Abstract
Over two years into the COVID-19 pandemic, the human immune response to SARS-CoV-2 during the active disease phase has been extensively studied. However, the long-term impact after recovery, which is critical to advance our understanding SARS-CoV-2 and COVID-19-associated long-term complications, remains largely unknown. Herein, we characterized single-cell profiles of circulating immune cells in the peripheral blood of 100 patients, including convalescent COVID-19 and sero-negative controls. Flow cytometry analyses revealed reduced frequencies of both short-lived monocytes and long-lived regulatory T (Treg) cells within the patients who have recovered from severe COVID-19. sc-RNA seq analysis identifies seven heterogeneous clusters of monocytes and nine Treg clusters featuring distinct molecular signatures in association with COVID-19 severity. Asymptomatic patients contain the most abundant clusters of monocytes and Tregs expressing high CD74 or IFN-responsive genes. In contrast, the patients recovered from a severe disease have shown two dominant inflammatory monocyte clusters featuring S100 family genes: one monocyte cluster of S100A8 & A9 coupled with high HLA-I and another cluster of S100A4 & A6 with high HLA-II genes, a specific non-classical monocyte cluster with distinct IFITM family genes, as well as a unique TGF-β high Treg Cluster. The outpatients and seronegative controls share most of the monocyte and Treg clusters patterns with high expression of HLA genes. Surprisingly, while presumably short-lived monocytes appear to have sustained alterations over 4 months, the decreased frequencies of long-lived Tregs (high HLA-DRA and S100A6) in the outpatients restore over the tested convalescent time (≥ 4 months). Collectively, our study identifies sustained and dynamically altered monocytes and Treg clusters with distinct molecular signatures after recovery, associated with COVID-19 severity.
Keywords: COVID-19, Sustained immunity, Monocytes, Tregs
1. Introduction
The COVID-19 pandemic caused by SARS-CoV-2 has infected near a billion people and caused multiple million deaths since its emergence in late 2019. New, evolving variants of concern and delays in vaccination (over 90% unvaccinated in low-income countries) mean that it will continue to disrupt the global society for extended time in the future. Shortly after COVID-19 became a global pandemic, post-acute health issues were reported [41]. Typical issues included shortness of breath, muscle fatigue, and prolonged loss of smell. Other reported symptoms include lingering headaches and cognitive difficulties, skin rashes and gastrointestinal discomfort. Combined, these constellation symptoms are now informally referred to as “long COVID” and likely represent a diverse group of post-infectious syndromes [8,13,47]. These symptoms have been reported to occur at a higher frequency in patients with severe initial infection [45,57]. In addition to these long-term ailments, COVID has frequently been characterized by high amounts of inflammation. Together, these suggest that long-term changes in the immune system even following recovery from COVID could be at the root of some of these long-term symptoms.
An efficient immune response against invading pathogens including SARS-CoV-2 requires the early activation of innate immunity, a nonspecific but quick frontline response able to control infection [7,11,36,53,58]. This effective innate immune response plays a critical role to mount the antigen-specific adaptive immunity. The latter contributes to clearing the infection and preventing reinfection by the same pathogen and more importantly, often sustains the memory response ready for future threatens by the same pathogen [37]. Numerous examinations of patient response to COVID-19 at the single-cell level have been performed since the disease became widespread. Multiple meta-analyses of single-cell RNA-sequencing (sc-RNA seq) data sets have defined the several immune dysregulations are involved in COVID-19 pathogenicity including lymphopenia, impaired IFN response, hyperactivation of myeloid cells and dysregulated macrophage and monocyte functions [3,5,6,20,24,28,29,36,38,43,51,54]. A common feature of SARS-CoV-2 infection in severe COVID-19 patients is lymphopenia with a drastic reduction in both T and B lymphocytes in the circulating blood. This lymphogenic response is often negatively associated with the viral load of SARS-CoV-2 as well as the disease severity [46,51]. In addition, Wilk et al. performed some of the earliest single-cell RNA sequencing of COVID-19 patient-derived cells, identifying changes in the myeloid compartment associated with severe disease states [51]. Despite these progresses, critical questions regarding to whether the dysregulated immune cell phenotypes observed during SARS-CoV-2 infection persist in COVID-19 patients after their full recovery, and if yes, how long the fingerprints in gene expression can be maintained, remain to be answered. In order to understand the post-COVID immune response, we determined the molecular signatures of monocytes and understudied regulatory T (Treg) cells in the circulating blood from convalescent COVID-19 patients (when they were unvaccinated), using single cell RNA sequencing and other approaches. Our studies revealed several unique clusters and molecular signatures in both populations sustained during the recovery phase after SARS-CoV-2 infection.
2. Results
2.1. Association of monocyte and Treg populations with COVID-19 pathogenicity
During the convalescent phase following SARS-CoV-2 infection, the immune system consists of two distinct cellular pools, the virus experienced and naïve cells. As a patient recovers, the SARS-CoV-2 experienced pool changes from a mixture of short lived myeloid, lymphoid and tissue resident cells to predominantly long-lived lymphocytes and tissue resident macrophages. Thus, in convalescent patients, the circulating monocytes and granulocytes are predominantly COVID-19 naïve. To assess whether and how SARS-CoV-2 infection maintains a sustained impact to immune function, we characterized the levels of circulating monocytes from convalescent COVID-19 patients. All patents had been enrolled during the early phase of COVID-19 pandemic from June to November 2020, who did not receive any COVID-19 vaccination (Table 1 ). We enrolled 100 convalescent patients (included healthy controls) for blood sampling at least three weeks after recovery from COVID-19 (within 50–145 days from the onset) and analyzed both plasma IgG antibodies specific to SARS-CoV-2 receptor binding domain (RBD) and cellular immunity profiling (Table 1, Fig. 1A). Based on severity of clinical symptoms and RBD-IgG levels in the plasma [15,27] (Supplementary Fig. 1A), patients were classified into 5 groups, including sero-negative, healthy control group (undetectable RBD-IgG and PCR negative/no PCR), asymptomatic (detectable RBD-IgG but symptom free during SARS-CoV-2 infection), outpatient (mild symptoms, detectable RBD-IgG), hospitalized group (non-ICU), and ICU subgroups. Patients in the groups of asymptomatic and outpatient did not receive any prescribed medication treatment, while some of hospitalized (non-ICU) and ICU groups were treated with vasopressors, high flow nasal cannula, or ventilation, but not any SARS-CoV-2 specific therapies, which were not available during the time (Table 1, Supplementary Table 1).
Table 1.
Data sheet for patients recruited in the study.
| Sero-negative (N = 5) |
Asymptomatic (N = 18) |
Outpatients (N = 56) |
Hospitalized (N = 12) |
ICU (N = 9) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mean / count | SD / % | mean / count | SD / % | mean / count | SD / % | mean / count | SD / % | mean / count | SD / % | |
| Age, years | 39.4 | 15.7 | 33.4 | 8.2 | 42.3 | 15.4 | 48.6 | 11.5 | 50.7 | 16.5 |
| Sex, male | 0 | 0.0% | 9 | 50.0% | 22 | 39.3% | 5 | 41.2% | 6 | 66.7% |
| Race | ||||||||||
| Black | 0 | 0.0% | 0 | 0.0% | 4 | 7.1% | 6 | 50.0% | 1 | 11.1% |
| White | 3 | 60.0% | 8 | 44.4% | 30 | 53.6% | 5 | 41.7% | 8 | 88.9% |
| Asian | 0 | 0.0% | 4 | 22.2% | 1 | 1.8% | 0 | 0.0% | 0 | 0.0% |
| Other | 2 | 40.0% | 6 | 33.3% | 21 | 37.5% | 1 | 8.3% | 0 | 0.0% |
| SOFA score* | NA | NA | NA | NA | NA | NA | NA | NA | 6.4 | 3.5 |
| Intermediate to advanced interventions | ||||||||||
| Vasopressor use | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 1 | 8.3% | 7 | 77.8% |
| High flow nasal cannula | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 5 | 55.6% |
| Non-invasive ventilation | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 1 | 11.1% |
| Mechanical ventilation | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | 7 | 77.8% |
| LOS, days | NA | NA | NA | NA | NA | NA | 6.8 | 5.3 | 29.6 | 19.2 |
| Onset to sampling, days | NA | NA | 80.7 | 51.4 | 82.6 | 51.1 | 145.1 | 53.8 | 119.6 | 39.1 |
Fig. 1.
