ABSTRACT
During cytokinesis, a series of coordinated events partition a dividing cell. Accurate regulation of cytokinesis is essential for proliferation and genome integrity. In fission yeast, these coordinated events ensure that the actomyosin ring and septum start ingressing only after chromosome segregation. How cytokinetic events are coordinated remains unclear. The GTPase Cdc42 promotes recruitment of certain cell wall-building enzymes whereas the GTPase Rho1 activates these enzymes. We show that Cdc42 prevents early Rho1 activation during fission yeast cytokinesis. Using an active Rho probe, we find that although the Rho1 activators Rgf1 and Rgf3 localize to the division site in early anaphase, Rho1 is not activated until late anaphase, just before the onset of ring constriction. We find that loss of Cdc42 activation enables precocious Rho1 activation in early anaphase. Furthermore, we provide functional and genetic evidence that Cdc42-dependent Rho1 inhibition is mediated by the Cdc42 target Pak1 kinase. Our work proposes a mechanism of Rho1 regulation by active Cdc42 to coordinate timely septum formation and cytokinesis fidelity.
Keywords: Cdc42, Rho1, Cytokinesis, Pak kinase
Highlighted Article: Using a bio-probe, we show that Rho1 is activated in late cytokinesis, just prior to septum ingression in fission yeast. We find that Cdc42 prevents premature Rho1 activation during early cytokinesis.
INTRODUCTION
During cytokinesis, a mother cell is physically partitioned to generate two daughter cells through a series of coordinated steps (Jordan and Canman, 2012; Pollard, 2010). Fission yeast cells, as in animal cells, divide via an actomyosin-based contractile ring, which is assembled in the medial region of the cell (Balasubramanian et al., 2004; Pollard, 2010). The cell wall is essential for survival and cytokinesis in fungi. Thus, during cytokinesis in fission yeast a new cell wall known as the septum is synthesized to form the new ends of daughter cells. The septum primarily consists of linear β (1-3), branched β (1-6) glucan and α-glucan components (Cortes et al., 2016; Munoz et al., 2013), and is formed in coordination with membrane ingression and ring constriction (Cortes et al., 2015, 2016; Onwubiko et al., 2019). Failure to form a septum leads to cytokinetic failure as these cells cannot undergo ring constriction and membrane ingression (Arasada and Pollard, 2014; Cortes et al., 2015). Cytokinesis begins with the assembly of protein nodes enriched in the formin Cdc12, type II myosin, the F-BAR protein Cdc15 and the IQGAP protein Rng2 at the division site (Padmanabhan et al., 2011; Wu et al., 2003). Under normal conditions, the ring is assembled as myosin heads that interact with nucleated actin filaments to condense nodes into an actomyosin-based contractile ring (Wu et al., 2006). In fission yeast, unlike animal cells, ring constriction does not immediately follow ring assembly. Instead, once the ring assembles, it dwells or ‘matures’ (Wu et al., 2003, 2006) while serving as a landmark for the recruitment of essential septum synthesizing proteins such as Bgs1 (Arasada and Pollard, 2014; Vjestica et al., 2008; Wei et al., 2016). Membrane trafficking enables the recruitment of septum-synthesizing enzymes and furrow formation (Hercyk et al., 2019b; Vjestica et al., 2008; Wang et al., 2016). The septum consists of three layers made up of a primary septum sandwiched between two secondary septa, with primary septum synthesis preceding secondary septum synthesis (Pérez et al., 2018). The process of septum synthesis is well coordinated, but not entirely understood. The enzyme Bgs1 is required for primary septum formation, and thus its recruitment is critical for cytokinesis (Cortes et al., 2002, 2007; Onwubiko et al., 2021; Pérez et al., 2018; Wei et al., 2016).
We have previously shown that Cdc42 promotes timely septum formation via ensuring timely Bgs1 delivery and membrane trafficking at the division site (Campbell et al., 2022; Onwubiko et al., 2021; Wei et al., 2016). Cdc42 is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), and is active when GTP bound and inactive in the GDP-bound state. GEFs promote GTP exchange to activate the GTPase, whereas the GAPs increase GTP hydrolysis, thus promoting GTPase inactivation. In fission yeast, Cdc42 is sequentially activated by the GEFs Gef1 and Scd1 at the division site (Wei et al., 2016). Gef1 first activates Cdc42 during early anaphase, whereas Scd1 activates Cdc42 in late anaphase at the onset of ring constriction (Hercyk and Das, 2019b; Hercyk et al., 2019b; Wei et al., 2016). In the absence of gef1, Bgs1 delivery and the initiation of septum formation and ring constriction are delayed (Wei et al., 2016).
In addition to Cdc42, other Rho GTPases also play a role in cytokinesis (Hercyk and Das, 2019a). The essential GTPase Rho1 is required for septum formation and cell wall integrity via the activation of cell-wall-building enzymes (Arellano et al., 1996). Septum synthesis and Rho1 activation occur once chromosome segregation completes. The timing of septum synthesis is regulated by the septation initiation network (SIN) pathway, a signal transduction cascade emanating from the microtubule-organizing center of the cell or the spindle pole body (SPB). The SIN pathway becomes active in early anaphase and is required for proper cytokinesis progression (Hou et al., 2000). The SIN is analogous to HIPPO signaling in mammalian cells, but the role of HIPPO in mammalian cytokinesis is yet to be clarified (Johnson et al., 2012). Genetic evidence indicates that the SIN pathway promotes Rho1 activation, which then allows septum synthesis (Alcaide-Gavilán et al., 2014; Hou et al., 2000). Thus, the SIN pathway ensures septum synthesis is activated once chromosome segregation is successful. The animal Rho1 homolog RhoA is required for ring formation and is essential for cytokinesis (Basant and Glotzer, 2018; Jantsch-Plunger et al., 2000). Although Rho1 is essential for septum formation in yeast, the current literature suggests that it is dispensable for ring formation (Onishi et al., 2013; Yoshida et al., 2009). In fission yeast, where both the actomyosin ring and the septum have important roles in cytokinesis, Rho1 has no reported roles in ring formation but is essential for septation (Balasubramanian et al., 2004).
Cytokinetic defects in yeasts have been linked to improper Cdc42 and Rho1 regulation (Onishi et al., 2013; Onwubiko et al., 2021). It is unclear how Cdc42 and Rho1 coordinate septation during cytokinesis. Previous work in budding yeast suggested antagonistic roles for Rho1 and Cdc42, where Cdc42 inactivation and Rho1 activation are required for the completion of cytokinesis (Onishi et al., 2013). Although Cdc42 promotes the delivery of Bgs1, Rho1 is the regulatory subunit required for the activation of Bgs1 (Arellano et al., 1996; Cabib et al., 1998).
In animal cells, the direct role of Cdc42 in cytokinesis remains undefined. In Xenopus embryos and mouse fibroblasts, for example, constitutively active Cdc42 impairs cytokinesis completion (Drechsel et al., 1997). However, in other cases such as in mouse embryonic stem cells, Cdc42 is only critical for development but not cytokinesis (Chen et al., 2000). RNA interference in animal cells demonstrates that whereas RhoA is required for cytokinesis, Cdc42 is not required for this process (Jantsch-Plunger et al., 2000). Cdc42 also promotes spindle positioning and polar body extrusion in mouse oocytes, but it is not known whether its localization at these spindles affects RhoA (Na and Zernicka-Goetz, 2006). Thus, the role of Cdc42 in the cytokinetic process may be cell type specific, and these data highlight the importance for more investigation to elucidate Cdc42 regulation in dividing cells (Jordan and Canman, 2012).
Here, we investigate how Cdc42 and Rho1 regulate cytokinetic events in fission yeast (Schizosaccharomyces pombe). With the respective molecular probes designed to bind active Cdc42 and Rho1, we thoroughly compared the activity of these GTPases during cytokinesis. We report that Cdc42 activation precedes Rho1 activation at the division site. Our results indicate that Gef1-dependent Cdc42 activation prevents Rho1 activation during early anaphase in a Pak1-dependent manner, likely via the regulation of the Rho1 GEFs and/or GAPs.
RESULTS
A Rho probe detects Rho1 activation at cell tips and the division site
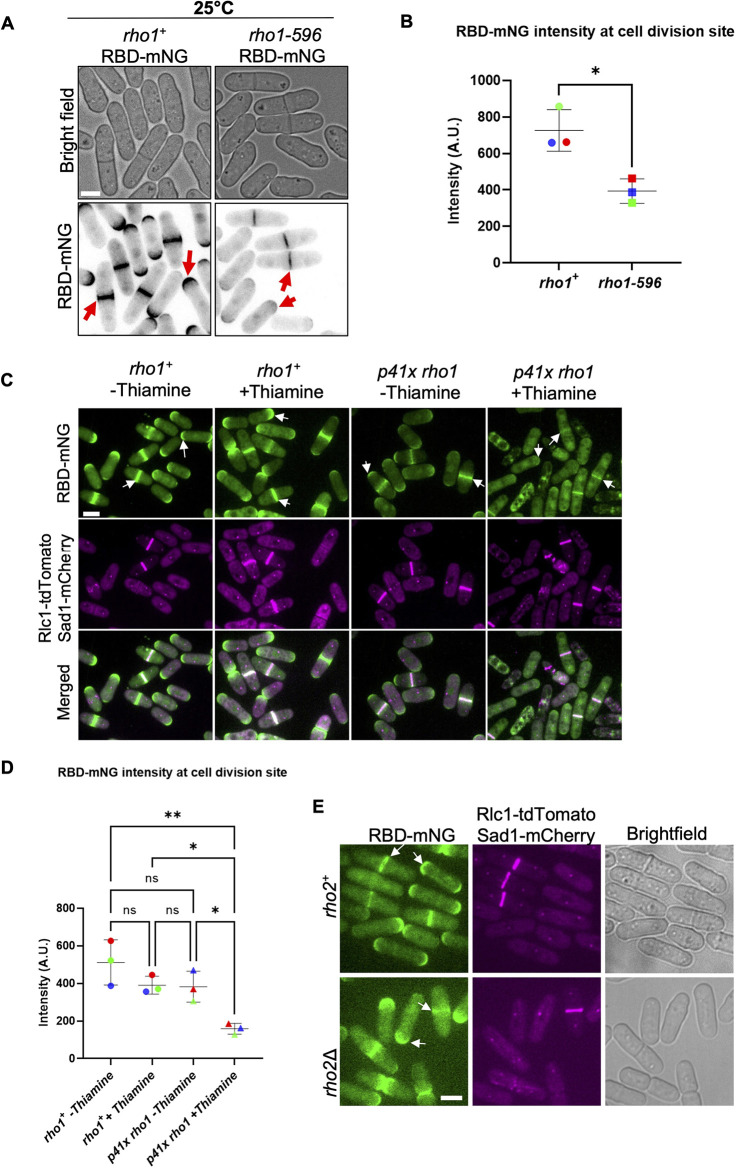
To study Rho1 activation, we designed a Rho probe using the Rho-binding domain (RBD) of the protein kinase C Pck2 modified from the RBD previously used in budding yeast (Kono et al., 2012; Yoshida et al., 2006). Briefly, we placed the promoter region of fission yeast pck2 upstream of the PCK2 RBD with a C-terminal fluorescent reporter and integrated the construct into the genome (Fig. S1Ai). Successful integration was confirmed by visualizing the localized Rho probe signal in transformed wild-type cells, noting that these cells showed normal morphology with no aberrant phenotypes (Fig. S1Aii). The Rho probe RBD–mNG localized to the growing tips of interphase cells, and the division site of dividing cells (Fig. S1Aii). To verify that the Rho probe detected Rho1 activation, we investigated the localization of the probe RBD–mNG in temperature-sensitive and in switch-off rho1 mutants. The rho1 gene is essential and the temperature-sensitive rho1-596 mutant is inviable at restrictive temperatures (Fig. S1B). We compared RBD–mNG localization in these mutants under permissive conditions to ensure that pleotropic effects and cell death did not mislead our observations. We found that even under permissive temperature (25°C) the RBD–mNG intensity at the division site was decreased in rho1-596 mutants compared to rho1+ cells (Fig. 1A,B). As expected, RBD–mNG intensity at the cell tips was also decreased in these mutants. Next, we compared RBD–mNG intensity at the division site in rho1 switch-off mutants. Under complete rho1 repression the cells are inviable. Thus, we partially repressed rho1 in these mutants with thiamine treatment for 18 h at 25°C. In rho1+ cells, RBD–mNG localized to the division site with assembled actomyosin ring labeled with Rlc1–tdTomato and to the cell tips (Fig. 1C, arrows). In rho1Δ mutants transformed with p41X rho1, in the presence of thiamine, RBD–mNG intensity at the division site and the cell tips was decreased (Fig. 1C,D). Together, these data indicate that the Rho probe RBD–mNG detects active Rho1. It was previously reported that the Pck2p–RBD interacts with Rho1 and Rho2 GTPases (Arellano et al., 1999). To ensure that our Rho probe reported active Rho1 and not Rho2 in cells, we expressed the RBD–mNG in rho2Δ cells (Fig. 1E). In rho2Δ mutants RBD–mNG intensity at the division site and the cell tips did not show any change compared to rho2+ cells. RBD–mNG localized to the division site with an assembled actomyosin ring and to the cell tips (Fig. 1E, arrows). Taken together, these results indicate that the Rho probe reports active Rho1 localization in cells.
