Abstract
Hypoxia-induced adenosine creates an immunosuppressive tumor microenvironment (TME) and dampens the efficacy of immune checkpoint inhibitors (ICIs). We found that hypoxia-inducible factor 1 (HIF-1) orchestrates adenosine efflux through two steps in hepatocellular carcinoma (HCC). First, HIF-1 activates transcriptional repressor MXI1, which inhibits adenosine kinase (ADK), resulting in the failure of adenosine phosphorylation to adenosine monophosphate. This leads to adenosine accumulation in hypoxic cancer cells. Second, HIF-1 transcriptionally activates equilibrative nucleoside transporter 4, pumping adenosine into the interstitial space of HCC, elevating extracellular adenosine levels. Multiple in vitro assays demonstrated the immunosuppressive role of adenosine on T cells and myeloid cells. Knockout of ADK in vivo skewed intratumoral immune cells to protumorigenic and promoted tumor progression. Therapeutically, combination treatment of adenosine receptor antagonists and anti–PD-1 prolonged survival of HCC-bearing mice. We illustrated the dual role of hypoxia in establishing an adenosine-mediated immunosuppressive TME and offered a potential therapeutic approach that synergizes with ICIs in HCC.
Hypoxia promotes adenosine efflux to dampen anti-tumor immune responses and contributes to immune checkpoint inhibitor resistance.
INTRODUCTION
Hepatocellular carcinoma (HCC), a major primary liver cancer transformed from hepatocytes, ranks the third deadliest cancer worldwide (1). High mortality of HCC is ascribable to late symptom presentation and its resistance against conventional chemotherapies. Most patients with HCC are deemed ineligible for surgical treatment at the time of diagnosis. Currently, there are only two types of U.S. Food and Drug Administration (FDA)–approved drugs for patients with advanced HCC, namely, tyrosine kinase inhibitors (TKIs) and immune checkpoint inhibitors (ICIs). TKIs such as sorafenib and lenvatinib extend patients’ survival for only around 3 months (2, 3). Nivolumab, an anti–PD-1 (programmed cell death protein 1) monoclonal antibody was recently approved by FDA as second-line HCC treatment with a response rate of 20% in patients with HCC (4). Nivolumab has demonstrated an unprecedented clinical benefit in a subset of patients with HCC; nonetheless, majority of patients with HCC remain unresponsive to nivolumab. Therefore, the investigations on combinatorial therapies to improve ICIs are actively underway. Recently, the combination treatment of bevacizumab, an anti–vascular endothelial growth factor monoclonal antibody, and atezolizumab, an anti–PD-L1 (programmed cell death ligand 1) monoclonal antibody, has been FDA-approved as a first-line treatment for patients with advanced HCC, and it is the first treatment that achieves a better survival outcome than the standard treatment sorafenib with a response rate of around 27% (5). While immunotherapies have emerged to be a major HCC therapy especially in a combination treatment approach, more thorough understanding of the resistance mechanisms against ICIs in HCC is crucial to provide a mechanistic rationale for combination treatment and to enhance therapeutic efficacy.
Tumor microenvironment (TME) plays a key role in mediating immune evasion and resistance to ICIs. TME is a complexed and interactive network with multiple components including cancer cells, immune cells, stromal cells, extracellular matrix, and vasculature (6, 7). A diversity of immune cells is found in the HCC microenvironment including antitumorigenic immune cells including dendritic cells (DCs), natural killer (NK) cells, and T cells, and protumorigenic immune cells such as myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs) (8, 9). HCC cells, taking advantage of this shared TME, favor the accumulation of protumorigenic cells while suppressing antitumorigenic immune cells through production of immunosuppressive chemokines and metabolites. One of the key contributors to this immunosuppressive TME is hypoxia, oxygen (O2) deprivation.
Hypoxia is a prominent feature in HCC due to abnormal tumor vasculature resulting in uneven O2 delivery. The major molecular mechanisms induced by hypoxia are mediated by the transcription factor, hypoxia-inducible factor (HIF). HIF comprises an O2-responsive HIF-1/2α subunit and a constitutively expressed HIF-1β subunit (10). In the presence of O2 (normoxia), HIF-1/2α is hydroxylated by prolyl hydroxylase (11), permitting E3 ubiquitin ligase von Hippel–Lindau protein (VHL) to bind to HIF-1/2α and mediate the proteasomal degradation of HIF-1/2α (12). When O2 level declines, stabilized HIF-1/2α protein translocates to the nucleus and forms dimeric complex with HIF-1β. The HIF complex initiates transcription by binding to genes enclosing consensus sequence –A/GCGTG-, hypoxia response elements (HREs) (13). HIF regulates a wide repertoire of genes that benefit essentially all steps in tumor development including angiogenesis, metabolic reprogramming, metastasis, chemo- and radioresistance, and immune evasion.
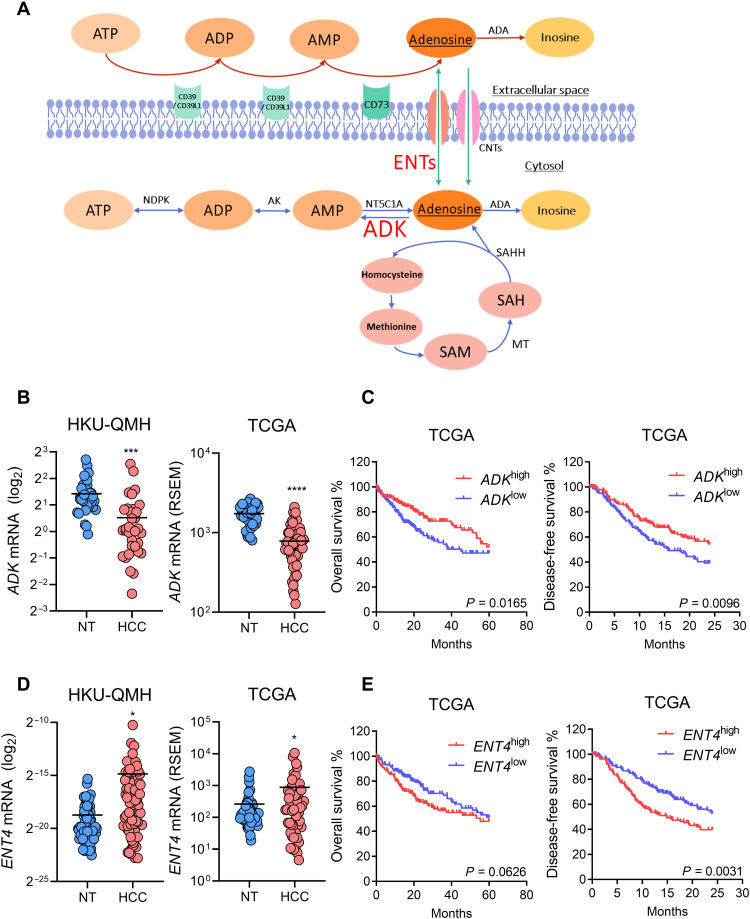
HIF promotes immune escape through multiple ways. HIF promotes immunosuppressive cell recruitment like MDSC and Treg by inducing cancer cell–secreted chemokines C-C motif chemokine ligand 26 (CCL26) and C-C motif chemokine ligand 28 (CCL28), respectively (14, 15). In breast cancer stem cells, HIF induces integrin CD47 which acts as a do-not-eat-me signal through interaction with receptor signal regulatory protein alpha (SIRPα) expressed on macrophages to prevent phagocytosis (16). HIF also induces the production of immunosuppressive metabolites such as adenosine through transcriptionally activating the ectoenzymes CD39/CD39L1 and CD73 (17–19). CD39/CD39L1 hydrolyzes extracellular adenosine triphosphate (ATP)/adenosine diphosphate (ADP) to ADP/adenosine monophosphate (AMP), while CD73 converts AMP to adenosine eventually (Fig. 1A) (20).
Fig. 1. ENT4 is overexpressed, while ADK is suppressed in human HCC.
(A) Schematic representation of pathways involved in adenosine metabolism. (B) ADK mRNA expression in paired HCC and NT (nontumorous) tissues from (left) University of Hong Kong-Queen Mary Hospital (HKU-QMH) patients and (right) The Cancer Genome Atlas (TCGA) database. (C) Kaplan-Meier curves showing the association of ADK mRNA expression with (left) overall and (right) disease-free survival in patients with HCC from TCGA database. (D) ENT4 mRNA expression in paired HCC and NT tissues from (left) HKU-QMH patients and (right) TCGA database. (E) Kaplan-Meier curves showing the association of ENT4 mRNA expression with (left) overall and (right) disease-free survival in patients with HCC from TCGA database. *P < 0.05, ***P < 0.001, and ****P < 0.0001 versus NT. (B) and (D) Student’s t test. (C) and (E) Kaplan-Meier followed by log-rank test. RSEM, RNA sequencing expression estimation by expectation maximization.
Extracellular adenosine exerts its immunosuppressive effect via interacting with four heterotrimeric guanine nucleotide-binding protein–coupled receptors [adesonsine A1 receptor (A1R), adenosine 2A receptor (A2AR), adenosine 2B receptor (A2BR), and A3R] (21). Adenosine has been shown to enrich Tregs via A2AR (22). Treg also expresses CD39, which, together with CD73 expressed on cancer cells and other stromal cells, produces adenosine to inhibit T cells and NK cells (23–25). It also promotes the expansion of MDSCs and maintains their immunosuppressive activities while inhibiting tumor antigen cross-presentation in DCs through A2BR (26, 27). Given the strong immunosuppressive roles of adenosine, blocking extracellular adenosine accumulation represents a tantalizing therapeutic strategy in cancer treatment for its potential effect to reinstate immunity. CD39 and CD73 have been hotly pursued therapeutic targets in treating cancers (28, 29). Although intracellular adenosine is an indispensable source of adenosine that can be exported to the surroundings, how intracellular adenosine contributes to extracellular adenosine accumulation and the establishment of immunosuppressive TME is underappreciated. Therefore, understanding the underlying molecular mechanisms of intracellular adenosine accumulation and its export sheds light on another important contributor to adenosine-rich TME.
