Summary
Although the differentiation of human induced pluripotent stem cells (hiPSCs) into various types of blood cells has been well established, approaches for clinical-scale production of multipotent hematopoietic progenitor cells (HPCs) remain challenging. We found that hiPSCs cocultured with stromal cells as spheroids (hematopoietic spheroids [Hp-spheroids]) can grow in a stirred bioreactor and develop into yolk sac-like organoids without the addition of exogenous factors. Hp-spheroid-induced organoids recapitulated a yolk sac-characteristic cellular complement and structures as well as the functional ability to generate HPCs with lympho-myeloid potential. Moreover, sequential hemato-vascular ontogenesis could also be observed during organoid formation. We demonstrated that organoid-induced HPCs can be differentiated into erythroid cells, macrophages, and T lymphocytes with current maturation protocols. Notably, the Hp-spheroid system can be performed in an autologous and xeno-free manner, thereby improving the feasibility of bulk production of hiPSC-derived HPCs in clinical, therapeutic contexts.
Keywords: human iPSC, stromal cell, HPC, yolk sac, organoid, hematopoiesis, bulk production, xeno-free condition
Graphical abstract

Highlights
-
•
Coculturing hiPSCs with stromal cells in 3D (Hp-spheroids) induces hematopoiesis
-
•
Hp-spheroids develop into yolk sac-like organoids without exogenous factors
-
•
Organoid-derived hematopoietic progenitor cells (HPCs) are of the definitive type
-
•
Hp-spheroid system allows for scalable generation of HPCs in a xeno-free condition
Motivation
The differentiation of blood cells from human induced pluripotent stem cells (hiPSCs) involves the generation of multipotent hematopoietic progenitor cells (HPCs) in advance and then directed maturation into target blood cell types. However, a scalable and simple approach to generate hiPSC-derived HPCs in a xeno-free manner has not been established, which significantly hinders clinical application of hiPSC-based blood cell therapy. We report a production protocol that overcomes these hurdles and that allows for mass production of HPCs from hiPSCs via the induction of yolk sac-like organoids without addition of exogenous factors.
Tamaoki et al. develop a simple and scalable method to generate multipotent hematopoietic progenitor cells (HPCs) from iPSCs by coculturing iPSCs with stromal cells as spheroids. This system produces HPCs in an autologous and xeno-free manner, helping to overcome current limitations for bulk production of iPSC-derived HPCs in clinical contexts.
Introduction
Generation of blood cells from patient-specific human induced pluripotent stem cells (hiPSC) has enormous potential for the treatment of malignant and nonmalignant hematologic disorders using cell-based therapy. The differentiation of blood cells from human pluripotent stem cells (hPSCs) requires the generation of multipotent hematopoietic progenitor cells (HPCs) and then directed maturation into target blood cell types. Even though various types of blood cells could be induced from hPSC-derived HPCs,1,2,3,4,5,6 the clinical use of hPSC-based blood cell therapy faces several challenges. One of the major obstacles is that current methods to produce HPCs are not particularly scalable, yet bulk production of HPCs is required to yield sufficient numbers of target blood cells for therapy. Currently, the most popular and well-established approach is the use of an embryoid body (EB)-mediated differentiation system in which hPSCs are cultured in 3D as aggregates and differentiated by adding exogenous factors specific to each developmental stage. As EBs are cultured in 3D, it is theoretically possible to scale up production using stirred bioreactors. However, the shell structure formed on the exterior surface of EBs prevents the effective diffusion of soluble biochemicals into the EBs,7,8 thereby making it more difficult to manipulate the delicate stages of differentiation using a complicated cocktail of cytokines/growth factors in a bioreactor setting. Furthermore, preparation of various GMP (good manufacturing practice) grade factors is a practical hurdle for cost-effective bulk production. The key to overcoming this hurdle is to avoid the use of exogenous factors. HPC formation can also be induced in a simple way by coculturing with mouse stromal cells such as OP9 cells,9,10 but this approach is laborious, and the use of selected fetal bovine serum (FBS) lots is critical for successful generation of HPCs.11 Moreover, autologous and xeno-free conditions are desirable for clinical use. Therefore, an ideal culture method would combine the benefits of both approaches.
The mammalian yolk sac, a double-layered organ composed of both endodermal and mesodermal layers, is the first site of hematopoiesis and supports the developing embryo by delivering oxygen and nutrients to the embryo during early embryogenesis.12,13,14 Previous studies have shown that spontaneous differentiation of EBs grown in 3D conditions can represent some yolk sac features and induce hematopoiesis.15,16 Moreover, the differentiation of hPSCs to hematopoietic cells recapitulates the hemato-vascular development in the yolk sac.16,17 Given that hematopoiesis of hPSCs proceeds through yolk sac development, providing yolk sac-specific microenvironments and signaling in the culture may facilitate the induction of HPCs from hPSCs. Recent advancements in 3D culture techniques have enabled the formation of hPSC-derived organoids, representing the remarkable complexity of organ-specific cell types and structural features that also function similarly to their in vivo counterparts.18 Although many types of organoids, such as brain, liver, and kidney, have been generated successfully from hPSCs,19,20,21 yolk sac organogenesis has not been reported. Importantly, the generation of yolk sac organoids potentially allows for hematopoietic cell production.
In this study, we explored the development of a simple and scalable system to generate multipotent HPCs from hiPSCs. We discovered that hematopoietic spheroid (Hp-spheroid), in which hiPSCs are cocultured with stromal cells in 3D, can develop into yolk sac-like organoids and generate HPCs. Hp-spheroid-derived organoids recapitulated not only human secondary yolk sac-specific cellular components and structures but also the functional ability to induce hematopoiesis from hiPSCs. Similar to the EB-mediated differentiation, a sequential hemato-vascular developmental process was observed during organoid formation. Moreover, organoid-induced HPCs possess a broad multipotency to generate erythroid, myeloid, and T cells. Importantly, our Hp-spheroid system is scalable and can be performed in an autologous and xeno-free condition. In summary, the Hp-spheroid platform allows for reproducible and cost-effective bulk production of HPCs from patient-specific hiPSCs and represents a new avenue for clinical application of hiPSC-based blood cell therapy.
Results
The Hp-spheroid system provides a simple and scalable approach to generate definitive HPCs from hiPSCs
Given that mouse bone marrow-derived OP9 stromal cells can present a hematopoietic niche-like microenvironment to induce hematopoiesis of hPSCs without the addition of exogenous growth factors,10,22 identifying a human cell type as an alternative to OP9 cells may provide a simple and clinically relevant protocol to generate HPCs. Previous studies have shown that there are many similarities between OP9 cells and human bone marrow stromal cells (hBMSCs),23 and considering that hBMSCs contribute to the bone marrow hematopoietic niche,24 we speculated that hBMSCs may possess the potency to induce the hematopoiesis of hiPSCs as well.
We first attempted to determine whether coculturing with healthy donor-derived hBMSCs25 in a 2D condition is able to generate CD34+CD43+ cells, which contain multipotent HPCs with lympho-myeloid potential.22,26 To track hiPSC-derived cells during coculture, we used a stable EGFP (enhanced green fluorescent protein)-expressing hiPSC line (NCRM5-AAVS1-CAG-EGFP, hereinafter referred to as NCRM5-EGFP).27 Contrary to the OP9 system, hiPSCs grown on hBMSCs formed flattened colonies and failed to develop into cystic structures after 13 days of coculture (Figure S1A). Flow cytometry analysis revealed that this approach mainly generates CD34+CD43− cells but not CD34+CD43+ cells (Figure S1B). Since hPSCs form cystic structures during their differentiation to HPCs using either the OP9 system or EB-mediated differentiation,28,29 we speculated that the formation of cystic structures may be a critical process. Therefore, we sought to engineer a consistent coculture system to facilitate this cystic formation.
As 3D culture techniques can provide improved physiological growth conditions and allow hiPSCs to organize multilayered tissue-like structures compared with 2D conditions,30 we attempted to convert the coculture system from 2D to 3D (Figure 1A). To promote hiPSC differentiation in 3D, we converted hiPSCs to EBs before coculture. Small EBs (approximately 100 cells/EB) were used to ensure enhanced cell-to-cell contact between hiPSCs and hBMSCs, improving intercellular signaling and cell-extracellular matrix interactions. We discovered that EBs and hBMSCs can form uniform-sized spheroids when they are mixed in ultra-low adherent microwells (Aggrewell plate) (Figure 1B). As EBs sink faster than hBMSCs in microwells, EBs and hBMSCs did not mix homogeneously in the forming spheroids, but this approach produced a mechanically resistant spheroid with firm interactions between hiPSCs and hBMSCs (Figures 1A and 1B).
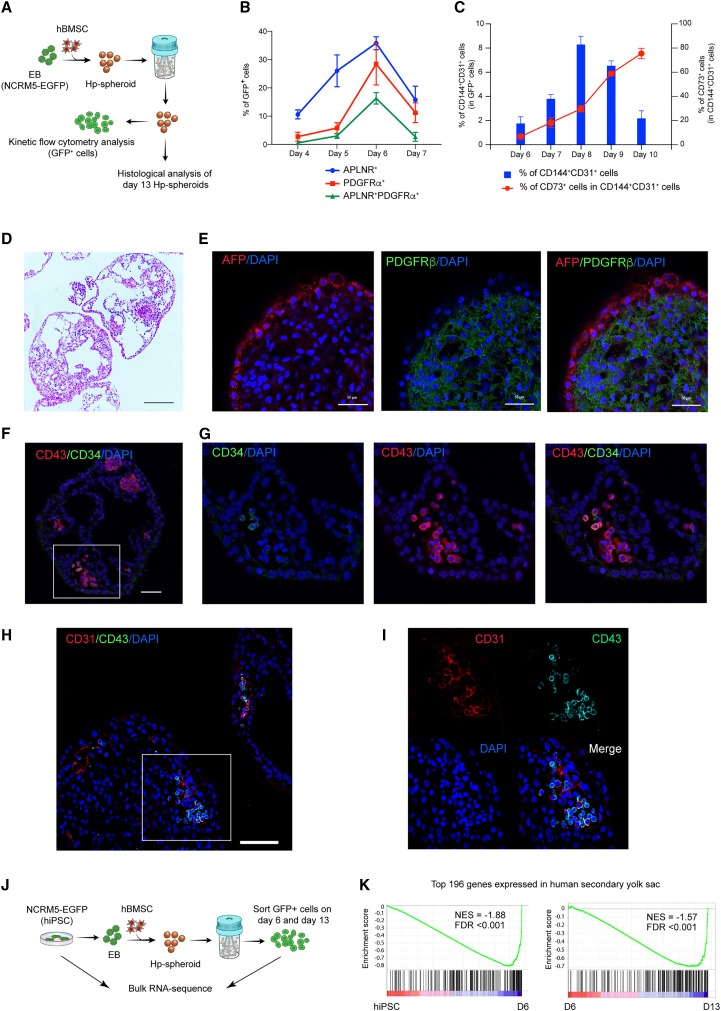
Figure 1.
Hp-spheroid system can induce hematopoiesis from hiPSCs without addition of exogenous factors
(A) Schematic of coculturing hiPSCs and hBMSCs as spheroids in 3D conditions.
(B) Representative GFP fluorescence microscopy image of Hp-spheroids on day 1. Scale bar: 300 μm.
(C) Light microscopy image of Hp-spheroids cultured in 3D culture plates (left) and in bioreactors (right) for 13 days. Scale bar: 600 μm. Red arrows indicate the outline of a single Hp-spheroid.
(D) Representative flow cytometry analysis of CD34+ and CD43+ cells in Hp-spheroids on day 13. GFP+ cells are gated for analysis.
(E) Yield comparison of 3D plate vs. bioreactor. Left: number of GFP+ cells in day 13 Hp-spheroids. Graph shows results when 1 × 106 hiPSCs were used for differentiation. Right: frequencies of CD34+CD43+ cells in GFP+ cells in day 13 Hp-spheroids. Values represent mean ± SD from 4 independent experiments, ∗∗∗∗p < 0.0001.
(F) Number (left) and the percentage (right) of GFP+ cells (hiPSC-derived cells) and GFP− cells (hBMSCs) in Hp-spheroids within 13 days of culture. Values represent mean ± SD from 3 independent experiments. Representative flow cytometry analysis is shown in Figure S3A.
(G) Fluorescence microscopy image of Hp-spheroids on day 7. Scale bar: 300 μm.
(H) NCRM5-EGFP cells were cocultured with hBMSCs or iSTCs (isolated after 8, 14, or 20 days of differentiation) to form Hp-spheroids. Spheroids were cultured in bioreactors for 13 days, and the percentage of CD34+CD43+ cells in GFP+ cells isolated from each type of Hp-spheroids was analyzed by flow cytometry. Values represent mean ± SD from 3 independent experiments; ns, not significant.
(I) Number of GFP+ cells (left) and the percentage of APLNR+ cells in GFP+ cells (right) detected in EBs cultured in conditioned medium or in Hp-spheroids on day 6. The same number of EBs was used for each condition. Horizontal bars represent mean value from 3 independent experiments, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. iSTC, hiPSC-derived stromal cell; CM, conditioned medium.
The cocultured spheroids were transferred from Aggrewell plates to ultra-low attachment 3D culture plates (Figure 1A) and were cultured in αMEM medium containing 20% FBS for 13 days. Notably, the spheroids developed into cystic structures (Figure 1C) containing GFP+CD34+CD43+ cells on day 13 (Figures S1C and S1D). Importantly, this process occurred in the absence of exogenous factors, which are required for EB-mediated differentiation (Figures S1C and S1D). Hereinafter, we refer to this cocultured spheroid, which can induce CD34+CD43+ cells, as Hp-spheroids. Similar to hBMSCs, we confirmed that OP9 cells can also be used to form Hp-spheroids and induce hematopoiesis, as they do in the conventional 2D culture system (Figures S1E–S1G). Our Hp-spheroid system provides a simple exogenous factor-free system to induce CD34+CD43+ cells from hiPSCs.
Given that one of the goals in this study was to establish a scalable system to induce HPCs, we sought to explore whether this system is scalable (Figure 1A). As the Hp-spheroid showed a strong resilience to mechanical stress, we were able to culture them in stirred suspension bioreactors after optimizing the rotation speed. Hp-spheroids grown in the bioreactors were homogeneous spheres with internalized cystic structures (Figure 1C). Remarkably, based on the size of the spheroids and the number of GFP+ cells, the use of bioreactors enhanced the growth of hiPSC and the fidelity of HPC induction (Figures 1C–1E). A kinetic analysis of CD34+CD43+ cells by flow cytometry showed that HPCs started to appear by day 9 and peaked on day 13–14 of culturing (Figure S2A).
