Key Points
-
•
Del(1p32) is an independent and important adverse prognostic factor in myeloma.
-
•
Biallelic deletion of 1p32 dramatically worsens the prognosis compared with a monoallelic loss.
Visual Abstract
Abstract
Cytogenetic abnormalities (CAs) are known to be the preponderant prognostic factor in multiple myeloma. Our team has recently developed a prognostic score based on 6 CAs, with which del(1p32) appears to be the second worst abnormality after del(17p). This study aimed to confirm the adverse effect of 1p32 deletion in patients with newly diagnosed multiple myeloma (NDMM). Among 2551 patients with newly diagnosed multiple myeloma, 11% were harboring del(1p32). Their overall survival (OS) was significantly inferior compared with patients without del(1p32) (median OS: 49 months vs 124 months). Likewise, progression-free survival was significantly shorter. More importantly, biallelic del(1p32) conferred a dramatically poorer prognosis than a monoallelic del(1p32) (median OS: 25 months vs 60 months). As expected, the OS of patients with del(1p32) significantly decreased when this abnormality was associated with other high-risk CAs [del(17p), t(4;14), or gain(1q)]. In the multivariate analysis, del(1p32) appeared as a negative prognostic factor; after adjustment for age and treatment, the risk of progression was 1.3 times higher among patients harboring del(1p32), and the risk of death was 1.9 times higher. At the dawn of risk-adapted treatment strategies, we have confirmed the adverse effect of del(1p32) in multiple myeloma and the relevance of its assessment at diagnosis.
In this month’s CME article, Schavgoulidze et al confirm the adverse impact of 1p32 deletion in patients newly diagnosed with multiple myeloma (NDMM). In a study of 2551 patients with NDMM, the 11% harboring del(1p32) have inferior progression-free and overall survival (OS). Biallelic del(1p32) confirmed a dramatically worse prognosis, with a median OS of only 25 months. Detection of biallelic del(1p32) at diagnosis defines an ultra–high-risk subgroup of NDMM.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited with commendation by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
Successful completion of this CME activity, which includes participation in the evaluation component, enables the participant to earn up to 1.0 MOC points in the American Board of Internal Medicine's (ABIM) Maintenance of Certification (MOC) program. Participants will earn MOC points equivalent to the amount of CME credits claimed for the activity. It is the CME activity provider's responsibility to submit participant completion information to ACCME for the purpose of granting ABIM MOC credit.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at https://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 1367.
Disclosures
Laurie Barclay, MD, freelance writer and reviewer, Medscape, LLC, has disclosed the following relevant financial relationships: stock, stock options, or bonds: AbbVie Inc. (former).
Learning Objectives
Upon completion of this activity, participants will have increased knowledge regarding the:
-
1.
Impact of monoallelic and biallelic del(1p32) on overall survival and progression-free survival among patients with newly diagnosed multiple myeloma (NDMM)
-
2.
Prognostic value of other high-risk cytogenetic abnormalities (CAs) in combination with del(1p32) among patients with NDMM, according to a study using a prognostic score developed from 6 CAs
-
3.
Clinical implications of the impact of del(1p32) on patients with NDMM
Release date March 16, 2023; Expiration date: March 16, 2024
Introduction
Multiple myeloma (MM) is the second most frequent hematological malignancy in Western countries. The exceptional development in patient treatments has led to a significant improvement in overall survival (OS), both for patients who are eligible for transplant and for those who are not.1 However, this improvement does not benefit patients at high risk who still represent an unmet medical need. It is unanimously recognized that cytogenetic abnormalities (CAs) displayed by malignant plasma cells (PCs) have a strong prognostic impact. Deletion 17p [del(17p)] is known to be the most unfavorable CA and affects ∼8% of patients with newly diagnosed MM (NDMM). In 2015, del(17p) was integrated into the criteria of the revised version of the International Staging System (ISS) score, as well as 2 other CAs, translocations t(4;14) and t(14;16).2
CAs affecting chromosome 1, gain(1q) and del(1p32), were not included in these new criteria despite their relatively high frequencies, 35% and 11%, respectively, in patients with NDMM. However, previous or subsequent studies have distinctly demonstrated their negative effect on patients.3, 4, 5 Moreover, Perrot et al have recently confirmed the significant prognostic impact of del(1p32) as being the second most adverse abnormality in myeloma, just after del(17p).6
This study aimed to update our data on the prognostic impact of del(1p32) in a large cohort of patients with NDMM, irrespectively of the therapeutic strategy used.
