Abstract
Objectives:
To employ different automated convolutional neural network (CNN)-based transfer learning (TL) methods for both binary and multiclass classification of Alzheimer’s disease (AD) using brain MRI.
Methods:
Herein, we applied three popular pre-trained CNN models (ResNet101, Xception, and InceptionV3) using a fine-tuned approach of TL on 3D T 1-weighted brain MRI from a subset of ADNI dataset (n = 305 subjects). To evaluate power of TL, the aforementioned networks were also trained from scratch for performance comparison. Initially, Unet network segmentedthe MRI scans into characteristic components of gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF). The proposed networks were trained and tested over the pre-processed and augmented segmented and whole images for both binary (NC/AD + progressive mild cognitive impairment (pMCI)+stable MCI (sMCI)) and 4-class (AD/pMCI/sMCI/NC) classification. Also, two independent test sets from the OASIS (n = 30) and AIBL (n = 60) datasets were used to externally assess the performance of the proposed algorithms.
Results:
The proposed TL-based CNN models achieved better performance compared to the training CNN models from scratch. On the ADNI test set, InceptionV3-TL achieved the highest accuracy of 93.75% and AUC of 92.0% for binary classification, as well as the highest accuracy of 93.75% and AUC of 96.0% for multiclass classification of AD on the whole images. On the OASIS test set, InceptionV3-TL outperformed two other models by achieving 93.33% accuracy with 93.0% AUC in binary classification of AD on the whole images. On the AIBL test set, InceptionV3-TL also outperformed two other models in both binary and multiclass classification tasks on the whole MR images and achieved accuracy/AUC of 93.33%/95.0% and 90.0%/93.0%, respectively. The GM segment as input provided the highest performance in both binary and multiclass classification of AD, as compared to the WM and CSF segments.
Conclusion:
This study demonstrates the potential of applying deep TL approach for automated detection and classification of AD using brain MRI with high accuracy and robustness across internal and external test data, suggesting that these models can possibly be used as a supportive tool to assist clinicians in creating objective opinion and correct diagnosis.
Advances in knowledge:
We used CNN-based TL approaches and the augmentation techniques to overcome the insufficient data problem. Our study provides evidence that deep TL algorithms can be used for both binary and multiclass classification of AD with high accuracy.
Introduction
Alzheimer’s disease (AD), the most frequent type of dementia, is an incurable and progressive brain disorder occurring in the elderly population, mostly for people aged 65 and older. 1 It represents one of the greatest challenges for healthcare systems in the 21st century and is the sixth leading cause of death in the United States. 1 AD destroys brain cells, resulting in loss of memory and mental functions. Initially, AD affects the hippocampus region, which controls language and memory. 2 Therefore, the early symptoms of AD are memory loss, confusion and difficulty in speaking, reading or writing. Taken together, AD has a significant negative effect on patients’ everyday lives. According to the World Alzheimer’s Report 2018, there are around 47 million people with AD throughout the world, and the number of patients with AD is estimated to increase to 152 million patients in 2050. 3
AD has three major stages: pre-clinical (normal cognitive (NC)), mild cognitive impairment (MCI), and Alzheimer’s dementia. AD is an irreversible, progressive neurodegenerative disease characterized by a decline in cognitive functioning with no effective disease-modifying treatment available today. 4 Therefore, it is very important to develop strategies for the detection of AD at its early or prodromal stage to prevent and/or slow its progression. 5 For example, MCI is a prodromal or transitional stage of AD where patients have the risk to develop AD. 6 Hence, over the past few decades, advanced neuroimaging technologies have been widely developed and used for AD and MCI diagnosis, such as magnetic resonance imaging (MRI) and positron emission tomography (PET). 7 MRI is a non-invasive imaging technology providing detailed 3D anatomical images of brain tissue, and has been widely used to identify AD-related structural and functional changes in the brain. 8 In particular, structural MRI scans can track the changes in brain structure and measure the inevitable cerebral atrophy, which is caused by the neurodegenerative aspect of AD pathology. The symptoms of AD typically progress slowly and gradually, and also patients may show various symptoms at cognitive and behavioral level; therefore, it can be difficult and complex to diagnose AD. Within this framework, developing innovative diagnostic tools to help diagnosing the disease at an earlier stage is a challenging task. In this context, there has been growing interest in using computer-aided diagnosis (CAD) systems for automatic detection of AD. 9,10
Over the past decade, the automated CAD of Alzheimer has employed machine learning (ML) approaches to analyze structural brain MRI for disease classification and detection. A large number of studies have used the MRI data to detect AD by means of conventional ML methods such as random forests (RF), 11 support vector machine (SVM), 12,13 and boosting algorithms. 14 ML-based classification typically involves four steps: feature extraction, feature selection, dimensionality reduction, and feature-based classification algorithm selection. There are several major problems with the aforementioned procedures. For example, feature extraction and feature selection usually depend on manual/semi-automated image segmentations, which is tedious and prone to inter- and intraobserver variability. 15 Moreover, these procedures require multiple stages of optimization; e.g., complex image pre-processing, which may be time-consuming and computationally demanding. 16 Also, another issue associated with these procedures is reproducibility. 16
To overcome the aforementioned issues, more recently, deep learning (DL), as a new ML technique, has emerged and shown promising results in the field of large-scale, high-dimensional medical imaging analysis. 17 Convolutional neural networks (CNNs), as the most widely used DL architecture, has attracted considerable attention owing to its great success in image classification, image segmentation and object detection. 18–21 CNNs are capable of performing ML tasks without manual functions. 22 DL methods and specifically CNN have outperformed traditional ML methods. 23 Numerous recent studies have used structural-MRI-based CNN models for automated diagnosis of AD. 15,24–26 However, the existing DL approaches train deep networks from scratch, which has some limitations 27,28 : (1) properly training a deep CNN architecture requires a huge amount of annotated medical imaging data, which is time-consuming and expensive to obtain owing to privacy and cost issues; (2)training a deep CNN architecture with huge amount of images requires substantial computational resources; and (3) training a deep network depends seriously on the careful tuning of many hyper-parameters, which is a tedious endeavor. An alternative solution to resolve these issues is fine tuning a deep CNN architecture through transfer learning (TL). 29 The concept behind TL is using and fine tuning the pre-trained CNNs built using large-scale datasets such as ImageNet on different problems with a smaller dataset. 30 Therefore, the purpose of the current study was to apply an automated CNN-based TL approach using three well-known pre-trained models (ResNet101, Xception, and Inception V3) for two classification tasks: (1) binary or 2-way classification (NC/AD+progressive MCI (pMCI)+stable MCI (sMCI)) and (2) 4-way classification (AD/pMCI/sMCI/NC) of 3D brain structural MRI scans. Furthermore, the aforementioned networks have been structured and trained from scratch to compare the effectiveness of TL and training from scratch approaches on the AD classification tasks. Also, we segmented brain MR images into gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) to evaluate the effect of different MRI segments in classifying AD.