Since both primitive and definitive types of hematopoiesis can be induced from hiPSCs, we sought to clarify the hematopoietic potential of HPCs generated by this system. The hematopoietic colony-forming unit (CFU) assay demonstrated that GFP+CD34+CD43+ cells sorted on day 13 could be induced into erythroid and myeloid lineage cells (Figure S2B). Furthermore, T cell differentiation potential was evaluated by coculturing on OP9/DLL1 in the presence of cytokines (stem cell factor [SCF], Flt3-L, and, interleukin-7 [IL-7]).4 CD5+CD7+ T cell progenitors were detected by day 17, and CD4+CD8+ double-positive T cells were detected by day 27 of T cell differentiation (Figure S2C). These data indicated that our Hp-spheroid system can induce definitive-type HPCs from hiPSCs.
To determine whether multipotent hematopoietic cells generated in Hp-spheroids contain engraftable hematopoietic stem cells (HSCs), we introduced NCRM5-EGFP-derived CD34+ cells into immunocompromised mice. In contrast to human adult mobilized CD34+ cells, we could not detect GFP+/human CD45+ cells at 3 months post-transfer (Figure S2D), suggesting that Hp-spheroid-derived HPCs were less differentiated than adult HSCs. Although our Hp-spheroid system cannot induce engraftable HSC-like cells, this system could provide a simple and scalable approach to generate definitive HPCs from hiPSCs in vitro.
hiPSC-derived stromal cells can be used for the Hp-spheroid system
Despite using similar numbers of NCRM5-EGFP cells and hBMSCs to establish Hp-spheroids, the number of GFP+ cells (hiPSCs) in day 1 Hp-spheroids was much lower than that of GFP− cells (stromal cells) due to the loss of hiPSCs during EB and spheroid formation. However, we found almost all cells in day 13 Hp-spheroids to be GFP+ cells (Figure S3A). To determine how long hBMSCs contributed to the hiPSC differentiation in Hp-spheroids, we tracked both GFP+ and GFP− cell populations through day 13 of coculture. Remarkably, hBMSCs decreased dramatically through day 4 of coculture, whereas GFP-labeled hiPSC-derived cells continued to grow to eventually form the entirety of the cystic structures by day 7 (Figures 1F, 1G, and S3A). The same result was obtained when OP9 cells were used (data not shown). Our observation suggests that hBMSCs and OP9 cells are not required for the entire process of hiPSC differentiation in the Hp-spheroid system but appear to play an important role during the early stage.
Using patients’ specific hBMSCs, our Hp-spheroid system can be performed in an autologous condition. However, the in vitro expansion of hBMSCs induces cell senescence and has limited scalability.31 Currently, generation of hiPSC-derived stromal cells (iSTCs) is well established.32,33 Given that hBMSCs do not directly contribute to hematopoiesis of hiPSCs, it would be beneficial to test whether iSTCs can be used in the Hp-spheroid system because they are easily accessible and have a great scalability to build a large-scale Hp-spheroid system in an autologous setting.
To test this, we generated iSTCs from NCRM5 cells (GFP− cells) according to our previous report33 (Figure S3B). Flow cytometry analysis demonstrated that iSTCs increased the expression of typical stromal cell markers such as CD44, CD105, and CD73 during differentiation. However, by day 20 of differentiation, the expression levels of CD105 and CD73 were much lower than CD44 (Figure S3C). Based on the expression pattern of cell surface markers, iSTCs at day 20 were still in a relatively immature state. Nevertheless, all types of iSTCs isolated from different time points (days 8, 14, and 20) could support the Hp-spheroid system with similar proficiency to using hBMSCs (Figures 1H and S3D). Our data indicated that various types of stromal cells can be used for the Hp-spheroid system. To exclude the possibility that this modified Hp-spheroid system using iSTCs (referred to as iSTC-Hp-spheroid) only worked for NCRM5-EGFP cells, we also tested another hiPSC line: a sickle cell disease-derived iPSC line (SCD-iPSCs).34 Similarly, iSTCs generated from SCD-iPSCs could contribute to the Hp-spheroid formation and support generation of HPCs with T lymphocyte potential (Figure S3E).
Next, we set out to determine whether early hiPSC differentiation is regulated by morphogens secreted from stromal cells or cell-cell contacts between hiPSCs and stromal cells. To this end, we tested if EBs cultured in conditioned medium from stromal cells can induce APLNR+ (apelin receptor) cells, which have been described as mesodermal progenitors with hemato-vascular potential.35 Notably, EBs cultured in contact with stromal cells as Hp-spheroids grew and induced APLNR+ cells, whereas EBs cultured in conditioned medium from OP9 cells or iSTCs for 6 days could not (Figure 1I). Our data suggested that direct contact signaling from stromal cells is critical for hiPSCs to grow in this 3D culture condition and is an important trigger to initiate their differentiation.
hiPSCs in the Hp-spheroids recapitulate the dynamics of hemato-vascular developmental processes and form yolk sac-like structures
Several studies have shown that the differentiation of hPSCs to hematopoietic cells recapitulates the embryonic hemato-vascular development in the yolk sac.17,36 We sought to understand whether the dynamics of hemato-vascular ontogenesis is also recapitulated in Hp-spheroids. To this end, Hp-spheroids were formed using NCRM5-EGFP cells and hBMSCs, and kinetic flow cytometry analyses were performed to observe sequential differentiation stages (Figure 2A). Embryonic hemato-vascular formation begins with the induction of multipotent mesodermal progenitor cells with hematoendothelial potential.36,37 These specific mesodermal progenitors can be detected by coexpression of APNLR+ platelet-derived growth factor receptor α (PDGFRα)+ during hPSC differentiation.35,38 In Hp-spheroids, GFP+APNLR+ PDGFRα+ cells were detected from day 5, then peaked on day 6 and declined toward day 7 (Figures 2B and S4A). The next developmental stage is the commitment of mesodermal progenitors to become hemogenic endothelium (HE) cells, which are common precursors of endothelial and hematopoietic cells, subsequently followed by endothelial-to-hematopoietic transition (EHT) to branch into both types of cells.36,38,39 To document this process, we analyzed various endothelial cell markers, CD144 (VE-cadherin), CD31, and CD73. When mesoderm progenitors become committed to bipotential HE cells, they start to express pan-endothelial markers CD144 and CD31 and then differentiate into mature endothelial cells (CD144+CD31+CD73+) and hematopoietic cells (CD144−CD73−) after the EHT process.38,39 Kinetic flow cytometry analysis revealed that GFP+ cells increased CD144 and CD31 expression from day 6 and peaked on day 8, followed by a decline toward day 10 (Figures 2C and S4B). Most of these CD144+CD31+ cells were CD73− until day 8 and then became CD73+, while CD144 expression decreased. Our observations suggested that the formation of HE cells peaked on day 8, and cells committed to the hematopoietic lineage then subsequently lost CD144 expression, while maturing endothelial cells acquired CD73 expression (Figures 2C and S4B). This was consistent with a dramatic increase of CD43+ hematopoietic cells from day 9 to 12 (Figure S2A). In summary, hiPSC differentiation in the Hp-spheroid system recapitulates embryonic hemato-vascular development.
Figure 2.
The Hp-spheroid system induces hiPSCs to self-organize into yolk sac-like organoids
(A) NCRM5-EGFP cells and hBMSCs were used for Hp-spheroid formation. The differentiation of GFP+ cells in Hp-spheroids was monitored by kinetic flow cytometry analyses. Histological analyses were performed on day 13 Hp-spheroids.
(B) Kinetic flow cytometry analysis of APLNR and PDGFRα expression on GFP+ cells in Hp-spheroids from days 4 through 7. Values represent mean ± SD from 3 independent experiments. A representative flow cytometry analysis is shown in Figure S4A.
(C) Kinetic flow cytometry analysis of CD144, CD31, and CD73 expression on GFP+ cells in Hp-spheroids from days 6 to 10. The percentages of CD144+CD31+ cells in GFP+ cells are shown by blue bars, and the percentages of CD73+ cells in GFP+CD144+CD31+ are shown by red dots. Values represent mean ± SD from 3 independent experiments. A representative flow cytometry analysis is shown in Figure S4B.
(D) Hematoxylin and eosin staining of day 13 Hp-spheroids. Scale bar: 200 μm.
(E) Whole-mount immunostaining analysis of AFP and PDGFRβ in a day 13 Hp-spheroid. Scale bar: 50 μm.
(F) Immunostaining of a day 13 Hp-spheroid for CD34 and CD43. Scale bar: 50 μm.
(G) Higher magnification of the boxed area in (F).
(H) Immunostaining of Hp-spheroids on day 13 for CD31 and CD43. Scale bar: 100 μm.
(I) Higher magnification of the boxed area in (H).
(J) Schematic outline of bulk RNA sequencing analysis.
(K) Gene set enrichment analysis for a human yolk sac gene set (top 196 yolk sac genes from Cindrova-Davies et al.12). NES, normalized enrichment score; FDR, false discovery rate.
We next explored whether hiPSCs patterned and formed yolk sac-like structures to induce hematopoiesis (Figure 2A). Hematoxylin and eosin staining suggested that day 13 Hp-spheroids are composed of an outer layer of cells encompassing multiple inner cystic structures (Figure 2D). Whole-mount immunostaining and confocal imaging revealed that the outer layer of the day 13 Hp-spheroid expressed AFP (alpha fetoprotein), whereas the PDGFRβ+ cells were located inside, indicating that an endodermal layer (AFP) and a mesodermal region (PDGFRβ) are separately organized in the day 13 Hp-spheroid (Figure 2E). To visualize the distribution of hematopoietic cells, we performed immunostaining for CD34 and CD43. Similar to the anatomical feature of hematopoietic cells in the blood island of the yolk sac, CD43+ hematopoietic cells were detected as cell clusters within the mesodermal region close to the endoderm layer with a subset of CD34+CD43+ putative HPCs (Figures 2F and 2G). Immunostaining for CD31 and CD43 revealed that CD31 single-positive endothelial cells were found to be located very close to CD31+CD43+ or CD31−CD43+ hematopoietic cells (Figures 2H and 2I), implying that the simultaneous emergence of hematopoietic and endothelial cells within blood islands is similarly observed in day 13 Hp-spheroids. These CD31+ endothelial cells, however, did not form a developed vascular plexus in day 13 Hp-spheroids. Our observations suggest that hematopoietic cells are generated through blood island formation but not directly from blood vessels as seen in the AGM (aorta-gonad-mesonephros) region.
To ascertain global gene expression signatures of hiPSCs during differentiation in the Hp-spheroid system, we performed bulk RNA sequencing of NCRM5-EGFP cells (hiPSCs) and GFP+ cells isolated from Hp-spheroids on days 6 and 13 (Figure 2J). Gene set enrichment analysis (GSEA) using a human secondary yolk sac gene set comprising the top 196 differentially expressed genes12 indicated that the gene expression signature of hiPSCs in Hp-spheroids became more similar to that of human secondary yolk sac as development progressed (Figure 2K; Table S1). Taken together, day 13 Hp-spheroids possessed functional and morphological features of the human secondary yolk sac, indicating that hiPSCs developed into yolk sac-like organoids using our Hp-spheroid system.
Single-cell RNA sequencing analysis reveals a day 6 Hp-spheroid cell state composition similar to that of the early human secondary yolk sac
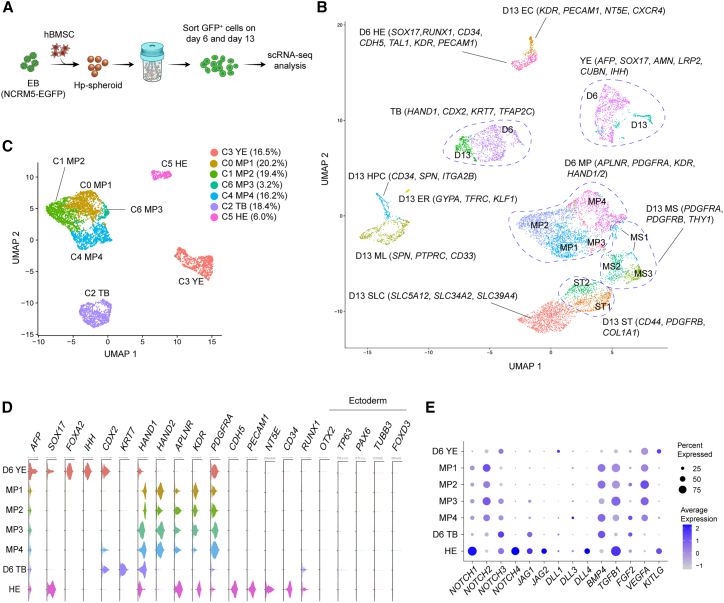
Since the yolk sac mesoderm dramatically changes its cell composition during hemato-vascular development, we sought to describe the cell complement of day 6 and 13 Hp-spheroids by single-cell RNA sequencing (scRNA-seq) analysis. We harvested GFP+ cells on days 6 and 13 for scRNA-seq analysis (Figure 3A). This yielded 4,895 day 6 cells and 4,401 day 13 cells with median counts of 5,292 genes and 30,096 transcripts (unique molecular identifiers) per cell after filtering (Figure S5A). We clustered cells from both days separately and in combination, identified biomarkers, and annotated the obtained clusters by using expression of previously described marker genes (Figures 3B, S5B, S5C, S5E, and S6A–S6C; Tables S2 and S3).12,38,40,41,42,43,44,45
Figure 3.
Single-cell RNA sequencing (RNA-seq) analysis reveals similarities between the day 6 Hp-spheroid cell complement and the early human secondary yolk sac
(A) Schematic outline of single-cell RNA-seq (scRNA-seq) analysis of GFP+ cells isolated from day 6 and 13 Hp-spheroids.
(B) UMAP embedding of 9,296 GFP+ cells isolated from both day 6 and 13 Hp-spheroids. Cluster annotations obtained when clustering day 6 and 13 cells separately are visualized.
(C) UMAP embedding for 4895 GFP+ cells isolated from day 6 Hp-spheroids.
(D) Violin plots showing the expression of representative lineage marker genes for day 6 clusters.
(E) Dot plot representing the expression of Notch signaling genes and key morphogens required for hemato-vascular development in the yolk sac in day 6 clusters. Circles are coded by color (average gene expression level) and size (proportion of cells in clusters expressing a gene). YE, yolk sac endoderm cell; MP, mesoderm progenitor cell; TP, trophoblast-like cell; HE, hemogenic endothelium cell; MS, early mesenchymal cell; ST, stromal cell; SLC, SLC transporter genes expressing stromal cells; EC, endothelial cell; HPC, hematopoietic progenitor cell; ML, myeloid cell; ER, erythroid cell.