Methods
The Toulouse Ethics Committee approved the study. Informed consent was obtained for all included patients. Clinical data were obtained from 2551 patients with NDMM recruited within hospitals involved in the Intergroupe Francophone du Myélome, followed up for ≥36 months or having died or progressed within 36 months after treatment. Diagnosis was established between 2010 and 2021, and 1258 patients were treated with intensive therapy. Progression was determined based on criteria defined by the International Myeloma Working Group.7
Bone marrow samples were obtained at diagnosis and shipped overnight to a central laboratory. Upon receipt, PCs were isolated using CD138+ magnetic cells sorting (Miltenyi Biotec, Paris, France). After sorting, purity was assessed by morphology, and only samples with a PC content of ≥70% after sorting were kept for the analysis. The mean purity was 97%. PCs were analyzed either by fluorescence in situ hybridization (FISH) (848 patients), single-nucleotide polymorphism (SNP) arrays (1395 patients), or by next-generation sequencing (NGS) (308 patients), depending on the date of receipt. We proved the equivalence of the sensitivity of all 3 techniques with a method validation imposed by our quality system. Moreover, for each positive del(17p) by SNP array or NGS, an additional FISH analysis was performed to assess the percentage of positive PCs. Del(1p32) was defined by the deletion of at least FAF1 or CDKN2C genes.
For FISH analysis, gain(1q), del(1p32), del(17p), and t(4;14) were detected using specific probes (Cytocell, Paris, France) for copy number, and Abbott Molecular (Paris, France) for translocations. Only del(17p) present in >55% of PCs was considered.8 For the other CAs, the positivity threshold was 30%. This threshold was defined in normal bone marrow PCs.
SNP arrays (Affymetrix, Santa Clara, CA) were performed using the Cytoscan HD Array Kit (Affymetrix), and NGS sequencing was performed using a panel of specific probes targeting regions of interest,9 as previously described.3,10,11
Categorical data were presented as percentages and compared using a chi-square test or Fisher exact test. Continuous variables were described by mean ± standard deviation and compared using the Student t test. Follow-up duration was estimated using the reverse Kaplan-Meier method. OS and progression-free survival (PFS) curves were estimated using the Kaplan-Meier method and were compared using the log-rank test. A univariate Cox model was performed to estimate the hazard ratios (HRs) for each variable along with 95% confidence intervals (95% CI). A multivariable Cox proportional hazard model was performed for all variables that were significant (P < .05) in the univariate analysis and which met the proportionality assumption. Tests were two-sided, and P < .05 was considered significant. All analyses were performed using R version 4.1.1.
Results
Characteristics of the patients are presented in Table 1. A flowchart is provided in supplemental Figure 1 (available on the Blood website). The median follow-up was 67.4 months. No significant differences were observed in terms of regimen repartition between patients according to del(1p32) status.