Methods and materials
Dataset
In this study, we used a publicly available dataset. The study’s data were obtained from the Alzheimer Disease Neuroimaging Initiative (ADNI) dataset (http://adni.loni.usc.edu/ (accessed on February 2021)). The ADNI was established in 2003 as a public–private partnership, initiated by Dr Michael W. Weiner. It has been designed as a multisite, longitudinal study to develop various biomarkers (clinical, imaging, genetic, etc.) for the early diagnosis of AD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
Herein, a subset of ADNI-1 and ADNI-2 datasets has been used. Standard 3T baseline 3D T 1-weighted structural MRI scans for 305 subjects (94 AD, 65 pMCI, 61 sMCI, and 85 NC) were considered. From the ADNI-1 dataset, images acquired with 3T scanners were included in the study. Structural T 1-weighted MRI images were acquired using 3T Siemens and Philips scanners using MPRAGE sequence with the typical 3T acquisition parameters, including repetition time (TR) = 2300 ms, minimum full echo time (TE), inversion time (TI) = 900 ms, flip angle = 8–9°, slice thickness = 1.2 mm without gap, field-of-view (FOV) = 256 × 256 mm2, matrix size = 256 × 256, and voxel size = 1 × 1 × 1.2 mm3. The demographic details of the ADNI dataset are outlined in Table 1. Imaging data were randomly divided into training, validation and test sets using a ratio of 80:10:10 respectively. We did not use the test data in the training or validation process. Our proposed models were trained and tested over whole MRI images from the ADNI dataset along with the segments of GM, WM, and CSF.
Table 1.
Demographic and clinical information of study participants from the ADNI, OASIS, and AIBL datasets
| dataset | Information | NC | sMCI | pMCI | AD | ||
|---|---|---|---|---|---|---|---|
| Internal | ADNI | Train | Gender (F/M) | 36/35 | 29/17 | 35/12 | 39/38 |
| Age (mean ± SD) | 66.9 ± 3.8 | 69.8 ± 5.3 | 71.3 ± 6.2 | 69.7 ± 5.5 | |||
| CDR (mean ± SD) | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.5 ± 0.0 | 0.8 ± 0.1 | |||
| Validation | Gender (F/M) | 3/3 | 3/4 | 5/5 | 3/6 | ||
| Age (mean ± SD) | 64.2 ± 1.1 | 74.8 ± 2.6 | 70.7 ± 6.2 | 69.5 ± 3.7 | |||
| CDR (mean ± SD) | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.5 ± 0.0 | 0.7 ± 0.5 | |||
| Test | Gender (F/M) | 2/6 | 4/4 | 3/5 | 7/1 | ||
| Age (mean ± SD) | 68.5 ± 3.6 | 67.4 ± 1.3 | 72.7 ± 2.2 | 68.6 ± 5.8 | |||
| CDR (mean ± SD) | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.5 ± 0.0 | 0.7 ± 0.5 | |||
| External | OASIS | Test | Gender (F/M) | 6/9 | - | - | 11/4 |
| Age (mean ± SD) | 67.3 ± 2.9 | - | - | 67.1 ± 9.2 | |||
| CDR (mean ± SD) | 0.0 ± .0.0 | - | - | 0.8 ± 0.2 | |||
| AIBL | Test | Gender (F/M) | 9/6 | 5/10 | 12/3 | 8/7 | |
| Age (mean ± SD) | 68.7 ± 3.2 | 71.3 ± 3.4 | 70.0 ± 6.9 | 69.6 ± 5.1 | |||
| CDR (mean ± SD) | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.6 ± 0.1 | 0.7 ± 0.3 | |||
F: Female; M: Male; CDR: Clinical dementia rating; NC: Normal cognitive; sMCI: Stable mild cognitive impairment; pMCI: Progressive mild cognitive impairment; AD: Alzheimer’s disease;
Moreover, we also included two additional test sets to externally evaluate the performance of the proposed algorithms for both binary and multiclass classification tasks on the whole MR images as well as segmented tissues including GM, WM, and CSF: 1) the Open Access Series of Imaging Studies (OASIS; oasis-brains.org) dataset for binary classification, and 2) Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL; https://aibl.csiro.au) dataset for both binary and multiclass classification. As outlined in Table 1, 30 (15 AD and 15 NC) and 60 (15 AD, 15 pMCI, 15sMCI, and 15 NC) subjects from the OASIS and AIBL datasets, respectively, were included, as two independent test sets, to evaluate the performance of the proposed algorithms outside of the ADNI dataset. Flowchart of the proposed methodology for binary and multiclass classification of AD using brain MR images is shown in Figure 1.
Figure 1.
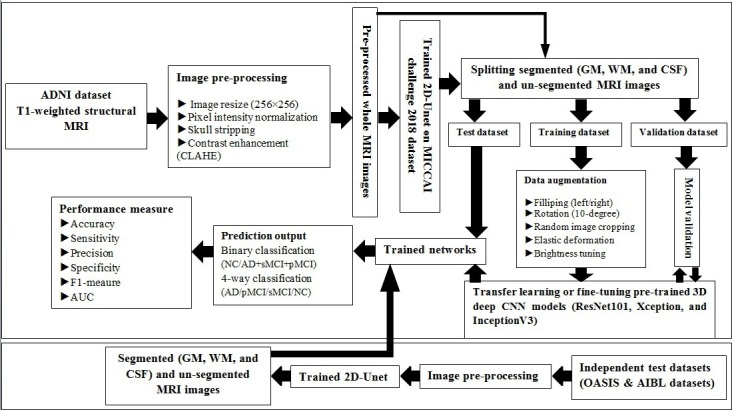
Flowchart of the proposed methodology for binary and multiclass classification of Alzheimer’s disease
Image preprocessing
As a first step of preprocessing, we converted the ADNI T 1-weighted MR images from the.raw to the.NIFTI format and then to.npy format. All images were resized to a size of 256 × 256. The images values were normalized to a range from 0 to 1. Also, a contrast limited adaptive histogram equalization (CLAHE) technique was used to enhance the contrast of MR images, as shown in Supplementary Material 1. Skull stripping was performed using a simple skull stripping algorithm, termed as S3, proposed by Roy and Maji. 31
Image segmentation
Segmentation of brain tissue into GM, WM, and CSF can help to detect AD. Hence, we adopted the standard 2D-Unet architecture 32 to automatically segment the whole brain into GM, WM, and CSF from MRI. Herein, for segmenting brain tissues, different preprocessing and augmentation strategies were used to train and test Unet model on MICCAI challenge 2018 dataset (https://mrbrains18.isi.uu.nl/) and compare the performance with Dice Coefficient Similarity as a metric. Schematic representation of the proposed 2D-Unet for the segmentation of brain tissue is depicted in Supplementary Material 1. The MRI preprocessing, including standardization and skull stripping, was performed for all T 1-weighted MRI images. We also applied a CLAHE algorithm for increasing the contrast level of the input images. Data augmentation was applied using filliping (left/right), rotation range 15 degree, random image cropping, and elastic deformation. The dataset was split randomly into three subsets: a training set (n = 300 images), a validation set (n = 18 images), and a testing set (n = 18 images). The Unet architecture was trained using a combined loss function (Dice-loss+categorical cross-entropy) and Adam optimizer. We used an initial learning rate of 0.01, batch size of 8, and epoch value of 400. Also, during training process, we applied learning rate scheduler for reducing the learning rate as the number of training epochs increases. The network was implemented in Python 3.8 using Pytorch 1.9. The training was performed on NVIDIA Tesla T4 GPU 8 GB and 12 GB RAM. The training time of Unet model was approximately 15 h. Then, the trained and tested U-Net architecture on MICCAI 2018 challenge dataset was used to segment brain MRI from the ADNI dataset into GM, WM, and CSF components with a visually evaluation and modification in part if necessary by an experienced radiologist.
