Abstract
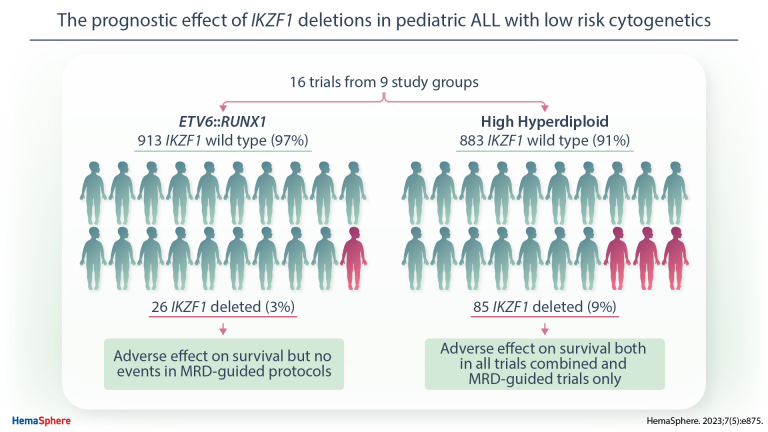
IKZF1 deletions are an established prognostic factor in childhood acute lymphoblastic leukemia (ALL). However, their relevance in patients with good risk genetics, namely ETV6::RUNX1 and high hyperdiploid (HeH), ALL remains unclear. We assessed the prognostic impact of IKZF1 deletions in 939 ETV6::RUNX1 and 968 HeH ALL patients by evaluating data from 16 trials from 9 study groups. Only 3% of ETV6::RUNX1 cases (n = 26) were IKZF1-deleted; this adversely affected survival combining all trials (5-year event-free survival [EFS], 79% versus 92%; P = 0.02). No relapses occurred among the 14 patients with an IKZF1 deletion treated on a minimal residual disease (MRD)-guided protocols. Nine percent of HeH cases (n = 85) had an IKZF1 deletion; this adversely affected survival in all trials (5-year EFS, 76% versus 89%; P = 0.006) and in MRD-guided protocols (73% versus 88%; P = 0.004). HeH cases with an IKZF1 deletion had significantly higher end of induction MRD values (P = 0.03). Multivariate Cox regression showed that IKZF1 deletions negatively affected survival independent of sex, age, and white blood cell count at diagnosis in HeH ALL (hazard ratio of relapse rate [95% confidence interval]: 2.48 [1.32-4.66]). There was no evidence to suggest that IKZF1 deletions affected outcome in the small number of ETV6::RUNX1 cases in MRD-guided protocols but that they are related to higher MRD values, higher relapse, and lower survival rates in HeH ALL. Future trials are needed to study whether stratifying by MRD is adequate for HeH patients or additional risk stratification is necessary.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most prevalent childhood malignancy and recent cure rates approach 90% on first-line therapy.1,2 This is mostly due to trials using increasingly precise risk stratification, but changes in drug doses and schedule as well as improved supportive care also play a role.1,2 Risk stratification is based on clinical and genetic parameters present at diagnosis2 and on early response to treatment as ascertained by minimal residual disease (MRD) analyses.3 Some study groups additionally use copy number alterations to adjust risk stratification.4–6 IKZF1 deletions have been reported as an unfavorable prognostic factor by various study groups,7–16 and some have consequently incorporated IKZF1 status in risk stratification.5,6 IKZF1 deletion is associated with older age,17 higher white blood cell count (WCC) at diagnosis,10 and higher MRD10,18 and is thus overrepresented in high-risk patients. In contrast, IKZF1 deletions are rare in ETV6::RUNX1 and high hyperdiploid (HeH) ALL,11,12 the 2 cytogenetic subgroups with the most favorable prognosis.19,20 Because of the rarity of IKZF1 deletions within the subset of patients with favorable cytogenetics, its prognostic effect remains unclear.10–12 Therefore, we assessed the prognostic impact of IKZF1 deletions in ETV6::RUNX1- and HeH-positive ALL by evaluating previously published data from 16 international trials.19,21–32
MATERIALS AND METHODS
Patients
We performed this retrospective analysis on data of children and adolescents of 1–18 years with B-cell precursor ALL diagnosed in 1991–2016 and treated on 1 of the 16 trials from 9 study groups, of which 6 were MRD guided (Table 1; Suppl. Table S1).33,34 Cytogenetic, fluorescence in-situ hybridization, and reverse transcription polymerase chain reaction (RT-PCR) analyses of pretreatment bone marrow samples to determine ploidy and fusion gene status were performed locally. Trials using MRD for risk stratification applied either PCR19,21,23,25 or flow cytometry analyses.22,24 The IKZF1 status was determined by multiplex ligation-dependent probe amplification (MLPA) by each individual study group (Suppl. Table S1). In addition, we classified cases according to the IKZF1plus profile described by Stanulla et al.16 Because we did not have data on ERG deletions, we used a modified definition of IKZF1plus, namely an IKZF1 deletion plus a deletion involving CDKN2A/B, PAX5, or PAR1. Intragenic ERG deletions are exclusively observed in cases with IGH::DUX4, which is widely considered mutually exclusive with ETV6::RUNX1 and high hyperdiploidy.35 All trials were approved by the local ethics committees and patients, parents, or guardians gave written consent.
Table 1.
Datasets Used for Assessing the Prognostic Value of IKZF1 Deletions in ETV6::RUNX1 and High Hyperdiploid Acute Lymphoblastic Leukemia
| Dataset | Genetic Subtype | IKZF1 Status | Inclusion Criteria From Original Study | |||
|---|---|---|---|---|---|---|
| Deleted | Wild-type | |||||
| n | n | Time Period | Trials/Protocols | |||
| A | ETV6::RUNX1 | 26 | 913 | Availability of MLPA data | 1991–2015 | BFM-ALL 2000, IC-BFM ALL 2002/2009, ANZCHOG ALL8, DCOG ALL8/9/10, NOPHO ALL-92/2000/2008, UKALL2003, MB-ALL-2002/2008, GBTLI LLA-2009, JACLS-ALL02, Other |
| B | High hyperdiploid | 85 | 883 | Availability of MLPA data | ||
| C | High hyperdiploid | 34 | 299 | EOI MRD >0% and <5% | 2003–2014 | CoALL07-03, DCOG ALL10, NOPHO ALL2008, UKALL2003 |
| D | High hyperdiploid | 29 | 276 | Participation in clinical trial | 2003–2013 | DCOG ALL10, UKALL2003 |
EOI MRD = end of induction minimal residual disease; MLPA = multiplex ligation-dependent probe amplification.
