Abstract
The proteins ZC3HC1 and TPR are structural components of the nuclear basket (NB), a fibrillar structure attached to the nucleoplasmic side of the nuclear pore complex (NPC). ZC3HC1 initially binds to the NB in a TPR-dependent manner and can subsequently recruit additional TPR polypeptides to this structure. Here, we examined the molecular properties of ZC3HC1 that enable its initial binding to the NB and TPR. We report the identification and definition of a nuclear basket-interaction domain (NuBaID) of HsZC3HC1 that comprises two similarly built modules, both essential for binding the NB-resident TPR. We show that such a bimodular construction is evolutionarily conserved, which we further investigated in Dictyostelium discoideum and Saccharomyces cerevisiae. Presenting ScPml39p as the ZC3HC1 homologue in budding yeast, we show that the bimodular NuBaID of Pml39p is essential for binding to the yeast NB and its TPR homologues ScMlp1p and ScMlp2p, and we further demonstrate that Pml39p enables linkage between subpopulations of Mlp1p. We eventually delineate the common NuBaID of the human, amoebic, and yeast homologue as the defining structural entity of a unique protein not found in all but likely present in most taxa of the eukaryotic realm.
INTRODUCTION
Nuclear pore complexes (NPCs) are gateways that connect the cytoplasm and nucleus in eukaryotes. The nuclear ring of the NPC serves as the attachment site for the nuclear basket (NB), a structure composed of thin fibrils that project rectilinearly toward the nuclear interior. These NB fibrils then bifurcate and interconnect laterally with their neighboring fibrils, resulting in a ring-like arrangement commonly called the terminal ring (TR).
The NB was possibly first described as a fish trap–like structure sporadically observed in electron micrographs of ultrathin sections of monkey kidney cells (Maul, 1976). Later, it was visualized as a three-dimensional structure in amphibian and avian oocytes (Ris, 1989, 1991; Jarnik and Aebi, 1991; Goldberg and Allen, 1992; Goldberg et al., 1997). NBs were also detected in the salivary gland cells of the midge Chironomus tentans (Kiseleva et al., 1996) and in the slime mold Dictyostelium discoideum (Beck et al., 2004). In addition, NB-like structures have been described for budding yeast (Kiseleva et al., 2004) and tobacco plant cells (Fiserova et al., 2009), indicating that this structure may exist in many eukaryotes and different cell types. However, while various functions have been attributed to the NB or some of its components in different cell types and species, a universal NB function that holds for all cells remains elusive (e.g., Strambio-De-Castillia et al., 2010; Niepel et al., 2013; Snow and Paschal, 2014; Ashkenazy-Titelman et al., 2020; Bensidoun et al., 2021).
Furthermore, a consensus on the complete protein inventory of the NB remains pending, even though several proteins have been localized at the NPC-attached intranuclear fibrils in different species. Among these proteins is TPR, a large, coiled-coil–forming vertebrate protein (e.g., Cordes et al., 1997; Hase et al., 2001; Frosst et al., 2002; Krull et al., 2004) and its budding yeast homologues, Mlp1p and Mlp2p (Strambio-de-Castillia et al., 1999; Kosova et al., 2000). Homologues of TPR and the Mlps have also been identified in insects and plants (Zimowska et al., 1997; Kuznetsov et al., 2002; Jacob et al., 2007; Xu et al., 2007) and in numerous other species across the eukaryotic realm (Holden et al., 2014). The vertebrate TPR polypeptides and their homologues in insects and yeasts have been proposed to form the backbone of the fibrillar NB scaffold (Krull et al., 2004; Soop et al., 2005; Niepel et al., 2013; Gunkel et al., 2021). Finally, TPR has been shown to be essential for the integrity of the NBs in human cells and budding yeast (Krull et al., 2010; Funasaka et al., 2012; Niepel et al., 2013; Duheron et al., 2014).
Various proteins have been described as binding partners of TPR or its homologues. Some of these proteins were found to colocalize with the NPC-attached TPR and reside at the nuclear envelope (NE) in a TPR-dependent manner. However, these proteins were considered neither to contribute to NB assembly nor to maintain its structural integrity. Instead, it was proposed that they use the NB as either an operational platform for conducting specific tasks or as an NPC-adjacent storage site (e.g., Zhao et al., 2004; Palancade et al., 2005; Scott et al., 2005; Xu et al., 2007; Lee et al., 2008; Ding et al., 2012; Schweizer et al., 2013; Umlauf et al., 2013; Aksenova et al., 2020; Ouyang et al., 2020).
Recently, we reported the identification of vertebrate ZC3HC1 as an NB-resident protein that occurs at the TR of the prototypic NB and is a binding partner of TPR (Gunkel et al., 2021). As with the other NB-appended proteins, NB localization of ZC3HC1 is TPR-dependent. Unlike the other NB proteins, though, ZC3HC1 functions as a structural element that enables the recruitment and linkage of large additional amounts of TPR to preexisting NPC-attached TPR (Gunkel et al., 2021; Gunkel and Cordes, 2022). However, it still needed to be determined how this reciprocally dependent NB positioning of ZC3HC1 and subpopulations of TPR occurs, and which parts of ZC3HC1 enable it to engage in possibly different types of interactions with TPR.
In the present study, we first investigated the molecular features of human ZC3HC1 that enable its initial binding to the NB. We show that ZC3HC1 is composed of two modules that form a functional entity, which we named the nuclear basket interaction domain (NuBaID). We demonstrate that the integrity of the NuBaID is essential for ZC3HC1 binding to the NB and those TPR polypeptides already anchored to the NPC. We defined a composite sequence signature for this bimodular arrangement and describe it as the evolutionarily conserved characteristic of a unique protein existing in many but not all species of the eukaryotic realm. We further demonstrate that the NuBaID signature–containing proteins of two model species, DDB0349234 of D. discoideum and Pml39p of Saccharomyces cerevisiae, are true homologues of HsZC3HC1. While ScPml39p was already known to be an Mlp-binding protein (Palancade et al., 2005) and recently declared the budding yeast homologue of ZC3HC1 (Gunkel et al., 2021), we now unveil that ScPml39p possesses a prototypic NuBaID whose integrity is required for the interactions with NBs and Mlps. Furthermore, we demonstrate that the NB association of Pml39p is a prerequisite for the positioning of additional amounts of Mlp1p at the yeast NBs, and that Pml39p even enables interconnections between Mlp1 polypeptides at sites remote from the NB. In addition, we show that structural similarities predicted by the artificial intelligence (AI) network–based program AlphaFold2 (Jumper et al., 2021) underscore the ZC3HC1 and Pml39p homology. Finally, we discuss why ZC3HC1 and its homologues stand out as unique.
RESULTS
ZC3HC1 contains a bimodular NB interaction domain essential for its binding to TPR
Full-length ZC3HC1 tagged with a fluorescent protein (FP) was known to preferentially associate with the NE, via binding to NBs, when ectopically expressed in different cell lines (Gunkel et al., 2021; Gunkel and Cordes, 2022). Furthermore, fluorescence-loss-in-photobleaching (FLIP) experiments in HeLa cells had revealed that such interactions between FP-tagged ZC3HC1 and the NBs lasted far longer than those between the NPCs and transiently interacting proteins (Supplemental Figure S1). Hence, to specify which parts of ZC3HC1 enable its initial binding to the NB, we also studied the subcellular distributions of FP-tagged ZC3HC1 mutants ectopically expressed in HeLa cells.
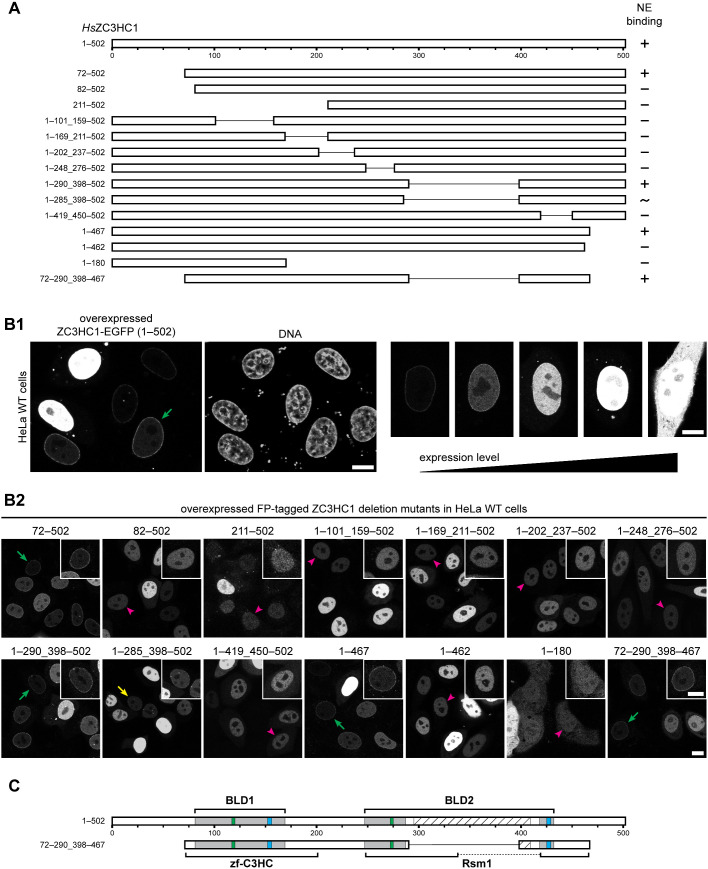
First, we generated stepwise truncations of HsZC3HC1, beginning from the termini of the 502-amino-acid (aa)-long protein. This approach defined two outer boundaries, one close to aa 72 and the other near aa 467, up to which truncations still allowed for NB binding, while further truncations beyond these boundaries did not.
Next, a series of internal deletions revealed additional regions that are non-essential for NB binding (Figure 1, A and B; for further deletions, see Supplemental Figure S2A). Of particular note, we could delete aa 291–397 without compromising NB binding. This latter part had been predicted to primarily represent a natively unfolded region when using tools like Predictor of Natural Disordered Regions (PONDR; e.g., Garner et al., 1999). Finally, we combined most of these tolerable deletions and created a mutant comprising aa 72–290_398–467 that could still bind to the NBs (Figure 1, A and B).
FIGURE 1:
A tandem arrangement of two predicted zinc ion binding modules is essential for the association of ZC3HC1 with NBs. (A) Schematic depiction, drawn to scale, of expression vector–encoded HsZC3HC1 and deletion mutants (see Supplemental Table 1). NB-binding ability is indicated (+, binding visible; −, no binding visible; ∼, unclear or only traces of binding visible). These ratings pertained to cells that expressed lower levels of the recombinant protein, as exemplified in B. (B) Fluorescence microscopy of wild-type (WT) HeLa P2 cells transiently transfected with the vectors shown in A. Bars, 10 µm. (B1) Examples of cells in which NE association of an NB binding–competent version of ZC3HC1, here represented by the WT protein, was discernible only at lower expression levels (green arrow). (B2) Examples of cells with ZC3HC1 mutants ectopically expressed in different amounts. Representative cells with low expression levels are marked, indicating apparent presence at the NE (green arrows), only trace amounts (yellow arrow), or no signal at the NE (magenta arrowheads). Insets show the magnified and signal-enhanced images of the marked cells. As an aside, note that mutant 1–180, lacking the nuclear localization signal (NLS) of ZC3HC1 (Ouyang et al., 2003), shows only slight nuclear enrichment. (C) Schematic depiction of the central regions of the two BLDs of ZC3HC1, relative to the simple schemes of the WT and mutant 72–290_398–467 in A. Green and blue boxes represent the positions of the C-X(2)-C and H-X(3)-C sequence elements, respectively. Gray regions represent the initially approximated expanse of each BLD (Higashi et al., 2005; Kokoszynska et al., 2008). The outer boundaries of BLD1 are defined by R81 and M169, the outer boundaries of BLD2 by T247 and V432, and the inner boundaries of BLD2 by I287 and F418. The hatched box, corresponding to D295–S409, represents an insertion predicted to be largely disordered, except for the P322–S329 region. Brackets represent the expanse of regions comprising the Pfam database motifs zf-C3HC and Rsm1, as specified in the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/). Note that ZC3HC1 mutants lacking any part of the C-X(2)-C or H-X(3)-C peptide sequences did not bind to the NE.
Inspecting those sequence segments capable of NB binding, we realized that others had discovered local sequence similarities between the family of inhibitor of apoptosis proteins (IAPs) and ZC3HC1 (Kokoszynska et al., 2008) and likewise between the IAPs and a new family of proteins considered IAP paralogues and thus called ILPs (IAP-like proteins; Higashi et al., 2005). Among these ILPs were HsILP1, which was actually identical to HsZC3HC1, and SpILP1, which was Schizosaccharomyces pombe Rsm1p, a protein reported to be involved in RNA export (Yoon, 2004).