Data augmentation
We also applied aggressive data augmentation techniques to artificially increase the size of training data because a lot of data is required to train deep neural networks. This approach can improve the classification accuracy and make the models more generalized, resulting in reduced overfitting. We used different augmentation methods such as left and right flipping, rotation range 10 degree, random image cropping, elastic deformation, and brightness tuning.
Network architecture
As stated earlier, training a deep CNN model with randomly initialized weights from scratch is a difficult task, especially in medical image analysis, owing to the lack of a massive amount of training data. Using pre-trained model can result in a significant reduction in the amount of training data. In TL, a CNN is pre-trained on large-scale dataset like ImageNet; the weights of the pre-trained deep CNNs are then adopted and fine-tuned to learn a new task.
In this study, we used the pre-trained weights of three different CNN architectures and retrained them on target dataset (i.e., a subset of ADNI) to classify AD. Three powerful and well-known pre-trained CNNs were used as backbone model for TL: ResNet101, 33 Xception, 34 and InceptionV3. 35 All these networks have been pre-trained on the ImageNet dataset. ResNet-101 is a CNN that has 101 layers with 33 three-layer residual blocks. The Xception model is a 71-layer deep CNN based on depth-wise separable convolution layers. InceptionV3 is a deep CNN architecture of 48 layers. The overall architecture of the aforementioned CNN models consists of multilayered structures including convolution, pooling, number of consecutive fully connected, and SoftMax layers. The convolution layers were followed by the exponential linear unit (ELU) activation function. During TL, we froze the three first residual blocks, two first separable blocks, and two first inception blocks for ResNet101, Xception, and InceptionV3 architectures, respectively, to achieve the best accuracy of the models. The other layers were open for modification. The transformed vectors in the flatten layer were fed to a dense layer with 512 neurons, followed by another dense layer with 256 neurons. A dropout with a threshold of 0.5 was implemented in the fully connected layer. A last dense layer with two or four neurons was used with SoftMax activation for binary or multiclass classification, respectively. Also, ResNet101, Xception, and InceptionV3 were regenerated with randomly initialized weights for all layers to perform the training from scratch approach for AD classification tasks.
Training details
Before starting the training, it is necessary to set hyper-parameters; i.e., all the training variables, manually. The optimization of hyper-parameters is performed with an iterative process using the validation loss, which is a guide to model performance. Validation loss indicates errors within a network and also proves how well a network is operating. We trained the networks until there was no further improvement in the validation loss. The networks with the best performance (i.e., lowest validation loss) were saved.
Herein, in order to make a meaningful comparison, we used the same training parameters for all experiments. The binary and categorical cross-entropy loss functions were applied for binary and multiclass Alzheimer’s classification, respectively. All proposed networks were trained using an Adaptive Moment Estimation (Adam) optimizer, with a learning rate of 0.01, which decayed by a factor of 0.1 every 10 epochs whenever loss plateaus, batch size of 16, and epoch value of 200. As stated earlier, data augmentation techniques were applied on images in order to create additional artificial images and consequently reduce overfitting. All deep TL models were implemented in Python 3.8 using Pytorch 1.9 and trained on a computing system with dedicated NVIDIA Tesla T4 GPU 8 GB and 12 GB RAM.
Performance evaluation
The performance of the proposed classification models was evaluated using unseen internal and external test datasets. Confusion matrices were calculated to assess the classification performance of the proposed models. For classification performance, various evaluation metrics, including accuracy, weighted-average based precision, sensitivity, specificity, and F1-meaure, were computed. The value of the area under the receiver operating characteristic (ROC) curve (AUC) was also calculated. Furthermore, we compared binary classification performance of different deep networks and training approaches on the ADNI test set in terms of AUC using the DeLong test. Statistical analyses were done using MedCalc, version 20.0.27 (MedCalc Software, bvba, Ostend, Belgium). A p-value less than 0.05 was considered statistically significant.
Where: TP: True Positive. FP: False Positive. TN: True Negative, and FN: False Negative.
Results
Figure 2 shows an example of auto-segmentation of input brain MR images into WM, GM, and CSF from the MICCAI challenge 2018 dataset and ADNI dataset. The proposed standard 2D-Unet architecture achieved Dice scores of 0.89, 0.88, and 0.92 for WM, GM, and CSF segmentation on the MICCAI challenge 2018 dataset, respectively.
Figure 2.
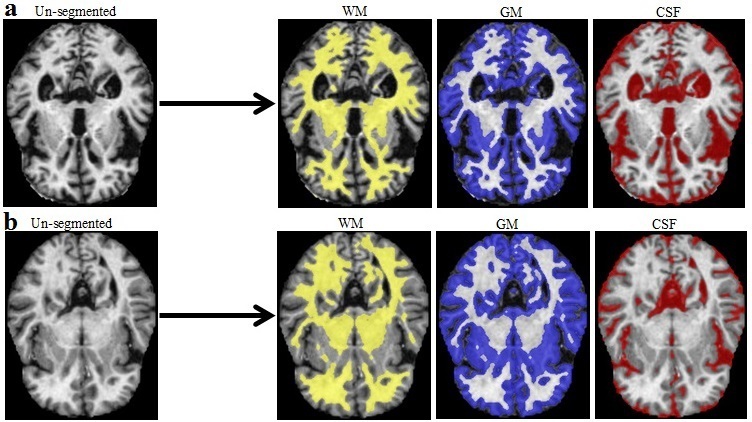
An example of the auto-segmentation of white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) in brain MRI from the MICCAI challenge 2018 dataset (a) and ADNI dataset (b) using 2D-Unet architecture.