Procedures
We used several previously curated datasets to define 4 datasets for our analysis based on genetics and the availability of MRD data (Table 1; Suppl. Table S1). Overlap in patients between datasets might be present and, therefore, datasets were analyzed separately. Because the largest datasets (A and B) did not contain MRD data, datasets C and D were acquired. Dataset C contains categorized MRD data, meaning MRD values have been divided into 5 categories between MRD positive but not quantifiable and 5%. Dataset D contains exact quantitative MRD values without categorization. ALL with both HeH and ETV6::RUNX1 was classified as the latter on the assumption that the fusion gene was the primary genetic abnormality.
Statistics
The survival analyses considered 3 end points: event-free survival (EFS), relapse rate (RR), and overall survival (OS). An event was defined as either relapse, second malignant neoplasm, or death. All end points were censored at last contact. Survival rates were calculated and compared using Kaplan-Meier curves, log-rank tests, and univariate and multivariate Cox regression models. Variables included in the models were sex, age < or ≥10 years, and WCC <50 or ≥50 × 109/L. All variables were linear and conformed to the proportional hazard assumption. Hazard ratios are reported with 95% confidence interval. Survival analyses presented in results were not stratified per trial because of small sample sizes (Suppl. Table S1) and resulting low number of events. However, where number of included patients permitted (>20 cases within a cytogenetic subgroup), survival analysis was performed per trial to assess differences in outcome among trials and these differences did not affect further analyses (Suppl. Table S2). Forest plots were drawn to depict variation in effect size of IKZF1 deletions on outcome among studies. Heterogeneity was tested using Higgins I2 test.36 An I2 statistic ≥50% was considered representing statistically significant heterogeneity.
Categorical variables were compared between groups with Fishers exact test continuous variables with Wilcoxon-rank sum test. To examine MRD as a continuous variable in dataset D, we log-transformed quantitative MRD values, assigned patient-cases with undetectable MRD a value of 1 × 10−6 (one log below the minimum detection level of 1 × 10−5) and assumed a maximum value of 0.99999.37 Normality was assessed by using the skewness, kurtosis, and Shapiro-Wilk test. Log normal distributions were compared by t test.
RESULTS
ETV6::RUNX1
Among the 939 ETV6::RUNX1 positive cases in dataset A (Table 1), 3% had an IKZF1 deletion (Table 2). There was no significant difference between IKZF1-deleted and wild-type cases in terms of age, sex and WCC (P > 0.168 for all, Table 2). When assessing all protocols, ETV6::RUNX1 positive cases with an IKZF1 deletion had a significantly worse outcome (5-year EFS, 79% versus 92%; RR, 18% versus 6%; OS, 87% versus 97%, respectively; P > 0.03 for all, Table 2; Suppl. Figure S1A). However, among the 14 patients with an IKZF1 deletion treated on and MRD-guided protocol (n = 646), no adverse events occurred; suggesting that MRD stratification negated the adverse prognostic impact of an IKZF1 deletion (5-year EFS, 100% versus 93%; RR, 0% versus 5%; OS, 100% versus 98%; P > 0.34 for all; Table 2 and Suppl. Figure S1B).
Table 2.
Features and Treatment Outcome of ETV6::RUNX1 Acute Lymphoblastic Leukemia
| ETV6::RUNX1 | All Protocols | MRD-guided Protocols Only | Non-MRD-guided Protocols Only | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IKZF1 | IKZF1 | IKZF1 | |||||||
| Deleted | Wild-type | Deleted | Wild-type | Deleted | Wild-type | ||||
| n = 939 | n = 26 | n = 913 | P-value | n = 14 | n = 632 | P-value | n = 12 | n = 281 | P-value |
| Age | |||||||||
| <10 | 22 (85%) | 837 (92%) | 0.27 | 13 (93%) | 585 (93%) | 1.00 | 9 (75%) | 252 (90%) | 0.132 |
| ≥10 | 4 (15%) | 76 (8%) | 1 (7%) | 47 (7%) | 3 (25%) | 29 (10%) | |||
| Median (IQR) | 5.00 (3.03–6.21) | 4.00 (3.00–6.00) | 0.434 | 4.50 (3.19–5.96) | 4.00 (3.00–6.00) | 0.741 | 5.35 (2.67–8.75) | 4.00 (3.00–6.00) | 0.486 |
| Sex | 0.844 | 1.00 | 1.00 | ||||||
| Male | 13 (50%) | 478 (52%) | 7 (50%) | 335 (53%) | 6 (50%) | 143 (51%) | |||
| Female | 13 (50%) | 435 (48%) | 7 (50%) | 297 (47%) | 6 (50%) | 138 (49%) | |||
| WCC | |||||||||