The human secondary yolk sac is composed of mesodermal and endodermal cells. Consistent with this, we detected mesodermal and endodermal gene expression signatures in day 6 Hp-spheroids (Figures 3C and 3D). We found that most hiPSC-derived cells on day 6 lacked expression of canonical ectoderm marker genes, such as OTX2, TP63, PAX6, TUBB3, and FOXD3 (Figure 3D). For day 6 cells, 4 distinct populations were evident in the uniform manifold approximation and projection (UMAP) (Figure 3C). Cells within a single cluster showed elevated expression of both endodermal markers (AFP, SOX17, and FOXA2) as well as genes known to be expressed in the human secondary yolk sac (AMN, LRP2, CUBN, TTR, and IHH) (Figures 3D and S5C),12 and this cluster was annotated as yolk sac endoderm (cluster YE). The main functions of the human yolk sac endoderm are absorption of nutrients from the exocoelomic cavity and transport to the developing embryo.14,46,47 Endocytic uptake of nutrients is mediated by LRP2-CUBN-AMN endovesicular complexes.48 Moreover, genes required for transport of nutrients (TTR, SLC39A5, and SLC39A14) and various apolipoprotein genes (APOA1, APOA2, APOA4, APOB, APOE, and APOM) were highly expressed in YE cluster (Figures S5C and S5D).12,13 Mouse yolk sac endoderm secretes soluble factors, such as IHH (Indian Hedgehog) and VEGF (vascular endothelial growth factor), to regulate the differentiation of mesoderm progenitor cells.49,50 Similarly, high expression of IHH and VEGFA was detected in cells of cluster YE (Figures 3D, 3E, and S5C). These expression data demonstrated that endodermal cells formed in day 6 Hp-spheroids showed expression of several genes identified as essential in murine and human yolk sac endoderm.
The most dominant population in day 6 cells comprised mesoderm progenitor cells (cluster MP; KDR, APLNR, and PDGFRA) (Figures 3D and S6A). In accordance with previous reports, MP cluster also enriched lateral plate/extraembryonic mesoderm genes (HAND1/2 and FOXF1) (Figures 3D and S6A).45 Remarkably, a subset of mesoderm progenitor cells appeared to be committed to HE cell fate, as indicated by the expression of endothelial lineage genes (cluster HE; CDH5, CD34, KDR, and PECAM1) (Figures 3D and S6C). Although our flow cytometry analysis demonstrated that most CD144+ cells on day 6 were CD73− (Figure 2C), cells in cluster HE were already found to express NT5E (CD73) (Figures 3D and S6C), suggesting that HE cells were primed to undergo the EHT process. Additionally, HE cells also expressed SOX17 and SOX7, which are key genes for the transition to hematopoietic cells42,51 (Figures 3D and S6C). Notch signaling plays a pivotal role in regulating the EHT process and induction of definitive hematopoiesis.44,52,53 Consistent with this, both Notch ligands (JAG1, JAG2, and DLL4) and receptors (NOTCH1, NOTCH3, and NOTCH4) were highly expressed in cluster HE (Figure 3E), suggesting that auto-activation of Notch signaling promotes the transition process and a role in the induction of definitive hematopoiesis.
The hemato-vascular specification was precisely controlled in growing Hp-spheroids despite the early loss of hBMSCs. We hypothesized that the critical morphogens required for this stage may be provided from hiPSC-derived cells. Genes encoding morphogens known to be essential for hemato-vascular development, such as BMP4 (BMP4), FGF2 (FGF2), VEGF (VEGFA), SCF (KITLG), and transforming growth factor β-1 (TGF-β-1; TGFB1), were highly expressed in hiPSC-derived cells54,55 (Figure 3E). Our observations raised the possibility that these important morphogens, and the signaling required for early embryonic development in the yolk sac, can be provided by hiPSC-derived cells reciprocally.
In addition to mesoderm progenitors and their derivatives, we identified a unique cell type within mesodermal lineage cells in day 6 Hp-spheroids. Cells in this cluster expressed early trophoblast-associated genes, such as HAND1, CDX2, KRT7, TFAP2A/2C, GATA3, CDH1, and CXCL12, and were annotated as trophoblast-like cells (cluster TB)43,56,57,58 (Figures 3D and S5E). Previous studies have shown that BMP4 can induce a trophoblast phenotype from hPSCs.40,59 Most mesoderm lineage cells show high expression of BMP4, raising the possibility that hiPSCs may be induced into trophoblast-like cells by exposure to BMP4 generated in Hp-spheroids. Indeed, we detected many genes that have been found to be upregulated in differentiated hPSCs with a short-term BMP4 treatment in the day 6 TB cluster (Table S2, yellow highlighted genes).43
scRNA-seq analysis of day 6 Hp-spheroids revealed a cell complement reminiscent of the cellular components of developing human secondary yolk sac. Gene expression profiling revealed that signaling pathways required for early hemato-vascular development were recapitulated in the Hp-spheroids.
Various hematopoietic cell subsets are detected in day 13 Hp-spheroids
Unsupervised clustering of day 13 cells revealed 12 transcriptionally distinct clusters (Figure 4A). Similar to day 6 Hp-spheroids, we detected yolk sac endoderm cells (cluster YE) and trophoblast-like cells (cluster TB). Approximately 70% of day 13 cells were stromal-like cells (CD44, PDGFRB, and COL1A1). Remarkably, these stromal-like cells expressed HAND1/2 and FOXF1, suggesting that they are derivatives of mesoderm progenitors in an early mesoderm stage (Figures 4B and S6A). Stromal-like cells were classified into three different groups: early mesenchymal cell (cluster MS; PDGFRA, PDGFRB, and THY1), stromal cell (cluster ST; CD44, PDGFRB, COL1A1, and DCN), and solute carrier (SLC) transporter genes expressing stromal cells (cluster SLC) (Figures 4B and S6B). SLCs are transmembrane transporters that are widely expressed in the human yolk sac to import various nutrients from the exocoelomic cavity.12 Notably, various SLC family genes were enriched in the top 10 of the SLC cluster specific genes (Figure S6B; Table S3).
Figure 4.
Various types of hematopoietic cells are present in the day 13 Hp-spheroid
(A) UMAP embedding for 4,401 GFP+ cells isolated from day 13 Hp-spheroids.
(B) Violin plots showing expression of representative lineage marker genes in day 13 clusters.
(C) Comparison of the gene expression levels of TAL1 and CXCR4 between day 6 HE cells and day 13 EC cells, ∗∗∗∗p < 0.0001.
(D) UMAP representation for a sub-cluster analysis of hematopoietic lineage cells present in day 13 Hp-spheroids.
(E) Coexpression of CD34 and SPN within hematopoietic sub-clusters. An expression cutoff of 2.5 is used for visualization.
(F) UMAP representation of hematopoietic sub-clusters colored by gene expression of various hematopoietic lineage genes. YE, yolk sac endoderm cell; TP, trophoblast-like cell; MS, early mesenchymal cell; ST, stromal cell; SLC, SLC transporter genes expressing stromal cell; EC, endothelial cell; HPC, hematopoietic progenitor cell; ML, myeloid cell; ER, erythroid cell; HE, hemogenic endothelium cell.
Three SPN (CD43)-expressing hematopoietic clusters and an endothelial cell cluster (cluster EC; KDR, PECAM1, CD34, and NT5E) were identified in day 13 Hp-spheroids (Figures 4B and S6C). Day 13 endothelial cells showed lower expression of TAL1 and higher CXCR4 expression when compared with HE cells from day 6 (Figure 4C), consistent with previous findings for this lineage.38,39 Hematopoietic lineage cells were tentatively classified into multipotent HPC (cluster HPC; expressing both CD34 and SPN), myeloid cell (cluster ML; SPN, CD33, and PTPRC), and erythroid cell (cluster ER; GYPA and TFRC) (Figure 4B). In EB-mediated differentiation, hematopoietic specification from HE cells requires various growth factors and cytokines, such as VEGF, fms-related tyrosine kinase 3 (Flt3), thrombopoietin (TPO), erythropoietin (EPO) and IL6. Interestingly, only expression of VEGFA was detected in day 13 cells (Figure S7A), suggesting that these factors are not required for HPC generation in Hp-spheroids. It has been reported that expression of HOXA family genes, such as HOXA7 and HOXA9, segregates yolk sac-like and AGM-like hematopoiesis.60,61 Consistent with this, Hp-spheroid-derived HPCs lack reconstitution ability in immunocompromised mice, and these genes were also not expressed in cells of the HPC cluster (Figure S6C).
In the murine yolk sac, the first definitive hematopoietic cells emerge as erythro-myeloid progenitor (EMP) cells, then followed by HPCs with lymphoid-myeloid potential.62,63 These two types of definitive HPCs can also be observed during hPSC differentiation. Generally, hPSC-derived EMP cells can be detected by coexpression of CD41a (ITGA2B) and CD235a (GYPA), while multipotent HPCs do not express these lineage markers.22,36 Even though myeloid and erythroid cell states were present in day 13 Hp-spheroids, cells with EMP-like transcriptional signatures were absent, and high expression of ITGA2B was detected solely in the HPC cluster, while GYPA expression was observed only in the erythroid cluster (Figure 4B). This was consistent with a kinetic flow cytometry analysis for CD41a and CD235a expression (Figure S7B). Since hPSC-derived multipotent hematopoietic cells have been reported as a heterogeneous population,64 we sought to explore the heterogeneity within the cells of the three hematopoietic clusters.
We performed a sub-cluster analysis, which recovered seven hematopoietic subsets (Figure 4D; Table S4). Coexpression of CD34 and SPN was mainly found in sub-cluster 2 (Figure 4E), and cells in this cluster also expressed HSC-specific genes, such as SPINK2, KIT, and SOX4 (Figures 4F and S7C).64,65 Previous reports demonstrated that early hematopoietic cells derived from HE cells express CD41a,66,67 and ITGA2B expression could be detected in cells of sub-clusters 2 and 3, with stronger expression in sub-cluster 3 (Figure 4F). Consistent with this, early hematopoiesis-specific genes, such as GATA2, KIT, MEIS1, and ANGPT1, were found expressed in sub-cluster 3 (Figures 4F and S7C).65,68,69,70,71 Interestingly, megakaryocytic lineage-specific genes, such ESAM, GP9, and ITGB3, were also expressed in cells of this cluster (Figure S7C). However, GYPA as well as other erythroid-specific genes were barely detected. Notably, an early T lymphoid lineage gene CD7 was detected in sub-clusters 2 and 3, suggesting that these less differentiated hematopoietic cells possess lymphoid potential (Figure S7C). Myeloid lineage genes, such as CD33, S100A9, and MNDA, were expressed in sub-clusters 0, 1, 4, and 6 (Figures 4F and S7C), suggesting myeloid lineage cell states. Sub-cluster 5 was positive for erythroid cell-associated genes including GYPA, KLF1, and hemoglobin genes HBZ, HBA1, HBE1, and HBG (Figures 4F and S7C). The gene expression patterns of hemoglobin implied that erythroid cells in day 13 Hp-spheroids were primitive or the fetal type since HBB expression was sparse (Figures 4F and S7C). Collectively, gene expression data indicated that day 13 Hp-spheroids contain myeloid and erythroid cells as well as HPCs in different states. In the mice yolk sac, a variety of hematopoietic cells can be identified during the transition from primitive to definitive hematopoiesis.72 Day 13 Hp-spheroids seems to recapitulate this developmental stage.
HPCs generated by a modified Hp-spheroid system using iSTCs give rise to various blood cell lineages
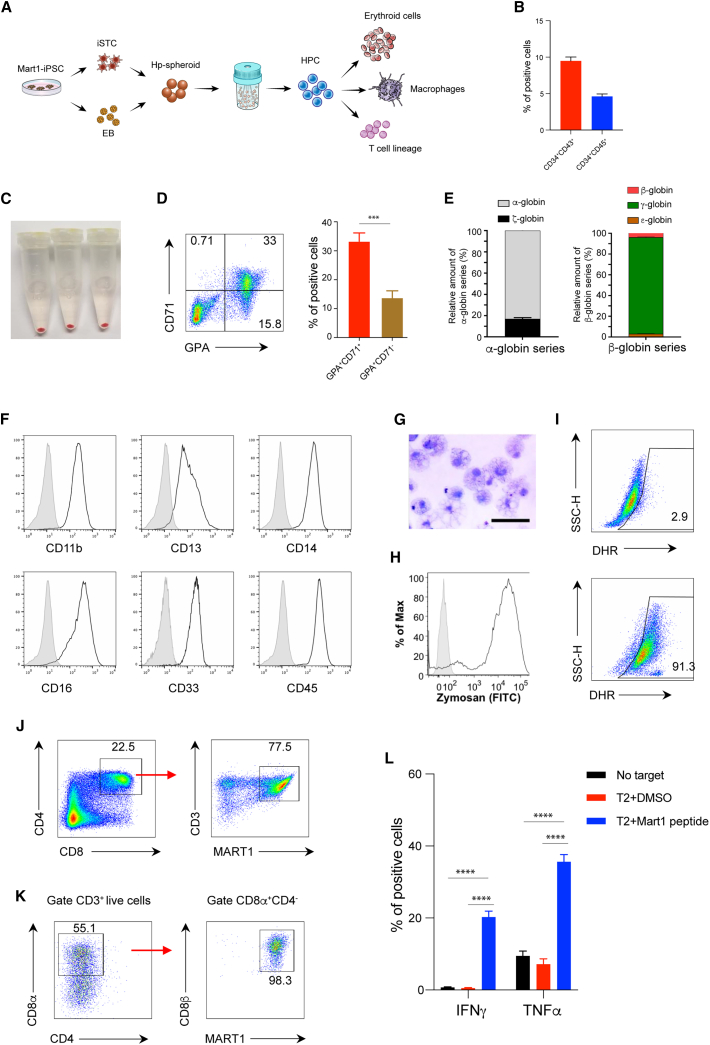
To further evaluate the potential of Hp-spheroid system for clinical application of iPSC-based blood cell therapy, we sought to determine whether HPCs produced by the iSTC-Hp-spheroid system could be differentiated to erythroid cells, macrophages, and T lymphocytes by current maturation protocols. To this end, we used a patient-specific hiPSC line, Mart1-iPSC, that was generated from a cytotoxic T cell possessing a T cell receptor (TCR) specific for the melanoma epitope Mart1.4 Mart1-iPSC-derived HPCs were harvested on day 13 and subjected to further differentiation toward these targeted blood cell lineages (Figure 5A). Similar to NCRM5-EGFP cells, Mart1-iPSC cells were differentiated into HPCs in the iSTC-Hp-spheroids (Figure 5B). For erythroid and macrophage differentiation, CD34+ cells were isolated with magnetic-activated cell sorting (MACS). After 15 days of erythroid differentiation,6,73 the cell pellets turned red (Figure 5C), and flow cytometry analysis of CD71 and glycophorin A (GPA) revealed that these cells contained CD71+GPA+ cells (erythroblasts) and CD71−GPA+ cells (mature erythroid cells) (Figure 5D). Globin gene expression patterns in Mart1-iPSC-derived erythroid cells were determined by quantitative RT-PCR. We found that the dominant globin expression pattern in Mart1-iPSC-derived erythroid cells included α-globin and γ-globin, indicating that most of them were fetal-type erythroid cells, not the primitive type (Figure 5E). Similarly, after culturing under macrophage differentiation conditions for 14 days,74 Mart1-iPSC-derived CD34+ cells generated cells with typical human macrophage cell markers (Figure 5F) and morphology (Figure 5G). Additionally, normal phagocyte activities including phagocytosis of zymosan particles and production of reactive oxygen species were observed in Mart1-iPSC-derived macrophages upon stimulation with phorbol 12-myristate 13-acetate (PMA) (Figures 5H and 5I). Finally, we confirmed that Mart1-iPSC-derived HPCs cocultured with OP9-DLL1 cells for 21 days differentiated into mature CD4+CD8+ double-positive T cells, with a high population of CD3+ T cells recognizing the Mart1-tetramer (Figure 5J). As reported in previous studies, antibody-driven TCR stimulation of these CD4+CD8+ double-positive T cells induced CD8+ single-positive T cells (Figure 5K).5 Function and antigen specificity of these Mart1-iPSC-derived CD8+ single-positive T cells were confirmed by cytokine release after coculture with Mart1 peptide-pulsed T2 cells4,5 (Figure 5L). Collectively, our data demonstrate that HPCs generated by our iSTC-Hp-spheroid system are able to successfully produce erythroid, myeloid, and T lymphoid lineage cells.
