Abstract
Pathogenic short tandem repeat (STR) expansions cause over 20 neurodegenerative diseases. To determine the contribution of STRs in sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), we used ExpansionHunter, REviewer, and polymerase chain reaction validation to assess 21 neurodegenerative disease-associated STRs in whole-genome sequencing data from 608 patients with sporadic ALS, 68 patients with sporadic FTD, and 4703 matched controls. We also propose a data-derived outlier detection method for defining allele thresholds in rare STRs. Excluding C9orf72 repeat expansions, 17.6% of clinically diagnosed ALS and FTD cases had at least one expanded STR allele reported to be pathogenic or intermediate for another neurodegenerative disease. We identified and validated 162 disease-relevant STR expansions in C9orf72 (ALS/FTD), ATXN1 [spinal cerebellar ataxia type 1 (SCA1)], ATXN2 (SCA2), ATXN8 (SCA8), TBP (SCA17), HTT (Huntington’s disease), DMPK [myotonic dystrophy type 1 (DM1)], CNBP (DM2), and FMR1 (fragile-X disorders). Our findings suggest clinical and pathological pleiotropy of neurodegenerative disease genes and highlight their importance in ALS and FTD.
Expanded segments of DNA known to cause neurodegenerative disease are present in up to one-quarter of ALS and FTD patients.
INTRODUCTION
Short tandem repeats (STRs, also known as microsatellites) are tracts of repetitive DNA units 2 to 6 nt in length that comprise at least 3% of the human genome (1–3). These repetitive sequences are inherently unstable and are prone to both germline and somatic variations in repeat number. Expansion of STRs can be pathogenic, with more than 40 monogenic disorders currently attributed to expanded STR loci (2). Most of these disorders primarily affect the nervous system with almost half leading to neurodegenerative disorders. Many pathogenic mechanisms have been proposed for STR expansions including loss of gene function, formation of repeat-containing RNA foci, polyglutamine aggregation, and repeat-associated non-AUG translation of toxic peptides, as previously reviewed (3). The pathogenic mechanism, and ultimately the phenotypic outcome, is dependent on the location of the STR expansion relative to the gene, the specific nucleotide composition, and, importantly, the number of repeated motifs. Disease-associated STRs are generally categorized into three repeat size ranges: normal (no disease phenotype), intermediate (or premutation), and pathogenic (disease phenotype). While intermediate (or premutation) STR expansions do not usually cause the disease associated with a pathogenic expansion at the locus, they can be more prone to meiotic expansion (genetic anticipation) and may cause or confer risk to related diseases and phenotypes (1, 2).
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two neurodegenerative disorders recognized to exist on a disease spectrum (4). ALS is characterized by the degeneration of upper and lower motor neurons resulting in progressive paralysis, culminating in death typically due to respiratory failure (5, 6). In FTD, the degeneration of the frontal and temporal lobes of the brain causes progressive changes in behavior, language, and movement (7). In most cases, abnormal aggregation of tar DNA binding protein 43 (TDP-43) is associated with the pattern of degeneration (8). Cognitive changes have been identified in more than 50% of patients with ALS, with around 15% demonstrating comorbidity with FTD (9). Conversely, up to 30% of patients with FTD demonstrate motor dysfunction, with 10 to 15% also developing ALS (10, 11). Accompanying the clinical and pathological overlap between ALS and FTD are shared genetic features, such as an intronic STR expansion in the C9orf72 gene, the most commonly known genetic cause of ALS and FTD (12–15). Although this is currently the only STR expansion proven to cause ALS/FTD, intermediate STR expansions in the spinocerebellar ataxia (SCA) genes, ATXN1 (SCA1) and ATXN2 (SCA2), have been associated with ALS risk (16, 17). Pathogenic STR expansions in the HTT gene, recognized to cause clinical and pathological Huntington’s disease (HD), were recently reported in individuals diagnosed with clinical and pathological ALS-FTD (18), with such cases overrepresented in HD brain banks (19). The emerging evidence suggests that STR expansions have shared pathophysiological mechanisms that can give rise to different clinical phenotypes (e.g., ALS or FTD) and, in certain circumstances, different underlying pathologies (e.g., HD or ALS-FTD). Because of technical limitations, the extent of pleiotropy in genes associated with repeat expansion disorders is yet to be assessed, although there is in silico evidence of higher frequencies in the population than previously thought (20).
The genetic etiologies of ALS and FTD are complex, with disease-linked mutations identified in more than 30 genes for ALS and 13 genes in FTD to date (7, 21). Several genes are clinically pleiotropic, causing both ALS and FTD (22). While 90% of patients with ALS are classified as sporadic, heritability estimates based on parent-offspring and twin studies in ALS suggest that genetic factors account for between 40 and 60% of sporadic ALS (sALS) risk respectively (23–25). For patients with FTD, at least 30% have a family history of disease, with behavioral variant FTD and ALS-FTD demonstrating the greatest heritability (up to 50%) (7). Despite this apportioned genetic contribution, a causal genetic variant has only been identified in about 10% of ALS and FTD cases (15). Excluding the large genetic contribution of STR expansions in C9orf72, most of the other known genetic factors include missense mutations, insertions, and deletions (7, 21). Other variants such as STRs, which until recently have been challenging to detect because of low throughput and cost of detection techniques, could represent a source of the missing heritability. The detection of STR expansions has traditionally relied on time-consuming laboratory-based methods that are not conducive to high-throughput screening, including fragment analysis, repeat-primed polymerase chain reaction (PCR) and Southern blot assays. However, the growing accessibility and availability of whole-genome sequencing (WGS) data, has seen a growth in bioinformatic tools capable of detecting STR expansions on a genome-wide scale. Bioinformatic tools such as ExpansionHunter (26, 27) have recently been used to systematically screen many known STR loci across disease cohorts including autism spectrum disorder (28) and broad groups of neurodegenerative disorders (29), resulting in the identification of rare disease-linked STR expansions. Online databases such as the Genome Aggregation Database (gnomAD) (30) now provide population frequencies of disease-relevant STR expansions, which are invaluable for estimating the prevalence of neurodegenerative and neurological expansions in the general population.
Here, we specifically selected 21 STR expansions known to cause motor phenotype–related neurodegenerative diseases. We used ExpansionHunter to genotype these 21 STR loci in WGS data from 608 patients with sALS, 68 patients with sporadic FTD (sFTD), and 4703 population-matched control participants and performed PCR-based validation of identified expansions in the clinically diagnosed patients with ALS and FTD. We identified intermediate and pathogenic repeat expansions in C9orf72, as well as genes implicated in HD (HTT), SCA (ATXN1, ATXN2, ATXN8, and TBP), myotonic dystrophy (DMPK and CNBP), and fragile X syndrome/fragile X-associated tremor/ataxia syndrome (FMR1).
RESULTS
sALS and sFTD patients showed phenotypic variability
WGS data were available for STR analysis of 608 patients with sALS and 68 patients with sFTD. Mutation and clinical summary data have been previously reported for 426 of 608 patients with sALS in the study by McCann et al. (21) and is reported here for all 608 patients with sALS. Forty-eight patients with sALS carried known disease-related mutations including SOD1 p.I114T (n = 3), TARDBP p.I383V (n = 2), TARDBP p.G287S (n = 1), and TARDBP p.G295C (n = 1) and pathogenic repeat expansions in C9orf72 (n = 41), representative of a typical sALS cohort recruited to a neurology clinic. Patients with sFTD did not harbor any known disease-related mutations in MAPT, GRN, or C9orf72.
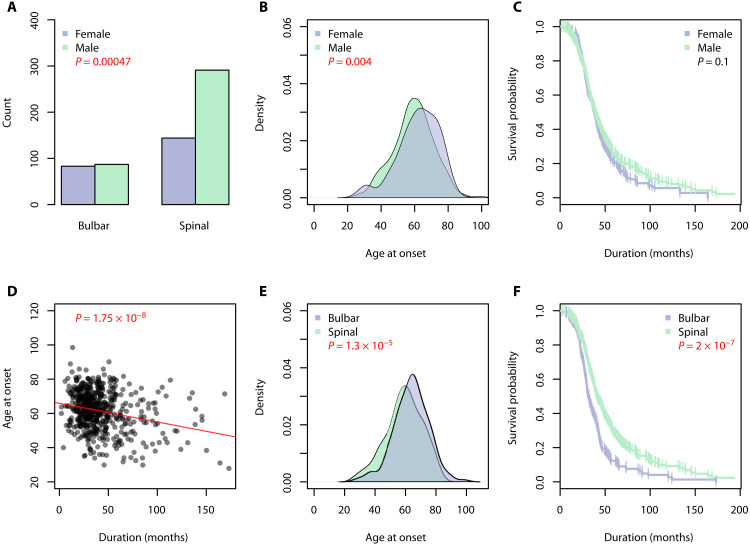
In the sALS cohort, around two-thirds of cases were male. Differences in the site of disease onset (P = 0.0005) and age of onset (P = 0.004) were observed between male and female patients with ALS (Fig. 1, A and B). Seventy-seven percent of males presented with spinal onset ALS compared to 63% of females. The average age of disease onset was 59 years in males and 62 years in females. The average disease duration for patients with sALS was 44 months, with no difference between males and females (Fig. 1C; P = 0.1). Individuals who presented with ALS at a younger age had a longer disease duration (Fig. 1D; P = 1.75 × 10−8). On average, patients with sALS diagnosed in their 40s lived for 52 months compared to 44 months and 38 months for individuals diagnosed with ALS in their 50s and 60s, respectively. Patients with sALS with spinal onset disease had a younger age of onset (mean difference of 4.7 years, P = 1.3 × 10−5) and slower disease progression (median difference of 7.8 months, P = 2 × 10−7) compared with bulbar onset cases (Fig. 1, E and F).
Fig. 1. Statistical analysis of clinical variables in sALS cohort (n = 608).
Significant associations are denoted by red P values. (A) There was a significant difference in the site of onset between males and females with ALS, with more males presenting with spinal onset ALS. (B) The age of onset was significantly older in females with ALS. (C) Males with ALS tended to have a longer disease duration, although not statistically significant. (D) Patients diagnosed with ALS at a younger age lived significantly longer than those diagnosed later in life. (E) Patients with bulbar onset ALS typically presented with disease at an older age. (F) A significant difference in ALS disease duration was observed between the site of onset, with bulbar onset having a worse prognosis.
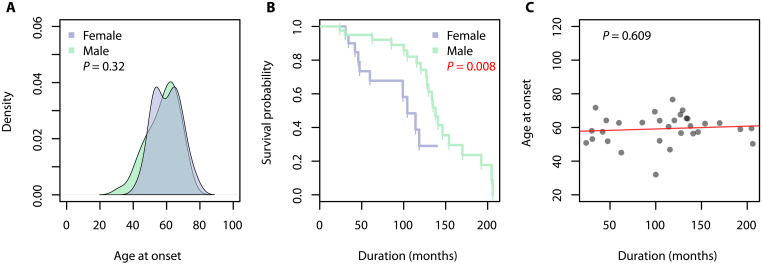
In the sFTD cohort, males comprised 70% of cases. The average age of disease onset in sFTD was 59 years with no difference between males and females (Fig. 2A; P = 0.32). A difference in disease duration was observed between males and females (Fig. 2B; P = 0.008), with females having a more rapid disease progression. On average, females with sFTD lived for 70 months following diagnosis while males lived for 129 months. There was no association between the age of disease onset and disease progression in patients with sFTD (Fig. 2C; P = 0.609).
Fig. 2. Statistical analysis of clinical variables in sFTD cohort (n = 68).
Significant associations are denoted by red P values. (A) There was no significant difference in the age of onset between males and females with FTD. (B) Females with FTD have a significantly worse prognosis compared to male with FTD. (C) No associations were found between the age of onset and the disease duration in patients with FTD.