| <50 × 109/L | 19 (73%) | 769 (84%) | 0.168 | 11 (79%) | 546 (86%) | 0.421 | 8 (67%) | 223 (79%) | 0.288 |
| ≥ 50 × 109/L | 7 (27%) | 143 (16%) | 3 (21%) | 85 (13%) | 4 (33%) | 58 (21%) | |||
| Median (IQR) | 16.35 (6.02–47.53) | 10.34 (5.20–30.30) | 0.268 | 16.35 (6.03–31.95) | 9.80 (5.02–25.65) | 0.572 | 16.00 (9.47–130.40) | 11.40 (5.60–40.80) | 0.34 |
| CNS disease | 1.00 | 1.00 | Unknown | Unknown | |||||
| Yes | 0 | 1 (<0.5%) | 0 (<0.5%) | 1 (<0.5%) | |||||
| No | 5 (19%) | 226 (25%) | 5 (36%) | 226 (36%) | |||||
| Clinical remission | 1.00 | 1.00 | 1.00 | ||||||
| Yes | 26 (100%) | 908 (99%) | 14 (100%) | 629 (100%) | 12 (100%) | 279 (99%) | |||
| No | 0 | 4 (<0.5%) | 0 | 2 (<0.5%) | 0 | 2 (1%) | |||
| Induction death | 0 | 1 (<0.5%) | 0 | 1 (<0.5%) | 0 | 0 | |||
| Outcome | |||||||||
| 5-y EFS | 79% (63%-97%) | 92% (90%-94%) | 0.021 | 100% | 93% (91%-95%) | 0.341 | 58% (36%-94%) | 91% (87%-94%) | <0.001 |
| 5-y RR | 18% (0%-33%) | 6% (4%-8%) | 0.026 | 0% | 5% (3%-7%) | 0.41 | 37% (0%-59%) | 7% (4%-10%) | <0.001 |
| 5-y OS | 87% (74%-100%) | 97% (96%-98%) | 0.013 | 100% | 98% (67%-99%) | 0.626 | 74% (53%-100%) | 95% (92%-98%) | 0.009 |
| Hazard ratio EFS | 2.80 (1.10–6.90) | Reference | 0.027 | No event | Reference | 5.10 (1.90–13.00) | Reference | <0.001 | |
| Follow-up in years, median (IQR) | 6.72 (4.65–8.71) | 7.40 (5.45–9.22%) | 6.28 (2.25–8.57) | 7.30 (5.71–9.13) | 7.28 (5.84–9.13) | 7.67 (5.08–9.73) | |||
Data are n (%), rates at 5 y (95% CI) or median (IQR).
WCC = white blood cell count; CNS = central nervous system; CI = confidence interval; EFS = event-free survival; IQR = inter-quartile range; OS = overall survival; RR = relapse rate.
Summarizing, dataset A shows that IKZF1 deletions do not affect survival in ETV6::RUNX1 ALL for patients treated on MRD-guided protocols.
High hyperdiploidy
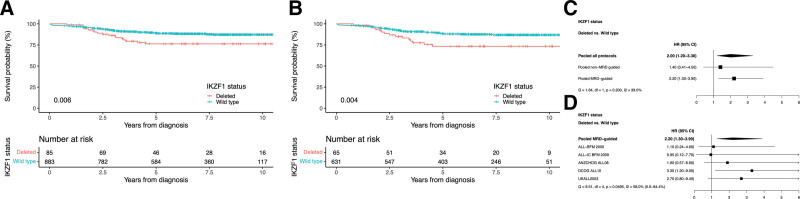
Among the 968 HeH ALL cases in dataset B (Table 1), 9% had an IKZF1 deletion (Table 3). IKZF1-deleted cases were significantly older (median of 5 versus 4 years at diagnosis, P = 0.005; Table 3), but sex and WCC were not significantly different between IKZF1-deleted and wild-type patients (P > 0.557; Table 3). There was no difference in the survival of HeH cases between non-MRD-guided and MRD-guided protocols (hazard ratio of EFS non-MRD-guided trials versus MRD-guided: 0.83 [0.55-1.30], P = 0.372; Table 3). However, the outcome of HeH patients was not equivalent across all the trials. In ALL-IC BFM 2002, ANZCHOG ALL8, and DCOG ALL10, patients with HeH showed a significantly higher hazard ratio for RR, EFS, and OS compared with patients treated on UKALL2003 (Suppl. Table S2). IKZF1-deleted HeH ALL had a significantly worse outcome than HeH with IKZF1 wild-type (5-year EFS, 76% versus 89%; RR, 20% versus 8%; OS, 88% versus 94%, respectively; P < 0.01 for all; Table 3 and Figure 1A) when examining the total cohort (dataset B). When we examined MRD-guided and non-MRD-guided protocols separately, we observed a lower and nonsignificant hazard ratio among the non-MRD-guided protocols but the test for heterogeneity was not significant (Figure 1C). IKZF1 deletions had a negative effect in MRD-guided protocols (n = 696): 5-year EFS, 73% versus 88%; RR, 23% versus 9%; OS, 88% versus 94%; P < 0.01 for all; Table 3 and Figure 1B. When adjusting for clinical parameters by including sex, age, and WCC in a multivariate Cox regression model, IKZF1 status still affected survival significantly in HeH cases treated on MRD-guided protocols (hazard ratio IKZF1-deleted versus wild-type EFS, 2.09 [1.19-3.65]; RR, 2.48 [1.32-4.66]; OS, 2.37 [1.17-4.79]; P < 0.03 for all). The individual MRD-guided protocols had percentages of 0%–4% of IKZF1-deleted cases (Suppl. Table S1). However, among the 5 MRD-guided protocols, there was an evidence of heterogeneity albeit with a marginal P-value (P = 0.0495) indicating that the prognostic impact of IKZF1 deletions may be protocol specific (Figure 1D). Applying the modified IKZF1plus profile in HeH cases did not show any additional effect on survival over IKZF1 deletions only (P > 0.18 for all, Suppl. Table S3). In summary, dataset B shows that IKZF1 deletions can lead to lower survival and higher RRs in HeH ALL cases treated on MRD-guided protocols and that IKZF1plus does not have additional prognostic effect over IKZF1 deletion alone.
Table 3.