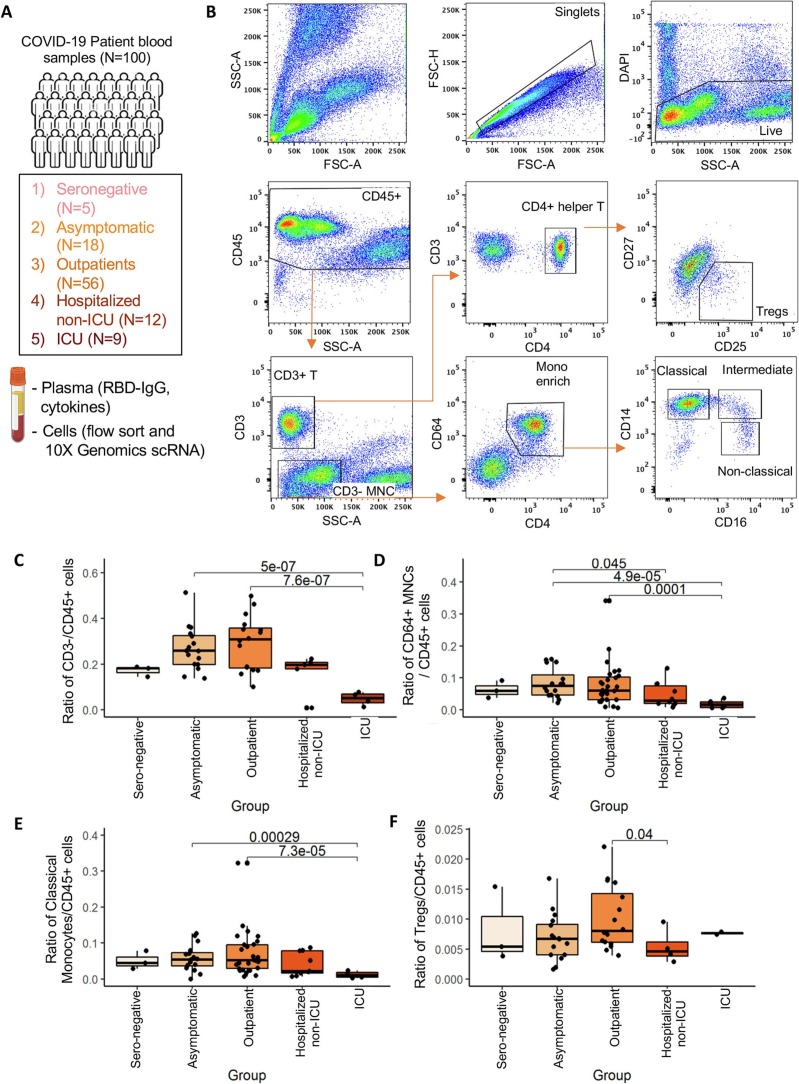
Flow profiles of circulating immune cells associated with COVID-19 patients (N = 100).
(A) Number of patients enrolled and cell isolation strategies for RBD-IgG, flow and single-cell sequencing analysis.
(B) Representative flow cytometry analysis showing the gating and sorting strategies for WBCs isolated from convalescent COVID-19 patients. CD45+DAPI− alive cells were gated into CD3+ and CD3− within the SSClow groups. CD3+ cells were further sub-divided into CD4+ Helper T cells, and Treg cells were sorted as CD4+CD25highCD127low population. CD3- cell fraction were sorted based on CD4dimCD64+ MNCs fraction (enriched for monocytes); these cells were further sub-divided into CD14+CD16− classical monocytes, CD14−CD16+ nonclassical, and CD14+CD16+ intermediate monocytes for analysis.
(C-E) Ratio of flow gated CD3− mononuclear (B), CD64+ monocytes (C), and CD14+ classical monocytes (D) within CD45+ WBCs among five groups of COVID-19 patients (sero-neg, asymptomatic, outpatients, hospitalized-no ICU, ICU), with elevation in mild disease groups (asymptomatic/outpatient) in comparison with severe ICU patients.
(F) Ratio of flow gated Treg cells within CD45+ WBCs among five groups of COVID-19 patients, elevated in outpatients in comparison with hospitalized patients.
As expected, severe COVID symptoms in hospitalized and ICU groups correlated with higher RBD-IgG levels (Supplementary Fig. 1A). Consistently, plasma capacity to block RBD binding to ACE2-expressing cells was positively correlated with RBD-IgG levels and disease severity (Supplementary Fig. 1B). While we expected that time from disease onset would likewise correlate due to eventual senescence of antibody producing cells, the plasma RBD-IgG levels and capacity to inhibit RBD binding in outpatients gradually decline by four months after their recovery (Supplementary Fig. 1C-D).
Initial flow cytometric analysis of the gated CD45+ peripheral white blood cells (WBCs) identified monocytes based on the smaller cell size and lower granularity than granulocytes, negative for expression of CD3, dim positive for CD4, and positive for CD64, a specific marker for human myeloid cells, particularly macrophages and monocytes (Fig. 1B). Based on their expression of CD14 and CD16, these CD45+CD3−CD4dimCD64+ cells were further sub-divided into CD14+CD16− classical monocytes, CD14+CD16+ intermediate monocytes, and CD14−CD16+ non-classical monocytes (Fig. 1B). Interestingly, the frequency of monocyte population showed a dynamic pattern in association with the COVID-19 disease severity. As shown in Fig. 1C-E, the ratio of gated monocytes (CD3− mononuclear and CD64+CD4dim) and the classical CD14+ monocyte levels relative to total CD45+ cells statistically reduced in the ICU group of patients recovered from the most severe COVID-19, compared to those recovered from less severe disease (asymptomatic and outpatient). Therefore, the decline in monocyte populations during the recovery phase appear to be a signature memory for severe disease.
One possible speculation for the long-term changes observed in circulating monocytes in convalescent patients from COVID-19 is global alterations to the immune system driven by long-lived adaptive immune cells. As CD4+CD25+FoxP3+ Tregs are known to modulate broad aspects of the immune response including maintenance of immune tolerance and global immunosuppression, we hypothesized that changes in the Treg compartment might persist following the recovery from COVID-19. To assess changes in Treg population in convalescent patients, CD45+CD3+ T lymphocytes were further gated on CD4 expression as helper T cell population, and Tregs were then defined as CD4+CD25+CD127low populations as reported [55] (Fig. 1B). Importantly, the Treg cell frequency during the convalescent phase relatively increased in less severe outpatients compared to hospitalized patients (Fig. 1F), suggesting that a possible association of Treg increase in those patients with a mild COVID-19.
2.2. Identification of persistent subsets of monocytes with distinct molecular signatures from convalescent COVID-19 patients
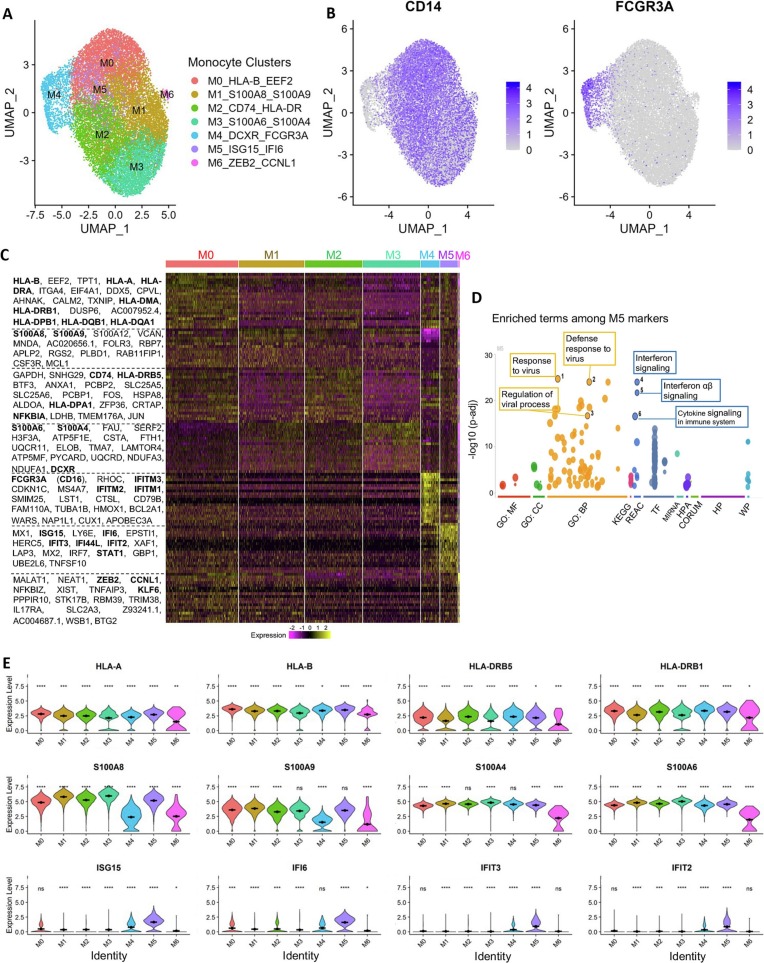
To further investigate the effect of COVID-19 severity on the molecular signature of convalescent monocyte phenotypes, we sorted 18,165 cells of CD45+CD3−CD64hiCD4dim monocytes on intermediate side scatter from 50 convalescent patients and sero-negative controls and subjected to the 10× Genomics single cell RNA sequencing (See Supplementary Table 2 for further demographic information). We first performed the alignment analysis using the 10× genomics CellRanger software with either human or SARS-CoV2 genomes, and our results show there were no viral genes detected from any of the monocytes (Fig. 1A). Clustering analysis identified seven distinct clusters (M0-M6), each of which is specified with top enriched genes, including five CD14+ classical/intermediate monocyte clusters (M0-M4) with inflammatory features, one distinct CD16+ non-classical monocyte cluster M5_ISG15_IFI6, and one minor dendritic cell cluster M6_ZEB2_CCNL1 showing lower expression of both HLA and S100 family genes (Fig. 2A-E).