IAPs contain at least one, but usually several, zinc ion–binding modules called BIR (baculovirus inhibitor of apoptosis protein repeat) domains (Verhagen et al., 2001; Silke and Vucic, 2014). The ILPs, in turn, were described as possessing two BIR-like domains (BLD1 and BLD2), each containing residues likely to form a zinc ion coordination sphere (Higashi et al., 2005). Furthermore, BLD1 had been noted (Kokoszynska et al., 2008) to correspond to a zinc finger (zf) motif listed in the Pfam database (Finn et al., 2006) as zf-C3HC (https://www.ebi.ac.uk/interpro/entry/pfam/PF07967). On the other hand, part of the HsZC3HC1 BLD2 had been assigned the so-called Rsm1 motif (Finn et al., 2008, 2010; https://www.ebi.ac.uk/interpro/entry/pfam/PF08600; for further details, see Supplemental Information 1). However, the IAPs lacked such zf-C3HC and Rsm1 motifs. Moreover, they also lacked another feature found in some ILPs: an extensive sequence insertion that disrupted the BLD2 sequence (Higashi et al., 2005; Kokoszynska et al., 2008). Remarkably, this sequence insertion largely corresponded to the intrinsically disordered region, also termed “the loop” further below, that we had identified as dispensable for NB binding of ZC3HC1.
When comparing the predicted BLDs with the NB binding–competent mutant 72–290_398–467 (Figure 1C; Supplemental Figure S2A1), we noted that the integrity of both BLD1 and BLD2, yet without the BLD2 loop, seemed to be essential for NB binding. Accordingly, deletion mutants that consisted of only BLD1 or BLD2, or those that even lacked only minor parts of the regions likely involved in zinc ion coordination, had failed to bind to the NB (Figure 1A).
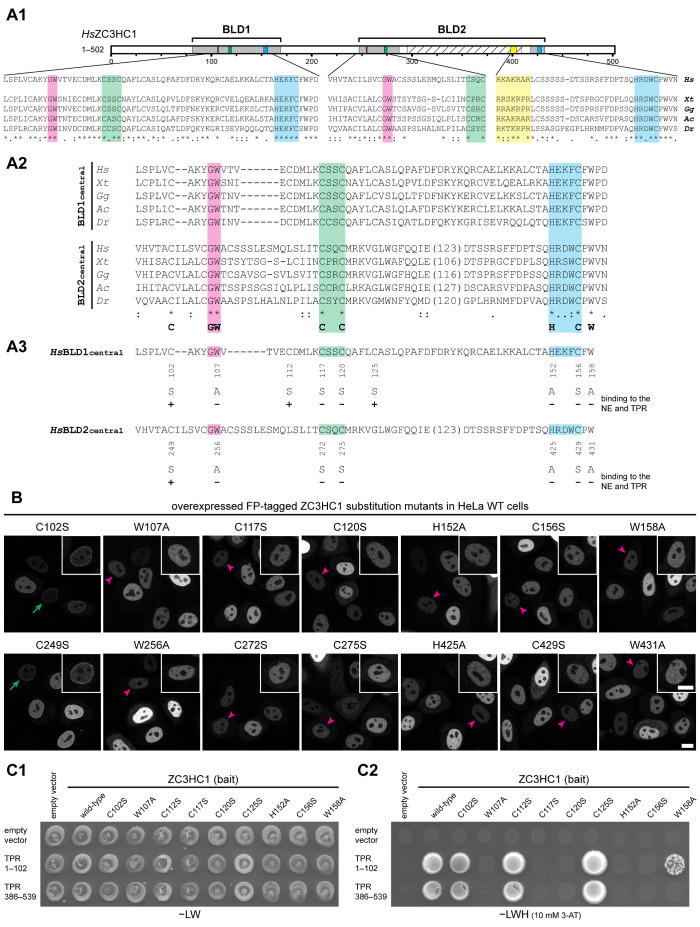
Next, we created HsZC3HC1 aa substitution mutants to investigate whether the predicted zinc ion–coordinating aa and some other seemingly evolutionarily conserved residues were required for NB binding. We focused on a minimal sequence signature of the central part of both BLDs in vertebrates, which reads C-X(3,5)-G-W-X(9,15)-C-X(2)-C-X(31,153)-H-X(3)-C-X-W (Figure 2A; Supplemental Information 2). We altered this signature in one or the other BLD by introducing single-aa substitutions, each one individually, into full-length ZC3HC1. Furthermore, we also substituted a few other single aa, including some present only within BLD1 or the loop of BLD2 (Figure 2A3; Supplemental Figure S2B1).
FIGURE 2:
Specific amino acids within both BLDs of ZC3HC1 are essential for the initial binding to TPR. (A) Schematic depiction of the two BLDs, sequence alignments of representative vertebrate homologues, and an overview of the single-aa-substitution mutants of FP-tagged HsZC3HC1. (A1) Schemes of the two HsZC3HC1 BLDs and alignment of the vertebrate homologue sequence segments corresponding to the minimal central region of each BLD, including the G-W, C-X(2)-C, and H-X(3)-C peptides, and some flanking residues. Sequences are from the human homologue (Hs), amphibians (Xenopus tropicalis, Xt), birds (Gallus gallus, Gg), reptiles (Anolis carolinensis, Ac), and fish (Danio rerio, Dr). Areas highlighted in addition to those in Figure 1C represent G-W dipeptides (magenta) and the NLS (yellow). (A2) Alignments between sequences representing the central BLD1 region and corresponding BLD2 segments but excluding the BLD2-specific sequence insertions (variable lengths in brackets). The bottom line provides a minimal sequence signature identical for both BLDs in these vertebrate homologues. The HsZC3HC1 BLD1 and BLD2 sequences shown represent L97–D160 and V244–N433, respectively. The inner boundaries flanking the BLD2 insertion correspond to E288 and D412. (A3) Individual aa substitutions within the BLD regions and their effects on NE binding and TPR interaction. (B) Fluorescence microscopy of WT HeLa cells transiently transfected with a selection of expression vectors encoding full-length HsZC3HC1 mutants, each carboxy-terminally tagged with EGFP and carrying one of the single-aa substitutions specified in A3. Representative cells with low expression levels are marked as in Figure 1B2 by green arrows and magenta arrowheads, with insets showing the magnified and signal-enhanced images of the marked cells. Bar, 10 µm. (C) Y2H experiments analyzing the interaction of the single-aa-substitution mutants of HsZC3HC1 with two HsTPR segments that include ZC3HC1 interaction domains. (C1) Representative colony growth of diploid cells expressing TPR segments together with WT ZC3HC1 or some of its mutants. Cells were grown on a selection medium lacking leucine and tryptophan (−LW). (C2) Visualization of Y2H interactions after replica-plating onto –LW selection medium also lacking histidine (−LWH) and supplemented with 3-AT. Note that those single-aa-substitution mutants of ZC3HC1 that did not impair NE association in HeLa cells (e.g., C102S, C112S, and C125S) allowed colony growth when paired with the ZC3HC1-binding domains of TPR. By contrast, no colony growth was observed for the mutants incapable of associating with the NE (e.g., C117S, C120S, and C156S). Further note that mutant W158A, which associated with the NE in ZC3HC1 KO cells (Supplemental Figure S2B2) but not in WT cells (as shown in B), was capable of an attenuated TPR interaction (see also Supplemental Figure S2E).
When they were ectopically expressed in HeLa cells, we observed that solitary C-to-S mutations at positions 117, 120, and 156 in BLD1 and 272, 275, and 429 in BLD2 abolished NB binding, as did H-to-A substitutions at positions 152 in BLD1 and 425 in BLD2. In addition, NB binding was abolished by solitary W-to-A substitutions at positions 107 and 158 in BLD1 and 256 and 431 in BLD2 (Figure 2B; Supplemental Figure S2B2).
Furthermore, since the BIR domains of the IAPs harbor other aromatic residues at the positions that correspond to HsZC3HC1 W107 and W256, we tested whether ZC3HC1 would tolerate W-to-Y and W-to-F substitutions at these sites, and found them not to notably impair NB binding (Supplemental Figure S2C). Moreover, solitary C-to-S substitutions of several other HsZC3HC1 cysteine residues (aa 102, 112, 125, and 249), conserved in vertebrates and a few other organisms, did not notably impact NB binding either (Figure 2A3; Supplemental Figure S2B). Of these substitutions, the one at position 102 was of particular interest because C102, together with C117 and C120, formed part of the original consensus sequence of the Pfam zf-C3HC motif (Supplemental Figure S2D), and these three cysteines may have been eponymic for the C3 in the motif name.
To examine whether any mutants that appeared NB binding-incompetent had any residual NB binding potential, we also tested them in later available ZC3HC1 knockout (KO) cells (Gunkel et al., 2021), where these mutants would not need to compete with the wild-type (WT) protein for binding sites (Supplemental Figure S2, A2, B2, and C, 2 and 3). These experiments corroborated almost all the results acquired with the ZC3HC1 WT cell line. The exceptions were mutants W158A and W431A, which could bind to the NB in the absence of WT ZC3HC1 (Supplemental Figure S2B2), indicating that residues W158 and W431 support NB binding but are not essential for this interaction.
Thus far, our data allowed us to conclude that ZC3HC1 uses the tandem arrangement of two BLDs for its binding to the NB, with the functionality of both domains depending on residues likely to be involved in zinc ion coordination. Furthermore, our experiments had revealed that signature residues at seemingly corresponding positions within both BLDs could be functionally equivalent.
To determine whether the NB binding of ZC3HC1 that depended on the integrity of its BLDs reflected genuine interactions with TPR, we used the yeast two-hybrid (Y2H) methodology (Fields and Song, 1989). We thereby tested whether the single-aa-substitution mutants of ZC3HC1 could bind to those regions of TPR that we had already found to interact with WT ZC3HC1 (Gunkel et al., unpublished data). These TPR segments comprised, for example, aa 1–102 and 386–539 (Figure 2C; Supplemental Figure S2E) and thus included some of the coiled-coil–forming regions of TPR (Hase et al., 2001).
The Y2H experiments revealed that all ZC3HC1 mutants capable of NB binding also showed a robust Y2H interaction with TPR (Figure 2C; Supplemental Figure S2E). Furthermore, those aa-substitution mutants that had failed to bind to the NB in the presence of WT ZC3HC1, while capable of NB binding in ZC3HC1 KO cells, also engaged, albeit attenuated, in interactions with TPR (Supplemental Figure S2E). Most strikingly, however, all aa-substitution mutants that failed to associate with the NBs in both ZC3HC1 WT and KO cells also did not exhibit any Y2H interaction with TPR (Figure 2C; Supplemental Figure S2E).
The strict correlation between the results in HeLa cells and those from the Y2H experiments argued that the impaired NB binding of a ZC3HC1 mutant reflected its impaired interaction with TPR. Furthermore, it was evident that both BLD1 and BLD2 were required to establish a functional TPR-binding interface. Thus, the two BLDs formed an experimentally validated functional entity, which we termed the NuBaID.
ZC3HC1 homologues with a NuBaID signature and capable of binding to corresponding TPR homologues are NB proteins in different eukaryotic phyla
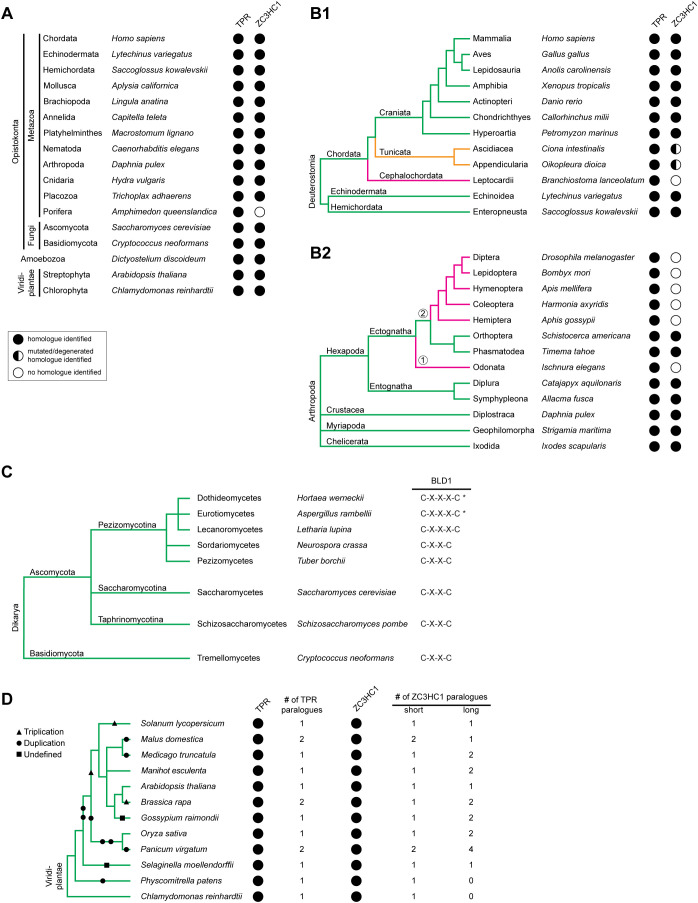
Having determined that the NuBaID and its tandem arrangement of predicted zinc fingers is an identifying feature of HsZC3HC1, we explored whether NuBaID signatures could identify NB and TPR interaction partners across phyla. We applied complementary approaches to seek such potential ZC3HC1 homologues, including signature-based and primary sequence end-to-end alignment searches (Supplemental Information 3).
This approach allowed the identification of potential ZC3HC1 homologues in all eukaryotic supergroups. A genuine homologue remained undetectable in only some phyla and classes (Figure 3, A and B1). The latter was true, for example, for most insect orders (Figure 3B2), which was in line with no ILP reported detectable in Drosophila (Higashi et al., 2005). Furthermore, the existing ZC3HC1 homologues in some groups of organisms appeared to be subject to various mutations (Figure 3B1), most of which would abolish binding to the NB and TPR if present at corresponding positions in the human protein.