Features and Treatment Outcome of High Hyperdiploid Acute Lymphoblastic Leukemia
| High Hyperdiploid | All Protocols | MRD-guided Protocols Only | Non-MRD-guided Protocols Only | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IKZF1 | IKZF1 | IKZF1 | |||||||
| Deleted | Wild-type | Deleted | Wild-type | Deleted | Wild-type | ||||
| n = 968 | n = 85 | n = 883 | P-value | n= 65 | n = 631 | P-value | n = 20 | n = 252 | P-value |
| Age | |||||||||
| <10 | 68 (80%) | 780 (88%) | 0.037 | 51 (78%) | 556 (88%) | 0.032 | 17 (85%) | 224 (89%) | 0.485 |
| ≥10 | 17 (20%) | 103 (12%) | 14 (22%) | 75 (12%) | 3 (15%) | 28 (11%) | |||
| Median (IQR) | 5.00 (3.09–7.83) | 4.00 (2.52–6.45) | 0.005 | 5.36 (3.28–7.92) | 3.94 (2.60–6.41) | 0.003 | 4.00 (2.91–6.55) | 4.00 (2.00–6.86) | 0.676 |
| Sex | 0.734 | 1.00 | 0.493 | ||||||
| Male | 42 (49%) | 457 (52%) | 33 (51%) | 322 (51%) | 9 (45%) | 135 (54%) | |||
| Female | 43 (51%) | 426 (48%) | 32 (49%) | 309 (49%) | 11 (55%) | 117 (46%) | |||
| WCC | |||||||||
| <50 × 109/L | 76 (89%) | 803 (91%) | 0.557 | 60 (92%) | 577 (91%) | 1.00 | 16 (80%) | 226 (90%) | 0.247 |
| ≥50 × 109/L | 9 (11%) | 79 (9%) | 5 (8%) | 54 (9%) | 4 (20%) | 25 (10%) | |||
| Median (IQR) | 7.20 (3.40–17.80) | 7.30 (3.50–18.35) | 0.922 | 7.10 (3.40–16.50) | 7.00 (3.40–17.30) | 0.781 | 15.95 (3.32–32.59) | 8.10 (3.89–21.80) | 0.599 |
| CNS disease | 1.00 | 1.00 | |||||||
| Yes | 0 | 2 (<0.5%) | 0 | 2 (0.3%) | Unknown | Unknown | |||
| No | 17 (20%) | 204 (23%) | 17 (26%) | 204 (32%) | |||||
| Clinical remission | 1.00 | 1.00 | |||||||
| Yes | 84 (99%) | 870 (99%) | 65 (100%) | 623 (99%) | 19 (95%) | 247 (98%) | 0.37 | ||
| No | 1 (1%) | 8 (1%) | 0 | 5 (1%) | 1 (5%) | 3 (1%) | |||
| Induction death | 0 | 5 (1%) | 0 | 3 (<0.5%) | 0 | 2 (1%) | |||
| Outcome | |||||||||
| 5-y EFS | 76% (67%-87%) | 89% (87%-91%) | 0.006 | 73% (63%-86%) | 88% (86%-91%) | 0.004 | 85% (70%-100%) | 90% (87-94%) | 0.62 |
| 5-y RR | 20% (10%-28%) | 8% (6%-10%) | 0.003 | 23% (10%-33%) | 9% (6%-11%) | 0.002 | 11% (0%-24%) | 6% (3-9%) | 0.576 |
| 5-y OS | 88% (81%-96%) | 94% (93%-96%) | 0.005 | 88% (79%-97%) | 94% (92%-96%) | 0.004 | 90% (78%-100%) | 94% (91-97%) | 0.541 |
| Hazard ratio EFS | 2.00 (1.20–3.30) | Reference | 0.007 | 2.20 (1.30%–3.90) | Reference | 0.005 | 1.40 (0.41%–4.50) | Reference | 0.621 |
| Follow-up in years, median (IQR) | 6.93 (4.48–9.83) | 7.19 (4.89–9.10) | 6.90 (4.48–8.96) | 6.95 (4.70–8.86) | 8.64 (4.68%–10.52) | 7.62 (5.68–10.43) | |||
Data are n (%), rates at 5 y (95% CI) or median (IQR).
WCC = white blood cell count; CNS = central nervous system; CI = confidence interval; EFS = event-free survival; IQR = inter-quartile range; OS = overall survival; RR = relapse rate.
Figure 1.
Outcome of patients with high hyperdiploid acute lymphoblastic leukemia. (A) Kaplan-Meier of event-free survival including both MRD-guided and non-MRD-guided trials. (B) Kaplan-Meier of event-free survival of MRD-guided trials. (C) Effect of IKZF1 deletions on hazard ratio of event-free survival: pooled data from MRD-guided and non-MRD-guided trials. (D) Effect of IKZF1 deletions on hazard ratio of event-free survival: data from each MRD-guided trial. P-values from log-rank test for comparing survival function estimates. MRD = minimal residual disease.
High hyperdiploidy in ALLTogether
The new European collaborative treatment protocol ALLTogether uses the UKALL-CNA profile5,34 along with other genetic abnormalities and MRD for risk stratification (AVM, personal communication, March 2, 2022). Patients are assigned at the end of induction to standard-, intermediate-, or high-risk groups based on MRD and the presence of selected high-risk features.
HeH patients allocated to the initial intermediate-risk group (detectable MRD <5% at the end of induction) are further stratified into the intermediate low-risk group if they fulfill either of the following criteria: (a) MRD <0.03% or (b) a good risk UKALL-CNA profile and MRD <0.05%. The presence of an IKZF1 deletion would not influence the first criterion but would prevent the patient qualifying on the basis of the second criterion, because the IKZF1 deletion would automatically assign them to a UKALL-CNA poor risk profile and would lead to assignment to the intermediate-risk-high group if their MRD was ≥0.03%.