Fig. 2.
Analysis of the sustained molecular signatures in monocytes from convalescent COVID-19 patients.
(A) Dimensional UMAP plot of monocytes from convalescent COVID-19 patients and sero-negative controls.
(B) Expression of CD14, indicating classical monocytes, and FCGR3A/CD16, indicating nonclassical monocytes, on the UMAP projection from A. Classical monocytes make up the majority of the cells and five of the seven clusters. Nonclassical monocytes are principally cluster M4 on the dimensional plot in A.
(C) Expression of selected genes across clusters. Class I HLA molecules (HLA-A and HLA-B) are strongly expressed in cluster M0, while class II HLA molecules (HLA-DRB5 and HLA-DRB1) are more strongly expressed in cluster M2. S100 genes (second row) are most highly expressed in clusters M1 and M3. Interferon-responsive genes (third row) are most present in cluster M5, which appears to be a unique interferon-responsive cluster of monocytes.
(D) Heatmap showing the expression levels of selected genes across all cells. Nonclassical monocyte associated genes are clearly visible in cluster M4, as are interferon-responsive genes in cluster M5.
Significance of gene expression difference in individual clusters evaluated by Wilcoxon comparison of cluster-cell expression levels with all-cell expression levels. Significance values: ns: p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Analysis of the cells by cluster and featured genes show 4 clusters M0-M3 containing the majority of monocytes (Fig. 2C). Both M1 and M3 clusters showed high but distinct S100 genes with high levels of S100A8 & A9 in M1, but high S100A6 & A4 expression in M3 (Fig. 2C, E). Cluster M0_HLA-B_EEF2_S100A8/A9 and cluster M2_CD74_HLA-DR shared relatively similar gene expression patterns; however, M0 displayed enhanced S100A8/9, STAT1, and ZEB2 genes within multiple high level heatmap blocks and appear to be intermediate populations. Also, the M0_HLA-B_EEF2 monocytes show uniquely high levels of class I HLA molecules whereas cluster M2_CD74_HLA-DR expressed predominantly HLA class II genes (Fig. 2C & D). Consistent with previous studies showing the altered IFN response during the acute SARS-CoV-2 infection is responsible for severe disease [34,53], we detected two clusters with high but distinct IFN responsive genes: the classical monocyte cluster M5_ISG15_IFI6 displayed a unique gene expression pattern characterized by high expression of interferon stimulated genes including ISG15, IFI6, IFIT3, and IFIT2. In contrast, the cluster M4_IFITM_FCGR3A (CD16), which belong to CD16+CD14− non-classical monocytes, expressed high levels of different IFN responsive genes, in particular the Interferon Induced Transmembrane Protein (IFITM) family genes (IFITM1–3) (Fig. 2C-E). Consistently, GO and KEGG pathways analyses revealed that the top differentially expressed genes in monocyte clusters, such as M5, are involved in response to viral infection, regulation of viral process and interferon and cytokine responses in monocytes even after recovery from COVID-19 (Fig. 2D, Supplementary Fig. 2A). Pseudotime analyses of monocytes using S100A8 as a rooting gene indicate alterations in many gene may contribute to the dynamics and differentiation of these cells (Supplementary Fig. 2B-C).
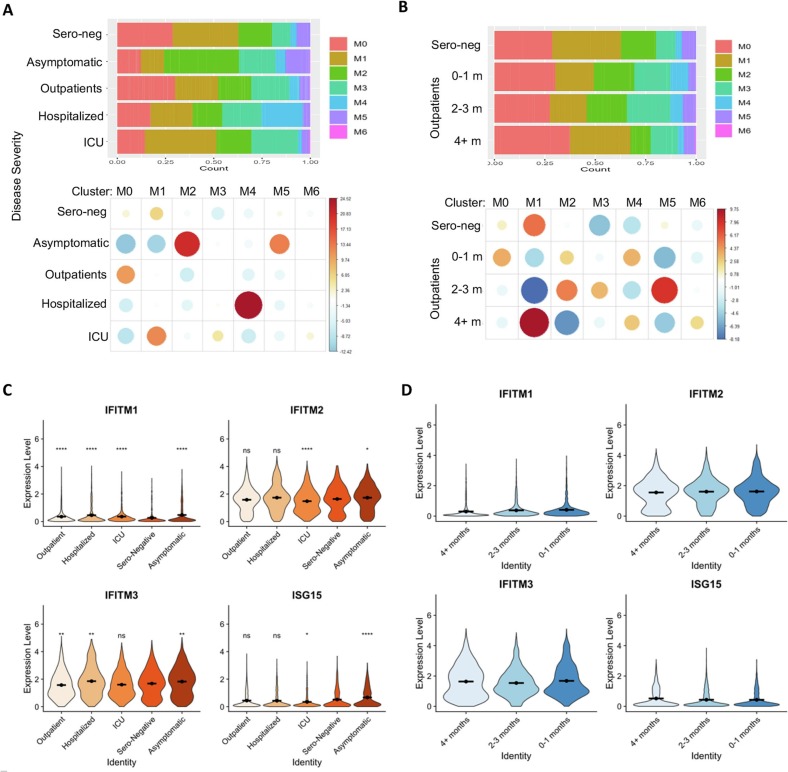
Importantly, these monocyte clusters with distinct molecular signatures were found to be associated with COVID-19 severity and/or time from the onset (Fig. 3A-B, Supplementary Fig. 3 and Table 3A). In the convalescent patients with asymptomatic COVID-19 but detectable RBD-IgG, M0_HLA-B_EEF2 cluster and S100 family gene-high cluster M1-S100A8_S100A9 were underrepresented but clusters M2-CD74_HLA-DR with high immune regulatory receptor CD74 levels and M5_ISG15_IFI6 with high IFN signature genes were enriched in these patients (Fig. 3A, Supplementary Fig. 3). Instead, both M2 and M5 clusters were relatively lost in other COVID-19 patients (Fig. 3A, Supplementary Fig. 3). Of note, among outpatients, while Cluster M0_HLA-B_EEF2_S100A8/A9 remained more abundant than other groups over 4 months, the dynamic changes in monocyte clusters M1-M3 appeared to be temporary for 2–3 months and partially restored after 4 months from the onset (Fig. 3B, Supplementary Fig. 3), reminiscent of the dynamic changes in plasma RBD-IgG levels.
Fig. 3.
Association of monocytes' signatures with COVID-19 disease severity and recovery.
(A) Proportion of monocyte cells from each disease severity category in each cluster. Cluster M5 (interferon-stimulated monocytes) appears enriched in asymptomatic patients, while cluster M4 (nonclassical monocytes) appears enriched in hospitalized patients.
(B) Proportion of monocytes in outpatients and sero-negative controls separated by time. Changes in monocyte proportions in these patients are largely not sensitive to time since disease.
(C) Interferon stimulated gene expression in cells separated by disease severity. Both genes appear to be most highly expressed among asymptomatic patients.
(D) Expression of interferon stimulated genes in cells derived from outpatients and sero-negative controls only separated by time from disease. These genes show very little change over time.
C-D: Significance of gene expression difference in individual groups evaluated by Wilcoxon comparison of group-cell expression levels with Sero-Negative-cell expression levels. Significance values: ns: p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Along with changes in the classical monocyte populations, the non-classical cluster M4_IFITM_FCGR3A appeared to be enriched specifically in recovered hospitalized patients over 4 months from the onset (Fig. 3A, Supplementary Fig. 3). Interestingly, unlike the classical monocyte cluster M5_ISG15_IFI6, the non-classical monocytes in Cluster M4_IFITM_FCGR3A/CD16 express distinct interferon-induced genes IFITM1–3. In contrast, these gene expression levels, in particular IFITM3, in outpatients were significantly reduced (Figs. 3C). Further longitudinal analysis shows the reduction in IFNTM3 levels sustained for >4 months in outpatients after COVID-19 recovery (Fig. 3D). The IFITM proteins have been identified as cell-autonomous proteins that suppress the early stages of viral replication [25], our data suggest that the increase of this non-classical cluster might be responsible for the severe COVID-19 pathogenesis.