Table 1.
Characteristics of patients according to deletion-1p32 status
| Patients with del(1p32) | Patients without del(1p32) | P value | |
|---|---|---|---|
| Patients, n | 282 | 2269 | |
| Age at diagnosis, mean (SD), y | 63.8 (10.8) | 63.6 (10.7) | .9876 |
| Sex, n (%) | .6655 | ||
| Male | 169 (59.9) | 1323 (58.4) | |
| Female | 113 (40.1) | 943 (41.6) | |
| Missing | 0 | 3 | |
| ISS, n (%) | .0177 | ||
| 1 | 48 (21.7) | 540 (29.2) | |
| 2 | 94 (42.5) | 795 (42.9) | |
| 3 | 79 (35.7) | 517 (27.9) | |
| Patients, n | 221 | 1852 | |
| Missing, n | 61 | 417 | |
| Del(17p), n (%) | <.0001 | ||
| No | 217 (77.5) | 2064 (91.2) | |
| Yes | 63 (22.5) | 199 (8.8) | |
| Missing | 2 | 6 | |
| t(4;14), n (%) | .8478 | ||
| No | 245 (88.4) | 1966 (87.8) | |
| Yes | 32 (11.6) | 272 (12.2) | |
| Missing | 5 | 31 | |
| Gain(1q), n (%) | <.0001 | ||
| No | 127 (45.0) | 1502 (66.2) | |
| Yes | 155 (55.0) | 766 (33.8) | |
| Missing | 0 | 1 | |
| t(14;16), n (%) | .0016 | ||
| No | 91 (87.5) | 689 (95.6) | |
| Yes | 13 (12.5) | 32 (4.4) | |
| Missing | 178 | 1,548 | |
| t(14;20), n (%) | .6969 | ||
| No | 67 (98.5) | 301 (96.8) | |
| Yes | 1 (1.5) | 10 (3.2) | |
| Missing | 214 | 1,958 | |
| TP53mutation, n (%) | .0010 | ||
| No | 55 (72.4) | 288 (88.1) | |
| Yes | 21 (27.6) | 39 (11.9) | |
| Missing | 206 | 1,942 | |
| Hyperdiploidy (>50 chromosomes), n (%) | .0065 | ||
| No | 188 (69.9) | 1336 (61.1) | |
| Yes | 81 (30.1) | 849 (38.9) | |
| Missing | 13 | 84 | |
| First-line therapy, n (%) | |||
| Patients eligible for transplant | |||
| IMiD or PI without anti-CD38 | 20 (14.9) | 178 (15.8) | .1061 |
| Triplet∗ | 104 (77.6) | 905 (80.6) | |
| Triplet + anti-CD38 | 10 (7.5) | 41 (3.6) | |
| Patients not eligible for transplant | |||
| IMiD or PI without anti-CD38 | 84 (59.2) | 716 (64.0) | .0819 |
| IMiD or PI with anti-CD38 | 11 (7.7) | 41 (3.7) | |
| Triplet | 44 (31.0) | 350 (31.3) | |
| Triplet + anti-CD38 | 3 (2.1) | 12 (1.0) | |
| Missing | 6 | 26 | |
| Maintenance therapy for patients eligible for transplant, n (%) | .3161 | ||
| No | 77 (57.5) | 696 (61.9) | |
| Yes | 57 (42.5) | 428 (38.1) | |
| Missing | 0 | 0 |
IMiDs, immunomodulatory drugs; ISS, International Staging System; PI, proteasome inhibitors; SD, standard deviation.
Triplet is defined by the association of 1 IMiD, 1 PI, and 1 corticosteroid (mostly dexamethasone).
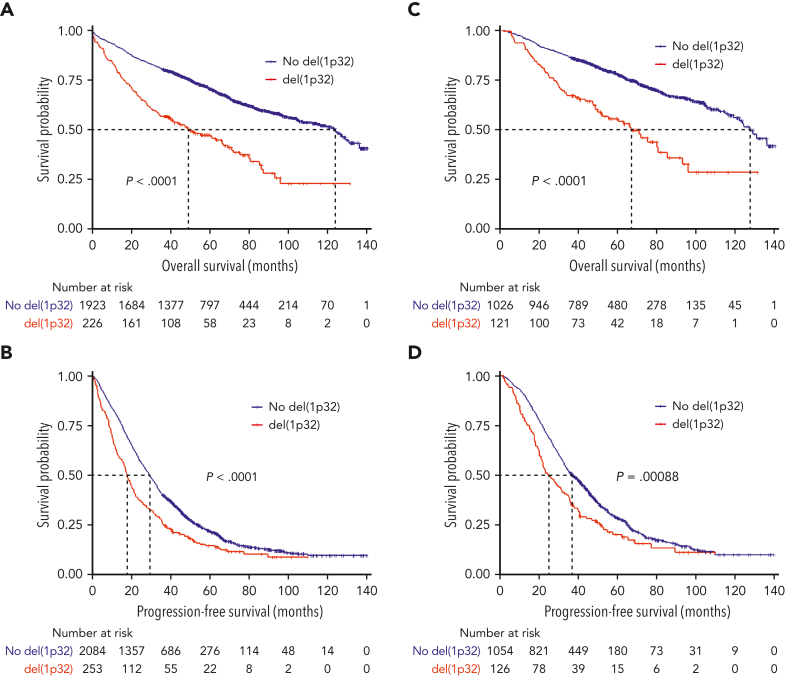
In the analyzed cohort, 11.1% of the patients displayed del(1p32), which was the expected proportion. The OS of patients harboring del(1p32) was significantly inferior compared with patients lacking the del(1p32) (median OS, 49.1 and 123.9 months, respectively; P < .0001) (Figure 1A). Likewise, PFS was significantly shorter in patients with del(1p32) (median PFS 17.7 and 29.2 months, respectively; P < .0001) (Figure 1B). These poorer outcomes were also observed when we focused on patients treated with an intensive therapy (del(1p32) vs no del(1p32); median PFS, 24.9 vs 36.8 months, P = .0009; median OS, 66.9 vs 127.4 months, P < .0001) (Figure 1C-D).