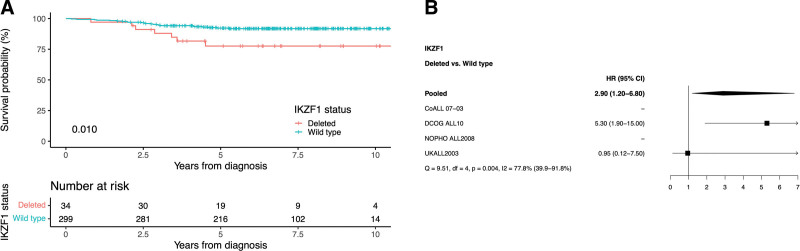
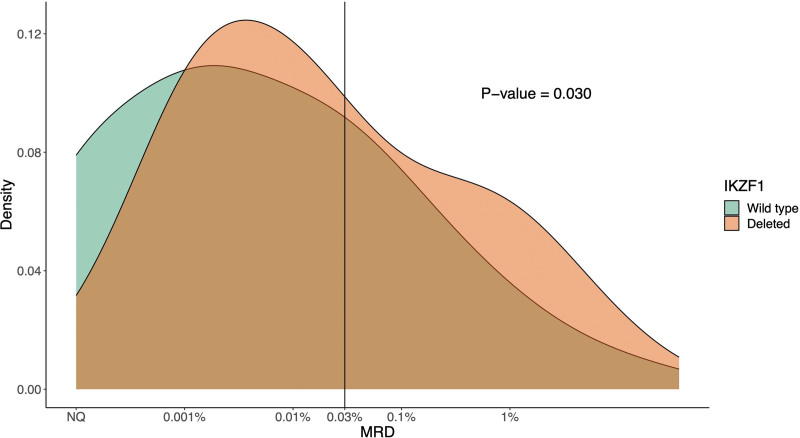
Therefore, we examined the prognostic effect of IKZF1 deletion in this specific subgroup in more detail by looking at the 333 HeH patients with detectable MRD <5% at the end of induction in dataset C33 (Table 1 and Suppl. Table S4). Among these cases, 10% carried an IKZF1 deletion and while the age difference between IKZF1-deleted and wild-type cases was not significant in this subgroup, a significant difference in sex distribution was seen (68% versus 44% males, respectively; P = 0.011; Table 4). Within dataset C, IKZF1-deleted cases also had a worse outcome compared with IKZF1 wild-type cases (5-year EFS, 78% versus 92%; RR, 18% versus 6%; OS, 88% versus 97%; P < 0.05 for all; Table 4 and Figure 2A). When adjusting for sex, age, and WCC in a multivariate Cox regression model, IKZF1 deletion still affected survival significantly (hazard ratio of IKZF1-deleted versus wild-type EFS, 2.96 [1.23-7.14]; RR, 3.03 [1.08-8.54]; OS, 4.20 [1.41-12.53]; P < 0.05 for all). The prognostic effect of IKZF1 deletion was again largest in the DCOG ALL10 trial (n = 95), while no events were observed in IKZF1-deleted cases treated on CoALL 07-03 (n = 2) or NOPHO ALL2008 (n = 5) (Figure 2B, Suppl. Table S5). There was strong evidence for statistical heterogeneity among studies (P = 0.004). When assessing the quantitative MRD data of 305 HeH patients from the UKALL2003 and DCOG ALL10 cohorts in dataset D (Table 1), cases with an IKZF1 deletion had significantly higher MRD values (P = 0.03; Figure 3) and 80% had an MRD ≥0.03%, which would already exclude them from the intermediate low-risk group, based on MRD levels only.
Table 4.
Features and Treatment Outcome of High Hyperdiploid Acute Lymphoblastic Leukemia and Minimal Residual Disease at the End of Induction >0% and <5%
| High Hyperdiploid | IKZF1 | P-value | |
|---|---|---|---|
| Deleted | Wild-type | ||
| n = 333 | n = 34 | n = 299 | |
| Age | |||
| <10 | 30 (88%) | 261 (87%) | 1.00 |
| ≥10 | 4 (12%) | 38 (13%) | |
| Median (IQR) | 5.13 (3.72–6.84) | 4.00 (2.66–6.12) | 0.072 |
| Sex | 0.011 | ||
| Male | 23 (68%) | 133 (44%) | |
| Female | 11 (32%) | 166 (56%) | |
| WCC | |||
| <50 × 109/L | 31 (91%) | 247 (83%) | 0.334 |
| ≥50 × 109/L | 1 (3%) | 25 (8%) | |
| Median (IQR) | 6.65 (3.40–13.72) | 7.70 (4.10–21.50) | 0.167 |
| CNS disease | 1.00 | ||
| Yes | 0 | 1 (<0.5%) | |
| No | 15 (44%) | 128 (43%) | |
| Clinical remission | 1.00 | ||
| Yes | 34 (100%) | 299 (100%) | |
| No | 0 | 0 | |
| Induction death | 0 | 0 | |
| Ouctome | |||
| 5-y EFS | 78% (64%-94%) | 92% (89%-95%) | 0.01 |
| 5-y RR | 18% (2%-31%) | 6% (3%-9%) | 0.041 |
| 5-y OS | 88% (77%-100%) | 97% (95%-99%) | 0.041 |
| Hazard ratio EFS | 2.90 (1.20–6.80) | Reference | 0.014 |
| Follow-up in years, median (IQR) | 6.74 (4.50–8.88) | 6.69 (5.31–8.29) | |
Data are n (%), rates at 5 y (95% CI) or median (IQR).
WCC = white blood cell count; CNS = central nervous system; CI = confidence interval; EFS = event-free survival; IQR = inter-quartile range; OS = overall survival; RR = relapse rate.
Figure 2.
Outcome of patients with high hyperdiploid acute lymphoblastic leukemia and minimal residual disease at the end of induction between >0% and <5%. (A) Kaplan-Meier of event-free survival. (B) Effect of IKZF1 deletion on hazard ratio of event-free survival. P-values from log-rank test for comparing survival function estimates. No hazard ratio for NOPHO and CoALL due to no events.
Figure 3.
Distribution of the log-transformed MRD value, τ(MRD), at the end of induction of 305 patients with HeH ALL treated on UKALL2003 or DCOGALL10 trials. Smoothed density plots of the log-transformed minimal τ(MRD) by IKZF1 status: not deleted (green) and deleted (orange); 0.03% is MRD cutoff at the end of induction for HeH ALL for placement in intermediate-risk-low or intermediate-risk-high in ALLTogether trial. HeH = high hyperdiploid; MRD = minimal residual disease.