Examining the individual gene expression patterns, we found that ISG15 was mostly depleted in the monocytes of ICU patients (Fig. 3C-D, Supplementary Fig. 3 and Table 2). It has been well established that the impairments in the IFN response is the key critical for anti-SARS-CoV-2 immune response [34,53], our data imply a possibility that the patients with a better expansion of monocytes in M5_ISG15_IFI6, as well as cluster M2_CD74_HLA-DR, have a favorable clinical outcome. Consistent with this notion, further analysis revealed a trend in reduction of IFN-responsive monocytes during the early recovery phase of outpatients (2–3 months), which gradually returned 4 months after acute infection (Fig. 3D). More importantly, while results from our trajectory analysis show that some inflammatory responsive genes such as S100 family genes A6, A8 and A9 gradually reduced back after disease recovery, surprisingly, multiple HLA family members and IFN-responsive genes show a trend of further increase along the time after disease recovery (Supplementary Fig. 2), implying a long-term impact of SARS-CoV2 infection to innate immune response.
To further validate our conclusion, we reanalyzed our sc-RNA seq data using the SCT/integrate workflow form Seurat, which yielded 10 monocyte clusters (m0-m9) (Supplementary Fig. 4A). Consistently, monocytes in three clusters, m0, m1 and m4, show high expression of S100 family genes including S100A8, S100A12 and S100A9. Clusters m3 m5 & m7 show uniquely high expression of HLA family genes, two of which, m3 and m7 are non-classical monocytes with high expression of FCGR3A as well as a group of IFN-inducible genes (Supplementary Fig. 4B & C). In addition, m6 monocytes are classical regulatory monocytes that maintain elevated expression of CD74 and IFN-inducible genes. Importantly, both analyses confirmed that the S100 family gene-high cluster M1 and m1 reduced but the immune regulatory receptor CD74-high and HLA-high cluster M2 and m3 enriched in the convalescent patients with asymptomatic COVID-19 vs other patient groups (Supplementary Fig. 4D & E, Supplementary Table 3B), confirming our initial conclusion that the dynamic changes of unique monocyte clusters with disease severity even in convalescent COVID-19 patients.
Circulating, classical monocytes are known to have a short lifespan of no more than a few days, while non-classical monocytes display longer survival, but still are unlikely to live for >1 or 2 weeks [31]. One possibility is that some, if not all of the monocyte molecular signatures are maintained by the altered circulating cytokines. We then utilized a multiple plex approach and analyzed the levels of total 20 inflammatory and anti-inflammatory cytokines in the circulating blood in parallel. As expected, the majority of cytokines analyzed are comparable between sero-negative and convalescent COVID-19 patients regardless their disease severities (Supplementary Fig. 5A). Interestingly, we detected a statistically significant increase in IP-10 and VEGF-A levels in ICU group (Supplementary Fig. 5B-C). High levels of both IP-10 and VEGF-A have been detected in critically ill COVID-19 patients during their active phase of the disease [10,23]. In addition, a high of IFN-β level was detected in hospitalized group of convalescent COVID-19 patients. However, this high level of IFN-β was not statistically significant in ICU patients, which is likely due the small sample size in this group (Supplementary Fig. 5D). Analysis of cytokine profiles using sPLS-DA does not appear to produce clear discrimination between disease severity among the analyzed cohort (Supplementary Fig. 5E & F). Nevertheless, these data imply that only few circulating cytokines involved in virial infection and tissue repairment remain elevated in convalescent COVID-19 patients with severe disease. One of the important questions is what are the cells that produce these cytokines to maintain high levels in circulating blood even several months after disease recovery. Interestingly, analysis of the expression levels of these VEGFA, IP10 and IFN-b in the sc-RNA seq data revealed a significant increase in VEGFA in monocytes, but not Tregs from ICU groups in comparison to Outpatient and Asymptomatic groups. In contract, IP-10 was comparable among the disease groups (Supplementary Fig. 5G & H), indicating that VEGFA produced by monocytes contributes, at least partially, to its increase in blood.
Collectively, the monocytes we observed in our analysis likely developed following clearance of SARS-CoV-2 and the restoration of some degree of immune homeostasis in these patients. In that context, our data demonstrated the long-term changes in monocyte phenotypes in patients recovered from COVID-19 is surprising, which suggests that acute SARS-CoV-2 infection induces immune system alteration even after elimination of replicating virus.
2.3. scRNA-Seq identifies unique Treg clusters and long lasting Treg cell signatures in patients recovered from COVID-19
Our discovery that the frequency of FoxP3+ Tregs is increased in the circulating blood from recovered COVID-19 outpatients but decreased in hospitalized patients (Fig. 1F), suggests that dynamic changes in circulating Treg frequency may impact both acute COVID-19 pathogenesis and long-term complications. We sorted the 7178 cells with CD45+CD3+CD4+CD25highCD127low expression, which are known to sufficiently define human Tregs from PBMC [55], from 27 convalescent COVID-19 patients and sero-negative controls (Supplemental Table 4 for further information), and subjected them to the 10× Genomics single cell sequencing platform. Similar to that in monocytes, the alignment analysis did not detect any viral genes in Tregs (Fig. 1A). Clustering analysis identified 10 clusters with 7 clusters T0–6 with the majority of Treg populations (Fig. 4A-C). indicating that Tregs in the circulating blood from both COVID-19 patients and healthy donors are highly heterogeneous.
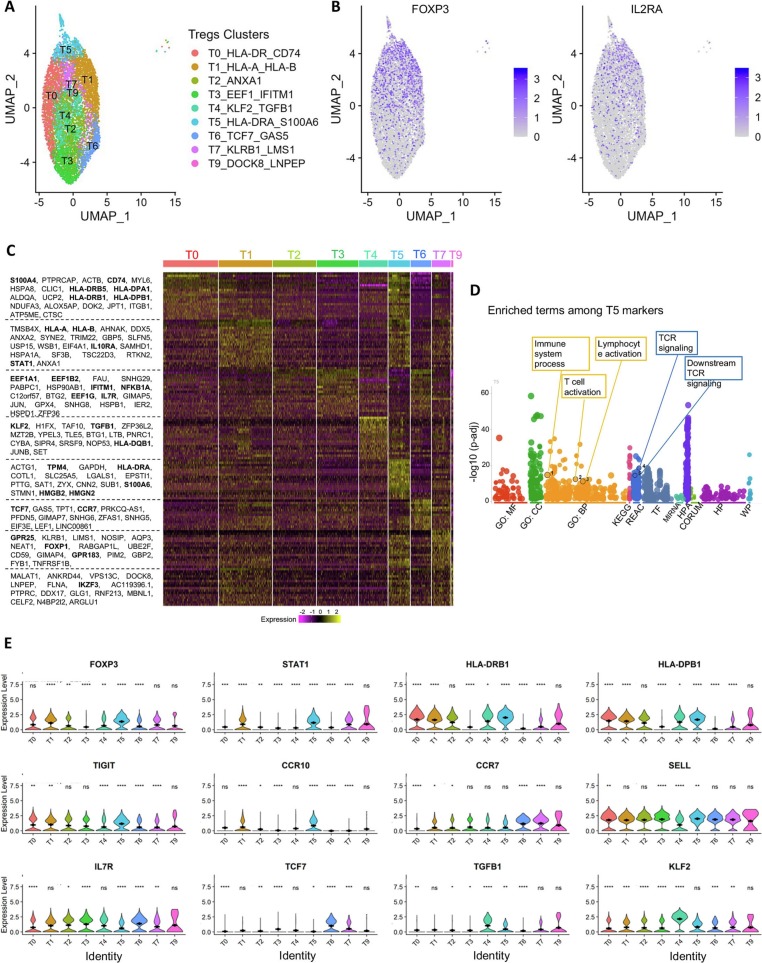
Fig. 4.
Analysis of the sustained Treg molecular signatures from convalescent COVID-19 patients.
(A) Dimensional UMAP plot of Tregs from convalescent COVID-19 patients and sero-negative controls.
(B) Expression of FOXP3 and CD25/IL2RA on the UMAP projection from A.
