Dear Editor,
Mutation on CEBPA (CEBPAmut) is one of the most common molecular abnormalities in acute myeloid leukemia (AML), especially in east Asian population [1]. As recently reported, in-frame mutations in bZIP domain of CEBPA (CEBPAbZIP-inf) exerted higher potency in favorable-risk prediction than biallelic mutated CEBPA (CEBPAbi), although cases were highly overlapped between the two categories [2–4]. However, about 30–50% CEBPAbi AML cases consolidated with chemotherapy alone suffered from disease relapse [5, 6], retaining the same CEBPAmut patterns as diagnosis [7, 8]. Hence, there might be clinically and biologically heterogeneous under current context of CEBPAmut grouping, and a comprehensive assessment of CEBPAmut AML prognosis remains to be established.
In this study, a total of 293 de novo CEBPAmut AML patients were enrolled, with biological data available in 124 patients (Supplementary Fig. 1A). Usually, CEBPAbZIP-inf AML patients were diagnosed at younger age, with higher white blood cell counts, hemoglobin levels and lower platelet counts compared with other CEBPAmut AML patients (CEBPAother); while risk classification of karyotypes according to the ELN 2022 showed no differences between CEBPAbZIP-inf and CEBPAother AML (Supplementary Table 1).
Consistence with previous reports [2, 3], CEBPAbZIP-inf AML correlated with higher CR rate (Supplementary Table 1), which could translate into improved overall survival (2-year OS: 86% vs. 53.1%, p = 0.0019) and event-free survival (2-year EFS: 64.7% vs. 37.5%, p = 0.01) (Supplementary Fig. 1B). However, the OS, EFS and relapse rate (Supplementary Table 1) of the patients who achieved CR showed no difference between CEBPAbZIP-inf and CEBPAother AML (2-year OS: 87% vs. 63.1%, p = 0.07; 2-year EFS: 65.5% vs. 47.7%, p = 0.19) (Supplementary Fig. 1C). It seemed that the current risk stratification based on CEBPAmut locus could not sufficiently distinguish certain patients who may develop disease progression.
Most patients with CEBPAbZIP-inf (85/89, 95.5%) displayed cross-lineage expression of CD7, while only 20/35 (57.1%) patients with CEBPAother harboring CD7-positive immunophenotype (p < 0.001) (Supplementary Table 1). CD7-positive cases showed distinct gene expression patterns compared with CD7 negative cases (Supplementary Fig. 2). Survival analysis further indicated CD7 could significantly distinguish the clinical outcome in the whole cohort of 293 CEBPAmut AML cases, with improved 2-year OS and EFS of 81.8% and 66.4% respectively in CD7-positive CEBPAmut AML vs. 48.8% and 33.0% respectively in CD7-negative CEBPAmut AML (p < 0.0001 and <0.0001; Supplementary Fig. 3A, B). Moreover, CD7-negative CEBPAmut AML patients also had a shorter disease-free survival compared with CD7-positive patients (2-year DFS: CD7-positive CEBPAmut AML 63.1% vs. CD7-negative CEBPAmut AML 39.4%, p < 0.0001; Supplementary Fig. 3C).
Given the prognostic significance of CD7 in CEBPAmut AML, survival analysis of 117 CR patients demonstrated that the combine of CD7 with the CEBPAmut locus could discriminate disease prognosis, with distinguished 2-year OS and EFS: CEBPAbZIP-inf/CD7 + AML, 90.4% and 68.8% vs. other CEBPAmut AML, 56.6% and 41.3%, respectively (p < 0.0001 and = 0.0076; Supplementary Fig. 3D, E). Besides, the 2-year DFS of CEBPAmut AML was as follows: CEBPAbZIP-inf/CD7 + AML 72.5% vs. other CEBPAmut AML 40.6%, (p = 0.0028; Supplementary Fig. 3F). Multivariable analysis further confirmed CEBPAbZIP-inf/CD7+ as an independent risk factor that favors the prognosis of CEBPAmut AML, with hazard ratio of 0.16 (p = 0.001), 0.45 (p = 0.034), and 0.39 (p = 0.018) in OS, EFS and DFS, respectively (Supplementary Table 2).
