Abstract
A strain of Streptococcus pyogenes resistant to multiple fluoroquinolones was isolated from the blood of an immunocompromised patient. Resistance to fluoroquinolones in S. pyogenes has not been previously studied. Compared to 10 sensitive strains of S. pyogenes, the fluoroquinolone-resistant clinical isolate of S. pyogenes presented point mutations in gyrA, predicting that serine-81 was changed to phenylalanine and that methionine-99 was changed to leucine, and in parC, predicting that serine-79 was changed to tyrosine. The mechanism of fluoroquinolone resistance in this isolate of S. pyogenes appears to be analogous to previously reported mechanisms for Streptococcus pneumoniae.
Development of penicillin resistance in Streptococcus pneumoniae has prompted a search for alternative effective therapy for infections caused by this organism (4, 12, 13). Newer fluoroquinolones have demonstrated excellent activity against penicillin-sensitive and penicillin-resistant S. pneumoniae strains. However, with the increasing use of fluoroquinolones, there have been reports of emergence of S. pneumoniae isolates with resistance to this class of antibiotics (3, 7, 8, 12, 16). In contrast to S. pneumoniae, Streptococcus pyogenes remains uniformly sensitive to penicillin despite intensive exposure to the agent, and penicillin remains the drug of choice for infections caused by S. pyogenes (15). For this reason, susceptibility testing of S. pyogenes isolates is not routinely performed. Resistance to fluoroquinolones among S. pyogenes isolates has not been reported previously, though slightly increased MICs of sparfloxacin (10) and ciprofloxacin and levofloxacin (2) have been described elsewhere. We report here a clinical strain of S. pyogenes (NIH-R01-GAS) isolated from an immunocompromised patient who had received repeated antibiotic treatment including levofloxacin for various infections. This isolate was found to be highly resistant to several fluoroquinolones, and analysis of gyrA and parC gene sequences from the isolate indicated that point mutations along the quinolone resistance-determining regions (QRDRs) were the probable mechanism for its resistance.
Case history.
The patient was an eighteen-year-old black male with hyper-immunoglobulin E recurrent infection (Job's syndrome) diagnosed at age four who has been previously described (6). He had had multiple recurrent pulmonary and sinus infections requiring multiple courses of long-term therapy and prophylactic antibiotics. One month prior to admission, he had extensive bilateral inguinal crease infections. Empiric therapy with levofloxacin, 500 mg orally daily, was initiated. Wound cultures subsequently grew S. pyogenes. One month later, while still on levofloxacin, he complained of headaches, fever (40.1°C), and purulent drainage from his right ear and nose. Blood cultures at this time grew S. pyogenes resistant to levofloxacin. He was admitted for 10 days for intravenous administration of vancomycin, and other antibiotics were discontinued. An echocardiogram was negative for any vegetations, and ophthalmic examination revealed no Roth spots. Blood cultures taken after completion of vancomycin therapy were negative.
All isolates of S. pyogenes (ATCC 700294, 12384, and 12344; the fluoroquinolone-resistant blood isolate; and seven fluoroquinolone-sensitive clinical isolates from a community hospital) were initially grown on 5% sheep blood plates (Remel, Lenexa, Kans.) in the presence of 5% CO2 at 35°C. The original isolate from the patient's wound cultures was unavailable for further investigation. Antimicrobial susceptibility was determined by a frozen microdilution MicroStrep panel (Dade Behring, Inc., West Sacramento, Calif.), the Etest (AB Biodisk, Solna, Sweden), or the Kirby-Bauer (KB) disk (Becton Dickinson, Cockeysville, Md.) diffusion methods following manufacturers' or NCCLS recommendations (5). Interpretation of susceptibility was made according to NCCLS standards whenever available (5). Quality control for all methods of susceptibility testing was performed using S. pneumoniae strain ATCC 49619, and results were within acceptable limits.