Adenosine is transported out from cells through energy-independent equilibrative nucleoside transporters (ENTs), while adenosine uptake is mediated by both ENTs and energy-dependent concentrative nucleoside transporters (CNTs) (30). ENT family comprises ENT1 to ENT4 family members. Although they all facilitate the transport of adenosine, they have distinct subcellular localization. ENT1 and ENT2 are mainly expressed in plasma membrane and could be found in the nuclear membrane. ENT1 could also be found in the mitochondrial membrane. ENT3 is exclusively expressed in endosomal/lysosomal and mitochondrial membranes. ENT4 is exclusively found in the plasma membrane (31). ENT1 and ENT4 are the major studied transporters that could efflux adenosine. ENT4 is more divergent from ENT1 to ENT3, and it is only specific to adenosine, while ENT1 to ENT3 members have affinities for other nucleosides (30). ENT4 has been reported to regulate the level of extracellular adenosine in heart under acidic condition (32). Direction of ENT-mediated adenosine transport is solely determined by concentration gradient, and thus, intracellular adenosine metabolism plays a key role in adenosine export. Intracellular metabolism of adenosine can be principally controlled by three major enzymes, S-adenosylhomocysteine hydrolase (SAHH), adenosine deaminase (ADA), and adenosine kinase (ADK). SAHH reversibly hydrolyzes S-adenosylhomocysteine to adenosine and homocysteine. ADA hydrolyzes adenosine to inosine. ADK phosphorylates intracellular adenosine to AMP (Fig. 1A). ADK has a high affinity for adenosine and is the major enzyme that clears intracellular adenosine rapidly to maintain low intracellular adenosine (33). Although it is demonstrated in endothelial cells that ADK was shown to be repressed by hypoxia in a HIF-1–dependent manner, how HIF mediated ADK suppression requires further delineation (34).
We found that ADK was repressed, while ENT4 was induced by hypoxia in HCC. At the regulatory level, we found that HIF-1 induced a transcriptional repressor MAX interacting protein 1 (MXI1) to inhibit ADK transcription, while HIF-1 directly activated ENT4 transcriptionally. We hypothesize that the reduction of ADK impaired the clearance of intracellular adenosine leading to its accumulation intracellularly. By concentration gradient, hypoxia-induced ENT4 facilitated the export of adenosine to the extracellular space, leading to substantial accumulation of extracellular adenosine. We demonstrated that ADK repression and ENT4 overexpression (OE) drove extracellular adenosine accumulation. Extracellular adenosine drove Treg differentiation and MDSC induction and suppressed T cell proliferation. We further showed that ADK knockout (KO) in HCC tumors skewed intratumoral immune cells to protumorigenic and promoted tumor progression. Blocking adenosine receptors on immune cells could work synergistically with anti–PD-1. We illustrated the significance of the dual role of hypoxia/HIF in establishing an adenosine-mediated immunosuppressive TME and offered a potential therapeutic approach that might synergize with ICIs in HCC.
RESULTS
ADK was down-regulated and ENT4 was up-regulated in human HCC
To understand the clinical relevance of adenosine metabolism in HCC, we investigated the expression levels in patients with HCC. We found that ADK was down-regulated in HCC tissues when compared to paired nontumorous (NT) liver tissues in both the University of Hong Kong-Queen Mary Hospital (HKU-QMH) cohort and The Cancer Genome Atlas (TCGA) database (Fig. 1B). TCGA data showed that lower expression of ADK was significantly correlated with poorer overall and disease-free survival (Fig. 1C). On the other hand, ENT4 was overexpressed in HCC tissues when compared to paired NT liver tissues in both HKU-QMH and TCGA cohort (Fig. 1D). Higher expression of ENT4 was remarkably correlated with shorter overall and disease-free survival (Fig. 1E). Among all the ENT family members, ENT4 was the only family member associated with poor prognosis in both overall and disease-free survival in TCGA cohort (fig. S1, A and B). Moreover, HCC samples from HKU-QMH with higher ENT4 expression were significantly associated with larger tumor size (>5 cm; table S1). Given the important role of these two genes in adenosine metabolism and transport, the unique clinical expression patterns of these two genes prompted us to further investigate the regulation of these two genes in HCC.
Hypoxia repressed ADK and induced ENT4 through HIF-1 in HCC
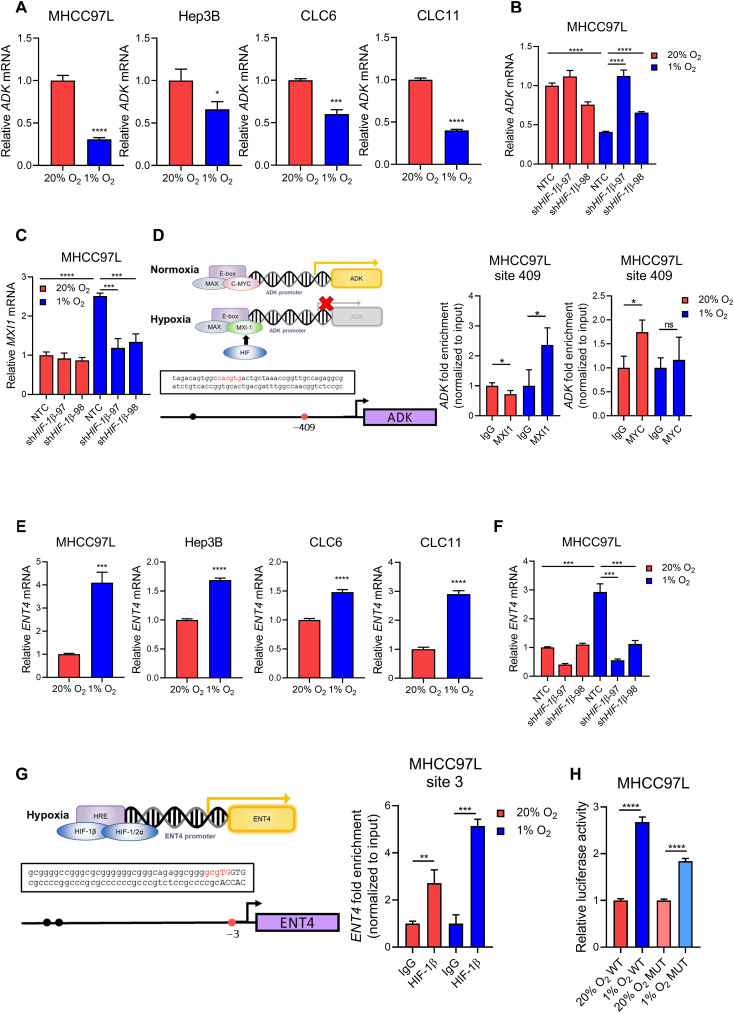
Next, we investigated how these two important genes in adenosine metabolism were being regulated in HCC. Hypoxia was previously reported to promote extracellular adenosine accumulation in TME via up-regulation of the ectoenzymes CD39 and CD73 to enhance ATP/AMP/adenosine conversion. Therefore, we explored whether hypoxia also regulated ADK and ENT4, orchestrating extracellular adenosine accumulation via regulating intracellular adenosine. We found that ADK was suppressed under hypoxia in multiple human HCC cell lines (Fig. 2A) and the effect could be abrogated by HIF-1β knockdown (Fig. 2B and fig. S2, A and B). As the direct binding of HIF to gene promoter regions usually enhances gene transcription, the suppression of ADK under hypoxia may be regulated in an indirect manner. MXI1 is a transcription repressor that has been reported to be induced by hypoxia and inhibited C-MYC activities (35). Under hypoxia, MXI1 is up-regulated and functions as a transcription repressor by competing with MYC to dimerize with MAX and bind to E-box element (5′-CACGTG-3′) at gene promoter region (36), thereby preventing the transcriptional activation mediated by MYC binding. We showed that MXI1 was induced in HCC cells and the effect was reversed by knockdown of HIF-1β (Fig. 2C and fig. S2C). In silico analysis revealed putative E-box element at ADK promoter region (Fig. 2D). Chromatin immunoprecipitation (ChIP) assay demonstrated that there was a significant enrichment of MXI1 binding to ADK promoter region under hypoxia, which was accompanied by the lack of MYC enrichment at the site (Fig. 2D). Meanwhile, ENT4 was up-regulated under hypoxia in multiple HCC cell lines (Fig. 2E), and the effect could be abrogated by HIF-1β knockdown (Fig. 2F and fig. S2, D and E). In silico analysis identified putative HRE at ENT4 promoter region (Fig. 2G). ChIP assay demonstrated that there was a significant enrichment of HIF-1β binding to ENT4 promoter region under hypoxia (Fig. 2G). Luciferase reporter assay further confirmed this putative HRE to be functional, as more prominent induction of luciferase activity by hypoxia was observed in HCC cells expressing wild-type (WT) HRE than those expressing a mutated (MUT) one (Fig 2H). Together, ADK was suppressed by hypoxia via HIF/MXI1 axis, while ENT4 was a direct transcriptional target of HIF.
Fig. 2. ADK and ENT4 are regulated by hypoxia via HIF.
(A) ADK mRNA expression in human HCC cell lines including MHCC97L, Hep3B, CLC6, and CLC11 exposed to 20 and 1% O2 for 24 hours. (B) ADK mRNA expression in MHCC97L-shHIF1β cells compared to NTC (nontargeting control) exposed to 20 and 1% O2 for 24 hours. (C) MXI1 mRNA expression in MHCC97L-shHIF-1β cells compared to NTC exposed to 20 and 1% O2 for 24 hours. (D) Left: Schematic representation of competition of MYC and MXI1 for E-box element binding under hypoxia and the binding site at ADK promoter region. Right: ChIP assay on MHCC97L cells exposed to 20 and 1% O2 using MXI1, MYC, and immunoglobulin G (IgG) antibodies. (E) ENT4 mRNA expression in human HCC cell lines including MHCC97L, Hep3B, CLC6, and CLC11 exposed to 20 and 1% O2 for 24 hours. (F) ENT4 mRNA expression in MHCC97L-shHIF-1β cells compared to NTC exposed to 20 and 1% O2 for 24 hours. (G) Left: Putative HRE at the promoter region of ENT4. Right: ChIP assay in MHCC97L cells exposed to 20 and 1% O2 using HIF-1β and IgG antibodies. (H) Relative luciferase activity in MHCC97L cells transfected with luciferase plasmids containing WT HRE or MUT HRE. (A) to (C), (E), and (F) mRNA expression was determined by quantitative real-time polymerase chain reaction (qRT-PCR), and values were normalized to 20% O2 or 20% O2 NTC. (D) and (G) Fold of enrichment was normalized to the corresponding IgG controls. (H) Relative luciferase activity was normalized to 20% O2 WT or 20% O2 MUT, respectively. Error bars indicate means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 versus 20% O2, 20% O2 NTC, IgG, 20% O2 WT, or 20% O2 MUT as indicated. Student’s t test.