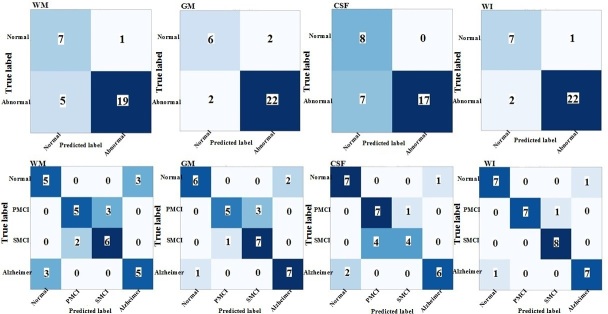
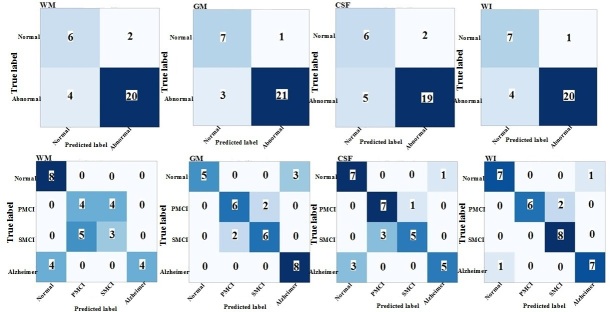
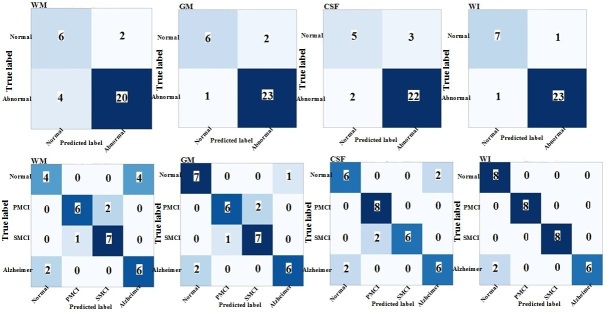
Table 2 summarizes binary (NC/AD+pMCI+ sMCI) and multiclass (AD/pMCI/sMCI/NC) classification performances of various deep networks (ResNet101, Xception, and InceptionV3) and training approaches (i.e., TL and training from scratch) on the ADNI test set. As shown in Table 2, ResNet101, Xception, and InceptionV3, when trained from scratch, resulted in poor performance for both classification tasks. The TL outperformed the training from scratch approach for all three ResNet101, Xception, and InceptionV3. As given in Table 2, InceptionV3 with TL achieved the highest accuracy of 93.75% and an AUC of 92.0% for binary classification of the whole MR images. For the multiclass classification task, again InceptionV3 with TL obtained the highest accuracy of 93.75% and an AUC of 96.0%, when the whole MR images were used as input (Table 2). When GM segmentations were provided as an input to deep networks, InceptionV3 with TL achieved the best performance in both binary and multiclass classification among the three models. As can be seen in Table 2, ResNet101-TL method outperformed the other proposed deep TL models in terms of AUC in binary classification of AD, when the WM and CSF segments were applied as the individual inputs. The GM segment of the brain MR images as input provided the highest performance in both binary and multiclass classification of AD, as compared to the WM and CSF segments (Table 2).
Table 2.
Performance metrics for different deep networks and training approaches on ADNI testing data for both binary and multiclass classification of Alzheimer’s (TL/TS)a
| Input | Classification type | AUC | Accuracy (%) | Sensitivity (%) | Precision (%) | Specificity (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| ResNet101 | |||||||
| ADNI (GM) | Binary | 0.83/0.79a | 87.50/81.25 | 87.50/81.25 | 87.50/83.18 | 79.17/77.08 | 87.50/81.89 |
| Multiclass | 0.85/0.81 | 78.14/71.88 | 78.12/71.88 | 79.20/72.86 | 92.71/90.62 | 77.89/71.62 | |
| ADNI (WM) | Binary | 0.83/0.67 | 81.25/75.00 | 81.25/75.00 | 85.83/75.00 | 85.40/58.33 | 83.47/75.00 |
| Multiclass | 0.77/0.73 | 65.62/59.38 | 65.62/59.38 | 65.78/59.43 | 88.54/86.46 | 65.57/59.29 | |
| ADNI (CSF) | Binary | 0.85/0.75 | 78.12/75.00 | 78.12/68.75 | 88.33/80.00 | 92.71/75.00 | 79.58/76.37 |
| Multiclass | 0.83/0.79 | 75.00/68.75 | 75.00/68.75 | 76.78/69.05 | 91.66/89.58 | 74.39/68.63 | |
| ADNI (whole image) | Binary | 0.90/0.81 | 90.62/84.38 | 90.63/84.38 | 91.18/85.14 | 88.54/78.12 | 90.80/84.67 |
| Multiclass | 0.94/0.89 | 90.63/84.37 | 90.62/84.38 | 90.97/84.62 | 96.87/94.79 | 90.61/84.34 | |
| Xception | |||||||
| ADNI (GM) | Binary | 0.88/0.83 | 87.50/81.25 | 87.50/81.25 | 89.09/85.83 | 87.50/85.42 | 87.92/82.28 |
| Multiclass | 0.85/0.83 | 78.13/75.00 | 78.12/75.00 | 80.68/75.40 | 92.71/91.67 | 77.78/74.90 | |
| ADNI (WM) | Binary | 0.79/0.85 | 81.25/78.12 | 81.25/78.12 | 83.18/88.33 | 77.08/92.71 | 81.89/79.58 |
| Multiclass | 0.72/0.69 | 59.37/53.13 | 71.88/53.12 | 50.99/53.07 | 82.88/84.38 | 58.43/53.02 | |
| ADNI (CSF) | Binary | 0.77/0.75 | 78.12/75.00 | 78.13/75.00 | 81.50/80.00 | 76.04/75.00 | 79.13/76.37 |
| Multiclass | 0.83/0.81 | 75.00/71.87 | 75.00/71.88 | 76.66/72.54 | 91.66/90.62 | 74.61/71.54 | |
| ADNI (whole image) | Binary | 0.85/0.73 | 84.38/78.12 | 84.37/78.12 | 87.34/79.11 | 86.46/67.71 | 85.09/78.54 |
| Multiclass | 0.92/0.87 | 87.50/81.25 | 87.50/81.25 | 88.75/83.33 | 95.83/93.76 | 87.40/82.27 | |
| InceptionV3 | |||||||
| ADNI (GM) | Binary | 0.85/0.77 | 90.62/84.38 | 90.62/84.38 | 90.43/83.86 | 80.21/69.79 | 90.41/84.02 |
| Multiclass | 0.88/0.85 | 81.25/78.12 | 81.25/78.12 | 81.74/81.51 | 93.99/92.71 | 81.18/77.58 | |
| ADNI (WM) | Binary | 0.79/0.77 | 81.25/78.12 | 81.25/78.13 | 83.18/81.50 | 77.08/76.04 | 81.89/79.13 |
| Multiclass | 0.85/0.83 | 78.12/75.00 | 78.12/75.00 | 82.54/75.40 | 92.71/91.67 | 77.26/74.90 | |
| ADNI (CSF) | Binary | 0.77/0.77 | 84.38/78.12 | 84.38/78.13 | 83.86/81.50 | 69.79/76.04 | 84.02/79.13 |
| Multiclass | 0.88/0.81 | 81.25/71.87 | 81.25/81.25 | 82.50/64.83 | 93.75/87.20 | 81.15/70.59 | |
| ADNI (whole image) | Binary | 0.92/0.92 | 93.75/87.50 | 93.75/87.50 | 93.75/91.67 | 89.58/95.83 | 93.75/88.18 |
| Multiclass | 0.96/0.93 | 93.75/90.62 | 93.75/90.62 | 95.00/90.87 | 97.92/96.88 | 93.65/90.59 | |
GM: Gray matter; WM: White matter; CSF: Cerebrospinal fluid; AUC: Area under curve
TL/TS: Transfer learning/Training from scratch
A comparison of the binary classification performance of the proposed models for different training approaches in terms of AUC on the ADNI test dataset using the DeLong test is given in Supplementary Material 1. Statistically significant differences in the AUCs between training from scratch and TL approaches were not found. Also, we compared binary classification performance of the proposed deep TL models on the ADNI test set in terms of AUC using the DeLong test, as shown in Supplementary Material 1. There were no statistically significant differences between the proposed deep TL models for binary classification with a same input.
The performance of the proposed TL models on the segmented MR images with three labels (i.e., WM, GM, and CSF) and whole MR images are presented in the form of confusion matrix in Figures 3–5 for both binary and multiclass classification. Confusion matrices for the proposed models with training from scratch are shown in Supplementary Material 1.
