Abstract
The enzyme nicotine oxidoreductase (NicA2) is a member of the flavoprotein amine oxidase family that uses a cytochrome c protein (CycN) as its oxidant instead of dioxygen, which is the oxidant used by most other members of this enzyme family. We recently identified a potential binding site for CycN on the surface of NicA2 through rigid body docking [J. Biol. Chem. (2022) 298(8), 102251]. However, this potential binding interface has not been experimentally validated. In this paper, we used unnatural amino acid incorporation to probe the binding interface between NicA2 and CycN. Our results are consistent with a structural model of the NicA2-CycN complex predicted by protein-protein docking and AlphaFold, suggesting that this is the binding site for CycN on NicA2’s surface. Based on additional mutagenesis of potentially redox active residues in NicA2, we propose that electron transfer from NicA2’s flavin to CycN’s heme occurs without the assistance of a protein-derived wire.
Graphical Abstract
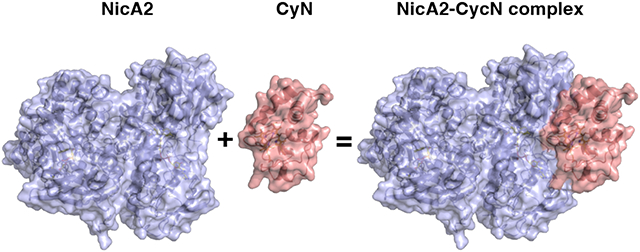
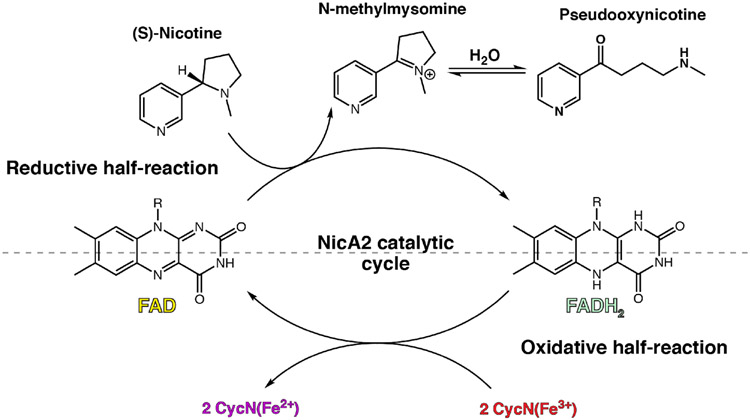
Nicotine oxidoreductase (NicA2) is an enzyme that facilitates the degradation of nicotine in the organism Pseudomonas putida S16. Commonly found in tobacco fields, this organism can use nicotine as its sole carbon and nitrogen source.1 This is possible due to NicA2’s flavin adenine dinucleotide (FAD) cofactor which is responsible for converting nicotine into N-methylmysomine, and subsequently psuedooxynicotine (Pon) after hydrolysis, in the first step of nicotine catabolism (Figure 1).2 Due to this nicotine degradation property, NicA2 has gained attention as a smoking cessation therapeutic.3 When given to nicotine-dependent mice, NicA2 has been shown to expunge nicotine from the blood, mitigate symptoms of withdrawal, and reduce compulsive nicotine intake.4,5 However, unlike other members of the flavoprotein amine oxidase (FAO) family, NicA2 does not react with molecular oxygen (O2) as an oxidant on a physiologically relevant timescale, leading to diminished efficacy as a therapeutic.5-10 Our lab has recently discovered that O2 is not the physiological electron acceptor of NicA2 but rather a cytochrome c protein (CycN) native to P. putida S16, meaning that NicA2 is a dehydrogenase unlike most other FAOs.11 When using CycN as an electron acceptor in vitro, NicA2 is oxidized as quickly as other flavoprotein amine oxidases that utilize O2 as their electron acceptor.
Figure 1.
The catalytic cycle for nicotine oxidoreductase (NicA2). Like many flavin-dependent enzymes, the catalytic cycle can be split into reductive and oxidative half-reactions that can be studied independently.