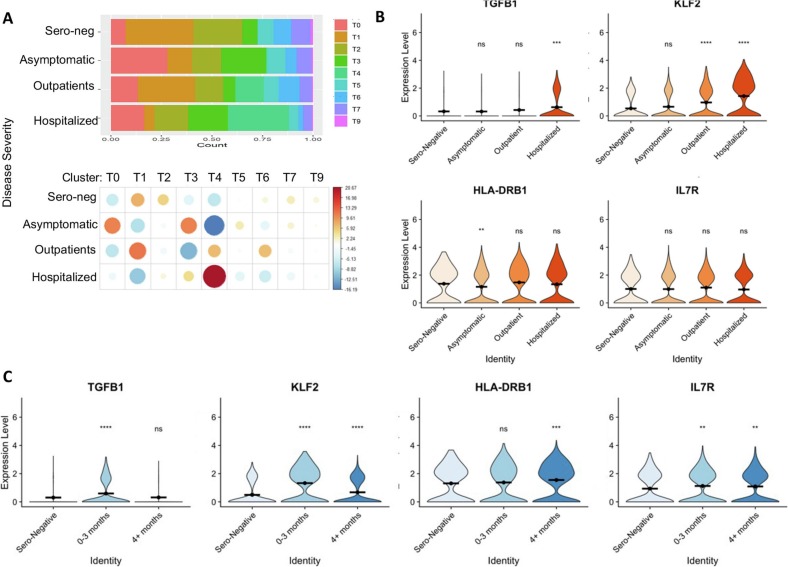
(C) Expression of selected genes across clusters. FOXP3 and STAT1 are strongly expressed in clusters T1, T5 and T7, indicating effector cells. Clusters T2, T3, T6 and T9 show lower levels of FOXP3 and STAT1 but higher levels of SELL and IL7R, indicating central/homeostatic Treg cells. Cluster T5 expresses high levels of CCR10, CCR7 and FOXP3, potentially indicating a tissue-migratory effector Treg phenotype. Cluster T4 is unique to patients recovering from COVID-19 and expresses high levels of TGFB1 and KLF2.
(D) Heatmap showing the expression levels of selected genes across all cells.
Significance of gene expression difference in individual clusters evaluated by Wilcoxon comparison of cluster-cell expression levels with all-cell expression levels. Significance values: ns: p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Further annotation of gene expression of the individual clusters identified related signatures between clusters, and each cluster is defined by a few top enriched genes (Fig. 4C). Treg clusters T1_HLA-A_HLA-B and T5_HLA-DRA_S100A6 displayed high expression of FOXP3 and STAT1, which are consistent with an activated/effector Treg phenotype. To support this designation, clusters T1 and T5 also express high levels of HLA class II molecules and TIGIT (Fig. 4C), which is thought to define a unique Treg subset that preferentially suppresses Th1 and Th17-driven inflammatory responses [22]. Tregs in T1_HLA-A_HLA-B and T5_HLA-DRA_S100A6 also show a tissue migratory Treg-signature with high expresses levels of CCR10, CCR7, and FOXP1. In contrast, Tregs in clusters T2_ANXA1 and T3_EEF1_IFITM1, have reduced expression of TIGIT and class two HLA molecules. In addition, the cluster T6_TCF7_GAS5 shows lower levels of FOXP3 and CD25, as well as HLA family genes, but displays higher levels of central/homeostatic Treg markers including TCF7, SELL and IL-7R. Cluster T7_KLRB1_LMS1 also showed increased expression of CCR7, TCF7 and KLF2 consistent with a less suppressive Treg phenotype. Interestingly, a Treg population cluster T4_KLF2_TGFB1 expressed elevated expression of TGF-β and KLF2 In addition to these 7 clusters with unique gene expression signatures, Tregs in cluster 0, T0_HLA-DR_CD74, share a largely common gene expression profile with all the rest clusters, implying a possibility that Tregs in this cluster are an intermediate population. Tregs in cluster 9, T9_DOCK8_LNPEP, which are rather minor, show reduced expression of genes enriched in cluster T3_EEF1_IFITM1 including EEF family and IFN responsive genes (Fig. 4C-E).
GO pathways analysis revealed that the top differentially expressed genes in Treg clusters, such as T5, are involved in T cell immunity including immune system processing, T cell activation and TCR signaling regulation, indicating a sustained Treg activation response even after recovery from COVID-19 (Fig. 4D, Supplementary Fig. 6A). Consistently, trajectory analysis revealed that a group of genes involved in cell proliferation, such as CCNA2, CCNB2, CDC20, EEF1A1 and EEF1B1 show a trend of increase along the time (Supplementary Fig. 6B & C), indicating a sustained Treg expansion induced by SARS-COV2 infection after COVID-19 recovery. In addition, similar to what we observed in monocytes, multiple HLA-associated genes including HLA-A and B2M, and inflammatory genes, IL-32 and show a trend of further increase along the time after disease recovery (Supplementary Fig. 6C), imply a long-term impact of SARS-CoV2 infection to Treg functions. Pseudotime analyses of Tregs using KLF2 as a rooting gene indicate alterations in many genes may contribute to the dynamics and differentiation of these cells (Supplementary Fig. 6B-C).
Similar to monocytes in convalescent COVID-19 patients, the relative proportions of the identified Treg clusters show differences that are correlated with initial disease severity (Fig. 5 , Supplementary Table 5A). Comparing to sero-negative control and convalescent COVID-19 patients with milder disease, the hospitalized patients had a dramatic reduction in the population of effector/activated Tregs (clusters T1_HLA-A_HLA-B and T5_HLA-DRA_S100A6), which are known important for protecting patients from tissue injury during respiratory virus infections including SARS-CoV-2 [50]. In contrast, the hospitalized patients had a relative expansion in TGF-β high cluster T4_KLF2_TGFB1 Tregs (Fig. 5A & B). Interestingly, patients recovered from an asymptomatic infection showed a unique expansion of cluster T0_HLA-DR_CD74 relative to any of the severity groups. This cluster demonstrates the highest expression of class II HLA molecules (Fig. 4C and 5A, Supplementary Table 5A), which has previously been suggested to mark a specific highly suppressive Treg subset that primarily inhibits CD4+ helper T responses through contact-mediated suppression [4]. In addition, our integrative analysis of single-cell Treg transcriptomes shows increased Cluster T0-HLA-DR_CD74 and Cluster T3-EEF1_IFITM1, but decreased Cluster T6-TCF7_GAS5, in asymptomatic and hospitalized patients. In contrast, Tregs in Clusters T2-ANXA1 and T7-KLRB1-LMS1 are decreased in all COVID groups (Fig. 5A). Cluster T4_KLF2_TGFB1 was only identified in a subset of convalescent COVID-19 patients, and not identified in sero-negative patients (Fig. 5A, Supplementary Table 5A). Further analysis showed that both TGF-β1 and IL-7R upregulation occurred during the early COVID-19 recovery phase (0–3 month), which gradually returned to normal during the later recovery phase whereas KLF2 increased and sustained over time in the Tregs from COVID-19 patients (Fig. 5C). The clinical significance of this cluster T4_KLF2_TGFB1 population is unclear. However, as Treg-specific production of TGF-β is known to have a variety of immunologic roles, the fact that this population is only observed in patients recovering from COVID-19 suggests a possible role in COVID-19 pathogenesis and recovery. Importantly, because this cluster was only identified in patients who had recovered from SARS-CoV-2 infection, these cells may identify a distinct SARS-CoV-2-associated immunological profile.
Fig. 5.
Association of Treg molecular signatures with COVID-19 disease severity and recovery.
(A) Proportion of treg cells from each patient type in each cluster. Cluster T4 is only present among symptomatic COVID patients, not in asymptomatic or sero-negative patients.
(B) Expression level of genes characteristic of cluster 4 separated by disease severity. KLF2 and TGFB1 appear particularly high in the most severe patients.
(C) Expression of cluster T4 characteristic genes across time in outpatients and sero-negative controls. TGFB and KLF2 are most highly expressed 0–3 months post disease.
(D) Cluster T5 (circulating Tregs) appears to decrease temporarily after COVID-19 illness in outpatients before recovering after 4+ months. Cluster T0 shows no changes after COVID-19 illness.
B-C: Significance of gene expression difference in individual groups evaluated by Wilcoxon comparison of group-cell expression levels with Sero-Negative-cell expression levels. D: Boxplots indicate quartiles. Individual comparisons made using Wilcoxon signed rank test. Significance values: ns: p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
We also noticed a significant decrease in Tregs of cluster T5_HLA-DRA_S100A6, which shows lower levels of FOXP3 in hospitalized patients, which is possibly due to the inflammation-induced downregulation of FOXP3 (Fig. 5A). Importantly, the frequency of effector/activated Tregs with high expression of FOXP3 and STAT1 in cluster T5_HLA-DRA_S100A6 were dramatically reduced during the early recovery phase in COVID-19 outpatients, which returned to a comparable level similar to that in sero-negative controls 4 months after disease recovery. As expected, the intermediate Cluster T0_HLA-DR_CD74, which occupy one of the major Treg populations, did not show significant changes (Fig. 5D).
