (3) | C8—C9—C10—C11 | 1.5 (4) |
| Rh1—O2—C1—C2 | −177.62 (16) | C14—C15—C16—C17 | −0.4 (4) |
| Rh1—O1—N1—C1 | 1.5 (3) | C14—C19—C18—C17 | −0.2 (4) |
| Rh1—O1—N1—C8 | 175.94 (16) | C20—C25—C24—C23 | −1.8 (6) |
| As—C26—C27—C28 | −178.1 (2) | C20—C21—C22—C23 | −1.2 (5) |
| As—C26—C31—C30 | 179.1 (2) | C15—C14—C19—C18 | −1.6 (4) |
| As—C14—C15—C16 | −175.2 (2) | C15—C16—C17—C18 | −1.4 (4) |
| As—C14—C19—C18 | 175.6 (2) | C7—C2—C3—C4 | 0.4 (4) |
| As—C20—C25—C24 | −176.8 (3) | C7—C6—C5—C4 | −0.1 (4) |
| As—C20—C21—C22 | 178.4 (3) | C27—C26—C31—C30 | 1.4 (4) |
| O2—C1—C2—C7 | 137.2 (2) | C27—C28—C29—C30 | 0.6 (5) |
| O2—C1—C2—C3 | −39.0 (3) | C13—C8—C9—C10 | −4.1 (4) |
| O1—N1—C1—O2 | −4.1 (3) | C2—C7—C6—C5 | −0.8 (4) |
| O1—N1—C1—C2 | 178.1 (2) | C2—C3—C4—C5 | −1.3 (4) |
| O1—N1—C8—C13 | −58.9 (3) | C19—C14—C15—C16 | 1.9 (4) |
| O1—N1—C8—C9 | 121.5 (2) | C19—C18—C17—C16 | 1.7 (4) |
| N1—C1—C2—C7 | −45.0 (4) | C6—C7—C2—C1 | −175.5 (2) |
| N1—C1—C2—C3 | 138.8 (3) | C6—C7—C2—C3 | 0.6 (4) |
| N1—C8—C13—C12 | −175.6 (2) | C28—C29—C30—C31 | 0.3 (5) |
| N1—C8—C9—C10 | 175.5 (2) | C9—C8—C13—C12 | 4.0 (4) |
| C1—N1—C8—C13 | 115.1 (3) | C3—C4—C5—C6 | 1.1 (4) |
| C1—N1—C8—C9 | −64.5 (3) | C31—C26—C27—C28 | −0.5 (4) |
| C1—C2—C3—C4 | 176.7 (2) | C12—C11—C10—C9 | 1.2 (4) |
| C26—C27—C28—C29 | −0.5 (4) | C10—C11—C12—C13 | −1.3 (4) |
| C26—C31—C30—C29 | −1.3 (5) | C25—C20—C21—C22 | −3.2 (5) |
| C8—N1—C1—O2 | −177.9 (2) | C25—C24—C23—C22 | −2.7 (5) |
| C8—N1—C1—C2 | 4.3 (4) | C24—C23—C22—C21 | 4.2 (5) |
| C8—C13—C12—C11 | −1.3 (4) | C21—C20—C25—C24 | 4.7 (5) |
References
- Brandenburg, K. & Putz, H. (2005). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Bruker (2012). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Elmakki, M. A., Alexander, O. T., Venter, G. J. S. & Venter, J. A. Z. (2017). Z. Kristallogr. New Cryst. Struct. 232, 831–833.
- Elmakki, M. A., Koen, R., Drost, R. M., Alexander, O. T., Venter, J. A. & Venter, J. A. (2016). Z. Kristallogr. New Cryst. Struct. 231, 781–783.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Leipoldt, J. C. & Grobler, E. C. (1982). Inorg. Chim. Acta, 60, 141–144.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314623003553/wm4184sup1.cif
CCDC reference: 2257230
Additional supporting information: crystallographic information; 3D view; checkCIF report