FIGURE 3:
Distribution of ZC3HC1 and its homologues among eukaryotes. (A) Selection of eukaryotic phyla and divisions with representative species in which ZC3HC1 homologues were identified by sequence database mining. The presence of TPR homologues is depicted for comparison. Taxonomy is mostly according to the NCBI taxonomy database (Schoch et al., 2020; https://www.ncbi.nlm.nih.gov/taxonomy). Note that ZC3HC1 homologues exist in most animal phyla and the divisions of fungi, amoeba, and green plants. Among the listed animal phyla, ZC3HC1 was undetectable only in the sponges. (B) Exemplifying cladograms of the clade Deuterostomia and the phylum Arthropoda, illustrating the evolutionary fate of ZC3HC1 in some subphyla. (B1) Cladogram of the Deuterostomia, based on phylogenetic trees and former cladograms (e.g., Delsuc et al., 2018), listing the phyla Echinodermata, Hemichordata, and Chordata, the chordate subphyla Craniata, Tunicata, and Cephalochordata, and a selection of classes and species. Note that ZC3HC1 homologues were undetectable in the Cephalochordata, while homologues in the Tunicata harbor various mutations that differ between different classes and orders, but would all prevent HsZC3HC1 from binding to the NB and TPR. (B2) Cladogram of the phylum Arthropoda, deduced from a phylogenetic tree (Sasaki et al., 2013), listing the subphyla Hexapoda, Crustacea, Myriapoda, and Chelicerata, and a selection of orders and species. Note that ZC3HC1 homologues were undetectable in the infraclass Paleoptera (①), here represented by the order Odonata, and are similarly absent in most orders of the infraclass Neoptera (②). However, ZC3HC1 homologues are present in the orders Orthoptera and Phasmatodea, suggesting that at least two independent events led to the disappearance of the ZC3HC1 gene, or its alteration beyond recognition, in the other insects. (C) Cladogram of the subkingdom Dikarya, representing an excerpt of a previous cladogram (Spatafora et al., 2017), with its divisions Basidiomycota and Ascomycota. For the Ascomycota, three subdivisions are listed, i.e. the Pezizomycotina, Saccharomycotina, and Taphrinomycotina, together with some of their classes and representative species. While most fungal ZC3HC1 homologues possess the C-X(2)-C tetrapeptide as part of their BLD1 zinc finger, some Pezizomycotina classes feature a C-X(3)-C instead. However, even though the C-X(3)-C pentapeptide is predominant in the classes Eurotiomycetes and Dothideomycetes, a few of their orders feature only sequences with C-X(2)-C. These classes are therefore marked with an asterisk. (D) Cladogram of the kingdom Viridiplantae, representing an excerpt of a phylogenetic tree (Panchy et al., 2016; Wang et al., 2020), including some green plant orders in which genome duplication events did or did not occur. Representative species and genome duplication (circles), triplication (triangles), and undefined polyploidization events (squares) are indicated. Several genome duplications led to six ZC3HC1 and two TPR paralogues in the order Poales, here represented by the switchgrass Panicum virgatum. By contrast, in other orders, the additional ZC3HC1 gene appears to have been inactivated again, for example, in the spreading earth moss Physcomitrella patens.
Other homologues had different spacing between the first two cysteines of the BLD1 zinc finger signature, reading C-X(3)-C instead of C-X(2)-C, which nevertheless allowed for NB binding (Figure 3C and Supplemental Figure S3). Furthermore, in many but not all species, we again found the BLD2 sequence disrupted by extensive sequence insertions (Supplemental Figure S4), sometimes far exceeding the length of the BLD2 insertion in HsZC3HC1. Most of these insertions did not share any conspicuous sequence similarity, sometimes not even between closely related species. Instead, they often appeared to only have in common that they were predicted to represent intrinsically disordered loops.
Of further note, we could identify two or more NuBaID-encoding genes per species only in organisms that had undergone whole-genome duplications (Figure 3D). Thus, species with nonduplicated genomes appeared to generally have only one gene encoding a NuBaID signature–containing protein (for further details, see Supplemental Information 4). Currently, most of the putative ZC3HC1 homologues that we detected and regarded as potentially NB binding-competent can be described by a NuBaID signature that reads G-[WYF]-X(6,24)-C-X(2,3)-C-X(17,82)-H-X(3)-C-X-[WYFML]-X(48,232)-G-[WYF]-X(8,140)-C-X(2)-C-X(14,994)-H-X(3)-C-X-[WYFRCV].
Next, we selected two potential ZC3HC1 homologues to investigate their relationship with HsZC3HC1 in further detail. One was the D. discoideum protein DDB0349234, here referred to as DdZC3HC1. The other was S. cerevisiae Pml39p, a nonessential protein and known binding partner of the TPR homologues Mlp1p and Mlp2p (Palancade et al., 2005). Back then, however, no human homologue of ScPml39p had been identified by sequence similarity searches. Correspondingly, previous sequence searches for ILP/ZC3HC1 homologues had not detected a homologue in S. cerevisiae (Higashi et al., 2005). Similarly, most of the search tools that we used for sequence database mining did not detect HsZC3HC1 from ScPml39p, nor vice versa (Supplemental Information 5).
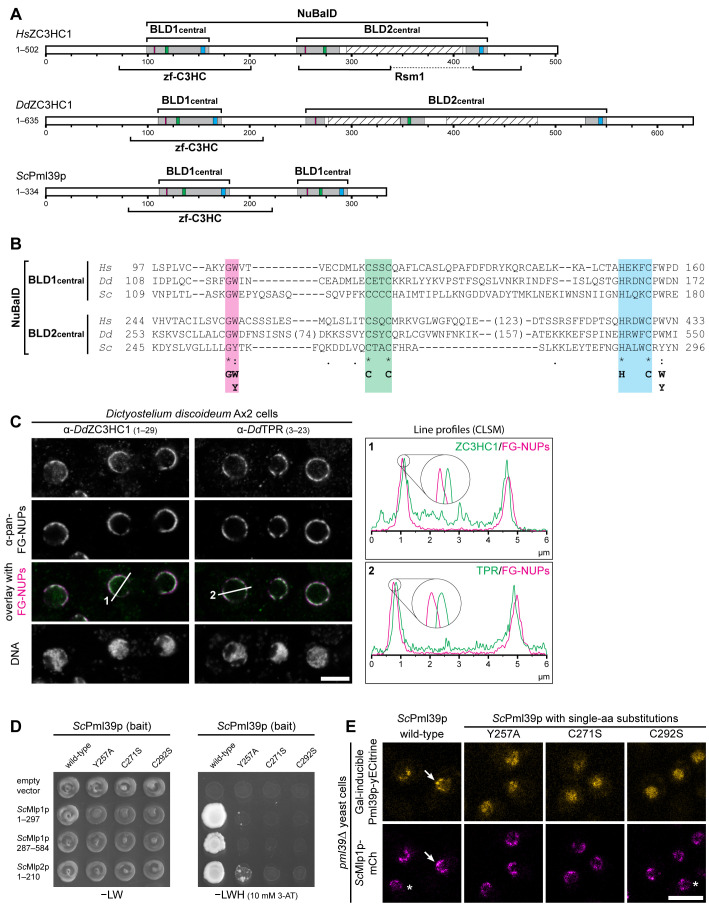
Apart from their low degree of sequence similarity, the diversity between HsZC3HC1, ScPml39p, and DdZC3HC1 manifested itself in various ways, including size, isoelectric point (pI), and aa composition. Specifically, while the 502-aa-long human homologue is a 55.3 kDa protein with a pI of 5.44, the 386-aa-long ScPml39p of 39.2 kDa has a pI of 9.13. DdZC3HC1 (accession number ON368701; see also Supplemental Figure S5A) is a 635-aa-long protein of 72.5 kDa with a pI of 8.38 and with an aa composition strikingly different from both ScPml39p and HsZC3HC1. Moreover, the linear sequence of the DdZC3HC1 BLD2 appeared disrupted by two sequence insertions predicted to be largely unstructured. ScPml39p, by contrast, evidently lacked such BLD2-embedded loops (Figure 4, A and B). However, all three proteins could be described by a NuBaID signature reading G-W-X(9,14)-C-X(2)-C-X(31,34)-H-X(3)-C-X-W-X(77,96)-G-[WY]-X(10,89)-C-X(2)-C-X(16,183)-H-X(3)-C-X-[WY].
FIGURE 4:
NB- and TPR/Mlp-interacting ZC3HC1 homologues with a conserved NuBaID signature exist in S. cerevisiae and D. discoideum. (A) Schematic depiction of DdZC3HC1 and ScPml39p, compared to HsZC3HC1. The highlighted regions of the two BLDs, together representing the bimodular NuBaID, correspond to those in Figures 1C and 2A1. The boxes in magenta now depict positions that can read either G-W or G-Y, the latter dipeptide part of the Pml39p BLD2. The known NLS of HsZC3HC1 and the unknown, only conjecturable NLS of the two other homologues appear differently positioned and are not depicted here. The expanse of the minimal central region of each HsZC3HC1 BLD is as specified in Figure 2A2. The central regions of DdZC3HC1 BLD1 and BLD2 shown here comprise I108–N172 and K253–I550, respectively. The inner boundaries flanking two apparent insertions within DdZC3HC1 BLD2 here correspond to S273 and D348, and to K371 and A529. Parts of these insertions were predicted to be mostly unstructured, except for V418–N433, and to range from N277 to K347 and N393 to S482 (hatched boxes). For ScPml39p, the central BLD1 and BLD2 regions presented here comprise V109–E180 and K245–N296, respectively. Brackets indicate regions to which a zf-C3HC or Rsm1 motif has been attributed to date. So far, no Rsm1 motif has been assigned to DdZC3HC1 and ScPml39p. (B) Sequence alignments of the central regions of the BLDs, according to those in Figure 2A2. The minimal sequence signature shared by the two BLDs in all three proteins is depicted. (C) Double-labeling IFM of D. discoideum Ax2 cells with pan-FG-NUPs antibodies and antibodies for either DdZC3HC1 or DdTPR, with the focal plane approximately at the equator of most nuclei. DNA staining is shown for reference. Section lines across the nuclei, marked 1 and 2 in the overlay micrographs, were analyzed by ImageJ, with line profiles plotted. Note that the 4× enlarged line profiles from both sides of the corresponding nuclei reveal the offset location of DdZC3HC1 and DdTPR toward the nuclear interior, relative to the immunolabeled FG-repeat nucleoporins of the NPCs. Bar, 5 µm. (D) Representative Y2H data, obtained by expressing segments of Mlp1p and Mlp2p with either intact Pml39p or a selection of Pml39p mutants with single-aa substitutions in the NuBaID. Experiments were performed as described in Figure 2C. Note that while a robust Y2H interaction occurred between intact Pml39p and distinct parts of the Mlps, no colony growth was observed for Pml39p mutants such as Y257A, C271S, and C292S. (E) Live-cell imaging of pml39∆ yeast cells endogenously expressing mCherry-tagged Mlp1p and, upon induced ectopic expression, either yECitrine-tagged WT Pml39p or a selection of the likewise tagged mutants with single-aa substitutions. Note that the newly synthesized intact Pml39p primarily accumulated at the NE (arrow). By contrast, the Pml39p mutants were distributed throughout the nuclear interior. Asterisks mark a few cells in which Pml39p expression was not detected. Bar, 5 µm.
We next examined the location of DdZC3HC1 relative to its NPC and NB. We had already found that D. discoideum possesses a TPR protein, namely DDB0308586, which shared characteristics with, e.g., metazoan TPR homologues (Supplemental Figure S5B, 1 and 2; Kuznetsov et al., 2002). Furthermore, we had localized DdTPR, with a sequence-deduced Mr of 235 kDa (accession number ON368702; Supplemental Figure S5B3), at the nuclear side of the NPC (see below). In addition, Y2H studies revealed that parts of the N-terminal domain of DdTPR robustly interacted with a loop-free DdZC3HC1 deletion mutant (Supplemental Figure S5, A and C). Finally, immunofluorescence microscopy (IFM) revealed that DdZC3HC1 was primarily associated with the NPC, with a location offset toward the nuclear interior, similar to DdTPR (Figure 4C; for antibody characterization, see Supplemental Figure S5D).
ScPml39p had previously been shown to interact with the Mlps and locate at the yeast NPC (Palancade et al., 2005). To examine whether the predicted NuBaID of Pml39p is essential for this interaction, we created Y2H constructs for (i) Mlp segments similar to those previously found to interact with ScPml39p (Palancade et al., 2005), (ii) WT Pml39p, and (iii) a collection of Pml39p single-aa-substitution mutants. The Y2H experiments then confirmed that WT Pml39p could interact with distinct Mlp regions (Supplemental Figure S6A). In contrast, mutations at distinct NuBaID signature positions, equivalent to those essential for TPR binding of HsZC3HC1, abolished or markedly impaired the binding of Pml39p to both Mlps (Figure 4D; Supplemental Figure S6B).