Adenosine is an immunosuppressive metabolite in HCC
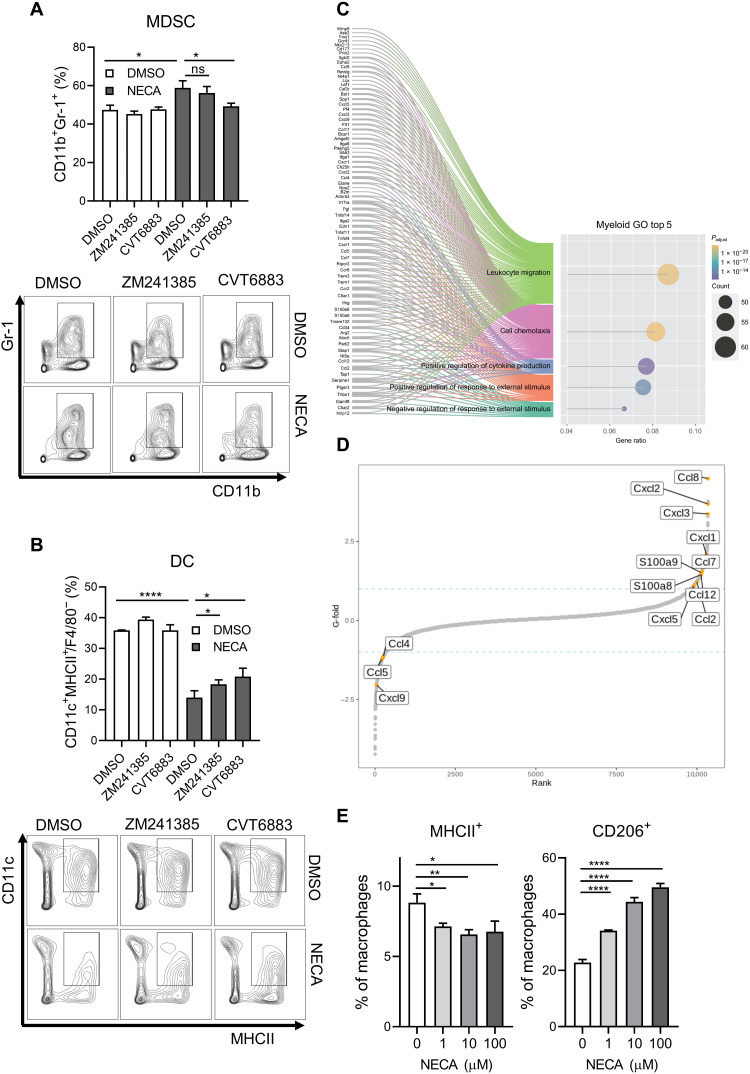
We demonstrated that hypoxia suppresses ADK, and we proposed that this promotes intracellular adenosine accumulation that creates a concentration gradient of adenosine favoring its efflux. Meanwhile, hypoxia elevates ENT4 that may facilitate the export of adenosine down the concentration gradient, leading to extracellular adenosine accumulation in TME. Therefore, we then explored how extracellular adenosine influences immune cells. As extracellular adenosine is degraded rapidly, we used a stable analog of adenosine, 5′-N-ethylcarboxamide adenosine (NECA) to study the immunomodulatory effect of adenosine. We cultured splenic T cells in the presence of CD3/CD28 Dynabeads and interleukin-2 (IL-2) for 3 days at indicated conditions. T cells were then harvested for flow cytometry analysis or bulk RNA sequencing. We showed that NECA suppressed both CD8+ and CD4+ T cell proliferation in a dose-dependent manner (Fig. 3A). We also observed a dose-dependent decrease of interferon-γ (IFN-γ) and granzyme B–expressing CD8+ T cells but an increase of cell death in both CD8+ and CD4+ T cells upon NECA treatment (Fig. 3, B and C). Moreover, NECA increased the percentage of Tregs (CD4+CD25+FOXP3+) after culturing for 3 days, and this could be abrogated by the addition of A2AR antagonist (ZM241385) but not A2BR antagonist (CVT6883; Fig. 3D). Gene set enrichment analysis (GSEA) on our RNA sequencing result showed that NECA-treated T cells were enriched with genes associated with Treg gene signature (Fig. 3, E and F) and negative regulation of T cell proliferation (Fig. 3G), echoing our observation in flow cytometry analysis. We also observed the up-regulation of genes commonly observed in Treg, such as Csf2, Foxp3, Tnfrsf18, Ccr4, Ccl22, and Ctla4, and a down-regulation of multiple cytotoxic genes, including Gzmb, Gzmm, Prf1, Gzma, and Gzmk (Fig. 3H), suggesting that NECA promotes the differentiation to Treg and dampens the cytotoxicity of T cells.
Fig. 3. Adenosine promotes Treg differentiation and cell death while suppressing T cell proliferation and functions.
(A) CellTrace Violet (CTV) T cell proliferation assay on (left) CD8+ and (right) CD4+ T cells treated with indicated doses of NECA (adenosine stable analog) for 3 days with IL-2. (B) Percentages of (left) IFN-γ+ and (right) granzyme B+ CD8+ T cells treated with indicated doses of NECA for 3 days with IL-2. (C) Percentage of cell death of (left) CD8+ and (right) CD4+ T cells treated with indicated doses of NECA for 3 days with IL-2. (D) Splenic T cells were cultured with 100 μM NECA in the presence of vehicle [dimethyl sulfoxide (DMSO)], 100 nM A2AR antagonist (ZM241385), or 100 nM A2BR antagonist (CVT6883) for 3 days with IL-2. The percentage of Tregs (CD4+ CD25+FOXP3+) was determined by flow cytometry. (E to H) Bulk RNA sequencing was performed on NECA-treated T cells compared to vehicle (DMSO). (E) to (G) GSEA showed the enrichment of selected gene sets in NECA-treated T cells. (H) Fold change of selected genes as compared to vehicle. Error bars indicate means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 versus 0 μM or DMSO as indicated. Student’s t test. ns, not significant.
To look at how NECA influences the differentiation of myeloid cells, we cultured bone marrow progenitor cells in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 for 5 days. Cells were harvested for flow cytometry analysis or bulk RNA sequencing. We demonstrated that NECA promoted MDSCs (CD11b+Gr-1+) accumulation, which could only be abrogated by A2BR blockade but not A2AR blockade (Fig. 4A). Meanwhile, NECA suppressed differentiation to DCs and could be rescued by both A2AR and A2BR blockade (Fig. 4B). Gene Ontology (GO) enrichment analysis showed that the top two GO terms were related to leukocyte migration and cell chemotaxis (Fig. 4C). Therefore, we further investigated the gene expression change of a panel of chemokines and cytokines. We observed that multiple chemokines that are well known to be recruiting immunosuppressive immune cells were up-regulated (Fig. 4D). Ccl2, Ccl7, Ccl8, and Ccl12 recruit MDSCs via CCR2 receptors (37). Cxcl1, Cxcl2, Cxcl3, and Cxcl5 recruit CXCR2+ cells, such as neutrophils and MDSCs (38). S100a8 and S100a9 promote MDSC accumulation (39, 40). On the other hand, there was a decrease in T cell chemoattractants including Cxcl9, Ccl4, and Ccl5 (Fig. 4D). We also studied the effect of NECA on macrophages. NECA treatment suppressed major histocompatibility complex II (MHCII) expression, which is important for antigen presentation by antitumorigenic macrophages, while inducing CD206 expression, an immunosuppressive marker on macrophages (Fig. 4E).
Fig. 4. Adenosine skews myeloid cells to immunosuppressive phenotypes.
(A and B) Bone marrow progenitor cells were cultured with 10 μM NECA in the presence of vehicle (DMSO), 100 nM A2AR antagonist (ZM241385), or 100 nM A2BR antagonist (CVT6883) for 5 days with IL-4 and GM-CSF. The percentages of (A) MDSC and (B) DC were determined by flow cytometry. (C and D) Bulk RNA sequencing was performed on NECA-treated bone marrow progenitor cells. (C) GO term enrichment analysis revealed the top five GO terms most enriched in NECA-treated bone marrow cells as compared to vehicle control. (D) Dot plot showed the fold change of a panel of chemokines as compared to vehicle. (E) Percentages of (left) MHCII+ and (right) CD206+ bone marrow–derived macrophages treated with indicated doses of NECA for 2 days with M-CSF. Error bars indicate means ± SD. *P < 0.05, **P < 0.01, and ****P < 0.0001 versus 0 μM or DMSO as indicated. Student’s t test.
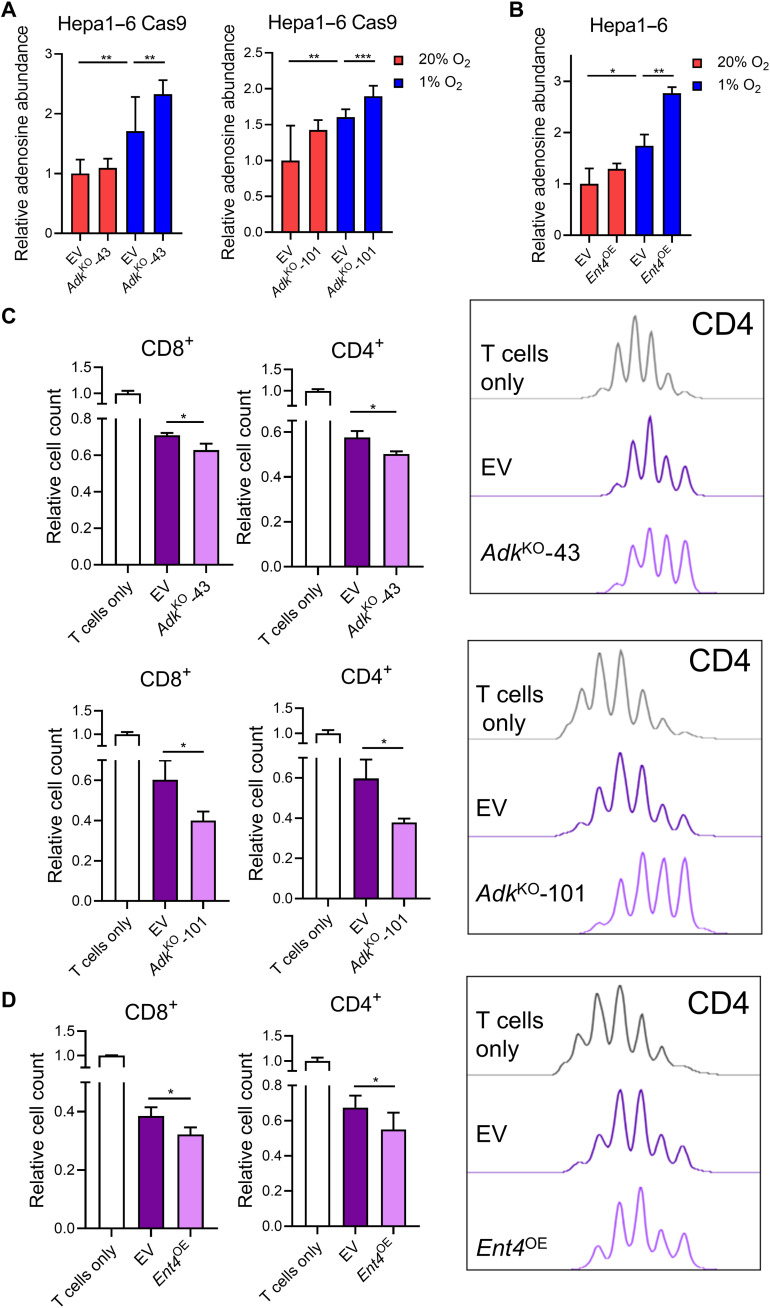
Upon confirming the immunosuppressive role of adenosine, we investigated how manipulation of ADK and ENT4 influence extracellular adenosine level. We first established stable Adk KO clones in mouse HCC cell line using the CRISPR-Cas9 system with two independent single-guide RNA (sgRNA) sequences (AdkKO-43 and AdkKO-101; fig. S3A). We harvested conditioned medium from these clones and performed liquid chromatography–mass spectrometry (LC-MS) to detect extracellular adenosine. We showed that extracellular adenosine level increased upon Adk ablation with a more obvious effect observed under hypoxia (Fig. 5A). As the endogenous expression level of Ent4 in mouse HCC cell line is low, we used an OE system to explore its function. We overexpressed the coding DNA sequence (CDS) of Ent4 in mouse HCC Hepa1–6 cells (fig. S3B). Gas chromatography–MS (GC-MS) revealed that extracellular adenosine level increased upon Ent4 OE, and this effect was more prominent under hypoxia, when the adenosine gradient was created (Fig. 5B). We further performed a coculture experiment to explore how ADK and ENT4 manipulation affects immune cells. Using the culturing conditions as above, we activated splenic T cells for 24 hours and cocultured T cells and mouse HCC cells in 1-to-1 ratio for 48 hours. Coculturing with HCC cells suppressed T cell proliferation, and we showed that KO of Adk or OE of Ent4 in HCC cells further suppressed T cell proliferation (Fig. 5, C and D).