Figure 1.
Kaplan-Meier survival of patients with NDMM according to del(1p32). The red curve corresponds to patients with del(1p32), the blue curve to patients without del(1p32). P values are determined by the log-rank test comparison. (A) Overall survival of patients with NDMM, irrespective of the treatment. (B) Progression-free survival of patients with NDMM, irrespective of the treatment. (C) Overall survival of patients treated with high-dose melphalan. (D) Progression-free survival of patients treated with high-dose melphalan.
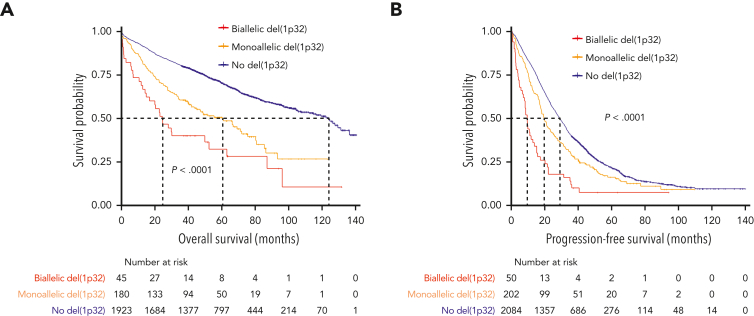
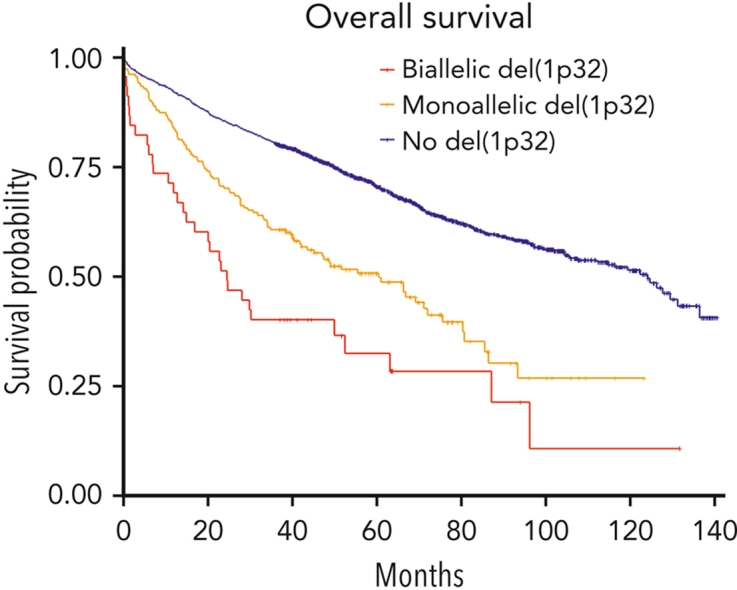
We then postulated that a biallelic del(1p32) should have a greater effect on prognosis than a monoallelic loss. Among 282 patients with del(1p32), a 1p32 copy number was available for 281 patients; we divided them into “biallelic del(1p32)” (0 copy) or “monoallelic del(1p32)” (1 copy). The OS of patients with a biallelic del(1p32) was significantly shorter than that of patients with a monoallelic del(1p32) (median OS, 24.6 and 60.4 months, respectively, P < .0001) (Figure 2A). The same phenomenon was observed for PFS (median PFS, 9.6 and 19.6, respectively, P < .0001) (Figure 2B). These data show that monoallelic loss of 1p32 still had an adverse effect on prognosis, although it was less unfavorable than biallelic loss.
Figure 2.
Kaplan-Meier survival of patients with NDMM according to del(1p32) status. The blue curve corresponds to patients without del(1p32), the yellow curve to patients with a monoallelic del(1p32), and the red curve to patients with a biallelic del(1p32). P values are determined by the log-rank test comparison. (A) Overall survival. (B) Progression-free survival.