Neurodegenerative disease STR expansions were identified in 22% of patients with sALS and sFTD
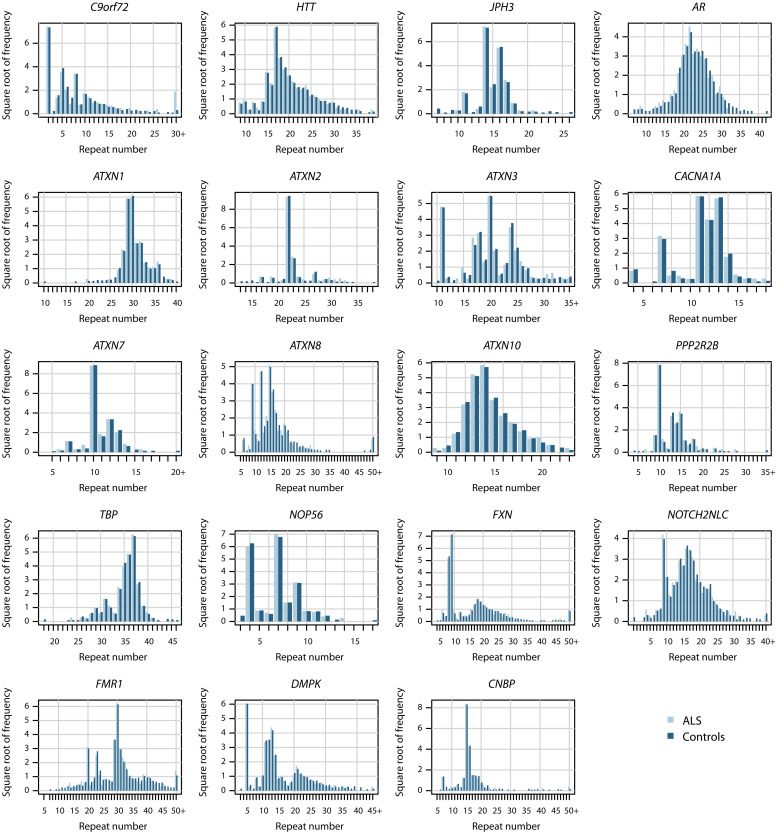
Individuals were screened for 21 repeat expansions known to cause motor phenotype–related neurodegenerative disorders using ExpansionHunter v4.0.1 (26, 27) (table S1). We examined repeat expansions with length estimates within the reported intermediate and pathogenic ranges for each neurodegenerative disease gene obtained from the literature. The accuracy of the repeat lengths estimated by ExpansionHunter was assessed for all 21 loci in 10 randomly selected samples for each STR locus by visualizing the alignment of reads in the tandem repeat regions using REViewer (31) (figs. S1 to S21). ExpansionHunter could not accurately identify STRs in BEAN (SCA31) or DAB1 (SCA37) (figs. S14 and S16); therefore, we removed these two loci from further analysis.
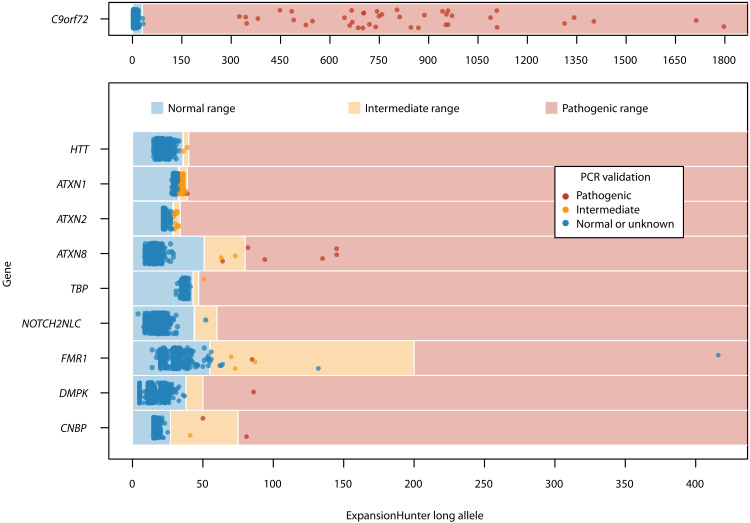
In patients with sALS and sFTD, we identified 276 repeat expansions above the intermediate and/or pathogenic thresholds for 10 neurodegenerative disease genes (Table 1). ExpansionHunter correctly identified all patients with sALS with a pathogenic expansion in C9orf72 previously identified through routine repeat-primed PCR screening. The remaining nine loci included expansions in ATXN1 and ATXN2, which have been previously implicated in sALS and/or sFTD, as well as HTT (HD), ATXN8 (SCA8), TBP (SCA17), NOTCH2NLC [neuronal intranuclear inclusion disease (NIID)], FMR1 [fragile X syndrome/fragile X-associated tremor/ataxia syndrome (FRAXA/FXTAS)], DMPK [myotonic dystrophy type 1 (DM1)] and CNBP [myotonic dystrophy type 2 (DM2)]. Repeat-primed PCR and fluorescence PCR genotyping were performed to validate ExpansionHunter results. All ATXN1 PCR genotypes were exactly one repeat unit shorter than the lengths estimated by ExpansionHunter. Visual inspection of aligned reads over the ATXN1 STR locus using REViewer showed that the repeat boundaries were out of frame, leading to the false inclusion of an additional codon in the ExpansionHunter allele length estimates (fig. S22). We adjusted ExpansionHunter ATXN1 allele lengths by subtracting one repeat unit from the size estimates to produce an allele frequency distribution that was consistent with PCR results (32–34). Of the remaining 173 repeat expansions above the intermediate and/or pathogenic thresholds estimated by ExpansionHunter (Fig. 3), 10 of 173 (5.78%) had PCR genotypes that were within the normal range [FMR1 (n = 7), C9orf72 (n = 2) and NOTCH2NLC (n = 1)]. One sample failed repeated PCR validation attempts for FMR1. Overall, 162 of 173 (93.6%) of pathogenic and intermediate expansions identified by ExpansionHunter were confirmed by PCR genotyping to be pathogenic or intermediate in 150 of 676 (22%) patients with sALS and sFTD (Table 1 and table S2). Excluding repeat expansions in C9orf72, 119 of 676 (17.6%) patients with sALS and sFTD harbored a PCR-validated pathogenic or intermediate repeat expansion in a gene known to cause other neurodegenerative diseases (table S2).
Table 1. Intermediate and pathogenic STR expansions identified in ALS, FTD, and control individuals using ExpansionHunter and PCR genotyping.
| ExpansionHunter v4 frequencies | PCR validation frequencies in ALS/FTD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Disease ID | Gene | Expansion type | Control | sALS | sFTD | sALS | sFTD | |||
| n = 4703* | n = 608* | n = 68* | ||||||||
| n | % | n | % | n | % | |||||
| FTDALS1 | C9orf72 | Pathogenic | 9 | 0.19 | 43 | 7.07 | 0 | 0 | 41 | 0 |
| HD | HTT | Intermediate | 6 | 0.13 | 2 | 0.33 | 0 | 0 | 2 | 0 |
| SCA1 | ATXN1 | Pathogenic | 5 | 0.11 | 1 | 0.16 | 0 | 0 | 1 | 0 |
| Intermediate | 564 | 12 | 81 | 13.3 | 8 | 11.8 | 81 | 8 | ||
| SCA2 | ATXN2 | Intermediate | 31 | 0.66 | 12 | 1.97 | 0 | 0 | 12 | 0 |
| SCA8 | ATXN8 | Pathogenic | 40 | 0.85 | 5 | 0.82 | 0 | 0 | 6 | 0 |
| Intermediate | 35 | 0.74 | 2 | 0.33 | 1/67 | 1.49 | 1 | 1 | ||
| SCA17 | TBP | Pathogenic | 0 | 0 | 0 | 0 | 1 | 1.47 | 0 | 0 |
| Intermediate | 6 | 0.13 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| NIID | NOTCH2NLC | Intermediate | 10/4690 | 0.21 | 1 | 0.16 | 0 | 0 | 0 | 0 |
| FRAXA/ FXTAS | FMR1 | Pathogenic | 0 | 0 | 1/607 | 0.16 | 0 | 0 | 1† | 0 |
| Intermediate | 56/4699 | 1.19 | 10/607 | 1.65 | 1 | 1.47 | 3 | 0 | ||
| DM1 | DMPK | Pathogenic | 1 | 0.02 | 1 | 0.16 | 0 | 0 | 1 | 0 |
| DM2 | CNBP | Pathogenic | 0 | 0 | 1 | 0.16 | 0 | 0 | 2 | 0 |
| Intermediate | 20 | 0.43 | 2 | 0.33 | 0 | 0 | 1 | 0 | ||
| Total | 783 | 16.65‡ | 162 | 26.64‡ | 11 | 16.18‡ | 152 | 10 | ||
*Total number of control/ALS/FTD individuals unless otherwise stated.
†Individual has both an intermediate and pathogenic STR expansion in FMR1 and has only been included in the pathogenic count.
‡Percentage calculated using the total number of repeat expansions divided by the total number of individuals within that group.
Fig. 3. ExpansionHunter STR expansion size estimates (number of repeats) for the long allele and PCR validation results for 608 patients with sALS and 68 patients with sFTD.
C9orf72 has been graphed separately to allow for a larger expansion range. The color of each point is reflective of the PCR validation results. Note that only intermediate and pathogenic expansions identified by ExpansionHunter were PCR-validated.
In patients with sALS and sFTD, PCR genotyped expansions exceeding the pathogenic thresholds for each neurodegenerative disease gene were observed in patients with sALS only. C9orf72 had the greatest number of individuals harboring pathogenic expansions (n = 41 of 608, 6.74%), consistent with C9orf72 expansion frequencies in Caucasian patients with sALS (35). Pathogenic repeat expansions were also confirmed in ATXN8 (n = 6, 0.99%), CNBP (n = 2, 0.33%), ATXN1 (n = 1, 0.16%), DMPK (n = 1, 0.16%), and FMR1 (n = 1, 0.16%) (Table 1). The patient with sALS harboring a pathogenic STR expansion in FMR1 was also found to harbor an intermediate expansion in FMR1 on the other allele. Intermediate repeat expansions were observed in ATXN1 (n = 81 sALS cases or 13.3%; n = 8 sFTD cases or 11.8%), ATXN2 (n = 12 sALS cases, 2.0%), FMR1 (n = 3 sALS cases, 0.49%), HTT (n = 2 sALS cases, 0.33%), ATXN8 (n = 1 sALS case or 0.16%; n = 1 sFTD case or 1.47%), TBP (n = 1 sFTD case, 1.47%), and CNBP (n = 1 sALS case, 0.16%).
For all 19 individuals with pathogenic or intermediate expansions in neurodegenerative disease genes not previously implicated in ALS or FTD (i.e., excluding C9orf72, ATXN1, or ATXN2), we assessed available clinical data and postmortem pathology records to confirm a diagnosis of ALS or FTD (available clinical summaries for these patients is provided in Supplementary Text).
Repeat expansions associated with neurodegenerative disorders were also present in controls
To compare the frequency of repeat expansions against a control population, ExpansionHunter v4.0.1 was run on 4703 neurologically healthy individuals (n = 2309 males; n = 2394 females) of population-matched ancestry (Tables 1 and 2, Fig. 4, and fig. S23). Expansions exceeding the pathogenic threshold were found in control participants in C9orf72, ATXN1, ATXN8, and DMPK but not TBP, FMR1, or CNBP (Table 1). Intermediate repeat expansions were found at the same loci in cases and controls (Table 1). Because DNA was not available for PCR-based validation of control participants, ExpansionHunter-identified expanded repeats could not be PCR genotyped in the control cohort. Hence, repeat expansion frequency comparisons use only ExpansionHunter repeat sizes. Following correction for multiple testing of 19 genes, pathogenic expansions in C9orf72 and intermediate expansions in ATXN2 were overrepresented in patients with sALS compared to controls (P = 2.25 × 10−32 and P = 0.0025, respectively; Table 2). Pathogenic expansions in C9orf72 were >30 times more common in patients with sALS compared to controls (7.07% versus 0.19%, P = 2.25 × 10−32; Tables 1 and 2). Intermediate ATXN2 expansions were three times more common in patients with sALS compared to controls (1.97% versus 0.66%, P = 0.0025; Tables 1 and 2), while intermediate ATXN1 expansions were present at similar frequencies in sALS, sFTD and control individuals (sALS: 13.3% versus 12%, P = 0.197; sFTD: 11.8% versus 12%, P = 0.574; Tables 1 and 2). Intermediate HTT expansions were more than twice as common in patients with sALS compared to controls (0.33% versus 0.13%, P = 0.231; Tables 1 and 2), and pathogenic expansions in TBP (SCA17), DMPK (DM1), and CNBP (DM2) were either absent from controls or in a single control (P = 0.014, P = 0.216, and P = 0.115, respectively; Table 2). Across all loci in Table 1, intermediate and pathogenic repeat expansions determined by ExpansionHunter were overrepresented in cases (n = 157 of 676 or 23.2%) compared to controls (n = 766 of 4703 or 16.3%, P = 7.7 × 10−6) (table S2). Excluding repeat expansions in C9orf72, a higher proportion of cases (n = 125 of 676 or 18.5%) had a suspected pathogenic or intermediate expansion in a neurodegenerative disease gene compared to controls (n = 758 of 4703 or 16.1%), although not statistically significant (P = 0.119).
Table 2. The odds ratios and P values for repeat expansions identified using ExpansionHunter in patients with sALS/sFTD versus controls.