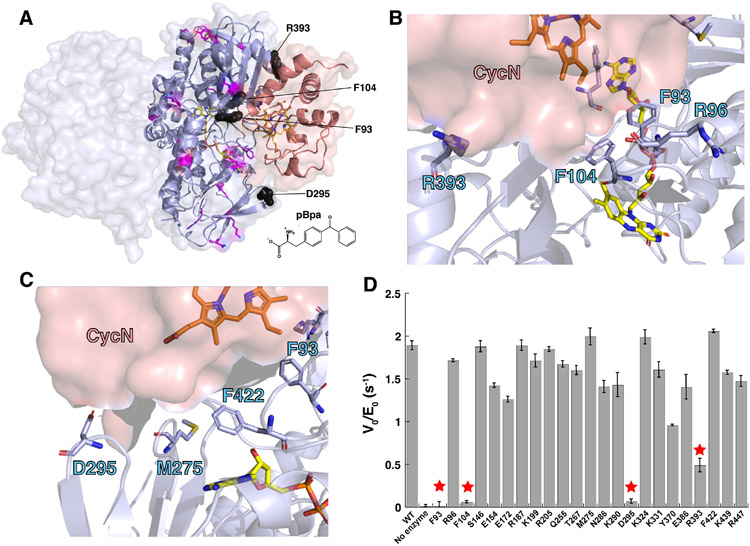
Our lab has also recently characterized the enzyme pseudooxynicotine amine dehydrogenase (Pnad),12 formerly called pseudooxynicotine amine oxidase,1 another FAO family enzyme that is responsible for the second step in nicotine catabolism in P. putida S16, where Pon is converted into 3-succinoylsemialdehyde-pyridine and methylamine. Pnad forms an operon with nicA2 and cycN in the genome of P. putida S16, and our biochemical characterization of Pnad indicated that it, like NicA2, is also a dehydrogenase that uses CycN as its natural electron acceptor. Rigid body docking of CycN on the structure of NicA2 and Pnad using ZDOCK13 identified a potential binding site that is conserved between these two proteins.12 AlphaFold2 Multimer14,15 of the NicA2 dimer with CycN also predicts that CycN binds in the same location predicted by ZDOCK, and this AlphaFold-predicted complex is shown in Figure 2A (the coordinates for this NicA2-CycN AlphaFold complex can be downloaded using the link in the Notes section).
Figure 2.
(A) Structure of the NicA2-CycN complex predicted by AlphaFold2 Multimer. The NicA2 homodimer is shown in light blue and CycN is shown in salmon. Residue positions replaced with pBpa that had minimal impact on the rate of electron transfer from nicotine to CycN are shown as sticks in magenta. Residue positions replaced with pBpa that substantial decreased the rate of electron transfer from nicotine to CycN are shown as black spheres. The chemical structure of pBpa is shown for reference. (B) and (C) Orientation of side chains for select residues in NicA2 at the predicted CycN binding site that were mutated to pBpa in this study. (D) Measured rate of electron transfer from nicotine to CycN by various pBpa-containing mutants of NicA2. Residue positions that dramatically decreased the rate of electron transfer upon replacement with pBpa are labeled with stars.
However, this potential binding site has not been experimentally validated, which was the primary objective of this study. Unfortunately, CycN binds to NicA2 with poor affinity,11 making it difficult to identify their binding interface through crystallography or other methods that rely on the formation of a stable complex. In this communication, we used an indirect approach to identify the binding interface between NicA2 and CycN by using unnatural amino acid incorporation followed by activity assays. The CycN binding site on NicA2 identified using this approach closely overlaps with the potential binding interface identified by ZDOCK and AlphaFold, suggesting that this is the true binding site for CycN. Furthermore, tryptophan and tyrosine residues in NicA2 adjacent to its FAD are not necessary for electron transfer from NicA2’s flavin to CycN’s heme on the surface of NicA2, suggesting that electron transfer between these two redox cofactors is unassisted by protein residues in the NicA2-CycN complex.
Our initial objective was to identify the CycN binding site on NicA2 using genetic code expansion to site-specifically incorporate the unnatural amino acid, p-benzoylphenylalanine (pBpa), at several different positions on the surface of NicA2, which can capture weak interactions through photoactivatable crosslinking.16,17 Our expectation was that NicA2 mutants containing pBpa at CycN binding sites would form crosslinks with CycN upon UV illumination whereas residues distant from the CycN binding site would not. Twenty-eight surface residue positions were selected, including eight in the region of the potential CycN docking site identified by AlphaFold Multimer (Ser86, Phe93, Arg96, Phe104, Met275, Asp295, Arg393, and Phe422), and site-directed mutagenesis was used to individually introduce the amber stop codon at these positions (Figure 2A-C). We were successfully able to express and purify 23 of the 28 pBpa-containing FAD bound variants via C-terminal his tags whereas five (Arg78, Ser86, Asp136, Lys346 and Phe405) were insoluble and were not studied further. Crosslinking between the pBpa-containing NicA2 mutants and CycN was attempted by exposing samples containing a singular NicA2 mutant and CycN to a 365 nm UV lamp followed by SDS-PAGE and heme staining.16,18 The heme staining for all variant samples showed a strong band at ~12 kDa for free CycN, but no higher molecular weight crosslinked species were observed for any of the pBpa-containing variants (Figure S1), indicating that crosslinking did not occur with any of the NicA2 variants.
