Abstract
The AmpC β-lactamase gene and a small portion of the regulatory ampR sequence of Enterobacter aerogenes 97B were cloned and sequenced. The β-lactamase had an isoelectric point of 8 and conferred cephalosporin and cephamycin resistance on the host. The sequence of the cloned gene is most closely related to those of the ampC genes of E. cloacae and C. freundii.
Several members of the family Enterobacteriaceae are naturally resistant to cephalosporins due to the production of an inducible, chromosomally encoded cephalosporinase (7). In the uninduced state, transcription of the structural β-lactamase gene, ampC, is repressed by the product of the linked ampR gene (6, 12, 16). Chromosomal ampC-ampR systems have been described for Citrobacter freundii (11, 13), Enterobacter cloacae (4, 6), Morganella morganii (2, 16), and Yersinia enterocolitica (21). The ampC of Escherichia coli (8) lacks the regulatory gene, possibly as the result of a deletion (6).
Recently discovered plasmid-borne AmpC cephalosporinases such as MIR-1, ACT-1, and LAT-1 appear to be genetic descendants of these chromosomally encoded AmpC enzymes and have been implicated in the spread of the cephalosporin-resistant phenotype to other genera such as Klebsiella (3). In this paper, we describe a hitherto unknown chromosomal ampC from E. aerogenes that was cloned accidentally while searching for a potential plasmid-mediated cephalosporinase.
Enterobacter aerogenes isolates 97A and 97B, identified with the Vitek Identification System (bioMérieux Vitek, Inc., Hazelwood, Mo.), were obtained from the urine of a 30-year-old female patient receiving antibiotic therapy (ampicillin, cefazolin, and gentamicin) for recurring urinary tract infections. The specimen containing 97B was collected 17 h after collection of the one containing 97A. Standard disk diffusion tests (14) and Etests (AB Biodisk North America, Inc., Piscataway, N.J.) were used to determine antibiograms and MICs, respectively. Isolate 97B showed greater resistance than 97A to cefuroxime, cefotetan, cefazolin, ceftriaxone, ceftazidime, and ampicillin (AMP)-sulbactam (Table 1). Both isolates exhibited cefoxitin (FOX) resistance, with MICs of >256 μg/ml.
TABLE 1.
β-Lactam disk diffusion zone sizes for E. aerogenes isolates, the cloning host, and recombinants clones
| Antibiotic | Zone size (mm)
|
||||
|---|---|---|---|---|---|
| E. aerogenes isolate 97A | E. aerogenes isolate 97B | E. coli DH5α | E. coli DH5α (pACM204) | E. coli DH5α (pACM206) | |
| AMP | 6 | 6 | 19 | 6 | 6 |
| AMP-Sulbactam | 15 | 6 | 24 | 6 | 6 |
| Pipericillin | 22 | 22 | 30 | 6 | 6 |
| Pipericillin-tazobactam | 21 | 21 | 29 | 17 | 17 |
| Cefazolin | 7 | 6 | 26 | 6 | 12 |
| Cefuroxime | 16 | 6 | 22 | 8 | 20 |
| FOX | 6 | 6 | 25 | 10 | 25 |
| Cefotetan | 22 | 6 | 32 | 22 | 31 |
| Ceftriaxone | 24 | 20 | 31 | 24 | 31 |
| Ceftazidime | 24 | 19 | 30 | 25 | 30 |
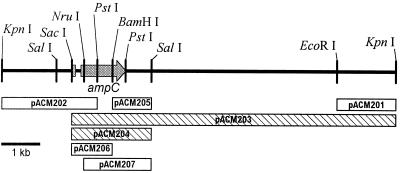
In a “shotgun” attempt to clone a possible plasmid-mediated cephamycinase, libraries of plasmid fragments from 97A and 97B were created by standard procedures (9, 19). Plasmid DNAs were combined with the vector pUC19, digested with PstI or KpnI, and ligated. E. coli DH5α (Gibco-BRL, Gaithersburg, Md.) transformants were selected on agar containing 16 μg of FOX per ml or 50 μg of AMP per ml. This experiment produced only one recombinant, pACM200, from the 97B KpnI-fragment library. The construct contained a 10-kb insert (Fig. 1).
FIG. 1.
Fragment of E. aerogenes chromosomal DNA cloned in recombinant pACM200. The fragments included in each subclone of pACM200 are indicated by boxes below the map; the hatched boxes indicate constructs that mediated FOX resistance. The coding sequence for ampC and the small portion of ampR that was sequenced are indicated by a gray arrow and a gray bar, respectively. Only relevant restriction sites are shown.