NA, not applicable.
| Disease ID | Gene | Expansion type | Control | sALS | sFTD | ||||
|---|---|---|---|---|---|---|---|---|---|
| n | n | Odds ratio | P value | n | Odds ratio | P value | |||
| FTDALS1 | C9orf72 | Pathogenic | 9 | 43 | 38.23 | 2.25 × 10−32 | 0 | NA | NA |
| HD | HTT | Intermediate | 6 | 2 | 2.58 | 0.2309 | 0 | NA | NA |
| SCA1 | ATXN1 | Pathogenic | 5 | 1 | 1.55 | 0.5179 | 0 | NA | NA |
| Intermediate | 564 | 81 | 1.09 | 0.2472 | 8 | 0.95 | 0.6117 | ||
| SCA2 | ATXN2 | Intermediate | 31 | 12 | 3.01 | 0.0025 | 0 | NA | NA |
| SCA8 | ATXN8 | Pathogenic | 40 | 5 | 0.97 | 0.5981 | 0 | NA | NA |
| Intermediate | 35 | 2 | 0.44 | 0.9360 | 1 | 4.06 | 0.0949 | ||
| SCA17 | TBP | Pathogenic | 0 | 0 | NA | NA | 1 | Inf | 0.0143 |
| NIID | NOTCH2NLC | Intermediate | 10 | 1 | 0.77 | 0.7380 | 0 | NA | NA |
| FRAXA/ FXTAS | FMR1 | Pathogenic | 0 | 1 | Inf | 0.1143 | 0 | NA | NA |
| Intermediate | 56 | 10 | 1.58 | 0.0795 | 1 | 1.88 | 0.2955 | ||
| DM1 | DMPK | Pathogenic | 1 | 1 | 7.74 | 0.2159 | 0 | NA | NA |
| DM2 | CNBP | Pathogenic | 0 | 1 | Inf | 0.1145 | 0 | NA | NA |
| Intermediate | 20 | 2 | 0.77 | 0.7353 | 0 | NA | NA | ||
Fig. 4. ExpansionHunter STR length frequency distributions in 608 patients with sALS and 4703 control participants for 19 neurodegenerative disease loci.
The y axis is the square root of the total number of alleles for each repeat length to allow better depiction of frequency variations.
Evidence of oligogenic expansions in patients with sALS
We identified 12 patients with sALS (12 of 608 = 1.97%) who harbored more than one disease-related variant (intermediate/pathogenic expansion or known mutation) in genes implicated in neurodegenerative disorders (Table 3). Of these, 10 cases had a pathogenic C9orf72 expansion in addition to either a pathogenic expansion in ATXN8 (n = 1) or FMR1 (n = 1) or an intermediate expansion in ATXN1 (n = 6), ATXN2 (n = 1), or CNBP (n = 1). A pathogenic CNBP expansion was observed in one case with both an intermediate ATXN1 and ATXN2 expansion. One sALS case with a SOD1 p.I114T mutation also carried an intermediate-sized expansion in FMR1. A multinomial logistic regression analysis was performed to assess whether the number of disease-related variants carried by a sALS case influenced their clinical presentation. No clinical variables were associated with the number of disease-related variants harbored by an individual.
Table 3. Summary of 12 patients with sALS with multiple variants detected following PCR validation.
Genetic variation known to be pathogenic are in bold. NA, not available.
| Variant one | Variant two | Variant three | Sex | Diagnosis | Age onset | Disease duration (months) |
|---|---|---|---|---|---|---|
| C9orf72 pathogenic (FTDALS1) | ATXN8 pathogenic (SCA8) | Male | sALS* | 53.0 | 25.7 | |
| C9orf72 pathogenic (FTDALS1) | FMR1 pathogenic (FRAXA) | Male | sALS*† | 57.1 | 19.3 | |
| C9orf72 pathogenic (FTDALS1) | CNBP intermediate (DM2) | Female | sALS* | 56.7 | 71.0 | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Male | sALS | 66.4 | 13.2 | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Male | sALS | 45.7 | 45.5 | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Male | sALS | 68.0 | 28.6 | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Female | sALS | 63.3 | 8.2 (alive) | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Male | sALS | 65.2 | 48.7 | |
| C9orf72 pathogenic (FTDALS1) | ATXN1 intermediate | Male | sALS | NA | NA | |
| C9orf72 pathogenic (FTDALS1) | ATXN2 intermediate | Male | sALS | 49.9 | 38.9 | |
| CNBP pathogenic (DM2) | ATXN2 intermediate | ATXN1 intermediate | Female | sALS* | 39.0 | 36 (alive) |
| SOD1 p.I114T (ALS) | FMR1 intermediate (FXTAS) | Female | sALS* | 54.5 | 17.4 (alive) |
*Detailed clinical data available in Supplementary Text.
†Postmortem neuropathology report confirmed ALS diagnosis.
Statistical detection of outlier allele length thresholds
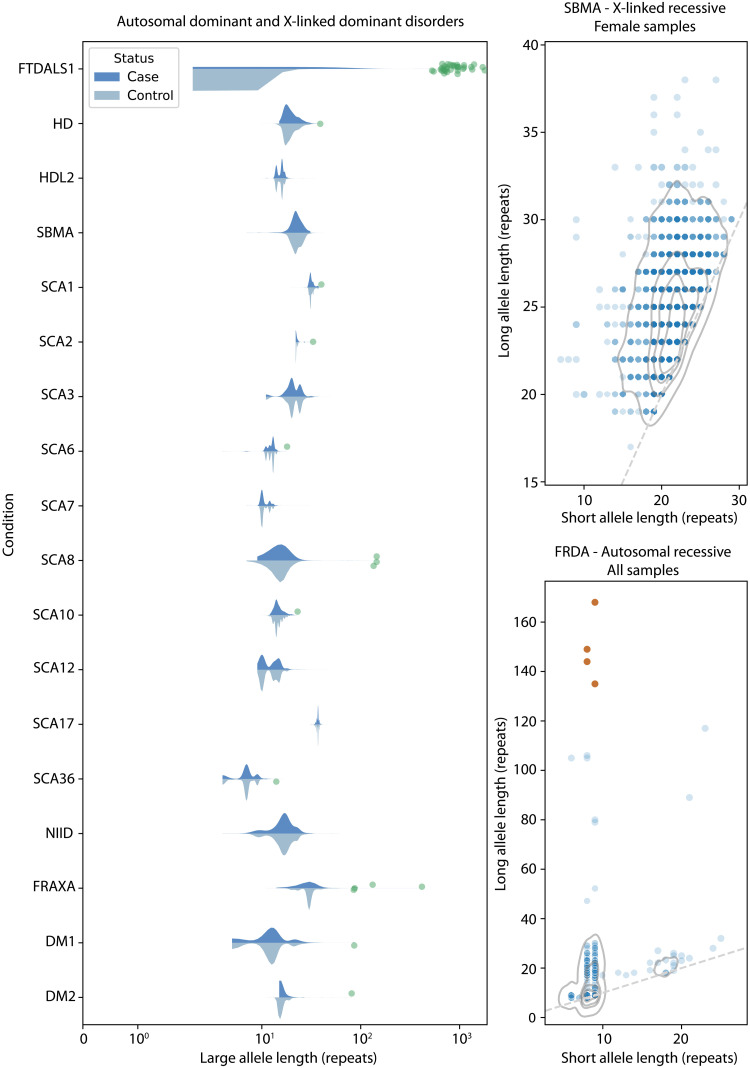
For each neurodegenerative disease gene, a data-derived outlier detection method was used to determine a threshold that 0.1% of allele lengths exceeded following 10,000 iterations of bootstrap sampling of ExpansionHunter allele lengths in the control cohort (Table 4). Samples from the ALS and FTD cohorts were considered outliers for a disease locus if their ExpansionHunter repeat length exceeded the defined outlier threshold (Fig. 5 and fig. S24).
Table 4. Data-derived repeat length thresholds using the 99.9th percentile statistical threshold and number of patients with sALS and sFTD exceeding the threshold.
| Gene | Disease ID | 99.9th percentile threshold | Repeat size difference compared to literature-defined pathogenic threshold | Patients with sALS and sFTD exceeding data-derived threshold (n) |
|---|---|---|---|---|
| C9orf72 | FTDALS1 | 512 | + 482 | 34 |
| HTT | HD | 39 | − 1 | 1 |
| JPH3 | HDL2 | 30 | − 12 | 0 |
| AR | SBMA | 38 | 0 | 0 |
| ATXN1 | SCA1 | 40 | + 1 | 1 |
| ATXN2 | SCA2 | 33 | − 1 | 1 |
| ATXN3 | SCA3/MJD | 43 | − 18 | 0 |
| CACNA1A | SCA6 | 18 | − 2 | 1 |
| ATXN7 | SCA7 | 20 | − 17 | 0 |
| ATXN8 | SCA8 | 122 | + 42 | 3 |
| ATXN10 | SCA10 | 23 | − 377 | 1 |
| PPP2R2B | SCA12 | 35 | − 16 | 0 |
| TBP | SCA17 | 45 | − 2 | 1 |
| NOP56 | SCA36 | 13 | − 637 | 1 |
| FXN | FRDA | 134 | + 64 | 5* |
| NOTCH2NLC | NIID | 55 | − 5 | 0 |
| FMR1 | FRAXA/FXTAS | 76 | − 124 | 4 |
| DMPK | DM1 | 43 | − 7 | 1 |
| CNBP | DM2 | 69 | − 6 | 1 |
*Patients have one expanded allele and are considered carriers for the autosomal recessive disorder.
Fig. 5. Outlier alleles in 608 patients with sALS using 99.9th percentile data-derived thresholds.
Distributions of repeat lengths in patients with sALS (dark blue) and controls (light blue) are shown for diseases inherited in a dominant pattern (either autosomal dominant, X-linked dominant, or male carriers of X-linked recessive STRs). Two recessive disorders [spinal bulbar muscular atrophy (SBMA), an X-linked STR; and Friedreich’s ataxia (FRDA), an autosomal recessive STR] are shown in scatter plots of patient allele lengths with control distributions rendered as contours at the quartiles of the distribution. Allele lengths detected as outliers in patients with sALS are indicated by green points. For recessive disorders, patients with only one allele detected as an outlier are indicated by orange points (i.e., carriers).
DISCUSSION
Heritability studies suggest a substantial genetic component of up to 60% underlies sALS and sFTD disease (23–25). Yet, few causal genes and risk alleles have been identified to date and single-nucleotide polymorphism (SNP)–based heritability estimates are only 8.5% (26). This suggests that complex genetic variation may contribute to the missing heritability. Notably, the most common known cause of ALS and FTD is the pathogenic STR expansion in an intron of C9orf72, which has been identified in around 7% of sALS cases and 6% of sFTD cases (28). There is no current SNP-based evidence that there are additional common pathogenic STRs in ALS genome-wide arising from a common founder (27). Until recently, there has been limited capacity to systematically screen large disease cohorts for STR expansions that may be associated with disease. Bioinformatics tools have been developed to screen WGS data for many STR loci simultaneously, and associations between rare repeat expansions and clinically distinct neurodegenerative disorders have been uncovered, resolving genetic heterogeneity (36–37). Of interest is whether these repeat expansions display pleiotropy; if patients have been misdiagnosed; or whether they are simply present without influencing the alternately diagnosed phenotype.
Using WGS data, ExpansionHunter identified rare pathogenic repeat expansions in sALS and sFTD cases that were completely absent from 4703 control participants in TBP (SCA17), FMR1 (FRAXA/FXTAS), and CNBP (DM2). The pathogenic FMR1 expansion was determined to be within the normal repeat range using PCR genotyping validation; however, expansions in TBP and CNBP were confirmed above intermediate or pathogenic thresholds following PCR genotyping. DNA was not available for control participants, and therefore, the frequency of intermediate and pathogenic expansions in cases versus controls could only be compared using ExpansionHunter results. Across 19 genes, repeat expansions from WGS data were overrepresented in cases compared to controls (23.2% versus 16.3%). Because repeat expansions in C9orf72 are causal for ALS/FTD and are significantly overrepresented in this cohort with an odds ratio of 38.2, even when omitting C9orf72 we were still able to demonstrate a larger proportion of cases with a repeat expansion in a neurodegenerative disease gene compared to controls (18.5% versus 16.1%). When considering all 19 individual genes, and after multiple testing correction, significant differences in the frequency of repeat expansions in patients with ALS versus controls were observed for pathogenic expansions in C9orf72 (P = 2.25 × 10−32) and intermediate-sized expansions in ATXN2 only (P = 0.0025), replicating previous studies (13, 14, 16). Intermediate expansions in ATXN1 were found in 12 to 13% of all cases and controls and were not associated with ALS or FTD disease risk in our study. A similar frequency of intermediate-sized ATXN1 expansions has been observed in other ALS cohorts where a statistical association with ALS disease risk was determined (38). Pathogenic repeat expansions in the HD-associated locus, HTT, were recently reported to be a risk factor for pathologically proven TDP-43–positive ALS and FTD, further evidence of pleiotropy in STR expansions (18). While no patients with sALS or sFTD harbored pathogenic HTT repeat expansions in our study, two patients with sALS carried intermediate expansions in HTT. This was more than twice the frequency seen in control individuals, although intermediate expansions in HTT were not significantly enriched in patients after correction for multiple testing of 19 genes.