Figure 3.
Confusion matrix of binary and multiclass classification of Alzheimer’s using ResNet101-transfer learning on the ADNI test set. WM: white matter; GM: gray matter; CSF: cerebrospinal fluid; WI: whole image
Figure 4.
Confusion matrix of binary and multiclass classification of Alzheimer’s using Xception-transfer learning on the ADNI test set. WM: white matter; GM: gray matter; CSF: cerebrospinal fluid; WI: whole image
Figure 5.
Confusion matrix of binary and multiclass classification of Alzheimer’s using InceptionV3-transfer learning on the ADNI test set. WM: white matter; GM: gray matter; CSF: cerebrospinal fluid; WI: whole image
The results for binary and multiclass classification performances of the proposed TL models (ResNet101, Xception, and InceptionV3) on the independent test sets; i.e., OASIS and AIBL testing data, are shown in Tables 3 and 4, and corresponding confusion matrices are given in Supplementary Material 1, respectively. As shown in Table 3, InceptionV3 obtained the highest overall performance with 93.33% accuracy and 93.0% AUC in binary classification of whole images on the OASIS test set among the three models. On the AIBL testing data, InceptionV3 model outperformed all other models in both binary and multiclass classification of the whole MR images and achieved an accuracy of 93.33% and an AUC of 95.0% in binary classification and an accuracy of 90.0% and an AUC of 93.0% in multiclass classification (Table 4).
Table 3.
Performance evaluation of the proposed transfer learning models (ResNet101, Xception, and InceptionV3) for binary classification on OASIS testing data
| Input | Model | AUC | Accuracy (%) | Sensitivity (%) | Precision (%) | Specificity (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| OASIS (GM) | ResNet101 | 0.83 | 83.33 | 83.33 | 83.48 | 83.33 | 83.40 |
| Xception | 0.80 | 80.00 | 80.00 | 80.54 | 80.00 | 80.26 | |
| InceptionV3 | 0.90 | 90.00 | 90.00 | 90.17 | 90.00 | 90.08 | |
| OASIS (WM) | ResNet101 | 0.87 | 86.67 | 86.67 | 86.67 | 86.67 | 86.67 |
| Xception | 0.77 | 76.67 | 76.67 | 76.78 | 75.00 | 76.72 | |
| InceptionV3 | 0.83 | 83.33 | 83.33 | 84.72 | 83.33 | 84.01 | |
| OASIS (CSF) | ResNet101 | 0.83 | 83.33 | 83.33 | 83.48 | 83.33 | 83.40 |
| Xception | 0.73 | 73.33 | 73.33 | 73.33 | 73.33 | 73.33 | |
| InceptionV3 | 0.83 | 83..33 | 83.33 | 84.72 | 83.33 | 84.01 | |
| OASIS (whole image) | ResNet101 | 0.90 | 90.00 | 90.00 | 91.67 | 90.00 | 90.82 |
| Xception | 0.80 | 80.00 | 80.00 | 80.54 | 80.00 | 80.26 | |
| InceptionV3 | 0.93 | 93.33 | 93.33 | 93.33 | 93.33 | 93.33 |
GM: Gray matter; WM: White matter; CSF: Cerebrospinal fluid; AUC: Area under curve
Table 4.
Performance evaluation of the proposed transfer learning models (ResNet101, Xception, and InceptionV3) for both binary and multiclass classification on the AIBL testing data
| Input | Classification type | AUC | Accuracy (%) | Sensitivity (%) | Precision (%) | Specificity (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|
| ResNet101 | |||||||
| AIBL (GM) | Binary | 0.82 | 83.33 | 83.33 | 85.30 | 81.11 | 83.92 |
| Multiclass | 0.83 | 75.00 | 75.00 | 75.71 | 91.66 | 75.35 | |
| AIBL (WM) | Binary | 0.79 | 78.33 | 78.34 | 82.72 | 79.44 | 79.47 |
| Multiclass | 0.81 | 71.67 | 71.66 | 72.22 | 90.56 | 71.43 | |
| AIBL (CSF) | Binary | 0.82 | 80.00 | 80.00 | 85.07 | 84.45 | 81.13 |
| Multiclass | 0.82 | 73.33 | 73.33 | 73.60 | 91.11 | 73.46 | |
| AIBL (whole image) | Binary | 0.91 | 90.00 | 90.00 | 91.59 | 92.22 | 90.35 |
| Multiclass | 0.91 | 86.67 | 86.67 | 87.33 | 95.56 | 86.61 | |
| Xception | |||||||
| AIBL (GM) | Binary | 0.89 | 90.00 | 90.00 | 90.63 | 87.78 | 90.20 |
| Multiclass | 0.90 | 85.00 | 85.00 | 85.07 | 95.00 | 85.03 | |
| AIBL (WM) | Binary | 0.84 | 86.67 | 86.67 | 87.41 | 82.22 | 86.93 |
| Multiclass | 0.74 | 61.67 | 61.67 | 62.45 | 87.22 | 61.15 | |
| AIBL (CSF) | Binary | 0.89 | 86.67 | 86.66 | 89.75 | 91.11 | 87.30 |
| Multiclass | 0.89 | 83.33 | 83.33 | 84.23 | 94.44 | 83.77 | |
| AIBL (whole image) | Binary | 0.86 | 86.67 | 86.67 | 88.44 | 86.67 | 87.54 |
| Multiclass | 0.91 | 86.67 | 86.67 | 86.99 | 95.55 | 86.82 | |
| InceptionV3 | |||||||
| AIBL (GM) | Binary | 0.92 | 91.67 | 91.66 | 92.66 | 92.78 | 91.90 |
| Multiclass | 0.91 | 86.67 | 86.67 | 86.83 | 95.56 | 86.65 | |
| AIBL (WM) | Binary | 0.74 | 75.00 | 75.00 | 79.60 | 73.89 | 76.31 |
| Multiclass | 0.88 | 81.67 | 81.67 | 81.74 | 93.89 | 81.66 | |
| AIBL (CSF) | Binary | 0.85 | 88.33 | 88.33 | 88.64 | 82.78 | 88.45 |
| Multiclass | 0.86 | 78.33 | 78.33 | 80.27 | 92.78 | 77.96 | |
| AIBL (whole image) | Binary | 0.95 | 93.33 | 93.33 | 94.74 | 97.78 | 93.57 |
| Multiclass | 0.93 | 90.00 | 90.00 | 90.33 | 96.67 | 90.16 | |
GM: Gray matter; WM: White matter; CSF: Cerebrospinal fluid; AUC: Area under curve
Discussion
The main purpose of this study was to exploit three different architectures of deep CNN models (ResNet101, Xception, and InceptionV3) for both automated binary and multiclass classification of AD using 3D brain MR images through a fine-tuned approach of TL. To assess power of TL, the aforementioned networks were also trained from scratch for performance comparison. Furthermore, the effect of different segments of the brain MRI scans (GM, WM, and CSF) was also evaluated in classifying AD. Herein, 2D-Unet, a type of CNN, was used to automatically segment the MR images into GM, WM, and CSF. Then, TL-based methods (ResNet101, Xception, and InceptionV3) were trained over the dataset of whole images along with the segmented components. We tested our proposed algorithms on two test datasets: 10% ADNI set as internal test set and independent test sets from the OASIS and AIBL datasets. From our data, it can be seen that the proposed TL-based CNN models achieved better performance than training deep CNN models from scratch. Using TL algorithms reveal promising results in terms of classification accuracy, sensitivity, specificity, and AUC. As a result, TL-based CNN models prevent the expensive training from scratch and achieve higher classification performance with a small amount of data.