Fig. 1.
RBD–mNG detects Rho1 activation at the cell tips and at the division site. (A) RBD–mNG localization at the cell tips and at the division site is decreased in rho1-596 mutant cells compared to rho1+ (red arrows) at permissive temperature. (B) Quantification of RBD–mNG intensity at the division site in rho1+ and rho1-596 cells (mean±s.d., three independent experiments, n≥26 cells each, P-value=0.0120, unpaired two-tailed Student's t-test). (C) RBD–mNG localization at the cell tips and at the division site (white arrows) is decreased in rho1-repressed cells (p41xrho1+ RBD–mNG Rlc1–tdTomato Sad1–mCherry cells) with thiamine for 18 h at 25°C as compared to cells expressing rho1 (rho1+ RBD–mNG Rlc1–tdTomato Sad1–mCherry and p41x rho1+ RBD-mNG-Rlc1 tdTomato-Sad1 mCherry) without thiamine. (D) Quantification of RBD–mNG intensity at the division site in the strains indicated (mean±s.d. three independent experiments, n≥24 cells each, *P<0.05, **P<0.01 one-way ANOVA with Tukey's multiple comparison test). (E) RBD–mNG localization in rho2+ and rho2Δ cells at the cell tips and division site (white arrows). Rlc1–tdTomato marks the actomyosin ring and Sad1–mCherry marks the SPBs. Quantification is provided in Fig. S2C. A.U., arbitrary units. Scale bars: 5 µm.
Given that Rho1 activation occurs at the division site during cytokinesis, we asked whether this required the actomyosin ring. The actomyosin ring can be disrupted in cells upon treatment with the drug latrunculin A (LatA). An asynchronous population of cells was treated with LatA, imaged and analyzed. Cells in cytokinesis with disrupted actomyosin rings still showed RBD–mNG localized to the division site. However, we did observe a loss of RBD–mNG at the growing ends of interphase cells. As cytokinesis also requires endocytosis via branched actin networks, we tested its requirement in Rho1 activation at the division site (Onwubiko et al., 2019). The drug CK666 has been shown to block the Arp2/3 complex and prevent branched actin assembly (Nolen et al., 2009). We found that in cells treated with CK666, RBD–mNG was not lost at the division site. However, we observed ectopic RBD–mNG in interphase cells (Fig. S1C, asterisks). We also depolymerized microtubules in cells by treating them with methyl benzimidazole-2yl carbamate (MBC). We did not observe any disruption in the localization pattern of RBD–mNG; however, the signal appeared dampened at the cell tips and division site compared to DMSO-treated control cells (Fig. S1C). These results indicate that although the actin cytoskeleton is not required for maintaining Rho1 activation at the division site, it is necessary at the growth sites of interphase cells. Our data also indicates that Rho1 activation at the division site is differentially regulated than at the growing cell ends.
Cdc42 is activated earlier whereas Rho1 is activated late in cytokinesis
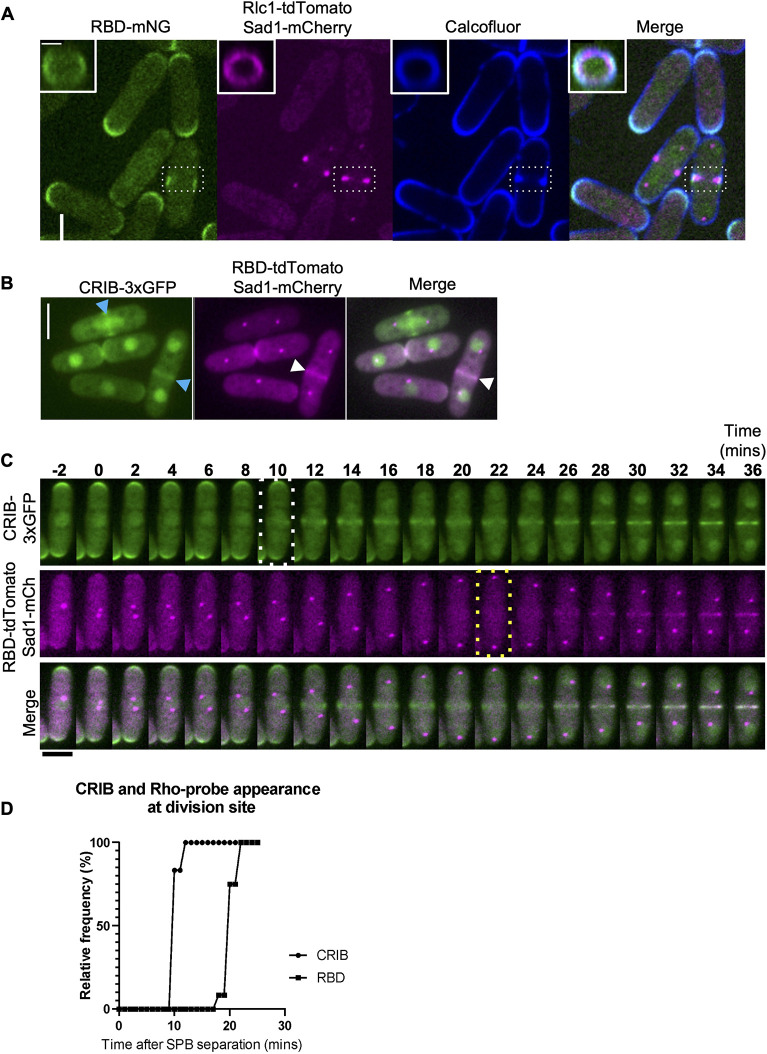
We found that RBD–mNG is only present at the division site of cells that also show ring constriction and septum deposition (dashed white box, Fig. 2A). 3D projections of the RBD–mNG, the ring marker Rlc1–tdTomato and the septum shows the spatial organization of the Rho probe at the division site (Fig. 2A, insets). RBD-mNG signal at the division site appears at the ring–membrane interface, overlapping with the Calcofluor-stained septum (Fig. 2A, 3D rings).
Fig. 2.
Cdc42 is activated earlier in cytokinesis whereas Rho1 is activated in late anaphase. (A) Middle of Z-plane image showing active Rho1 (RBD–mNG), the ring (Rlc1–tdTomato), and septum (Calcofluor White) at the division site in representative cells during cytokinesis (white dashed box). Scale bar: 5 µm. Insets, 3D projections of areas from the dashed white box of the division site show concentric rings of active Rho (green), the actomyosin ring (magenta), and the septum (blue). Scale bar: 2 µm. (B) Localization of active Cdc42 (CRIB–3xGFP, blue arrowheads), and active Rho probe in cells (RBD–mNG, white arrowheads). Scale bars: 5 µm. (C) Time-lapse series of a representative cell, time of Cdc42 activation (white dotted box) and Rho activation (yellow dotted box) during cytokinesis. Scale bar: 5 µm. (D) Quantification of the timing of Cdc42 and Rho activation at the division site during cytokinesis (n=12 cells). P<0.0001 (one way ANOVA followed by Tukey's HSD test).
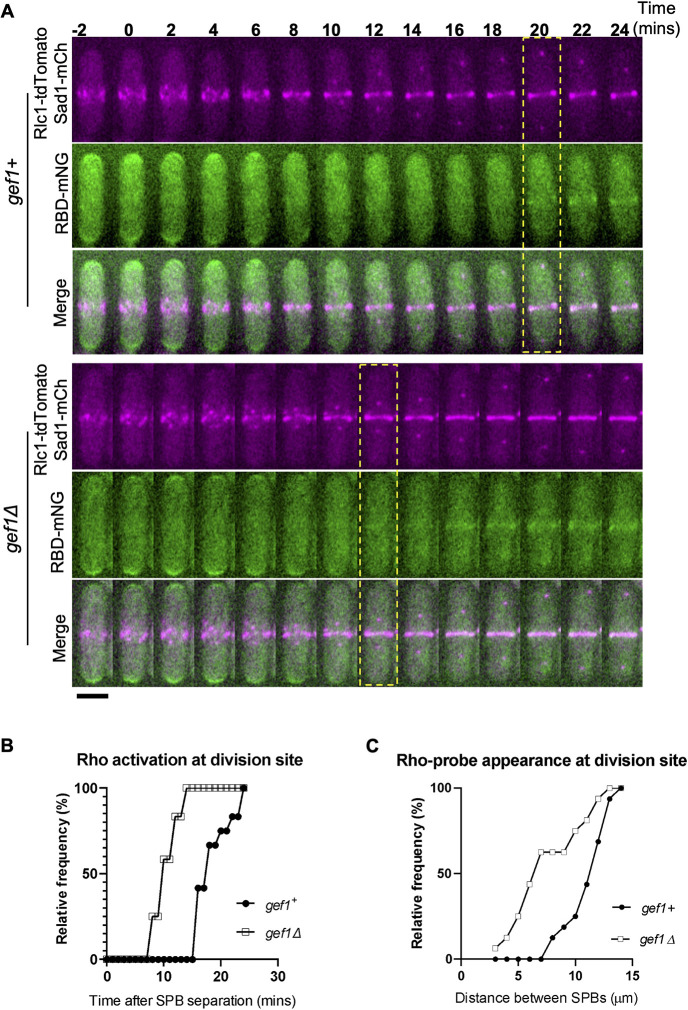
Fig. 3.