Fig. 5. ADK deficiency and ENT4 overexpression (OE) elevate extracellular adenosine level and suppress T cell proliferation.
(A and B) Conditioned media of (A) Hepa1–6 Cas9–AdkKO cells and (B) Hepa1–6–Ent4OE cells exposed to 20 and 1% O2 for 48 hours were harvested, and extracellular adenosine levels were determined by MS. (C and D) CTV T cell proliferation assay on (left) CD8+ and (right) CD4+ T cells cocultured with (C) Hepa1–6 Cas9–AdkKO cells or (D) Hepa1–6–Ent4OE cells for 3 days with IL-2. (A) and (B) Relative adenosine level was normalized to 20% O2 empty vector (EV). Error bars indicate means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 versus EV or 20% O2 EV. Student’s t test.
KO of Adk promoted HCC development and suppressed antitumor immunity
We then investigated how altering intracellular adenosine level changes the TME in vivo. Using hydrodynamic tail vein injection (HDTVi) model, we simultaneously knocked out Trp53 and Adk while overexpressing c-Myc in liver to induce HCC tumors in immunocompetent mice. KO of Adk markedly promoted tumor growth (Fig. 6A). We further studied the tumor-infiltrating immune cells to understand how ADK deficiency influenced the tumor immune landscape. We observed a reduction of CD8+ T cells, cytotoxic T cells that are responsible for cancer cell killing (Fig. 6, B and E, and fig. S4A), but there was no difference in the proportion of memory T and naïve T cell subsets (fig. S4B). These T cells were more exhausted with higher expression of multiple inhibitory receptors (Fig. 6C). There was also a decrease in CD4+ T cells (Fig. 6, D and E, and fig. S4C) and DCs (Fig. 6F), suggesting that ADK ablation suppressed multiple types of immune cells via creating an adenosine-rich TME.
Fig. 6. ADK deficiency promotes HCC progression and creates an immunosuppressive TME.
(A) In vivo HCC tumors were induced via HDTVi of plasmids carrying Trp53KO/c-MycOE (EV) or Trp53KOAdkKO/c-MycOE (-43 and -101) in C57BL/6N mice. (Left) Liver mass and (right) representative picture of harvested livers with tumors. (B to F) Flow cytometry analysis was performed to determine tumor infiltration of immune cells. (B) Left: Quantification of CD8+ T cells. Right: Representative contour plots. (C) Percentage of effector T cells (CD8+CD44+CD62L−) expressing multiple exhaustion markers. (D) Left: Quantification of CD4+ T cells. Right: Representative contour plots. (E) Quantification of (top) CD8+ and (bottom) CD4+ T cells in tumor by immunohistochemistry staining. (F) Left: Quantification of DCs. Right: Representative contour plots. Error bars indicate means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 versus empty vector (EV). Student’s t test. Scale bar, 1 cm.
Blockade of extracellular adenosine sensing enhanced the efficacy of anti–PD-1 in HCC
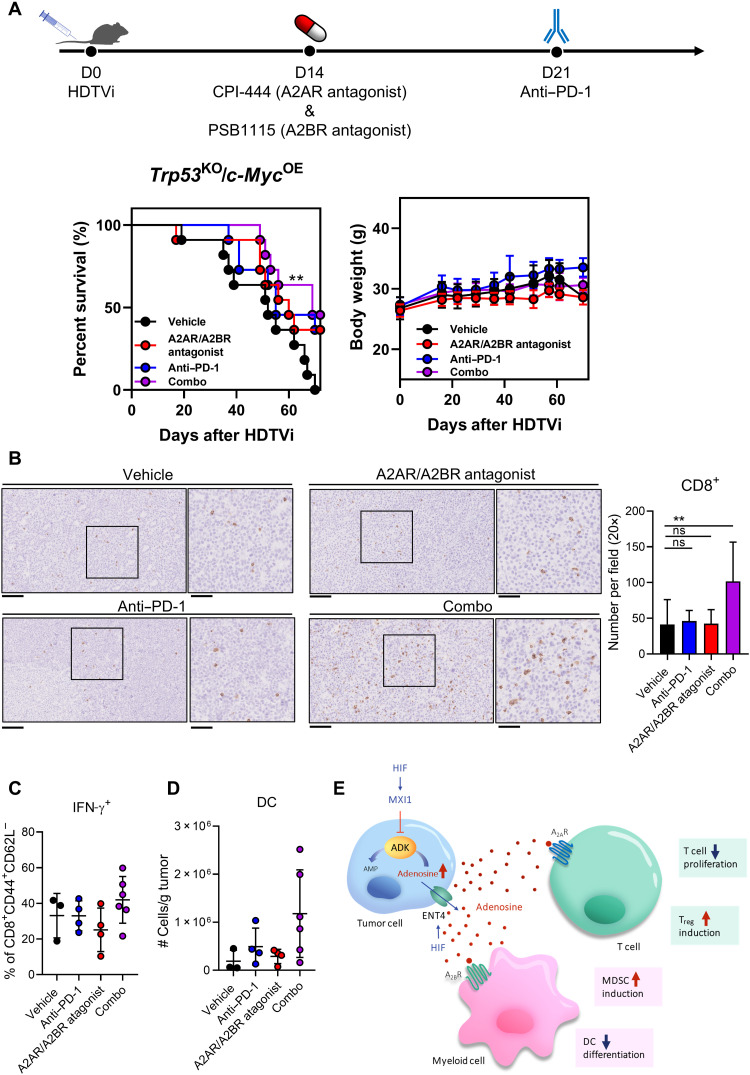
We previously showed that adenosine acts on A2AR and A2BR on immune cells to mediate their suppressive effect and the addition of adenosine receptor antagonists can reverse the suppressive effect of adenosine on immune cells in vitro. Therefore, we explored the therapeutic potential of the use of adenosine receptor antagonists to reverse the immunosuppressive effect caused by adenosine. We induced Trp53KO/c-MycOE HCC tumors by HDTVi and treated the mice using adenosine receptor antagonists in combination with anti–PD-1 monoclonal antibody, a current second line treatment for patients with HCC. The A2AR antagonist used in our in vivo study is CPI-444, which is currently used in clinical trials in other cancer types (NCT04280328 and NCT02655822). The A2BR antagonist used PSB1115, delayed tumor growth, and reduced MDSC accumulation in a melanoma mice model (41). We observed that single treatment of anti–PD-1 or the cotreatment of adenosine receptor antagonists did not bring significant survival benefit to HCC-bearing mice. We showed that combination treatment of anti–PD-1 and cotreatment of A2AR and A2BR antagonists significantly prolonged the survival of HCC-bearing mice (Fig. 7A). These treatments were well tolerated, as there were no significant changes in body weight between vehicle group and treatment groups. Accompanying the improvement in survival rate, we observed elevated tumor infiltration of CD8+ T cells with increased IFN-γ secretion in combination treatment group (Fig. 7, B and C). There was also a trend of increase in DC population in combination treatment group (Fig. 7D).
Fig. 7. Adenosine receptors blockade synergizes with anti–PD-1 treatment.
(A) In vivo HCC tumors were induced via HDTVi of plasmids carrying Trp53KO/c-MycOE in C57BL/6N mice. (Left) Survival plot and (right) body weight of HCC-bearing mice treated with vehicle, adenosine receptor antagonists, anti–PD-1 monoclonal antibodies, or the combination of both. (B) Representative pictures (left) and quantification (right) of CD8+ T cells in tumor by immunohistochemistry staining. (C) Percentage of effector T cells (CD8+CD44+CD62L−) expressing IFN-γ. (D) Quantification of DCs in tumor by flow cytometry. (E) Schematic summary of the role of HIF in promoting intracellular adenosine efflux and mediating immunosuppressive TME. Error bars indicate means ± SD. **P < 0.01 versus vehicle. (A) Kaplan-Meier followed by log-rank test. (B) Student’s t test. Original, 20× magnification (scale bars, 100 μm); Inset, 40× magnification (scale bars, 50 μm).
DISCUSSION
In our study, we identified the dual role of hypoxia in establishing an adenosine-rich immunosuppressive TME that favors HCC progression. Hypoxia suppressed ADK, which is important for intracellular adenosine clearance. Hypoxia also up-regulated ENT4, which is a passive transporter of adenosine. Together, this promotes extracellular adenosine accumulation. The contributions of ADK and ENT4 in cancers are not clearly understood. The role of ADK in cancers varies in different cancer types. Although previous studies showed that ADK was overexpressed in tumor tissues in patients with breast cancer and glioma (42, 43), it was shown in patients with HCC that ADK was down-regulated, consistent with what we have observed (44). ENT4 was reported to be up-regulated in desmoplastic small round cell tumor (DSRCT) (45). Here, we revealed a key mechanism by which hypoxia contributes to extracellular adenosine accumulation via regulation of ADK and ENT4, creating an immunosuppressive TME. Given that hypoxia is a common phenomenon in solid tumors, it would be meaningful to study whether the HIF/ADK/ENT4 axis in extracellular adenosine accumulation is also involved in other cancer types.
