Abstract
A collection of rifampin-resistant mutants of Staphylococcus aureus with characterized RNA polymerase β-subunit (rpoB) gene mutations was cross-screened against a number of other RNA polymerase inhibitors to correlate susceptibility with specific rpoB genotypes. The rpoB mutants were cross-resistant to streptolydigin and sorangicin A. In contrast, thiolutin, holomycin, corallopyronin A, and ripostatin A retained activity against the rpoB mutants. The second group of inhibitors may be of interest as drug development candidates.
Bacterial DNA-dependent RNA polymerase is an attractive drug target because RNA chain elongation is essential for bacterial growth (6, 16). Among those antibiotics discovered in the last 50 years, there are several known, or suspected, inhibitors of bacterial DNA-dependent RNA polymerase that have not been developed and could be candidates for new drugs. These agents include thiolutin (18), holomycin (B. Oliva, A. O'Neill, J. M. Wilson, P. J. O'Hanlon and I. Chopra, submitted for publication), streptolydigin (6, 16), the ripostatins, corallopyronins and sorangicins (10–12, 23) (Fig. 1). However, bacterial resistance has already developed to the rifamycins, the only class of RNA polymerase inhibitor that is in use clinically (21). Therefore, before considering whether other RNA polymerase inhibitors might be developed, it is important to establish whether resistance to rifamycins, such as rifampin, also confers cross-resistance to the other agents. Some attempts to address this issue have been made (9, 12, 13, 15, 24). However, the data are incomplete and the genetic basis of resistance to rifamycins in those strains used for cross-screening has rarely been determined. Furthermore, some data are contradictory; e.g., cross-resistance between rifampin and streptolydigin has been observed by some authors (13) but not by others (9, 15).
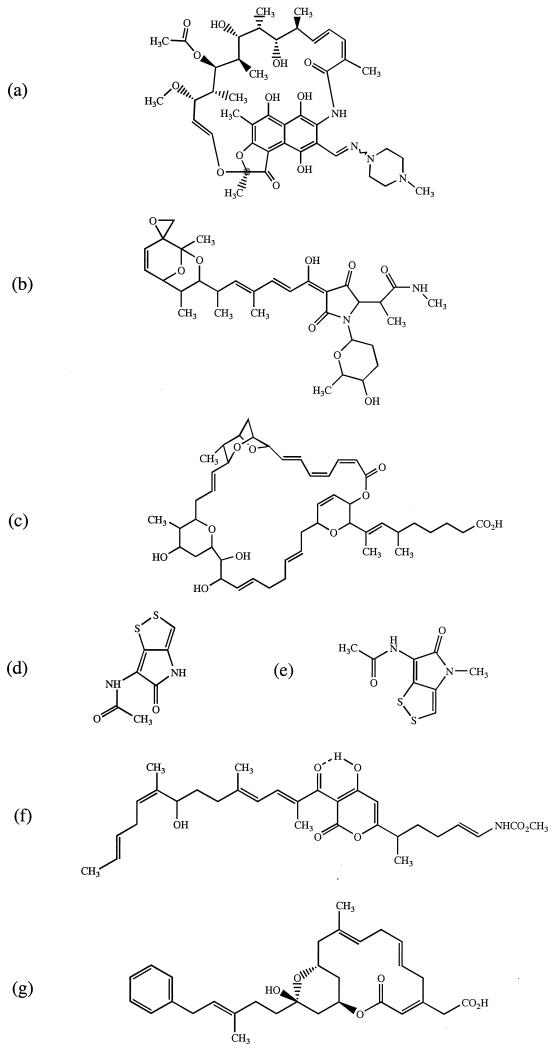
FIG. 1.
Structures of rifampin (a), streptolydigin (b), sorangicin A (c), holomycin (d), thiolutin (e), corallopyronin A (f), and ripostatin A (g).
To assist the evaluation of these older agents we cross-screened them against a collection of rifampin-resistant mutants of Staphylococcus aureus, generated in an isogenic background, with defined RNA polymerase β-subunit (rpoB) gene mutations. These S. aureus strains, which provide a model for rpoB mutations occurring in naturally occurring isolates of staphylococci and other organisms (1, 7, 8, 15, 22, 28, 29), have allowed us to correlate susceptibility with specific rpoB genotypes.