Together, dataset B, C, and D show that IKZF1 deletions can lead to lower survival and higher RRs in HeH ALL with intermediate MRD at the end of induction and to higher MRD levels in general.
DISCUSSION
IKZF1 deletions are an established poor prognostic factor in childhood ALL, but their value in the favorable cytogenetic subgroups ETV6::RUNX1 and HeH is unclear. The present comprehensive analysis of 16 trials assesses the prognostic effect of IKZF1 deletions in these 2 subgroups. Our results show that IKZF1 deletions can predict significantly lower survival in HeH ALL, even when treated on MRD-adapted protocols and when adjusting for other clinical parameters. However, IKZF1 deletions do not have a prognostic effect in ETV6::RUNX1 ALL when treated on MRD-adapted protocols.
Previous DCOG trials showed that IKZF1 deletions do not have prognostic value in patients stratified as standard risk18 or in ETV6::RUNX1 as a subgroup.11 However, it did have prognostic value in HeH patients6,11 and in patients stratified as medium risk, independent of the MRD level within the medium risk arm.6,18 These findings led to the design of the DCOG ALL11 trial in which having an IKZF1 deletion resulted in longer treatment (3 years) in the medium risk arm.6 However, in the current trial used by many European countries, ALLTogether, IKZF1 status alone is not used for treatment stratification of ETV6::RUNX1 and HeH cases. For these 2 subgroups, specific optimal MRD thresholds at the end of induction have been established and are the main parameters used for stratification to intermediate-risk-low or intermediate-risk-high treatment arms.37
MRD was previously shown to be a very accurate prognostic parameter for treatment outcome in childhood ALL,19,38,39 also in combination with IKZF1 status.11,18 Unfortunately, we could not take MRD values into account for the largest datasets A and B. Therefore, we only compared quantitative values of the first time point (end of induction) of 2 recent trials (dataset D). Our analysis of quantitative MRD data of HeH patients showed that IKZF1-deleted cases showed higher MRD levels than wild-type cases at the end of induction. To determine how HeH cases with IKZF1 deletions would be stratified in the new ALLTogether trial, we examined how cases assigned to the initial intermediate-risk group would distribute across the intermediate-risk-low and intermediate-risk-high arms based on quantitative end of induction MRD. The vast majority of these HeH cases (80% in dataset D) would be allocated to intermediate-risk-high group of the current ALLTogether trial and would not be eligible for any treatment reduction. This is the same treatment arm non-HeH cases with IKZF1 deletions would be placed in, based on the copy number alteration profile. In this scenario, stratification by primary genetic subtype (HeH) and MRD appears to have the same effect as stratifying by primary and secondary genetic abnormality. The pattern of chromosomal gain in HeH has been linked to both MRD and outcome. A recent study identified that the pattern of gain of 4 chromosomes (5, 17, 18, and 20) could defined low- and high-risk subtypes of HeH ALL.40 Interestingly, the proportion of cases with an IKZF1 deletion was higher in the UKALL-HeH high-risk group compared with the low-risk group: 11% versus 6%, P = 0.66.
One of the limitations of this study is that it is based on data from 16 trials spanning 25 years and during this period OS for ALL has increased. However, IKZF1 deletions are rare in ETV6::RUNX1 and HeH ALL; hence, analysis of large retrospective multitrial datasets are the only practical source of information. All retrospective studies spanning long periods are limited by the fact that patients are treated on different, often improving protocols. To address this issue, we present data by trial and also by 2 major eras—pre and post the advent of MRD risk stratification. Survival analysis of patients with HeH ALL treated on different protocols showed several differences in RR, EFS, and OS, although the trials were recent and MRD-guided. It is not clear why this difference in survival occurs. All trials use similar drugs but in different doses and regimens. In our analysis, the prognostic effect of IKZF1 deletion was largest in the DCOG ALL10 trial and even more pronounced when we examined intermediate-risk patients as defined by the ALLTogether trial (Dataset C). This difference cannot be explained by difference in methodology or classification. MLPA was used to detect all IKZF1 deletions, and we use the same definition of HeH, that is, cytogenetic presence of an abnormal clone with 51–67 chromosomes. Furthermore, we re-examined the DCOG cases to ensure that there was no misclassification or inclusion of masked near-haploidy. Because ALL with IKZF1 deletions has been shown to be more resistant to therapeutic drugs, this mechanism of drug resistance might underlie the difference in prognostic effect of IKZF1 deletions among trials.6
Stanulla et al16 described the prognostic effect of the IKZF1plus profile characterized by a co-occurrence of IKZF1 deletions with deletions of CDKN2A, CDKN2B, or PAX5 or the PAR1 region in the absence of ERG deletion. We did not have data on ERG status and could, therefore, not assess the IKZF1plus profile, although the numbers for this co-occurrence are expected to be small in the ETV6::RUNX1 and HeH subtypes. Our analysis using the modified profile without ERG status did not show an additional prognostic effect of IKZF1plus over assessing IKZF1 status only. In addition, due to slight differences in IKZF1 status calling between trials and in concordance with previous reports on the prognostic value of copy number alterations, single-exon deletions have not been categorized as IKZF1-deleted in this dataset. Although single-exon deletions can have a prognostic effect,41 they only comprise <10% of IKZF1 deletions11 and are, therefore, unlikely to influence our conclusions.
In conclusion, our analysis of a large composite cohort consisting of 16 trials shows no evidence to suggest that IKZF1 deletions affect outcome in the small number of ETV6::RUNX1 cases in MRD-guided protocols. In contrast, our data show that in HeH ALL, IKZF1 deletions are associated with lower survival rates, higher RRs, and higher MRD values. Future results of current trials such as the ALLTogether will likely reveal whether risk stratification predominantly reliant on MRD is adequate for HeH patients or whether stratification by copy number alteration profile, including IKZF1 status, or by other methods, would be more suitable.