Hypoxia in cancers is evident, and it promotes cancer progression mediated by HIF-1. Hypoxia induces transcriptional activation of multiple genes involved in tumor development including cancer cell proliferation, angiogenesis, and migration (46). While most of the studies focused on the hypoxia-induced gene activation, a significant number of genes are suppressed by hypoxia, which is HIF-1 dependent in an indirect manner (47, 48). Hypoxia induced multiple transcription repressors to suppress gene expression indirectly. For example, hypoxia induced transcription factor 3 (TCF3), zinc finger homeobox protein 1A (ZFHX1A), and ZFHX1B, resulting in the down-regulation of E-cadherin in hypoxic cells, reducing cell-cell adhesion (49). We previously showed that hypoxia induced hairy/enhancer-of-split related with YRPW motif protein 1 (HEY1), another transcription suppressor that repressed PINK1 to reduce reactive oxygen species production, benefiting HCC cell survival (50). In our study, we showed that ADK, a key enzyme converting adenosine to AMP, was suppressed under hypoxia. We demonstrated that this was mediated by hypoxia-induced MXI1, which competed with MYC for the binding at ADK promoter region, thereby suppressing its expression. This is consistent with a previous study demonstrating that MXI1 was induced under hypoxia and suppressed MYC activities (35). A study showed that there were enriched binding sites of multiple components of the SIN3 transcription regulator family member A (SIN3A) histone deacetylase (HDAC) co-repressor complex together with MXI1 at hypoxia-suppressed genes and suggested that MXI1 interacted with SIN3A/HDAC co-repressor complex to mediate gene suppression under hypoxia (51). The down-regulation of ADK, together with the transcriptional induction of ENT4, promoted extracellular adenosine accumulation and suppressed antitumor immune responses in HCC. Together, these illustrated that hypoxia promoted cancer growth in a complexed and coordinated manner by inducing global transcriptional changes to establish a protumorigenic niche.
Adenosine is a well-studied immunosuppressive metabolite in promoting immune evasion. Multiple studies have demonstrated that adenosine impairs DCs and cytotoxic functions of T cells and NK cells while promoting Treg and MDSC development, establishing an immunosuppressive TME (52). We observed that ADK mediated extracellular adenosine accumulation and reduced the tumor infiltration of CD4+ T cells, CD8+ T cells, and DCs, which all play a key role in suppressing tumor progression. While CD4+ T cells secrete proinflammatory cytokines and DCs present tumor antigens to T cells, both mediate their tumor cell elimination indirectly through CD8+ T cells; thus, we believe that CD8+ T cells, being the major cell type that mediates direct cancer cell killing, play the most important role among these cell types. The CD39/CD73 axis has been identified as the key extracellular adenosine-producing enzymes; therefore, blocking these enzymes becomes an attractive therapeutic target. Ongoing clinical trials aim to study the efficacy of monoclonal antibodies targeting CD39 and CD73 in combination with ICIs (NCT02503774 and NCT04261075). However, CD39/CD73 is not the sole pathway that generates adenosine. Here, we showed that intracellular adenosine also contributes to the extracellular adenosine accumulation in TME. We demonstrated that hypoxia suppressed ADK, leading to intracellular adenosine accumulation, creating a concentration gradient favoring adenosine efflux. Meanwhile, hypoxia up-regulated ENT4, an adenosine transporter that carries adenosine down the concentration gradient. Together, they contributed to extracellular adenosine accumulation in TME and suppressed T cell proliferation and DC population while promoting Treg and MDSC population accumulation in vitro. Furthermore, ADK deficiency promoted tumor growth and impaired infiltration of T cell and DC in vivo. ENT4 overexpression also supported tumor progression (fig. S5). The adenosine transporter could be a potential therapeutic target to block adenosine export to TME. Although, currently, there is no commercially available inhibitor to ENT4, cotargeting adenosine pumping and CD39/CD73 ectoenzymes might enhance efficacy of ICIs.
The immunosuppressive effects of adenosine are mediated by adenosine receptors. Among the four subtypes of adenosine receptors, A2A receptors are the most well characterized for their role in promoting cancer immune evasion. It has been shown that A2AR deficiency in host mice led to complete tumor rejection, and adenosine suppressed both T cells and NK cells cytotoxicity via A2AR (53–55). Therefore, A2AR blockade is being pursued as a therapeutic target to reverse immunosuppressive effect mediated by adenosine. CPI-444 used in this study is one of the most commonly studied small-molecule inhibitors highly selective against A2AR. It demonstrated antitumor efficacy and worked synergistically with ICIs in preclinical models (56). CPI-444 has been used in phase 1 clinical trials on patients with renal cell cancer or prostate cancer as monotherapy or in combination with atezolizumab (anti–PD-L1) in which signs of efficacy were observed in the combination group (NCT02655822). In our study, we demonstrated that the immunosuppressive effects of adenosine on immune cell types were mediated by different adenosine receptors. While the effect on T cells was mediated by A2AR, the effect on myeloid cells was mediated by A2BR, consistent with previous studies (26, 27). Therefore, to further enhance the effect of blocking adenosine signaling, we treated HCC-bearing mice with coadministration of A2AR and A2BR receptor antagonists and showed that it worked synergistically with anti–PD-1 to prolong the survival of mice. Recently, a dual adenosine receptor antagonist has been developed and tested in an ongoing clinical trial to study its efficacy in combination with chemotherapy or anti–PD-1 (NCT03719326). Our study highlights the scientific basis for targeting both A2AR and A2BR to enhance ICI efficacy.
We demonstrated that ADK repression and ENT4 OE promoted extracellular adenosine accumulation. HIF-1 induced a transcriptional repressor MXI1 to inhibit ADK transcription, while HIF-1 directly activated ENT4 transcriptionally. The reduction of ADK impaired the removal of intracellular adenosine and established a concentration gradient of adenosine. Meanwhile, hypoxia-induced ENT4 mediated the export of adenosine, leading to substantial accumulation of extracellular adenosine that suppressed multiple immune cells (Fig. 7E). We further showed that ADK deficiency in HCC tumors skewed intratumoral immune cells to protumorigenic and promoted tumor growth. Blocking adenosine receptors on immune cells could work synergistically with anti–PD-1. We delineated the dual role of hypoxia/HIF in creating an immunosuppressive TME and offered a potential therapeutic approach that might improve ICIs in HCC.
MATERIALS AND METHODS
Study approval
Approval was obtained for the use of clinical HCC tumor samples and paired NT liver tissues from the Institutional Review Board of The University of Hong Kong and the Hospital Authority of Hong Kong. All animal procedures performed throughout this study were approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong and adhered to the Animals (Control of Experiments) Ordinance of Hong Kong. All animal procedures were performed under the U.K. National Cancer Research Institute (NCRI) PMID: 20502460 Guidelines for the Welfare and Use of Animals in Cancer Research to minimize suffering on the animals throughout the experiments.
Statistical analysis
Data were analyzed by Student’s t test unless otherwise specified and were expressed as means ± SD. P value <0.05 were considered statistically significant. GraphPad Prism 8.0 or above software (GraphPad Software Inc., La Jolla, CA, USA) is used for data analysis.
The Cancer Genome Atlas
Transcriptome sequencing was performed on 49 pairs of human HCC and the corresponding NT liver tissues, and the results were retrieved from TCGA through cBioPortal (www.cbioportal.org/) to examine the mRNA expression levels of ADK, ENT1, ENT2, ENT3, and ENT4. Survival test was done by Kaplan-Meier curve and log-rank test.
Clinical samples
Paired clinical HCC tumor and NT liver tissues were collected from patients with HCC admitted to Queen Mary Hospital and used in this study. Approval was obtained from the Institutional Review Board of The University of Hong Kong and the Hospital Authority of Hong Kong. Signed consent forms were obtained from patients as acknowledgement before the collection and usage of resected tissues for research purpose. The expression level of ENT4 in patients with HCC was correlated with multiple clinicopathological parameters (tumor microsatellite formation, direction liver invasion, tumor encapsulation, venous invasion, tumor stage, tumor size, and Edmondson grading) using SPSS 20.0 software as described (57).
Immunohistochemistry staining
Mouse liver tissues fixed in 4% formalin and 75% ethanol were embedded in paraffin. Paraffin sections were first dewaxed with xylene and rinsed with ethanol. Antigen retrieval was performed by boiling in 1 mM EDTA buffer of pH 7.8 or 10 mM tris and 1 mM EDTA buffer of pH 9.0. The samples were stained at 4°C overnight with primary antibody (table S2). Samples were then incubated at room temperature for 30 min with horseradish peroxidase–conjugated secondary antibody (Dako, Glostrup, Denmark) followed by development with 3,3′-diaminobenzidine (Sigma-Aldrich, St. Louis, MO, USA) and counterstaining with hematoxylin.
Chromatin immunoprecipitation
HCC cells were fixed in 3.7% formaldehyde for DNA cross-linking and lysed with SDS buffer, followed by sonication. Sheared DNA fragments were then incubated with antibodies against MYC, MXI1, HIF-1β, or immunoglobulin G (IgG) control (table S2) together with Protein A Agarose/Salmon Sperm DNA Beads (Merck Millipore, Burlington, MA, USA). After the incubation step, DNA-protein-antibody complexes were washed with salt buffer with gradient concentrations and then eluted in 1% SDS/NaHCO3. ChIP DNA samples were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) using primers flanking the putative E-box element or HRE (table S3).
Cell culture and hypoxia
Human HCC cell MHCC97L was a gift from Z. Y. Tang of Fudan University (Shanghai, China). CLC6 and CLC11 are gifts from L. Hui of Chinese Academy of Sciences (Shanghai, China). Human HCC cell Hep3B and mouse HCC cell Hepa1–6 were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines within this study were authenticated with the AuthentiFiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). Cell cultures were routinely tested for mycoplasma infection. MHCC97L, Hep3B, and Hepa1–6 were cultured in Dulbecco’s modified Eagle’s medium (DMEM)–high glucose media with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) supplementations (all reagents are from Gibco by Life Technologies, Grand Island, NY, USA). CLC6 and CLC11 were cultured in RPMI 1640 medium (Gibco) with 10% FBS and 1% PS supplemented with epidermal growth factor (40 ng/ml; PeproTech, Cranbury, NJ, USA) and 1% Insulin-Transferrin-Selenium (Gibco). For in vitro culture of immune cell assays, cells were cultured in RPMI 1640 media with 10% FBS and 1% PS supplemented with 25 mM Hepes buffer (Gibco) and 50 μM 2-mercaptoethanol (Sigma-Aldrich). Cells were cultured in a 37°C humidified incubator supplied with 5% CO2. Hypoxic environment was created by culturing HCC cells in 1% O2 in a hypoxia incubator chamber for 24 or 48 hours as indicated.