Further reanalysis of the Treg sc-Seq data using the SCT/integrate workflow form Seurat confirmed the HLA and CD74 high t0 cluster as one of the major Treg population. Similar to T6 clusters, Tregs in t3 cluster display higher levels of central/homeostatic Treg markers including TCF7 and IL-7R, and with decreased expression of HLA family genes and reduced inflammatory response. In contrast, Tregs in cluster t4, similar to that in T5, appear to be effector/activated Tregs with high expression of FOXP3, HLAs, STAT1 as well as cell proliferative genes (Supplementary Fig. 7–8, Supplementary Table 5B). When the frequencies of monocyte and Tregs were examined based on both severity and time from onset, most of the changes observed in patients recovered from severe disease show a long period of immune memories.
Collectively, our study identified a dynamic association of frequencies in clusters of monocytes and Tregs and their immune regulatory and tissue injury protective molecular signatures with disease severity identified in convalescent COVID-19 patients.
3. Discussion
Our study enrolled 100 convalescent patients with different disease severities including asymptomatic, outpatient, hospitalized and ICU as well as healthy controls. All patients were enrolled at least three weeks after recovery from COVID-19 confirmed by negative in nucleic acid tests and complete disappearance in clinical symptoms. Their history with SARS-CoV-2 infection was all confirmed by the presence of abundant RBD-specific IgG in their circulating blood. We focused on the analysis of the monocytes, an innate immune cell population that are largely short-lived in the circulating blood, and the understudied regulatory T cells for dissecting the experienced immune responses of COVID-19 patients at the single-cell resolution.
Surprisingly, flow cytometry analysis revealed that the frequency of the short-lived monocyte and Treg populations display a sustained and dynamic association with disease severity in convalescent COVID-19 patients. Consistent with their dynamic frequency changes, we discovered several COVID-19-associated immune signatures such as the elevated IFN responsive genes in asymptomatic COVID-19 patients, which appears to be downregulated in patients with severe disease in particular the ICU patients. Many studies through single cell transcriptome analysis of PBMCs obtained during the active phase of the disease, have shown that the impairments in the IFN response is the key mediators of severe disease [34,53]. Our study here confirms that this dynamic association of IFN response with COVID-19 disease severity is sustained after full recovery. Interestingly, further longitudinal analysis showed that the elevated IFN responsive genes in monocytes could be sustained for >4 months of the studied time. In addition to IFN response, the altered HLA expression on myeloid cells during active SARS-CoV-2 infection, which is presumably due to the virus-induced inflammatory cytokines, have been identified [16,51]. Our study here demonstrated that this HLA high signature is also sustained after COVID-19 recovery. Importantly, pseudo-time analysis revealed that the expression both IFN-responsive genes and HLA family members show an increase trend along the time after disease recovery. As the current study collected PBMCs in convalescent COVID-19 patients up to 4 months after disease recovery, it will be interesting to define a time point when the expression of IFN-responsive genes and HLA family members return back to a normal level.
During the convalescent phase following COVID-19 infection, the immune system consists of two distinct pools of cells, COVID-19 experienced, such as memory T and B cells specific to SARS-CoV-2 antigens, and COVID-19 unexperienced cells. It has been well documented that the half-life of classical and non-classical monocytes in both mice and human is estimated at half- to 2.2 days [30,32,44]. Therefore, in convalescent patients, the circulating monocytes and granulocytes are presumably unexperienced directly to SARS-CoV-2-specific antigens as well as the associated inflammatory environment during the acute phase of infection. In addition, our data suggest that the sustained COVID-19 molecular signatures are likely independent of the inflammatory cytokines in the circulating blood, because their levels are largely indistinguishable in convalescent phase of the patients from healthy controls. Therefore, it is reasonable to speculate that the COVID-19-associated molecular signatures are largely inherited from their precursors and/or even the hematopoietic stem cells experienced with SARS-CoV-2 infection. Indeed, a recent study has suggested that the CD34+ hematopoietic stem/progenitor cells were primed toward megakaryopoiesis, accompanied by expanded megakaryocyte-committed progenitors and increased platelet activation by SARS-CoV-2 infection [43].
Tregs cells are known to modulate broad aspects of both innate and adaptive immune responses to maintain immune tolerance and global immunosuppression and consequently protect the host from autoimmunity [14]. It has been well established that Tregs play a critical role in protecting tissue injury during microbial infections including influenza viruses and SARS-CoV-2 [2,49]. Consistent with those observations note, our study here demonstrated that compared to sero-negative healthy controls, the Treg cell frequency was increased in COVID-19 patients with milder disease severity and this increase was not observed in hospitalized and ICU patients. As expected, our in-depth single cell transcriptome analysis of the sorted Treg populations identified 10 distinct clusters, confirming that human circulating Treg populations are highly heterogenous regardless with or without microbial infection [12,19,39,40,52]. Interestingly, a Treg population (cluster T4_KLF2_TGFB1) that was characterized by elevated expression of TGF-β and KLF2, was only identified in some COVID-19 patients in particular the ICU patients, but not in sero-negative healthy controls. Further longitudinal analysis shows that the upregulation of both TGF-β and KLF2 sustained only during the first three months after full recovery, which was not observed in these convalescent COVID-19 patients 4 months after their full recovery. The clinical significance of this TGF-β and KLF2 high population is unclear. Since Treg-specific production of TGF-β is critical for a variety of immunologic roles, the fact that this population is only observed in patients recovering from COVID-19 suggests a possible role in COVID-19 pathogenesis and recovery. Consistent to our observation, it has been recently observed that SARS-CoV-2 in severe COVID-19 patients induces TGF-β expression in plasma blasts and possibly involved in IgA production for improving the lung mucosal immunity [17]. A recent interesting study observed that Tregs from patients with severe disease produce a substantial amount of interleukin (IL)-6 and IL-18 [18], which was not observed in COVID-19 patients in their recovery phase. Tregs are often depolarized or instable in the inflammatory environment, which consequently triggers a Treg expansion to maintain the immune homeostasis [26]. Interestingly, our trajectory study demonstrated a trend of sustained increase in a group of genes involved in cell proliferation and gene transcription along the time after disease recovery. It will be interesting to study how Treg frequencies and their COVID-19-associated molecular signatures dynamically change from active infection to recovery phase.
4. Methods
4.1. Human subject study and biosafety approvals for blood draws
All research activities with human blood specimens of pre-COVID-19, sero-negative (healthy) donors and convalescent COVID-19 patients were implemented under NIH guidelines for human subject studies and the protocols approved by the Northwestern University Institutional Review Board (STU00205299) as well as the Institutional Biosafety Committee for COVID-19 research. For collecting human blood specimens, patients and donors were recruited at Northwestern Memorial Hospital based on their availability and willingness to consent and participate in the research. Participants were compensated $20.00 and the blood was collected prior to COVID-19 vaccination.
All convalescent COVID-19 patients enrolled in our study meet the following criteria: testing positive for SARS-CoV-2, at least three weeks following recovery; some patients were also enrolled if they had a positive RBD-specific IgG in a community screening study. 20 mL of blood was drawn into EDTA tubes and transported on ice blocks to the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility at Northwestern University.
4.2. Analysis of sera RBD-specific IgG levels in the plasma and its neutralization capacity
Plasma was centrifuged at 1200 xg for 10 min at 4 °C (brake off) after blood samples were diluted 1:1 with PBS during B cell isolation using RosetteSep Human B-Cell Enrichment Cocktail (Stem Cell Technologies). SARS-CoV-2 RBD-specific IgG ELISA tests with the plasma were performed in the lab of Dr. Alexis Demonbreun using enzyme-linked immunoassay (ELISA) as previously described [1]. In brief, plasma was run in one or two duplicates and reported as the average. Results were normalized to the CR3022 antibody with known affinity to the receptor binding domain of SARS-CoV2 (Creative Biolabs, MRO-1214LC) [27,56]. Anti-RBD IgG concentration (μg/mL) was calculated from the 4PL regression of the CR3022 calibration curve.