To investigate whether these Pml39p mutations also impair binding to the NB in vivo, we generated a yeast strain stably expressing Mlp1p as a mCherry-tagged polypeptide in a pml39∆ background. We then used this PML39 deletion strain for the galactose-inducible expression of yECitrine-tagged intact and mutant versions of Pml39p. Live-cell imaging of the transformed cells confirmed that the intact Pml39p colocated with Mlp1p at the NEs. In contrast, Pml39p mutants carrying single-aa substitutions that attenuated or abolished Mlp binding in Y2H experiments were either impaired in NE binding or not located at the NE at all. Instead, these mutants appeared to be distributed throughout the nuclear interior (Figure 4E; Supplemental Figure S6C). Thus, these outcomes resembled the effects of equivalent HsZC3HC1 mutations that abolished binding to the NB and TPR in human cells.
Together, these findings indicated that an intact prototypic NuBaID signature can identify proteins that are genuine ZC3HC1 homologues capable of binding to their corresponding TPR homologues.
Pml39p is required for positioning Mlp1p subpopulations at the NB and enables interlinkage of Mlp1 polypeptides in nuclear foci
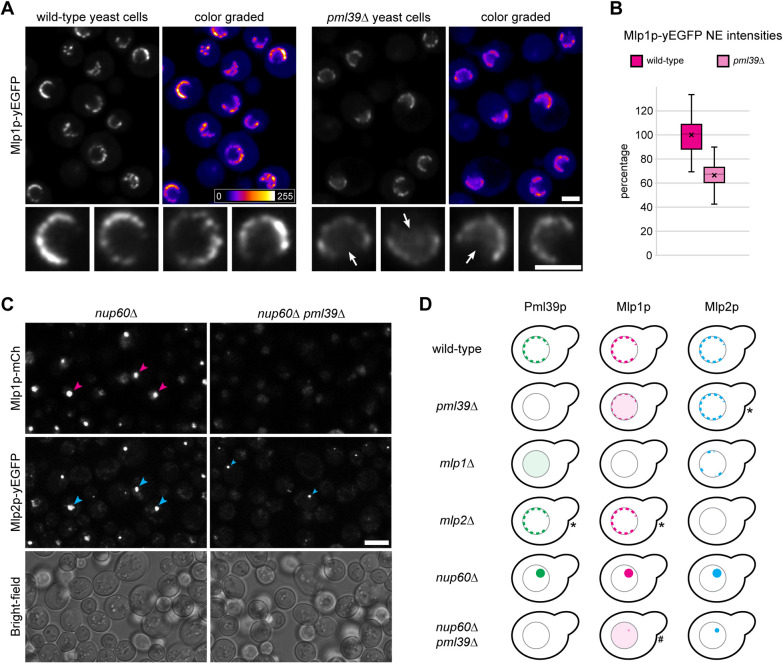
While Pml39p is located at the NE through its binding to the Mlps, in particular to Mlp1p (Palancade et al., 2005; Supplemental Figure S7A1), the deletion of PML39 had been noted to neither affect the NE localization of Mlp1p nor Mlp2p (Palancade et al., 2005). On the other hand, HsZC3HC1 was known to be required for the NB positioning of TPR subpopulations (Gunkel et al., 2021). Therefore, although it seemed possible that Pml39p and ZC3HC1 do not share all functional properties because their sequences differ significantly, we nonetheless examined whether Pml39p plays a role in the NE positioning of the Mlps.
In line with the former study on Pml39p (Palancade et al., 2005), we noticed hardly any reduction in the NE-associated signal intensities for Mlp2p-yEGFP when comparing a PML39 wild-type (wt) and a pml39∆ yeast strain (Supplemental Figure S7A2). Similarly, the NE localization of other NB-associated proteins, specifically Mad1p, Sac3p, and Ulp1p, was not notably or only moderately affected in the pml39∆ cells (Supplemental Figure S7B).
By contrast, in pml39∆ yeast strains expressing Mlp1p-yEGFP, the NE association of Mlp1p was notably reduced compared to PML39wt cells grown in parallel (Figure 5A; Supplemental Figure S7A3). Moreover, in many pml39∆ cells, some yEGFP-tagged Mlp1p was diffusely distributed throughout the nucleus, a feature generally not noted within PML39wt cells (Figure 5A). Furthermore, the comparison of PML39wt and pml39∆ strains expressing all Mlp1p tagged with mCherry confirmed these observations (Supplemental Figure S7A4).
FIGURE 5:
Absence of Pml39p results in reduced amounts of NE-associated Mlp1p in pml39∆ cells and prevents Mlp1p from accumulating in nuclear clusters in nup60∆ pml39∆ cells. (A) Live-cell fluorescence microscopy of PML39wt and pml39∆ yeast cells endogenously expressing all Mlp1p as yEGFP-tagged polypeptides. The two strains, grown as asynchronous populations, were analyzed in parallel with identical microscope settings. Representative overview images are also shown color graded, together with a color lookup table. Some nuclei of the overviews are also shown at higher magnification. Note that the yEGFP signal intensities at the NEs of the pml39∆ cells were, on average, conspicuously reduced compared to the PML39wt cells. Often, some Mlp1p-yEGFP appeared instead distributed throughout the nuclear interior of the pml39∆ cells (arrows). Bars, 2 µm. (B) Quantification of yEGFP signals from tagged Mlp1p at the NEs of PML39wt and pml39∆ cells. The data represent the mean results of two separate experiments conducted on different days and evaluated independently. For each experiment (n = 50 nuclei per strain), PML39wt and pml39∆ cell populations were grown in parallel under identical conditions (see Supplemental Figure S7C1 for the individual data sets). Box plots display the relative signal intensity values, with the arithmetic means marked by x and the means for the PML39wt cells set to 100%. SDs are provided. Note that the mean Mlp1p-yEGFP signal yields for the NEs of the pml39∆ cells were generally reduced significantly. While the signal intensities between individual nuclei differed notably, such variation was less pronounced in the pml39∆ cells than in the PML39wt cells. (C) Live-cell imaging of nup60∆ and nup60∆ pml39∆ cells endogenously expressing mCherry-tagged Mlp1p and yEGFP-tagged Mlp2p. Bright-field micrographs are shown as a reference. Note that both Mlps were no longer attached to the NPCs in the nup60∆ cells. Instead, the Mlps were often focally accumulated, together with Pml39p (Supplemental Figure S7E), within prominent nuclear clusters (magenta and large blue arrowheads). By contrast, in the nup60∆ pml39∆ cells, Mlp1p was hardly or no longer detectable as part of such nuclear clusters, while Mlp2p could still be found in foci (small blue arrowheads) that were then typically much smaller. Bar, 5 µm. (D) Schematic depiction of the subcellular distribution of FP-tagged Mlp and Pml39 polypeptides in yeast KO strains. Contrary to most strains, the outcome for a few strains, marked here by asterisks, appeared to vary moderately between experimental replicates. Such ambiguity pertained to some minor reduction in the amounts of (i) NE-associated Mlp2p in the pml39∆ cells, (ii) NE-associated Pml39p in the mlp2∆ cells, and (iii) NE-associated Mlp1p in the mlp2∆ cells. However, such reductions were noted in only some replicates and occasionally appeared to correlate with cell culture growth phases. A hash marks the nup60∆ pml39∆ strain, as we technically could not unequivocally exclude the possibility that trace amounts of Mlp1p, apart from the bulk of Mlp1p distributed throughout the nucleoplasm, may still occur associated with the small Mlp2p foci in this strain.
To approximate the extent to which the NE-associated amounts of Mlp1p were diminished in the absence of Pml39p, we determined the mean signal yields for Mlp1p-yEGFP at the NEs of PML39wt and pml39∆ cells. These quantifications revealed that the mean total amount of NE-associated Mlp1p was reduced by up to one third in the pml39∆ cells (Figure 5B; Supplemental Figure S7C).
We next investigated whether Pml39p would enable the interconnection of Mlp1 polypeptides even at sites remote from the NPC, similar to ZC3HC1 in human cells. In the latter, knockdown of the NPC protein NUP153 was known to cause TPR accumulation in cytoplasmic and nuclear foci (e.g., Hase and Cordes, 2003). These foci were mostly no longer detectable when both NUP153 and ZC3HC1 were absent (Gunkel and Cordes, 2022). Furthermore, NUP153 had been considered homologous to the yeast protein Nup60 as both proteins contribute similarly to recruiting TPR/Mlp1 polypeptides to the NPC (e.g., Hase and Cordes, 2003). We thus examined whether the nuclear clusters of Mlp1p and Mlp2p, which appear in the absence of Nup60p (e.g., Feuerbach et al., 2002), would be affected by a concurrent removal of Pml39p.
Confirming earlier results (e.g., Feuerbach et al., 2002), we found conspicuous nuclear clusters of Mlp1p and Mlp2p in nup60∆ cells (Figure 5C; Supplemental Figure S7, D and E1). Furthermore, Pml39p colocated in such foci with the Mlps (Supplemental Figure S7E1), also consistent with previous observations (Palancade et al., 2005). However, as a novel result, we found that Mlp1p foci were barely or not detectable within nup60∆ pml39∆ cells (Figure 5C). Instead, the Mlp1 polypeptides then often appeared diffusely distributed throughout the nuclei. We thus concluded that Pml39p could keep Mlp1 polypeptides interconnected even at sites other than the NB.
The focal accumulations of Mlp2p appeared less affected by Pml39p deficiency (Figure 5, C and D), indicating that the interactions between Pml39p and Mlp2p differ from the likely more complex arrangements between Pml39p and Mlp1p. Along these lines, the loss of Mlp2p in nup60∆ mlp2∆ cells did not prevent the formation of conspicuous foci in which Mlp1p and Pml39p were colocated (Supplemental Figure S7E). In contrast, the notably smaller-sized and less frequently observed Mlp2p foci in nup60∆ mlp1∆ cells appeared to attract only minor amounts of Pml39p (Supplemental Figure S7F).
Overall, we rated the absence of Mlp1p foci in nup60∆ pml39∆ cells as equivalent to the absence of TPR foci in NUP153- and ZC3HC1-deficient cells. Furthermore, because degron-mediated rapid degradation of ZC3HC1 had proven that ZC3HC1 holds the TPR foci together as a structural element (Gunkel and Cordes, 2022), we considered it reasonable to also regard Pml39p as a protein that can act as a linker between Mlp1 polypeptides.
Even though neither Pml39p nor ZC3HC1 interacted with each other’s TPR/Mlp homologues (Supplemental Figure S8), we considered such findings likely explainable by binding interfaces for Mlp and TPR that share too little sequence similarity. Therefore, even without an ultimate proof based on homologue interchangeability, we were confident that HsZC3HC1 and ScPml39p are genuine homologues that share at least one similar task. However, because these homologues have diverged considerably at the primary sequence level, apart from their NuBaID signature, we wondered which structural features they would have in common.
ZC3HC1 structure predictions illustrate an evolutionarily conserved NuBaID construction and allow the redefinition of BLD boundaries
We used the neural network–based deep-learning program AlphaFold2 (Jumper et al., 2021; Tunyasuvunakool et al., 2021) in combination with the ColabFold platform (Mirdita et al., 2022) to compare the computationally predicted structures of the ZC3HC1 homologues. Beforehand, we scrutinized how the information packages representing the input materials for such predictions (Supplemental Information 6) would affect the computed outcome. On the basis of these preparatory analyses (Supplemental Figures S9 and S11A), we considered the predictions for the structured parts of the ZC3HC1 homologues to be conclusive within the range of accuracy deemed sufficient in the context of this study.
In addition to features presented in more detail (Supplemental Figures S9–S12), the characteristics of the predicted structures can be outlined as follows. When comparing the homologues in their entirety, the loop-like insertions within the human and amoebic ZC3HC1, and their absence from ScPml39p, made these proteins appear different at first glance (Supplemental Figure S10A). However, upon blinding out these insertions, a compact arrangement of the two BLDs as two adjoining modules became apparent for all three homologues (Supplemental Figure S10B1). Furthermore, closer inspection revealed an evolutionarily conserved BLD1:BLD2 binding interface, underscoring that the NuBaID is not only a functional unit but also a structural entity composed of two modules (Supplemental Figure S10B, 2 and 3).
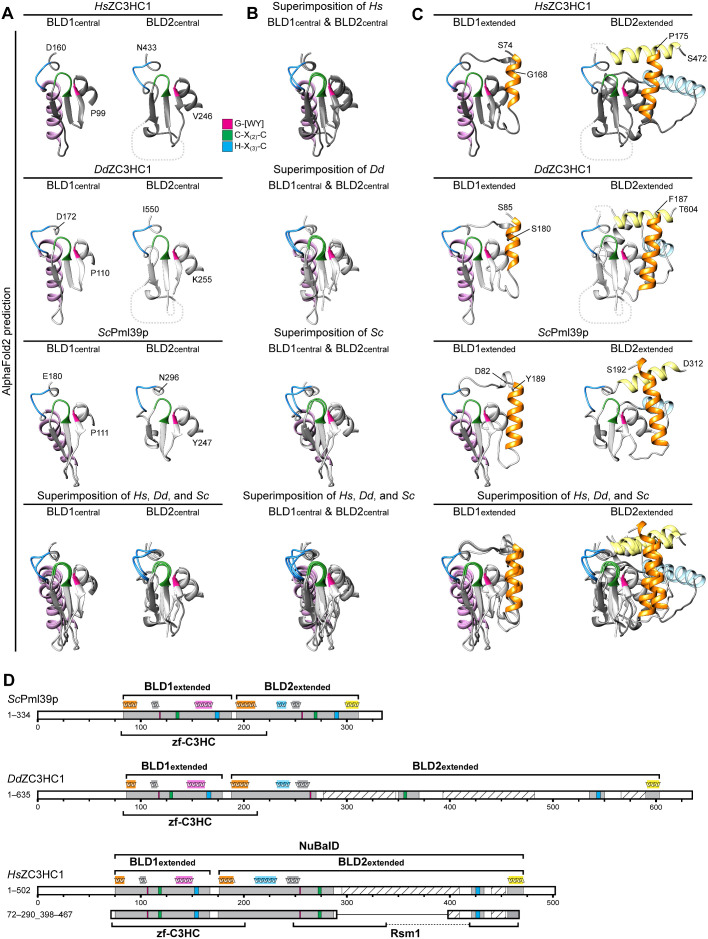
We next focused on the central regions of the BLDs, essentially corresponding to the parts indicated in Figure 4A. Again, we blinded out the large loop-like insertions within the human and amoebic BLD2 to expose only the structured elements, which revealed the structural similarity between the elements that contribute to the zinc coordination spheres (Figure 6, A and B). While the resemblance was most evident when the corresponding BLDs of the three homologues were compared (Figure 6A), comparison of the BLD1 and BLD2 of each homologue also revealed similarities. Nevertheless, both modules were also distinguishable by BLD1- and BLD2-specific features, including, for example, a conspicuous BLD1-specific α-helix (Figure 6, A and B; for further information on the BLD1 and BLD2 α-helices, see Supplemental Information 7).