Establishment of stable cell lines with knockdown, KO, or OE
All HCC cell lines were established with the lentiviral-mediated approach. For HIF-1β knockdown HCC cell lines, short hairpin RNAs (shRNAs) targeting HIF-1β, or nontargeting control was inserted into pLKO.1-puro vectors and was stably transfected into MHCC97L and CLC11 cells (58), followed by puromycin selection. Mouse Adk stable KO cell lines were established by CRISPR gene editing system (lentiCRISPR v2) obtained as a gift from Sanjana and colleagues (59). Cas9 and sgRNAs targeting Adk were sequentially transfected into Hepa1–6 cells. For mouse Ent4 OE cell lines, plasmid containing the CDS of Ent4 (GenBank accession NM_146257) was purchased from GenScript. CDS of Ent4 was amplified using Pfu DNA Polymerase (Promega, Madison, WI, USA) and inserted into pLenti6-V5 vector using pGEM-T Easy Vector Systems (Promega). The vector was then stably transfected into Hepa1–6 cells and selected using blasticidin. Sequences of all shRNAs and sgRNAs used are provided in table S4.
Luciferase reporter assay
Using pGL4 firefly luciferase promoter reporter vector as the backbone, oligonucleotides containing the WT or MUT HREs of ENT4 were inserted into this vector. These plasmids containing WT or MUT HREs were transfected together with pRL-CMV into MHCC97L cells. The cells were then exposed to 1% or 20% O2 for 24 hours. Cells were harvested, and luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Insert sequences were provided in table S4.
Animal studies
To study the effect of Adk KO on tumor immune landscape, we use HDTVi model. Briefly, plasmids diluted in sterile saline with a total volume of 10% of the mouse body weight were injected into the lateral tail vein of 8- to 10-week-old male C57BL/6N mice in 6 to 8 s. A total of 20 μg of CRISPR-Cas9 vector system carrying sgRNA targeting Trp53 and Adk simultaneously, together with the transposon system carrying c-Myc vector, was injected into the lateral tail vein of mice. The sgRNA sequences are provided in table S3. HCC tumors were harvested 5 weeks after HDTVi for tumor dissociation and study of immune landscape.
To study the therapeutic effect of A2AR and A2BR antagonists in combination with anti–PD-1 in HCC, we also used HDTVi model. For this experiment, 20 μg of CRISPR-Cas9 vector system carrying sgRNA targeting Trp53 and the transposon system carrying c-Myc were injected. Fourteen days after HDTVi, mice were treated with CPI-444 (10 mg/kg; A2AR antagonist; MedChemExpress, Monmouth Junction, NJ, USA) administered via daily oral gavage and PSB1115 (1 mg/kg; A2BR antagonist; Tocris Bioscience, Bristol, UK) administered via intraperitoneal injection every other day. Treatment of anti–PD-1 monoclonal antibody (clone RMP1.14; Bio X Cell, Lebanon, NH, USA) started 21 days after HDTVi. Mice were treated with anti–PD-1 (10 mg/kg) twice weekly via intraperitoneal injection. Mice were euthanized upon reaching the humane end point. For orthotopic implantation, 4 × 106 Hepa1–6 cells were resuspended in 100% Matrigel (BD Biosciences, San Jose, CA, USA) and implanted into left lobes in 100% of livers of 5- to 7-week-old, male C57BL/6N mice. Tumors were harvested 14 days after operation.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted from clinical samples (snap-frozen with liquid nitrogen and stored in −80°C), HCC cell lines, or primary immune cells with TRIzol reagent (Ambion by Life Technologies, Austin, TX, USA). cDNA was prepared by reverse transcription using the GeneAmp RNA PCR Core Kit (Applied Biosystems). For clinical samples, qRT-PCR amplifications of ADK or ENT4 and internal control 18S were performed using the TaqMan Gene Expression Assay (Applied Biosystems). For HCC cell lines, qRT-PCR amplifications of ADK, ENT4, or MXI1 and internal control 18S were performed using SYBR Green PCR Master Mix (Applied Biosystems) with the respective qRT-PCR primers (table S3). All qRT-PCR amplifications were performed with StepOne Real-Time PCR System (Applied Biosystems).
Immunofluorescent staining
HCC cells were seeded on glass coverslips and then exposed to 1 or 20% O2 for 48 hours. Cells were fixed with 4% formalin and followed by washing with 0.5% Triton X-100 at room temperature for 10 min. Cells were then blocked in 3% bovine serum albumin (BSA) in 1× phosphate-buffered saline (PBS) at room temperature for 20 min before overnight incubation with primary antibodies at 4°C. Cells were washed with 1× PBS and incubated with secondary antibody [Donkey anti-Rabbit IgG (H+L), Alexa Fluor 488; Invitrogen, Carlsbad, CA, USA] at room temperature for 2 hours in dark. Cells were then mounted with ProLong Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (Invitrogen). Images were captured with an LSM 800 confocal microscope (Carl Zeiss).
Protein extraction and Western blotting
Radioimmunoprecipitation assay lysis buffer with cOmplete protease and PhosSTOP phosphatase inhibitors (Roche Diagnostics, Mannheim, Germany) was used to extract total protein lysates from HCC cell lines. Equal amounts of protein lysates were separated by SDS–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane (GE HealthCare, Little Chalfont, UK). The membrane was incubated with corresponding primary antibodies overnight at 4°C. The dilution of the antibodies is listed in table S2. The membrane was then washed with 1× Tris buffered saline with Tween 20 (TBST) and incubated with secondary antibody at room temperature for 2 hours. The protein expression was detected with the ECL detection system (GE HealthCare).
Adenosine quantification by MS
Sample preparation
Media were changed to phenol red–free media (Gibco) before exposing HCC cells to 1 or 20% O2 for 48 hours. For the metabolite extraction from cultured medium, 1 ml of sample was mixed with 4 ml of chilled 80% methanol and incubated at −80°C for 15 min. The mixture was spun down at 4°C for 15 min with a speed of 13,500 rpm, and the supernatant were transferred to another tube and dried with a vacuum centrifuge.
Liquid chromatography–mass spectrometry
The dried samples were resuspended in 50% methanol containing 13C5 Ribose Adenosine (Cambridge Isotope Laboratories, Tewksbury, MA, USA) [M + 5] as internal standard. For preparation of standard solution, adenosine (Sigma-Aldrich) was dissolved in methanol and diluted into a series of solutions for qualitative and quantitative analysis. The standard mobile phase was prepared as follows: A = water with 4 mM ammonium bicarbonate and B = 80% methanol with 4 mM ammonium bicarbonate. The flow rate was 0.3 ml/min; the sample injection volume was 10 μl, and the column temperature was 37°C. The analytes were separated by hypercarbon column (5-μm particles, 2.1 mm by 150 mm; Thermo Fisher Scientific, Waltham, MA, USA). The LC solvent gradient was as follows: From 0 to 10 min, B was linearly changed from 5 to 62.5%; from 10 to 11 min, B was linearly changed from 62.5 to 40%, and from 11 to 14 min, B was maintained at 5%. The electrospray ionization source was operated in positive mode. Its conditions were set as follows: Ion source gas 1 was 40 psi. Ion source gas 2 was 40 psi. Curtain gas was 30 psi. Temperature was 500°C. IonSpray voltage floating was ±5500 V. Declustering potential was 80 V, and collision energy was 30 eV. Multiple reaction monitoring experiment was selected to MS acquisition. The transition from precursor ion to product ions was referred to as an ion transition. Analyst Software (AB SCIEX) was used for peak identification and integration. Adenosine was identified by exact mass and retention time matched to adenosine standards. Adenosine level was normalized with internal standard and cell number.
Gas chromatography–mass spectrometry
To increase the volatility of adenosine for GC-MS analysis, the cell extracts were derivatized with a two-chemical reaction approach. Briefly, the samples were initially mixed with 30 μl of methoxyamine in pyridine (201 mg/ml), which also contained an internal standard (13C5-adenosine, 2 μg/ml). Samples were incubated at 37°C for 60 min and cooled to room temperature. Next, 35 μl of BSTFA with 1% of TMCS (Sigma-Aldrich) was added to each sample, followed by incubation at 65°C for 90 min and cooling to room temperature. The adenosine was analyzed on an Agilent GC-MS system, comprising of an Agilent 7890b GC connected to an Agilent 5975C mass selective detector. The GC was set in splitless mode, maintaining a constant helium gas velocity at 1 ml/min. The derivatized samples were introduced into a DB-5MS column (Agilent, Santa Clara, CA, USA), and the GC oven program was optimized as follows: Start at 80°C with a hold for 1 min; then, increase by 10°C per 1 min to 320°C, followed by a final hold at 320°C for 5 min. Intensity of adenosine was extracted, and ion fragment of 136.06 mass/charge ratio was integrated by using MassHunter software (Agilent).
Cell staining for flow cytometry
For analysis of cells from tumor tissues, cells were stained with LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen) first. For cell surface staining for both in vitro and in vivo studies, cells were stained with primary antibodies conjugated with fluorochromes at 4°C for 30 min in the dark after a 10-min blocking step with anti-mouse CD16/32 antibody (BioLegend, San Diego, CA, USA) at room temperature. Cells were then washed and resuspended in 300 μl of cell staining buffer [0.5% (w/v) BSA and 2 mM EDTA in PBS] and analyzed by flow cytometry. For studying Tregs or T cell cytokine production, after cell surface staining, T cells were subjected to intracellular staining using the eBioscience Foxp3/Transcription Factor Staining Buffer Set (Invitrogen) according to the manufacturer’s instruction. Cells were resuspended in 300 μl of cell staining buffer in the end and analyzed with the analyzer BD LSRFortessa (BD Biosciences). Primary antibodies used and the dilution were indicated in table S2. Software FlowJo was used for data analysis.
T cell differentiation assay
Splenic T cells from 5- to 7-week-old C57BL/6N male mice were obtained with Mouse T Cell Enrichment Columns (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instruction. Isolated T cells were activated with Dynabeads Mouse T-Activator CD3/CD28 (Gibco) and cultured in the presence IL-2 (R&D Systems). NECA (Tocris Bioscience) was added at indicated concentrations for 3 days in the presence of vehicle [dimethyl sulfoxide (DMSO)], ZM241385 (A2AR antagonist; Tocris Bioscience), or CVT6883 (A2BR antagonist; Tocris Bioscience). T cells were harvested for transcriptome sequencing or subjected to steps for intracellular staining as described above, followed by flow cytometry analysis. Tregs were defined by CD4+ CD25+ FOXP3+.
T cell proliferation assay
Splenic T cells were obtained as above. Isolated T cells were labeled with 5 μM CellTrace Violet (CTV; Invitrogen) for 15 min at 37°C with mixing at regular interval. Labeled T cells were cultured in the presence of NECA at indicated concentrations or cocultured with Hepa1–6 Cas9 or Hepa1–6 cells [empty vector (EV) and AdkKO or Ent4OE] in 1:1 ratio for 3 days in the presence of activation beads and IL-2. The CTV signal would be halved after each cell division cycle. T cells were harvested and analyzed by flow cytometry.
T cell cytokine production
For in vitro study, splenic T cells were extracted as above. Isolated T cells were cultured in the presence of NECA at indicated concentrations for 3 days in the presence of activation beads and IL-2. For in vivo study, single-cell suspensions were obtained from murine tumor tissues. Cells were incubated for 4 hours in the presence of cell activation cocktail (with Brefeldin A; BioLegend) according to the manufacturer’s suggestion before harvesting for flow cytometry analysis.