To ensure that this effect was not biased by a potential cooccurrence with del(17p), which is known to increase genetic instability, we performed a Fisher exact test. There was no difference in terms of del(17p) frequency between biallelic and monoallelic del(1p32) (27.6% vs 21.3%, P = .49).
Conversely, we had noticed that the proportion of patients harboring gain(1q) was significantly higher in the biallelic del(1p32) group compared with the monoallelic group (76.3% vs 49.1%, P = .0002). Nevertheless, when we focused only on patients harboring gain(1q), there was still a clear discrepancy between biallelic and monoallelic del(1p32) (supplemental Figure 2).
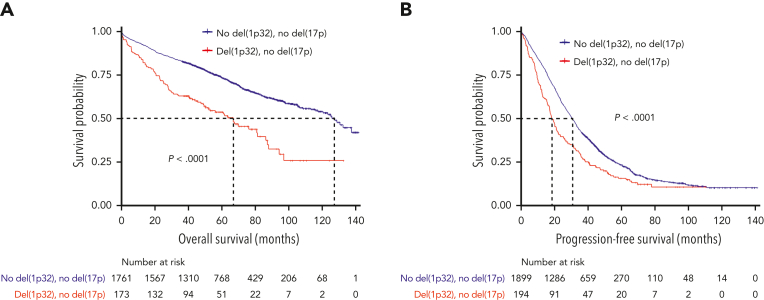
Next, we went back to the whole del(1p32) cohort. Of patients displaying a del(1p32), 22.5% also harbored a del(17p), which is twice the amount seen in the general NDMM population. To confirm that the poor survival was not simply because of this higher level of association, we studied survival according to del(17p) status. In patients without del(17p), outcomes were comparable with what we described in the entire cohort, with significantly decreased PFS and OS for patients with del(1p32) vs those without del(1p32) (median PFS, 18.3 vs 30.4 months, P < .0001; median OS, 66.3 vs 126.0 months, P < .0001) (Figure 3).
Figure 3.
Kaplan-Meier survival of patients with del(17p)-negative NDMM according to del(1p32). The red curve corresponds to patients with del(1p32) and the blue curve to patients without del(1p32). P values are determined by the log-rank test comparison. (A) Overall survival in patients without del(17p). (B) Progression-free survival in patients without del(17p).
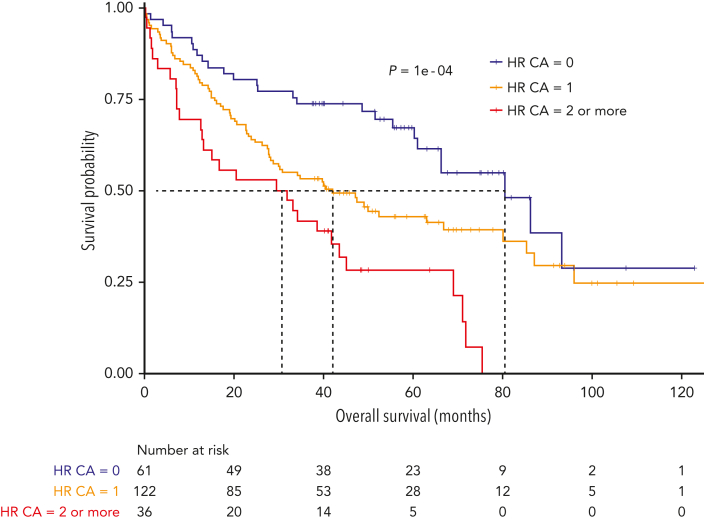
A higher association was also observed with 1q gain. To check whether the poor survival was not simply because of the higher level of association with known adverse cytogenetics, we focused on patients with del(1p32) without the main high-risk (HR) CAs. HR CAs were defined by the presence of del(17p), t(4;14), and/or gain(1q). Patients without HR CAs had a lower PFS and OS when they carried del(1p32) [del(1p32) vs no del(1p32); median PFS, 23.0 vs 34.9 months, P = .0003; median OS, 80.6 vs 136.1 months, P = .0002] (supplemental Figure 3). As previously described, accumulating HR CAs worsen the prognosis.3,6,12, 13, 14 Thus, we have assessed the effect of additional HR CAs on the prognosis of patients with del(1p32). Not surprisingly, the OS of patients with del(1p32) significantly decreased when this abnormality was associated with other HR CAs (median OS: del(1p32) alone, 80.6 months; del(1p32) with 1 HR CA, 42.2 months; del(1p32) with ≥2 HR CAs, 30.8 months; P = .0001) (Figure 4).