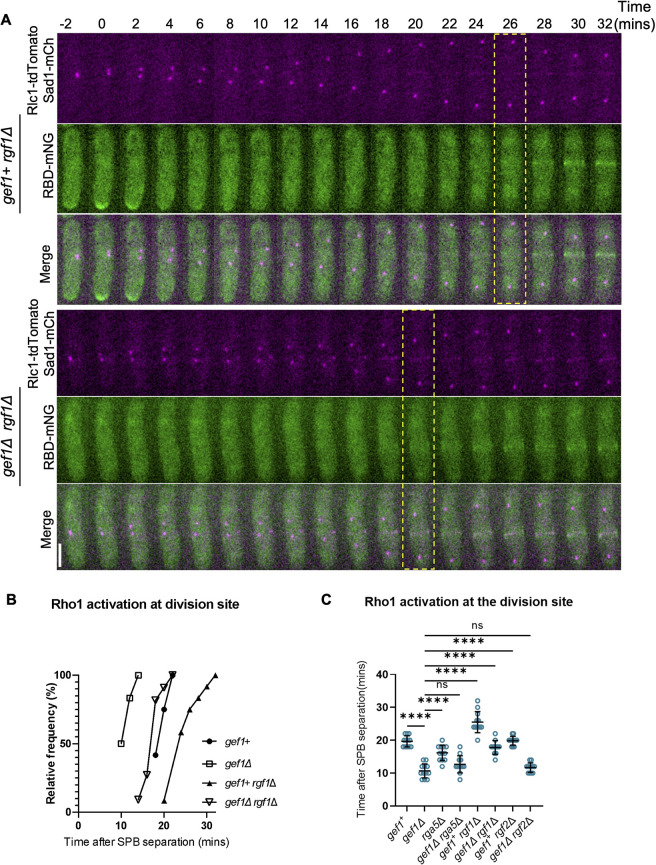
Loss of gef1 results in early Rho1 activation in cytokinesis. (A) Time-lapse montage showing the time of Rho activation (RBD–mNG) at the division site in representative gef1+ and gef1Δ cells (yellow dotted box), Time=0 marks the time of SPB separation, and onset of cytokinetic events. Scale bar: 5 µm. (B) Outcome plot showing frequency of Rho-activation over time during cytokinesis from time-lapse movies of strains as indicated (n=12 cells from time-lapse movies). (C) Outcome plot shows frequency distribution of SPB distances at which Rho-activation at the division site is observed in acquired still images of gef1+ and gef1Δ cells (n=16 cells per indicated strain).
To investigate Rho1 and Cdc42 dynamics during cytokinesis, we performed live imaging of cells simultaneously expressing the active Cdc42 probe CRIB–3xGFP (Das et al., 2012; Tatebe et al., 2008; Wei et al., 2016) and the Rho probe RBD–tdTomato in wild-type cells (Fig. 2B). We observed that although some cells displayed RBD–tdTomato and CRIB–3xGFP simultaneously at the division site, others only displayed CRIB–3xGFP (Fig. 2B). Time-lapse imaging of cells simultaneously expressing CRIB–3xGFP and RBD–tdTomato revealed that Cdc42 was activated at the division site ∼10 min after the onset of SPB separation, whereas Rho1 was activated ∼20 min after SPB separation (Fig. 2C). Following the previously described timeline of cytokinetic events in fission yeast (Wu et al., 2003), we observed Cdc42 activation in early anaphase, at the time of ring assembly. In contrast, we found that the Rho probe RBD–tdTomato localized to the division site in late anaphase, immediately preceding the onset of ring constriction and septum ingression (Fig. 2C,D; Fig. S2A). These observations are consistent with the fact that septum formation is activated by Rho1 and occurs in late anaphase.
Rho1 is activated by the GEFs Rgf1, Rgf2 and Rgf3 (Mutoh et al., 2005; Tajadura et al., 2004), all of which promote cell wall integrity (Fig. S3A). We asked whether the delay in Rho1 activation was due to a delay in the localization of its GEFs at the division site. Previously, it has been reported that loss of rgf1 or rgf2 does not disrupt cell viability, but rgf1Δ cells display severe cell wall defects, and rgf1Δ rgf2Δ is lethal (Mutoh et al., 2005). Rgf3 is the primary cytokinetic GEF for Rho1, and loss of rgf3 is lethal (Tajadura et al., 2004). We imaged live cells expressing fluorescently labeled Rgf3 and Rgf1. Both Rgf1–GFP and Rgf3–eGFP localize to the division site in early anaphase (Fig. S3B,C) at the time when active Rho1 is absent from the division site. Although we could not acquire a time-lapse series to assess the exact timing of Rho1 GEF localization, we quantified Rgf1 and Rgf3 localization during cytokinesis by measuring the distance between the mitotic SPBs. SPB separation and movement from mitosis onset serve as a conventional timer for cytokinesis (Nabeshima et al., 1998; Pollard and Wu, 2010). Cytokinesis progression is therefore proportional to the distance between mitotic spindles up until late anaphase, when ring constriction and septum formation begin. Here, we labeled the SPBs with Sad1–mCherry in cells expressing the fluorescently tagged GEFs and computed SPB distances at which the GEF localizes to the division site (Fig. S3B–D). Using the distance between SPBs as a proxy for timing of mitosis and cytokinesis, we found that in most cells, Rgf1–GFP and Rgf3–eGFP was localized to the division site at early stages in anaphase (Fig. S3D,E). This could be observed by the short distance between the SPBs of ∼2 µm (Fig. S3D). We also measured the distance for which active Rho1 appeared at the division site, and found that at the distance between SPBs of ∼10 µm, active Rho1 was present at the division site in ∼50% of the population of control cells (Fig. S3E). A comparison of the cumulative frequency distribution of the localization of the two GEFs with increasing SPB distance suggests that the GEFs localize to the division site simultaneously (Fig. S3E). However, although the GEFs localize to the division site earlier, the Rho probe was visible only at a greater SPB distance, suggesting a time delay between GEF localization and Rho1 activation. Thus, although Rho1 GEFs Rgf1 and Rgf3 localize to the division site in early anaphase, they are unable to activate Rho1 at this stage.
Cdc42 inhibits Rho1 activation in early anaphase
Next, we asked what prevents early Rho1 activation. Given that Cdc42 is activated before Rho1, we asked whether active Cdc42 prevents early Rho1 activation. As previously shown, Cdc42 activation in early anaphase is dependent upon its GEF Gef1 (Wei et al., 2016). To test whether active Cdc42 prevents Rho1 activation during cytokinesis, we looked at RBD–mNG localization in gef1Δ cells. Time-lapse imaging of gef1+ and gef1Δ cells expressing RBD–mNG reveal that gef1Δ causes premature Rho1 activation in early anaphase (Fig. 3A). We found that ∼100% of gef1Δ cells display early RBD–mNG localization at the division site at ∼12 min after SPB separation compared to ∼20 min in gef1+ control cells (Fig. 3B). Furthermore, we found that RBD–mNG normally localized to the division site at SPB distances of greater than ∼7 µm in gef1+ control cells, whereas in gef1Δ cells they appeared early at an SPB distance of ∼4 µm or less (Fig. 3C). This suggests that Gef1 prevents premature Rho1 activation at the division site.
Given that Gef1 is a Cdc42 GEF, we posit that Gef1-dependent Cdc42 activation prevents premature Rho1 activation in early cytokinesis. To test this, we assessed the localization of the Rho probe in cells expressing the constitutively active cdc42G12V allele. Previously, we have shown that cytokinetic defects in gef1 mutants can be rescued by moderate expression of constitutively active cdc42G12V (Wei et al., 2016). With the medium-strength thiamine-repressible promoter nmt41, we moderately expressed cdc42G12V in gef1+ and gef1Δ cells expressing RBD–tdTomato. Experimental controls were gef1+ and gef1Δ cells expressing the empty pjk148 vector with RBD–tdTomato. All cells expressed the ring marker Rlc1–GFP and the SPB marker Sad1–mCherry. As described above, the distance between the SPBs was used to identify the stages of cytokinesis at which RBD–tdTomato was detected at the division site. We found that RBD–tdTomato localized to the division site in late anaphase in gef1+ pjk148 empty control cells and was present earlier in gef1Δ pjk148 empty cells (Fig. S4A,B). However, the expression of cdc42G12V in gef1Δ cells reverted RBD–TdTomato localization at the division site to late anaphase, similar to the timing seen in gef1+ controls (Fig. S4A,B). We quantified the percentage of non-constricting actomyosin rings with RBD–TdTomato localization and found a significant increase in gef1Δ cells compared to gef1+ controls, and this was rescued by cdc42G12V expression in gef1Δ cells (Fig. S4D). We also observed that the RBD–TdTomato signal was diminished in cdc42G12V-expressing cells at the cell tips and division site (Fig. S4A). We quantified the mean fluorescence intensity of the RBD–mNG signal at the division site and confirmed that it was indeed significantly reduced in cdc42G12V-expressing cells (Fig. S4C). Together, these results indicate that gef1 inhibits Rho1 via Cdc42 activation.
Even though the Rho probe detects Rho1 activation, we questioned whether early Rho probe localization in gef1Δ cells during cytokinesis could also be due to Rho2 activity. To verify this, we assessed the localization of the RBD–mNG in rho2Δ cells (Fig. S2B). We found that whereas RBD–mNG localized in late anaphase in gef1+ rho2+ control cells, it still localizes earlier in gef1Δ rho2Δ double mutants (Fig. S2B,C). Hence, early RBD–mNG localization in gef1Δ cells is indeed due to early Rho1 activation. Collectively, these data indicate that active Cdc42 prevents premature Rho1 activation in early anaphase.
Genetic evidence indicates that Rho1 activation requires the SIN pathway (Jin et al., 2006). We asked whether early Rho1 activation in gef1Δ mutants was sufficient to bypass the requirement for SIN-dependent activation. Using RBD–mNG, we assessed Rho1 activation in a temperature-sensitive sid2 mutant (sid2-250), which disrupts SIN function at the restrictive temperature (36°C). As previously shown (Feoktistova et al., 2012), SIN inactivation resulted in elongated, non-dividing cells (Fig. S5A). We found that under permissive conditions (25°C) RBD–mNG localized to the division site in both gef1+ sid2-250 and gef1Δ sid2-250 cells (Fig. S5A,B). However, SIN inactivation at 36°C abolished RBD–mNG localization from the division site in gef1+ sid2-250 and gef1Δ sid2-250 cells (Fig. S5A,B), suggesting that the SIN is required for Rho1 activation during cytokinesis regardless of gef1. Interestingly, these mutants were still able to activate Rho1 at the cell tips, suggesting that the SIN pathway is only required for Rho1 activation at the division site.
Disruption of Pak1 kinase function results in early Rho1 activation during cytokinesis
The p21-activated kinase (Pak1) is a known downstream effector of Cdc42 in fission yeast (Magliozzi et al., 2020; Ottilie et al., 1995). In animal cells, PAK kinases differentially regulate the RhoA GEFs (Alberts et al., 2005; Zenke et al., 2004). Pak1 localizes to the division site in early cytokinesis (Magliozzi et al., 2020), via its interaction with active Cdc42. Given the early localization of Pak1 to the division site, we wondered whether Cdc42-dependent inhibition of Rho1 activation was due to Pak1 function. We reasoned that if Pak1 was responsible for blocking Rho1 activation, then loss of pak1 function should allow early Rho1 activation at the division site. To evaluate this idea, we assessed Rho1 activation in the temperature-sensitive pak1 mutant orb2-34 (Verde et al., 1995), hereafter denoted pak1-ts for clarity. We first quantified the appearance of RBD–mNG at the division site in the temperature-sensitive pak1 mutants. At the permissive temperature, RBD–mNG at the division site appeared as normal in late anaphase in pak1+ cells (Fig. 4A). In pak1-ts mutants at the permissive temperature, RBD–mNG appeared a bit earlier than in pak1+ cells as determined by the distance between SPBs and the appearance of assembling rings (Fig. 4B). At restrictive temperature (35.5°C), RBD–mNG localized to the division site in early anaphase in most pak1-ts mutant cells (Fig. 4A,B). To assess the timing of Rho1 activation during cytokinesis, we measured the distance between the Sad1–mCherry-labeled SPBs in pak1+ and pak1-ts cells (Fig. 4B). At 25°C, pak1+ and pak1-ts cells localize RBD-mNG late in anaphase as determined by longer SPB distances (Fig. 4A,B). In contrast, at 35.5°C pak1-ts mutants displayed early RBD–mNG localization at the division site as determined by the short SPB distances. Thus, disruption of pak1 function enables early Rho1 activation in cytokinesis. Although we observed RBD–mNG at a shorter SPB distance in pak1 mutants under restrictive conditions, we also note that these mutants are smaller in size. However, the small cell size does not impact the SPB distance at which RBD–mNG appears at the division site. This is because even under permissive conditions, pak1 mutants are smaller in size, and in these cells, RBD–mNG does not appear at the division site at a relatively short SPB distance (Fig. 4B).