ACKNOWLEDGMENTS
Authors thank all the study groups and their representatives who contributed data to the 2 original studies: (1) The international BFM study group report34: UKALL study group (Lina Hamadeh, Amir Enshaei, Claire Schwab, Christine J Harrison, Anthony V. Moorman, and Ajay Vora), NOPHO, Nordic Society for Paediatric Haematology and Oncology (Gisela Barbany, Ingegerd I. Öfverholm, and Mats M. Heyman), DCOG, Dutch Childrens Oncology Group (Monique L. den Boer, Judith M. Boer, Roland P. Kuiper, and Rob Pieters), ANZCHOG, Australian and New Zealand Children Haematology/Oncology Group (Luciano Dalla Pozza, Rosemary Sutton, and Nicola Venn), EMiLI, Brazilian Group for the treatment of ALL (Maria S Pombo-de-Oliveira and Mariana Emerenciano), Austrian BFM study group (Andishe Attarbaschi, Karin Nebral, and Sabine Strehl), ALL-IC-2009 study group (Cristina N Alonso, Marcin Braun, Eva Fronkova, Gábor Kovács, Wojciech Mlynarski, Irén Haltrich, Sarah Elitzur, Agata Pastorczak, Henriett Piko, Patricia Rubio, Marta Jeison, Maria Sara Felice, Jan Trka, and Jan Stary), Russian Childhood ALL study group (Larisa Fechina and Grigory Tsaur), Japan Association of Childhood Leukaemia Study (Keizo Horibe, Toshihiko Imamura, and Mio Yano); (2) The UKALL prognostic index study33: UKALL study group (Amir Enshaei, David O’Connor, Jack Bartram, Jeremy Hancock, Christine J. Harrison, Rachael Hough, Sujith Samarasinghe, Ajay Vora, John Moppett, and Anthony V Moorman), NOPHO, Nordic Society for Paediatric Haematology and Oncology (Hanne V. Marquart, Ulrika Norén-Nyström, Kjeld Schmiegelow, Mats M Heyman), DCOG, Dutch Childhood Oncology Group (Monique L den Boer, Judith M Boer, Hester A. de Groot-Kruseman, and Rob Pieters), German Co-operative Study Group (CoALL)–07-03 (Martin A. Horstmann and Gabriele Escherich).
AUTHOR CONTRIBUTIONS
FvL, RK, and RP conceived and designed the study. AV, MAH, GE, BJ, MH, MLdB, and KS supplied data. AE and AVM collected and assembled the data. AØ and AE analyzed the data. AØ, AE, PH, MLdB, RP, AVM, FNvL, and JB interpreted the data. AØ wrote the original draft. All authors reviewed and approved the final article.
DATA AVAILABILITY
The data used in this study are available from the corresponding author.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
AØ received funding from the Princess Máxima Center Foundation, Talent Programme. AVM received funding from Blood Cancer UK. MdB and JB received funding from the Oncode Institute.
Supplementary Material
Footnotes
AØ and AE shared first author.
AVM, JMB, and FNvL shared last author.
Ethical statement: The study was conducted in accordance with the Declaration of Helsinki. All research protocols were approved by the local Ethics Committees of the included trials. Informed consent was obtained from all participants and/or their parents or caretakers.
Supplemental digital content is available for this article.
REFERENCES
- 1.Pui CH, Yang JJ, Hunger SP, et al. Childhood acute lymphoblastic leukemia: progress through collaboration. J Clin Oncol. 2015;33:2938–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vrooman LM, Silverman LB. Treatment of childhood acute lymphoblastic leukemia: prognostic factors and clinical advances. Curr Hematol Malig Rep. 2016;11:385–394. [DOI] [PubMed] [Google Scholar]
- 3.Contreras Yametti GP, Ostrow TH, Jasinski S, et al. Minimal residual disease in acute lymphoblastic leukemia: current practice and future directions. Cancers (Basel). 2021;13:1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125:3977–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moorman A, Enshaei A, Schwab C, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444. [DOI] [PubMed] [Google Scholar]
- 6.Steeghs EMP, Boer JM, Hoogkamer AQ, et al. Copy number alterations in B-cell development genes, drug resistance, and clinical outcome in pediatric B-cell precursor acute lymphoblastic leukemia. Sci Rep. 2019;9:4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuiper RP, Waanders E, Van Der Velden VHJ, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. [DOI] [PubMed] [Google Scholar]
- 8.Asai D, Imamura T, Suenobu SI, et al. IKZF1 deletion is associated with a poor outcome in pediatric B-cell precursor acute lymphoblastic leukemia in Japan. Cancer Med. 2013;2:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita Y, Shimada A, Yamada T, et al. IKZF1 and CRLF2 gene alterations correlate with poor prognosis in Japanese BCR-ABL1-negative high-risk B-cell precursor acute lymphoblastic leukemia. Pediatr Blood Cancer. 2013;60:1587–1592. [DOI] [PubMed] [Google Scholar]
- 10.Dörge P, Meissner B, Zimmermann M, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clappier E, Grardel N, Bakkus M, et al. IKZF1 deletion is an independent prognostic marker in childhood B-cell precursor acute lymphoblastic leukemia, and distinguishes patients benefiting from pulses during maintenance therapy: results of the EORTC Children’s Leukemia Group study 58951. Leukemia. 2015;29:2154–2161. [DOI] [PubMed] [Google Scholar]
- 13.Olsson L, Ivanov Öfverholm I, Norén-Nyström U, et al. The clinical impact of IKZF1 deletions in paediatric B-cell precursor acute lymphoblastic leukaemia is independent of minimal residual disease stratification in Nordic Society for Paediatric Haematology and Oncology treatment protocols used between 1992 and 2013. Br J Haematol. 2015;170:847–858. [DOI] [PubMed] [Google Scholar]
- 14.Sutton R, Venn NC, Law T, et al. A risk score including microdeletions improves relapse prediction for standard and medium risk precursor B-cell acute lymphoblastic leukaemia in children. Br J Haematol. 2018;180:550–562. [DOI] [PubMed] [Google Scholar]