FIGURE 6:
Tertiary structure predictions by AlphaFold2 uncover striking similarities between the BLDs of HsZC3HC1, DdZC3HC1, and ScPml39p and allow the redefinition of their boundaries. (A) Structures predicted for the central regions of the BLD1 and BLD2 of HsZC3HC1, DdZC3HC1, and ScPml39p. The outer boundaries of the central part of BLD1 shown here are P99 and D160 for HsZC3HC1, P110 and D172 for DdZC3HC1, and P111 and E180 for ScPml39p. The outer boundaries of the central part of BLD2 here correspond to V246 and N433 for HsZC3HC1, K255 and I550 for DdZC3HC1, and Y247 and N296 for ScPml39p. Having blinded out the AlphaFold2 prediction for the major loop-like BLD2 insertion of HsZC3HC1 and the two major insertions of DdZC3HC1 BLD2, the inner BLD2 boundaries shown here correspond to I287 and F418 for HsZC3HC1, and to I270 and K350, and I370 and E535, respectively, for DdZC3HC1. However, the relative positions of the blinded-out loops are depicted as dashed lines (not to scale). The HsZC3HC1 and ScPml39p structures, here and in B and C, were obtained from the AlphaFold database. The structure for the sequence-corrected version of DdZC3HC1 (accession number ON368701) was determined using the AlphaFold2 source code. The sequence elements C-X(2)-C and H-X(3)-C, assumed to be involved in zinc ion coordination, and the G-[WY] dipeptide are colored as in Figure 4A. Note the similarities between the central BLD1 structures of the homologues and those between the central BLD2 structures. Also note the BLD1-specific α-helix, colored in light pink. (B) Superimposition of the central parts of BLD1 and BLD2 onto each other. Aside from the evolutionarily conserved BLD1-specific α-helix, the structural similarity between the other central parts of both BLDs, which are considered involved in zinc ion coordination, appears evident. (C) Structural predictions for essentially the entire BLD1 and BLD2 modules as newly defined in our study. An additional residue was appended to each boundary to facilitate recognition. The BLD1 of HsZC3HC1, comprising K75–F167, is thus presented as S74–G168. The HsZC3HC1 BLD2, comprising A176–S471, is shown as P175–S472, yet with the major loop and now also a smaller second loop between I434 and E455 blinded out as in A. Accordingly, the BLD1 of DdZC3HC1 is presented as S85–S180, instead of N86–F179, and its BLD2 as F187–T604, instead of Q188–S603. Again, the two major loops within the amoebic BLD2 have been blinded out, as was a smaller loop between V551 and I589. BLD1 and BLD2 of ScPml39p, defined as L83–E188 and S193–E311, are presented correspondingly as D82–Y189 and S192–D312. The BLD1-specific α-helix is again colored in light pink, while the α-helices specific for the BLD2 of all three homologues are shown in light yellow and light blue. The α-helix common to the N-terminal boundary of both BLDs is highlighted in orange. As an aside, note that all segments shown here primarily comprise residues for which AlphaFold2 assigned, with only a few exceptions, a high per-residue confidence score of at least 70, mostly exceeding 90. (D) Schematic depiction of HsZC3HC1, DdZC3HC1, and ScPml39p with the newly defined BLD boundaries. These schemes depict an additional minor insertion within the BLD2 of HsZC3HC1 (I434–E455) and DdZC3HC1 (V551–I589), with the predicted unstructured regions (S440–A454 of HsZC3HC1, G566–S590 of DdZC3HC1) again shown as hatched. For simplification, other potentially unstructured regions beyond the outer BLD boundaries and found in all three homologues are not highlighted. The schematic indications of the α-helices above the scheme for each homologue represent the relative positions of the α-helices correspondingly colored in C. According to the novel BLD delineations, the zf-C3HC motif of the Pfam database would now comprise sequences encompassing the entire BLD1 and part of BLD2. The Rsm1 motif, assigned to HsZC3HC1 and not to DdZC3HC1 or ScPml39p, corresponds only to parts of the BLD2 and its loop-like insertions. Finally, note that the minimal NE binding–competent HsZC3HC1 mutant 72–290_398–467, schematically depicted here for comparison, comprises, with the exception of the four residues 468–471, the newly defined BLD regions in their entirety.
The common feature of all BLDs was an antiparallel arrangement of several β-sheets and their tetrahedral arrangement of three cysteines and one histidine, forming the likely zinc coordination sphere. Moreover, relative to its CCHC arrangement, each BLD also featured essentially the same positioning of (i) its G-W/G-Y dipeptide that is part of the G-[WY]-X(9,89)-C-X(2)-C peptide and (ii) its aromatic residue that resides at the C-terminal side of its H-X(3)-C-X-[WY] peptide (Figure 6, A and B; Supplemental Figure S11C).
These structure predictions also attested that the residues we had defined as the minimal sequence signature of the NuBaID are unlikely to engage in direct intermolecular interactions with TPR as part of a yet-to-be-defined TPR-binding interface. Instead, the NuBaID signature residues, and most of the small number of other residues evolutionarily conserved across different phyla, contribute to establishing or maintaining the central structures of the BLDs. Apart from those directly involved in zinc ion coordination, some of these conserved residues engage in other intra-BLD interactions required to establish the BLD core structure and a stable zinc coordination sphere (Supplemental Figure S11, F and G).
Of further note, the predicted structural features of the most central parts of the BLDs resembled those of the BIR domains of IAPs (Supplemental Figure S11, A–C), consistent with formerly proposed structural similarities between BLD1 of HsILP1/ZC3HC1 and the BIR domain of human survivin/BIRC5 (Higashi et al., 2005). Moreover, we noted that the BIR domains and vertebrate BLD1 also share another feature, i.e., a conspicuous groove exposed on the surface of these domains. For HsZC3HC1, this groove is formed by residues between L128 and L144 (Supplemental Figure S11D), which to some extent resembles the so-called IBM (IAP binding motif) groove of the type II BIR domains (e.g., Cossu et al., 2019; also see Supplemental Discussion 1).
Inspection of the structural elements flanking the central BLD regions revealed some additional, evolutionarily conserved BLD α-helices (Figure 6C), some even extending beyond the regions initially defined as the BLDs. For example, while one of these α-helices had not been considered part of BLD2 before, we now found it to be equivalent to another α-helix that we had assigned to BLD1. Both of these α-helices are colored in orange in Figure 6C (see also Supplemental Information 7).
Considering the outcome of our deletion experiments, the AlphaFold2 structure predictions, and the structure-based alignments of the homologue sequences, we defined the BLD1 of HsZC3HC1 as ranging from at least K75 to F167. Correspondingly, the BLD2 of HsZC3HC1 would start at A176 and extend to S471, even though we had found the last residues dispensable for NE association (Figure 1). These few BLD2 residues were part of a carboxy-terminal α-helix, shown in light yellow in Figure 6C. Dispensability of these residues in humans appeared to be in line with the corresponding α-helix of ScPml39p being naturally shorter (Figure 6C) and nevertheless sufficient for NB binding in yeast.
A schematic depiction of the newly defined BLD regions relative to the primary sequences of HsZC3HC1, DdZC3HC1, and ScPml39p illustrates how closely BLD1 and BLD2 are positioned next to each other in each homologue (Figure 6D). Furthermore, our redefinition of the boundaries indicated that both BLDs actually differ in size, with BLD1 ranging from 93 to 107 aa across the three homologues and BLD2 ranging from about 119 to 144 aa when excluding the loop-like insertions. Of further note, it is now evident that the Pfam motif zf-C3HC actually encompasses residues corresponding to BLD1 plus part of BLD2. On the other hand, the Rsm1 motif, so far only assigned to HsZC3HC1 but not to DdZC3HC1 and ScPml39p, applies to only some parts of the BLD2 while missing others (Figure 6D). Finally, with this more precise delineation of the NuBaID and its BLDs, it is now even more apparent that their boundaries are close to those of the minimal NB binding–competent HsZC3HC1 mutant we had experimentally determined (Figure 1C). The structure predicted for this 289-aa-long mutant reveals a compact polypeptide mainly composed of the two BLDs, including the newly assigned α-helices, plus only a few residual unstructured regions (Supplemental Figure S12). The now adapted schematic depiction of this mutant illustrates that it is practically the entirety of the structural regions forming the NuBaID that is both indispensable and sufficient for the initial NB binding of ZC3HC1 (Figure 6D).
DISCUSSION
The NuBaID of ZC3HC1 as the conserved bimodular TPR interaction domain of a unique protein present in numerous eukaryotes
In this study, we identified and characterized the domain of HsZC3HC1 that enables its initial interaction with the NB and NPC-anchored TPR. We demonstrate that this bimodular NuBaID represents an evolutionarily conserved functional and structural entity. As such, it also functions as the NB- and Mlp-binding domain of ScPml39p, which we present as the budding yeast homologue of ZC3HC1.
Our finding of a modularly built NuBaID is consistent with a study (Higashi et al., 2005) in which database mining for potential IAP homologues in Arabidopsis uncovered proteins with two BIR domainresembling modules in plants and other organisms. These proteins were termed ILPs and included a human ILP now known as ZC3HC1. The similarities and differences between the IAPs and ZC3HC1 are addressed in Supplemental Discussion 1, including a comparison of the NuBaID and the human BIR domains with their minimal consensus sequence C-X(2)-C-X(16)-H-X(6)-C.
Our database searches identified additional likely ZC3HC1 homologues in a wide range of organisms across the eukaryotic realm. Furthermore, apart from some rare ambiguous exceptions, we found that species with a ZC3HC1 homologue also have a TPR/Mlp homologue, suggesting that the presence of TPR might be mandatory for a TPR-binding ZC3HC1 to persist within a taxon. By contrast, such coexistence does not appear required for the TPR homologues, which we also found in organisms without an evident ZC3HC1.
Moreover, in each ZC3HC1-positive species with a nonduplicated genome, we found only one NuBaID signature–containing protein. This finding suggested early on that the ZC3HC1 homologues in different species may have at least one common function, which is defined by the NuBaID and needs to be performed together with TPR. However, as the NuBaID signature apparently does not describe the TPR-binding interface, it remains to be determined whether the two or more ZC3HC1 paralogues in species with genome duplications represent solely TPR-binding proteins or whether some have adopted other binding partners and tasks. These questions hold, for example, for the Arabidopsis ILP/ZC3HC1 proteins At1g17210 and At1g48950 (e.g., Higashi et al., 2005; Lu et al., 2020; Wang et al., 2022; for further considerations, see Supplemental Discussion 2).
However, the NuBaID of HsZC3HC1 currently appears specific for only one stable binding partner, i.e., HsTPR. Other proteins previously stated to be regular ZC3HC1 binding partners, which included SCF complex components, CCNB1, and FANCD2 (e.g., Bassermann et al., 2005; Kreutmair et al., 2020), have been ruled out (Gunkel et al., 2021) or assessed as unlikely binding partners (Supplemental Figure S13). However, we cannot exclude that some ZC3HC1 paralogues might have established binding interfaces for stably interacting partners other than TPR that are yet unknown. Therefore, as long as it remains uncertain whether every ZC3HC1 paralogue with a current version of the NuBaID signature is an NB-interacting protein, one could imagine an alternative name for this signature that does not assign a function to it. In addition, one could also consider merging the NuBaID signature and the Pfam motifs zf-C3HC and Rsm1 into a single, all characteristics–encompassing novel motif, as the individual signatures represent parts of the same protein (see also Supplemental Discussion 3).
Furthermore, it remains to be determined whether the commonality shared by all ZC3HC1 homologues with an intact NuBaID is confined to the initial NB binding, or whether all are capable of also recruiting further TPR polypeptides. We consider it possible that some homologues with an NB binding–competent NuBaID might lack the potential to attract more TPR. In this context, it is noteworthy that the HsZC3HC1 mutant 72–290_398–467, while competent to bind to the NB and TPR already positioned there, cannot attract additional TPR polypeptides. Such TPR recruitment requires additional HsZC3HC1 features that are not readily identifiable in every ZC3HC1 homologue (Gunkel et al., unpublished data).