Deletion subclones of pACM200 were selected on AMP agar and were subsequently tested for growth on FOX agar. FOX resistance was localized to the 2-kb SacI-SalI fragment in pACM204 (Fig. 1). Further subcloning of pACM204 by cutting the BamHI or NruI sites (pACM205, pACM206, and pACM207) eliminated FOX resistance (Table 1).
Probe 206 was made from the insert of pACM206 by using the Non-Radioactive Labeling and Detection Kit (Roche, Indianapolis, Ind.). Probe 206 failed to hybridize at 42°C with plasmid DNAs from either 97A or 97B (data not shown), even though the cloned fragment originated, presumably, in plasmid DNA from 97B. The probe was then applied to a Southern blot (19) of XbaI-digested total DNAs from a pulsed-field gel electrophoresis gel (20) (data not shown). The blot included DNAs from E. aerogenes strains ATCC 35028 and ATCC 35029 (American Type Culture Collection [ATCC], Rockville, Md.), isolates 97A and 97B, E. coli DH5α, and clinical isolates of E. cloacae and Enterobacter agglomerans (one isolate each). Probe 206 hybridized with a single large fragment (485 kb or larger) in each of three E. aerogenes lanes; in the fourth lane (with isolate ATCC 35028), the DNA was degraded to a smear that ran near the bottom of the gel, but the probe hybridized with the smeared DNA as well. The probe did not bind to the DNA of E. coli DH5α or E. agglomerans, and only a very faint band was present in the E. cloacae lane. These observations led to the conclusion that the cloned fragment had originated in residual chromosomal DNA in the plasmid preparation for 97B and serve as a reminder that the origin of any cloned fragment must be confirmed by hybridization with donor DNA.
A collection of 43 E. aerogenes strains dating back to the 1980s was screened by in situ colony hybridization (19) with probe 206 (data not shown). All E. aerogenes colonies were positive. Other enteric organisms were negative (Klebsiella oxytoca, E. coli, C. freundii, E. agglomerans) or only weakly positive (E. cloacae). These results indicate that the cloned gene is specific to E. aerogenes.
The isoelectric point of the cloned β-lactamase was determined by focusing, as described previously (22), crude protein extracts prepared with the B-PER II Bacterial Protein Extraction Reagent (Pierce, Rockford, Ill.). A sonicated preparation of K. oxytoca β-lactamases (22) provided reference enzymes with known isoelectric points. Focused β-lactamases (data not shown) were detected with the chromogenic cephalosporin pyridinium-2-azo-p-dimethylaniline chromophore (Calbiochem, La Jolla, Calif.) (10). The cloned β-lactamase had an isoelectric point of approximately 8 and comigrated with the single β-lactamase produced by each of four E. aerogenes strains (ATCC and clinical strains), including the DNA donor 97B. Since there is no evidence that 97B produced any β-lactamase other than the one that was cloned, we ruled out a novel plasmidic β-lactamase gene as the source of increased cephalosporin resistance.
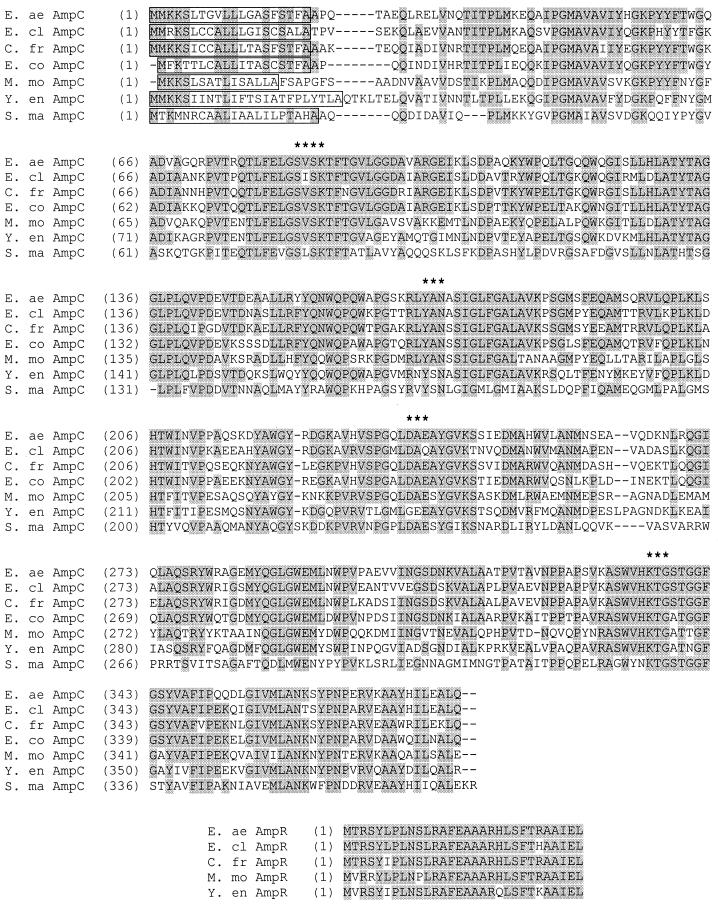
The nucleotide sequence of the insert of pACM204 was determined as described previously (17). The sequence was analyzed with Vector NTI Suite software (InforMax, North Bethesda, Md.) and compared to sequences in the GenBank database through BLAST (1). The insert of pACM204 (Fig. 2) has an open reading frame (ORF) that encodes 381 amino acids. The coding sequence has a 73% identity to the ampC of E. cloacae P99, a 72% identity to the ampC of C. freundii OS60, and a 70% identity to the ampC of E. coli K-12 (Table 2). The ampC ORF is preceded by an incomplete ORF (30 codons) on the complementary strand that corresponds to the beginning of the ampR regulatory genes of E. cloacae and C. freundii. Putative promoters and ribosomal binding sites of the intercistronic region are indicated. There are two possible alternate start codons preceding and in frame with the indicated start of the ampC coding sequence. On the basis of a comparison with other ampC sequences, the indicated start codon is predicted to be the actual start codon. There is no evidence of a transcript terminator sequence following ampC. The nucleotide sequence following ampC has no significant similarity to any other nucleotide sequences in the GenBank database; translations of short ORFs in this region have no protein homologs.