Neutralization assays with 10 μL or 80 μL of plasma were performed using HEK293 cells with stable expression of human ACE2. Commercially available SARS-CoV-2 spike receptor binding domain (RBD) (Raybiotech catalog no. 230–20,407) was biotinylated using EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific catalog no. 21335) and desalted with a Zeba quick spin column (Thermo Scientific catalog no. 89877). 5 μg biotinylated RBD was pre-coupled with 0.6 ul Streptavidin-AF647 (Invitrogen catalog no. S21374), used as RBD-647. 0.5 μL RBD-647 was then pre-incubated with PBS as a control or 80 μL patient-derived plasma for 45 min on ice, after which it was combined with 106 HEK/ACE2 cells. These cells were incubated on ice for 45 min, washed and stained with DAPI. The levels of RBD-647 binding to HEK293/ACE2 cells were then assessed BD FACSAria Cell Sorter and analyzed using FlowJo v10software.
4.3. Flow cytometry analysis of PBMCs and cell sorting of monocytes and Tregs
Total white blood cells from 1 to 2 mL freshly collected blood in EDTA tubes were enriched through ammonium chloride lysis (BD Bioscience, catalog no. 555899), followed by centrifugation (300 ×g for 10 min). Supernatant was discarded, cells were washed twice using MACS buffer (Miltenyi, catalog no. 130–091-221); and pellet resuspended in 200uls MACS + Fc-block (ThermoFisher, catalog no. 14–9161-73). Cells were stained for surface antigens using fluorescence-conjugated monoclonal Abs specific to human CD45, CD3, CD4, CD127 and CD25 antibodies for Treg identification and sorting as well as CD45, CD3, CD64, CD14 and CD16 for monocyte characterization and sorting. Cells were also incubated with a patient-specific hashtag oligo (Biolegend TotalSeq-C0251–260 hashtag antibodies) as per manufacturer instruction. Post staining for 30 min on ice, cells were washed with MACS buffer and pellet resuspended in 1 mL MACS buffer for sorting. Sorting was carried out on BD FACSAria SORP cell sorter housed in an independent negative pressure lab space in Baker BioProtect IV Biological Safety Cabinet. Sorted cell fractions were collected in MACS buffer and processed immediately for 10× library preparation. All of the sample preparation and cell sorting were carried out using BSL-2+ practices per Institutional Biosafety Committee approved protocols. Monocytes and Treg analyses with respective gating strategies are shown in Fig. 1B. Detailed information of all fluorescent-conjugated monoclonal Abs used for this study are shown in supplemental Table 5.
4.4. 10× library preparation
The concentration and viability of the single cell suspension were measured using a Nexcelom Cellometer Auto 2000 with ViaStain AOPI staining solution (Nexcelom, CS2–0106). Cells were loaded onto a 10× Chromium Controller for GEM generation followed by single cell library construction using 10× Chromium Next GEM Single Cell 5’ Library and gel bead kit v1.1 (10× Genomics, PN-1000165) according to manufacturer's protocol. Quality control of the libraries was performed using an Invitrogen Qubit DNA high sensitivity kit (ThermoFisher Scientific, Q32851) and Agilent Bioanalyzer high sensitivity DNA kit (Agilent, 5067–4626). The libraries were pooled in equal molar ratio and sequenced on an Illumina HiSeq 4000 using sequencing parameters indicated by the manufacturer (Read 1: 26 cycles, i7 index: 8 cycles; Read 2: 91 cycles).
4.5. scRNA-seq data acquisition and analysis
Data from scRNA-seq was demultiplexed and mapped to hg38 (refdata-gex-GRCh38–2020-A) using Cell Ranger software version 4.0.0 (10× genomics). Matrix files were analyzed using the Seurat R package (Seurat v4.0.3, R verson 4.1.0) [21]. Libraries were loaded individually and log-normalized to correct for batch effects. Cells with mitochondrial reads >5% of total and fewer than 200 mRNAs were filtered out. Patient-specific hashtags were demultiplexed from each library. Alignment was done using the 10× genomics CellRanger software with either human or SARS-CoV2 genomes. Libraries were then merged together and scaled across all samples. Monocytes and Treg clusters were identified by expression of CD14, CD16, FOXP3 and CD25 (Supplemental Fig. S9). Monocyte and Treg clusters were split from one another and analyzed separately. Principal component analysis was performed on each data set, and Uniform Manifold Approximation and Projections (UMAP) were made from the top 10 principal components. Positive markers for each cluster were identified with a minimum log-FC threshold of 0.25. Statistically significant markers with the highest log-FC were used in the creation of heatmaps. Pseudotime analysis created using monocle3 [9,33,42,48]. The single cell RNA sequences are deposited in the GEO repository (GSE232523).
4.6. Multiplex analysis of cytokines in huma sera/plasma
Patient plasma samples were isolated from blood samples during cellular analysis and stored at -80C. After all of the patient samples for the study had been collected, plasma samples were analyzed using a U-PLEX Viral Combo 1 Human cytokine array (Meso Scale Diagnositics Cat No. K15343K-1). The cytokine array plate was read at the Northwestern University Immunotherapy Assessment Core.
4.7. Statistical analyses
Kruskal-Wallis tests were used to identify overall significant differences at the population level. Wilcoxon signed-rank tests were used to identify significant differences between a sub-group and the overall population (Figs. 2C and 4C) or to compare individual subgroups against one another (Fig. 1, Fig. 3, Fig. 5). All statistics were computed on R v 4.1.0. Correlation plots were created using R package corrplot (Wei T, Simko V (2021). R package ‘corrplot’: Visualization of a Correlation Matrix. (Version 0.92), https://github.com/taiyun/corrplot.). GO analysis and Manhattan plots created using R package gprofiler2 (Uku Raudvere, Liis Kolberg, Ivan Kuzmin, Tambet Arak, Priit Adler, Hedi Peterson, Jaak Vilo: g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) [35].
Author contributions statement
A.D·H, and S.E.W. designed and led the bench experiments, analyzed data, prepared figures, and contributed to writing. S.S., S.C., H.F.M, M.J.S., C.M., X.W., L.E., N.K.D., J.W., P.J.M., L.J.S., C.M.W., C.O., Y.J., P.D., and N.R.W. provided technical support or conducted bench experiments, and analyzed data. Y.L., A.R.D., and M.G.I. provided critical resources and supervised the research project. D.F., and H.L designed experiments, analyzed data, wrote the manuscript, and supervised the work.
Boxplots in B-E indicate quartiles. Comparison p-values calculated using Wilcoxon signed rank test.
Declaration of Competing Interest
Northwestern University and H. L., D. F., L. E., A. D. H., and N. K. D. hold issued and/or provisional patents in the area of exosome therapeutics and COVID-19 therapeutics. H.L., D. F., and A.D·H are scientific co-founders in ExoMira Medicine Inc.
Acknowledgments
We are thankful to the team of Northwestern COVID-19 Antibody and Cancer Collaborative Group and advisory members, especially Drs. Alfred L. George Jr., Richard D'Aquila, Leonidas C. Platanias, Rex L. Chishom, and William A. Muller for their scientific input and resourceful support for the project. The work was partially funded by The National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) R01AI167272 and Chicago Biomedical Consortium Accelerator Award A-017 (H.L. and D.F.), Northwestern University Feinberg School of Medicine Emerging and Re-emerging Pathogens Program (EREPP) (H.L.), Department of Pharmacology Start-up fund (H.L.), and the R.H. Lurie Comprehensive Cancer Center Blood Biobank fund and Northwestern University Clinical & Translational Sciences Institute (NUCATS) grant UL1TR001422 (M.I.). We gratefully acknowledge the support from the NUseq Core Facility. This work was supported by the Northwestern University RHLCCC Flow Cytometry Facility and a Cancer Center Support Grant National Cancer Institute (NCI) (CA060553). Flow Cytometry Cell Sorting was performed on a BD FACSAria SORP system and BD FACSymphony S6 SORP system, purchased through the support of NIH 1S10OD011996-01 and 1S10OD026814-01.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clim.2023.109634.
Appendix A. Supplementary data
1. Supplemental Figures 1-9 and legends; 2. Supplemental Tables 1-6
Data availability
The single cell RNA sequences are deposited in the GEO repository (GSE232523).