The antibiotics used here were either purchased from Sigma (rifampin and streptolydigin) or were gifts from H. Reichenbach, Gesellschaft für Biotechnologische Forschung, Braunschweig, Germany (corallopyronin A, ripostatin A, and sorangicin A); P. O'Hanlon, SmithKline Beecham Pharmaceuticals, Harlow, United Kingdom (holomycin and thiolutin); and Pharmacia & Upjohn (rifabutin). Spontaneous rifampin-resistant mutants of S. aureus 8325-4 (20) were isolated by plating approximately 108 CFU onto Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) containing 0.032 μg of rifampin/ml (four times the MIC). A number of rifampin-resistant mutants were picked at random, and their MICs of rifampin were determined by agar dilution in Iso-Sensitest agar using an inoculum of 106 CFU/spot (2). This resulted in the identification of a series of mutants for which the MICs of rifampin were in the range 0.25 to 1024 μg/ml.
The rpoB gene mutations were determined in three low-level-resistant mutants (MIC, 0.25 μg/ml), three intermediate-level-resistant mutants (MIC, 8 to 16 μg/ml), and three high-level-resistant mutants (MIC, >500 μg/ml). Total DNA was prepared (25) from the mutants and the parental strain 8325-4 and was subjected to PCR amplification of rpoB using the primers F3 and F4 (1) (Table 1). The amplification products were visualised by agarose gel electrophoresis (25) and then extracted from gels by solubilization in QG buffer (Qiagen, Crawley, United Kingdom). DNA was purified using the QIAquick PCR purification kit (Qiagen) and then sequenced from both F3 and F4 using an Applied Biosystems 377 DNA sequencer. This procedure resulted in the identification of mutations in all strains apart from Rif21, Rif22, and Rif26. Additional primers (rif1 and rif6) (Table 1) were used to amplify the whole of rpoB in these mutants and all primers (Table 1) used for sequencing of the amplified products.
TABLE 1.
Primers used for PCR amplification and sequencing of regions of rpoB from rifampin-resistant mutants of S. aureus 8325-4
| Primera | Nucleotide sequence (5′-3′) | Position (bp) in rpoB (direction) |
|---|---|---|
| F3 | AGTCTATCACACCTCAACAA | 1325–1344 (sense) |
| F4 | TAATAGCCGCACCAGAATCA | 2026–2007 (antisense) |
| rif1 | ATCTGTTTGGCAGGTCAAGTTGTC | 1–24 (sense) |
| rif2 | ACAGATGCTAAAGATGTTGTATAC | 526–549 (sense) |
| rif3 | TCAATTAAAGTATATGTTCCTAAC | 1042–1065 (sense) |
| rif4 | ACCAATATAAACGATACCACGATC | 2505–2482 (antisense) |
| rif5 | ATCACGAGCCATACCAGCTTCTTC | 3045–3022 (antisense) |
| rif6 | AAATTGCGTATTAATCAGTAACTC | 3569–3546 (antisense) |
Primers rif1 to rif6 were based on existing rpoB sequence data (GenBank accession no. X64172).
Nine mutational changes were found in the rifampin-resistant mutants occurring at seven positions from amino acid 137 to 486 (Table 2). With the exception of the mutation at amino acid 137, the other mutations were all located in cluster I of rpoB (15, 16) and are either identical to those previously reported for rifampin resistance in S. aureus (1, 28) or involve different amino acid substitutions (e.g., Asp471→Glu and His481→Asp [at sites 471 and 481]) where other mutational changes are already known to confer rifampin resistance (1, 28). The mutation at position 137 (Gln137→Leu) in mutant Rif21 has not previously been reported in S. aureus. Furthermore, it does not appear to represent a mutational site which exactly corresponds to those found in the rpoB genes of other organisms (16). However, we observed an identical mutation in two other independent mutants (Rif22 and Rif26) that also displayed low-level resistance to rifampin, and mutations conferring rifampin resistance in Escherichia coli (19) and Rickettsia typhi (27) have been reported at the amino terminus of the β-subunit, corresponding to positions 135 and 125 in S. aureus. The S. aureus rifampin-resistant mutants studied here displayed cross-resistance to streptolydigin and sorangicin A (Table 2). However, cross-resistance was not observed with thiolutin, holomycin, corralopyronin A, or ripostatin A (Table 2). For control purposes we also screened the set of rpoB mutants for cross-resistance to another member of the rifamycin class, rifabutin. In all cases cross-resistance was observed (data not shown).