Figure 5.
HPCs generated in iSTC-Hp-spheroids can undergo maturation to erythroid cells, macrophages, or T lymphocytes using established cell maturation protocols
(A) Schematic outline illustrating the use of Mart1-iPSC cells for iSTC-Hp-spheroid formation and subsequent erythroid cell, macrophage, and T lymphocyte differentiation.
(B) Frequencies of CD34+CD43+ and CD34+CD45+ cells in Mart1-iPSC-derived iSTC-Hp-spheroids on day 13. Values represent mean ± SD from 3 independent experiments.
(C–E) Mart1-iPSC-derived HPCs cultured in the erythroid differentiation condition for 15 days were harvested to characterize erythroid cell properties. The color of cell pellets changed to red (C), and typical erythroid lineage markers, such as CD71 and GPA, could be detected by flow cytometry analysis (D). Globin expression patterns were determined by relative RNA expression of α- and ζ-globin (α-globin series) and ε-, γ-, and β-globin (β-globin series) (E). Values represent mean ± SD from 3 independent experiments.
(F–I) Macrophage differentiation from Mart1-iPSC-derived CD34+ cells.
(F) Macrophage surface markers were analyzed by flow cytometry (gray; isotype control).
(G) Giemsa stain of macrophage cytospin. Scale bar: 50 μm.
(H) Phagocytosis assay of fluorescein isothiocyanate (FITC)-labeled zymosan A particles (gray; isotype control).
(I) DHR assay of reactive oxygen species production by Mart1-iPSC-derived macrophages in response to stimulation with PMA (bottom panel); unstimulated macrophage control is also shown (top panel).
(J–L) T lymphocytes generated from Mart1-iPSC-derived HPCs.
(J) After 22 days of T cell differentiation, CD4+CD8+ double-positive T cells could be detected by flow cytometry analysis, most of which were CD3+Mart1-tetramer+ cells.
(K) Mart1-iPSC-derived CD4+CD8+ double-positive T cells could be induced to CD8αβ single-positive T cells by stimulation with Mart1 peptide-primed T2 cells.
(L) Mart1-iPSC-derived CD8+ single-positive T cells released interferon γ (INFγ) and tumor necrosis factor α (TNF-α) in response to Mart1 peptide-pulsed T2 cells. T cell responses without T2+Mart1 peptide (no target) or without Mart1 peptide (T2+DMSO) are also shown. Values represent mean ± SD from 3 independent experiments, ∗∗∗∗p < 0.0001.
The iSTC-Hp-spheroid system can be performed in a xeno-free condition
For the therapeutic use of iPSC-based cell therapies, production of cells under xeno-free conditions is desirable due to biosafety issues. Consequently, we attempted to develop a xeno-free medium for our iSTC-Hp-spheroid system (Figure 6A). To this end, we used αMEM as a basal medium and screened various commercial FBS replacements. We found that αMEM containing 2.5% PLTGold (heparin-free human platelet lysate) and StemFit C02 (an FBS replacement kindly provided by Ajinomoto) supported the iSTC-Hp-spheroids in generating CD34+CD43+ cells in bioreactors similar to using FBS (Figures 6B and 6C). To assess the definitive hematopoietic potential of HPCs generated in this setting, we conducted T cell differentiation as described above. Similar to results obtained using FBS, HPCs generated from NCRM5-EGFP (non-T cell-derived hiPSCs) under xeno-free conditions differentiated into T cell progenitors and CD4+CD8+ double-positive T cells (Figures 6D and 6E). However, CD3 expression on NCRM5-EGFP-derived CD4+CD8+ T cells was lower on day 22 when compared with Mart1-iPSC-derived cells (Figures 5J, 6F, and 6G). During T cell development, CD3 expression is concomitant with TCR expression after TCR gene rearrangements. Accordingly, CD4+CD8+ T cells generated from non-T cell-derived hiPSCs need to complete the rearrangement process prior to the onset of CD3 expression.4 We observed a significant increase of CD3+ cells in NCRM5-EGFP-derived CD4+CD8+ T cells after day 27 (Figures 6F and 6G).4,28 Importantly, there was no significant difference in CD3 expression between xeno-free or FBS-containing medium on day 32 (Figure 6H). Measuring TCR diversity in CD3+CD4+CD8+ T cells on day 32 indicated successful TCRβ rearrangement in NCRM5-EGFP-derived T lineage cells (Figure 6I). Taken together, the iSTC-Hp-spheroid system can provide a scalable approach to generate definitive HPCs in an autologous and simple xeno-free condition.
Figure 6.
The iSTC-Hp-spheroid system can be performed in a xeno-free condition
(A) Schematic outline of screening a xeno-free condition for the iSTC-Hp-spheroid system.
(B) Representative flow cytometry analysis of CD34, CD43, and CD45 on GFP+ cells in iSTC-Hp-spheroids cultured in the xeno-free medium for 13 days. GFP+ cells are gated for analysis.
(C) Comparison of the frequency of CD34+CD43+ cells and CD34+CD45+ cells in iSTC-Hp-spheroids between culturing in FBS-based medium vs. xeno-free medium. GFP+ cells are gated for analysis. Values represent mean ± SD from 3 independent experiments; ns, not significant.
(D) Representative flow cytometry analysis of T lineage cell markers, including CD5, CD7, CD4, and CD8, on cells differentiated from HPCs generated in xeno-free condition. Cells were harvested on day 22 of T cell differentiation. GFP+ cells are gated for analysis.
(E) Quantifications of flow cytometry analysis in (D). Values represent mean ± SD from 3 independent experiments.
(F) Representative flow cytometry analysis of CD3+ cells in CD4+CD8+ cells during the T cell differentiation. GFP+ cells are gated for analysis.
(G) Quantifications of flow cytometry analysis in (F). Values represent mean ± SD from 3 independent experiments.
(H) Frequencies of CD3+ cells in CD4+CD8+ cells on day 32 of T cell differentiation. GFP+ cells are gated for analysis. Horizontal bars represent mean value from 3 independent experiments; ns, not significant.
(I) TCR sequence analysis showing the diversity of rearranged TCR genes in NCRM5-EGFP-derived CD4+CD8+CD3+ T cells isolated from day 32 of the T cell differentiation.
Discussion
Here, we report a Hp-spheroid system that can induce yolk sac-like organoids from hiPSCs in a simple and scalable manner. We demonstrated that day 13 Hp-spheroid mimicked the unique structural features and the cellular complement of the human secondary yolk sac and showed the functional capability to generate hematopoietic cells from hiPSCs. Single-cell transcriptional profiling revealed that endodermal cells in Hp-spheroids expressed many genes related to the functions of the human/mouse yolk sac endoderm. Several studies have shown that the developmental process of yolk sac hematopoiesis can be observed during the hPSC differentiation into HPCs16,17; however, the generation of yolk sac organoids presenting both yolk sac-specific endoderm and mesoderm has not been reported. Our Hp-spheroid system may provide a new in vitro platform to investigate the signaling crosstalk between mesoderm and endoderm during human embryonic hematopoiesis.
It was intriguing to find the disappearance of cocultured stromal cells at the early differentiation stages since they were expected to support the entire differentiation process in the Hp-spheroid. It is therefore conceivable that the primary role of stromal cells is to initiate self-organization in the early spheroid. Considering that the conditioned medium from stromal cells was not sufficient to induce a similar process, it is more likely that cell contacts with the stromal cells in a 3D setting are required rather than secreted factors. Previous studies have demonstrated that mechanic stimulations, such as physical contact or cell-adhesion tension, can induce mesoderm specification from embryonic stem cells.75,76 As EBs form firm contacts with stromal cells, this can foster mesoderm formation at the contact side and result in symmetry breaking, which is a critical prerequisite for the formation of embryonic tissues.
Current hiPSC-derived organoid systems have been shown to possess self-organization capacities; however, extrinsic biochemicals were necessary to induce organ-specific stem cells as well as niche components that regulate the stem cell fate.18,77 In our Hp-spheroid system, mesoderm progenitor cells and yolk sac endodermal cells could be induced through short-term coculture with stromal cells. Notably, subsequent hemato-vascular ontogenesis was recapitulated without the addition of stage-specific factors. These observations suggest that a niche-like microenvironment is created in the day 6 Hp-spheroid to control the differentiation of mesoderm progenitor cells. Indeed, scRNA-seq analysis revealed that cells in the day 6 Hp-spheroid expressed genes encoding important morphogens for the early hemato-vascular development.
Although blood island-like structures were observed, a developed vascular plexus was not identified in day 13 Hp-spheroids, implying that hematopoietic cells were not directly generated from blood vessels, which can be observed in the AGM region or in the murine yolk sac.78,79,80 There are two possible explanations for the lack of well-established vasculature in Hp-spheroids. One is that the endogenous production of VEGF in Hp-spheroids may not be sufficient for vascular remodeling, as angiogenesis in organoids generally requires higher doses of VEGF than those used for hematopoietic induction from hPSCs.81,82 Another is that it may be too early to detect vascular remodeling in day 13 Hp-spheroids. It would be of interest to explore whether exogenous VEGF can promote vascularization in Hp-spheroids after day 13.
Mass production of HPCs from hiPSCs is a major obstacle for hiPSC-based blood cell therapy. Traditionally, generation of HPCs in a xeno-free condition needs a stepwise tuning of exogenous biochemicals for each developmental stage. This requires complicated handling steps and many GMP-grade supplemental factors and therefore is severely limited in establishing cost-effective bulk production. Recently, a cytokine-free approach has been published,83 allowing production of HPCs in a simple and low-cost manner. However, this method still relies on the 2D planar culture format, which generally limits scalability because a large culture area is necessary for high cell yields. Furthermore, the process can be more complex and laborious if cell passaging is necessary to prevent cell overgrowth during the differentiation.
Conversely, generation of HPCs from Hp-spheroids can overcome many restrictions for clinical-scale manufacturing. We demonstrated that Hp-spheroids can grow in stirred bioreactors in a xeno-free condition without supplement factors, and this allows them to be developed in a simple medium in large-scale tank bioreactors. As hiPSC-derived cells grow in a 3D condition, our method does not require monitoring of cell confluency and is more scalable than 2D culture approaches. Importantly, despite no exogenous factors being used, definitive HPCs can be generated robustly across experiments and different hiPSC lines. Moreover, HPCs isolated from Hp-spheroids can be differentiated into various types of blood cells with current published protocols, indicating that our system is adaptable to most current maturation methods. Several recent studies highlighted the promise of hiPSC-derived T and natural killer (NK) cells for cancer immunotherapy due to their potential to overcome aging and exhaustion and to restore stemness.84,85,86 Scalable production of hiPSC-derived HPCs would enhance the output of mature hematopoietic lineage cells with clinical potential.
In conclusion, we developed a simple, scalable system to generate HPCs from yolk sac-like organoids that offers a new avenue to the clinical application of hiPSC-based blood cell therapy.