FIG. 2.
Nucleotide sequence of the insert of pACM204 and deduced amino acid sequences for two ORFs corresponding to ampR and ampC. The standard single-letter code for amino acids is used; the stop codon is marked with an asterisk. Putative ribosomal binding sites (rbs), promoters (−10, −35), and selected restriction sites are indicated. Two in-frame alternate start codons for ampC are double underlined. The putative active-site serine is boxed; amino acids in boldface are characteristic motifs of serine β-lactamases. The predicted cleavage point for the signal peptide is marked with an arrowhead.
TABLE 2.
Sequences discussed in the text and included in alignment figures
| Species and strain | Abbreviation in Fig. 3 to 5 | Nucleotide sequence accession no(s). | Reference |
|---|---|---|---|
| Enterobacter aerogenes 97B | E. ae | AF211348 | This work |
| Enterobacter cloacae P99 | E. cl | X07247 | 4 (ampC) |
| Enterobacter cloacae MHN1 | X04730 | 6 (ampR, i-cisa) | |
| Escherichia coli K-12 | E. co | J01611 | 8 |
| Citrobacter freundii OS60 | C. fr | X03866, M27222 | 11 (ampC) |
| 13 (ampR, i-cis) | |||
| Morganella morganii SLM01 | M. mo | Y10283 | 2 |
| Serratia marcescens SR50 | S. ma | X52964 | 15 |
| Yersinia enterocolitica IP97 | Y. en | X63149 | 21 |
i-cis, intercistronic region.
The deduced amino acid sequence has characteristics typical of AmpC enzymes from other enteric organisms. The first 20 amino acids fit predictions for a putative signal peptide (23). The predicted mature peptide of 361 amino acids has a calculated molecular mass and isoelectric point of 39.5 kDa and 7.99, respectively. The mature peptide includes four “signature” motifs [SXXK, Y(A or S)N, D/E, and KTG] that are involved in the formation of the active site in the “active-site serine” enzymes (5). The putative active-site serine occurs at amino acid 64 in the mature peptide sequence.
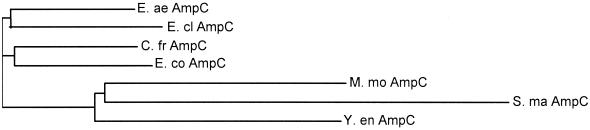
The alignments of the deduced amino acid sequences for seven AmpC sequences and five partial AmpR sequences (AmpR is absent in E. coli and unknown in S. marcescens) are shown in Fig. 3. The dendrogram (Fig. 4) depicts the relationship between the AmpC sequences calculated by the neighbor joining method (18).
FIG. 3.
Alignment of deduced amino acid sequences for seven chromosomal AmpC β-lactamases and the first 30 amino acids of five regulatory AmpR proteins (Table 2). The AmpC signal peptides (predicted or experimentally determined by each author) are boxed, and motifs involved in the formation of the active site are marked with asterisks. Amino acids conserved in more than half the sequences are shaded. E. ae, E. aerogenes; E. cl, E. cloacae; C. fr, C. freundii; M. mo, M. morganii; Y. en, Y. enterocolitica; S. ma, Serratia marcescens.
FIG. 4.
A dendrogram calculated by the neighbor joining method (18) shows the relatedness of the AmpC amino acid sequences; branch lengths are proportional to the number of amino acid changes between sequences. E. co, E. coli; see the legend to Fig. 3 for definitions of the other abbreviations.