TABLE 2.
Susceptibility of S. aureus 8325-4 rpoB mutants to various antibiotics
| Strain | Mutation in RpoB | MIC (μg/ml) ofa:
|
||||||
|---|---|---|---|---|---|---|---|---|
| RIF | STL | SOR A | HOL | THL | COR A | RIP A | ||
| 8325-4 | 0.008 | 16 | 0.032 | 8 | 8 | 0.5 | 8 | |
| Rif21 | Gln137→Leu | 0.25 | 32 | 0.016 | 8 | 8 | 0.5 | 8 |
| Rif37 | Asp471→Glu | 0.25 | 32 | 1 | 8 | 8 | 0.5 | 8 |
| Rif39 | Leu466→Ser | 0.25 | 32 | 1 | 8 | 8 | 0.5 | 8 |
| Rif28 | His481→Asn | 8 | 64 | 16 | 8 | 4 | 0.5 | 8 |
| Rif34 | Asp471→Tyr | 8 | 64 | 2 | 8 | 8 | 0.5 | 8 |
| Rif35 | Ser464→Pro | 16 | 64 | 1 | 4 | 4 | 1 | 8 |
| Rif38 | Ala477→Asp | 512 | 64 | 2 | 8 | 4 | 1 | 8 |
| Rif23 | Ser486→Leu | 1,024 | 64 | 32 | 8 | 8 | 0.5 | 4 |
| Rif40 | His481→Asp | 1,024 | >128 | >64 | 4 | 8 | 0.5 | 4 |
Abbreviations: COR A, corallopyronin A; HOL, holomycin; RIF, rifampin; RIP A, ripostatin A; SOR A, sorangicin A; STL, streptolydigin; THL, thiolutin.
The emergence of bacterial resistance to antimicrobial agents is a serious threat to human health (3–5). One approach to the problem is the discovery and development of new antibacterial agents with novel targets that will be effective against organisms resistant to current agents (3, 4). However, since this process is complex and lengthy (3, 4) an alternative strategy is the reevaluation of older unexploited antibiotic classes that have not so far been developed for human use (4, 30).
When choosing earlier agents for development it is desirable to select compounds that are structurally unrelated to current agents so that problems of cross-resistance mediated by existing mechanisms are minimized (4). Apart from the fact that the structures of rifampin, ripostatin A, and sorangicin A all contain cyclic frameworks (25, 14, and 30 membered, respectively) (Fig. 1), there is essentially very little structural similarity between rifampin and the earlier RNA polymerase inhibitors. This is particularly the case when comparing the macrocyclic naphthol-based structure in rifampin with the tetramic acid structure of streptolydigin and with the structurally simple thiolutin and holomycin systems (Fig. 1). Thus, none of the earlier RNA polymerase inhibitors described here has any obvious pharmacophoric similarity to rifampin. In view of their structural novelty these agents might therefore be expected to retain activity against rifampin-resistant isolates. However, there have been reports of cross-resistance between some of the older compounds described here and rifampin (13, 24). We examined this in more detail by establishing whether the non-rifamycin-type RNA polymerase inhibitors were active against genetically defined rifampicin-resistant (rpoB) mutants of S. aureus.
On the basis of the staphylococcal cross-resistance patterns we conclude that streptolydigin and sorangicin A are not potential drug candidates. In contrast, holomycin, thiolutin, corallopyronin A, and ripostatin A are worthy of further study since they overcome rifampin-resistant genotypes. There are also no reports that these agents exhibit cross-resistance with other antibiotic resistance determinants. Our data suggest that this second group of inhibitors could have an application in the treatment of infections caused by rifampin-resistant and -sensitive staphylococci. However, since these agents have a spectrum of activity that also encompasses other bacterial species (10, 12, 17, 24, 26; Oliva et al., submitted for publication), they may have applications which could extend beyond their use simply as antistaphylococcal agents. With the exception of corallopyronin A, the other three agents are less active as antimicrobial agents (MICs, 8 to 4 μg/ml) (Table 2). Therefore, it will probably be necessary to derive more-active analogs of these agents for administration as antibacterial drugs.