Mutational alterations in the QRDRs of gyrA and parC of NIH-R01-GAS were investigated by PCR using Ready-To-Go PCR Beads (Pharmacia Biotech, Piscataway, N.J.) with chromosomal DNA as template and subsequent DNA sequencing (Perkin-Elmer, Applied Biosystems, Foster City, Calif.). For amplification of a 614-bp fragment of gyrA containing the QRDR, a pair of primers (5′ GCAAGATCGAAATTTAATTGACGTC, nucleotides 3 to 27, and 5′ ACTCTCTTGTTGTACAGTCTGG, nucleotides 595 to 616) was used. For the amplification of the QRDR of parC of S. pyogenes, primers 5′ ATGTCAAACATTCAAAACATGTCC, nucleotides 1 to 24, and 5′ AGCCTGCGGAAATACCAGAAG, nucleotides 500 to 520, were used to amplify a 520-bp fragment.
The levofloxacin-resistant isolate NIH-R01-GAS was sensitive to other antibiotics in the MicroStrep panel (azithromycin, ceftriaxone, chloramphenicol, clindamycin, penicillin, tetracycline, and vancomycin) according to NCCLS criteria (5). Susceptibility testing by Etest, however, demonstrated that the NIH-R01-GAS isolate was resistant to trovafloxacin, levofloxacin, and grepafloxacin as defined by NCCLS criteria (5) (Table 1). High MICs of ciprofloxacin, sparfloxacin, and norfloxacin were also found, and there were no zones of inhibition around the KB disks for enrofloxacin, lomefloxacin, and ofloxacin (Table 1). A MIC of 1.0 μg/ml suggested that the isolate was sensitive to clinafloxacin. Low MICs (≤2.0 μg/ml) and/or large KB disk zone sizes (≥19 mm) for all fluoroquinolones tested, indicative of sensitivity, were found for the three ATCC strains and the seven additional clinical isolates that were tested.
TABLE 1.
Fluoroquinolone susceptibilities of the ATCC strain and clinical isolate of S. pyogenes determined by disk diffusion and Etest
| Quinolone | Result for straina:
|
|||
|---|---|---|---|---|
| ATCC 700294
|
NIH-R01-GAS
|
|||
| KB disk (mm) | Etest (μg/ml) | KB disk (mm) | Etest (μg/ml) | |
| Ciprofloxacin | 21 | 0.50 | No zone | >32 |
| Clinafloxacin | 23 | 0.10 | 19 | 1.00 |
| Enrofloxacin | 13 | ND | No zone | ND |
| Grepafloxacin | 21 | 0.50 | No zone | >32 |
| Levofloxacin | 20 | 0.50 | No zone | >32 |
| Lomefloxacin | 16 | ND | No zone | ND |
| Norfloxacin | 20 | 3.00 | No zone | >256 |
| Ofloxacin | 18 | ND | No zone | ND |
| Sparfloxacin | 21 | 0.50 | No zone | >32 |
| Trovafloxacin | 21 | 0.13 | No zone | >32 |
Results for disk diffusion are expressed as diameters of zones of inhibition. Results for Etest are expressed as MICs of quinolones. ND, not determined.
The gyrA and parC genes of S. pneumoniae, encoding DNA gyrase A and topoisomerase IV subunit C, respectively, have been well characterized elsewhere (1, 9, 11). The genome of S. pyogenes ATCC 700294 is currently being sequenced at the University of Oklahoma (Streptococcus pyogenes Genome Project). For defining gyrA in S. pyogenes, the nucleic acid sequences of gyrA (1) and parC (11) from S. pneumoniae were used to search the Streptococcus pyogenes Genome Project Database. Based on homology with the counterpart gyrA genes of S. pneumoniae, Staphylococcus aureus, and Escherichia coli, the putative open reading frame of gyrA of S. pyogenes was defined as a gene of 2,487 bp, encoding a protein of 829 residues. The putative promoter of gyrA of S. pyogenes has a striking similarity to that of gyrA of S. pneumoniae (1). Extended putative −10 (TATGGTATAAT) (1) and −35 (CTGATAA) regions were identified upstream of the start codon ATG. The deduced amino acid sequence of the gyrase subunit A of S. pyogenes demonstrated 79% identity with GyrA of S. pneumoniae (1). However, identity was 88% when the first 400 amino acids in the N-terminal region were compared based on the genes from these two species (data not shown). The open reading frame of parC of S. pyogenes is a gene of 2,460 bp encoding a protein of 820 residues. The deduced amino acid sequence of subunit C of topoisomerase IV had 82% identity among the first 620 residues between S. pyogenes and S. pneumoniae (data not shown).