Figure 4.
Kaplan-Meier overall survival of patients with NDMM with del(1p32) according to the association with other HR CA. HR CAs are defined by the presence of del(17p), t(4;14), and/or gain(1q). The blue curve corresponds to patients with del(1p32) without other HR CAs, the yellow curve to patients with ≥1 CA, and the red curve to patients with ≥2 other HR CAs. P value is determined by the log-rank test comparison.
In the univariate analysis, the risk of progression and risk of death were both increased for patients with del(1p32), del(17p), gain(1q), or t(4;14) (Table 2). Likewise, the risk was significantly higher when we compared patients in stage ISS II vs ISS I and stage ISS III vs ISS I.
Table 2.
Univariate and multivariate analyses for PFS and OS
| Variable | Univariate analysis |
Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| PFS (n = 1860) |
OS (n = 1721 ) |
PFS (n = 1860) |
OS (n = 1721) |
|||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | 1.0 (1.0-1.0) | <.0001 | 1.0 (1.0-1.0) | <.0001 | ||||
| High-dose melphalan | 1.7 (1.6-1.9) | <.0001 | 1.6 (1.4-1.) | <.0001 | ||||
| ISS II vs I | 1.4 (1.3-1.6) | <.0001 | 1.9 (1.5-2.4) | <.0001 | 1.2 (1.1-1.4) | .0016 | 1.5 (1.2-1.8) | .001 |
| ISS III vs I | 2.0 (1.7-2.3) | <.0001 | 2.9 (2.3-3.6) | <.0001 | 1.7 (1.5-2.0) | <.0001 | 2.1 (1.6-2.6) | <.0001 |
| del(1p32) | 1.5 (1.2-1.7) | <.0001 | 2.5 (2.0-3.1) | <.0001 | 1.3 (1.1-1.5) | .0065 | 1.9 (1.5-2.40) | <.0001 |
| del(17p) | 1.9 (1.6-2.2) | <.0001 | 3.2 (2.6-3.9) | <.0001 | 1.8 (1.5-2.1) | <.0001 | 2.9 (2.3-3.8) | <.0001 |
| t(4;14) | 1.6 (1.4-1.8) | <.0001 | 2.2 (1.8-2.7) | <.0001 | 1.4 (1.2-1.7) | <.0001 | 1.7 (1.4-2.2) | <.0001 |
| Gain(1q) | 1.5 (1.3-1.6) | <.0001 | 1.9 (1.7-2.3) | <.0001 | 1.3 (1.2-1.5) | <.0001 | 1.6 (1.3-1.9) | <.0001 |
CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Finally, in the multivariate analysis, del(1p32) appeared as a factor of poor prognosis. The risk of progression was 1.3 times higher among patients with del(1p32) (P = .0065), and the risk of death was 1.9 higher (P < .0001), after adjustment for age and type of treatment (Table 2).
Discussion
In conclusion, this study confirms the adverse impact of del(1p32) in MM in, to the best of our knowledge, the largest cohort of patients with NDMM ever evaluated. Indeed, we analyzed 282 patients with del(1p32) compared with 85 and 34 patients with del(1p32) in the Hebraud et al and Wang et al studies, respectively.4,5
Most of the studies do not specify the locus of deletion 1p. In our study, we have deliberately focused on locus 1p32. Our previous work,6 done for the construction of a cytogenetic prognostic index (linear predictive score), studied the most recurrent cytogenetic aberrations, including 1p12, 1p22, and 1p32; only del(1p32) was found to be predictive of survival. The locus 1p32 contains important genes such as FAF1, involved in the initiation of apoptosis, or CDKN2C, which prevents cell cycle G1 progression. Deleting these genes could promote tumorigenesis but it still has to be proved in myeloma.
In the case of del(17p), it is well known that the biallelic inactivation of TP53 is more unfavorable than an isolated del(17p). To the best of our knowledge, no study has compared biallelic deletion and monoallelic loss of the 1p32 locus. In our study, we have shown that the biallelic deletion of 1p32 dramatically worsens the prognosis compared with a monoallelic loss. Although the biallelic inactivation of TP53 is mostly owing to the association of a deletion on 1 allele and a mutation on the other allele, we have only seen double-deletion in patients with del(1p32).