Other aspects of these compounds, e.g., their toxicity, will need to be addressed before they can be considered as development candidates. Preliminary toxicity studies have been performed on some of these agents. Corallopyronin and ripostatin are only weakly active against eukaryotic cells in vitro (10, 23), and corallopyronin is reported to show no toxicity for mice when administered subcutaneously (10). Thiolutin appears to possess activity against eukaryotic cells since it has antifungal activity (14, 26) and is reported to be moderately toxic in mice (26). Holomycin appears to lack antifungal activity (Oliva et al., submitted for publication), but there are no published animal toxicity data for this compound. Some of the toxicity studies on these compounds were conducted more than 50 years ago (26), and clearly more-detailed studies are required to determine the specificities of these agents for prokaryotic organisms.
Although studies on cross-resistance between rifampin, and holomycin, thiolutin, and corallopyronins have not been reported, the lack of cross-resistance between rifampin and ripostatin A that we observed is consistent with earlier findings (12). Similarly, we have confirmed an earlier report of cross-resistance between sorangicins and rifampin (24). Our results with streptolydigin are only in partial agreement with earlier reports. Iwakura et al. (13) demonstrated cross-resistance in E. coli between rifampin and streptolydigin at the level of rpoB. However, although mutations conferring resistance to streptolydigin are known to occur in rpoB (between clusters I and II in E. coli) (9, 15) others have reported no relationship between these mutations and resistance to rifampin (9). It has therefore been concluded that each drug has a separate binding site in the β-subunit of RNA polymerase (9, 15). We suggest, however, that the streptolydigin binding site, at least in S. aureus, may extend into cluster I or is influenced by the nature of the amino acids present in this region.
Acknowledgments
This work was supported by grants to I. Chopra from Smith and Nephew Research, Hull, United Kingdom, and Intrabiotics Pharmaceuticals, Mountain View, Calif.
We thank H. Reichenbach and P. O'Hanlon for the provision of antibiotics.
REFERENCES
- 1.Aubry-Damon H, Soussy C-J, Courvalin P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:2590–2594. doi: 10.1128/aac.42.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.British Society for Antimicrobial Chemotherapy. A guide to sensitivity testing. Report of the working party on antibiotic sensitivity testing of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother. 1991;27(Suppl. D):1–50. [PubMed] [Google Scholar]
- 3.Chopra I, Hodgson J, Metcalf B, Poste G. New approaches to the control of infections caused by antibiotic-resistant bacteria. An industry perspective. JAMA. 1996;275:401–403. [PubMed] [Google Scholar]
- 4.Chopra I, Hodgson J, Metcalf B, Poste G. The search for antimicrobial agents effective against bacteria resistant to multiple antibiotics. Antimicrob Agents Chemother. 1997;41:497–503. doi: 10.1128/aac.41.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen M L. Epidemiology of drug resistance: implications for a postantimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 6.Das A, DeVito J, Sparkowski J, Warren F. RNA synthesis in bacteria: mechanism and regulation of discrete biochemical events at initiation and termination. In: Sutcliffe J, Georgopapadakou N H, editors. Emerging targets in antibacterial and antifungal chemotherapy. New York, N.Y: Chapman and Hall; 1992. pp. 68–116. [Google Scholar]
- 7.Drancourt M, Raoult D. Characterization of mutations in the rpoB gene in naturally rifampin-resistant Rickettsia species. Antimicrob Agents Chemother. 1999;43:2400–2403. doi: 10.1128/aac.43.10.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright M, Zawadski P, Pickerill P, Dowson C G. Molecular evolution of rifampicin resistance in Streptococcus pneumoniae. Microb Drug Res. 1998;4:65–70. doi: 10.1089/mdr.1998.4.65. [DOI] [PubMed] [Google Scholar]