Fig. 4.
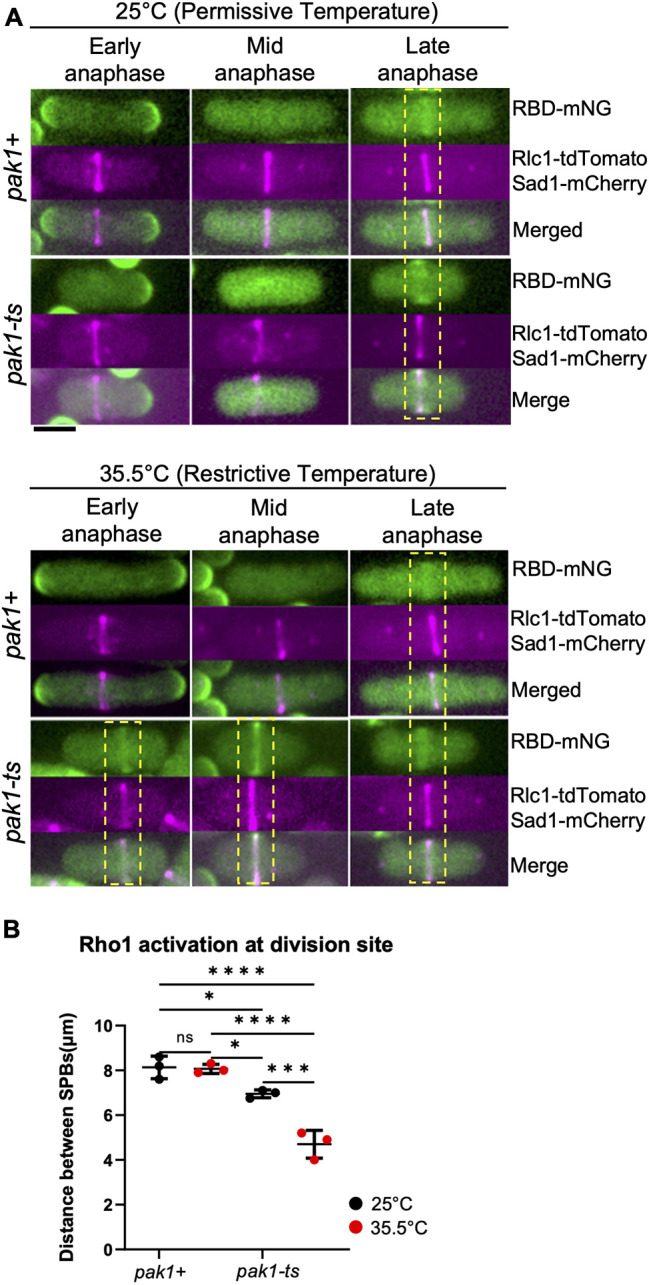
A hypomorphic pak1 mutant displays early Rho1 activation in cells during cytokinesis. (A) Rho1 activation (RBD–mNG) in pak1+ (orb2+) and pak1-ts (orb2-34) strains at the permissive temperature (25°C), and restrictive temperature (35.5°C). Yellow dotted boxes highlight the stage of cytokinesis in which Rho1 activation is observed at the division site in the indicated strains. Scale bar: 5 μm. (B) Outcome plot showing frequency distribution of the quantified distance between the SPBs at which Rho1 activation at the division site is observed in all conditions shown (mean±s.d; n=3 representative experiments). Data points on the graph represent the first quartile of SPBs distances measured in the indicated strains. *P≤0.03; ***P≤0.0008; ****P≤0.0001; ns, not statistically significant between strains (one-way ANOVA followed by Tukey's HSD test).
Next, to further confirm that inhibition of Rho1 is indeed dependent on Pak1 kinase, we tested whether pak1 overexpression disrupts Rho1 activation in dividing cells even in the absence of gef1. Utilizing the high-strength thiamine repressible promoter nmt1 (Javerzat et al., 1996), we assessed the effect of pak1 overexpression on Rho1 activation during cytokinesis. The nmt1-3xHA-pak1 allele (MBY3451) was either expressed or repressed in gef1+ and gef1Δ cells expressing RBD–mNG, Sad1–mCherry and Rlc1–tdTomato (Fig. S6A). Cells were grown in the presence (pak1-repressed) and absence (pak1OE) of thiamine. Under repressing conditions, the RBD–mNG appeared at late anaphase, similar to what was seen in pak1+ cells (Fig. S6B). This is due to the previously shown leaky expression of pak1 due to the high-strength nmt1 promoter even in the presence of thiamine (Javerzat et al., 1996; Wei et al., 2016). Thus gef1+ pak1-repressed cells behaved similarly to wild-type and localized RBD–mNG in late anaphase (Fig. S6B,C). However, early RBD–mNG localization at the division site observed in gef1Δ cells was rescued upon pak1OE, restoring it to late anaphase (Fig. S6A,B). We also observed an overall decrease in mean intensity of RBD–mNG in pak1OE cells compared to pak1-repressed cells (Fig. S6B,C). These results indicate that overexpression of pak1 blocks Rho1 activation even in the absence of gef1. Together, these data suggest that Gef1 mediates Rho1 inhibition in early anaphase via the Pak1 kinase.
Loss of gef1 rescues rgf3 repression-induced lethality
Based on our observation that Rgf1 and Rgf3 localize to the division site in early anaphase, even before the ring is fully assembled (Fig. S3B,C), we wondered whether the localization of either of these Rho GEFs was either enhanced or early in gef1 mutants. However, when comparing Rgf3–eGFP and Rgf1–GFP localization in gef1+ and gef1Δ cells using the distance between SPBs as the marker for cytokinesis progression, we saw that both GEFs localize in early cytokinesis in gef1+ and gef1Δ cells (Fig. S7A,B). The mean intensity of Rgf1 and Rgf3 at the division site in gef1+ and gef1Δ cells also remained similar (Fig. S7C,D). Thus neither the timing of localization nor the intensity of Rgf3 and Rgf1 is changed in the absence of gef1. It is possible that regulation of Rho1 GEF activity prevents Rho1 activation in the early anaphase.
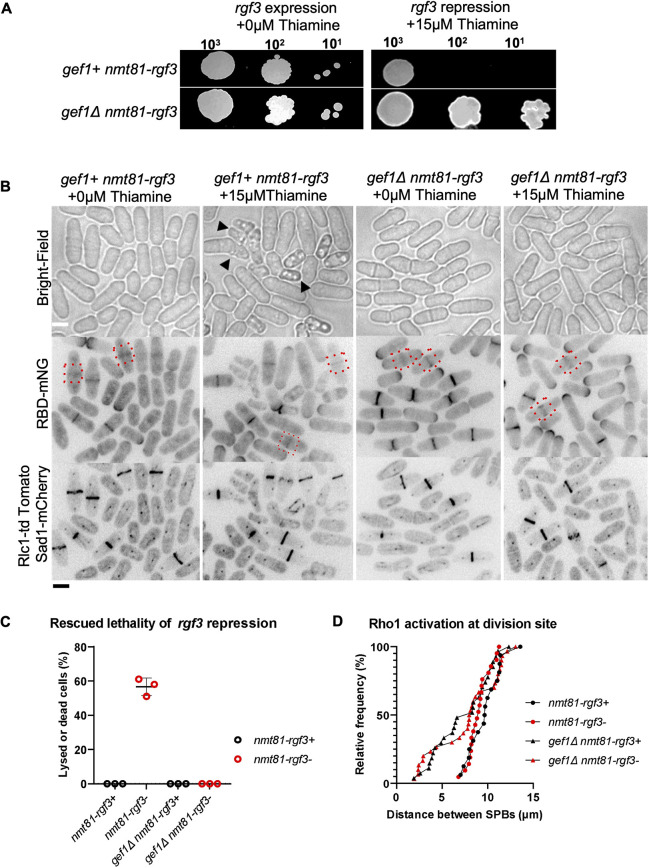
Given that Rgf3 is the only Rho1 GEF reported to localize specifically to the division site but not the cell tips, (Morrell-Falvey et al., 2005; Mutoh et al., 2005), we asked whether active Cdc42 inhibits Rgf3-dependent Rho1 activation in early cytokinesis. Loss of rgf3 results in cell lysis and death (Tajadura et al., 2004). To test whether Rgf3 was responsible for early Rho1 activation in gef1Δ cells, we repressed rgf3 expression via the low-strength thiamine-repressible promoter nmt81 in gef1+ and gef1Δ cells. We first assessed the viability of rgf3-depleted (nmt81-rgf3) cells in the presence and absence of gef1 by performing a growth assay of gef1+ nmt81-rgf3, and gef1Δ nmt81-rgf3 cells (Fig. 5A). Cells were subjected to rgf3-expressing or -repressing (+ thiamine) conditions in growth media at 25°C. As expected, gef1+ nmt81-rgf3 cells showed lethality under rgf3-repressing conditions; however, gef1Δ nmt81-rgf3 cells survived under these conditions (Fig. 5A). To corroborate our observations, we analyzed Rho1 activity at the division site in gef1Δ nmt81-rgf3 cells. Repression of nmt81-rgf3 resulted in lysed and dying cells, as shown previously (Tajadura et al., 2004). In gef1+ cells, repression of nmt81-rgf3 resulted in lysis of ∼58% of cells in an asynchronous population (Fig. 5B, black arrowheads, C). Loss of gef1 rescued the lethality of rgf3 repression, thus corroborating our growth assay results (Fig. 5B,C). Moreover, rgf3 repression did not disrupt the localization of RBD–mNG at the division site in gef1+ and gef1Δ cells (Fig. 5B,D). This indicates that the loss of gef1 rescues the lethality of rgf3 repression, and early Rho1 activation in gef1 mutants is not Rgf3-dependent. This suggests that Gef1-mediated repression of Rho1 activity occurs via another mechanism.
Fig. 5.
Loss of gef1 rescues the lethality of rgf3-repressed strains. (A) Rescue of rgf3-repression lethality in gef1Δ cells as shown by a spot assay on supplemented EMM with or without 15 µM thiamine for the strains as indicated. Representative of three repeats. (B) Rho1 activation (RBD–mNG) at the division site (red dotted boxes) under rgf3-repressed and expressing conditions in the indicated strains. Black arrowheads point to dying or lysed rgf3-depleted cells. Scale bar: 5 µm. (C) Quantification of cell lysis in the indicated strains. Mean±s.d, n=3 experiments. (D) Outcome plot showing the distance between SPBs at which Rho1 activation is observed at the division site of the indicated strains and conditions. n=30 cells per indicated strain.