After PCR-based validation, we confirmed repeat expansions in 22% (17.6% excluding C9orf72) of 676 patients with sALS and sFTD identified using ExpansionHunter. The repeat expansions were larger than the reported pathogenic and intermediate thresholds for nine respective neurodegenerative disease genes. These included C9orf72 (6.74% as expected for ALS/FTD (21, 28)) but also HTT, ATXN1, ATXN2, ATXN8, TBP, DMPK, CNBP, and FMR1, where expansions above the pathogenic thresholds typically cause ALS/FTD, HD, SCA types 1, 2, 8, and 17, myotonic dystrophy types 1 and 2, and fragile X syndrome/fragile X-associated tremor/ataxia syndrome, respectively. Such clinical pleiotropy has been identified previously in individuals diagnosed with other neurodegenerative disorders (18, 39–41), and, in considering the alternate possibility of misdiagnosis of patients harboring pathogenic repeat expansions, careful examination of the clinical records indicated that misdiagnosis was unlikely in our study.
All available clinical reports for the patients with sALS with pathogenic repeat expansions in ATXN1 (SCA1, n = 1), ATXN8 (SCA8, n = 6), FMR1 (FRAXA, n = 1), DMPK (DM1, n = 1), and CNBP (DM2, n = 2) were consistent with a diagnosis of ALS (Supplementary Text). Patients had classic limb or bulbar onset ALS, with progressive upper and lower motor neuron weakness as well as widespread denervation using electromyography and normal magnetic resonance imaging (MRI) scans of the brain and spinal cord. There were no signs of cognitive impairment, speech and language difficulties, or mood disturbances in one patient with sALS harboring a pathogenic FMR1 repeat expansion, as may be expected in fragile X syndrome (FRAXA). A postmortem report confirmed a neuropathological diagnosis of ALS in this patient. Pathogenic FMR1 expansions give rise to FRAXA through hypermethylation of the FMR1 promotor region, resulting in gene silencing and the absence of the FMR1 encoded protein, FMRP (42, 43). FMRP is known to directly interact with TDP-43 and Staufen to form a functional complex involved in mRNA dendritic transport and translation (44). The recruitment of FMRP to TDP-43–positive stress granules reduces TDP-43 aggregation and restores translation of specific mRNAs in motor neurons (44). PCR genotyping showed two signals for FMR1 in this male patient with sALS, indicating one intermediate expansion in FMR1, which usually causes fragile X-associated tremor/ataxia syndrome (FXTAS), and one pathogenic expansion in FMR1 (FRAXA). In contrast to pathogenic expansions, intermediate expansions of FMR1 are typically unmethylated and produce FMRP. Studies of mosaicisms of FMR1 expansions in males have shown that nonzero expression of FMRP, due to shorter unmethylated FMR1 expansions, may be sufficient to positively affect cognitive function in individuals with pathogenic FMR1 expansions (42). Some production of FMRP with such mosaicism of FMR1 may explain the lack of cognitive impairment in the patient with sALS harboring both a pathogenic and intermediate FRM1 expansion, but the potentially reduced quantities of FMRP would allow TDP-43 to abnormally aggregate.
An intermediate repeat expansion in TBP, which is clinically associated with SCA17, was recently found to segregate with disease in a family with partially penetrant FTD and cerebellar atrophy (40). We identified one patient with sFTD with an intermediate repeat expansion in TBP who was originally diagnosed with behavioral variant FTD but was later diagnosed with progressive supranuclear palsy (PSP) (Supplementary Text). While this patient showed some clinical signs consistent with SCA (gait disturbances and problems with eye movements), these phenotypes are consistent with PSP and no cerebellar abnormalities were seen in MRI scans. Pathogenic and intermediate repeat expansions in TBP are extremely rare and have been associated with unique clinical features, including dementia and behavioral changes (40), demonstrating a possible link to disease in this patient.
In DM1, the age of onset and disease severity is generally inversely correlated with the repeat length (45). Congenital DM1 typically has DMPK repeat sizes >1000, whereas childhood DM1 (onset age, 1 to 10 years) and classic DM1 (onset age, 10 to 30 years) typically have between 50 and 1000 repeats. Mild DM1 cases (50 to 100 repeats; age onset, 20 to 70 years) typically display cataracts and mild myotonia symptoms and can have a full life expectancy (45, 46). One patient with sALS harbored a pathogenic expansion in DMPK comprising 81 repeats and was diagnosed with progressive bulbar palsy (PBP) at age 54. Clinical notes briefly mentioned development of early cataracts with removal in his mid-30s. No other symptoms of classic DM1 were reported in this patient. This patient likely had mild undiagnosed DM1 in his early 30s, developing comorbid PBP in his 50s.
Two patients with sALS have repeat expansions beyond the pathogenic threshold in CNBP; causal for DM2, where the onset of DM2 is in the third to fourth decade (20 to 40 years) of life (47). One patient was diagnosed with ALS-FTD at age 69 and died at age 73 following disease progression typical of ALS-FTD with no comorbid diagnosis of DM2. The second patient was clinically diagnosed with ALS at age 39 with no reported symptoms suggestive of DM2, nor a prior diagnosis of DM2. Clinical notes were only available for this patient at one time-point following ALS diagnosis at age 39, so it is plausible that this patient may have developed comorbid myotonic dystrophy after their last recorded presentation at the neurology clinic.
Six patients with sALS were found to harbor a pathogenic repeat expansion in the SCA8-associated gene, ATXN8, yet were clinically diagnosed with ALS. A similar percentage of control participants were also identified as having an expanded repeat in ATXN8. Large repeat expansions in ATXN8 have been identified in asymptomatic relatives of both familial and sporadic SCA8 patients, suggesting that genetic modifiers or environmental factors may influence disease onset (48).
Two patients with pathogenic expansions in ATXN8 or FMR1 also harbored a pathogenic C9orf72 repeat expansion. These patients were diagnosed with ALS in their mid-50s and had disease progression typical of C9orf72 mutation carriers (49). A multistep hypothesis has been proposed in ALS, whereby the cumulation of multiple genetic variants accounts for a greater number of steps in the model, requiring fewer disease-relevant factors before ALS is established (50). McCann et al. (21) recently demonstrated that patients with sALS who harbored more disease-implicated variants had an earlier age of onset. We identified 11 oligogenic patients with sALS who harbored multiple repeat expansions within the intermediate or pathogenic range and 1 patient with sALS with a SOD1 p.I114T mutation and an intermediate expansion. No significant associations were found between clinical variables, including age of onset, and the number of variants, suggesting that the presence of multiple repeat expansions that cause neurodegenerative disorders does not influence the ALS multistep hypothesis; however, this is based on a small sample size.
ExpansionHunter uses reads aligned to the reference genome to estimate the size of STR expansions. Several factors influence the capacity of ExpansionHunter to accurately size repeat expansions, including the read length, locus coverage, and complexity of the repeat motif (29, 51). Two loci, BEAN (SCA31) and DAB1 (SCA37), were removed from analysis as ExpansionHunter sized the incorrect repeat motif (figs. S14 and S16). Both BEAN and DAB1 STR expansions are complex pentanucleotide repeats that contain the disease-motif (TGGAA and TGAAA, respectively) flanked by a benign pentanucleotide TAAAA repeat. For both loci, the TAAAA repeat was incorrectly sized as the disease-motif. Because ExpansionHunter relies on aligned reads to estimate the size of a repeat, it may not be accurate for repeats whose lengths extend close to or beyond the read length. Intermediate and pathogenic expansions in ATXN8 (SCA8), TBP (SCA17), CNBP (DM2), and FMR1 (FRAXA/FXTAS) may be difficult to size using ExpansionHunter given the 150-bp sequencing read length, leading to reclassification of expansions within the normal, intermediate, or pathogenic range following PCR validation. For example, intermediate STR expansions in ANTX8 (n = 1) and CNBP (n = 1) were PCR-genotyped within the pathogenic range, while a pathogenic expansion in TBP (n = 1) was PCR-genotyped within the intermediate range. Seven patients with intermediate FMR1 expansions were PCR-genotyped in the normal range; however, three of these patients harbored expansions in the “gray-zone” range (45 to 54 repeats), while three other patients had FMR1 expansions ≥40 repeats.
ExpansionHunter overestimated the repeat size of ATXN1 by a single repeat. Inspection of aligned reads over ATXN1 (using REViewer) established that the repeat boundaries were out of frame, leading to the inclusion of an additional codon in the ExpansionHunter allele length estimates (fig. S22). Corrected ATXN1 repeat lengths produced an allele frequency distribution that was consistent with previously reported PCR genotyped cohorts (35, 38, 52). The distribution of ATXN1 STR alleles prior to correction was similar to the distribution of ATXN1 STR alleles on gnomAD (33) and WebSTR, two web-based platforms that use bioinformatics tools to infer STR allele lengths in WGS data. While bioinformatics tools are useful for estimating STR allele lengths, the sequence immediately flanking STR loci may influence the accuracy of genotypes estimated from bioinformatics tools. Gold-standard PCR genotyping should be used for accurate sizing STR expansions.
Classification of neurodegenerative disorders, including ALS and FTD, is currently based on clinicopathological phenotypes. However, these disorders do not always present as a distinct clinical phenotype and can display marked variability including presenting features, in age at disease onset, disease duration and clinical progression. While disorders such as ALS, FTD, SCA, HD, and DM are relatively rare and have distinct clinical classifications, phenotypic overlap is observed. Symptoms typically manifest in adulthood and include progressive degeneration with some degree of muscular atrophy. In rare instances, patients genetically diagnosed with HD or SCA have pathological evidence of ALS (53, 54). Understanding the underlying biological mechanisms that allow a genetic variant to cause either pathological ALS, FTD, or another neurodegenerative disease phenotype is crucial for patients as it directs genetic counselling (55) and prognosis and allows them to join suitable clinical trials (56). In addition, it reduces genetic heterogeneity for studies into the remaining missing heritability for both ALS and FTD. Rare repeat expansions, such as these identified in this study, may be truly associated with disease risk; however, larger ALS and FTD patient cohorts will be required to determine their significance to disease.
MATERIALS AND METHODS
sALS and sFTD cohort
A total of 635 Australian patients with sALS and 78 Australian patients with sFTD were recruited for analysis from the Macquarie University Neurodegenerative Disease Biobank, Australian MND DNA Bank (Royal Prince Alfred Hospital), and Brain and Mind Centre (The University of Sydney). Each participant provided informed written consent as approved by the human research ethics committees of Macquarie University (5201600387) or The University of Sydney. Each patient with sALS was clinically diagnosed by a neurologist according to El Escorial criteria (57) or on the basis of a progressive upper and lower motor neuron weakness, with electrophysiology confirming widespread muscle denervation, and no other diagnosis being found on neuroimaging. Each patient with sFTD was diagnosed according to the modified version of the Goldman score (58). Patients with sALS were prescreened for disease-related mutations in SOD1, TARDBP, and FUS and for the pathogenic C9orf72 repeat expansion, while patients with sFTD were prescreened for mutations in MAPT and GRN and for the pathogenic C9orf72 repeat expansion.
Statistical analysis of clinical data
Clinical records were examined for phenotypic features: sex, age at disease onset, duration of disease from onset (until death or last known date of survival), and, for sALS patients only, site of onset (bulbar or spinal). All statistical analyses were performed in R (V.3.5.1). A chi-squared analysis was performed between sex and site of onset, while Welch’s two sample t tests were performed between age of onset versus both sex and site of onset. Kaplan-Meier survival analyses were performed between disease duration and both sex and site of onset. Additionally, a linear regression model was fitted between age at onset and duration (for deceased cases only). Multiple testing was accounted for using a Bonferroni-corrected significance threshold of P < 0.008 for sALS cases (α = 0.05 and 6 comparisons) and P < 0.017 for sFTD cases (α = 0.05 and 3 comparisons). A multinomial logistic regression analysis was performed for each clinical variable to compare patients with sALS carrying two or more disease-implicated genetic variants (repeat expansions or known ALS-linked mutations in SOD1, TARDBP, and FUS) with patients with sALS carrying none, or only one, disease-implicated genetic variant.