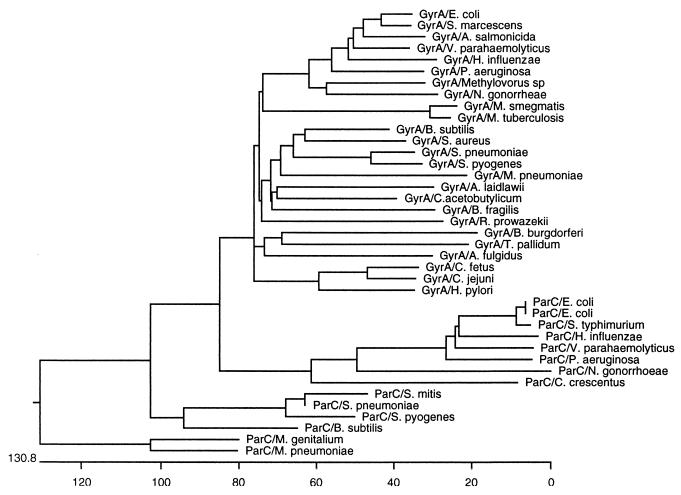
A phylogenetic protein tree was constructed based on amino acid sequences available from GenBank using an unbalanced method provided by the computer program MegAlign (DNAStar, Inc. Madison, Wis.). Both GyrA and ParC of S. pyogenes were most closely related to those of S. pneumoniae (Fig. 1). Based on the available data, gyrase A of S. pyogenes is next most closely related to those of S. aureus and Bacillus subtilis; ParC is next most closely related to that of Streptococcus mitis. These data are in agreement with but expand the data from the phylogenetic comparisons of these two genes reported by Balas et al. (1).
FIG. 1.
Protein tree for the full-length gyrase A and ParC subunit of topoisomerase IV of several bacterial species. The protein sequences are from GenBank. The phylogenetic distance was determined using the unbalanced method provided by the computer program MegAlign. Lengths of branches correspond to sequence divergence. The scale underneath the tree represents the actual number of amino acid substitutions.
All 10 fluoroquinolone-sensitive ATCC and clinical isolates demonstrated identical amino acid sequences for the QRDRs of both gyrA and parC (data not shown). In contrast, mutations were identified in both gyrA and parC in the isolate NIH-R01-GAS. Specifically, two point mutations within the QRDR were identified in gyrA, with codon TCT (Ser-81, location designation for S. pyogenes ATCC 700294) being replaced by TTT (Phe) and ATG (Met-99) being replaced by CTG (Leu). Only a single point mutation was found in the QRDR of parC, in which TAC (Tyr) replaced the codon TCC (Ser-79). Resistance to fluoroquinolones usually results from mutations in the QRDRs of either gyrA or parC, or both genes, particularly at the highly conserved residues Ser-83 and Asp-87 (positions refer to those of E. coli) (14, 18). Munoz and De La Campa (11) demonstrated that most ciprofloxacin-resistant S. pneumoniae strains in their study had alterations at Ser-79 (analogous to Ser-83 of E. coli or Ser-81 of S. pyogenes), and the amino acid replacing the serine residue was either phenylalanine or tyrosine. This observation has also been reported by other investigators studying fluoroquinolone resistance in S. pneumoniae (9, 12). Therefore, the quinolone-resistant isolate of S. pyogenes has developed mutational alterations of key topoisomerases analogous to those reported for quinolone resistance of S. pneumoniae.