Loss of rgf1 rescues premature Rho1 activation in gef1 mutants
Rho1 GEFs differ in their spatial dynamics during ring constriction. Unlike Rgf3, which localizes at, and constricts with the actomyosin ring, Rgf1 and Rgf2 ingress with the membrane furrow and septum (Morrell-Falvey et al., 2005; Mutoh et al., 2005). Active Rho1 at the division site appears diffuse and condenses into a band-like appearance as constriction begins (Fig. 2A,C). Rho1 might therefore be differentially activated by its GEFs at the division site. Our observation that Rho1 activation remained early in gef1Δ cells depleted of rgf3 (Fig. 5B,D) suggests that another GEF is responsible for this activation in early cytokinesis. Therefore, we tested whether the other Rho1 GEFs rgf1 and rgf2 were required for early Rho1 activation in gef1Δ cells. Although rgf1Δ cells are viable, they display severe cell wall and morphology defects (Tajadura et al., 2004). On the other hand, rgf2Δ cells do not show any obvious defect, but the rgf1Δ rgf2Δ double mutant is not viable (Morrell-Falvey et al., 2005; Mutoh et al., 2005). This indicates that Rgf1 and Rgf2 are redundant, with Rgf1 being the primary Rho1 GEF. We asked whether early Rho1 activation in gef1Δ cells was Rgf1 dependent. We assessed RBD–mNG localization at the division site of rgf1Δ cells in the presence and absence of gef1 via time-lapse confocal microscopy. RBD–mNG localized normally at the onset of ring constriction in gef1+ rgf1+ control cells (Fig. 6B). As expected, RBD–mNG also appeared early in gef1Δ cells (Fig. 6B). RBD–mNG localization at the division site was delayed in rgf1Δ cells compared to the gef1+ controls (Fig. 6A,B). Moreover, loss of rgf1 prevents premature RBD–mNG localization at the division site in gef1Δ cells restoring it to normal in late anaphase (Fig. 6A,B). This suggests that Rgf1 is responsible for early Rho1 activation in gef1Δ cells. We observed that Rho1 activation in rgf1Δ cells was delayed much more than in rgf1Δ gef1Δ cells (Fig. 6A–C), indicating that in gef1Δ, in addition to Rgf1, another regulator is responsible for early Rho1 activation. We asked whether this regulator could be the other GEF Rgf2, which has been shown to behave similarly to Rgf1 (Morrell-Falvey et al., 2005). We found that rgf2Δ did not alter Rho1 activation at the division site (Fig. S7F; Fig. 6C), likely due to the presence of Rgf1. Accordingly, gef1Δ rgf2Δ double mutants showed early Rho1 activation at the division site, similar to what was seen for gef1Δ cells (Fig. S7F,G). We were unable to assess Rho1 activation in the rgf1Δ rgf2Δ mutants as these double mutants were lethal.
Fig. 6.
Loss of rgf1 rescues premature Rho1 activation in gef1 mutants. (A) Time-lapse of representative gef1+ rgf1Δ and gef1Δ rgf1Δ cells showing the time of Rho1 activation at the division site during cytokinesis (yellow dotted box). Scale bar: 5 µm. (B) Outcome plot showing the frequency of the time at which Rho1 activation (RBD-mNG) occurs at the division site during cytokinesis as determined from time-lapse movies as shown in A. Time=0 marks the onset of cytokinetic events (n=12 cells per strain). (C) Quantification of the timing of Rho1 activation during cytokinesis from movies of the strains indicated (mean±s.d; n=12 cells per strain). ****P≤0.0001; ns, not statistically significant (one-way ANOVA followed by Tukey's HSD test).
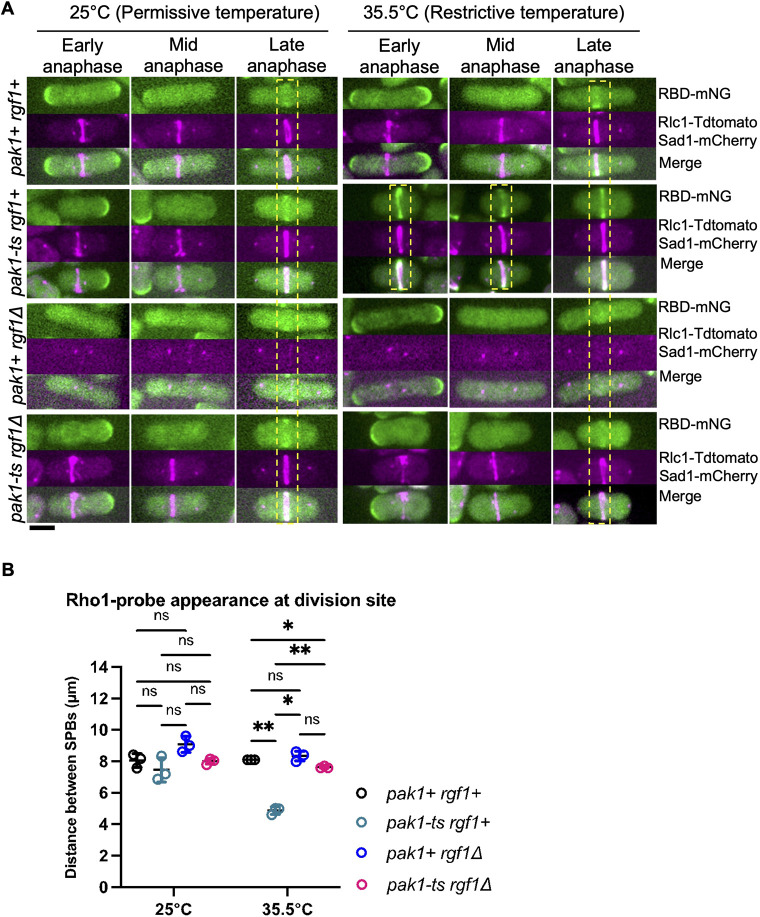
Given that Rho1 activation at the division site occurred early in pak1-ts cells at the restrictive temperature, we posit that Pak1 prevents Rgf1-dependent Rho1 activation in early anaphase. We tested this hypothesis by analyzing RBD–mNG localization in the rgf1Δ pak1-ts double mutant. If indeed Pak1 regulates Rgf1, then the loss of rgf1 in pak1-ts mutants should rescue early RBD-mNG localization at the division site. We imaged the rgf1+, pak1-ts, rgf1Δ, and pak1-ts rgf1Δ cells expressing RBD-mNG at permissive (25°C) and restrictive (35.5°C) temperatures. Cytokinesis progression was determined by measuring the distance between the mitotic SPBs labeled with Sad1–mCherry. We also labeled the actomyosin rings with Rlc1–tdTomato to discern whether the rings were constricting. The timing of RBD–mNG localization at the division site occurs late in cytokinesis during late anaphase as depicted by longer SPB distances in pak1+ rgf1+, pak1-ts rgf1+, and pak1+ rgf1Δ cells at 25°C (Fig. 7B). As previously shown, RBD–mNG localizes to the division site in early anaphase in pak1-ts rgf1+ cells at the restrictive temperature (35.5°C, Fig. 7A,B). In agreement with our reasoning, early RBD–mNG localization in pak1-ts mutants at 35.5°C was rescued in the absence of rgf1 (Fig. 7A,B). These data suggest that similar to what is seen in gef1Δ mutants, early Rho1 activation in the pak1-ts mutant is Rgf1 dependent.
Fig. 7.
Loss of rgf1 rescues early Rho1 activation in mutants with disrupted Pak1 function. (A) Rho1 activation (RBD–mNG) in rgf1Δ mutants in pak1-ts functional (25°C) and pak1-ts (orb2-34) hypomorphic (35.5°C) conditions. Yellow dashed boxes highlight stage of cytokinesis in which Rho1 activation is observed in representative cells for each indicated genotype. Scale bar: 5 µm. (B) Quantification of the distance between the SPBs at which Rho1 activation is observed in all conditions shown. The data points on the graph represent first quartile computations of measurements obtained (mean±s.d; n=3 replicate experiments). *P≤0.01; **P≤0.004; ns, not statistically significant (one-way ANOVA followed by Tukey's HSD test).
Although Rho1 is activated by the GEFs Rgf1–Rgf3, it is inactivated by the GAPs Rga1, Rga5 and Rga8 (Calonge et al., 2003; Nakano et al., 2001). Of these Rho1 GAPs, Rga5 has been implicated in Rho1 function during cytokinesis (Calonge et al., 2003). Loss of rga5 function has been reported to rescue septation defects in cells where Rho1 activity is impaired and is lethal in cells where Rho1 is overexpressed (Alcaide-Gavilan et al., 2014; Calonge et al., 2003). To address whether Cdc42 inhibited Rho1 activation by promoting Rga5 GAP function, we compared Rho1 activation at the division site in gef1+, rga5Δ, gef1Δ, and gef1Δ rga5Δ cells. We expect that if Rga5 is responsible for Cdc42-dependent Rho1 inhibition in early cytokinesis, then rga5Δ should phenocopy gef1Δ. Indeed, we found that in rga5Δ mutants RBD–mNG localizes to the division site earlier than in gef1+ cells (Fig. 6C); however, it was not as early as in gef1Δ cells. Thus, Rga5 alone cannot account for the Cdc42-dependent Rho1 inhibition observed in early anaphase, and other regulators such Rgf1 and Rgf2 could be involved in this process.
Early Rho1 activation leads to cytokinetic defects
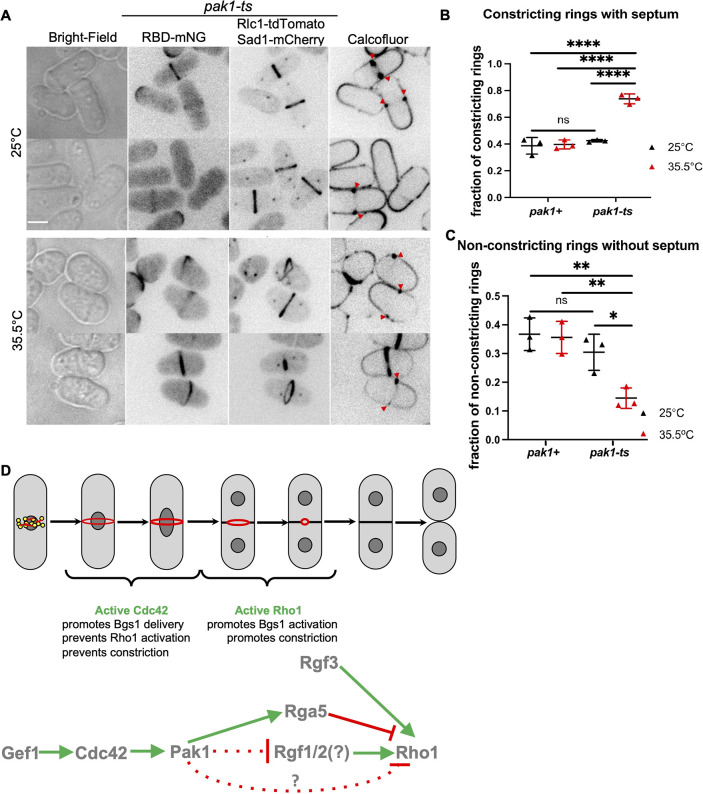
What are the implications of early Rho1 activation during cytokinesis? Normally during cytokinesis, the septum is formed in late anaphase, after the actomyosin ring assembles and matures. Furthermore, ring constriction is only initiated when septum deposition starts (Balasubramanian et al., 2004; Proctor et al., 2012). We reported previously that septum ingression and ring constriction begin ∼30 min after SPB separation, whereas the recruitment of the primary septum-synthesizing enzyme Bgs1 to the division site occurs ∼16 min after SPB separation (Wei et al., 2016). The Bgs1 enzyme becomes active and builds the septum only when it binds active Rho1 (Arellano et al., 1996). Thus, the time lag between Bgs1 localization and septum ingression likely occurs due to a delay in Bgs1 activity as a result of Rho1 inhibition in early cytokinesis. We have shown that in gef1Δ mutants Bgs1 recruitment to the division site is delayed and thus septum ingression is delayed in these mutants (Onwubiko et al., 2021; Wei et al., 2016). We postulate that early Rho1 activation in pak1-ts mutants will induce early Bgs1 activation, and consequently early septum deposition and ring constriction. In these cells, the time lag between ring assembly and constriction should be alleviated as septum synthesis initiates early. In an asynchronous population, cells undergoing cytokinesis with an assembled actomyosin ring are at one of the following stages – a non-constricting actomyosin ring with no septum deposits, a not-yet-constricting ring with early septum deposits, or with a constricting ring with mature septum deposits. We expect that in cells prematurely activating Rho1, the fraction of non-constricting rings without septum deposits would be lower, as in these cells, septum deposition will initiate early. In contrast, the fraction of constricting actomyosin rings with septum deposits would increase in these cells. Indeed, we found that pak1-ts mutants under restrictive conditions displayed a higher number of constricting rings with septum as compared to the controls (Fig. 8A,B; Fig. S8). Furthermore, in these mutants, the fraction of non-constricting rings without septum deposits is decreased (Fig. 8A,C; Fig. S8). Taken together, this suggests that ring constriction initiates early in the absence of pak1 kinase likely due to early septum deposition.