WGS of sALS and sFTD cases
Genomic DNA extraction was performed using whole blood according to standard protocols. All samples underwent library preparation using the TruSeq PCR free library preparation kit (Illumina, v2.5). Prepared libraries underwent multiplex 150-bp paired-end WGS on an Illumina HiSeq X Ten instrument (Kinghorn Centre for Clinical Genomics, Sydney, Australia).
Database of Genotypes and Phenotypes control cohort
A set of 5664 nontumor and non-neurological samples with WGS data available were obtained from National Heart, Lung and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) studies to be used as population-matched control participants. The ages of control participants were not available for all TOPMed studies; however, the average age was provided for some studies with most control participants aged 57 years or older. A total of 3541 samples aged 65 years or older were obtained from the Cardiovascular Health Study (phs001368.v3.p2), 1351 samples with a mean age of 57 years from the Mayo Clinic Venous Thromboembolism Study (phs001402.v3.p1), 640 samples from the Groningen Genetics of Atrial Fibrillation Study (phs001725.v2.p1), 128 samples with a mean age of 49 years from the Partners HealthCare Biobank (phs001024.v5.p1), 2 from the Malmo Preventive Project (phs001544.v2.p1), and 2 from the Johns Hopkins University School of Medicine Atrial Fibrillation Genetics Study (phs001598.v2.p1). All samples underwent 150 to 151-bp paired-end WGS on an Illumina HiSeq X Ten instrument with PCR-free library preparation methods.
WGS data processing and sample quality control filtering
Sequencing data was mapped to the 1000 Genomes GRCh38 reference genome (GRCh38_full_analysis_set_plus_decoy_hla.fa) using the joint Centers for Common Disease Genomics/TOPMed functionally equivalent read mapping pipeline (59). This pipeline standardizes read alignment processing to resolve batch effects from different genome centers generating functionally equivalent cohorts for integrated data analysis. Genotype call sets for the TOPMed control cohort (data freeze 9) were produced by the TOPMed variant calling pipeline (https://github.com/statgen/topmed_variant_calling). Genotype calls for the ALS/FTD cohort were generated using the GATK Best Practices pipeline as previously described (60).
Genotype call sets for ALS/FTD cases and TOPMed controls were merged with the combined HapMap Phase II and III dataset consisting of 11 populations (61). A principal components analysis was performed using KING v2.2.7 (62) to identify samples of European ancestry and PLINK v1.9 (63) was used to identify close relatives. Duplicate samples (including unaffected cotwins) were excluded from the ALS/FTD cohort, while control samples identified as third-degree relatives or closer were removed. The final population-matched cohort comprised 608 sALS, 68 sFTD, and 4703 control individuals.
Alignment-based repeat expansion detection, visualization, and statistical association analysis
Samples were screened for 21 repeat expansions known to cause motor phenotype–related neurodegenerative diseases using ExpansionHunter v4.0.1 (29, 30) (table S1) using custom repeat expansion catalogues. To assess the accuracy of the repeat lengths estimated by ExpansionHunter for each of the 21 STR loci, read alignments over the 21 repeat regions were visualized for 10 randomly selected samples using REViewer (34). Loci with the correct repeat motif aligned to the reference genome and appropriate ExpansionHunter repeat size estimates in all 10 samples were included for additional screening or analysis. Intermediate repeat size estimates and pathogenic thresholds were sourced from literature (table S1). A one-tailed Fisher’s exact test was used to evaluate associations between repeat sizes generated from ExpansionHunter and case-control status for all alleles, with a Bonferroni-corrected threshold of P < 0.0026 (α = 0.05 and 19 comparisons). A chi-square test was used to determine whether there was a difference in the total number of cases and controls with a repeat expansion compared to the total numbers excluding expansions in C9orf72.
PCR genotyping
To confirm ExpansionHunter-determined repeat sizes, PCR genotyping was performed for patients with ALS/FTD predicted to harbor repeat expansions in the intermediate or pathogenic range. STR expansions in C9orf72, ATXN1, ATXN2, ATXN8, TBP, NOTCH2NLC, DMPK, and CNBP were validated in-house. PCR primers and conditions are described in table S3. Standard PCRs underwent Sanger sequencing (repeat sizing) and fragment analysis (high-throughput genotyping) (Macrogen, Seoul, South Korea). Repeat-primed PCRs (C9orf72 and CNBP) underwent fragment analysis and were used to determine the presence or absence of a repeat expansion. GeneScan 500 LIZ size standard (Applied Biosystems, MA, USA) was used for all fragment analyses, and results were analyzed on Peak Scanner Software 2 (Thermo Fisher Scientific, MA, USA). HTT and FMR1 genotyping was performed by an accredited diagnostic pathology laboratory.
Outlier repeat expansion detection
To detect samples harboring an outlier repeat allele length without the bias of a preselected literature-defined allele-length threshold, we ran a data-derived outlier detection method on each gene. We used a statistical threshold of the 99.9th percentile (equivalent to a minor allele frequency; MAF = 0.001) on our control cohort, using 10,000 iterations of bootstrap sampling of allele lengths estimated by ExpansionHunter. This statistical threshold was selected because of the size of the control cohort and the expected rarity of pathogenic expanded alleles.
Acknowledgments
We thank D. Zhu from Molecular Medicine Laboratory, Concord Hospital, Sydney, Australia for providing PCR protocols to genotype STRs in ATXN1, ATXN2, TBP, DMPK, and CNBP. We thank A. Smith from the Macquarie University Centre for Motor Neuron Disease Research, Sydney, New South Wales, Australia for creating a repository on Zenodo to store code and data related to this study. Biospecimens and related clinical data used in this research were obtained from the Neurodegenerative Disease Biobank, Macquarie University, New South Wales, Australia and the Australian MND DNA Bank and the ForeFront research team at the Brain and Mind Centre, University of Sydney, New South Wales, Australia. This research was undertaken with the assistance of resources and services from the National Computational Infrastructure (NCI), which is supported by the Australian Government. This work was also supported by the Victorian Government’s Operational Infrastructure Support Program and the NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS). Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the NHLBI. Genome sequencing for “NHLBI TOPMed: Cardiovascular Health Study” (phs001368.v3.p2) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN2682016000331). Funded in part by grants from the National Institutes of Health, NHLBI (HL66216 and HL83141), and the National Human Genome Research Institute (HG04735). Genome sequencing for “NHLBI TOPMed: Mayo Clinic Venous Thromboembolism Study” (phs001402.v3.p1) was performed at the Baylor College of Medicine Human Genome Sequencing Center (3U54HG003273-12S2, HHSN268201500015C). Genome sequencing for “NHLBI TOPMed: Groningen Genetics of Atrial Fibrillation Study” (phs001725.v2.p1) was performed at the Baylor College of Medicine Human Genome Sequencing Center (3UM1HG008898-01S3). This funding source was an NHLBI supplement to NHGRI’s Centers for Common Disease Genomics (CCDG). Genome sequencing for “NHLBI TOPMed: Partners HealthCare Biobank Study” (phs001024.v5.p1) was performed at the Broad Institute Genomics Platform (3R01HL092577-06S1). Genome sequencing for “NHLBI TOPMed: Malmo Preventive Project” (phs001544.v2.p1) was performed at the Broad Institute Genomics Platform (3UM1HG008895-01S2). This funding source was an NHLBI supplement to NHGRI’s CCDG. Genome sequencing for “NHLBI TOPMed: Johns Hopkins University School of Medicine Atrial Fibrillation Genetics Study” (phs001598.v2.p1) was performed at the Broad Institute Genomics Platform (3UM1HG008895-01S2). This funding source was an NHLBI supplement to NHGRI’s CCDG. Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity QC, general program, and coordination were provided by the TOPMed Administrative Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Funding: This study was supported by National Health and Medical Research Council of Australia grants 1095215 and 1176913 (I.P.B.); National Health and Medical Research Council of Australia grants 1037746, 1095127, and 1132524 (G.M.H. and M.C.K.); National Health and Medical Research Council of Australia grant 1153439 and fellowship 1156093 (M.C.K.); National Health and Medical Research Council of Australia grant 1195236 (M.B.); National Health and Medical Research Council of Australia fellowship 1092023 (K.L.W.); National Health and Medical Research Council of Australia fellowship 1176607 (G.M.H.); National Health and Medical Research Council of Australia fellowship 1138223 (C.D.-S.); DHB Foundation Centenary Postdoctoral Fellowship (L.G.F.); and FightMND. Motor Neurone Disease Research Australia.
Author contributions: Conceptualization: L.He., L.G.F., M.B., K.L.W., and I.P.B. Software: L.G.F. Formal analysis: L.He. and L.G.F. Visualization: L.He. and L.G.F. Validation: N.G., C.D.-S., E.P.M., S.C.M.F., L.F., K.F., and L.Ho. Resources: D.B.R., J.B.K., G.M.H., M.C.K., and R.P. Data curation: L.He., L.G.F., C.D.-S., G.M.H., D.B.R., S.D., S.M., H.C.T., M.Z., L.A., and K.L.W. Supervision: K.L.W., I.P.B., and M.B. Writing—original draft: L.He., L.G.F., N.G., M.B., I.P.B., and K.L.W. Writing—review and editing: L.He., L.G.F., N.G., E.P.M., C.D.-S., L.F., K.F., L.Ho., S.C.M.F., D.B.R., S.D., J.B.K., G.M.H., M.C.K., S.M., H.C.T., M.Z., R.P., L.A., M.B., I.P.B., and K.L.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Data and code needed to evaluate the conclusions in the paper are available in a Zenodo repository (https://doi.org/10.5281/zenodo.7343770). Raw and processed sequencing data for the ALS and FTD patients are not publicly available because of ethics and patient consent constraints. To access the ALS and FTD patient data for academic research purposes, contact the corresponding author (K.L.W., kelly.williams@mq.edu.au).