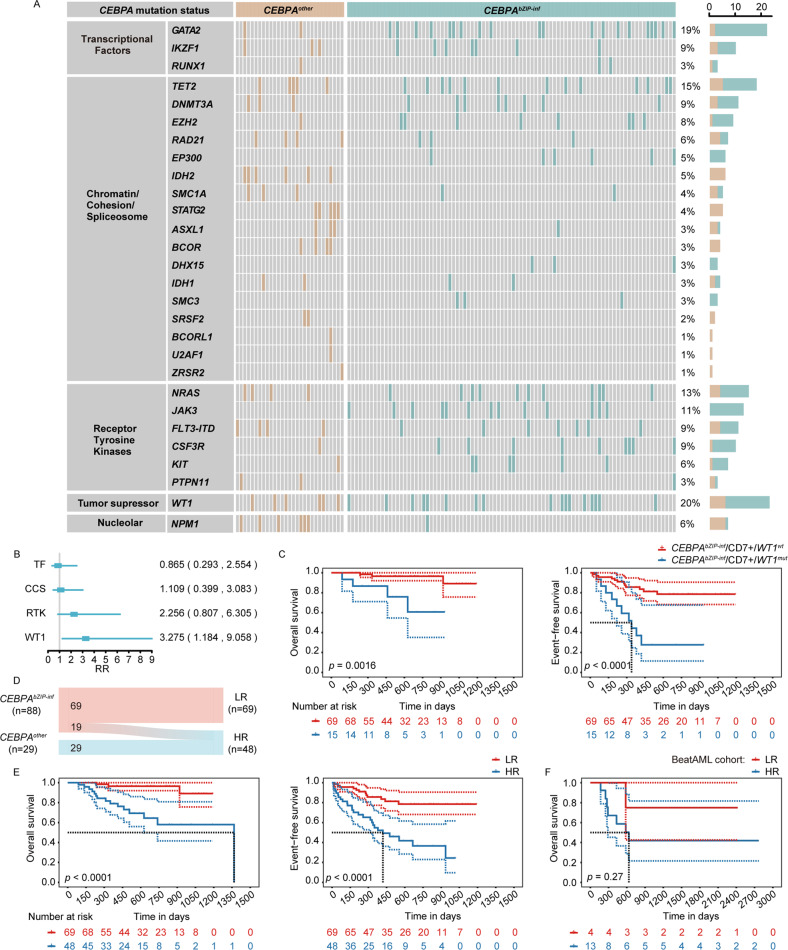
The distribution of co-mutations was illustrated in Fig. 1A. The co-mutations of the 117/124 CR patients were categorized into Transcriptional Factors (TFs, 31/117, 26.5%), Chromatin/Cohesion/Spliceosome (CCS, 54/117, 46.2%), Receptor Tyrosine Kinases (RTKs, 49/117, 41.9%), Tumor Suppressor (TS, only WT1 mutation in this group, 23/117, 19.7%) and Nucleolar (only NPM1 mutation in this group, 7/117, 6.0%). Notably, CEBPAbZIP-inf/CD7 + AML were more frequently accompanied with mutations in TFs than other CEBPAmut AML (32.1% vs. 12.1%, p = 0.027). Whereas mutations in CCS were highly enriched in other CEBPAmut AML compared to CEBPAbZIP-inf/CD7 + AML (60.6% vs. 40.5%, p = 0.049). No Nucleolar (NPM1) mutations were found in CEBPAbZIP-inf/CD7 + AML, while 21.2% of the rest CEBPAmut patients harboring NPM1 mutations (p < 0.001; Supplementary Table 3).
Fig. 1. Integrating CD7 expression and WT1 mutation status for revised risk stratification of CEBPAbZIP-inf AML patients.
A The distribution of co-mutations within the cohort of 124 CEBPAmut AML patients. Genes were categorized into groups as labeled on the left. B Cox-proportional hazard regression analysis for the categorized co-mutations independently affecting OS of 117 CR-achieved CEBPAmut AML patients. TF transcriptional factor, CCS chromatin/cohesion/spliceosome, RTK receptor tyrosine kinase, TS tumor suppressor. C Kaplan–Meier curves for the survival of 84 CR-achieved CEBPAbZIP-inf/CD7+AML patients according to WT1 mutation status. D Sankey plot for reclassification of 117 CR-achieved CEBPAmut AML patients from CEBPAbZIP-inf/CEBPAother grouping to the revised risk stratification. E Kaplan–Meier curves for the survival of 117 CR-achieved CEBPAmut AML patients according to the revised risk stratification. F Kaplan–Meier survival curves for OS of 17 CEBPAmut AML patients within the BeatAML cohort according to the revised risk stratification.
The corresponding clinical impacts of co-mutations were involved into prognosis evaluation. We categorized the co-mutations into groups (CCS, RTKs, TS, Nucleolar) to avoid the interference of low-frequency mutations as independent variables for hazard analysis. Multivariate Cox regression analysis showed TS (WT1 mutations, WT1mut) significantly affected the OS of CEBPAmut AML, with risk ratio (RR) of 3.275, p = 0.0223 (Fig. 1B). Further analysis indicated WT1mut could significantly shorten the survival of CEBPAbZIP-inf/CD7 + AML, with 2-year OS and EFS of 96.6% and 78.6% vs. 60.7% and 27.8%, respectively (p = 0.0016 and <0.0001, respectively) (Fig. 1C).