Our study also confirms the cumulative risk induced by the association of multiple CAs. Indeed, del(17p), t(4;14), and gain(1q) worsen the prognosis of patients with del(1p32). This was already well described in several studies.6,13,15, 16, 17, 18 The association of del(1p32) with other genetic features such as t(14;16), t(14;20), 1q amplification, or recurrent mutations like TP53 mutation still have to be investigated with larger study populations.
Previously, Hebraud et al focused only on patients eligible for transplant who had received older induction regimens (bortezomib only or vincristine-doxorubicin (Adriamycin)-dexamethasone).4 In our study, we confirmed del(1p32) adverse effects in both patients who are eligible for transplant and those who are not, of whom the latter group had mostly been treated with triplet therapy, which is the current standard of care. However, treatment recommendations are evolving quickly. Currently, we do not have enough data to study the effect of new generations of immunomodulatory drugs and proteasome inhibitors, and more importantly, the effect of immunotherapies such as anti-CD38. This should be investigated in further studies especially after the results of a post hoc analysis of the Endurance trial presented by Kapoor et al at the 2021 International Myeloma Workshop.19 They have shown that patients without del(17p), t(14;16), t(14;20), or increased LDH levels had poorer outcomes with bortezomib/lenalidomide/dexamethasone induction when the patients harbored del(1p), whereas the triplet carfilzomib/lenalidomide/dexamethasone appeared to abrogate its adverse effect.19 This trend has to be confirmed in a larger cohort. Nevertheless, our results are still of interest because we show the impact of del(1p32) on prognosis in a heterogeneous cohort, which is similar to real-life settings.
One limitation of our retrospective study is the unavoidable missing data. There are increasing trials designed according to cytogenetic features and it could be interesting to validate our results in such prospective studies.
Despite the proof of its poor prognosis, there are no official guidelines recommending the systematic research of del(1p32) at diagnosis, and it was not integrated into the revised ISS. However, this is changing with a recent paper, in which a panel of experts gave new recommendations, including the detection of chromosome 1 abnormalities (del(1p) and 1q gain), to improve the outcomes definition for high-risk disease.20 To support this idea, Giri et al have also confirmed the association between chromosome 1 abnormalities and inferior survival, independent of other HR CAs.21 They worked on a cohort of patients recruited in the same time period as our cohort (recruitment between 2011 and 2018), benefiting mostly from novel agents. Although they did not separate 1p deletion from 1q gain, they argue for inclusion of chromosome 1 abnormalities in risk assessment for clinical trials. Along the same line, other authors have recommended the inclusion of gain(1q) for the assessment of prognosis.14,22
FISH 1p1q probes designed to detect gains and deletions in the 1q21 and 1p32 regions are available, which make analysis of del(1p32) and gain(1q) easily applicable in laboratories that already perform FISH.
In conclusion, our results again demonstrate the importance of detecting del(1p32) at diagnosis because of its significant effect on the prognosis. Moreover, the number of copies involved in the del(1p32) matters, with a prognosis that is greatly inferior for patients who are double hit. In this era of risk-adapted treatment strategies, we cannot afford to miss these patients with del(1p32) who face a dismal prognosis. Thus, we argue for the systematic detection of del(1p32) at diagnosis for risk assessment.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
This work was financially supported by a National Institutes of Health (NIH), National Cancer Institute PO1 grant (NCI P01-155258), an NIH Specialized Programs of Research Excellence grant (NCI 5P50CA100707), the Fondation ARC (grant PGA1∗20160203788), and the Fondation Toulouse Cancer Santé.
Authorship
Contribution: A.S., A.T., J.C., and H.A.-L. designed the research and analyzed data; A.S. and J.C. wrote the manuscript; A.S. performed the statistical analysis; and all other authors provided study samples and clinical data and reviewed and edited the manuscript.
Footnotes
The data reported in this article have been deposited in the Gene Expression Omnibus database under accession number GSE216574.