Supplementary Materials
This PDF file includes:
Figs. S1 to S24
Tables S1 to S3
Supplementary Text
References
REFERENCES AND NOTES
- 1.S. R. Chintalaphani, S. S. Pineda, I. W. Deveson, K. R. Kumar, An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics. Acta Neuropathol. Commun. 9, 98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. Depienne, J. L. Mandel, 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? Am. J. Hum. Genet. 108, 764–785 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.I. Malik, C. P. Kelley, E. T. Wang, P. K. Todd, Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 22, 589–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. R. Burrell, G. M. Halliday, J. J. Kril, L. M. Ittner, J. Götz, M. C. Kiernan, J. R. Hodges, The frontotemporal dementia-motor neuron disease continuum. Lancet 388, 919–931 (2016). [DOI] [PubMed] [Google Scholar]
- 5.A. Shatunov, A. Al-Chalabi, The genetic architecture of ALS. Neurobiol. Dis. 147, 105156 (2021). [DOI] [PubMed] [Google Scholar]
- 6.J. M. Shefner, A. Al-Chalabi, M. R. Baker, L.-Y. Cui, M. de Carvalho, A. Eisen, J. Grosskreutz, O. Hardiman, R. Henderson, J. M. Matamala, H. Mitsumoto, W. Paulus, N. Simon, M. Swash, K. Talbot, M. R. Turner, Y. Ugawa, L. H. van den Berg, R. Verdugo, S. Vucic, R. Kaji, D. Burke, M. C. Kiernan, A proposal for new diagnostic criteria for ALS. Clin. Neurophysiol. 131, 1975–1978 (2020). [DOI] [PubMed] [Google Scholar]
- 7.C. V. Greaves, J. D. Rohrer, An update on genetic frontotemporal dementia. J. Neurol. 266, 2075–2086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.E. M. J. de Boer, V. K. Orie, T. Williams, M. R. Baker, H. M. De Oliveira, T. Polvikoski, M. Silsby, P. Menon, M. van den Bos, G. M. Halliday, L. H. van den Berg, L. Van Den Bosch, P. van Damme, M. C. Kiernan, M. A. van Es, S. Vucic, TDP-43 proteinopathies: A new wave of neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry 92, 86–95 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.G. M. Ringholz, S. H. Appel, M. Bradshaw, N. A. Cooke, D. M. Mosnik, P. E. Schulz, Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology 65, 586–590 (2005). [DOI] [PubMed] [Google Scholar]
- 10.R. M. Ahmed, E. M. Devenney, C. Strikwerda-Brown, J. R. Hodges, O. Piguet, M. C. Kiernan, Phenotypic variability in ALS-FTD and effect on survival. Neurology 94, e2005–e2013 (2020). [DOI] [PubMed] [Google Scholar]
- 11.J. R. Burrell, M. C. Kiernan, S. Vucic, J. R. Hodges, Motor neuron dysfunction in frontotemporal dementia. Brain 134, 2582–2594 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Y. A. Abramzon, P. Fratta, B. J. Traynor, R. Chia, The overlapping genetics of amyotrophic lateral sclerosis and frontotemporal dementia. Front. Neurosci. 14, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. DeJesus-Hernandez, I. R. Mackenzie, B. F. Boeve, A. L. Boxer, M. Baker, N. J. Rutherford, A. M. Nicholson, N. A. Finch, H. Flynn, J. Adamson, N. Kouri, A. Wojtas, P. Sengdy, G.-Y. R. Hsiung, A. Karydas, W. W. Seeley, K. A. Josephs, G. Coppola, D. H. Geschwind, Z. K. Wszolek, H. Feldman, D. S. Knopman, R. C. Petersen, B. L. Miller, D. W. Dickson, K. B. Boylan, N. R. Graff-Radford, R. Rademakers, Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. E. Renton, E. Majounie, A. Waite, J. Simón-Sánchez, S. Rollinson, J. R. Gibbs, J. C. Schymick, H. Laaksovirta, J. C. van Swieten, L. Myllykangas, H. Kalimo, A. Paetau, Y. Abramzon, A. M. Remes, A. Kaganovich, S. W. Scholz, J. Duckworth, J. Ding, D. W. Harmer, D. G. Hernandez, J. O. Johnson, K. Mok, M. Ryten, D. Trabzuni, R. J. Guerreiro, R. W. Orrell, J. Neal, A. Murray, J. Pearson, I. E. Jansen, D. Sondervan, H. Seelaar, D. Blake, K. Young, N. Halliwell, J. B. Callister, G. Toulson, A. Richardson, A. Gerhard, J. Snowden, D. Mann, D. Neary, M. A. Nalls, T. Peuralinna, L. Jansson, V.-M. Isoviita, A.-L. Kaivorinne, M. Hölttä-Vuori, E. Ikonen, R. Sulkava, M. Benatar, J. Wuu, A. Chiò, G. Restagno, G. Borghero, M. Sabatelli; ITALSGEN Consortium, D. Heckerman, E. Rogaeva, L. Zinman, J. D. Rothstein, M. Sendtner, C. Drepper, E. E. Eichler, C. Alkan, Z. Abdullaev, S. D. Pack, A. Dutra, E. Pak, J. Hardy, A. Singleton, N. M. Williams, P. Heutink, S. Pickering-Brown, H. R. Morris, P. J. Tienari, B. J. Traynor, A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. R. Turner, A. Al-Chalabi, A. Chio, O. Hardiman, M. C. Kiernan, J. D. Rohrer, J. Rowe, W. Seeley, K. Talbot, Genetic screening in sporadic ALS and FTD. J. Neurol. Neurosurg. Psychiatry 88, 1042–1044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. C. Elden, H.-J. Kim, M. P. Hart, A. S. Chen-Plotkin, B. S. Johnson, X. Fang, M. Armakola, F. Geser, R. Greene, M. M. Lu, A. Padmanabhan, D. Clay-Falcone, L. McCluskey, L. Elman, D. Juhr, P. J. Gruber, U. Rüb, G. Auburger, J. Q. Trojanowski, V. M.-Y. Lee, V. M. Van Deerlin, N. M. Bonini, A. D. Gitler, Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Lattante, M. G. Pomponi, A. Conte, G. Marangi, G. Bisogni, A. K. Patanella, E. Meleo, C. Lunetta, N. Riva, L. Mosca, P. Carrera, M. Bee, M. Zollino, M. Sabatelli, ATXN1 intermediate-length polyglutamine expansions are associated with amyotrophic lateral sclerosis. Neurobiol. Aging 64, 157.e1–157.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 18.R. Dewan, R. Chia, J. Ding, R. A. Hickman, T. D. Stein, Y. Abramzon, S. Ahmed, M. S. Sabir, M. K. Portley, A. Tucci, K. Ibáñez, F. N. U. Shankaracharya, P. Keagle, G. Rossi, P. Caroppo, F. Tagliavini, M. L. Waldo, P. M. Johansson, C. F. Nilsson; American Genome Center (TAGC); FALS Sequencing Consortium; Genomics England Research Consortium; International ALS/FTD Genomics Consortium (iAFGC); International FTD Genetics Consortium (IFGC); International LBD Genomics Consortium (iLBDGC); NYGC ALS Consortium; PROSPECT Consortium, J. B. Rowe, L. Benussi, G. Binetti, R. Ghidoni, E. Jabbari, C. Viollet, J. D. Glass, A. B. Singleton, V. Silani, O. A. Ross, M. Ryten, A. Torkamani, T. Tanaka, L. Ferrucci, S. M. Resnick, S. Pickering-Brown, C. B. Brady, N. Kowal, J. A. Hardy, V. Van Deerlin, J. P. Vonsattel, M. B. Harms, H. R. Morris, R. Ferrari, J. E. Landers, A. Chiò, J. R. Gibbs, C. L. Dalgard, S. W. Scholz, B. J. Traynor, Pathogenic huntingtin repeat expansions in patients with frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 109, 448–460.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R. A. Hickman, R. Dewan, E. Cortes, B. J. Traynor, K. Marder, J.-P. Vonsattel, Amyotrophic lateral sclerosis is over-represented in two Huntington’s disease brain bank cohorts: Further evidence to support genetic pleiotropy of pathogenic HTT gene expansion. Acta Neuropathol. 143, 105–108 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F. Akçimen, J. P. Ross, C. Liao, D. Spiegelman, P. A. Dion, G. A. Rouleau, Expanded CAG repeats in ATXN1, ATXN2, ATXN3, and HTT in the 1000 Genomes Project. Mov. Disord. 36, 514–518 (2021). [DOI] [PubMed] [Google Scholar]
- 21.E. P. McCann, L. Henden, J. A. Fifita, K. Y. Zhang, N. Grima, D. C. Bauer, S. C. M. Fat, N. A. Twine, R. Pamphlett, M. C. Kiernan, D. B. Rowe, K. L. Williams, I. P. Blair, Evidence for polygenic and oligogenic basis of Australian sporadic amyotrophic lateral sclerosis. J. Med. Genet. 58, 87–95 (2021). [DOI] [PubMed] [Google Scholar]
- 22.R. Ranganathan, S. Haque, K. Coley, S. Shepheard, J. Cooper-Knock, J. Kirby, Multifaceted genes in amyotrophic lateral sclerosis-frontotemporal dementia. Front. Neurosci. 14, 684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. Al-Chalabi, F. Fang, M. F. Hanby, P. N. Leigh, C. E. Shaw, W. Ye, F. Rijsdijk, An estimate of amyotrophic lateral sclerosis heritability using twin data. J. Neurol. Neurosurg. Psychiatry 81, 1324–1326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Ryan, M. Heverin, R. L. Mclaughlin, O. Hardiman, Lifetime risk and heritability of amyotrophic lateral sclerosis. JAMA Neurol. 76, 1367–1374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.T. S. Wingo, D. J. Cutler, N. Yarab, C. M. Kelly, J. D. Glass, The heritability of amyotrophic lateral sclerosis in a clinically ascertained United States research registry. PLOS ONE 6, e27985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.W. van Rheenen, A. Shatunov, A. M. Dekker, R. L. McLaughlin, F. P. Diekstra, S. L. Pulit, R. A. A. van der Spek, U. Võsa, S. de Jong, M. R. Robinson, J. Yang, I. Fogh, P. T. van Doormaal, G. H. P. Tazelaar, M. Koppers, A. M. Blokhuis, W. Sproviero, A. R. Jones, K. P. Kenna, K. R. van Eijk, O. Harschnitz, R. D. Schellevis, W. J. Brands, J. Medic, A. Menelaou, A. Vajda, N. Ticozzi, K. Lin, B. Rogelj, K. Vrabec, M. Ravnik-Glavač, B. Koritnik, J. Zidar, L. Leonardis, L. D. Grošelj, S. Millecamps, F. Salachas, V. Meininger, M. de Carvalho, S. Pinto, J. S. Mora, R. Rojas-García, M. Polak, S. Chandran, S. Colville, R. Swingler, K. E. Morrison, P. J. Shaw, J. Hardy, R. W. Orrell, A. Pittman, K. Sidle, P. Fratta, A. Malaspina, S. Topp, S. Petri, S. Abdulla, C. Drepper, M. Sendtner, T. Meyer, R. A. Ophoff, K. A. Staats, M. Wiedau-Pazos, C. Lomen-Hoerth, V. M. Van Deerlin, J. Q. Trojanowski, L. Elman, L. McCluskey, A. N. Basak, C. Tunca, H. Hamzeiy, Y. Parman, T. Meitinger, P. Lichtner, M. Radivojkov-Blagojevic, C. R. Andres, C. Maurel, G. Bensimon, B. Landwehrmeyer, A. Brice, C. A. M. Payan, S. Saker-Delye, A. Dürr, N. W. Wood, L. Tittmann, W. Lieb, A. Franke, M. Rietschel, S. Cichon, M. M. Nöthen, P. Amouyel, C. Tzourio, J.-F. Dartigues, A. G. Uitterlinden, F. Rivadeneira, K. Estrada, A. Hofman, C. Curtis, H. M. Blauw, A. J. van der Kooi, M. de Visser, A. Goris, M. Weber, C. E. Shaw, B. N. Smith, O. Pansarasa, C. Cereda, R. Del Bo, G. P. Comi, S. D’Alfonso, C. Bertolin, G. Sorarù, L. Mazzini, V. Pensato, C. Gellera, C. Tiloca, A. Ratti, A. Calvo, C. Moglia, M. Brunetti, S. Arcuti, R. Capozzo, C. Zecca, C. Lunetta, S. Penco, N. Riva, A. Padovani, M. Filosto, B. Muller, R. J. Stuit; PARALS Registry; SLALOM Group; SLAP Registry; FALS Sequencing Consortium; SLAGEN Consortium; NNIPPS Study Group, I. Blair, K. Zhang, E. P. McCann, J. A. Fifita, G. A. Nicholson, D. B. Rowe, R. Pamphlett, M. C. Kiernan, J. Grosskreutz, O. W. Witte, T. Ringer, T. Prell, B. Stubendorff, I. Kurth, C. A. Hübner, P. N. Leigh, F. Casale, A. Chio, E. Beghi, E. Pupillo, R. Tortelli, G. Logroscino, J. Powell, A. C. Ludolph, J. H. Weishaupt, W. Robberecht, P. Van Damme, L. Franke, T. H. Pers, R. H. Brown, J. D. Glass, J. E. Landers, O. Hardiman, P. M. Andersen, P. Corcia, P. Vourc’h, V. Silani, N. R. Wray, P. M. Visscher, P. I. W. de Bakker, M. A. van Es, R. J. Pasterkamp, C. M. Lewis, G. Breen, A. Al-Chalabi, L. H. van den Berg, J. H. Veldink, Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.W. van Rheenen, R. A. A. van der Spek, M. K. Bakker, J. J. F. A. van Vugt, P. J. Hop, R. A. J. Zwamborn, N. de Klein, H.