- 9.Heisler L M, Suzuki H, Landick R, Gross C A. Four contiguous amino acids define the target for streptolydigin resistance in the β subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 10.Irschik H, Jansen R, Hofle G, Gerth K, Reichenbach H. The corralopyronins, new inhibitors of bacterial RNA synthesis from myxobacteria. J Antibiot. 1985;38:145–152. doi: 10.7164/antibiotics.38.145. [DOI] [PubMed] [Google Scholar]
- 11.Irschik H, Jansen R, Gerth K, Hofle G, Reichenbach H. The sorangicins, novel and powerful inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J Antibiot. 1987;40:7–13. doi: 10.7164/antibiotics.40.7. [DOI] [PubMed] [Google Scholar]
- 12.Irschik H, Augustiniak H, Gerth K, Hofle G, Reichenbach H. The ripostatins, novel inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J Antibiot. 1995;48:787–792. doi: 10.7164/antibiotics.48.787. [DOI] [PubMed] [Google Scholar]
- 13.Iwakura Y, Ishihama A, Yura T. RNA polymerase mutants of Escherichia coli. II. Streptolydigin resistance and its relation to rifampicin resistance. Mol Gen Genet. 1973;121:181–196. doi: 10.1007/BF00277531. [DOI] [PubMed] [Google Scholar]
- 14.Jiminez A, Tipper D J, Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 16.Jin D J, Zhou Y N. Mutational analysis of structure-function relationship of RNA polymerase in Escherichia coli. Methods Enzymol. 1996;273:300–319. doi: 10.1016/s0076-6879(96)73027-6. [DOI] [PubMed] [Google Scholar]
- 17.Kenig M, Reading C. Holomycin and an antibiotic related to tunicamycin, metabolites of Streptomyces clavuligerus. J Antibiot. 1979;32:549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 18.Khachatourians G G, Tipper D J. Inhibition of messenger ribonucleic acid synthesis in Escherichia coli by thiolutin. J Bacteriol. 1974;119:795–804. doi: 10.1128/jb.119.3.795-804.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisitsyn N A, Sverdlov E D, Moiseyeva E P, Danilevskaya O N, Nikiforov V G. Mutation to rifampin resistance at the beginning of the RNA polymerase beta subunit gene in Escherichia coli. Mol Gen Genet. 1984;196:173–174. doi: 10.1007/BF00334112. [DOI] [PubMed] [Google Scholar]
- 20.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 21.Parenti F, Lancini G. Rifamycins. In: O'Grady F, Lambert H P, Finch R G, Greenwood D, editors. Antibiotic and chemotherapy. 7th Edition. New York, N.Y: Churchill Livingstone; 1997. pp. 453–459. [Google Scholar]
- 22.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis. Tuberc Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 23.Reichenbach H, Hofle G. Myxobacteria as producers of secondary metabolites. In: Grabley S, Thiericke R, editors. Drug discovery from nature. Berlin, Germany: Springer-Verlag; 1999. pp. 149–178. [Google Scholar]
- 24.Romele G, Wirz G, Solf R, Vosbeck K, Gruner J, Wehrli W. Resistance of Escherichia coli to rifampicin and sorangicin A - a comparison. J Antibiot. 1990;43:88–91. doi: 10.7164/antibiotics.43.88. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- 26.Seneca H, Kane J H, Rockenbach J. Bactericidal, protozoicidal and fungicidal properties of thiolutin. Antibiot Chemother. 1952;2:357–360. [PubMed] [Google Scholar]
- 27.Troyer J M, Radulovic S, Andersson S G E, Azad A F. Detection of point mutations in rpoB gene of rifampin-resistant Rickettsia typhi. Antimicrob Agents Chemother. 1998;42:1845–1846. doi: 10.1128/aac.42.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wichelhaus T A, Schafer V, Brade V, Boddinghaus B. Molecular characterisation of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:2813–2816. doi: 10.1128/aac.43.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams D L, Spring L, Collins L, Miller L P, Heifets L B, Gangadharam P R J, Gillis T P. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahner H, Fiedler H-P. The need for new antibiotics: possible ways forward. In: Hunter P A, Darby G K, Russell N J, editors. Fifty years of antimicrobials: past perspectives and future trends. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 67–84. [Google Scholar]