These observations raise the question of why evolution would have allowed some species to possess a ZC3HC1 homologue with a prototypic NuBaID capable of NB binding but then be unfit to recruit more TPR. We recently speculated (Gunkel et al., 2021) that ZC3HC1 might also act as an adjustable insulator of TPR segments, with such a function even uncoupled from the recruitment of additional TPR. Future work will now need to scrutinize whether some ZC3HC1 homologues lack the ability to recruit TPR, and whether the NB binding of ZC3HC1 on its own already has specific functional relevance.
Computational predictions indicating common and specific features of BLD domain structures
AlphaFold2 excluded certain topologies for zinc ion coordination in ZC3HC1 (Supplemental Figure S14) and revealed the likely structures of different ZC3HC1 homologues. Despite their low sequence identity, which held even for the corresponding BLDs alone (Supplemental Figure S12B), the structured parts of these homologues were predicted to be strikingly similar. In addition to our experimental insights, these predictions underscored that intact ZC3HC1 homologues possess two zinc ion coordination modules that together constitute the NuBaID as a conserved structural entity. Furthermore, the predictions allowed us to redefine the extent of each BLD. This also revealed that the Pfam motif zf-C3HC refers to BLD1 plus an N-terminal part of BLD2, while the Rsm1 motif corresponds to a C-terminal BLD2 segment.
These predictions also confirmed that the minimal sequence signature of the NuBaID represents only residues involved in establishing the central structures of the BLDs. Therefore, the integrity of both BLD core regions is only a prerequisite for forming a fully functional TPR-binding interface, whose residues are positioned elsewhere in ZC3HC1 and still need to be specified. Notably, only some of the few other ZC3HC1 residues that appear relatively well conserved across phyla were predicted to be surface-exposed (Supplemental Figure S11, D and H). Thus, it is possible that the TPR-binding interfaces of different ZC3HC1 homologues from distant phyla may share only a few conserved residues at most.
We also noted that conspicuous α-helices surround each BLD core region. The outwardly exposed parts of some of these α-helices appear suitably positioned for contributing to a TPR-binding interface. We can picture a scenario in which some of them align with the coiled-coil homodimers of TPR, and that these ZC3HC1 α-helices would need to be in a specific arrangement for such interactions. Indeed, the positioning of these α-helices appears to be evolutionarily conserved and in some cases defined by interactions with the BLD cores. Thus, some aa substitutions in this study may have led to a damaged BLD core that no longer allowed the correct placement of these outward-facing a-helices, resulting in a loss of TPR-binding competence.
Other BLD-specific structural features included a conspicuous surface groove identified in the BLD1 of vertebrates. Such a groove appears to be absent or altered in BLD2, which instead exhibits a few relatively well-conserved surface-exposed residues in this position (Supplemental Figure S11H; also see Supplemental Discussion 1). In addition to the α-helices, these other single-BLD-specific features also need to be examined further to clarify which regions of ZC3HC1 constitute its TPR-binding interfaces.
Intrinsically disordered regions as a characteristic feature of many ZC3HC1 homologues
HsZC3HC1 contains several segments that are dispensable for NB binding. The most prominent one comprises more than 100 residues embedded within BLD2 and predicted to be largely unstructured. ZC3HC1 homologues of numerous other eukaryotes also harbor a large insertion at the corresponding position. Except for a few conserved residues, which are part of the Pfam Rsm1 motif (e.g., Supplemental Figure S11H) and located at the transitions between the structured part of BLD2 and its loop, these insertions do not share any obvious sequence conservation across phyla.
Some of these unstructured loops comprise more than 500 aa, and they might even be far longer in several stick insect homologues (Supplemental Figure S4). These findings raise the question of why evolution has favored and expanded such loops in these insects, while most insects appear to lack any detectable ZC3HC1 homologue (Supplemental Discussion 4). In further striking contrast, the genuine ZC3HC1 homologues of other species lack such additional insertions in BLD2, as exemplified by ScPml39p.
Notably, such large insertions are generally absent from the BLD1 in the different homologues. Such absence suggests that this domain, when located at the NB, faces a binding interface or local environment that cannot tolerate extensive, space-filling insertions. By contrast, the possibility of loops within BLD2 suggests free spaces that can be occupied there, for example, when these loops would project from the NB and surround the TR. One could further speculate that such loops would then contribute to flexible barriers or the demarcation of distinct cargo pathways. Alternatively, these loops could function as spacer elements involved in defining distances between the NB and the neighboring chromatin. Furthermore, while such loops are dispensable for initial NB binding, they may contribute to the regulated formation of higher-order cylindrical arrangements between TPR and ZC3HC1 of the type discussed recently (Gunkel et al., 2021; Gunkel and Cordes, 2022).
Also of note, these BLD2-embedded loops appear to be targets of cellular signaling processes. For example, the loop of HsZC3HC1 harbors many sites that appear to be also phosphorylated in the interphase after different stimuli (e.g., Christensen et al., 2010; Moritz et al., 2010; Yu et al., 2011; see also Supplemental Figure S15). Such posttranslational modifications might adjust the performance of the loop at the NB perimeter, with a more or less phosphorylated loop perhaps repelling specific molecules more or less efficiently (see also Supplemental Discussion 5).
Future dissection of the function of this loop might also reveal why a distinct HsZC3HC1 polymorphism has been associated with atherosclerosis of coronary arteries, ischemic stroke, and distinct blood pressure profiles. Such pathological phenotypes have been assigned to an arginine at aa 363, centrally located within the loop, while histidine at this position was classified as a noneffective or protective residue (e.g., Schunkert et al., 2011; Deloukas et al., 2013; Wirtwein et al., 2016; Jafaripour et al., 2019). Some studies speculated that these disease-associated phenotypes reflect a dysfunction of ZC3HC1 in its formerly proposed role (Bassermann et al., 2005) as an SCF component ensuring CCNB1 degradation (e.g., Jones et al., 2016; Linseman et al., 2017; see also López-Mejías et al., 2013; Kunnas and Nikkari, 2015). However, since ZC3HC1 is neither an SCF component nor involved in some direct regulation of cellular CCNB1 amounts (Gunkel et al., 2021), we can now rule out such a causal relationship. Therefore, it remains to be determined how R363, within a region not even sequence-conserved among vertebrates, would provoke the phenotypes assigned to this residue, especially since neither the R363 nor H363 variant affects initial NB binding notably (Supplemental Figure S2B). One could speculate, as others have (Jones et al., 2016; Linseman et al., 2017; Wang et al., 2019), that the two residues differentially impact the phosphorylation of nearby serine or threonine residues, perhaps by altering kinase binding preferences (e.g., Ren et al., 2010). The phenotypes assigned to this polymorphism would then be caused by changes in phosphorylation, affecting the yet unknown function of this disordered loop.
Pml39p as the budding yeast homologue of ZC3HC1
The BLD2 loop is missing in ScPml39p, and we found that Pml39p cannot stably bind to HsTPR, nor did HsZC3HC1 stably interact with the ScMlps. Nevertheless, as stated recently (Gunkel et al, 2021; Gunkel and Cordes, 2022), we regard Pml39p as the unequivocal and only homologue of HsZC3HC1 in S. cerevisiae. Why these proteins have not been considered homologues or structural NB proteins earlier may have had several reasons, outlined in Supplemental Discussion 6.
Our reasoning that ZC3HC1 and Pml39p are homologues is based not only on our finding that the nonessential Pml39p has a prototypic NuBaID signature, but also on our experimental results. Some of them confirmed that Pml39p locates at the yeast NE (Huh et al., 2003) via binding to the Mlp proteins (Palancade et al., 2005), just as the nonessential ZC3HC1 binds to vertebrate NBs by binding to TPR (Gunkel et al., 2021; Gunkel and Cordes, 2022). Moreover, previous work had found that Pml39p binds to distinct parts of the Mlps (Palancade et al., 2005), which correspond to those TPR regions that we found binding ZC3HC1. In addition, we validated Pml39p as the ZC3HC1 homologue by showing that the integrity of its NuBaID is required for both NB association and Mlp binding.
Furthermore, our finding that the NE-attached amounts of Mlp1p are significantly reduced in pml39∆ cells, in line with recent similar observations (Bensidoun et al., 2022), indicates that specific arrangements between Mlp subpopulations at the nuclear periphery are Pml39p-dependent. Again, this result is reminiscent of ZC3HC1 being required for tethering TPR subpopulations to NBs in human cells (Gunkel et al., 2021; Gunkel and Cordes, 2022). Moreover, our finding that Pml39p is a prerequisite for the focal accumulation of Mlp1 in nup60∆ cells is equivalent to our data showing that ZC3HC1 is responsible for keeping TPR accumulated in NPC-remote foci in NUP153-deficient cells (Gunkel and Cordes, 2022). Furthermore, an observation hinting at perhaps yet another potential commonality between HsZC3HC1 and ScPml39p is also outlined in Supplemental Information 8.
Finally, AlphaFold2 predicted striking similarities between the NuBaID constructions of HsZC3HC1 and ScPml39p, demonstrating the relationship between these proteins also at the structural level. This finding was further underscored by the recent ScPml39p crystal structure (Hashimoto et al., 2022), which appears nearly identical to the AlphaFold2 prediction (Supplemental Figure S16).
Given the commonalities between HsZC3HC1 and ScPml39p, the question arises whether a function previously assigned to Pml39p might also apply to ZC3HC1. Pml39p has been described as an upstream effector of the Mlps, involved in the nuclear retention of incompletely spliced mRNAs (Palancade et al., 2005; Bonnet et al., 2015). In this model, Pml39p uses the NB and Mlps only as a platform to prevent intron-containing pre-mRNAs from exiting the nucleus, by interacting directly, and in principle also autonomously, with distinct mRNA-binding proteins and potentially also components of the splicing machinery (Palancade et al., 2005; for further considerations, see Supplemental Discussion 7). However, regarding human cells, none of our currently available data suggests that HsZC3HC1 would effectively execute autonomous functions when uncoupled from TPR. Furthermore, neither ZC3HC1 nor ZC3HC1-appended or NPC-anchored TPR seemed to play a universal role in monitoring mRNAs for splice sites and retained introns in HeLa cells (Iino, 2017, and our unpublished data). While these findings conflicted with one report (Rajanala and Nandicoori, 2012), they are consistent with TPR not promoting nuclear retention of 5′splice site–containing transcripts in human U-2 OS cells (Lee et al., 2020). However, human cells have a far larger nucleus than yeast cells, an abundance of introns, and, also unlike yeast, an open mitosis. Most quality control of splicing events in the proliferating human cell is therefore likely to occur already early during transcript processing (e.g., Schmid and Jensen, 2013; Garland and Jensen, 2020), and thus far from the NB. Such early quality controlling in humans could allow for fewer unfulfilled quality control tasks at the onset of mitosis and NE breakdown. Given these differences between humans and yeasts, we do not regard our human cell data as necessarily in conflict with those arguing for specific types of mRNA quality control at the yeast NBs (e.g., Schmid and Jensen, 2010).
To have or have not: trade-offs underlying the absence of a ZC3HC1 homologue in some species and its presence in others?
ZC3HC1 homologues are nonessential in different species, including budding and fission yeast (Yoon, 2004; Palancade et al., 2005), nematodes (Rual et al., 2004; Sönnichsen et al., 2005), and mice (e.g., Illert et al., 2012; Aherrahrou et al., 2021; our unpublished data). Furthermore, with ZC3HC1 homologues also appearing naturally absent in other organisms, like in Dipteran insects, these findings raise questions (see also Supplemental Discussions 8–10, the latter an extended version of this chapter). For example, what species-spanning general advantage does ZC3HC1 provide in the numerous organisms in which selection pressure ensured its persistence, and why did it become dispensable or perhaps even disadvantageous for those organisms that lost it during evolution? These questions inevitably come with another question, namely which advantages come with those TPR polypeptides that ZC3HC1 appends to the NB? Since ZC3HC1 is dispensable, NB appendage of such additional amounts of TPR would not be essential either. On the other hand, TPR is an essential gene in some organisms, including mammals and insects, indicating that those TPR polypeptides anchored independently of ZC3HC1 to the NPC are indispensable. This NPC-anchored TPR subpopulation is even essential for human tumor cells, as we could not produce TPR KO cell lines using CRISPR/Cas9n technology. Furthermore, organisms naturally lacking ZC3HC1 can still have TPR-containing NBs, as in insects of the order Diptera (Kiseleva et al., 1996; Soop et al., 2005). While these insects were initially considered ZC3HC1-deficient based only on sequence searches, a potential Dipteran ZC3HC1 homologue could also not be detected with recent structure search tools such as Foldseek (van Kempen et al., 2022; Supplemental Figure S17).