Limitations of the study
Two aspects of the protocol need further optimization to allow for clinical application of the Hp-spheroid system. The first challenge is the loss of hiPSCs during Hp-spheroid formation. A large number of uniform Hp-spheroids can be obtained with minimal manipulation by using Aggrewell plates. However, some EBs fail to establish strong adhesion with the sphere-forming stromal cells during overnight coculture in the plates, and these EBs detach from the forming spheroid during the transfer to bioreactors. Optimizing the timing of the transfer may allow more EBs to form firm interactions with stromal cells. The second challenge is the efficiency of the organoid dissociation process. In this study, we used exclusively enzymatic dissociation to obtain HPCs from organoids; however, the dissociation process remains time-consuming and treatment with enzymes for an extended time can impair cell viability. Combinations of mechanical (e.g., involving the gentleMACS dissociator) and enzymatic dissociation should be explored to improve this process. Further optimization for the dissociation process would be necessary for industrial-scale mass production.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-Cy7 Mouse anti-Human CD34 (Clone 581) | BD Biosciences | Catalog No: 560710, RRID AB_1727470 |
| APC Mouse anti-Human CD43 (Clone 1G10) | BD Biosciences | Catalog No: 560198, RRID AB_1645460 |
| Brilliant Violet 711 Mouse anti-Human CD45 (Clone HI30) | Biolegend | Catalog No: 304050, RRID AB_2563466 |
| APC Mouse anti-Human APNLR (APJ) (Clone 72133) | R&D system | Catalog No: FAB856A RRID AB_ 2044604 |
| PE-Cy7 Mouse anti-Human CD73 (Clone AD2) | Biolegend | Catalog No: 344010, RRID AB_2561542 |
| APC Mouse anti-Human CD44 (Clone G44-26) | BD Biosciences | Catalog No: 559942, RRID AB_398683 |
| PE Mouse anti-Human CD140a (PDGFRα) (Clone 16A1) | Biolegend | Catalog No: 323506, RRID AB_2268113 |
| PE Mouse anti-Human CD105 (Clone 43A3) | Biolegend | Catalog No: 323206, RRID AB_755958 |
| PE Mouse anti-Human CD140b (PDGFRβ) (Clone 18A2) | Biolegend | Catalog No: 323606, RRID AB_2268134 |
| PE Mouse anti-Human SSEA-4 (Clone MC-813-70) |
Biolegend | Catalog No: 330406, RRID AB_1089206 |
| Brilliant Violet 605 Mouse anti-Human CD31 (Clone WM59) | Biolegend | Catalog No: 303122, RRID AB_2562149 |
| Brilliant Violet 605 Mouse anti-Human CD5 (Clone UCHT2) | BD Biosciences | Catalog No: 563945, RRID AB_2738500 |
| PE Mouse anti-Human CD7 (Clone M-T701) | BD Biosciences | Catalog No: 555361, RRID AB_395764 |
| PE-Cy7 Mouse anti-Human CD4 (Clone SK3) | BD Biosciences | Catalog No: 557852, RRID AB_396897 |
| APC Mouse anti-Human CD8 (Clone RPA-T8) | BD Biosciences | Catalog No: 555369, RRID AB_398595 |
| Brilliant Violet 421 Mouse anti-Human CD8b (Clone 2ST8.5H7) | BD Biosciences | Catalog No: 742390, RRID AB_AB_2740746 |
| V450 Mouse anti-Human CD3 (Clone UCHT1) | BD Biosciences | Catalog No: 561812, RRID AB_1645570 |
| APC Mouse Anti-Human CD71 (Clone L01.1) | BD Biosciences | Catalog No: 341028, RRID AB_400560 |
| PE Mouse Anti-Human CD235a | BD Biosciences | Catalog No: 555570, RRID AB_395949 |
| PE Mouse Anti-Human CD235a (Clone HI264) | Biolegend | Catalog No: 349106, RRID AB_10640739 |
| Brilliant Violet 421 Mouse anti-human CD41a (Clone HIP8) | Biolegend | Catalog No: 303730, RRID AB_2629627 |
| FITC Mouse Anti-Human CD14 (Clone M5E2) | BD Biosciences | Catalog No: 555397, RRID AB_395798 |
| PE Mouse Anti-Human CD45 (Clone HI30) | BD Biosciences | Catalog No: 555483, RRID AB_395875 |
| APC Mouse Anti-Human CD11b (Clone ICRF44) | BD Biosciences | Catalog No: 550019, RRID AB_398456 |
| PE Mouse Anti-Human CD16 (Clone 3G8) | BD Biosciences | Catalog No: 555407, RRID AB_395807 |
| APC Mouse Anti-Human CD13 (Clone WM15) | BD Biosciences | Catalog No: 557454, RRID AB_398624 |
| APC Mouse Anti-Human CD33 (Clone WM53) | BD Biosciences | Catalog No: 551378, RRID AB_398502 |
| Brilliant Violet 605 Mouse anti-Human TNF-α (Clone MAb11) | Biolegend | Catalog No: 502932, RRID AB_10898321 |
| PE-Cy7 Mouse anti-human IFN-γ (Clone 4S.B3) | Biolegend | Catalog No: 502536, RRID AB_11125368 |
| eBioscience™ Fixable Viability Dye eFluor™ 450 | Thermo Fisher Scientific | Catalog No: 65-0863-14 |
| CD4 Micro-Bead Kit, human | Miltenyi Biotec | Catalog No: 130-045-101 |
| CD34 Micro-Bead Kit, human | Miltenyi Biotec | Catalog No: 130-046-702 |
| Peptide HLA-A∗02:01 Mart-1 ELAGIGILTV | MBL | Catalog No: SP0009 |
| iTAg Tetramer/PE – HLA-A∗02:01 Mart-1 | MBL | Catalog No: TB-0009-1 |
| Mouse anti-alpha-Fetoprotein/AFP Monoclonal Antibody (1:200) (Clone 189502) | R&D system | Catalog No: MAB1368, RRID AB_357658 |
| Rabbit anti-PDGFRb Monoclonal Antibody (1:200) (Clone 42G12) | Abcam | Catalog No: ab69506, RRID AB_1269704 |
| Rabbit anti-CD31 Monoclonal Antibody (1:100) (Clone EPR3094) | Abcam | Catalog No: ab76533, RRID AB_1523298 |
| Rabbit anti-CD34 Monoclonal Antibody (1:100) (Clone EP373Y) | Abcam | Catalog No: ab81289, RRID AB_1640331 |
| Mouse Anti-CD43 Monoclonal Antibody (1:300) (W3/13) | Abcam | Catalog No: ab22351, RRID AB_447013 |
| Alexa 488-conjugated anti-Mouse IgG (H + L) | Thermo Fisher Scientific | Catalog No: A11001, RRID AB_2534069 |
| Alexa 555-conjugated anti-Rabbit | Thermo Fisher Scientific | Catalog No: A21429, RRID AB_2535850 |
| Alexa 647-conjugated anti-Mouse | Abcam | Catalog No: ab150115, RRID AB_2687948 |
| Alexa 488-conjugated anti-Rabbit | Abcam | Catalog No: ab150081, RRID AB_2734747 |
| TotalSeq™-A0251 anti-human Hashtag 1 Antibody | Biolegend | Catalog No: 394601, RRID AB_2750015 |
| TotalSeq™-A0252 anti-human Hashtag 2 Antibody | Biolegend | Catalog No: 394603, RRID AB_2750016 |
| Chemicals, peptides, and recombinant proteins | ||
| Hoechst 33342 trihydrochloride trihydrate | Thermo Fisher Scientific | Catalog No: H3570 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | Catalog No: P36930 |
| Fixation/Permeabilization Solution Kit with BD GolgiStop™ | BD Cytofix/Cytoperm | Catalog No: 554715 |
| Recombinant Human BMP-4 | R&D systems | Catalog No: 314-BP |
| Recombinant Human FGF2 | R&D Systems | Catalog No: 233-FB |
| Recombinant Human IL3 | R&D Systems | Catalog No: 203-IL |
| Recombinant Human IL6 | R&D Systems | Catalog No: 206-IL |
| Recombinant Human IL7 | R&D Systems | Catalog No: BT-007 |
| Recombinant Human Flt3-Ligand | R&D Systems | Catalog No: 427-FL |
| Recombinant Human stem cell factor (SCF) | R&D Systems | Catalog No: 255-SC 050/CF |
| Recombinant Human VEGF | PeproTech | Catalog No: 100-20 |
| Recombinant Human Thrombopoietin (TPO) | R&D Systems | Catalog No: 288-TP |
| Recombinant Human PDGF-BB | R&D Systems | Catalog No: 220-BB |
| Erythropoietin | Amgen | EPOGEN® |
| Dexamethasone | VETone | Catalog No: 501012 |
| Estradiol | Pfizer | DEPO®-ESTRADIOL |
| Insulin | Lilly | Humulin® R U-100 |
| holo-Transferrin human | Sigma-Aldrich | Catalog No: T4132-500MG |
| Bovine Serum Albumin (Roche) | Millipore Sigma | Catalog No: 10735078001 |
| CHIR99021 | Tocris Bioscience | Catalog No: 4953 |
| Activin A | PeproTech Inc. | Catalog No: 120–14P |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Catalog No: P8139 |
| Zymosan An S. cerevisiae BioParticles, FITC conjugate | Thermo Fisher Scientific | Catalog No: Z2841 |
| Dihydrorhodamine 123 (DHR) | Thermo Fisher Scientific | Catalog No: D632 |
| Giemsa stain Solution | Thermo Fisher Scientific | Catalog No: 10092-013 |
| Gibco MEM α, nucleosides, powder (α MEM) | Thermo Fisher Scientific | Catalog No: 11900024 |
| Iscove’s Modified Dulbecco’s Medium (for Macrophage differentiation) | Thermo Fisher Scientific | Catalog No: 12440-053 |
| Iscove’s Modified Dulbecco’s Medium (for erythroid cell differentiation) | Sigma-Aldrich | Catalog No: I3390 |
| Fetal Bovine Serum (FBS), Embryonic Stem Cell Qualified | R&D Systems | Catalog No: S10250H |
| Recombinant Human Macrophage Colony Stimulating Factor (M-CSF) | PeproTech | Catalog No: 300-25 |
| Interferon γ (IFN-γ) | Horizon Therapeutics | Actimmune® |
| Busulfan | Otsuka America Pharmaceutical | Busulfex® IV |
| Y-27632 dihydrochloride | R&D Systems | Catalog No: 1254 |
| DMEM/F12 - Dulbecco’s Modified Eagle Medium | Thermo Fisher Scientific | Catalog No: 11320033 |
| Granulocyte-Colony Stimulating Factor (G-CSF) | Amgen | Neupogen® Filgrastim |
| Corning Matrigel matrix | Corning | Catalog No: 354230 |
| PLTGold® Human Platelet Lysate (heparin-free) | Mill Creek Life Sciences | Catalog No: PLTGold100R |
| StemFit C02 | Ajinomoto Co., Inc | Kindly provided from Ajinomoto Co., Inc |
| StemFit for differentiation | Nacalai USA | Catalog No: AS401 |
| Stemfit Basic02 | Nacalai USA | Catalog No: BASIC02 |
| CELLBANKER | Amsbio | Product code: 11910 |
| TrypLE™ Express Enzyme | Thermo Fisher Scientific | Catalog No: 12605010 |
| Liberase™ TM Research | Roche | Catalog No: 5401127001 |
| DNase I | Roche | Catalog No: 10104159001 |
| 0.1% Gelatin in Water | STEMCELL Technologies | Catalog No: 07903 |
| Able Bioreactor System Controller and Motor 30 mL | REPROCELL | ABBWDW-1013 |
| Able Bioreactor Magnetic Stir System Base 30mL | REPROCELL | ABBWBP03N0S-6 |
| Ultra-low attachment plates: 96 well, U bottom | S-BIO | Catalog No: MS-9096UZ |
| Anti-Adherence Rinsing Solution | STEMCELL Technologies | Catalog No: 07010 |
| AggreWell™400 (6-well plate) | STEMCELL Technologies | Catalog No: 34425 |
| AggreWell™800 (24-well plate) | STEMCELL Technologies | Catalog No: 34815 |
| Critical commercial assays | ||
| MethoCult GF H4034 Starter Kit | STEMCELL Technologies | Catalog No: 04064 |
| RNeasy Plus Mini Kit | Qiagen | Catalog No: 74134 |
| TruSeq Stranded Total RNA Library Prep | Illumina | Catalog No: 20020596 |
| Chromium Next GEM Single Cell 3′ Kit | 10X Genomics | Catalog No: 1000268 |
| Deposited data | ||
| Raw Bulk RNA-seq and scRNA-seq data, count matrices | This paper | GEO: GSE157140 |
| Experimental models: Cell lines | ||
| OP9/N-DLL1 | Riken Bioresource center | RRID:CVCL_B220 |
| OP9/N | Riken Bioresource center | RRID:CVCL_B219 |
| NCRM5-AAVS1-CAG-EGFP | iPSC core, NHLBI, NIH | https://www.nhlbi.nih.gov/science/ipsc-core |
| NCRM5 | iPSC core, NHLBI, NIH | https://www.nhlbi.nih.gov/science/ipsc-core |
| Mart1-iPSC | Kawamoto Lab | https://www.infront.kyoto-u.ac.jp/en/laboratory/lab14/ |
| SCD-iPSC | Tisdale Lab | https://www.nhlbi.nih.gov/science/cellular-and-molecular-therapeutics |
| Experimental models: Organisms/strains | ||
| NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice | The Jackson Laboratory | Strain #: 005557 |
| Software and algorithms | ||
| Graphpad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo software | FlowJo, LCC | https://www.flowjo.com/ |
| Cutadapt 1.18 | Martin, 201187 | https://github.com/marcelm/cutadapt/ |
| STAR 2.6.1 | Dobin et al., 201388 | https://github.com/alexdobin/STAR |
| RSEM 1.2.31 | Li and Dewey, 201189 | https://github.com/deweylab/RSEM |
| edgeR 3.28.1 | Robinson et al., 201090 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GSEA 4.0.3 | Mootha et al., 200391; Subramanian et al., 200592 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Cell Ranger 3.1.0 | 10X Genomics, Inc. | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat_4.0.5 | Butler et al., 201893; Stuart et al., 201994 | https://satijalab.org/seurat/ |
| Other | ||
| CliniMACS CD34 GMP Micro-Beads | Miltenyi Biotec | Catalog No: 170-076-711 |
| CliniMACS Plus Instrument | Miltenyi Biotec | Catalog No: 151-01 |
| ImmunoSEQ (TCR seq) | Adaptive Biotechnologies | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Naritaka Tamaoki (naritaka0102@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture of hiPSCs
NCRM5 and NCRM5-AAVS1-CAG-EGFP (NCRM5 iPSCs with CAG-EGFP integrated at Chr.19 AAVS1 safe harbor locus) were established by the iPSC core facility of the National Heart, Lung, and Blood Institute in NIH. Mart1-iPSC were kindly provided by Riken BBC, in Japan. SCD-iPSC line was established from bone marrow stromal cells isolated from a sickle cell disease patient.34 All hiPSCs were cultured on Matrigel (Corning) or iMatrix-511 (Nippi) coated dishes in xeno-free hiPSC medium Essential 8 (Invitrogen) or StemFit Basic02 (Ajinomoto Co., Inc). They were routinely passaged as small clumps/single cells using 0.5 mM EDTA in phosphate buffered saline (PBS) with the split ratio of 1:6 to 1:10 every 3 to 4 days after reaching 65%–80% confluence. After EDTA treatment, hiPSCs were transferred to new Matrigel or iMatrix-511 coated dishes in hiPSC medium supplemented with ROCK inhibitor Y-27632 (10 μM, R&D Systems Inc). Next day, the medium was changed to hiPSC medium without ROCK inhibitor.
Approval for human samples and animal use
Bone marrow was collected and hBMSCs produced from healthy human donors after written informed consent under the auspices of National Institutes of Health Clinical Center Review Board-approved protocol NCT01071577. Mobilized CD34+ hematopoietic stem/progenitor cells were obtained from healthy human donors after written informed consent under the auspices of National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board-approved protocol 94-I-0073. The conduct of these studies conforms to the Declaration of Helsinki protocols and all United States federal regulations required for protection of human subjects. The use of immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) for human cell transplant studies was approved by the NIAID Institutional Animal Care and Use Committee under animal use protocol LCIM-1E. Four-month-old male NSG mice were maintained under specific pathogen-free conditions at an American Association for the Accreditation of Laboratory Animal Care (AAALAC) accredited animal facility and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. The conduct of these studies conforms to AAALAC International guidelines and all US federal regulations required for protection of research animals.