-J. Westra, O. B. Bakker, P. Deelen, G. Shireby, E. Hannon, M. Moisse, D. Baird, R. Restuadi, E. Dolzhenko, A. M. Dekker, K. Gawor, H.-J. Westeneng, G. H. P. Tazelaar, K. R. van Eijk, M. Kooyman, R. P. Byrne, M. Doherty, M. Heverin, A. Al Khleifat, A. Iacoangeli, A. Shatunov, N. Ticozzi, J. Cooper-Knock, B. N. Smith, M. Gromicho, S. Chandran, S. Pal, K. E. Morrison, P. J. Shaw, J. Hardy, R. W. Orrell, M. Sendtner, T. Meyer, N. Başak, A. J. van der Kooi, A. Ratti, I. Fogh, C. Gellera, G. Lauria, S. Corti, C. Cereda, D. Sproviero, S. D’Alfonso, G. Sorarù, G. Siciliano, M. Filosto, A. Padovani, A. Chiò, A. Calvo, C. Moglia, M. Brunetti, A. Canosa, M. Grassano, E. Beghi, E. Pupillo, G. Logroscino, B. Nefussy, A. Osmanovic, A. Nordin, Y. Lerner, M. Zabari, M. Gotkine, R. H. Baloh, S. Bell, P. Vourc’h, P. Corcia, P. Couratier, S. Millecamps, V. Meininger, F. Salachas, J. S. M. Pardina, A. Assialioui, R. Rojas-García, P. A. Dion, J. P. Ross, A. C. Ludolph, J. H. Weishaupt, D. Brenner, A. Freischmidt, G. Bensimon, A. Brice, A. Durr, C. A. M. Payan, S. Saker-Delye, N. W. Wood, S. Topp, R. Rademakers, L. Tittmann, W. Lieb, A. Franke, S. Ripke, A. Braun, J. Kraft, D. C. Whiteman, C. M. Olsen, A. G. Uitterlinden, A. Hofman, M. Rietschel, S. Cichon, M. M. Nöthen, P. Amouyel; SLALOM Consortium; PARALS Consortium; SLAGEN Consortium; SLAP Consortium, B. J. Traynor, A. B. Singleton, M. M. Neto, R. J. Cauchi, R. A. Ophoff, M. Wiedau-Pazos, C. Lomen-Hoerth, V. M. van Deerlin, J. Grosskreutz, A. Roediger, N. Gaur, A. Jörk, T. Barthel, E. Theele, B. Ilse, B. Stubendorff, O. W. Witte, R. Steinbach, C. A. Hübner, C. Graff, L. Brylev, V. Fominykh, V. Demeshonok, A. Ataulina, B. Rogelj, B. Koritnik, J. Zidar, M. Ravnik-Glavač, D. Glavač, Z. Stević, V. Drory, M. Povedano, I. P. Blair, M. C. Kiernan, B. Benyamin, R. D. Henderson, S. Furlong, S. Mathers, P. A. McCombe, M. Needham, S. T. Ngo, G. A. Nicholson, R. Pamphlett, D. B. Rowe, F. J. Steyn, K. L. Williams, K. A. Mather, P. S. Sachdev, A. K. Henders, L. Wallace, M. de Carvalho, S. Pinto, S. Petri, M. Weber, G. A. Rouleau, V. Silani, C. J. Curtis, G. Breen, J. D. Glass, R. H. J. Brown, J. E. Landers, C. E. Shaw, P. M. Andersen, E. J. N. Groen, M. A. van Es, R. J. Pasterkamp, D. Fan, F. C. Garton, A. F. McRae, G. D. Smith, T. R. Gaunt, M. A. Eberle, J. Mill, R. L. McLaughlin, O. Hardiman, K. P. Kenna, N. R. Wray, E. Tsai, H. Runz, L. Franke, A. Al-Chalabi, P. Van Damme, L. H. van den Berg, J. H. Veldink, Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 53, 1636–1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. Majounie, A. E. Renton, K. Mok, E. G. P. Dopper, A. Waite, S. Rollinson, A. Chiò, G. Restagno, N. Nicolaou, J. Simon-Sanchez, J. C. van Swieten, Y. Abramzon, J. O. Johnson, M. Sendtner, R. Pamphlett, R. W. Orrell, S. Mead, K. C. Sidle, H. Houlden, J. D. Rohrer, K. E. Morrison, H. Pall, K. Talbot, O. Ansorge; Chromosome 9-ALS/FTD Consortium; French research network on FTLD/FTLD/ALS; ITALSGEN Consortium, D. G. Hernandez, S. Arepalli, M. Sabatelli, G. Mora, M. Corbo, F. Giannini, A. Calvo, E. Englund, G. Borghero, G. L. Floris, A. M. Remes, H. Laaksovirta, L. McCluskey, J. Q. Trojanowski, V. M. Van Deerlin, G. D. Schellenberg, M. A. Nalls, V. E. Drory, C.-S. Lu, T.-H. Yeh, H. Ishiura, Y. Takahashi, S. Tsuji, I. Le Ber, A. Brice, C. Drepper, N. Williams, J. Kirby, P. Shaw, J. Hardy, P. J. Tienari, P. Heutink, H. R. Morris, S. Pickering-Brown, B. J. Traynor, Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 11, 323–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.E. Dolzhenko, J. J. F. A. van Vugt, R. J. Shaw, M. A. Bekritsky, M. Van Blitterswijk, G. Narzisi, S. S. Ajay, V. Rajan, B. R. Lajoie, N. H. Johnson, Z. Kingsbury, S. J. Humphray, R. D. Schellevis, W. J. Brands, M. Baker, R. Rademakers, M. Kooyman, G. H. P. Tazelaar, M. A. Van Es, R. Mclaughlin, W. Sproviero, A. Shatunov, A. Jones, A. Al Khleifat, A. Pittman, S. Morgan, O. Hardiman, A. Al-Chalabi, C. Shaw, B. Smith, E. J. Neo, K. Morrison, P. J. Shaw, C. Reeves, L. Winterkorn, N. S. Wexler, D. E. Housman, C. W. Ng, A. L. Li, R. J. Taft, L. H. Van Den Berg, D. R. Bentley, J. H. Veldink, M. A. Eberle, Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 27, 1895–1903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.E. Dolzhenko, V. Deshpande, F. Schlesinger, P. Krusche, R. Petrovski, S. Chen, D. Emig-Agius, A. Gross, G. Narzisi, B. Bowman, K. Scheffler, J. J. F. A. van Vugt, C. French, A. Sanchis-Juan, K. Ibáñez, A. Tucci, B. R. Lajoie, J. H. Veldink, F. L. Raymond, R. J. Taft, D. R. Bentley, M. A. Eberle, ExpansionHunter: A sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics 35, 4754–4756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. Trost, W. Engchuan, C. M. Nguyen, B. Thiruvahindrapuram, E. Dolzhenko, I. Backstrom, M. Mirceta, B. A. Mojarad, Y. Yin, A. Dov, I. Chandrakumar, T. Prasolava, N. Shum, O. Hamdan, G. Pellecchia, J. L. Howe, J. Whitney, E. W. Klee, S. Baheti, D. G. Amaral, E. Anagnostou, M. Elsabbagh, B. A. Fernandez, N. Hoang, M. E. S. Lewis, X. Liu, C. Sjaarda, I. M. Smith, P. Szatmari, L. Zwaigenbaum, D. Glazer, D. Hartley, A. K. Stewart, M. A. Eberle, N. Sato, C. E. Pearson, S. W. Scherer, R. K. C. Yuen, Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature 586, 80–86 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. Ibáñez, J. Polke, R. T. Hagelstrom, E. Dolzhenko, D. Pasko, E. R. A. Thomas, L. C. Daugherty, D. Kasperaviciute, K. R. Smith; WGS for Neurological Diseases Group, Z. C. Deans, S. Hill, T. Fowler, R. H. Scott, J. Hardy, P. F. Chinnery, H. Houlden, A. Rendon, M. J. Caulfield, M. A. Eberle, R. J. Taft, A. Tucci; Genomics England Research Consortium , Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: A retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol. 21, 234–245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski; Genome Aggregation Database Consortium, B. M. Neale, M. J. Daly, D. G. MacArthur, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.E. Dolzhenko, B. Weisburd, K. Ibañez, I.-S. Rajan-Babu, C. Anyansi, M. F. Bennett, K. Billingsley, A. Carroll, S. Clamons, M. C. Danzi, V. Deshpande, J. Ding, S. Fazal, A. Halman, B. Jadhav, Y. Qiu, P. A. Richmond, C. T. Saunders, K. Scheffler, J. J. F. A. van Vugt, R. R. A. J. Zwamborn; Genomics England Research Consortium, S. S. Chong, J. M. Friedman, A. Tucci, H. L. Rehm, M. A. Eberle, REViewer: Haplotype-resolved visualization of read alignments in and around tandem repeats. Genome Med. 14, 84 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S. L. Gardiner, M. W. Boogaard, S. Trompet, R. de Mutsert, F. R. Rosendaal, J. Gussekloo, J. W. Jukema, R. A. C. Roos, N. A. Aziz, Prevalence of carriers of intermediate and pathological polyglutamine disease-associated alleles among large population-based cohorts. JAMA Neurol. 76, 650–656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.H. Rafehi, D. J. Szmulewicz, M. F. Bennett, N. L. M. Sobreira, K. Pope, K. R. Smith, G. Gillies, P. Diakumis, E. Dolzhenko, M. A. Eberle, M. G. Barcina, D. P. Breen, A. M. Chancellor, P. D. Cremer, M. B. Delatycki, B. L. Fogel, A. Hackett, G. M. Halmagyi, S. Kapetanovic, A. Lang, S. Mossman, W. Mu, P. Patrikios, S. L. Perlman, I. Rosemergy, E. Storey, S. R. D. Watson, M. A. Wilson, D. S. Zee, D. Valle, D. J. Amor, M. Bahlo, P. J. Lockhart, Bioinformatics-based identification of expanded repeats: A non-reference intronic pentamer expansion in RFC1 causes CANVAS. Am. J. Hum. Genet. 105, 151–165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Ishiura, S. Shibata, J. Yoshimura, Y. Suzuki, W. Qu, K. Doi, M. A. Almansour, J. K. Kikuchi, M. Taira, J. Mitsui, Y. Takahashi, Y. Ichikawa, T. Mano, A. Iwata, Y. Harigaya, M. K. Matsukawa, T. Matsukawa, M. Tanaka, Y. Shirota, R. Ohtomo, H. Kowa, H. Date, A. Mitsue, H. Hatsuta, S. Morimoto, S. Murayama, Y. Shiio, Y. Saito, A. Mitsutake, M. Kawai, T. Sasaki, Y. Sugiyama, M. Hamada, G. Ohtomo, Y. Terao, Y. Nakazato, A. Takeda, Y. Sakiyama, Y. Umeda-Kameyama, J. Shinmi, K. Ogata, Y. Kohno, S.-Y. Lim, A. H. Tan, J. Shimizu, J. Goto, I. Nishino, T. Toda, S. Morishita, S. Tsuji, Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 51, 1222–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 38.G. H. P. Tazelaar, S. Boeynaems, M. De Decker, J. J. F. A. van Vugt, L. Kool, H. S. Goedee, R. L. McLaughlin, W. Sproviero, A. Iacoangeli, M. Moisse, M. Jacquemyn, D. Daelemans, A. M. Dekker, R. A. van der Spek, H.-J. Westeneng, K. P. Kenna, A. Assialioui, N. Da Silva; Project MinE ALS Sequencing Consortium, M. Povedano, J. S. M. Pardina, O. Hardiman, F. Salachas, S. Millecamps, P. Vourc’h, P. Corcia, P. Couratier, K. E. Morrison, P. J. Shaw, C. E. Shaw, R. J. Pasterkamp, J. E. Landers, L. Van Den Bosch, W. Robberecht, A. Al-Chalabi, L. H. van den Berg, P. Van Damme, J. H. Veldink, M. A. van Es, ATXN1 repeat expansions confer risk for amyotrophic lateral sclerosis and contribute to TDP-43 mislocalization. Brain Commun. 2, fcaa064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.G. Koutsis, G. Karadima, A. Pandraud, M. G. Sweeney, R. Paudel, H. Houlden, N. W. Wood, M. Panas, Genetic screening of Greek patients with Huntington’s disease phenocopies identifies an SCA8 expansion. J. Neurol. 259, 1874–1878 (2012). [DOI] [PubMed] [Google Scholar]
- 40.D. A. Olszewska, E. M. Fallon, G. M. Pastores, K. Murphy, A. Blanco, T. Lynch, S. M. Murphy, Autosomal dominant gene negative frontotemporal dementia-think of SCA17. Cerebellum 18, 654–658 (2019). [DOI] [PubMed] [Google Scholar]
- 41.D. Eratne, A. Schneider, E. Lynch, M. Martyn, D. Velakoulis, M. Fahey, P. Kwan, R. Leventer, H. Rafehi, B. Chong, Z. Stark, S. Lunke, D. G. Phelan, M. O’Keefe, K. Siemering, K. West, A. Sexton, A. Jarmolowicz, J. A. Taylor, J. Schultz, R. Purvis, E. Uebergang, H. Chalinor, B. Creighton, N. Gelfand, T. Saks, Y. Prawer, Y. Smagarinsky, T. Pan, I. Goranitis, Z. Ademi, C. Gaff, A. Huq, M. Walsh, P. A. James, E. I. Krzesinski, M. Wallis, C. A. Stutterd, M. Bahlo, M. B. Delatycki, S. F. Berkovic, The clinical utility of exome sequencing and extended bioinformatic analyses in adolescents and adults with a broad range of neurological phenotypes: An Australian perspective. J. Neurol. Sci. 420, 117260 (2021). [DOI] [PubMed] [Google Scholar]