Fig. 8.
Disruption of Pak1 function results in early septum formation. (A) Rho1 activation (RBD–mNG) and septum formation in pak1-ts (orb2-34) strains grown at permissive and restrictive temperatures. Septum deposition at the division site (red arrowheads) is visualized with calcofluor staining. Scale bar: 5 µm. (B) Quantification of the fraction of constricting rings for cells undergoing cytokinesis in the indicated strains and conditions (mean±s.d; n=3 representative experiments). ****P<0.0001; ns, not statistically significant (one-way ANOVA with Tukey's HSD test). (C) Quantification of the fraction of non-constricting rings without the septum in the indicated strains and conditions (mean±s.d; n=3 representative experiments). *P≤0.025; **P≤0.005; ns, not statistically significant (one-way ANOVA with Tukey's HSD test). (D) Cdc42 prevents premature Rho1 activation during cytokinesis. Schematic describing how Cdc42 prevents early Rho1 activation and thus promotes proper cytokinesis fidelity. In early cytokinesis, active Cdc42 promotes recruitment of the septum-synthesizing enzyme Bgs1, but prevents Rho1 activation, thus inhibiting Bgs1 catalytic activity. In late cytokinesis, Rho1 is activated and this allows septum synthesis. Gef1 activates Cdc42, which in turn activates the Pak1 kinase. Rho1 is activated by the Rgf1 and/or Rgf2 (Rgf1/2) and Rgf3, and inactivated by the GAP Rga5. Rgf3 is an essential GEF and functions independently of the Cdc42 pathway. The data presented here suggest that Pak1 kinase prevents Rho1 activation, however the molecular details of this regulation is not known. Pak1 likely inhibits Rho1 activation either via inhibition of the GEFs Rgf1/2 or via activation of the GAPs Rga5. Alternatively, it is possible that Pak1 regulates Rho1 activation via an as yet unknown pathway.
DISCUSSION
Cytokinesis encompasses events that partition the cytoplasm of dividing cells at the end of the cell cycle. In fission yeast, cytokinesis is accomplished via septum formation and actomyosin ring constriction. The GTPase Rho1 is required for septum formation during cytokinesis and cell wall integrity (Arellano et al., 1996, 1997; Pérez et al., 2018). Previous work has indicated that Cdc42 promotes cytokinesis through its roles in septation, membrane trafficking and concentric furrow formation (Hercyk et al., 2019a; Onwubiko et al., 2019, 2021; Wei et al., 2016). Here, we investigated the relationship between Cdc42 and the essential GTPase Rho1 in the regulation of cytokinesis. We find that although Cdc42 is activated in early anaphase during ring assembly, Rho1 activation occurs in late anaphase, and this is immediately followed by ring constriction. Late Rho1 activation occurs in spite of the fact that the Rho1-specific GEFs Rgf1 and Rgf3 localize early to the division site, immediately after ring assembly, suggesting that either the GEFs were unable to activate Rho1 in early cytokinesis or Rho1 is tightly inhibited such that the GEFs are ineffective at this stage. Our data demonstrate that Cdc42 is responsible for the inhibition of Rho1 activity during early anaphase. In mutants lacking gef1, Cdc42 activation at the division site is delayed. In these mutants, Rho1 is activated in early anaphase immediately after the actomyosin ring assembles. We find that constitutively active Cdc42 prevents Rho1 activation at the division site and also at the ends of interphase cells. Together these observations suggest that Cdc42 globally inhibits Rho1 activation both at the division site and the site of cell growth.
The SIN pathway is required for coupling mitosis and cytokinetic events in cells, and has been reported to be upstream of Rho1 activation during cytokinesis (Alcaide-Gavilán et al., 2014). Using the Rho probe, we showed that the SIN is required for Rho1 activation at the division site and loss of Cdc42 activity cannot bypass this. We also show that the SIN pathway is not required for Rho1 activation at the cell ends. Given that Cdc42 inhibits Rho1 both at the division site and the cell ends, it further highlights that the SIN and Cdc42 pathways regulate Rho1 via independent mechanisms. Cdc42 is activated at the division site when the actomyosin ring assembles whereas Rho1 is activated just prior to ring constriction. The SIN pathway kinase Sid2 also localizes to the division site just prior to ring constriction (Alcaide-Gavilan et al., 2014). Thus, although Cdc42 prevents Rho1 activation during early cytokinesis, it is possible that Sid2 localizes to the division site in late cytokinesis to activate Rho1. It is unknown how the SIN activity overcomes Cdc42-mediated Rho1 inhibition during late cytokinesis. Further investigation will explain the molecular details of Rho1 activation in late cytokinesis.
Although our findings demonstrate Cdc42-mediated inhibition of Rho1 activation, the molecular details of how this is brought about is unclear. We find that early Rho1 activation in gef1Δ mutants is disrupted only in the absence of the GEF Rgf1. In contrast, absence of the GAP rga5 leads to premature Rho1 activation regardless of the presence of gef1. Thus, it is possible that Cdc42 inhibits Rho1 activation during early cytokinesis either via Rgf1 inhibition or Rga5 activation. Alternatively, it is possible that Cdc42 regulates the Rho1 protein itself and prevents it from being activated. Although Rgf1 is the primary GEF for Rho1 activation both at the division site and the cell ends, it is not essential (Morrell-Falvey et al., 2005; Mutoh et al., 2005; Tajadura et al., 2004). In contrast, Rgf3 an essential Rho1 GEF only localizes to the division site (Morrell-Falvey et al., 2005; Tajadura et al., 2004). It is not clear why different Rho1 GEFs show different activation patterns at the division site. Our findings suggest that in the absence of active Cdc42, Rgf3 is no longer essential likely due to Rgf1-mediated Rho1 activation.
Reports indicate that regulators of the Rho1 homolog RhoA respond to Pak-mediated regulation in animal cells (Alberts et al., 2005; DerMardirossian et al., 2004; Tiedje et al., 2008; Zenke et al., 2004). In fission yeast, the p21-activated kinase Pak1 localizes to the division site in early anaphase (Magliozzi et al., 2020). Our observations suggest that Gef1 mediates Rho1 inhibition in early anaphase via Pak1 kinase. It is possible that Pak1 phosphorylates Rgf1, Rga5, Rho1 or an intermediate protein to block Rho1 activation. Further biochemical analysis will determine the mechanism by which Pak1 blocks Rho1 activation.
We have previously shown that Cdc42 is activated in a Gef1-dependent manner to promote Bgs1 recruitment and for the timely onset of ring constriction (Wei et al., 2016). Thus, although Cdc42 activation promotes the delivery of the first septum-synthesizing enzyme, Bgs1, to the division site, it also ensures that this enzyme is not prematurely activated by inhibiting Rho1 (Fig. 8D). It is unclear why septum synthesis is tightly regulated during cytokinesis. One potential explanation is to ensure that septum ingression and cell partitioning occur only after completion of nuclear division. Indeed, a recent paper shows that, in fission yeast, septum ingression initiates during anaphase B, but at a much slower rate (Garcia Cortes et al., 2018). The rate of septum ingression increases only after the completion of anaphase B. Inhibition of Rho1 activation likely allows this careful coordination of mitosis and septum ingression. In gef1Δ cells, the delay in Bgs1 recruitment in the absence of active Cdc42, even as Rho1 is activated early, ensures a delay in septum ingression. In contrast, the pak1 mutants appear to initiate septum ingression early, likely due to proper Bgs1 delivery and premature Rho1 activation. Our findings might explain the previously reported observation that pak1 kinase defective mutants prematurely initiate ring constriction. In mutants undergoing slower anaphase, Pak1 played an important role in delaying cytokinesis and preventing chromosome segregation defects (Loo and Balasubramanian, 2008). Thus, active Cdc42 at the division site in anaphase enforces spatiotemporal regulation of β-glucan septum synthesis. Cdc42 therefore functions as a cellular quality control protein at the division site to facilitate time-dependent organization of cytokinetic events.
In animal cells, RhoA activation is essential for the formation of an actomyosin ring (Basant and Glotzer, 2018). RhoA is activated at the division site and this leads to actomyosin ring formation at that site (Wagner and Glotzer, 2016). However, in fission yeast Rho1 is activated at the division site only after the ring is fully assembled and ready to constrict. This activation pattern does not support a role for Rho1 in ring assembly but supports a role in septum formation. Our work further provides new details of Cdc42 and Rho1 crosstalk during fission yeast cytokinesis, where these GTPases localize in concentric zones to regulate essential cytokinetic steps. Although we show that Cdc42 inhibits Rho1 activation in early anaphase, this inhibition is finally removed in late anaphase to enable septum formation. Concentric zones of active Cdc42 and RhoA have also been reported to drive wound healing in Xenopus oocytes (Benink and Bement, 2005). Crosstalk between Cdc42 and RhoA maintains these zones of GTPase activity and constriction dynamics. Indeed, it has been shown that dominant-negative Cdc42 eliminates RhoA activation during constriction of the actomyosin array during wound healing, whereas constitutively active Cdc42 broadens the RhoA activity zone (Benink and Bement, 2005). The specific zones of activity for Rho GTPases might be required for regulating distinct cellular processes and could be determined via regulation of their GEFs and GAPs to maintain GTPase distinct zones. It will be interesting to assess whether Rho1-dependent effectors regulate active Cdc42 zones during cytokinesis. During growth in fission yeast, Cdc42 is restricted to growing tips primarily via the activity of its GAP Rga4 (Das et al., 2007; Rich-Robinson et al., 2021). At the onset of ring constriction during cytokinesis, Cdc42 GAPs also localize to the division site (Campbell et al., 2022; Rich-Robinson et al., 2021). These GAPs might establish distinct zones that are devoid of active Cdc42 where Rho1 activation can occur to finally allow septum formation. In budding yeast, antagonism between Cdc42 and Rho1 regulates secondary septum formation, and inactivation of Cdc42 is required for proper cytokinesis completion (Atkins et al., 2013; Onishi et al., 2013). In-depth biochemical analysis and mathematical models that probe toggles between GTPase regulators in concentric zones will provide insights into the mechanisms of crosstalk between these Rho GTPases.
MATERIALS AND METHODS
Strains and cell culture
Strains used in this study are listed in Table S1. The fission yeast strains used in this study are isogenic to PN972. Unless mentioned, cells were cultured in yeast extract (YES) medium (Sunrise Science; Petersen and Russell, 2016) and grown exponentially at 25°C. All genetic manipulations of strains were carried out using standard techniques (Moreno et al., 1991). Cells were grown exponentially for at least three rounds of eight generations before experiments were performed.
Microscopy
Image acquisition was performed at room temperature (23–25°C) on a spinning disk confocal system that uses a Nikon Eclipse inverted microscope with a 100×1.49NA objective, a CSU-22 spinning disk system, and a Photometrics EM-CCD camera from Visitech International. Images were acquired using Metamorph (Molecular Devices) and analyzed using ImageJ/FIJI Bio-Formats plugins (National Institutes of Health). Microscopy was also performed at room temperature using a 3i spinning disc confocal microscope with integrated Yokogawa spinning disk (Yokogawa CSU-X1 A1 spinning disk scanner) confocal on a Zeiss Axio Observer fully automated inverted microscope with a 100× −1.40 NA oil immersion objective and a Prime 95B sCMOS camera (Photometrics). Images were acquired on Slide Book digital microscopy software. For still images, cells were mounted on glass slides with a #1.5 coverslip (Thermo Fisher Scientific, Waltham, MA) and imaged right away. All Z-series images were acquired with a depth interval of 0.4 µm for a total of 6.2 µm. In time-lapse image acquisition, cells were placed in a 3.5-mm glass-bottom culture dish and covered with YES medium with 0.6% agar. Ascorbic acid (100 µM vitamin C) was added to the cell culture to minimize fluorescence toxicity, as previously reported (Wei et al., 2017). All images analyzed for mean intensity were acquired with a Z-series and sum-projected unless noted otherwise. Statistical analysis was performed using one-way ANOVA, followed by Tukey's honestly significant difference (HSD) post hoc test or unpaired, two-tailed Student's t-test where appropriate. Comparisons between experimental groups were considered significant when P≤0.05.