In this study, three popular TL architectures; i.e., ResNet101, Xception, and InceptionV3 were implemented. As given in Tables 2–4, using InceptionV3 with TL was shown to be the best-performing individual architecture in classifying AD across internal and external test sets, when whole MR images were used as input. On the internal test set, InceptionV3 with TL approach outperformed two other models in both binary and multiclass classification of AD, when the GM segments and whole images were used as individual inputs. As observable in Table 2, InceptionV3-TL method achieved the highest classification performance in 4-class (AD/pMCI/sMCI/NC) classification, regardless of the type of input MR images (i.e., WM, GM, CSF, or whole image). Several previous studies focused only on the specific GM region because GM segmentation of brain MRI is more useful in predicting AD’s early diagnosis. 15,36,37 As can be seen in Tables 2–4, the GM segments as input resulted in higher classification performance than WM and CSF as the individual segmented components. However, from our data, it can be seen that the proposed models achieved their best classification performance on the whole MRI scan as input. As a consequence, the cumulative information of the segmented components (i.e., WM, GM, and CSF) revealed to be distinctive enough for the better binary or multiclass classification of Alzheimer’s (Tables 2–4).
Herein, the DeLong test was used to statistically compare binary classification performance of TL and training from scratch approaches, as well as the proposed deep TL models on the ADNI test set in terms of AUC. Although multiclass classification has a higher value than binary classification because it can distinguish between different stages of the disease, significance of AUCs for multiclass classification cannot determine by the Delong test. As observable in Table 2 and Supplementary Material 1, although using TL algorithms resulted in better AUC, as compared to training from scratch, no statistically significant differences in the AUCs between two training approaches were found. However, from our data (Table 2 and Figures 3–5), it is evident that TL approaches lead to better classification performance compared to training from scratch. From a clinical point of view, the improvement in outcomes observed with TL approach is significant because we are able to train models in a short time with a small dataset and achieve better performance, as compared to training from scratch. Also, we found no statistically significant differences in the AUCs between the proposed deep TL models for binary classification with a same input (Supplementary Material 1). One possible reason for these non-significant values could be due to small sample size, and cross-validation would be suggested to tackle this for further studies.
According to the accuracy metric, we compared the classification performance of our proposed deep TL models with other state-of-the-art models reported in the literature for both binary and multiclass classification of AD on the ADNI dataset, as outlined in Table 5. It is worthwhile to mention that the direct comparison of our proposed TL approaches with the reviewed methods for automated AD diagnosis is not possible, as different ADNI databases with different dataset sizes and different partitions of training and testing sets were used in each study. Also, different MRI modalities, including functional imaging, were used, as shown in Table 5. Furthermore, various classification problems were challenged. The majority of studies focused on the 2-way AD/NC classification 15,24,36,38,39,41 and 3-way classification (AD/MCI/NC). 24,38–41 However, some studies considered 4-way classification of AD. 44–46 A limited number of studies used functional MRI (fMRI) and Diffusion Tensor Imaging (DTI) data for 4-class AD/early MCI (EMCI)/late MCI (LMCI)/NC classification. 42,43 For 4-way classification (AD/pMCI/sMCI/NC), the proposed fine-tuned InceptionV3 and ResNet101 achieved the highest accuracy of 93.75% and the four-highest accuracy of 90.63%, respectively, as shown in Table 5. The model proposed by Odusami et al achieved the second-highest accuracy for 4-class (AD/LMCI/EMCI/NC) classification 46 ; however, they stated that overfitting may have occurred. 46 Hence, Odusami et al suggested that a larger dataset is needed to drastically decrease overfitting. 46 Compared to the study by Odusami et al, 46 we applied a relatively large dataset (three-fold larger) and data augmentation techniques to avoid overfitting. It should be noted; however, that our results are not comparable in a straightforward fashion given that the other studies considered EMCI and LMCI as two distinct phases of MCI, while we used sMCI and pMCI, which seems to be different in literature. 16 Therefore, owing to overlapping features of different stages of AD, classification of various stages of AD is a challenging task. In 4-class classification of AD, our proposed InceptionV3 model outperformed all other state-of-the-art methodologies by achieving an accuracy of 93.75%. Of note, Song et al 42 and Harshit et al 43 used DTI and fMRI modalities with a small size of dataset for 4-class classification, respectively, while other studies applied structural MR images and had a relatively larger dataset. Compared to other studies which applied 3D structural MR images as input, the proposed ResNet101 and Xception achieved the third-highest and four-highest accuracy for 4-way classification of AD, respectively. In this research, 2-way classification was used to separate NC from abnormal cases (AD, pMCI, and sMCI), whereas most of the studies aimed to perform a binary classification of two AD stages that include NC and AD (AD vs NC), as shown in Table 5. However, the classification of sMCI and pMCI is the most challenging task because these classes have similar features. From Table 5, it is evident that our proposed models achieved comparable results to previous studies for binary classification (i.e., NC vs abnormal case). It should be noted that we discriminate NC from AD + pMCI+ sMCI, whereas Hosseini-Asl et al 24,39 classified NC vs MCI + AD. These empirical comparison studies proved that our proposed architectures outperform other competing methods for 4-way AD/pMCI/sMCI/NC classification and demonstrate competitive performance for NC/AD + pMCI+ sMCI classification. As a consequence, the use of deep TL approach helped to achieve better performance.
Table 5.