Preparation of hBMSCs for coculturing with hiPSCs
Healthy donor derived hBMSCs were kindly provided from the Cell Processing Section, Department of Transfusion Medicine in NIH. Establishment of hBMSCs is described in our previous study.25 hBMSCs in cryobags (100 × 106 cells/bag) were pre-aliquoted into cryotubes (5 × 106 cells/tube) with CELLBANKER (AMSBIO) for use in the Hp-spheroid system. Based on our experience of using the classical OP9 coculture system, we reasoned that the gradual loss of stemness in expanded hBMSCs may be detrimental. Thus, we limited the expansion of hBMSCs to 4–5 passages prior to coculture.
Adult human CD34+ cell collection
Healthy donors underwent CD34+ cell mobilization with granulocyte-colony stimulating factor (G-CSF, 15 mg/kg daily for 5 days, Amgen) followed by leukapheresis. After collection, CD34+ cells were purified by CliniMACS CD34 cell separation (Miltenyi Biotec) in the Cell Processing Section of the Department of Transfusion Medicine at the NIH Clinical Center and were cryopreserved prior to use in mouse transplant studies.
Method details
EBs differentiation to HPCs with a cytokine/growth factor cocktail
Semi-confluent hiPSC were dissociated with 0.5 mM EDTA in PBS and transferred to Aggrewell 400 (6 well plate type, 1 × 106 cells/well, STEMCELL Technologies) for EB formation (100–150 cells/EB) in hiPSC medium supplemented with ROCK inhibitor Y-27632 (10 μM) according to the manufacturer’s protocol. To compare with the Hp-spheroid system, the differentiation of EBs with exogenous factors was performed according to the previous publication with some modifications.54 In brief, EBs were transferred from Aggrewell 400 plates to ultra-low attachment 3D culture plates (96 well U bottom, S-BIO) and were cultured in αMEM medium (Thermo Fisher Scientific) with 20% fetal bovine serum (FBS) and exogenous factors. Human cytokines and growth factors were added as follows: bone morphogenic protein 4 (BMP4, 10 ng/mL, R&D Systems) and fibroblast growth factor 2 (FGF2, 20 ng/mL, R&D Systems) from day 0 to day 3, FGF2 (20 ng/mL) from day 3 to day 5, FGF2 (20 ng/mL), vascular endothelial growth factor (VEGF, 10 ng/mL, PeproTech), interleukin 3 (IL3, 10 ng/mL, R&D Systems), interleukin 6 (IL6, 10 ng/mL, R&D Systems), Flt3-Ligand (Flt3-L, 10 ng/mL, R&D Systems) and stem cell factor (SCF, 100 ng/mL, R&D Systems) from day 5 to day 8, VEGF (10 ng/mL), IL3 (10 ng/mL), IL6 (10 ng/mL), Flt3-L (10 ng/mL), SCF (100 ng/mL), and thrombopoietin (TPO, 10 ng/mL, R&D Systems) from day 8 to day 13. Medium was changed every day.
HPCs generation from hiPSCs in 2D
The classic OP9 co-culture system in 2D for HPC generation was performed according to previous publication with a slight modification.4 In brief, OP9 cells were cultured on gelatin coated 10 cm dishes for 7–8 days before coculturing with hiPSCs. To place similar sizes of hiPSC clumps on OP9 dishes, we harvested hiPSC and formed EBs (2.5 × 103 cells/EB) in Aggrewell 800 plates (24 well plate type, STEMCELL Technologies) in hiPSC medium supplemented with ROCK inhibitor Y-27632 in advance. Next day, we transferred EBs onto gelatinized OP9 dishes in OP9 medium consisting of αMEM with 20% FBS. On day 13 of the differentiation, hiPSC-derived sac-like structures were enzymatically dissociated for further analysis. To prepare gelatinized hBMSC dishes, we transferred hBMSCs onto gelatin-coated 10 cm dishes within one passage after thawing and cultured them in OP9 medium for 7–8 days before coculture with hiPSCs.
Generation of yolk sac-like organoids from hiPSCs with Hp-spheroids system in bioreactors
We transferred 4 × 106 hiPSCs into 4 wells of an Aggrewell 400 plate to form small EBs. Next day, pooled all EBs, mixed them with the thawed 5 × 106 stromal cells (hBMSCs or OP9 cells or iSTCs) in αMEM with 20% FBS and distributed the suspension across 8 wells of an Aggrewell 800 plate (24 well plate type) to form Hp-spheroids. Using centrifuge (800rpm, 3min) to settle EBs and stromal cells into microwells of Aggrewell 800 and incubated overnight. About 8000 stromal cells are cocultured with 7–10 EBs in a Hp-spheroid. Next day, Hp-spheroids were transferred to bioreactors (ABLE Bioreactor Magnetic Stir System 30 mL, REPROCELL), stirring at 65 rpm, in αMEM with 20% FBS. Medium was changed every 2–3 days. Generally, we cultured 2400–2800 Hp-spheroids in one ABLE 30 mL Disposable Bioreactor. We used αMEM containing 2.5% PLTGold (Mill Creek Life Sciences) and 1x StemFit C02 (kindly provided by Ajinomoto Co., Inc) as xeno-free medium for the iSTC-Hp-spheroid system. EBs and thawed iSTCs were mixed in the xeno-free medium supplemented with ROCK Inhibitor Y-27632 (10 μM) for Hp-spheroids formation. The next day, Hp-spheroids were transferred to bioreactors in the xeno-free medium without ROCK inhibitor.
Dissociation of yolk sac-like organoids (day 13 Hp-spheroids)
To dissociate day 13 Hp-spheroids, we washed them with Hank’s Balanced Salt Solution (HBSS) once and then treated with HBSS containing Liberase TM (Roche, 250 μg/mL) and DNase I (Roche, 500 μg/mL) at 37°C for 15–20 min, followed by pipetting for 1 min with a P1000 disposable plastic tip to break Hp-spheroids. Cells were then washed with PBS once and treated with PBS containing 10% TrypLE Express (Gibco) and DNase I (500 μg/mL) at 37°C for 10–15 min, followed by pipetting for 1 min with a P1000 disposable plastic tip to make a single cell suspension.
Induction of stromal cells from hiPSCs
Generation of iSTCs was performed according to our previous study with a slight modification33 as summarized in Figure S3B. In brief, hiPSCs were plated on iMatrix-511 coated dishes in hiPSC medium supplemented with ROCK inhibitor Y-27632 (10 μM) before differentiation. We used DMEM/F12 (Gibco) containing 1x StemFit for Differentiation (AS401) (Ajinomoto Co., Inc) and 1% Glutamax as a base medium. For mesoderm induction stage, hiPSCs were cultured in the base medium supplemented with CHIR99021 (8 μM, Tocris Bioscience) and BMP4 (25 ng/mL). Medium was changed every day until day 3 of induction. On day 4, medium was changed to stromal cell induction medium consisting of the base medium supplemented with Activin A (2 ng/mL, PeproTech Inc.) and PDGF-BB (10 ng/mL, R&D Systems). From day 8, the medium was changed to stromal cell maturation medium consisting of the base medium supplemented with PDGF-BB (10 ng/mL). After the stromal cell induction stage, the medium was changed every 2 days.
Hematopoietic colony-forming unit (CFU) assay
HPCs were harvested from day 13 from Hp-spheroids and then NCRM5-EGFP derived CD34+CD43+ cells were sorted by a FACSAria II (BD Biosciences) for hematopoietic CFU assays. 1,500 CD34+CD43+ cells were seeded per 35mm-dish in MethoCult H4434 (STEMCELL Technologies) methylcellulose-based medium with recombinant cytokines for CFU assay according to the manufacturer’s protocol, and colonies were identified and counted after 14 days of culture.
Erythroid cell differentiation from Hp-spheroid derived HPCs
Erythroid cell differentiation from Hp-spheroid derived HPCs was performed as described in our previous publications with a slight modification.6,73 In brief, CD34+ cells isolated from Hp-spheroids on day 13 were enriched by human CD34 Micro-Bead Kit (Miltenyi Biotec) and were cultured on irradiated OP9 feeder cells for 2 days in Iscove’s Modified Dulbecco’s Medium (IMDM) (Sigma-Aldrich) containing VEGF (20 ng/mL), SCF (50 ng/mL), Flt3-L (50 ng/mL), TPO (50 ng/mL), IL3 (5 mg/mL), BMP4 (10 ng/mL), erythropoietin (EPO, 5 U/mL, Amgen), and 15% FBS. Then, the suspension cells were collected and transferred onto fresh irradiated OP9 dishes for 5 days in IMDM medium containing SCF (10 ng/mL), IL3 (1 ng/mL), EPO (2 U/ml), dexamethasone (1 mM, VETone), estradiol (1 mM, Pfizer), and 20% FBS. For erythroid maturation, the medium was replaced with IMDM media containing EPO (2 U/ml), insulin (10 ng/mL, Lilly), transferrin (0.56 mg/mL, Sigma-Aldrich), 2% bovine serum albumin (BSA) (Roche), 2mM L-glutamine, and 20% FBS. The cells were cultured in the maturation medium for 8 days hiPSC-derived erythroid cells on day 15 of differentiation were harvested for flow cytometry analysis and evaluation of globin-expression patterns by quantitative PCR assay and RP-HPLC.73
Macrophage differentiation from Hp-spheroid derived HPCs
Macrophage differentiation from Hp-spheroid derived HPCs was performed according to our previous report with minor modifications.74 In brief, CD34+ cells isolated from Hp-spheroids on day 13 were enriched by human CD34 Micro-Bead Kit and cultured for 14 days in macrophage differentiation medium consisting of IMDM (Thermo Fisher Scientific) medium containing 10% FBS and macrophage colony stimulating factor (M-CSF, 100 ng/mL, PeproTech). Medium was changed two times per week, with suspended cells centrifuged and re-plated until the majority of the cells became adherent. Macrophages were cultured with interferon γ (IFN-γ, 65 U/ml, Horizon Therapeutics) for 3 days prior to functional assays. Cells were analyzed by cytospin Giemsa stain for macrophage morphology, and color images of stained macrophages were acquired using an EVOS XL Core system (Thermo Fisher Scientific); whole image adjustments of brightness, color balance, and contrast were performed using Adobe Photoshop software (Adobe) without additional image processing. Phagocytosis ability of hiPSC-derived macrophages was evaluated as previously described.95 Briefly, macrophages were incubated for 30 min at 37°C while shaking (300 rpm) with zymosan A BioParticles isolated from Saccharomyces cerevisiae (Thermo Fisher Scientific) at a multiplicity of infection (MOI) of 10:1. The suspension was then transferred in a FACS tube on ice and the same volume of 2 mg/mL trypan blue was added to stop the reaction and quench the fluorescence of membrane-bound not-internalized particles. Flow cytometry analysis was performed by FACSCanto flow cytometer (BD Biosciences). Dihydrorhodamine-123 (DHR) assay of reactive oxygen species production was performed as previously described.74 Briefly, cells were incubated at 37°C for 5 min in 400 μL of HBSS containing 130 μM of DHR (Molecular Probes, Thermo Fisher Scientific) and 500 U of catalase (Sigma-Aldrich). Cells were then stimulated for reactive oxygen species production by addition of 100 μL of 400 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) in HBSS containing calcium and magnesium (Thermo Fisher Scientific), and incubated at 37°C for 14 min, after which flow cytometry was performed using a FACSCalibur system (BD Biosciences).
T cell differentiation from Hp-spheroid derived HPCs
T cell differentiation from Hp-spheroid derived HPCs was described in our previous reports.4,5 In brief, Hp-spheroids were dissociated, and single cells were transferred onto OP9/DLL1 dishes with T cell differentiation medium consisting of OP9 medium supplemented with interleukin 7 (IL7, 5 ng/mL, R&D Systems), Flt3-L (5 ng/mL), and SCF (10 ng/mL). After 3 days, semi adherent cells were transferred to new OP9/DLL1 dishes, followed passaging semi-adherent onto new OP9/DLL1 dishes every 5–7 days. For cytokine release analysis of Mart1-iPSC derived T cells, we harvested floating cells from OP9/DLL1 dishes on day 22 of the T cell differentiation and enriched CD4+CD8+ double-positive T cells with human CD4 Micro-Bead Kit (Miltenyi Biotec).5 CD4+CD8+ double-positive T cells were stimulated by Mart-1 peptide pulsed T2 cells to be induced to CD8+ single positive T cells. After one week of post stimulation, Mart1-iPSC derived CD8+ single positive T cells were cocultured with Mart-1 peptide or DMSO pulsed T2 cells for cytokines release assay. TCR-Vβ deep sequencing was performed by ImmunoSEQ (Adaptive Biotechnologies) on genomic DNA isolated from NCRM5-EGFP derived (GFP+) CD4+CD8+CD3+ cells on day 32 of T cell differentiation.
Transplantation of CD34+ cells into NSG mice
Four-month-old male NSG mice were given busulfan (20 mg/kg) for myelosuppressive conditioning via intraperitoneal injections 24 h before transplant. Mice were anesthetized by isoflurane inhalation prior to receiving 0.5 or 1.0 × 106 human CD34+ cells (from human donor G-CSF mobilized peripheral blood apheresis or from Hp-spheroids) via femur injection. After busulfan treatment, NSG mice received neomycin-supplemented water for prophylaxis. At 3 months post-engraftment, mice were euthanized and bone marrow from the transplanted femur was harvested and stained with antibody to human CD45 for flow cytometry analysis of engraftment using a FACSCalibur system.
Flow cytometry analysis
Antibodies used for cell surface staining or intracellular staining are listed in the key resources table. To analyze cell surface markers, cells were stained with antibodies against cell surface antigens and Propidium Iodide (PI) in FACS buffer (PBS with 2% FBS and 0.1% sodium azide). To determine intracellular cytokine expression, Mart1-iPSC derived CD8+ single positive cells were cocultured with Mart-1 peptide or DMSO pulsed T2 cells for 6 h in the presence of GolgiStop (BD Cytofix/Cytoperm). After stimulation, cells were stained for surface markers and a fixable live/dead stain (Invitrogen). Cells stained for viability and cell surface markers were fixed and permeabilized with Fixation/Permeabilization Solution Kit (BD Cytofix/Cytoperm), and then they were stained with intracellular anti-cytokine antibodies. Stained cells were washed three times by the wash buffer and analyzed by a LSRFortessa (BD Biosciences). All flow cytometry data was analyzed using FlowJo 10.6.1 software (TreeStar).
Histology
Day 13 Hp-spheroids were fixed in 4% paraformaldehyde for 15 min at room temperature. The fixed Hp-spheroids were paraffin embedded and sliced into 20 μm thick sections for hematoxylin and eosin (H&E) staining (performed in Histoserv) or immunostaining.