Designing the active Rho probe
The Rho probe was designed using the rho-binding domain of the budding yeast protein kinase C (PKC1) as described in Davidson et al. (2015) and Kono et al. (2012). From the budding yeast PKC1, the base pairs 1–1173 were tagged with a fluorescent probe and this was expressed under the fission yeast pkc2 promoter region from −464 to −1 upstream of the start codon. We stitched three fragments – the pkc2 promoter sequence, the RBD, and mNG-pjk148 or tdTomato-pjk210, using restriction sites in our primers. The C-terminal sequence of the RBD contains a glycine linker with either mNeonGreen or tdTomato in a Pjk148 or Pjk210 vector. Cells were transformed via lithium acetate transformation (Okazaki et al., 1990) to integrate constructs into the leu, or ura locus of PN975/YMD493. Primer sequences used for the pkc2 promoter region were: Fwd: 5′-AAGCTTGATATCGAATTCCTGCAGCCCGGGAATGAACTGTTCTATTAATTGGTC-3′ and Rev, 5′-GTGAAATCATTACTTTAAGCCTAATCC-3′.
Cytoskeleton disruptions
To block Arp2/3 complex-dependent branched actin assembly, cells were treated with 100 µM CK666 (Sigma-Aldrich, SML006-5MG) in DMSO (Sigma-Aldrich, D8418-250ML). To disassemble all F-actin, cells were treated with 100 µM Latrunculin A (LatA; EMD Millipore) dissolved in DMSO in YES medium for 30 min before imaging. To depolymerize microtubules, cells were treated with 25 µg/ml MBC dissolved in DMSO and incubated for 45 min before imaging. For all these experiments, control cells were treated with 0.1% DMSO in YES medium.
Expressing constitutively active Cdc42
The cdc42G12V fragment qA cloned into the pJK148 vector under the thiamine-repressible promoter nmt41 and integrated into the genome of gef1+ and gef1Δ cells, as previously described in Wei et al. (2016). Cells were initially grown in Edinburgh minimal medium (EMM; Sunrise Science; Petersen and Russell, 2016) with 15 µM thiamine. Partial induction of cdc42G12V expression was performed by harvesting strains via low-speed centrifugation (5000 g for 5 min), rinsing four times with deionized water, and then grown in EMM with 0.05 µM thiamine for 34 h before imaging at 25°C. The experimental controls were gef1+ and gef1Δ cells transformed with the empty pJK148 vector.
Quantification of fluorescence intensity
Fluorescence intensity was measured in images via ImageJ software. All images were sum-projected and mean intensities were reported. A box was positioned to measure the signal at the division site. The cytoplasm of the cell with little to no signal was used for background subtraction. Mean intensity measurements were collected after background subtractions.
Estimation of timing of cytokinetic progression
To determine the timing of cytokinetic progression, we measure the SPB distance in cells undergoing mitosis. Mitotic progression as measured via SPB distance functions as an internal clock for cytokinetic events. The timepoint at which the two spindle poles can be first distinguished is considered zero time. Z-series images of cells expressing the SPB protein Sad1–mCherry and the cytokinetic ring marker Rlc1–tdTomato (see Table S1) were acquired for cells in all conditions measured. The Line tool in ImageJ was used to measure the distance in microns (µm) between the SPBs in cells. Z-series helped visualize spindles that were on different focal planes. The Rlc1–tdTomato signal was also helpful for the clarification of constricting rings. Given that SPB measurements were obtained in an asynchronous population of cells, we only plotted the 25th percentile of our SPB data measurements which represents the smallest SPB measurements for each strain in each data set. All experiments were performed in triplicate.
Rho probe localization analysis
The onset of Rho probe localization was assessed by researchers who were not aware of the experimental conditions. The time point at which the Rho probe was first detected at the division site in a time-lapse experiment was recorded in the different strains and conditions used. For the temperature-sensitive rho1 allele, rho1+ and rho1-596 cells were grown at 25°C to log phase in YES culture medium. For restrictive-temperature experiments, cells were shifted to 36°C for 4 h. Post incubation, cells were concentrated by centrifugation (5000 g for 5 min) and imaged.
For rho1 switch-off, rho1::ura4/p41xRho1-RBD mNG-Rlc1-tdTomato and control cells were grown to log phase at 25°C in EMM. The log-phase culture was then transferred to EMM with 15 µM thiamine for 18 h at 25°C. Post incubation, cells were concentrated by centrifugation as above and imaged.
SIN inactivation via sid2-250-ts
sid2-250 temperature-sensitive cells were cultured in YES medium until healthy. Cells were grown to an optical density (OD) at 595 nm of 0.2 at 25°C, and the culture split into two halves, for each strain. One half was kept at the permissive temperature of 25°C for 4 h prior and then imaged. The other half was shifted to the restrictive temperature of 36°C for 4 h to inactivate Sid2 and then imaged as previously performed (Feoktistova et al., 2012).
rgf3 repression
To repress rgf3, nmt-81-rgf3 cells (VT88) were grown in EMM supplemented with thiamine. Weak suppression of rgf3 was achieved by growing nmt-81-rgf3 cells in EMM supplemented with 15 µM thiamine for six generations as previously performed (Tajadura et al., 2004). For controls, cells were grown in supplemented EMM with no thiamine for the same amount of time.
Spot growth assay
Cells were grown to OD 0.5 (107cells/μl), in EMM supplemented with adenine, leucine, histidine and uridine, (A, L, U, H) at 25°C. Serial dilutions were set up from 104 to 101 cells, which were spotted for each strain on both EMM-ALUH+0μM thiamine, and EMM-ALUH+15μM thiamine media plates and incubated at 25°C. Cell growth was assessed after 7 days.
Disruption of Pak1 kinase function via temperature-sensitive allele orb2-34
Pak1 kinase was inactivated via the orb2-34 (pak1-ts) temperature-sensitive mutation. Cells were grown till healthy in YES medium. On the day of the experiment, cells were cultured to an OD of 0.2. A subset of these cells were incubated at the permissive temperature of 25°C, while the other set was incubated at the restrictive temperature of 35.5°C for 4 h. Cells were imaged after 4 h.
Pak1 overexpression and repression
In experiments with nmt1-3HA-pak1 (MBY), cells were grown in YES medium for at least three generations. Cells were washed thoroughly four times with thiamine-free EMM, switching to a new tube during the fourth wash. Washed cells were divided into two groups. For repression of nmt1-3HA-pak1, washed cells were transferred to EMM with 15 μM thiamine and grown for 48 h then imaged. For pak1OE, washed cells were transferred to EMM containing no thiamine and grown for 48 h, and imaged immediately.
Calcofluor staining
To stain the septum and cell wall, live cells were stained in YE liquid with 50 μg/ml Calcofluor White M2R (Sigma-Aldrich) at room temperature and imaged.
Supplementary Material
Acknowledgements
We thank Yolanda Sanchez, Kathy Gould, Mohan Balasubramanian, and Pilar Perez for providing strains and Bret Judson at Boston College for confocal imaging support.
Footnotes
Author contributions
Conceptualization: U.N.O., M.D.; Methodology: U.N.O., D.K., M.D.; Validation: U.N.O., D.K.; Formal analysis: U.N.O., D.K., E.K., S.P., H.R.; Investigation: U.N.O., D.K., E.K., S.P., H.R., A.M.; Resources: M.D.; Data curation: U.N.O., D.K.; Writing - original draft: U.N.O.; Writing - review & editing: U.N.O., D.K., M.D.; Visualization: U.N.O., D.K.; Supervision: M.D.; Project administration: M.D.; Funding acquisition: M.D.
Funding
This work is supported by the National Science Foundation (NSF; 1616495, 1941367). U.N.O. was supported by National Institutes of Health IMSD (R25GM086761) and is currently supported by an NSF GRFP (DGE-1452154). Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/lookup/doi/10.1242/jcs.261160.reviewer-comments.pdf.
References
- Alberts, A. S., Qin, H., Carr, H. S. and Frost, J. A. (2005). PAK1 negatively regulates the activity of the Rho exchange factor NET1. J. Biol. Chem. 280, 12152-12161. 10.1074/jbc.M405073200 [DOI] [PubMed] [Google Scholar]
- Alcaide-Gavilán, M., Lahoz, A., Daga, R. R. and Jimenez, J. (2014). Feedback regulation of SIN by Etd1 and Rho1 in fission yeast. Genetics 196, 455-470. 10.1534/genetics.113.155218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada, R. and Pollard, T. D. (2014). Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep. 8, 1533-1544. 10.1016/j.celrep.2014.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano, M., Duran, A. and Perez, P. (1996). Rho 1 GTPase activates the (1-3)beta-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 15, 4584-4591. 10.1002/j.1460-2075.1996.tb00836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano, M., Duran, A. and Perez, P. (1997). Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J. Cell Sci. 110, 2547-2555. 10.1242/jcs.110.20.2547 [DOI] [PubMed] [Google Scholar]
- Arellano, M., Valdivieso, M. H., Calonge, T. M., Coll, P. M., Duran, A. and Perez, P. (1999). Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J. Cell Sci. 112, 3569-3578. 10.1242/jcs.112.20.3569 [DOI] [PubMed] [Google Scholar]
- Atkins, B. D., Yoshida, S., Saito, K., Wu, C. F., Lew, D. J. and Pellman, D. (2013). Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 202, 231-240. 10.1083/jcb.201301090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, M. K., Bi, E. and Glotzer, M. (2004). Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14, R806-R818. 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Basant, A. and Glotzer, M. (2018). Spatiotemporal regulation of RhoA during Cytokinesis. Curr. Biol. 28, R570-R580. 10.1016/j.cub.2018.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benink, H. A. and Bement, W. M. (2005). Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 168, 429-439. 10.1083/jcb.200411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., Drgonová, J. and Drgon, T. (1998). Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67, 307-333. 10.1146/annurev.biochem.67.1.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge, T. M., Arellano, M., Coll, P. M. and Perez, P. (2003). Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 47, 507-518. 10.1046/j.1365-2958.2003.03312.x [DOI] [PubMed] [Google Scholar]
- Campbell, B. F., Hercyk, B. S., Williams, A. R., San Miguel, E., Young, H. G. and Das, M. E. (2022). Cdc42 GTPase activating proteins Rga4 and Rga6 coordinate septum synthesis and membrane trafficking at the division plane during cytokinesis. Traffic 23, 478-495. 10.1111/tra.12864 [DOI] [PubMed] [Google Scholar]
- Chen, F., Ma, L., Parrini, M. C., Mao, X., Lopez, M., Wu, C., Marks, P. W., Davidson, L., Kwiatkowski, D. J., Kirchhausen, T.et al. (2000). Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr. Biol. 10, 758-765. 10.1016/S0960-9822(00)00571-6 [DOI] [PubMed] [Google Scholar]
- Cortes, J. C., Ishiguro, J., Duran, A. and Ribas, J. C. (2002). Localization of the (1,3)beta-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 115, 4081-4096. 10.1242/jcs.00085 [DOI] [PubMed] [Google Scholar]
- Cortes, J. C., Konomi, M., Martins, I. M., Munoz, J., Moreno, M. B., Osumi, M., Duran, A. and Ribas, J. C. (2007). The (1,3)beta-D-glucan synthase subunit Bgs1p is responsible for the fission yeast primary septum formation. Mol. Microbiol. 65, 201-217. 10.1111/j.1365-2958.2007.05784.x [DOI] [PubMed] [Google Scholar]