- 42.D. Pretto, C. M. Yrigollen, H.-T. Tang, J. Williamson, G. Espinal, C. K. Iwahashi, B. Durbin-Johnson, R. J. Hagerman, P. J. Hagerman, F. Tassone, Clinical and molecular implications of mosaicism in FMR1 full mutations. Front. Genet. 5, 318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. I. Pretto, G. Mendoza-Morales, J. Lo, R. Cao, A. Hadd, G. J. Latham, B. Durbin-Johnson, R. Hagerman, F. Tassone, CGG allele size somatic mosaicism and methylation in FMR1 premutation alleles. J. Med. Genet. 51, 309–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. N. Coyne, S. B. Yamada, B. B. Siddegowda, P. S. Estes, B. L. Zaepfel, J. S. Johannesmeyer, D. B. Lockwood, L. T. Pham, M. P. Hart, J. A. Cassel, B. Freibaum, A. V. Boehringer, J. P. Taylor, A. B. Reitz, A. D. Gitler, D. C. Zarnescu, Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 24, 6886–6898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.C. Turner, D. Hilton-Jones, The myotonic dystrophies: Diagnosis and management. J. Neurol. Neurosurg. Psychiatry 81, 358–367 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Z. Musova, R. Mazanec, A. Krepelova, E. Ehler, J. Vales, R. Jaklova, T. Prochazka, P. Koukal, T. Marikova, J. Kraus, M. Havlovicova, Z. Sedlacek, Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am. J. Med. Genet. A 149A, 1365–1374 (2009). [DOI] [PubMed] [Google Scholar]
- 47.G. Meola, R. Cardani, Myotonic Dystrophy Type 2: An Update on Clinical Aspects, Genetic and Pathomolecular Mechanism. J. Neuromuscul. Dis. 2, S59–S71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B. A. Perez, H. K. Shorrock, M. Banez-Coronel, T. Zu, L. El Romano, L. A. Laboissonniere, T. Reid, Y. Ikeda, K. Reddy, C. M. Gomez, T. Bird, T. Ashizawa, L. J. Schut, A. Brusco, J. A. Berglund, L. F. Hasholt, J. E. Nielsen, S. H. Subramony, L. P. Ranum, CCG•CGG interruptions in high-penetrance SCA8 families increase RAN translation and protein toxicity. EMBO Mol. Med. 13, e14095 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.E. P. McCann, K. L. Williams, J. A. Fifita, I. S. Tarr, J. O’Connor, D. B. Rowe, G. A. Nicholson, I. P. Blair, The genotype-phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin. Genet. 92, 259–266 (2017). [DOI] [PubMed] [Google Scholar]
- 50.A. Chiò, L. Mazzini, S. D’Alfonso, L. Corrado, A. Canosa, C. Moglia, U. Manera, E. Bersano, M. Brunetti, M. Barberis, J. H. Veldink, L. H. van den Berg, N. Pearce, W. Sproviero, R. McLaughlin, A. Vajda, O. Hardiman, J. Rooney, G. Mora, A. Calvo, A. Al-Chalabi, The multistep hypothesis of ALS revisited: The role of genetic mutations. Neurology 91, e635–e642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. Bahlo, M. F. Bennett, P. Degorski, R. M. Tankard, M. B. Delatycki, P. J. Lockhart, Recent advances in the detection of repeat expansions with short-read next-generation sequencing. F1000Res. 7, 736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.I. Rosas, C. Martínez, J. Clarimón, A. Lleó, I. Illán-Gala, O. Dols-Icardo, B. Borroni, M. R. Almeida, J. van der Zee, C. Van Broeckhoven, A. C. Bruni, M. Anfossi, L. Bernardi, R. Maletta, M. Serpente, D. Galimberti, E. Scarpini, G. Rossi, P. Caroppo, L. Benussi, R. Ghidoni, G. Binetti, B. Nacmias, S. Sorbi, I. Piaceri, S. Bagnoli, A. Antonell, R. Sánchez-Valle, B. De la Casa-Fages, F. Grandas, M. Diez-Fairen, P. Pastor, R. Ferrari, V. Álvarez, M. Menéndez-González, Role for ATXN1, ATXN2, and HTT intermediate repeats in frontotemporal dementia and Alzheimer's disease. Neurobiol. Aging 87, 139.e1–139.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 53.M. Tada, E. A. Coon, A. P. Osmand, P. A. Kirby, W. Martin, M. Wieler, A. Shiga, H. Shirasaki, M. Tada, T. Makifuchi, M. Yamada, A. Kakita, M. Nishizawa, H. Takahashi, H. L. Paulson, Coexistence of Huntington’s disease and amyotrophic lateral sclerosis: A clinicopathologic study. Acta Neuropathol. 124, 749–760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. Nanetti, R. Fancellu, C. Tomasello, C. Gellera, D. Pareyson, C. Mariotti, Rare association of motor neuron disease and spinocerebellar ataxia type 2 (SCA2): A new case and review of the literature. J. Neurol. 256, 1926–1928 (2009). [DOI] [PubMed] [Google Scholar]
- 55.K. Salmon, M. C. Kiernan, S. H. Kim, P. M. Andersen, A. Chio, L. H. van den Berg, P. Van Damme, A. Al-Chalabi, P. Lillo, J. A. Andrews, A. Genge, The importance of offering early genetic testing in everyone with amyotrophic lateral sclerosis. Brain 145, 1207–1210 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. C. Kiernan, S. Vucic, K. Talbot, C. J. McDermott, O. Hardiman, J. M. Shefner, A. Al-Chalabi, W. Huynh, M. Cudkowicz, P. Talman, L. H. Van den Berg, T. Dharmadasa, P. Wicks, C. Reilly, M. R. Turner, Improving clinical trial outcomes in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 17, 104–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.B. R. Brooks, R. G. Miller, M. Swash, T. L. Munsat; World Federation of Neurology Research Group on Motor Neuron Diseases , El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1, 293–299 (2000). [DOI] [PubMed] [Google Scholar]
- 58.J. D. Rohrer, R. Guerreiro, J. Vandrovcova, J. Uphill, D. Reiman, J. Beck, A. M. Isaacs, A. Authier, R. Ferrari, N. C. Fox, I. R. A. Mackenzie, J. D. Warren, R. de Silva, J. Holton, T. Revesz, J. Hardy, S. Mead, M. N. Rossor, The heritability and genetics of frontotemporal lobar degeneration. Neurology 73, 1451–1456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. A. Regier, Y. Farjoun, D. E. Larson, O. Krasheninina, H. M. Kang, D. P. Howrigan, B.-J. Chen, M. Kher, E. Banks, D. C. Ames, A. C. English, H. Li, J. Xing, Y. Zhang, T. Matise, G. R. Abecasis, W. Salerno, M. C. Zody, B. M. Neale, I. M. Hall, Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9, 4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.L. Henden, N. A. Twine, P. Szul, E. P. McCann, G. A. Nicholson, D. B. Rowe, M. C. Kiernan, D. C. Bauer, I. P. Blair, K. L. Williams, Identity by descent analysis identifies founder events and links SOD1 familial and sporadic ALS cases. NPJ Genom. Med. 5, 32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.The International HapMap Consortium , The International HapMap Project. Nature 426, 789–796 (2003). [DOI] [PubMed] [Google Scholar]
- 62.A. Manichaikul, J. C. Mychaleckyj, S. S. Rich, K. Daly, M. Sale, W.-M. Chen, Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Purcell, B. Neale, K. Todd-Brown, L. Thomas, M. A. R. Ferreira, D. Bender, J. Maller, P. Sklar, P. I. W. De Bakker, M. J. Daly, P. C. Sham, PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R. H. Myers, Huntington’s disease genetics. NeuroRx. 1, 255–262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.P. K. Todd, H. L. Paulson, RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 67, 291–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. D. Figley, A. Thomas, A. D. Gitler, Evaluating noncoding nucleotide repeat expansions in amyotrophic lateral sclerosis. Neurobiol. Aging 35, 936.e1–936.e4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.A. Lund, B. Udd, V. Juvonen, P. M. Andersen, K. Cederquist, M. Davis, C. Gellera, C. Kölmel, L. O. Ronnevi, A. D. Sperfeld, S. A. Sörensen, L. Tranebjaerg, L. Van Maldergem, M. Watanabe, M. Weber, L. Yeung, M. L. Savontaus, Multiple founder effects in spinal and bulbar muscular atrophy (SBMA, Kennedy disease) around the world. Eur. J. Hum. Genet. 9, 431–436 (2001). [DOI] [PubMed] [Google Scholar]
- 68.W. Sproviero, A. Shatunov, D. Stahl, M. Shoai, W. van Rheenen, A. R. Jones, S. Al-Sarraj, P. M. Andersen, N. M. Bonini, F. L. Conforti, P. Van Damme, H. Daoud, M. Del Mar Amador, I. Fogh, M. Forzan, B. Gaastra, C. Gellera, A. D. Gitler, J. Hardy, P. Fratta, V. La Bella, I. Le Ber, T. Van Langenhove, S. Lattante, Y.-C. Lee, A. Malaspina, V. Meininger, S. Millecamps, R. Orrell, R. Rademakers, W. Robberecht, G. Rouleau, O. A. Ross, F. Salachas, K. Sidle, B. N. Smith, B.-W. Soong, G. Sorarù, G. Stevanin, E. Kabashi, C. Troakes, C. van Broeckhoven, J. H. Veldink, L. H. van den Berg, C. E. Shaw, J. F. Powell, A. Al-Chalabi, ATXN2 trinucleotide repeat length correlates with risk of ALS. Neurobiol. Aging 51, 178.e1–178.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.C. Bettencourt, M. Lima, Machado-Joseph Disease: From first descriptions to new perspectives. Orphanet J. Rare Dis. 6, 35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.G. Stevanin, P. Giunti, G. D. Belal, A. Dürr, M. Ruberg, N. Wood, A. Brice, De novo expansion of intermediate alleles in spinocerebellar ataxia 7. Hum. Mol. Genet. 7, 1809–1813 (1998). [DOI] [PubMed] [Google Scholar]
- 71.J. S. Kim, T. O. Son, J. Youn, C.-S. Ki, J. W. Cho, Non-Ataxic Phenotypes of SCA8 Mimicking Amyotrophic Lateral Sclerosis and Parkinson Disease. J. Clin. Neurol. 9, 274–279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.K. Nakamura, S. Y. Jeong, T. Uchihara, M. Anno, K. Nagashima, T. Nagashima, S. Ikeda, S. Tsuji, I. Kanazawa, SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 10, 1441–1448 (2001). [DOI] [PubMed] [Google Scholar]
- 73.N. Sato, T. Amino, K. Kobayashi, S. Asakawa, T. Ishiguro, T. Tsunemi, M. Takahashi, T. Matsuura, K. M. Flanigan, S. Iwasaki, F. Ishino, Y. Saito, S. Murayama, M. Yoshida, Y. Hashizume, Y. Takahashi, S. Tsuji, N. Shimizu, T. Toda, K. Ishikawa, H. Mizusawa, Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 85, 544–557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.A. I. Seixas, J. R. Loureiro, C. Costa, A. Ordóñez-Ugalde, H. Marcelino, C. L. Oliveira, J. L. Loureiro, A. Dhingra, E. Brandão, V. T. Cruz, A. Timóteo, B. Quintáns, G. A. Rouleau, P. Rizzu, Á. Carracedo, J. Bessa, P. Heutink, J. Sequeiros, M. J. Sobrido, P. Coutinho, I. Silveira, A Pentanucleotide ATTTC Repeat Insertion in the Non-coding Region of DAB1, Mapping to SCA37, Causes Spinocerebellar Ataxia. Am. J. Hum. Genet. 101, 87–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.A. Cook, P. Giunti, Friedreich’s ataxia: Clinical features, pathogenesis and management. Br. Med. Bull. 124, 19–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Y. Yuan, Z. Liu, X. Hou, W. Li, J. Ni, L. Huang, Y. Hu, P. Liu, X. Hou, J. Xue, Q. Sun, Y. Tian, B. Jiao, R. Duan, H. Jiang, L. Shen, B. Tang, J. Wang, Identification of GGC repeat expansion in the NOTCH2NLC gene in amyotrophic lateral sclerosis. Neurology 95, e3394–e3405 (2020). [DOI] [PubMed] [Google Scholar]
- 77.E. J. N. Groen, W. van Rheenen, M. Koppers, P. T. C. van Doormaal, L. Vlam, F. P. Diekstra, D. Dooijes, R. J. Pasterkamp, L. H. van den Berg, J. H. Veldink, CGG-repeat expansion in FMR1 is not associated with amyotrophic lateral sclerosis. Neurobiol. Aging 33, 1852.e1–1852.e3 (2012). [DOI] [PubMed] [Google Scholar]
- 78.L. L. Bachinski, T. Czernuszewicz, L. S. Ramagli, T. Suominen, M. D. Shriver, B. Udd, M. J. Siciliano, R. Krahe, Premutation allele pool in myotonic dystrophy type 2. Neurology 72, 490–497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S24
Tables S1 to S3
Supplementary Text
References