References
- 1.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpaia N., Green J.A., Moltedo B., Arvey A., Hemmers S., Yuan S., Treuting P.M., Rudensky A.Y. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162:1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baecher-Allan C., Wolf E., Hafler D.A. MHC class II expression identifies functionally distinct human regulatory T cells. J. Immunol. 2006;176:4622–4631. doi: 10.4049/jimmunol.176.8.4622. [DOI] [PubMed] [Google Scholar]
- 5.Bergamaschi L., Mescia F., Turner L., Hanson A.L., Kotagiri P., Dunmore B.J., Ruffieux H., De Sa A., Huhn O., Morgan M.D., et al. Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–1275.e1258. doi: 10.1016/j.immuni.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e1412. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyton R.J., Altmann D.M. Risk of SARS-CoV-2 reinfection after natural infection. Lancet. 2021;397:1161–1163. doi: 10.1016/S0140-6736(21)00662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callard F., Perego E. How and why patients made long Covid. Soc. Sci. Med. 2021;268 doi: 10.1016/j.socscimed.2020.113426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Wang J., Liu C., Su L., Zhang D., Fan J., Yang Y., Xiao M., Xie J., Xu Y., et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 12.Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., Sirven P., Magagna I., Fuhrmann L., Bernard C., et al. Fibroblast heterogeneity and immunosuppressive environment in human breast Cancer. Cancer Cell. 2018;33:463–479.e410. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re’em Y., Redfield S., Austin J.P., Akrami A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejaco C., Duftner C., Grubeck-Loebenstein B., Schirmer M. Imbalance of regulatory T cells in human autoimmune diseases. Immunology. 2006;117:289–300. doi: 10.1111/j.1365-2567.2005.02317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demonbreun A.R., McDade T.W., Pesce L., Vaught L.A., Reiser N.L., Bogdanovic E., Velez M.P., Hsieh R.R., Simons L.M., Saber R., et al. Patterns and persistence of SARS-CoV-2 IgG antibodies in Chicago to monitor COVID-19 exposure. JCI. Insight. 2021;6 doi: 10.1172/jci.insight.146148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evren E., Ringqvist E., Tripathi K.P., Sleiers N., Rives I.C., Alisjahbana A., Gao Y., Sarhan D., Halle T., Sorini C., et al. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity. 2021;54:259–275.e257. doi: 10.1016/j.immuni.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira-Gomes M., Kruglov A., Durek P., Heinrich F., Tizian C., Heinz G.A., Pascual-Reguant A., Du W., Mothes R., Fan C., et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat. Commun. 2021;12:1961. doi: 10.1038/s41467-021-22210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galván-Peña S., Leon J., Chowdhary K., Michelson D.A., Vijaykumar B., Yang L., Magnuson A.M., Chen F., Manickas-Hill Z., Piechocka-Trocha A., et al. Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2111315118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giganti G., Atif M., Mohseni Y., Mastronicola D., Grageda N., Povoleri G.A., Miyara M., Scottà C. Treg cell therapy: how cell heterogeneity can make the difference. Eur. J. Immunol. 2021;51:39–55. doi: 10.1002/eji.201948131. [DOI] [PubMed] [Google Scholar]
- 20.Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., Abbott D.A., Donnelly H.K., Donayre A., Goldberg I.A., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e3529. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joller N., Lozano E., Burkett P.R., Patel B., Xiao S., Zhu C., Xia J., Tan T.G., Sefik E., Yajnik V., et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40:569–581. doi: 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong Y., Han J., Wu X., Zeng H., Liu J., Zhang H. VEGF-D: a novel biomarker for detection of COVID-19 progression. Crit. Care. 2020;24:373. doi: 10.1186/s13054-020-03079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer K.J., Wilfong E.M., Voss K., Barone S.M., Shiakolas A.R., Raju N., Roe C.E., Suryadevara N., Walker L., Wall S.C., et al. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat Commun. 2022;13:3466. doi: 10.1038/s41467-022-31142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucca L.E., Dominguez-Villar M. Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat. Rev. Immunol. 2020;20:680–693. doi: 10.1038/s41577-020-0296-3. [DOI] [PubMed] [Google Scholar]
- 27.McDade T.W., McNally E.M., Zelikovich A.S., D’Aquila R., Mustanski B., Miller A., Vaught L.A., Reiser N.L., Bogdanovic E., Fallon K.S., et al. High seroprevalence for SARS-CoV-2 among household members of essential workers detected using a dried blood spot assay. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183:1340–1353.e1316. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., Katsyv I., Rendeiro A.F., Amin A.D., Schapiro D., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohri H., Perelson A.S., Tung K., Ribeiro R.M., Ramratnam B., Markowitz M., Kost R., Hurley A., Weinberger L., Cesar D., et al. Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med. 2001;194:1277–1287. doi: 10.1084/jem.194.9.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel A.A., Ginhoux F., Yona S. Monocytes, macrophages, dendritic cells and neutrophils: an update on lifespan kinetics in health and disease. Immunology. 2021;163:250–261. doi: 10.1111/imm.13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel A.A., Zhang Y., Fullerton J.N., Boelen L., Rongvaux A., Maini A.A., Bigley V., Flavell R.A., Gilroy D.W., Asquith B., et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randolph H.E., Fiege J.K., Thielen B.K., Mickelson C.K., Shiratori M., Barroso-Batista J., Langlois R.A., Barreiro L.B. Genetic ancestry effects on the response to viral infection are pervasive but cell type specific. Science. 2021;374:1127–1133. doi: 10.1126/science.abg0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. G:profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e1423. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuurman A.R., Reijnders T.D.Y., Saris A., Ramirez Moral I., Schinkel M., de Brabander J., van Linge C., Vermeulen L., Scicluna B.P., Wiersinga W.J., et al. Integrated single-cell analysis unveils diverging immune features of COVID-19, influenza, and other community-acquired pneumonia. Elife. 2021;10 doi: 10.7554/eLife.69661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevyrev D., Tereshchenko V. Treg heterogeneity, function, and homeostasis. Front. Immunol. 2019;10:3100. doi: 10.3389/fimmu.2019.03100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi H., Chi H. Metabolic control of Treg cell stability, plasticity, and tissue-specific heterogeneity. Front. Immunol. 2019;10:2716. doi: 10.3389/fimmu.2019.02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Special Expert Group for Control of the Epidemic of C-otCPMA Consideration on the strategies during epidemic stage changing from emergency response to continuous prevention and control. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:297–300. doi: 10.3760/cma.j.issn.0254-6450.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Srivatsan S.R., Regier M.C., Barkan E., Franks J.M., Packer J.S., Grosjean P., Duran M., Saxton S., Ladd J.J., Spielmann M., et al. Embryo-scale, single-cell spatial transcriptomics. Science. 2021;373:111–117. doi: 10.1126/science.abb9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tak T., Drylewicz J., Conemans L., de Boer R.J., Koenderman L., Borghans J.A.M., Tesselaar K. Circulatory and maturation kinetics of human monocyte subsets in vivo. Blood. 2017;130:1474–1477. doi: 10.1182/blood-2017-03-771261. [DOI] [PubMed] [Google Scholar]
- 45.Taquet M., Dercon Q., Luciano S., Geddes J.R., Husain M., Harrison P.J. Incidence, co-occurrence, and evolution of long-COVID features: a 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavakolpour S., Rakhshandehroo T., Wei E.X., Rashidian M. Lymphopenia during the COVID-19 infection: what it shows and what can be learned. Immunol. Lett. 2020;225:31–32. doi: 10.1016/j.imlet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The L. Facing up to long COVID. Lancet. 2020;396:1861. doi: 10.1016/S0140-6736(20)32662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H., Wang Z., Cao W., Wu Q., Yuan Y., Zhang X. Regulatory T cells in COVID-19. Aging Dis. 2021;12:1545–1553. doi: 10.14336/AD.2021.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Zheng J., Islam M.S., Yang Y., Hu Y., Chen X. The role of CD4(+)FoxP3(+) regulatory T cells in the immunopathogenesis of COVID-19: implications for treatment. Int. J. Biol. Sci. 2021;17:1507–1520. doi: 10.7150/ijbs.59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wing J.B., Tanaka A., Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and Cancer. Immunity. 2019;50:302–316. doi: 10.1016/j.immuni.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 53.Wong L.R., Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy? Nat. Rev. Immunol. 2021:1–10. doi: 10.1038/s41577-021-00656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida M., Worlock K.B., Huang N., Lindeboom R.G.H., Butler C.R., Kumasaka N., Conde C.D., Mamanova L., Bolt L., Richardson L., et al. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature. 2022;602:321–327. doi: 10.1038/s41586-021-04345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu N., Li X., Song W., Li D., Yu D., Zeng X., Li M., Leng X., Li X. CD4(+)CD25 (+)CD127 (low/−) T cells: a more specific Treg population in human peripheral blood. Inflammation. 2012;35:1773–1780. doi: 10.1007/s10753-012-9496-8. [DOI] [PubMed] [Google Scholar]
- 56.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X., Wang F., Shen Y., Zhang X., Cen Y., Wang B., Zhao S., Zhou Y., Hu B., Wang M., et al. Symptoms and health outcomes among survivors of COVID-19 infection 1 year after discharge from hospitals in Wuhan, China. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.27403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Supplemental Figures 1-9 and legends; 2. Supplemental Tables 1-6
Data Availability Statement
The single cell RNA sequences are deposited in the GEO repository (GSE232523).