Net reclassification improvement (NRI) was then performed and indicated that the outcome was significantly improved after the integration of CD7 expression and WT1 status into the clinical nomogram, with the value of 39.2% improvement [95% CI: 0.000–1.059]. Thus, we defined CEBPAbZIP-inf AML patients characterized by immunophenotypic CD7-positive and wild-type WT1 as low-risk group (LR group: CEBPAbZIP-inf/CD7 + /WT1wt); correspondingly, 19/89 patients of CEBPAbZIP-inf AML were re-stratified into the high-risk group (HR group, Fig. 1D). With the revised stratification, patients in LR group had a superior outcome than the patients in HR group (2-year OS: 96.6% vs. 64.4%, p < 0.0001; 2-year EFS: 78.6% vs. 36.6%, p < 0.0001; Fig. 1E). We also validate the revised stratification in patients from BeatAML cohort. Improved OS was observed in LR patients (n = 4) compared with HR patients (n = 13), although the difference was not significant due to the limited sample size (n = 17, Fig. 1F).
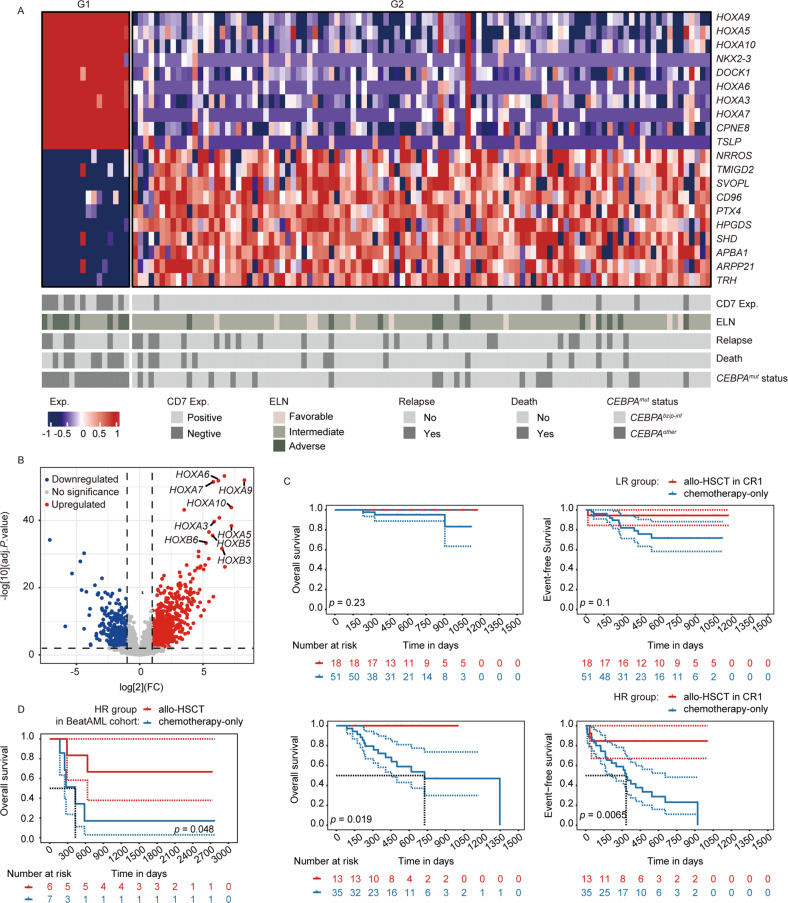
The transcriptomic data was available in 122 (data of 2 patients were missing) CEBPAmut patients from our cohort. With unsupervised cluster analysis, HOXA/B family genes were identified to be highly associated with poor prognosis in CEBPAmut AML (Fig. 2A). Besides, the differentially expressed genes (DEGs) analysis also showed that HOXA/B family genes were highly enriched in the up-regulated gene patterns of HR patients compared with LR patients (Fig. 2B); HR patients were usually accompanied with remarkably higher expression of HOXA/B family genes compared with LR patients (Supplementary Fig. 4).
Fig. 2. Differentially expressed genes and survival analysis of two distinct sub-cohorts of patients with CEBPAmut AML.
A Top ten DEGs identified as up- (red) or down- (blue) regulated were ranked by the the magnitude of expression value change. B Volcano plot showing DEGs according to the two distinct sub-cohorts clustered by unsupervised hierarchy. C Kaplan–Meier curves for the survival of LR (upper panel) and HR (lower panel) CEBPAmut AML patients according to treatment of allo-HSCT in CR1 or chemotherapy-only. D Kaplan–Meier curves for the OS of HR CEBPAmut AML patients within the BeatAML cohort according to treatment of allo-HSCT or chemotherapy-only.