Performance comparison of our transfer learning approaches with the state-of-the-art models in both binary and multiclass classification of Alzheimer’s disease on the ADNI dataset
| Study/ year | Architecture | Subjects | Modality | Classification accuracy | |||||
|---|---|---|---|---|---|---|---|---|---|
| NC | MCI | AD | AD vs NC | NC vs AC | 3-way classification | 4-way classification | |||
| Payan et al./ 2015 38 | 3D-CNN | 755 | 755 | 755 | MRI | 95.39% | - | 89.47% | - |
| Hosseini-Asl et al./ 2016 39 | 3D-ACNN | 70 | 70 | 70 | sMRI | 97.6% | 90.3% | 89.1% | - |
| Hosseini-Asl et al./ 2018 24 | 3D-DSA-CNN | 70 | 70 | 70 | sMRI | 99.3% | 95.7% | 94.8% | - |
| Khvostikov et al./ 2018 40 | 3D Inception-based CNN | 250 | 228 | 53 | sMRI, DTI | 93.3% | - | 68.9% | - |
| Sahumbaiev et al./ 2018 26 | 3D-CNN HadNet | 160 | 185 | 185 | MRI | - | - | 88.31% | - |
| Jain et al./ 2019 41 | 2D transfer learning-based CNN | 50 | 50 | 50 | sMRI | 99.14% | - | 95.73% | - |
| Song et al./ 2019 42 | Graph CNN | 12 | 12(E) 12(L) |
12 | DTI | - | - | - | 89.0% |
| Basaia et al./ 2019 15 | 3D deep CNN | 352 | 253(c) 510(s) |
294 | sMRI | 98.0% | - | - | - |
| Harshit et al./ 2020 43 | Modified 3D-CNN | 30 | 30(E) 30(L) |
30 | 4D fMRI | - | - | - | 93.0% |
| Abrol et al./ 2020 44 | 3D ResNet | 237 | 245(s) 189(p) |
157 | sMRI | - | - | - | 83.01% |
| Ruiz et al./ 2020 45 | 3D DenseNet ensemble | 120 | 120(E) 120(L) |
120 | MRI | - | - | - | 83.33% |
| Mehmood et al./ 2021 36 | Layer-wise transfer learning approach | 85 | 70(E) 70(L) |
75 | MRI | 98.73% | - | - | - |
| Odusami et al./ 2022 46 | Resnet18 and DenseNet121 with Randomized weight | 25 | 25(E) 25(L) |
25 | MRI | - | - | - | 93.06 |
| Present study | ResNet101 | 85 | 61(s) 65(p) |
94 | sMRI | - | 90.78% | - | 90.63% |
| Xception | - | 84.38% | - | 87.50% | |||||
| Inception v3 | - | 93.75% | - | 93.75% | |||||
NC: Normal cognitive; MCI: Mild cognitive impairment; AD: Alzheimer’s disease; AC: Abnormal case (e.g., MCI, sMCI, pMCI, AD); E: Early MCI; L: Late MCI; c: Convertor MCI; s: Stable MCI; p: Progressive MCI; DTI: Diffusion tensor imaging; 4D fMRI: Four-dimensional functional MRI
To investigate the robustness, we tested our proposed models on two independent test sets (i.e., external test set from the perspective of the algorithms), which contains T 1-weighted MR images from the OASIS (15 AD and 15 NC) and AIBL (15 AD, 15 pMCI, 15 sMCI, and 15 NC) datasets. From Tables 2–4, it can be seen that the performance of the proposed models does not fluctuate remarkably when tested on the external test sets. It is essential to point out that our external test sets were small and collected from public datasets. Thus, the performance of our proposed models on a more general patient population remains unproven. It should be noted, however, that a robust CAD model should be able to detect and classify AD in a normal patient population in presence or absence of other brain disorders. We applied TL approaches with the augmentation techniques to increase the size of dataset for improving performance accuracy. Also, the augmentation techniques can resolve the overfitting issue on a small dataset. Herein, we tested the proposed models on the public datasets. The pre-trained CNN models achieved strong performance on small test sets. In the light of these results, our proposed TL-based methods can be a promising supplementary diagnostic approach.
Conclusion
This study demonstrates the potential of application of deep TL approach for the automated detection and classification of AD from MRI studies of the brain with high accuracy and robustness across internal and external test data. In this study, we proposed three popular pre-trained networks; i.e., ResNet101, Xception, and InceptionV3, and fine-tuned the CNNs for both binary and multiclass classification of AD. Our models were fine-tuned over both segmented (i.e., WM, GM, and CSF) and whole images. We compared the performance classification of the proposed models, among which InceptionV3 with TL achieved the best performance with an accuracy of 93.75% on the internal test for both 2-class and 4-class classification, an accuracy of 93.33% on the OASIS test set in the 2-class classification, and an accuracy of 90.0% on the AIBL test set in the 4-class classification of AD, when the whole images were used as input. Furthermore, the pre-trained TL-based CNN models achieved higher classification performance with limited number of dataset compared to the training CNN models from scratch. The performance and robustness of our models cannot yet be guaranteed on real-life scenario patient cohorts. Hence, further large-scale studies with multiinstitutional data will be required to our proposed models integrate into clinical workflow and serve as a computer-assisted decision support system to aid physicians in detecting AD from MRI studies. Currently, our proposed TL-based methods can possibly be employed to provide diagnostic support.
Footnotes
Competing interests: The authors declare that they have no conflict of interests.
Patient consent: Consent is not required for this type of study.
Contributor Information
Hamed Ghaffari, Email: hamedghaffari@yahoo.com.
Hassan Tavakoli, Email: tavakoli@bmsu.ac.ir.
Gila Pirzad Jahromi, Email: g_pirzad_jahromi@yahoo.com.
REFERENCES
- 1. 2021 alzheimer’s disease facts and figures. Alzheimers Dement 2021; 17: 327–406. doi: 10.1002/alz.12328 [DOI] [PubMed] [Google Scholar]
- 2. Bachstetter AD, Van Eldik LJ, Schmitt FA, Neltner JH, Ighodaro ET, Webster SJ, et al. Disease-related microglia heterogeneity in the hippocampus of alzheimer’s disease, dementia with lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol Commun 2015; 3: 32. doi: 10.1186/s40478-015-0209-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christina P. World alzheimer report 2018– the state of the art of dementia research: new frontiers. 2018.
- 4. De Strooper B, Karran E. The cellular phase of alzheimer’s disease. Cell 2016; 164: 603–15. doi: 10.1016/j.cell.2015.12.056 [DOI] [PubMed] [Google Scholar]
- 5. Galvin JE. Prevention of alzheimer’s disease: lessons learned and applied. J Am Geriatr Soc 2017; 65: 2128–33. doi: 10.1111/jgs.14997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silveira M, Marques J. 20th International Conference on Pattern Recognition (ICPR). Istanbul, Turkey; 2010. pp. 2556–59. doi: 10.1109/ICPR.2010.626 [DOI] [Google Scholar]
- 7. Veitch DP, Weiner MW, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Understanding disease progression and improving alzheimer’s disease clinical trials: recent highlights from the alzheimer’s disease neuroimaging initiative. Alzheimers Dement 2019; 15: 106–52. doi: 10.1016/j.jalz.2018.08.005 [DOI] [PubMed] [Google Scholar]
- 8. Jack CR Jr, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the national institute on aging-alzheimer’s association workgroups on diagnostic guidelines for alzheimer’s disease. Alzheimers Dement 2011; 7: 257–62. doi: 10.1016/j.jalz.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaves R, Ramírez J, Górriz JM, López M, Salas-Gonzalez D, Alvarez I, et al. SVM-based computer-aided diagnosis of the alzheimer’s disease using t-test NMSE feature selection with feature correlation weighting. Neurosci Lett 2009; 461: 293–97. doi: 10.1016/j.neulet.2009.06.052 [DOI] [PubMed] [Google Scholar]
- 10. Bron EE, Smits M, van der Flier WM, Vrenken H, Barkhof F, Scheltens P, et al. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: the caddementia challenge. Neuroimage 2015; 111: 562–79. doi: 10.1016/j.neuroimage.2015.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tripoliti EE, Fotiadis DI, Argyropoulou M. A supervised method to assist the diagnosis and monitor progression of alzheimer’s disease using data from an fmri experiment. Artif Intell Med 2011; 53: 35–45. doi: 10.1016/j.artmed.2011.05.005 [DOI] [PubMed] [Google Scholar]