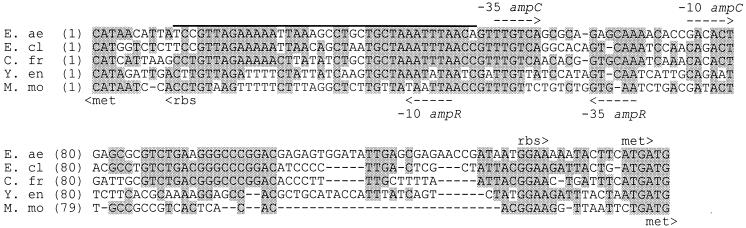
Figure 5 shows the alignment of the ampR-ampC intercistronic regions for six enteric organisms. Promoters, the ampC ribosomal binding site, and the AmpR binding region (determined experimentally for C. freundii by Lindquist et al. [13]) occur in highly conserved areas of the sequence. The putative ampR ribosomal binding site is less well conserved, which might reflect the observed weak expression of this gene (6, 13). Because of the close relationship between the E. aerogenes sequence and others of its type, the intercistronic region is assumed to function in a manner similar to those of C. freundii (13) and E. cloacae (6).
FIG. 5.
Alignment of the intercistronic regions of five ampR-ampC nucleotide sequences (Table 2). Nucleotides conserved in more than half the sequences are shaded. Putative start codons (met), promoters (−10, −35), and ribosomal binding sites (rbs) are indicated. The overlined sequence corresponds to the 38-bp AmpR binding region experimentally determined for C. freundii (13). See the legend to Fig. 3 for definitions of organism abbreviations.
Although cloned unintentionally, the E. aerogenes ampC sequence adds another branch to the phylogenetic tree of species-specific chromosomally encoded enzymes. As new sequences are added, a better understanding of evolutionary relationships among species is achieved.
Nucleotide sequence accession number. The nucleotide sequence of the insert of pACM204 has been deposited in GenBank under accession number AF211348.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnaud G, Arlet G, Danglot C, Philippon A. Cloning and sequencing of the gene encoding the AmpC β-lactamase of Morganella morganii. FEMS Microbiol Lett. 1997;148:15–20. doi: 10.1111/j.1574-6968.1997.tb10260.x. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galleni M, Lindberg F, Normark S, Cole S, Honoré N, Joris B, Frere J. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their products. Biochem J. 1988;250:753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghuysen J M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 6.Honoré N, Nicolas M H, Cole S T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986;5:3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones R N. Important and emerging β-lactamase-mediated resistances in hospital-based pathogens: the AmpC enzymes. Diagn Microbiol Infect Dis. 1998;31:461–466. doi: 10.1016/s0732-8893(98)00029-7. [DOI] [PubMed] [Google Scholar]
- 8.Juarin B, Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of the β-lactamases of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Arai S, Hayashi S, Sakaguchi T. Simple assay of β-lactamase with agar medium containing a chromogenic cephalosporin, pyridinium-2-azo-p-dimethylaniline chromophore (PADAC) Antimicrob Agents Chemother. 1988;32:1040–1045. doi: 10.1128/aac.32.7.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindberg F, Normark S. Sequence of the Citrobacter freundii OS60 chromosomal ampC β-lactamase gene. Eur J Biochem. 1986;156:441–445. doi: 10.1111/j.1432-1033.1986.tb09601.x. [DOI] [PubMed] [Google Scholar]
- 12.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc Natl Acad Sci USA. 1985;82:4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindquist S, Lindberg F, Normark S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J Bacteriol. 1989;171:3746–3753. doi: 10.1128/jb.171.7.3746-3753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 15.Nomura K, Yoshida T. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC β-lactamase gene. FEMS Microbiol Lett. 1990;70:295–300. doi: 10.1111/j.1574-6968.1990.tb13992.x. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preston K E, Radomski C C A, Venezia R A. The cassettes and 3′ conserved segment of an integron from Klebsiella oxytoca plasmid pACM1. Plasmid. 1999;42:104–114. doi: 10.1006/plas.1999.1418. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol. 1992;30:1491–1498. doi: 10.1128/jcm.30.6.1491-1498.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seoane A, Francia M V, García Lobo J M. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother. 1992;36:1049–1052. doi: 10.1128/aac.36.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venezia R A, Scarano F J, Preston K E, Steele L M, Root T P, Limberger R, Archinal W, Kacica M A. Molecular epidemiology of an SHV-5 extended-spectrum β-lactamase in Enterobacteriaceae isolated from infants in a neonatal intensive care unit. Clin Infect Dis. 1995;21:915–923. doi: 10.1093/clinids/21.4.915. [DOI] [PubMed] [Google Scholar]
- 23.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]