Myeloid cell differentiation assay
Bone marrow progenitor cells were isolated from the femur and tibia of male C57BL/6N mice and cultured in the presence of GM-CSF (R&D Systems) and IL-4 (R&D Systems) to drive myeloid cell differentiation. To study the effect of adenosine on differentiation of myeloid cells, 10 μM NECA was added to the culture in the presence of vehicle (DMSO), ZM241385, or CVT6883. The immune cell populations were harvested for transcriptome sequencing or determined by flow cytometry after 5 days of culture. DC population was defined by F4/80−CD11c+ MHCII+. MDSCs were defined by CD11b+ Gr-1+ population.
Macrophage polarization assay
Bone marrow progenitor cells were isolated as above and differentiated into macrophages in the presence of M-CSF (R&D Systems) for 6 days. Macrophages were then treated with NECA at indicated concentrations for 2 days and harvested for analysis of the expression of CD206 (immunosuppressive macrophage marker) and MHCII (stimulatory macrophage marker).
Preparation of single-cell suspension from tumor tissues
Single-cell suspensions were prepared from mouse tumors. Tumors harvested from HCC-bearing mice were first diced into small pieces. They were then put into gentleMACS C tubes (Miltenyi Biotec, Bergisch Gladbach, Germany). These C tubes contained 10 ml of DMEM/F12 medium with 30 μl of Liberase (2.5 mg/ml; Roche Diagnostics) and 60 μl of deoxyribonuclease I (10 mg/ml; Roche Diagnostics). Using the gentleMACS Dissociator (Miltenyi Biotec), the samples were further processed according to the manufacturer’s instructions. The cell suspensions were then passed through MACS SmartStrainers (Miltenyi Biotec), followed by centrifugation at 300g for 5 min. Red blood cells were removed from the cell suspensions using 10 ml of ACK lysing buffer (155 mM ammonium chloride, 10 mM potassium bicarbonate, and 0.1 mM EDTA in water) with a 5-min incubation at room temperature. After adding 20 ml of cell staining buffer to stop the lysis reaction, the cell suspensions were filtered again and centrifuged at 300g for 5 min. The cell pellets were resuspended in suitable volume of cell staining buffer and were ready for cell counting.
Transcriptome sequencing and data analysis
Transcriptome sequencing was performed on mouse splenic T cells treated with 100 μM NECA or vehicle and bone marrow cells treated with 10 μM NECA or vehicle. The library preparation and sequencing were done by Novogene (Beijing, China). Raw RNA sequencing data were trimmed and mapped to the reference mouse genome (mm10) by “Trim Galore” and “HISAT2.” Fragments per kilobase of transcript per million mapped reads (FPKM) and counts for gene were calculated by “StringTie” and the Python script “htseq-count,” respectively. To determine differential genes, “G-FOLD” was applied to compare the expression of each gene, and genes with absolute GFOLD value >1 were considered as differentially expressed genes. GO term enrichment analysis was performed by clusterProfiler. P values were adjusted by Benjamini-Hochberg method to control the false discovery rate (FDR), and threshold of FDR was set to 0.05. GSEA was performed by R package “fgsea.”
Acknowledgments
We thank the Centre for Comparative Medicine Research (CCMR) of The University of Hong Kong in support of our animal experiments and the School of Biological Sciences, The University of Hong Kong for technical support in our MS experiment. We also thank the Centre for PanorOmic Sciences (CPOS) from the Li Ka Shing Faculty of Medicine, The University of Hong Kong for the technical support.
Funding: This study is supported by the Hong Kong Research Grant Council (RGC) General Research Fund (17110220), National Natural Science Foundation of China (NSFC) Excellent Young Scientists Fund (Hong Kong and Macau) (82022077, C.C.-L.W.), Croucher Innovation Award, and The University of Hong Kong Outstanding Young Researcher Award. C.C.-L.W. is the funding recipient of the above funds. This study is also supported by RGC Theme-based Research Scheme (T12-716-22R). I.O.-L.N. is the funding recipient of this grant.
Author contributions: Conceptualization and methodology: J.W.-S.C., D.K.-C.C., and C.C.-L.W. Investigation and data acquisition: J.W.-S.C., D.K.-C.C., K.K.-L.K., C.Y., V.W.-H.Y., C.C.G., N.N.-Q.C., M.S.Z., M.H.-R.B., B.P.-Y.W., C.Y.-K.C., C.X.L., G.F.-W.S., Z.Y.O., H.D., A.P.-W.T., and C.C.-L.W. Data analysis: J.W.-S.C., D.K.-C.C., K.K.-L.K., C.Y., W.S., C.-T.L., and Q.L. Writing (original draft): J.W.-S.C. and C.C.-L.W. Writing (review and editing): J.W.-S.C. and C.C.-L.W. Clinical specimen procurement: I.O.-L.N. Supervision: C.C.-L.W. Funding acquisition: I.O.-L.N. and C.C.-L.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S1 to S4
REFERENCES AND NOTES
- 1.H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.J. M. Llovet, S. Ricci, V. Mazzaferro, P. Hilgard, E. Gane, J.-F. Blanc, A. C. De Oliveira, A. Santoro, J.-L. Raoul, A. Forner, M. Schwartz, C. Porta, S. Zeuzem, L. Bolondi, T. F. Greten, P. R. Galle, J.-F. Seitz, I. Borbath, D. Häussinger, T. Giannaris, M. Shan, M. Moscovici, D. Voliotis, J. Bruix; SHARP Investigators Study Group , Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008). [DOI] [PubMed] [Google Scholar]
- 3.M. Kudo, R. S. Finn, S. Qin, K.-H. Han, K. Ikeda, F. Piscaglia, A. Baron, J.-W. Park, G. Han, J. Jassem, J. F. Blanc, A. Vogel, D. Komov, T. R. J. Evans, C. Lopez, C. Dutcus, M. Guo, K. Saito, S. Kraljevic, T. Tamai, M. Ren, A.-L. Cheng, Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173 (2018). [DOI] [PubMed] [Google Scholar]
- 4.A. B. El-Khoueiry, B. Sangro, T. Yau, T. S. Crocenzi, M. Kudo, C. Hsu, T.-Y. Kim, S.-P. Choo, J. Trojan, T. H. Welling III, T. Meyer, Y.-K. Kang, W. Yeo, A. Chopra, J. Anderson, C. D. Cruz, L. Lang, J. Neely, H. Tang, H. B. Dastani, I. Melero, Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.R. S. Finn, S. Qin, M. Ikeda, P. R. Galle, M. Ducreux, T.-Y. Kim, M. Kudo, V. Breder, P. Merle, A. O. Kaseb, D. Li, W. Verret, D.-Z. Xu, S. Hernandez, J. Liu, C. Huang, S. Mulla, Y. Wang, H. Y. Lim, A. X. Zhu, A.-L. Cheng; IMbrave150 Investigators , Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382, 1894–1905 (2020). [DOI] [PubMed] [Google Scholar]
- 6.X. Jiang, J. Wang, X. Deng, F. Xiong, J. Ge, B. Xiang, X. Wu, J. Ma, M. Zhou, X. Li, Y. Li, G. Li, W. Xiong, C. Guo, Z. Zeng, Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 18, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Binnewies, E. W. Roberts, K. Kersten, V. Chan, D. F. Fearon, M. Merad, L. M. Coussens, D. I. Gabrilovich, S. Ostrand-Rosenberg, C. C. Hedrick, R. H. Vonderheide, M. J. Pittet, R. K. Jain, W. Zou, T. K. Howcroft, E. C. Woodhouse, R. A. Weinberg, M. F. Krummel, Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Q. Zhang, Y. He, N. Luo, S. J. Patel, Y. Han, R. Gao, M. Modak, S. Carotta, C. Haslinger, D. Kind, G. W. Peet, G. Zhong, S. Lu, W. Zhu, Y. Mao, M. Xiao, M. Bergmann, X. Hu, S. P. Kerkar, A. B. Vogt, S. Pflanz, K. Liu, J. Peng, X. Ren, Z. Zhang, Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179, 829–845.e20 (2019). [DOI] [PubMed] [Google Scholar]
- 9.B. Ruf, B. Heinrich, T. F. Greten, Immunobiology and immunotherapy of HCC: Spotlight on innate and innate-like immune cells. Cell. Mol. Immunol. 18, 112–127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.G. L. Semenza, Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. C. Epstein, J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, P. J. Ratcliffe, C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001). [DOI] [PubMed] [Google Scholar]
- 12.W. G. Kaelin Jr., P. J. Ratcliffe, Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008). [DOI] [PubMed] [Google Scholar]
- 13.G. L. Wang, B. H. Jiang, E. A. Rue, G. L. Semenza, Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. K.-C. Chiu, I. M.-J. Xu, R. K.-H. Lai, A. P.-W. Tse, L. L. Wei, H.-Y. Koh, L. L. Li, D. Lee, R. C.-L. Lo, C.-M. Wong, I. O.-L. Ng, C. C.-L. Wong, Hypoxia induces myeloid-derived suppressor cell recruitment to hepatocellular carcinoma through chemokine (C-C motif) ligand 26. Hepatology 64, 797–813 (2016). [DOI] [PubMed] [Google Scholar]
- 15.A. Facciabene, X. Peng, I. S. Hagemann, K. Balint, A. Barchetti, L. P. Wang, P. A. Gimotty, C. B. Gilks, P. Lal, L. Zhang, G. Coukos, Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature 475, 226–230 (2011). [DOI] [PubMed] [Google Scholar]
- 16.H. Zhang, H. Lu, L. Xiang, J. W. Bullen, C. Zhang, D. Samanta, D. M. Gilkes, J. He, G. L. Semenza, HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E6215–E6223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.H. K. Eltzschig, D. Köhler, T. Eckle, T. Kong, S. C. Robson, S. P. Colgan, Central role of Sp1-regulated CD39 in hypoxia/ischemia protection. Am. J. Hematol. 113, 224–232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D. K.-C. Chiu, A. P.-W. Tse, I. M.-J. Xu, J. Di Cui, R. K.-H. Lai, L. L. Li, H.-Y. Koh, F. H.-C. Tsang, L. L. Wei, C.-M. Wong, I. O.-L. Ng, C. C.-L. Wong, Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 8, 517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.K. Synnestvedt, G. T. Furuta, K. M. Comerford, N. Louis, J. Karhausen, H. K. Eltzschig, K. R. Hansen, L. F. Thompson, S. P. Colgan, Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J. Clin. Invest. 110, 993–1002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. Allard, M. S. Longhi, S. C. Robson, J. Stagg, The ectonucleotidases CD 39 and CD 73: Novel checkpoint inhibitor targets. Immunol. Rev. 276, 121–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.G. Haskó, J. Linden, B. Cronstein, P. Pacher, Adenosine receptors: Therapeutic aspects for inflammatory and immune diseases. Nat. Rev. Drug Discov. 7, 759–770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. Ohta, R. Kini, A. Ohta, M. Subramanian, M. Madasu, M. Sitkovsky, The development and immunosuppressive functions of CD4+ CD25+ FoxP3+ regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front. Immunol. 3, 190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Deaglio, K. M. Dwyer, W. Gao, D. Friedman, A. Usheva, A. Erat, J. F. Chen, K. Enjyoji, J. Linden, M. Oukka, V. K. Kuchroo, T. B. Strom, S. C. Robson, Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 204, 1257–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. Stagg, U. Divisekera, H. Duret, T. Sparwasser, M. W. L. Teng, P. K. Darcy, M. J. Smyth, CD73-deficient mice have increased antitumor immunity and are resistant to experimental metastasis. Cancer Res. 71, 2892–2900 (2011). [DOI] [PubMed] [Google Scholar]