In addition, survival analysis revealed LR patients may not benefit from allo-HSCT in CR1, with 2-year OS and EFS of 100% and 94.4% vs. 95.2% and 71.7% in chemotherapy-only, respectively (p = 0.23 and 0.10, respectively); whereas allo-HSCT in CR1 could significantly improve the outcome of HR patients, with 2-year OS and EFS of 100% and 84.6% vs. 53.6% and 23.1% in chemotherapy-only, respectively (p = 0.019 and 0.0065, respectively) (Fig. 2C). The therapeutic efficacy of allo-HSCT was also validated in patients from BeatAML cohort. There were 13 patients eligible for the criteria of HR CEBPAmut AML as we defined. The survival curves were different although the small sample size limited the statistical significance (p = 0.048, Fig. 2D). Therefore, not only CEBPAother AML patients, CEBPAbZIP-inf AML patients with negative CD7 expression or WT1mut may also be recommended for allo-HSCT as soon as CR achieved.
Conclusively, CD7 immunophenotype and WT1mut status is convenient for clinicians to acquire for the identification of CEBPAbZIP-inf AML patients who are in risk of disease relapse (Supplementary Fig. 5). Evidences in our cohort are provided to support the necessity of allo-HSCT in CR1 for high-risk cases, with further validation in an independent cohort from BeatAML. For the limitation of the retrospective nature in this study, relative clinical trial may be conducted in the future to validate our results and explore the therapeutic efficacy of allo-HSCT in CEBPAmut AML.
Supplementary information
Acknowledgements
The authors thank all the members of the Shanghai Institute of Hematology and National Research Center for Translational Medicine at Shanghai, and HOME for Researchers (https://www.home-for-researchers.com) for online analysis tools. This study was funded by the National Natural Science Foundation of China (82270116, 82000143), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161406) and Innovative research team of high-level local universities in Shanghai. All authors consent to publication.
Author contributions
SY, CXJ conceptualized the study; CXJ, ADY collected the original data. CXJ, ADY, ZYL, LJF, LXJ designed and performed the statistical analyses; SY, CXJ, ZYL, LJ, ZHM wrote the first draft of the manuscript. All authors participated in revised and approved the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable requests. More details are provided in Supplementary Information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xinjie Chen, Diyaer Abuduaini, Yuliang Zhang.
Contributor Information
Hongming Zhu, Email: zhm11931@rjh.com.cn.
Jianfeng Li, Email: ljf12500@rjh.com.cn.
Yang Shen, Email: yang_shen@sjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-023-00838-2.
References
- 1.Su L, Shi YY, Liu ZY, Gao SJ. Acute myeloid leukemia with CEBPA mutations: current progress and future directions. Front Oncol. 2022;12:806137. doi: 10.3389/fonc.2022.806137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Rollig C, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139:87–103. doi: 10.1182/blood.2020009680. [DOI] [PubMed] [Google Scholar]
- 3.Wakita S, Sakaguchi M, Oh I, Kako S, Toya T, Najima Y, et al. Prognostic impact of CEBPA bZIP domain mutation in acute myeloid leukemia. Blood Adv. 2022;6:238–47. doi: 10.1182/bloodadvances.2021004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77. doi: 10.1182/blood.2022016867. [DOI] [PubMed] [Google Scholar]
- 5.Schlenk RF, Taskesen E, van Norden Y, Krauter J, Ganser A, Bullinger L, et al. The value of allogeneic and autologous hematopoietic stem cell transplantation in prognostically favorable acute myeloid leukemia with double mutant CEBPA. Blood. 2013;122:1576–82. doi: 10.1182/blood-2013-05-503847. [DOI] [PubMed] [Google Scholar]
- 6.Su L, Tan Y, Lin H, Liu X, Yu L, Yang Y, et al. Mutational spectrum of acute myeloid leukemia patients with double CEBPA mutations based on next-generation sequencing and its prognostic significance. Oncotarget. 2018;9:24970–9. doi: 10.18632/oncotarget.23873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pulikkan JA, Tenen DG, Behre G. C/EBPalpha deregulation as a paradigm for leukemogenesis. Leukemia. 2017;31:2279–85. doi: 10.1038/leu.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih LY, Liang DC, Huang CF, Wu JH, Lin TL, Wang PN, et al. AML patients with CEBPalpha mutations mostly retain identical mutant patterns but frequently change in allelic distribution at relapse: a comparative analysis on paired diagnosis and relapse samples. Leukemia. 2006;20:604–9. doi: 10.1038/sj.leu.2404124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable requests. More details are provided in Supplementary Information.