Data are available on request from the corresponding author, Jill Corre (corre.jill@iuct-oncopole.fr).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Corre J, Perrot A, Hulin C, et al. Improved survival in multiple myeloma during the 2005-2009 and 2010-2014 periods. Leukemia. 2021;35(12):3600–3603. doi: 10.1038/s41375-021-01250-0. [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A, Avet-Loiseau H, Oliva S, et al. Revised International Staging System for Multiple Myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015;33(26):2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebraud B, Magrangeas F, Cleynen A, et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: the IFM experience. Blood. 2015;125(13):2095–2100. doi: 10.1182/blood-2014-07-587964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hebraud B, Leleu X, Lauwers-Cances V, et al. Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia. 2014;28(3):675–679. doi: 10.1038/leu.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Meng H, Wang J, et al. Clinical characteristics and prognostic values of 1p32.3 deletion detected through fluorescence in situ hybridization in patients with newly diagnosed multiple myeloma: a single-center study in China. Front Med. 2020;14(3):327–334. doi: 10.1007/s11684-019-0712-x. [DOI] [PubMed] [Google Scholar]
- 6.Perrot A, Lauwers-Cances V, Tournay E, et al. Development and validation of a cytogenetic prognostic index predicting survival in multiple myeloma. J Clin Oncol. 2019;37(19):1657–1665. doi: 10.1200/JCO.18.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajkumar SV, Harousseau J-L, Durie B, et al. Consensus recommendations for the uniform reporting of clinical trials: report of the International Myeloma Workshop Consensus Panel 1. Blood. 2011;117(18):4691–4695. doi: 10.1182/blood-2010-10-299487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thakurta A, Ortiz M, Blecua P, et al. High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood. 2019;133(11):1217–1221. doi: 10.1182/blood-2018-10-880831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolli N, Biancon G, Moarii M, et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia. 2018;32(12):2604–2616. doi: 10.1038/s41375-018-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corre J, Cleynen A, Robiou du Pont S, et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia. 2018;32(12):2636–2647. doi: 10.1038/s41375-018-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolli N, Li Y, Sathiaseelan V, et al. A DNA target-enrichment approach to detect mutations, copy number changes and immunoglobulin translocations in multiple myeloma. Blood Cancer J. 2016;6(9):e467. doi: 10.1038/bcj.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33(33):3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baysal M, Demirci U, Umit E, et al. Concepts of double hit and triple hit disease in multiple myeloma, entity and prognostic significance. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-62885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinhold N, Salwender HJ, Cairns DA, et al. Chromosome 1q21 abnormalities refine outcome prediction in patients with multiple myeloma - a meta-analysis of 2,596 trial patients. Haematologica. 2021;106(10):2754–2758. doi: 10.3324/haematol.2021.278888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker BA, Mavrommatis K, Wardell CP, et al. A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33(1):159–170. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schavgoulidze A, Lauwers-Cances V, Perrot A, et al. Heterogeneity in long-term outcomes for patients with Revised International Staging System stage II, newly diagnosed multiple myeloma. Haematologica. 2022 doi: 10.3324/haematol.2021.280566. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sergentanis TN, Kastritis E, Terpos E, Dimopoulos MA, Psaltopoulou T. Cytogenetics and survival of multiple myeloma: isolated and combined effects. Clin Lymphoma Myeloma Leuk. 2016;16(6):335–340. doi: 10.1016/j.clml.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 18.D’Agostino M, Cairns DA, Lahuerta JJ, et al. Second Revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: a European Myeloma Network (EMN) Report within the HARMONY Project. J Clin Oncol. 2022;40(29):3406–3418. doi: 10.1200/JCO.21.02614. [DOI] [PubMed] [Google Scholar]
- 19.Kapoor P, Schmidt T, Jacobus S, et al. OAB-052: impact of chromosome 1 abnormalities on newly diagnosed multiple myeloma treated with proteasome inhibitor, immunomodulatory drug, and dexamethasone: analysis from the ENDURANCE ECOG-ACRIN E1A11 trial. Clin Lymphoma Myeloma Leuk. 2021;21:S33–S34. [Google Scholar]
- 20.Davies FE, Pawlyn C, Usmani SZ, et al. Perspectives on the risk-stratified treatment of multiple myeloma. Blood Cancer Discov. 2022;3(4):OF1–OF12. doi: 10.1158/2643-3230.BCD-21-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giri S, Huntington SF, Wang R, et al. Chromosome 1 abnormalities and survival of patients with multiple myeloma in the era of novel agents. Blood Adv. 2020;4(10):2245–2253. doi: 10.1182/bloodadvances.2019001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95(5):548–567. doi: 10.1002/ajh.25791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.