- 25.T. Raskovalova, X. Huang, M. Sitkovsky, L. C. Zacharia, E. K. Jackson, E. Gorelik, Gs protein-coupled adenosine receptor signaling and lytic function of activated NK cells. J. Immunol. 175, 4383–4391 (2005). [DOI] [PubMed] [Google Scholar]
- 26.S. Ryzhov, S. V. Novitskiy, A. E. Goldstein, A. Biktasova, M. R. Blackburn, I. Biaggioni, M. M. Dikov, I. Feoktistov, Adenosinergic regulation of the expansion and immunosuppressive activity of CD11b+Gr1+ cells. J. Immunol. 187, 6120–6129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. V. Novitskiy, S. Ryzhov, R. Zaynagetdinov, A. E. Goldstein, Y. Huang, O. Y. Tikhomirov, M. R. Blackburn, I. Biaggioni, D. P. Carbone, I. Feoktistov, M. M. Dikov, Adenosine receptors in regulation of dendritic cell differentiation and function. Blood 112, 1822–1831 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.I. Perrot, H.-A. Michaud, M. Giraudon-Paoli, S. Augier, A. Docquier, L. Gros, R. Courtois, C. Dejou, D. Jecko, O. Becquart, H. Rispaud-Blanc, L. Gauthier, B. Rossi, S. Chanteux, N. Gourdin, B. Amigues, A. Roussel, A. Bensussan, J.-F. Eliaou, J. Bastid, F. Romagné, Y. Morel, E. Narni-Mancinelli, E. Vivier, C. Paturel, N. Bonnefoy, Blocking antibodies targeting the CD39/CD73 immunosuppressive pathway unleash immune responses in combination cancer therapies. Cell Rep. 27, 2411–2425.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 29.S. F. M. Häusler, I. M. Del Barrio, J. Diessner, R. G. Stein, J. Strohschein, A. Hönig, J. Dietl, J. Wischhusen, Anti-CD39 and anti-CD73 antibodies A1 and 7G2 improve targeted therapy in ovarian cancer by blocking adenosine-dependent immune evasion. Am. J. Transl. Res. 6, 129–139 (2014). [PMC free article] [PubMed] [Google Scholar]
- 30.J. D. Young, S. Y. M. Yao, J. M. Baldwin, C. E. Cass, S. A. Baldwin, The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Asp. Med. 34, 529–547 (2013). [DOI] [PubMed] [Google Scholar]
- 31.J. D. Young, The SLC28 (CNT) and SLC29 (ENT) nucleoside transporter families: A 30-year collaborative odyssey. Biochem. Soc. Trans. 44, 869–876 (2016). [DOI] [PubMed] [Google Scholar]
- 32.K. Barnes, H. Dobrzynski, S. Foppolo, P. R. Beal, F. Ismat, E. R. Scullion, L. Sun, J. Tellez, M. W. L. Ritzel, W. C. Claycomb, C. E. Cass, J. D. Young, R. Billeter-Clark, M. R. Boyett, S. A. Baldwin, Distribution and functional characterization of equilibrative nucleoside transporter-4, a novel cardiac adenosine transporter activated at acidic pH. Circ. Res. 99, 510–519 (2006). [DOI] [PubMed] [Google Scholar]
- 33.D. Boison, G. G. Yegutkin, Adenosine metabolism: Emerging concepts for cancer therapy. Cancer Cell 36, 582–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. C. Morote-Garcia, P. Rosenberger, J. Kuhlicke, H. K. Eltzschig, HIF-1-dependent repression of adenosine kinase attenuates hypoxia-induced vascular leak. Blood 111, 5571–5580 (2008). [DOI] [PubMed] [Google Scholar]
- 35.H. Zhang, P. Gao, R. Fukuda, G. Kumar, B. Krishnamachary, K. I. Zeller, C. V. Dang, G. L. Semenza, HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11, 407–420 (2007). [DOI] [PubMed] [Google Scholar]
- 36.P. C. Fernandez, S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, B. Amati, Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Fei, X. Ren, H. Yu, Y. Zhan, Targeting the CCL2/CCR2 axis in cancer immunotherapy: One stone, three birds? Front. Immunol. 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Y. Cheng, X.-l. Ma, Y.-q. Wei, X.-W. Wei, Potential roles and targeted therapy of the CXCLs/CXCR2 axis in cancer and inflammatory diseases. Biochim. Biophys. Acta Rev. Cancer 1871, 289–312 (2019). [DOI] [PubMed] [Google Scholar]
- 39.P. Cheng, C. A. Corzo, N. Luetteke, B. Yu, S. Nagaraj, M. M. Bui, M. Ortiz, W. Nacken, C. Sorg, T. Vogl, J. Roth, D. I. Gabrilovich, Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205, 2235–2249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. Sinha, C. Okoro, D. Foell, H. H. Freeze, S. Ostrand-Rosenberg, G. Srikrishna, Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 181, 4666–4675 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R. Iannone, L. Miele, P. Maiolino, A. Pinto, S. Morello, Blockade of A2b adenosine receptor reduces tumor growth and immune suppression mediated by myeloid-derived suppressor cells in a mouse model of melanoma. Neoplasia 15, 1400–1409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.B. Shamloo, N. Kumar, R. H. Owen, J. Reemmer, J. Ost, R. S. Perkins, H.-Y. Shen, Dysregulation of adenosine kinase isoforms in breast cancer. Oncotarget 10, 7238–7250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.G.-W. Huang, L.-Y. Yang, W.-Q. Lu, Expression of hypoxia-inducible factor 1alpha and vascular endothelial growth factor in hepatocellular carcinoma: Impact on neovascularization and survival. World J Gastroenterol: WJG 11, 1705–1708 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R. El-Kharrag, R. Owen, D. Boison, Adenosine kinase deficiency increases susceptibility to a carcinogen. J. Caffeine Adenosine Res. 9, 4–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. Li, G. A. Smolen, L. F. Beers, L. Xia, W. Gerald, J. Wang, D. A. Haber, S. B. Lee, Adenosine transporter ENT4 is a direct target of EWS/WT1 translocation product and is highly expressed in desmoplastic small round cell tumor. PLOS ONE 3, e2353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.A. L. Harris, Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002). [DOI] [PubMed] [Google Scholar]
- 47.D. J. Manalo, A. Rowan, T. Lavoie, L. Natarajan, B. D. Kelly, S. Q. Ye, J. G. N. Garcia, G. L. Semenza, Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669 (2005). [DOI] [PubMed] [Google Scholar]
- 48.D. R. Mole, C. Blancher, R. R. Copley, P. J. Pollard, J. M. Gleadle, J. Ragoussis, P. J. Ratcliffe, Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 284, 16767–16775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.B. Krishnamachary, D. Zagzag, H. Nagasawa, K. Rainey, H. Okuyama, J. H. Baek, G. L. Semenza, Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor–null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 66, 2725–2731 (2006). [DOI] [PubMed] [Google Scholar]
- 50.D. K.-C. Chiu, A. P.-W. Tse, C.-T. Law, I. M.-J. Xu, D. Lee, M. Chen, R. K.-H. Lai, V. W.-H. Yuen, J. W.-S. Cheu, D. W.-H. Ho, C.-M. Wong, H. Zhang, I. O.-L. Ng, C. C.-L. Wong, Hypoxia regulates the mitochondrial activity of hepatocellular carcinoma cells through HIF/HEY1/PINK1 pathway. Cell Death Dis. 10, 934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. Tiana, B. Acosta-Iborra, L. Puente-Santamaría, P. Hernansanz-Agustin, R. Worsley-Hunt, N. Masson, F. García-Rio, D. Mole, P. Ratcliffe, W. W. Wasserman, B. Jimenez, L. del Peso, The SIN3A histone deacetylase complex is required for a complete transcriptional response to hypoxia. Nucleic Acids Res. 46, 120–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. Vijayan, A. Young, M. W. L. Teng, M. J. Smyth, Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 17, 709–724 (2017). [DOI] [PubMed] [Google Scholar]
- 53.A. Ohta, E. Gorelik, S. J. Prasad, F. Ronchese, D. Lukashev, M. K. K. Wong, X. Huang, S. Caldwell, K. Liu, P. Smith, J.-F. Chen, E. K. Jackson, S. Apasov, S. Abrams, M. Sitkovsky, A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 13132–13137 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. Ohta, A. Ohta, M. Madasu, R. Kini, M. Subramanian, N. Goel, M. Sitkovsky, A2A adenosine receptor may allow expansion of T cells lacking effector functions in extracellular adenosine-rich microenvironments. J. Immunol. 183, 5487–5493 (2009). [DOI] [PubMed] [Google Scholar]
- 55.P. A. Beavis, U. Divisekera, C. Paget, M. T. Chow, L. B. John, C. Devaud, K. Dwyer, J. Stagg, M. J. Smyth, P. K. Darcy, Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc. Natl. Acad. Sci. U.S.A. 110, 14711–14716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R. D. Leone, I.-M. Sun, M.-H. Oh, I.-H. Sun, J. Wen, J. Englert, J. D. Powell, Inhibition of the adenosine A2a receptor modulates expression of T cell coinhibitory receptors and improves effector function for enhanced checkpoint blockade and ACT in murine cancer models. Cancer Immunol. Immunother. 67, 1271–1284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.I. O. L. Ng, E. C. S. Lai, S. T. Fan, M. M. T. Ng, M. K. P. So, Prognostic significance of pathologic features of hepatocellular carcinoma a multivariate analysis of 278 patients. Cancer 76, 2443–2448 (1995). [DOI] [PubMed] [Google Scholar]
- 58.C. C.-L. Wong, D. M. Gilkes, H. Zhang, J. Chen, H. Wei, P. Chaturvedi, S. I. Fraley, C.-M. Wong, U.-S. Khoo, I. O.-L. Ng, D. Wirtz, G. L. Semenza, Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc. Natl. Acad. Sci. U.S.A. 108, 16369–16374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. E. Sanjana, O. Shalem, F. Zhang, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S1 to S4