Immunostaining and confocal imaging
The antibodies used in this study were listed in the key resources table. For analysis of AFP and PDGFRβ, we used whole mount immunostaining. Hp-spheroids harvested on day 13 were permeabilized using PBS supplemented with 0.5% Triton X-100 for 15 min at room temperature. Blocking was performed for 1 h at room temperature using a blocking buffer (PBS supplemented with 1% BSA and 0.1% Triton X-100). Subsequently, samples were incubated with primary antibodies overnight at 4°C. Samples were then washed three times with PBS and followed by 1-h incubation with secondary antibodies at room temperature. Samples were then washed three times with PBS. Confocal images were collected with a 20x plan-apochromat (N.A. 0.75) objective lens using a Nikon Ti2 microscope (Nikon Instruments, Inc.) equipped with a Yokogawa CSU-W1 spinning disk confocal unit and Photometrics BSI sCMOS camera. For analysis of CD31, CD34, and CD43, we used section immunostaining. The sections were deparaffinized with xylene followed by different concentrations of ethanol. The immunofluorescence staining procedures were performed according to our previous publication.96 In brief, the sections were treated with glycine and then with 1% BSA and 10% normal donkey serum in PBS. Afterward, slides were incubated with the primary antibody overnight at 4°C. On the next day, slides were thoroughly washed in PBS, incubated with secondary antibodies for 1 h, and then thoroughly washed again. Coverslips were mounted on slides with Pro-Long Gold antifade reagent (Thermo Scientific), and confocal images were captured using a Zeiss LSM880 laser scanning confocal microscope equipped with a 20x plan-apochromat (N.A. 0.8) objective lens (Carl Zeiss Microscopy, LLC.).
RNA sequencing and analysis
RNA-seq was performed on day 0 iPSC (NCRM5-EGFP) and NCRM5-EGFP derived cells isolated from Hp-spheroids on day 6 and day 13 with three biological replicates for each time point. After dissociation of Hp-spheroids, GFP+ cells were sorted by Sony FX500 (Sony) prior to total RNA extraction using the RNeasy Plus Mini Kit (Qiagen). Libraries were pooled and sequenced in two lanes of a HiSeq4000 (Illumina). Raw reads were trimmed for adapters and low-quality bases using Cutadapt 1.1887 (-j 8 -b file:adapters.fa -B file:adapters.fa --trim-n -m 20 -o trimmed_R1.fq -p trimmed_R2.fq input_R1.fq input_R2.fq) prior to alignment to the reference genome (Human genome – hg38, annotation Gencode_v24) using STAR 2.6.1.88 Gene expression quantification analysis was performed using RSEM 1.2.31.89 Differential gene expression analysis was performed using edgeR 3.28.1.90 The analyses are available as a supplementary file (Data S1. Sup analysis 1).
Gene set enrichment analysis
Gene Set Enrichment Analysis was performed using the GSEAPreranked module within GSEA 4.0.3.91,92 Enrichment was tested using a custom yolk sac gene set which comprised the top 200 yolk sac genes (sorted by RPKM) as identified by Cindrova-Davies et al. (2017, supplemental Dataset S1). 196 of these genes were expressed in our dataset and were considered in the enrichment analysis (Table S1. GSEA using a human yolk sac gene set).
Single-cell RNA-seq library preparation and sequencing
Day 6 and day 13 Hp-spheroids were dissociated into single cells (see above) and GFP+ cells were sorted by Sony FX500. Cell suspensions from each day were labeled using oligonucleotide-coupled antibodies (Biolegend, TotalSeq-A0251 (barcode: GTCAACTCTTTAGCG) and TotalSeq-A0252 (barcode: TGATGGCCTATTGGG)) to allow for sample multiplexing on one capture lane and cells were mixed at 1:1 ratio. The sample concentration and viability were assessed using the LunaFL fluorescent cell counter (Logos Biosystems). A total recovery of 12,000 cells was targeted. Cell loading, library preparation and quality control were performed as described in the 10x Genomics 3′ user guide (v3.0). Three sequencing runs were performed on an Illumina NextSeq 500 (28 × 8 × 0 X 98 base pair (bp) read configuration). The standard 10x Genomics cellranger pipeline (version 3.1.0) was used to convert raw sequencing data to Fastq format. scRNA-seq reads were aligned to the GRCh38 human reference genome and gene expression counts and feature barcode counts were determined using Cell Ranger.
Single-cell data analysis
We used the R package Seurat for cluster analyses and exploration of the dataset.93,94 The complete analysis is available as a supplemental file (Data S1. Sup analysis 02). In the analyses we considered cells with nFeature_RNA (genes) > 1000 and <8000 and nCount_RNA (Unique molecular identifiers (UMI)) >2000 and <80000. Cells with mitochondrial read counts >20% were excluded. This filtering left 4895 days 6 cells and 4401 days 13 cells for analyses with a mean gene count of 5292 and a mean UMI count of 30,069. The data were normalized and scaled, and genes that varied more than expected for their expression level were identified. Principal components (PCs) were calculated on these variable genes which were then used in graph-based clustering followed by UMAP dimensionality reduction. PCs 1:30 were considered in the clustering of cells from both days. PCs 1:21 or 1:25 were used in separate clusterings of D6 cells or D13 cells respectively. The cluster annotations from both the day 6 and the day 13 clusterings were mapped to the resulting UMAP after cells from both days were clustered together. Sub-cluster analysis for the subset of hematopoietic cells considered PCs 1:40. Biomarkers for the day 6, day 13 and the hematopoietic cell clusterings were identified using Seurat’s FindAllMarkers.94 We annotated cluster identities using previously characterized genes.
Quantification and statistical analysis
The data are presented as the mean values ±SD. Statistical analyses were performed using unpaired Student’s t-tests (Figures 1E, 1H, 1I, 5D, 5L, 6H, S1B, S1D, and S3D) with GraphPad Prism 9.1.0 (GraphPad software). "The two-group comparison in Figure 4C was performed using function stat_compare_means and default method “wilcox.test” from R package ggpubr 0.40." p < 0.05 was considered statically significant.
Acknowledgments
For technical support, we thank Zhiya Yu, Rafiqul Islam, Ken-ichi Hanada, Francis Flomerfelt, Sherif Badr, Minh Tran, and Chengyu Liu. We thank Arnold Mixon and Shawn Farid for flow cytometry support. We thank Hiroshi Kawamoto and Kyoko Masuda for kindly providing the OP9/DLL1 cell line. We thank Erina He and Maria Romanova for graphic support and Celina Juliano for access to computational resources. This research was supported by the Intramural Research Programs of the NCI (ZIA BC010763), NIH/NCI (K08CA197966), NIAID (Z01 AI000644), and NINDS (ZIA NS003034). This work was also supported by the Tiens Charitable Foundation, the NIH Center of Regenerative Medicine, the Cancer Moonshot Program for the Center for Cell-Based Therapy at the NCI, NIH, the Milstein Family Foundation, and the Melanoma Research Alliance.
Author contributions
N.T. and R.V. conceived the project, designed the experiments, collected and analyzed the data, and wrote the manuscript. S.S. conducted bioinformatics analysis and edited the manuscript. J.J.H.-M. and N.U. performed erythroid cell differentiation with expert advice from J.F.T. T.M. and M.L.G. performed T cell differentiation. C.L.S., U.C., J.B., and S.K. performed macrophage differentiation and mouse transplants with expert advice from H.L.M. Y.H. and R.I. performed imaging analysis with expert advice from D.S.G. and M.J.K. N.-H.H., S.K.V., Y.Y., M.K., T.T., J.Z., D.F.S., and P.G.R. helped to interpret experimental results and provided advice regarding the research strategy. R.V. and N.P.R. supervised the project.
Declaration of interests
N.T., M.L.G., R.V., and N.P.R. are inventors on international patent (WO 2019/094614A1), published on May 16, 2019, entitled “Methods of preparing hematopoietic progenitor cells in vitro.” J.J.H.-M. is currently an employee of GeneDx. N.T., S.S., T.M., N.-H.H., Y.H., S.K.V., Y.Y., N.P.R., and R.V. are currently employees of Lyell Immunopharma.
Published: April 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100460.
Contributor Information
Naritaka Tamaoki, Email: naritaka0102@gmail.com.
Nicholas P. Restifo, Email: drnickrestifo@gmail.com.
Raul Vizcardo, Email: poh_de_arras@hotmail.com.
Supplemental information
Data and code availability
-
•
The raw and processed data reported in this publication are archived at NCBI GEO (accession number GSE157140).
-
•
Analysis code is available as supplemental files (Data S1).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Choi K.D., Vodyanik M.A., Slukvin I.I. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J. Clin. Invest. 2009;119:2818–2829. doi: 10.1172/jci38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dias J., Gumenyuk M., Kang H., Vodyanik M., Yu J., Thomson J.A., Slukvin I.I. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011;20:1639–1647. doi: 10.1089/scd.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senju S., Haruta M., Matsumura K., Matsunaga Y., Fukushima S., Ikeda T., Takamatsu K., Irie A., Nishimura Y. Generation of dendritic cells and macrophages from human induced pluripotent stem cells aiming at cell therapy. Gene Ther. 2011;18:874–883. doi: 10.1038/gt.2011.22. [DOI] [PubMed] [Google Scholar]
- 4.Vizcardo R., Masuda K., Yamada D., Ikawa T., Shimizu K., Fujii S.I., Koseki H., Kawamoto H. Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8(+) T cells. Cell Stem Cell. 2013;12:31–36. doi: 10.1016/j.stem.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Maeda T., Nagano S., Ichise H., Kataoka K., Yamada D., Ogawa S., Koseki H., Kitawaki T., Kadowaki N., Takaori-Kondo A., et al. Regeneration of CD8alphabeta T cells from T-cell-derived iPSC imparts potent tumor antigen-specific cytotoxicity. Cancer Res. 2016;76:6839–6850. doi: 10.1158/0008-5472.CAN-16-1149. [DOI] [PubMed] [Google Scholar]
- 6.Fujita A., Uchida N., Haro-Mora J.J., Winkler T., Tisdale J. Beta-globin-expressing definitive erythroid progenitor cells generated from embryonic and induced pluripotent stem cell-derived sacs. Stem Cell. 2016;34:1541–1552. doi: 10.1002/stem.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratt-Leal A.M., Nguyen A.H., Hammersmith K.A., Singh A., McDevitt T.C. A microparticle approach to morphogen delivery within pluripotent stem cell aggregates. Biomaterials. 2013;34:7227–7235. doi: 10.1016/j.biomaterials.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachlos E., Auguste D.T. Embryoid body morphology influences diffusive transport of inductive biochemicals: a strategy for stem cell differentiation. Biomaterials. 2008;29:4471–4480. doi: 10.1016/j.biomaterials.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Vodyanik M.A., Bork J.A., Thomson J.A., Slukvin I.I. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 10.Nakano T., Kodama H., Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A., D'Souza S.S., Thakur A.S. Understanding the journey of human hematopoietic stem cell development. Stem Cells Int. 2019;2019 doi: 10.1155/2019/2141475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cindrova-Davies T., Jauniaux E., Elliot M.G., Gong S., Burton G.J., Charnock-Jones D.S. RNA-seq reveals conservation of function among the yolk sacs of human, mouse, and chicken. Proc. Natl. Acad. Sci. USA. 2017;114:E4753–E4761. doi: 10.1073/pnas.1702560114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross C., Boroviak T.E. Origin and function of the yolk sac in primate embryogenesis. Nat. Commun. 2020;11:3760. doi: 10.1038/s41467-020-17575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zohn I.E., Sarkar A.A. The visceral yolk sac endoderm provides for absorption of nutrients to the embryo during neurulation. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:593–600. doi: 10.1002/bdra.20705. [DOI] [PubMed] [Google Scholar]
- 15.Poon E., Clermont F., Firpo M.T., Akhurst R.J. TGFbeta inhibition of yolk-sac-like differentiation of human embryonic stem-cell-derived embryoid bodies illustrates differences between early mouse and human development. J. Cell Sci. 2006;119:759–768. doi: 10.1242/jcs.02788. [DOI] [PubMed] [Google Scholar]
- 16.Zambidis E.T., Peault B., Park T.S., Bunz F., Civin C.I. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkins M.H., Scarfò R., McGrath K.E., Yang D., Palis J., Ditadi A., Keller G.M. Modeling human yolk sac hematopoiesis with pluripotent stem cells. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCauley H.A., Wells J.M. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development. 2017;144:958–962. doi: 10.1242/dev.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M., et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low J.H., Li P., Chew E.G.Y., Zhou B., Suzuki K., Zhang T., Lian M.M., Liu M., Aizawa E., Rodriguez Esteban C., et al. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell. 2019;25:373–387.e9. doi: 10.1016/j.stem.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 22.Vodyanik M.A., Thomson J.A., Slukvin I.I. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J., Yan X.L., Li R., Liu Y., He W., Sun S., Zhang Y., Liu B., Xiong J., Mao N. Characterization of OP9 as authentic mesenchymal stem cell line. J Genet Genomics. 2010;37:475–482. doi: 10.1016/s1673-8527(09)60067-9. [DOI] [PubMed] [Google Scholar]
- 24.Kfoury Y., Scadden D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Sabatino M., Ren J., David-Ocampo V., England L., McGann M., Tran M., Kuznetsov S.A., Khuu H., Balakumaran A., Klein H.G., et al. The establishment of a bank of stored clinical bone marrow stromal cell products. J. Transl. Med. 2012;10:23. doi: 10.1186/1479-5876-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guibentif C., Rönn R.E., Böiers C., Lang S., Saxena S., Soneji S., Enver T., Karlsson G., Woods N.B. Single-cell analysis identifies distinct stages of human endothelial-to-hematopoietic transition. Cell Rep. 2017;19:10–19. doi: 10.1016/j.celrep.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 27.Luo Y., Liu C., Cerbini T., San H., Lin Y., Chen G., Rao M.S., Zou J. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl. Med. 2014;3:821–835. doi: 10.5966/sctm.2013-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmermans F., Velghe I., Vanwalleghem L., De Smedt M., Van Coppernolle S., Taghon T., Moore H.D., Leclercq G., Langerak A.W., Kerre T., et al. Generation of T cells from human embryonic stem cell-derived hematopoietic zones. J. Immunol. 2009;182:6879–6888. doi: 10.4049/jimmunol.0803670. [DOI] [PubMed] [Google Scholar]