Assuming that the progenitors of such ZC3HC1-deficient eukaryotes once had a functional ZC3HC1 homologue, the question remains how and why they lost it. In one scenario, ZC3HC1 became disadvantageous in certain situations and environments, with evolutionary forces then expediting its elimination. This idea of a trade-off between advantages and disadvantages under different conditions is supported by several systematic studies in S. cerevisiae. Homozygous pml39∆ cells in these studies were viable in various growth conditions considered approximations of typical environments experienced by wild, domesticated, and laboratory yeast strains. Under these conditions, Pml39p deficiency did not affect or only minimally affected the competitive fitness of the KO cells compared to the WT strains (e.g., Breslow et al., 2008; Qian et al., 2012). However, when exposed to a plethora of chemical, physical, or nutritional stress conditions, the competitive fitness of the PML39wt and pml39∆ cells differed significantly under some of these conditions. For example, the pml39∆ cells were more sensitive to certain types of acute stress but more tolerant of nutritional deficiencies and other types of stress than the PML39wt cells (Brown et al., 2006; Hillenmeyer et al., 2008). These screening data thus suggest that the existence of Pml39p in free-living yeasts could reflect a dynamic balance between advantages and disadvantages. Furthermore, one could imagine that such a balancing act between pros and cons in some situations and life phases also applies to the ZC3HC1 homologues in other species. ZC3HC1 would thus conform to the current definition of a gene with multiple opposing effects on fitness and hence with antagonistic pleiotropy (e.g., Kirkwood, 2002; Elena and Lenski, 2003; Mitchell-Olds et al., 2007; Anderson et al., 2011; Qian et al., 2012; Austad and Hoffman, 2018).
In conclusion, while future research might naturally focus on elucidating the advantages that ZC3HC1, and the TPR it attaches to the NB, will provide to those species with a ZC3HC1 homologue, there may also be a dark side to the possession of this protein and the ZC3HC1-dependent TPR at the NB. However, even if ZC3HC1 were the product of a trade-off gene that also imposes disadvantages, future work would still have to unveil how the structural arrangements of ZC3HC1 and TPR at the NB could cause both beneficial and adverse effects.
MATERIALS AND METHODS
Antibodies, expression vectors, cell lines, and yeast strains
Antibodies and expression vectors used in the present study are listed in the Supplemental Materials and Methods and in Supplemental Table 1, respectively. The human HeLa cell line P2 and its CRISPR/Cas9n-generated ZC3HC1 KO progeny line were previously described along with growth conditions (Gunkel et al., 2021). The axenic D. discoideum strains Ax2, Ax3, and Ax4 (e.g., Bloomfield et al., 2008) were kindly provided by Martin Kollmar (Georg-August-University Göttingen, Göttingen, Germany) and Katarina Gunkel (Max Planck Institute for Dynamics and Self-Organization, Göttingen, Germany). The budding yeast strains used in this study, isogenic to S288c, are listed in Supplemental Table 2.
Transfections and transformations for live-cell imaging
HeLa P2 cells were transfected with expression vectors encoding FP-tagged polypeptides by using PolyJet (SignaGen Laboratories, Rockville, MD) and inspected within 24 h of transfection, as previously described (Gunkel et al., 2021). The lithium acetate method (Gietz and Schiestl, 2007) was used to transform Mlp1p-mCherry–expressing pml39∆ yeast cells with Gal-inducible expression vectors encoding yECitrine-tagged WT and mutant versions of Pml39p. The transformed yeast cells were first grown in a noninducing medium with raffinose and then transferred into a galactose-containing medium to induce expression of the yECitrine-tagged proteins, followed by live-cell imaging about 30 min later.
Live-cell imaging and IFM
Live-cell imaging and IFM of cultured HeLa P2 cells were performed as described (Gunkel et al., 2021; Gunkel and Cordes, 2022). A Leica TCS SP5 or SP8 confocal laser-scanning microscope equipped with a 63× Lambda objective (Leica Microsystems, Wetzlar, Germany) was used for image acquisition. The Leica TCS SP5 was also used for FLIP of HeLa cells transiently expressing FP-tagged polypeptides. Selected areas were pulsed with full laser power for 2 min, followed by the acquisition of post-bleach images. Additional brightness enhancement of a subset of images (Figures 1B2 and 2B, and Supplemental Figure S2) was carried out after the image acquisition, as described previously (Gunkel and Cordes, 2022). For live-cell imaging of yeast, cultured cells in logarithmic growth were transferred into 18-well slides (µ-Slide 18 Well Flat ibiTreat, ibidi GmbH, Gräfelfing, Germany). Once the cells had settled, they were imaged at room temperature with a 63× Lambda objective using the Leica SP5 confocal microscope or with a 63× NA1.4 objective on the Zeiss LSM880 Fast Airyscan confocal microscope (Carl Zeiss, Oberkochen, Germany). NE signal intensities were quantified with Fiji/ImageJ (versions 1.50i–1.51t, National Institutes of Health; Schneider et al., 2012) using yeast cell micrographs obtained by live-cell imaging, applying the quantification procedure described previously (Gunkel et al., 2021). In brief, PML39wt and pml39∆ cell populations grown in parallel were harvested in the exponential growth phase and then imaged as live cells using the same microscope settings. Signal intensities of NE segments were quantified from essentially all NEs that could be seen in an equatorial focus plane in several randomly chosen overview micrographs for each population of the two strains. NE-flanking GFP signals from cytoplasmic and nucleoplasmic areas were subtracted. Suspended Dictyostelium Ax2 cells were grown in HL5 medium with glucose (HLG01; Formedium, Hunstanton, United Kingdom) for 2–3 d to reach confluence in a culture dish. After being detached by gentle pipetting, the cells were collected in a 15 ml tube and centrifuged at 300 × g for 3 min. The sedimented cells were washed twice in 10 ml of phosphate-buffered saline (PBS) with 10 mM freshly added MgCl2. Next, 400 µl of cell suspension per well was pipetted onto 12 mm #1.5H coverslips (Gerhard Menzel B.V. & Co. KG, Braunschweig, Germany) already placed in a 24-well plate. The plate was centrifuged in a swing-out rotor at 300 × g for 3 min, allowing cells to settle onto the coverslips homogeneously. The PBS solution was aspirated and replaced with fixation solution (1% formaldehyde and 0.54% methanol in PBS; i.e., Formalin solution diluted 1:37) and 0.1% Triton X-100. After incubation for 15 min, the fixed and permeabilized samples were quenched (50 mM NH4Cl in PBS, 5 min), blocked (1% bovine serum albumin in PBS, 30 min), and immunolabeled as previously described for human cells (Gunkel et al., 2021). Fiji/ImageJ was used to generate line profiles across immunolabeled Dictyostelium nuclei.
Cloning of Dictyostelium discoideum cDNAs
For total RNA isolation, D. discoideum Ax4 cells were homogenized in TRIzol (Invitrogen, Carlsbad, CA), followed by RNA purification and on-column digestion of genomic DNA (Direct-zol RNA Miniprep kit; Zymo Research, Freiburg, Germany). The Superscript III First-Strand Synthesis System (Invitrogen) was used for cDNA synthesis with oligo(dT) primers and for the final RNA digestion, according to the manufacturer’s instructions. Primers used for PCR, for subcloning, and for sequencing of DdTPR and DdZC3HC1 sequences are given in Supplemental Table 3. cDNA sequences were deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers ON368701 (DdZC3HC1) and ON368702 (DdTPR).
Editing of yeast genes
Genetic manipulations were performed in yeast strains that were isogenic to S. cerevisiae S288C (for the list of strains, see Supplemental Table 2). Chromosomal integration of PCR-amplified cassettes, for the tagging of yeast genes with codon-optimized ORFs for yEGFP or mCherry to obtain carboxy-terminally tagged proteins, was performed as described (Janke et al., 2004). Null mutants were generated by replacing whole genes with integration cassettes. In brief, an integration cassette, including an ORF for a tag or solely a stop codon, followed by a selection marker under the control of the Ashbya gossypii TEF-promoter, was amplified using primers of up to ∼100 nucleotides (nt) in length. These primers included 55–78-nt-long overhangs complementary to the 5′ and 3′ sequences of the genomic target location. The lithium acetate method (Gietz and Schiestl, 2007) was used to transform the WT or auxotrophic yeast cells with the purified PCR product, followed by colony selection via antibiotic resistance or on dropout plates. Isolated strains were further analyzed (i) by live-cell imaging, when purposeful, and (ii) by PCR, using genomic DNA isolated as described (Harju et al., 2004).
Yeast two-hybrid experiments
Haploid S. cerevisiae MATa strain CG-1945 or AH109 and MATα strain Y187 (Harper et al., 1993; Feilotter et al., 1994; James et al., 1996) were used for single transformations (Dohmen et al., 1991) with bait or prey constructs, respectively (based on vectors from Clontech Laboratories; further plasmid details in Supplemental Table 1). After selection on either −Trp (prey) or −Leu (bait) plates, single colonies were picked and grown as liquid cultures. Mating was first done in solution in a 96-well plate, with subsequent plating onto adenine-containing yeast extract-peptone-dextrose agar (YPDA) agar plates. After 2 d of growth at 25°C, the cells were replica-plated onto agar plates with selective medium (−Trp/−Leu; −LW) for diploid cells, to monitor the success of mating. After 3 d of growth at 25°C, cells were replica-plated onto −Leu/−Trp/−His (−LWH) dropout plates, without and with increasing concentrations of 3-AT, a competitive inhibitor of the HIS3 gene product IGP dehydratase (Brennan and Struhl, 1980; Durfee et al., 1993). Cells were further cultured at 25°C for up to 10 d to allow the growth of cells expressing interacting protein pairs. In addition, each bait construct was tested for self-activation by mating the corresponding MATa strain with a MATα strain transformed with the empty prey vector.
Sequence database mining, protein structure predictions, and molecular graphics tools
Sequence database mining and the generation of sequence logos are described in the Supplemental Materials and Methods, complemented by Supplemental Information 2 and 3. Predicted structures were either retrieved as PDB files from the structure database of AlphaFold (https://www.alphafold.ebi.ac.uk) or predicted using the platform ColabFold (AlphaFold2 using MMseqs2; https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb). ColabFold uses the AlphaFold2 source code but uses a different tool for generating the input MSAs (multiple sequence alignments), a feature that we, in turn, exploited for assessing possible limitations in the BLD structure predictions (Supplemental Information 6). Most structures presented here were further processed using the UCSF Chimera system (e.g., Pettersen et al., 2004; https://www.rbvi.ucsf.edu/chimera) for coloring, structural alignments, superimpositions, and in silico truncations (for further details, see the Supplemental Materials and Methods). Fast structural comparisons using Foldseek (https://search.foldseek.com/search) are described in the Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge Frank Schwarz and Kerstin Mohr at the Core Facility for Genomics and Proteomics at the German Cancer Research Center (DKFZ) for contributions to Y2H experiments and Gabriele Hawlitscheck at the Department for Cellular Logistics at the Max Planck Institute for Multidisciplinary Sciences for contributions to cloning. Furthermore, we thank Dirk Görlich for generous support and Eberhard Bodenschatz, Steffen Frey, Dirk Görlich, Katharina Gunkel, Martin Kollmar, Arturo Vera Rodriguez, and Frank Schwarz for kindly providing research materials. In addition, we appreciate Thomas Güttler, Michael Ridders, and Arturo Vera Rodriguez for technical advice and Sabbi Lall, Karsten Weis, Helen Pickersgill, Andrea Devlin, and James Allen for contributions to text editing and proofreading the manuscript.
Abbreviations used:
- aa
amino acid
- AI
artificial intelligence
- BIR
baculovirus inhibitor of apoptosis protein repeat
- BLD
BIR-like domain
- FLIP
fluorescence loss in photobleaching
- FP
fluorescent protein
- IAP
inhibitor of apoptosis protein
- IFM
immunofluorescence microscopy
- ILP
IAP-like protein
- KO
knockout
- NB
nuclear basket
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- NuBaID
nuclear basket interaction domain
- NUP
nucleoporin
- TR
terminal ring
- WT
wild-type
- Y2H
yeast two hybrid
- zf
zinc finger.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-09-0402) on March 1, 2023.