Quinolone resistance in S. pneumoniae arises through mutations of parC (and/or parE) before changes in gyrA occur, suggesting that topoisomerase IV is the primary target for the fluoroquinolones in this organism (8, 12). In the quinolone-resistant isolate of S. pyogenes in this study, mutations were identified in both gyrA and parC, which may explain its high level of resistance to fluoroquinolones. Because the resistant strain in the current study presented mutations at both sites at the time of isolation, we cannot determine the sequence of genetic transition from quinolone sensitive to resistant for these two target genes. The resistant isolate in this study demonstrated no sensitivity to all available fluoroquinolones tested, except to clinafloxacin. The superior activity of clinafloxacin has also been previously observed for S. pneumoniae (9, 13). Clinafloxacin is a novel C-8-substituted fluoroquinolone and is highly active against S. pneumoniae (17). Pan and Fisher have demonstrated that, in S. pneumoniae, neither gyrA nor parC quinolone-resistance-conferring mutants alone confer increased resistance to clinafloxacin (13). Laboratory experiments have shown that four consecutive mutational steps are required to induce significant resistance to clinafloxacin, while only two steps are required to achieve the same level of resistance for ciprofloxacin and three steps are required for sparfloxacin resistance (13). Compared to other tested quinolones, the mutations identified in the gyrA and parC genes of the resistant clinical isolate of S. pyogenes had less effect on the activity of clinafloxacin, suggesting a potential clinical advantage for clinafloxacin.
Nucleotide sequence accession number.
The DNA sequences obtained from this study were submitted to GenBank under accession no. AF220945, AF220946, AF222013, and AF223159.
Acknowledgments
We thank Michael L. Pendrak for his assistance in the construction of the phylogenetic tree of gyrase A and ParC proteins and Julie E. Niemela and Jodie M. Keary for assistance in nucleotide sequencing.
REFERENCES
- 1.Balas D, Fernandez-Moreira E, De La Campa A G. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J Bacteriol. 1998;180:2854–2861. doi: 10.1128/jb.180.11.2854-2861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondeau J M. A review of the comparative in-vitro activities of 12 antimicrobial agents, with a focus on five new respiratory quinolones. J Antimicrob Chemother. 1999;43(Suppl. B):1–11. doi: 10.1093/jac/43.suppl_2.1. [DOI] [PubMed] [Google Scholar]
- 3.Chen D K, McGeer A, de Azavedo J C, Low E E the Canadian Bacterial Surveillance Network. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian Bacterial Surveillance Network. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 4.Drlica K. Mechanism of fluoroquinolone action. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 5.Ferraro M J, Craig W A, Dudley M N, Eliopoulos G, Hecht D W, Hindler J, Reller L B, Sheldon A T, Swenson J M, Tenover F C, Testa R T, Weinstein M P, Wikler M A. NCCLS (ed.), Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th ed. M7-A5 and M100-S10. Wayne, Pa: NCCLS; 2000. [Google Scholar]
- 6.Grimbacher B, Holland S, Gallin J, Greenberg F, Hill S, Malech H, Miller J, O'Connell A, Puck J. Hyper-IgE syndrome with recurrent infections—an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 7.Ho P L, Que T L, Tsang D N, Ng T K, Chow K H, Seto W H. Emergence of fluoroquinolone resistance among multiply resistant strains of Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1999;43:1310–1313. doi: 10.1128/aac.43.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janoir C, Zeller V, Kitzis M D, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen J H, Weigel L M, Ferraro M J, Swenson J M, Tenover F C. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob Agents Chemother. 1999;43:329–334. doi: 10.1128/aac.43.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louie A, Baltch A L, Ritz W J, Smith R P. In vitro activity of sparfloxacin and six reference antibiotics against gram-positive bacteria. Chemotherapy. 1991;37:275–282. doi: 10.1159/000238867. [DOI] [PubMed] [Google Scholar]
- 11.Munoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan X S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens D L. Invasive group A streptococcus infections. Clin Infect Dis. 1992;14:2–11. doi: 10.1093/clinids/14.1.2. [DOI] [PubMed] [Google Scholar]
- 16.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waites K, Brookings E, Nix S, Robinson A, Gray B, Swiatlo E. Comparative in vitro activities of four new fluoroquinolones against Streptococcus pneumoniae determined by Etest. Int J Antimicrob Agents. 1998;9:215–218. doi: 10.1016/s0924-8579(97)00053-8. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]