- 29.Woods N.B., Parker A.S., Moraghebi R., Lutz M.K., Firth A.L., Brennand K.J., Berggren W.T., Raya A., Izpisúa Belmonte J.C., Gage F.H., Verma I.M. Brief report: efficient generation of hematopoietic precursors and progenitors from human pluripotent stem cell lines. Stem Cell. 2011;29:1158–1164. doi: 10.1002/stem.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Y., Izpisua Belmonte J.C. Design approaches for generating organ constructs. Cell Stem Cell. 2019;25:447. doi: 10.1016/j.stem.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Jiang T., Xu G., Wang Q., Yang L., Zheng L., Zhao J., Zhang X. In vitro expansion impaired the stemness of early passage mesenchymal stem cells for treatment of cartilage defects. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frobel J., Hemeda H., Lenz M., Abagnale G., Joussen S., Denecke B., Sarić T., Zenke M., Wagner W. Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells. Stem Cell Rep. 2014;3:414–422. doi: 10.1016/j.stemcr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takebe T., Sekine K., Kimura M., Yoshizawa E., Ayano S., Koido M., Funayama S., Nakanishi N., Hisai T., Kobayashi T., et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Uchida N., Haro-Mora J.J., Fujita A., Lee D.Y., Winkler T., Hsieh M.M., Tisdale J.F. Efficient generation of beta-globin-expressing erythroid cells using stromal cell-derived induced pluripotent stem cells from patients with sickle cell disease. Stem Cell. 2017;35:586–596. doi: 10.1002/stem.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Q.C., Hirst C.E., Costa M., Ng E.S., Schiesser J.V., Gertow K., Stanley E.G., Elefanty A.G. APELIN promotes hematopoiesis from human embryonic stem cells. Blood. 2012;119:6243–6254. doi: 10.1182/blood-2011-12-396093. [DOI] [PubMed] [Google Scholar]
- 36.Slukvin I.I. Hematopoietic specification from human pluripotent stem cells: current advances and challenges toward de novo generation of hematopoietic stem cells. Blood. 2013;122:4035–4046. doi: 10.1182/blood-2013-07-474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uenishi G., Theisen D., Lee J.H., Kumar A., Raymond M., Vodyanik M., Swanson S., Stewart R., Thomson J., Slukvin I. Tenascin C promotes hematoendothelial development and T lymphoid commitment from human pluripotent stem cells in chemically defined conditions. Stem Cell Rep. 2014;3:1073–1084. doi: 10.1016/j.stemcr.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi K.D., Vodyanik M.A., Togarrati P.P., Suknuntha K., Kumar A., Samarjeet F., Probasco M.D., Tian S., Stewart R., Thomson J.A., Slukvin I.I. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ditadi A., Sturgeon C.M., Tober J., Awong G., Kennedy M., Yzaguirre A.D., Azzola L., Ng E.S., Stanley E.G., French D.L., et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernardo A.S., Faial T., Gardner L., Niakan K.K., Ortmann D., Senner C.E., Callery E.M., Trotter M.W., Hemberger M., Smith J.C., et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krendl C., Shaposhnikov D., Rishko V., Ori C., Ziegenhain C., Sass S., Simon L., Müller N.S., Straub T., Brooks K.E., et al. GATA2/3-TFAP2A/C transcription factor network couples human pluripotent stem cell differentiation to trophectoderm with repression of pluripotency. Proc. Natl. Acad. Sci. USA. 2017;114:E9579–E9588. doi: 10.1073/pnas.1708341114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lilly A.J., Costa G., Largeot A., Fadlullah M.Z.H., Lie-A-Ling M., Lacaud G., Kouskoff V. Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development. 2016;143:4341–4351. doi: 10.1242/dev.140970. [DOI] [PubMed] [Google Scholar]
- 43.Marchand M., Horcajadas J.A., Esteban F.J., McElroy S.L., Fisher S.J., Giudice L.C. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol. Reprod. 2011;84:1258–1271. doi: 10.1095/biolreprod.110.086413. [DOI] [PubMed] [Google Scholar]
- 44.Uenishi G.I., Jung H.S., Kumar A., Park M.A., Hadland B.K., McLeod E., Raymond M., Moskvin O., Zimmerman C.E., Theisen D.J., et al. NOTCH signaling specifies arterial-type definitive hemogenic endothelium from human pluripotent stem cells. Nat. Commun. 2018;9:1828. doi: 10.1038/s41467-018-04134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vodyanik M.A., Yu J., Zhang X., Tian S., Stewart R., Thomson J.A., Slukvin I.I. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton G.J., Hempstock J., Jauniaux E. Nutrition of the human fetus during the first trimester--a review. Placenta. 2001;22:S70–S77. doi: 10.1053/plac.2001.0639. [DOI] [PubMed] [Google Scholar]
- 47.Pereda T J., Motta P.M. New advances in human embryology: morphofunctional relationship between the embryo and the yolk sac. Med. Electron. Microsc. 1999;32:67–78. doi: 10.1007/s007950050011. [DOI] [PubMed] [Google Scholar]
- 48.Burke K.A., Jauniaux E., Burton G.J., Cindrova-Davies T. Expression and immunolocalisation of the endocytic receptors megalin and cubilin in the human yolk sac and placenta across gestation. Placenta. 2013;34:1105–1109. doi: 10.1016/j.placenta.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldie L.C., Nix M.K., Hirschi K.K. Embryonic vasculogenesis and hematopoietic specification. Organogenesis. 2008;4:257–263. doi: 10.4161/org.4.4.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyer M.A., Farrington S.M., Mohn D., Munday J.R., Baron M.H. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development. 2001;128:1717–1730. doi: 10.1242/dev.128.10.1717. [DOI] [PubMed] [Google Scholar]
- 51.Nakajima-Takagi Y., Osawa M., Oshima M., Takagi H., Miyagi S., Endoh M., Endo T.A., Takayama N., Eto K., Toyoda T., et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood. 2013;121:447–458. doi: 10.1182/blood-2012-05-431403. [DOI] [PubMed] [Google Scholar]
- 52.Jang I.H., Lu Y.F., Zhao L., Wenzel P.L., Kume T., Datta S.M., Arora N., Guiu J., Lagha M., Kim P.G., et al. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood. 2015;125:1418–1426. doi: 10.1182/blood-2014-04-568170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung A., Zulick E., Skvir N., Vanuytsel K., Morrison T.A., Naing Z.H., Wang Z., Dai Y., Chui D.H.K., Steinberg M.H., et al. Notch and aryl hydrocarbon receptor signaling impact definitive hematopoiesis from human pluripotent stem cells. Stem Cell. 2018;36:1004–1019. doi: 10.1002/stem.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy M., Awong G., Sturgeon C.M., Ditadi A., LaMotte-Mohs R., Zúñiga-Pflücker J.C., Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Ledran M.H., Krassowska A., Armstrong L., Dimmick I., Renström J., Lang R., Yung S., Santibanez-Coref M., Dzierzak E., Stojkovic M., et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Dong C., Beltcheva M., Gontarz P., Zhang B., Popli P., Fischer L.A., Khan S.A., Park K.M., Yoon E.J., Xing X., et al. Derivation of trophoblast stem cells from naive human pluripotent stem cells. Elife. 2020;9 doi: 10.7554/eLife.52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kubaczka C., Senner C.E., Cierlitza M., Araúzo-Bravo M.J., Kuckenberg P., Peitz M., Hemberger M., Schorle H. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell. 2015;17:557–568. doi: 10.1016/j.stem.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. doi: 10.1016/j.stem.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Xu R.H., Chen X., Li D.S., Li R., Addicks G.C., Glennon C., Zwaka T.P., Thomson J.A. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 60.Ivanovs A., Rybtsov S., Ng E.S., Stanley E.G., Elefanty A.G., Medvinsky A. Human haematopoietic stem cell development: from the embryo to the dish. Development. 2017;144:2323–2337. doi: 10.1242/dev.134866. [DOI] [PubMed] [Google Scholar]
- 61.Dou D.R., Calvanese V., Sierra M.I., Nguyen A.T., Minasian A., Saarikoski P., Sasidharan R., Ramirez C.M., Zack J.A., Crooks G.M., et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nat. Cell Biol. 2016;18:595–606. doi: 10.1038/ncb3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palis J., Robertson S., Kennedy M., Wall C., Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 63.Palis J., Yoder M.C. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp. Hematol. 2001;29:927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 64.Fidanza A., Stumpf P.S., Ramachandran P., Tamagno S., Babtie A., Lopez-Yrigoyen M., Taylor A.H., Easterbrook J., Henderson B.E.P., Axton R., et al. Single-cell analyses and machine learning define hematopoietic progenitor and HSC-like cells derived from human PSCs. Blood. 2020;136:2893–2904. doi: 10.1182/blood.2020006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng Y., He J., Bai Z., Li Z., Gong Y., Liu C., Ni Y., Du J., Ma C., Bian L., et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 2019;29:881–894. doi: 10.1038/s41422-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Alegria E., Menegatti S., Fadlullah M.Z.H., Menendez P., Lacaud G., Kouskoff V. Early human hemogenic endothelium generates primitive and definitive hematopoiesis in vitro. Stem Cell Rep. 2018;11:1061–1074. doi: 10.1016/j.stemcr.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitjavila-Garcia M.T., Cailleret M., Godin I., Nogueira M.M., Cohen-Solal K., Schiavon V., Lecluse Y., Le Pesteur F., Lagrue A.H., Vainchenker W. Expression of CD41 on hematopoietic progenitors derived from embryonic hematopoietic cells. Development. 2002;129:2003–2013. doi: 10.1242/dev.129.8.2003. [DOI] [PubMed] [Google Scholar]
- 68.Azcoitia V., Aracil M., Martínez-A C., Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 2005;280:307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Castaño J., Aranda S., Bueno C., Calero-Nieto F.J., Mejia-Ramirez E., Mosquera J.L., Blanco E., Wang X., Prieto C., Zabaleta L., et al. GATA2 promotes hematopoietic development and represses cardiac differentiation of human mesoderm. Stem Cell Rep. 2019;13:515–529. doi: 10.1016/j.stemcr.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sitnicka E., Buza-Vidas N., Larsson S., Nygren J.M., Liuba K., Jacobsen S.E.W. Human CD34+ hematopoietic stem cells capable of multilineage engrafting NOD/SCID mice express flt3: distinct flt3 and c-kit expression and response patterns on mouse and candidate human hematopoietic stem cells. Blood. 2003;102:881–886. doi: 10.1182/blood-2002-06-1694. [DOI] [PubMed] [Google Scholar]
- 71.Suri C., Jones P.F., Patan S., Bartunkova S., Maisonpierre P.C., Davis S., Sato T.N., Yancopoulos G.D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 72.Yamane T. Mouse yolk sac hematopoiesis. Front. Cell Dev. Biol. 2018;6:80. doi: 10.3389/fcell.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haro-Mora J.J., Uchida N., Demirci S., Wang Q., Zou J., Tisdale J.F. Biallelic correction of sickle cell disease-derived induced pluripotent stem cells (iPSCs) confirmed at the protein level through serum-free iPS-sac/erythroid differentiation. Stem Cells Transl. Med. 2020;9:590–602. doi: 10.1002/sctm.19-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merling R.K., Sweeney C.L., Chu J., Bodansky A., Choi U., Priel D.L., Kuhns D.B., Wang H., Vasilevsky S., De Ravin S.S., et al. An AAVS1-targeted minigene platform for correction of iPSCs from all five types of chronic granulomatous disease. Mol. Ther. 2015;23:147–157. doi: 10.1038/mt.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sagy N., Slovin S., Allalouf M., Pour M., Savyon G., Boxman J., Nachman I. Prediction and control of symmetry breaking in embryoid bodies by environment and signal integration. Development. 2019;146 doi: 10.1242/dev.181917. [DOI] [PubMed] [Google Scholar]
- 76.Muncie J.M., Ayad N.M.E., Lakins J.N., Xue X., Fu J., Weaver V.M. Mechanical tension promotes formation of gastrulation-like nodes and patterns mesoderm specification in human embryonic stem cells. Dev. Cell. 2020;55:679–694.e11. doi: 10.1016/j.devcel.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin X., Mead B.E., Safaee H., Langer R., Karp J.M., Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasaai B., Caolo V., Peacock H.M., Lehoux S., Gomez-Perdiguero E., Luttun A., Jones E.A.V. Erythro-myeloid progenitors can differentiate from endothelial cells and modulate embryonic vascular remodeling. Sci. Rep. 2017;7 doi: 10.1038/srep43817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palis J., Yoder M.C. Endothelial cells transition to blood cells but probably not back again. Circ. Res. 2020;127:1233–1235. doi: 10.1161/circresaha.120.318113. [DOI] [PubMed] [Google Scholar]
- 80.Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 81.Motazedian A., Bruveris F.F., Kumar S.V., Schiesser J.V., Chen T., Ng E.S., Chidgey A.P., Wells C.A., Elefanty A.G., Stanley E.G. Multipotent RAG1+ progenitors emerge directly from haemogenic endothelium in human pluripotent stem cell-derived haematopoietic organoids. Nat. Cell Biol. 2020;22:60–73. doi: 10.1038/s41556-019-0445-8. [DOI] [PubMed] [Google Scholar]
- 82.Wimmer R.A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hämmerle M., Esk C., Bagley J.A., et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Philonenko E.S., Tan Y., Wang C., Zhang B., Shah Z., Zhang J., Ullah H., Kiselev S.L., Lagarkova M.A., Li D., et al. Recapitulative haematopoietic development of human pluripotent stem cells in the absence of exogenous haematopoietic cytokines. J. Cell Mol. Med. 2021;25:8701–8714. doi: 10.1111/jcmm.16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawamoto H., Masuda K., Nagano S. Regeneration of antigen-specific T cells by using induced pluripotent stem cell (iPSC) technology. Int. Immunol. 2021;33:827–833. doi: 10.1093/intimm/dxab091. [DOI] [PubMed] [Google Scholar]
- 85.Jing R., Scarfo I., Najia M.A., Lummertz da Rocha E., Han A., Sanborn M., Bingham T., Kubaczka C., Jha D.K., Falchetti M., et al. EZH1 repression generates mature iPSC-derived CAR T cells with enhanced antitumor activity. Cell Stem Cell. 2022;29:1181–1196.e6. doi: 10.1016/j.stem.2022.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goldenson B.H., Hor P., Kaufman D.S. iPSC-derived natural killer cell therapies - expansion and targeting. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.841107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 88.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 92.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brault J., Vigne B., Stasia M.J. Ex vivo models of chronic granulomatous disease. Methods Mol. Biol. 2019;1982:587–622. doi: 10.1007/978-1-4939-9424-3_35. [DOI] [PubMed] [Google Scholar]
- 96.Isonaka R., Sullivan P., Jinsmaa Y., Corrales A., Goldstein D.S. Spectrum of abnormalities of sympathetic tyrosine hydroxylase and alpha-synuclein in chronic autonomic failure. Clin. Auton. Res. 2018;28:223–230. doi: 10.1007/s10286-017-0495-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw and processed data reported in this publication are archived at NCBI GEO (accession number GSE157140).
-
•
Analysis code is available as supplemental files (Data S1).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.