- Cortes, J. C., Pujol, N., Sato, M., Pinar, M., Ramos, M., Moreno, B., Osumi, M., Ribas, J. C. and Perez, P. (2015). Cooperation between Paxillin-like Protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet. 11, e1005358. 10.1371/journal.pgen.1005358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes, J. C., Ramos, M., Osumi, M., Perez, P. and Ribas, J. C. (2016). Fission yeast septation. Commun. Integr. Biol. 9, e1189045. 10.1080/19420889.2016.1189045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Wiley, D. J., Medina, S., Vincent, H. A., Larrea, M., Oriolo, A. and Verde, F. (2007). Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell 18, 2090-2101. 10.1091/mbc.e06-09-0883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Drake, T., Wiley, D. J., Buchwald, P., Vavylonis, D. and Verde, F. (2012). Oscillatory Dynamics of Cdc42 GTPase in the Control of Polarized Growth. Science 337, 239-243. 10.1126/science.1218377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, R., Laporte, D. and Wu, J. Q. (2015). Regulation of Rho-GEF Rgf3 by the arrestin Art1 in fission yeast cytokinesis. Mol. Biol. Cell 26, 453-466. 10.1091/mbc.E14-07-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian, C., Schnelzer, A. and Bokoch, G. M. (2004). Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 15, 117-127. 10.1016/j.molcel.2004.05.019 [DOI] [PubMed] [Google Scholar]
- Drechsel, D. N., Hyman, A. A., Hall, A. and Glotzer, M. (1997). A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr. Biol. 7, 12-23. 10.1016/S0960-9822(06)00023-6 [DOI] [PubMed] [Google Scholar]
- Feoktistova, A., Morrell-Falvey, J., Chen, J.-S., Singh, N. S., Balasubramanian, M. K. and Gould, K. L. (2012). The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol. Biol. Cell 23, 1636-1645. 10.1091/mbc.e11-09-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Cortes, J. C., Ramos, M., Konomi, M., Barragan, I., Moreno, M. B., Alcaide-Gavilan, M., Moreno, S., Osumi, M., Perez, P. and Ribas, J. C. (2018). Specific detection of fission yeast primary septum reveals septum and cleavage furrow ingression during early anaphase independent of mitosis completion. PLoS Genet. 14, e1007388. 10.1371/journal.pgen.1007388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S. and Das, M. E. (2019a). Rho Family GTPases in fission yeast cytokinesis. Commun. Integr. Bio.l 12, 171-180. 10.1080/19420889.2019.1678453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S. and Das, M. E. (2019b). F-BAR Cdc15 promotes Gef1-mediated Cdc42 activation during cytokinesis and cell polarization in S. pombe. Genetics 213, 1341-1356. 10.1534/genetics.119.302649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S., Onwubiko, U. N. and Das, M. E. (2019a). Coordinating septum formation and the actomyosin ring during cytokinesis in Schizosaccharomyces pombe. Mol. Microbiol. 112, 1645-1657. 10.1111/mmi.14387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S., Rich-Robinson, J., Mitoubsi, A. S., Harrell, M. A. and Das, M. E. (2019b). A novel interplay between GEFs orchestrates Cdc42 activity during cell polarity and cytokinesis in fission yeast. J. Cell Sci. 132, jcs236018. 10.1242/jcs.236018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, M.-C., Salek, J. and McCollum, D. (2000). Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10, 619-622. 10.1016/S0960-9822(00)00492-9 [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger, V., Gonczy, P., Romano, A., Schnabel, H., Hamill, D., Schnabel, R., Hyman, A. A. and Glotzer, M. (2000). CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149, 1391-1404. 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javerzat, J. P., Cranston, G. and Allshire, R. C. (1996). Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 24, 4676-4683. 10.1093/nar/24.23.4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q. W., Zhou, M., Bimbo, A., Balasubramanian, M. K. and McCollum, D. (2006). A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2-250 suppressors. Genetics 172, 2101-2112. 10.1534/genetics.105.050955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. E., McCollum, D. and Gould, K. L. (2012). Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 69, 686-699. 10.1002/cm.21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, S. N. and Canman, J. C. (2012). Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken) 69, 919-930. 10.1002/cm.21071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, K., Saeki, Y., Yoshida, S., Tanaka, K. and Pellman, D. (2012). Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell 150, 151-164. 10.1016/j.cell.2012.05.030 [DOI] [PubMed] [Google Scholar]
- Loo, T. H. and Balasubramanian, M. (2008). Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J. Cell Biol. 183, 785-793. 10.1083/jcb.200806127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliozzi, J. O., Sears, J., Cressey, L., Brady, M., Opalko, H. E., Kettenbach, A. N. and Moseley, J. B. (2020). Fission yeast Pak1 phosphorylates anillin-like Mid1 for spatial control of cytokinesis. J. Cell Biol. 219, e201908017. 10.1083/jcb.201908017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A. and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. 10.1016/0076-6879(91)94059-l [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey, J. L., Ren, L., Feoktistova, A., Haese, G. D. and Gould, K. L. (2005). Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J. Cell Sci. 118, 5563-5573. 10.1242/jcs.02664 [DOI] [PubMed] [Google Scholar]
- Munoz, J., Cortes, J. C., Sipiczki, M., Ramos, M., Clemente-Ramos, J. A., Moreno, M. B., Martins, I. M., Perez, P. and Ribas, J. C. (2013). Extracellular cell wall beta(1,3)glucan is required to couple septation to actomyosin ring contraction. J. Cell Biol. 203, 265-282. 10.1083/jcb.201304132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh, T., Nakano, K. and Mabuchi, I. (2005). Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells 10, 1189-1202. 10.1111/j.1365-2443.2005.00908.x [DOI] [PubMed] [Google Scholar]
- Na, J. and Zernicka-Goetz, M. (2006). Asymmetric positioning and organization of the meiotic spindle of mouse oocytes requires CDC42 function. Curr. Biol. 16, 1249-1254. 10.1016/j.cub.2006.05.023 [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Nakagawa, T., Straight, A. F., Murray, A., Chikashige, Y., Yamashita, Y. M., Hiraoka, Y. and Yanagida, M. (1998). Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9, 3211-3225. 10.1091/mbc.9.11.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K., Mutoh, T. and Mabuchi, I. (2001). Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells 6, 1031-1042. 10.1046/j.1365-2443.2001.00485.x [DOI] [PubMed] [Google Scholar]
- Nolen, B. J., Tomasevic, N., Russell, A., Pierce, D. W., Jia, Z., McCormick, C. D., Hartman, J., Sakowicz, R. and Pollard, T. D. (2009). Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031-1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, K., Okazaki, N., Kume, K., Jinno, S., Tanaka, K. and Okayama, H. (1990). High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 18, 6485-6489. 10.1093/nar/18.22.6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi, M., Ko, N., Nishihama, R. and Pringle, J. R. (2013). Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol. 202, 311-329. 10.1083/jcb.201302001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwubiko, U. N., Mlynarczyk, P. J., Wei, B., Habiyaremye, J., Clack, A., Abel, S. M. and Das, M. E. (2019). A Cdc42 GEF, Gef1, through endocytosis organizes F-BAR Cdc15 along the actomyosin ring and promotes concentric furrowing. J. Cell Sci. 132, jcs223776. 10.1242/jcs.223776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwubiko, U. N., Rich-Robinson, J., Mustaf, R. A. and Das, M. E. (2021). Cdc42 promotes Bgs1 recruitment for septum synthesis and glucanase localization for cell separation during cytokinesis in fission yeast. Small GTPases 12, 257-264. 10.1080/21541248.2020.1743926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottilie, S., Miller, P. J., Johnson, D. I., Creasy, C. L., Sells, M. A., Bagrodia, S., Forsburg, S. L. and Chernoff, J. (1995). Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 14, 5908-5919. 10.1002/j.1460-2075.1995.tb00278.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, A., Bakka, K., Sevugan, M., Naqvi, N. I., D'Souza, V., Tang, X., Mishra, M. and Balasubramanian, M. K. (2011). IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr. Biol. 21, 467-472. 10.1016/j.cub.2011.01.059 [DOI] [PubMed] [Google Scholar]
- Pérez, P., Cortés, J. C. G., Cansado, J. and Ribas, J. C. (2018). Fission yeast cell wall biosynthesis and cell integrity signalling. The Cell Surface 4, 1-9. 10.1016/j.tcsw.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, J. and Russell, P. (2016). Growth and the Environment of Schizosaccharomyces pombe. Cold Spring Harb Protoc. 2016, pdb.top079764. 10.1101/pdb.top079764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D. (2010). Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22, 50-56. 10.1016/j.ceb.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D. and Wu, J.-Q. (2010). Understanding cytokinesis: lessons from fission yeast. Nature reviews. Molecular cell biology 11, 149-155. 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, S. A., Minc, N., Boudaoud, A. and Chang, F. (2012). Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr. Biol. 22, 1601-1608. 10.1016/j.cub.2012.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich-Robinson, J., Russell, A., Mancini, E. and Das, M. (2021). Cdc42 reactivation at growth sites is regulated by local cell-cycle-dependent loss of its GTPase-activating protein Rga4 in fission yeast. J. Cell Sci. 134, jcs259291. 10.1242/jcs.259291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura, V., Garcia, B., Garcia, I., Garcia, P. and Sanchez, Y. (2004). Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J. Cell Sci. 117, 6163-6174. 10.1242/jcs.01530 [DOI] [PubMed] [Google Scholar]
- Tatebe, H., Nakano, K., Maximo, R. and Shiozaki, K. (2008). Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18, 322-330. 10.1016/j.cub.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje, C., Sakwa, I., Just, U. and Höfken, T. (2008). The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol. Biol. Cell 19, 2885-2896. 10.1091/mbc.e07-11-1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, F., Mata, J. and Nurse, P. (1995). Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131, 1529-1538. 10.1083/jcb.131.6.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica, A., Tang, X.-Z. and Oliferenko, S. (2008). The Actomyosin Ring Recruits Early Secretory Compartments to the Division Site in Fission Yeast. Mol. Biol. Cell 19, 1125-1138. 10.1091/mbc.e07-07-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. and Glotzer, M. (2016). Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213, 641-649. 10.1083/jcb.201603025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Lee, I. J., Rask, G. and Wu, J. Q. (2016). Roles of the TRAPP-II complex and the exocyst in membrane deposition during fission yeast cytokinesis. PLoS Biol. 14, e1002437. 10.1371/journal.pbio.1002437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B., Hercyk, B. S., Habiyaremye, J. and Das, M. (2017). Spatiotemporal analysis of cytokinetic events in fission yeast. J. Vis. Exp. 120, 55109. 10.3791/55109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B., Hercyk, B. S., Mattson, N., Mohammadi, A., Rich, J., DeBruyne, E., Clark, M. M. and Das, M. (2016). Unique spatiotemporal activation pattern of Cdc42 by Gef1 and Scd1 promotes different events during cytokinesis. Mol. Biol. Cell 27, 1235-1245. 10.1091/mbc.E15-10-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. Q., Kuhn, J. R., Kovar, D. R. and Pollard, T. D. (2003). Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell 5, 723-734. 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu, J. Q., Sirotkin, V., Kovar, D. R., Lord, M., Beltzner, C. C., Kuhn, J. R. and Pollard, T. D. (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174, 391-402. 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Kono, K., Lowery, D. M., Bartolini, S., Yaffe, M. B., Ohya, Y. and Pellman, D. (2006). Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313, 108-111. 10.1126/science.1126747 [DOI] [PubMed] [Google Scholar]
- Yoshida, S., Bartolini, S. and Pellman, D. (2009). Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 23, 810-823. 10.1101/gad.1785209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke, F. T., Krendel, M., DerMardirossian, C., King, C. C., Bohl, B. P. and Bokoch, G. M. (2004). p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J. Biol. Chem. 279, 18392-18400. 10.1074/jbc.M400084200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.