- 12. Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Alzheimer’s disease neuroimaging initiative.multimodal classification of alzheimer’s disease and mild cognitive impairment. Neuroimage 2011; 55: 856–67. doi: 10.1016/j.neuroimage.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rathore S, Habes M, Iftikhar MA, Shacklett A, Davatzikos C. A review on neuroimaging-based classification studies and associated feature extraction methods for alzheimer’s disease and its prodromal stages. Neuroimage 2017; 155: 530–48. doi: 10.1016/j.neuroimage.2017.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC, et al. Spatially augmented lpboosting for AD classification with evaluations on the ADNI dataset. Neuroimage 2009; 48: 138–49. doi: 10.1016/j.neuroimage.2009.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Basaia S, Agosta F, Wagner L, Canu E, Magnani G, Santangelo R, et al. Automated classification of alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. Neuroimage Clin 2019; 21: 101645. doi: 10.1016/j.nicl.2018.101645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samper-González J, Burgos N, Bottani S, Fontanella S, Lu P, Marcoux A, et al. Reproducible evaluation of classification methods in alzheimer’s disease: framework and application to MRI and PET data. Neuroimage 2018; 183: 504–21. doi: 10.1016/j.neuroimage.2018.08.042 [DOI] [PubMed] [Google Scholar]
- 17. Plis SM, Hjelm DR, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, et al. Deep learning for neuroimaging: a validation study. Front Neurosci 2014; 8: 229. doi: 10.3389/fnins.2014.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM 2017; 60: 84–90. doi: 10.1145/3065386 [DOI] [Google Scholar]
- 19. Salehi M, Mohammadi R, Ghaffari H, Sadighi N, Reiazi R. Automated detection of pneumonia cases using deep transfer learning with paediatric chest X-ray images. Br J Radiol 2021; 94(1121): 20201263. doi: 10.1259/bjr.20201263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. R M, M S, H G, A A R, R R. Transfer learning-based automatic detection of coronavirus disease 2019 (COVID-19) from chest X-ray images. J Biomed Phys Eng 2020; 10: 559–68. doi: 10.31661/jbpe.v0i0.2008-1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017; 36: 61–78. doi: 10.1016/j.media.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 22. Nie D, Wang L, Gao Y, Shen D. FULLY convolutional networks for multi-modality isointense infant brain image segmentation. Proc IEEE Int Symp Biomed Imaging 2016; 2016: 1342–45. doi: 10.1109/ISBI.2016.7493515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–44. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 24. Hosseini-Asl E, Ghazal M, Mahmoud A, Aslantas A, Shalaby AM, Casanova MF, et al. Alzheimer’s disease diagnostics by a 3D deeply supervised adaptable convolutional network. Front Biosci (Landmark Ed) 2018; 23: 584–96. doi: 10.2741/4606 [DOI] [PubMed] [Google Scholar]
- 25. Bae JB, Lee S, Jung W, Park S, Kim W, Oh H, et al. Identification of alzheimer’s disease using a convolutional neural network model based on T1-weighted magnetic resonance imaging. Sci Rep 2020; 10(1): 22252. doi: 10.1038/s41598-020-79243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sahumbaiev I, Popov A, Ramirez J, Gorriz JM, Ortiz A. 3D-CNN HadNet classification of MRI for Alzheimer’s Disease diagnosis. 2018 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC); Sydney, Australia; November 2018. pp. 1–4. 10.1109/NSSMIC.2018.8824317 [DOI] [Google Scholar]
- 27. Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016; 35: 1299–1312. doi: 10.1109/TMI.2016.2535302 [DOI] [PubMed] [Google Scholar]
- 28.] Dumitru E, Pierre-Antoine M, Yoshua B, Samy B, Pascal V.. The Difficulty of Training Deep Architectures and the Effect of Unsupervised Pre-Training. PMLR; 2009. p. 153-60. [Google Scholar]
- 29. Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks. 2014.
- 30. Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015; 115: 211–52. doi: 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 31. Roy S, Maji P. A simple skull stripping algorithm for brain MRI. 2015 Eighth International Conference on Advances in Pattern Recognition (ICAPR); Kolkata, India. ; January 2015. pp. 1–6. doi: 10.1109/ICAPR.2015.7050671 [DOI] [Google Scholar]
- 32. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer; 2015. pp. 234–41. doi: 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 33. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV, USA. ; June 2016. pp. 770–78. doi: 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 34. Chollet F. Xception: Deep Learning with Depthwise Separable Convolutions. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Honolulu, HI. ; July 2017. pp. 1800–1807. doi: 10.1109/CVPR.2017.195 [DOI] [Google Scholar]
- 35. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, NV, USA. ; June 2016. pp. 2818–26. 10.1109/CVPR.2016.308 [DOI] [Google Scholar]
- 36. Mehmood A, Yang S, Feng Z, Wang M, Ahmad AS, Khan R, et al. A transfer learning approach for early diagnosis of alzheimer’s disease on MRI images. Neuroscience 2021; 460: 43–52. doi: 10.1016/j.neuroscience.2021.01.002 [DOI] [PubMed] [Google Scholar]
- 37. Nanni L, Interlenghi M, Brahnam S, Salvatore C, Papa S, Nemni R, et al. Comparison of transfer learning and conventional machine learning applied to structural brain MRI for the early diagnosis and prognosis of alzheimer’s disease. Front Neurol 2020; 11: 576194. doi: 10.3389/fneur.2020.576194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Payan A, Montana G. Predicting alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. ArXiv 2015; 1–9. [Google Scholar]
- 39. Hosseini-Asl E, Keynton R, El-Baz A. Alzheimer’s disease diagnostics by adaptation of 3D convolutional network. 2016 IEEE International Conference on Image Processing (ICIP); Phoenix, AZ, USA. ; September 2016. pp. 126–30. doi: 10.1109/ICIP.2016.7532332 [DOI] [Google Scholar]
- 40. Khvostikov A, Aderghal K, Krylov A, Catheline G, Benois-Pineau J. 3D inception-based CNN with smri and MD-DTI data fusion for alzheimer’s disease diagnostics. 2018.
- 41. Jain R, Jain N, Aggarwal A, Hemanth DJ. Convolutional neural network based alzheimer’s disease classification from magnetic resonance brain images. Cognitive Systems Research 2019; 57: 147–59. doi: 10.1016/j.cogsys.2018.12.015 [DOI] [Google Scholar]
- 42. Chowdhury SR, Yang F, Jacobs H, Fakhri GE. Graph Convolutional Neural Networks For Alzheimer’s Disease Classification. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI). ; 2019. pp. 414–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harshit P, Brian N, Rodney L, Sameer A, Sunanda M. Spatiotemporal feature extraction and classification of alzheimer’s disease using deep learning 3D-CNN for fmri data. Journal of Medical Imaging 2020; 7: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abrol A, Bhattarai M, Fedorov A, Du Y, Plis S, Calhoun V, et al. Deep residual learning for neuroimaging: an application to predict progression to alzheimer’s disease. J Neurosci Methods 2020; 339: 108701. doi: 10.1016/j.jneumeth.2020.108701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz J, Mahmud M, Modasshir M, Shamim Kaiser M. et al. Alzheimer’s Disease Neuroimaging Initiative ft. 3D DenseNet Ensemble in 4-Way Classification of Alzheimer’s Disease. In: Mahmud M, Vassanelli S, Kaiser MS, et al., eds. Brain Informatics. Cham: Springer International Publishing; 2020, pp. 85–96. [Google Scholar]
- 46. Odusami M, Maskeliūnas R, Damaševičius R. An intelligent system for early recognition of alzheimer’s disease using neuroimaging. Sensors (Basel) 2022; 22(3): 740. doi: 10.3390/s22030740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.