REFERENCES
- Aherrahrou R, Reinberger T, Werner J, Otto M, Al-Hasani J, Munoz-Venegas ML, Civelek M, Schunkert H, Kessler T, Erdmann J, et al. (2021). Deficiency of ZC3HC1 increases vascular smooth muscle cell migration, proliferation and neointima formation following injury. bioRxiv 10.1101/2021.09.29.462212. [DOI] [Google Scholar]
- Aksenova V, Smith A, Lee H, Bhat P, Esnault C, Chen S, Iben J, Kaufhold R, Yau KC, Echeverria C, et al. (2020). Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat Commun 11, 4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T (2011). Evolutionary genetics of plant adaptation. Trends Genet 27, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH (2020). Into the basket and beyond: the journey of mRNA through the nuclear pore complex. Biochem J 477, 23–44. [DOI] [PubMed] [Google Scholar]
- Austad SN, Hoffman JM (2018). Is antagonistic pleiotropy ubiquitous in aging biology? Evol Med Public Health 2018, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F, von Klitzing C, Münch S, Bai R-Y, Kawaguchi H, Morris SW, Peschel C, Duyster J, von Klitzing C, Münch S, et al. (2005). NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell 122, 45–57. [DOI] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O (2004). Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390. [DOI] [PubMed] [Google Scholar]
- Bensidoun P, Reiter T, Montpetit B, Zenklusen D, Oeffinger M (2022). Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pores in budding yeast. Mol Cell 82, 3856–3871.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensidoun P, Zenklusen D, Oeffinger M (2021). Choosing the right exit: how functional plasticity of the nuclear pore drives selective and efficient mRNA export. Wiley Interdiscip Rev RNA 12, e1660. [DOI] [PubMed] [Google Scholar]
- Bloomfield G, Tanaka Y, Skelton J, Ivens A, Kay RR (2008). Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum. Genome Biol 9, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A, Bretes H, Palancade B (2015). Nuclear pore components affect distinct stages of intron-containing gene expression. Nucleic Acids Res 43, 4249–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MB, Struhl K (1980). Mechanisms of increasing expression of a yeast gene in Escherichia coli. J Mol Biol 136, 333–338. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS (2008). A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods 5, 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Sherlock G, Myers CL, Burrows NM, Deng C, Wu HI, McCann KE, Troyanskaya OG, Brown JM (2006). Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol Syst Biol 2, 2006.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GL, Kelstrup CD, Lyngsø C, Sarwar U, Bøgebo R, Sheikh SP, Gammeltoft S, Olsen JV, Hansen JL (2010). Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics 9, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW (1997). Identification of protein p270/TPR as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol 136, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu F, Milani M, Mastrangelo E, Lecis D (2019). Targeting the BIR domains of Inhibitor of Apoptosis (IAP). Proteins in cancer treatment. Comput Struct Biotechnol J 17, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, et al. (2013). Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 45, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Philippe H, Tsagkogeorga G, Simion P, Tilak M-K, Turon X, López-Legentil S, Piette J, Lemaire P, Douzery EJP (2018). A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol 16, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Muthuswamy S, Meier I (2012). Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol Biol 79, 203–216. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AW, Höner CB, Hollenberg CP (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7, 691–692. [DOI] [PubMed] [Google Scholar]
- Duheron V, Chatel G, Sauder U, Oliveri V, Fahrenkrog B (2014). Structural characterization of altered nucleoporin NUP153 expression in human cells by thin-section electron microscopy. Nucleus 5, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE (2003). Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4, 457–469. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D (1994). Construction of an improved host strain for two hybrid screening. Nucleic Acids Res 22, 1502–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin J-C, Scherthan H, Nehrbass U (2002). Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol 4, 214–221. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res 34, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. (2010). The Pfam protein families database. Nucleic Acids Res 38, D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. (2008). The Pfam protein families database. Nucleic Acids Res 36, D281–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW (2009). Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J 59, 243–255. [DOI] [PubMed] [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, Gerace L (2002). TPR is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol 156, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funasaka T, Tsuka E, Wong RW (2012). Regulation of autophagy by nucleoporin TPR. Sci Rep 2, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland W, Jensen TH (2020). Nuclear sorting of RNA. Wiley Interdiscip Rev RNA 11, e1572. [DOI] [PubMed] [Google Scholar]
- Garner R, Dunker B, Obradovic (1999). Predicting binding regions within disordered proteins. Genome Inform Ser Workshop Genome Inform 10, 41–50. [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2, 31–34. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD (1992). High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol 119, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Solovei I, Allen TD (1997). Nuclear pore complex structure in birds. J Struct Biol 119, 284–294. [PubMed] [Google Scholar]
- Gunkel P, Cordes VC (2022). ZC3HC1 is a structural element of the nuclear basket effecting interlinkage of TPR polypeptides. Mol Biol Cell 33, ar82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel P, Iino H, Krull S, Cordes VC (2021). ZC3HC1 is a novel inherent component of the nuclear basket, resident in a state of reciprocal dependence with TPR. Cells 10, 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harju S, Fedosyuk H, Peterson KR (2004). Rapid isolation of yeast genomic DNA: bust n’ grab. BMC Biotechnol 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hase ME, Cordes VC (2003). Direct interaction with NUP153 mediates binding of TPR to the periphery of the nuclear pore complex. Mol Biol Cell 14, 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Kuznetsov NV, Cordes VC (2001). Amino acid substitutions of coiled-coil protein TPR abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell 12, 2433–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Ramirez DH, Lautier O, Pawlak N, Blobel G, Palancade B, Debler EW (2022). Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules. Sci Rep 12, 17691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Takasawa R, Yoshimori A, Goh T, Tanuma S, Kuchitsu K (2005). Identification of a novel gene family, paralogs of inhibitor of apoptosis proteins present in plants, fungi, and animals. Apoptosis 10, 471–480. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St.Onge RP, Tyers M, Koller D, et al. (2008). The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden JM, Koreny L, Kelly S, Chait BT, Rout MP, Field MC, Obado SO (2014). Touching from a distance. Nucleus 5, 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Iino H (2017). Functional Analysis of the Nuclear Basket and Protein Tpr, Göttingen, Germany: Cuvillier Verlag. [Google Scholar]
- Illert AL, Kawaguchi H, Antinozzi C, Bassermann F, Quintanilla-Martinez L, von Klitzing C, Hiwatari M, Peschel C, de Rooij DG, Morris SW, et al. (2012). Targeted inactivation of nuclear interaction partner of ALK disrupts meiotic prophase. Development 139, 2523–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Mongkolsiriwatana C, Veley KM, Kim SY, Michaels SD (2007). The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol 144, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafaripour S, Sasanejad P, Dadgarmoghaddam M, Sadr-Nabavi A (2019). ADAMTS7 and ZC3HC1 share genetic predisposition to coronary artery disease and large artery ischemic stroke. Crit Rev Eukaryot Gene Expr 29, 351–361. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Jarnik M, Aebi U (1991). Toward a more complete 3-D structure of the nuclear pore complex. J Struct Biol 107, 291–308. [DOI] [PubMed] [Google Scholar]
- Jones PD, Kaiser MA, Ghaderi Najafabadi M, McVey DG, Beveridge AJ, Schofield CL, Samani NJ, Webb TR (2016). The coronary artery disease-associated coding variant in zinc finger C3HC-type containing 1 (ZC3HC1). affects cell cycle regulation. J Biol Chem 291, 16318–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TBL (2002). Evolution of ageing. Mech Ageing Dev 123, 737–745. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Allen TD, Rutherford S, Bucci M, Wente SR, Goldberg MW (2004). Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J Struct Biol 145, 272–288. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Daneholt B, Allen TD (1996). RNP export is mediated by structural reorganization of the nuclear pore basket. J Mol Biol 260, 304–311. [DOI] [PubMed] [Google Scholar]
- Kokoszynska K, Rychlewski L, Wyrwicz LS (2008). The mitotic entry regulator NIPA is a prototypic BIR domain protein. Cell Cycle 7, 2073–2075. [DOI] [PubMed] [Google Scholar]
- Kosova B, Panté N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E (2000). Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with Nic96p. J Biol Chem 275, 343–350. [DOI] [PubMed] [Google Scholar]
- Kreutmair S, Erlacher M, Andrieux G, Istvanffy R, Mueller-Rudorf A, Zwick M, Rückert T, Pantic M, Poggio T, Shoumariyeh K, et al. (2020). Loss of the Fanconi anemia–associated protein NIPA causes bone marrow failure. J Clin Invest 130, 2827–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC (2010). Protein TPR is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29, 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Björkroth B, Rackwitz H-R, Cordes VC (2004). Nucleoporins as components of the nuclear pore complex core structure and TPR as the architectural element of the nuclear basket. Mol Biol Cell 15, 4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnas T, Nikkari ST (2015). Association of zinc finger, C3HC-type containing 1 (ZC3HC1). rs11556924 genetic variant with hypertension in a Finnish population, the TAMRISK study. Medicine 94, e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NV, Sandblad L, Hase ME, Hunziker A, Hergt M, Cordes VC (2002). The evolutionarily conserved single-copy gene for murine TPR encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111, 236–255. [DOI] [PubMed] [Google Scholar]
- Lee ES, Wolf EJ, Ihn SSJ, Smith HW, Emili A, Palazzo AF (2020). TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res 48, 11645–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F (2008). TPR directly binds to MAD1 and MAD2 and is important for the MAD1-MAD2-mediated mitotic spindle checkpoint. Genes Dev 22, 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman T, Soubeyrand S, Martinuk A, Nikpay M, Lau P, McPherson R (2017). Functional validation of a common nonsynonymous coding variant in ZC3HC1 associated with protection from coronary artery disease. Circ Cardiovasc Genet 10, e001498. [DOI] [PubMed] [Google Scholar]
- López-Mejías R, Genre F, García-Bermúdez M, Corrales A, González-Juanatey C, Llorca J, Miranda-Filloy JA, Rueda-Gotor J, Blanco R, Castañeda S, et al. (2013). The ZC3HC1 rs11556924 polymorphism is associated with increased carotid intima-media thickness in patients with rheumatoid arthritis. Arthritis Res Ther 15, R152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Dai J, Yang L, La Y, Zhou S, Qiang S, Wang Q, Tan F, Wu Y, Kong W, et al. (2020). Involvement of MEM1 in DNA demethylation in Arabidopsis. Plant Mol Biol 102, 307–322. [DOI] [PubMed] [Google Scholar]
- Maul GG (1976). Fibrils attached to the nuclear pore prevent egress of SV40 particles from the infected nucleus. J Cell Biol 70, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB (2007). Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet 8, 845–856. [DOI] [PubMed] [Google Scholar]
- Moritz A, Li Y, Guo A, Villén J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, et al. (2010). Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 3, ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C (2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell 24, 3920–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang T, Bai R-Y, Bassermann F, von Klitzing C, Klumpen S, Miething C, Morris SW, Peschel C, Duyster J, von Klitzing C, et al. (2003). Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA).. J Biol Chem 278, 30028–30036. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Guo P, Takeda K, Fu Q, Fang H, Frucht DM (2020). ERK1/2 inactivation promotes a rapid redistribution of COP1 and degradation of COP1 substrates. Proc Natl Acad Sci USA 117, 4078–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Zuccolo M, Loeillet S, Nicolas A, Doye V (2005). Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol Biol Cell 16, 5258–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H (2016). Evolution of gene duplication in plants. Plant Physiol 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Qian W, Ma D, Xiao C, Wang Z, Zhang J (2012). The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep 2, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanala K, Nandicoori VK (2012). Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS One 7, e29921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Jiang C, Gao X, Liu Z, Yuan Z, Jin C, Wen L, Zhang Z, Xue Y, Yao X (2010). PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics 9, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris H (1989). Three-dimensional imaging of cell ultrastructure with high resolution low-voltage SEM. Int Phys Conf Ser 98, 657–662. [Google Scholar]
- Ris H (1991). The three-dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull 21, 54–56. [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14, 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki G, Ishiwata K, Machida R, Miyata T, Su Z-H (2013). Molecular phylogenetic analyses support the monophyly of Hexapoda and suggest the paraphyly of Entognatha. BMC Evol Biol 13, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2010). Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip Rev RNA 1, 474–485. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2013). Transcription-associated quality control of mRNP. Biochim Biophys Acta 1829, 158–168. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. (2020). NCBI taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, et al. (2011). Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N, Ferrás C, Kern DM, Logarinho E, Cheeseman IM, Maiato H (2013). Spindle assembly checkpoint robustness requires TPR-mediated regulation of MAD1/MAD2 proteostasis. J Cell Biol 203, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW (2005). Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell 16, 4362–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Vucic D (2014). IAP family of cell death and signaling regulators. Methods Enzymol 545, 35–65. [DOI] [PubMed] [Google Scholar]
- Snow CJ, Paschal BM (2014). Roles of the nucleoporin TPR in cancer and aging. Adv Exp Med Biol 773, 309–322. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume A-M, Artelt J, Bettencourt P, Cassin E, et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469. [DOI] [PubMed] [Google Scholar]
- Soop T, Ivarsson B, Björkroth B, Fomproix N, Masich S, Cordes VC, Daneholt B (2005). NUP153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol Biol Cell 16, 5610–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Aime MC, Grigoriev IV, Martin F, Stajich JE, Blackwell M (2017). The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectr 5, DOI: 10.1128/microbiolspec.FUNK-0053-2016. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP (1999). Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol 144, 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11, 490–501. [DOI] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A, et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L (2013). The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126, 2656–2667. [DOI] [PubMed] [Google Scholar]
- van Kempen M, Kim SS, Tumescheit C, Mirdita M, Söding J, Steinegger M (2022). Foldseek: fast and accurate protein structure search. bioRxiv 10.1101/2022.02.07.479398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen AM, Coulson EJ, Vaux DL (2001). Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol 2, reviews3009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu J, Sun P, Li C, Song X, Lei T, Li Y, Yuan J, Sun S, Ding H, et al. (2020). Paleo-polyploidization in Lycophytes. Genomics Proteomics Bioinf 18, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, La Y, Xia H, Zhou S, Zhai Z, La H (2022). Roles of MEM1 in safeguarding Arabidopsis genome against DNA damage, inhibiting ATM/SOG1-mediated DNA damage response, and antagonizing global DNA hypermethylation. J Integr Plant Biol 64, 87–104. [DOI] [PubMed] [Google Scholar]
- Wang X, Mo X, Zhang H, Zhang Y, Shen Y (2019). Identification of phosphorylation associated SNPs for blood pressure, coronary artery disease and stroke from genome-wide association studies. Curr Mol Med 19, 731–738. [DOI] [PubMed] [Google Scholar]
- Wirtwein M, Melander O, Sjőgren M, Hoffmann M, Narkiewicz K, Gruchala M, Sobiczewski W (2016). The relationship between gene polymorphisms and dipping profile in patients with coronary heart disease. Am J Hypertens 29, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I (2007). Nuclear pore anchor, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH (2004). Schizosaccharomyces pombe rsm1 genetically interacts with spmex67, which is involved in mRNA export. J Microbiol 42, 32–36. [PubMed] [Google Scholar]
- Yu Y, Yoon S-O, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. (2011). Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu C-Y, Blobel G (2004). Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol 167, 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR (1997). A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110 (Pt 8), 927–944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.