- 15.Vrooman LM, Blonquist TM, Harris MH, et al. Refining risk classification in childhood B acute lymphoblastic leukemia: results of DFCI ALL Consortium Protocol 05-001. Blood Adv. 2018;2:1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanulla M, Dagdan E, Zaliova M, et al. IKZF1 plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric b-cell precursor acute lymphoblastic leukemia. J Clin Oncol. 2018;36:1240–1249. [DOI] [PubMed] [Google Scholar]
- 17.Palmi C, Valsecchi MG, Longinotti G, et al. What is the relevance of Ikaros gene deletions as a prognostic marker in pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia? Haematologica. 2013;98:1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waanders E, Van Der Velden VHJ, Van Der Schoot CE, et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 2011;25:254–258. [DOI] [PubMed] [Google Scholar]
- 19.Pieters R, De Groot-Kruseman H, Van Der Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol. 2016;34:2591–2601. [DOI] [PubMed] [Google Scholar]
- 20.Moorman AV, Ensor HM, Richards SM, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. [DOI] [PubMed] [Google Scholar]
- 21.Möricke A, Zimmermann M, Valsecchi MG, et al. Dexamethasone vs prednisone in induction treatment of pediatric ALL: results of the randomized trial AIEOP-BFM ALL 2000. Blood. 2016;127:2101–2112. [DOI] [PubMed] [Google Scholar]
- 22.Zawitkowska J, Lejman M, Romiszewski M, et al. Results of two consecutive treatment protocols in Polish children with acute lymphoblastic leukemia. Sci Rep. 2020;10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karsa M, Dalla Pozza L, Venn NC, et al. Improving the identification of high risk precursor B acute lymphoblastic leukemia patients with earlier quantification of minimal residual disease. PLoS One. 2013;8:e76455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toft N, Birgens H, Abrahamsson J, et al. Results of NOPHO ALL2008 treatment for patients aged 1-45 years with acute lymphoblastic leukemia. Leukemia. 2018;32:606–615. [DOI] [PubMed] [Google Scholar]
- 25.Vora A, Goulden N, Wade R, et al. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2013;14:199–209. [DOI] [PubMed] [Google Scholar]
- 26.Stary J, Zimmermann M, Campbell M, et al. Intensive chemotherapy for childhood acute lymphoblastic leukemia: results of the randomized intercontinental trial ALL IC-BFM 2002. J Clin Oncol. 2014;32:174–184. [DOI] [PubMed] [Google Scholar]
- 27.Meleshko AN, Savva NN, Fedasenka UU, et al. Prognostic value of MRD-dynamics in childhood acute lymphoblastic leukemia treated according to the MB-2002/2008 protocols. Leuk Res. 2011;35:1312–1320. [DOI] [PubMed] [Google Scholar]
- 28.Kamps WA, Bökkerink JPM, Hakvoort-Cammel FGAJ, et al. BFM-oriented treatment for children with acute lymphoblastic leukemia without cranial irradiation and treatment reduction for standard risk patients: results of DCLSG protocol ALL-8 (1991-1996). Leukemia. 2002;16:1099–1111. [DOI] [PubMed] [Google Scholar]
- 29.Veerman AJ, Kamps WA, van den Berg H, et al. Dexamethasone-based therapy for childhood acute lymphoblastic leukaemia: results of the prospective Dutch Childhood Oncology Group (DCOG) protocol ALL-9 (1997-2004). Lancet Oncol. 2009;10:957–966. [DOI] [PubMed] [Google Scholar]
- 30.Rocha JMC, Xavier SG, Souza ME de L, et al. Comparison between flow cytometry and standard PCR in the evaluation of MRD in children with acute lymphoblastic leukemia treated with the GBTLI LLA - 2009 protocol. Pediatr Hematol Oncol. 2019;36:287–301. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto K, Imamura T, Kihira K, et al. Low incidence of osteonecrosis in childhood acute lymphoblastic leukemia treated with ALL-97 and ALL-02 study of Japan association of childhood leukemia study group. J Clin Oncol. 2018;36:900–907. [DOI] [PubMed] [Google Scholar]
- 32.Schmiegelow K, Forestier E, Hellebostad M, et al. Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia. 2010;24:345–354. [DOI] [PubMed] [Google Scholar]
- 33.Enshaei A, O’Connor D, Bartram J, et al. A validated novel continuous prognostic index to deliver stratified medicine in pediatric acute lymphoblastic leukemia. Blood. 2020;135:1438–1446. [DOI] [PubMed] [Google Scholar]
- 34.Hamadeh L, Enshaei A, Schwab C, et al. Validation of the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv. 2019;3:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab CJ, Murdy D, Butler E, et al. Genetic characterisation of childhood B-other-acute lymphoblastic leukaemia in UK patients by fluorescence in situ hybridisation and Multiplex Ligation-dependent Probe Amplification. Br J Haematol. 2022;196:753–763. [DOI] [PubMed] [Google Scholar]
- 36.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. [DOI] [PubMed] [Google Scholar]
- 37.O’Connor D, Enshaei A, Bartram J, et al. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2018;36:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton R, Venn NC, Tolisano J, et al. Clinical significance of minimal residual disease at day 15 and at the end of therapy in childhood acute lymphoblastic leukaemia. Br J Haematol. 2009;146:292–299. [DOI] [PubMed] [Google Scholar]
- 39.Bartram J, Wade R, Vora A, et al. Excellent outcome of minimal residual disease-defined low-risk patients is sustained with more than 10 years follow-up: results of UK paediatric acute lymphoblastic leukaemia trials 1997-2003. Arch Dis Child. 2016;101:449–454. [DOI] [PubMed] [Google Scholar]
- 40.Enshaei A, Vora A, Harrison CJ, et al. Defining low-risk high hyperdiploidy in patients with paediatric acute lymphoblastic leukaemia: a retrospective analysis of data from the UKALL97/99 and UKALL2003 clinical trials. Lancet Haematol. 2021;8:e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boer JM, Van Der Veer A, Rizopoulos D, et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia. 2016;30:32–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are available from the corresponding author.