Upon reflection, we hypothesized that due to the bulky size of pBpa relative to naturally occurring amino acids, incorporating pBpa at the binding interface could interfere with CycN’s ability to bind to NicA2 due to steric hindrance, which would prevent crosslinking from occurring; however, this should also result in a reduced rate of electron transfer between NicA2 and CycN. To test this hypothesis, we measured the rate of CycN reduction in the presence of nicotine for each of the pBpa variants using the increase in absorbance at 550 nm that accompanies reduction of CycN’s heme from the ferric (3+) to ferrous (2+) state as a readout.19 The rate of CycN reduction by most of the pBpa-containing variants was comparable to WT NicA2 (Figure 2D). However, mutants containing pBpa at positions Phe93, Phe104, and Asp295 showed a dramatically reduced rate of CycN reduction to a level comparable to that of background, indicating that the ability to transfer electrons from nicotine to CycN is severely compromised in these variants. Variants containing pBpa at positions Arg393 and Tyr370 also showed more modest reductions in relative activity, but still had activity above that of background, indicating that electron transfer to CycN is partially compromised in these two variants. Interestingly, Phe93, Phe104, Asp295 and Arg393 are in the same region as the predicted CycN binding site (Figure 2), consistent with this location being the true CycN binding site on NicA2.
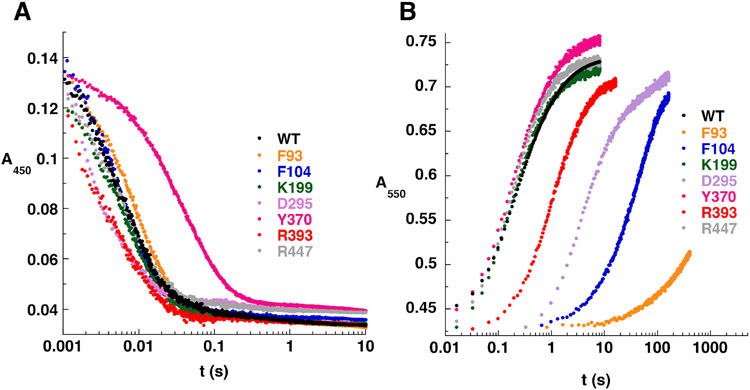
Although the above data indicate that pBpa incorporation at positions Phe93, Phe104, Asp295 and Arg393 inhibits NicA2 catalyzed turnover between nicotine and CycN, it does not indicate whether the electron transfer between nicotine and NicA2 or the subsequent electron transfer between NicA2 and CycN is impaired in these variants. To disentangle these two possibilities, we kinetically characterized the two individual half reactions for each pBpa-containing variant using anaerobic stopped-flow experiments (Figure 3).11,20 The mutants analyzed by stopped-flow included the five mutants with impaired relative activity as well as control mutants containing pBpa at positions Lys199 and Arg447. These two mutants were chosen at random from those that showed no impairment in enzyme activity relative to WT. Each pBpa-containing NicA2 mutant was anaerobically mixed with 100 μM nicotine in the stopped-flow to characterize the reductive half-reaction and the reaction monitored by following flavin reduction at 450 nm (Figure 3A). Reaction traces fit best to three exponentials, as observed previously with WT NicA2.11 All mutants reacted with nicotine in under 0.1 s on a similar timescale and had similar observed rate constants (kobs) as WT, except for mutant Tyr370, which took nearly 1 s to react and had rate constants ~2-4 fold lower than WT (Table S1). This indicates that the modest decrease in relative activity during turnover observed for this variant was likely due to a decreased rate constant(s) of electron transfer between nicotine and NicA2 and not the rate constant(s) of electron transfer to CycN. Tyr370 is on the surface of NicA2 more than 15 Å from the nicotine binding site and there may be some long-range effect on catalysis such as a change in dynamics upon substitution of this residue with pBpa. Notably, the rest of the pBpa-containing variants react with nicotine on a similar timescale as WT, indicating that pBpa incorporation at these positions does not impair the reductive half reaction for these enzymes.
Figure 3.
(A) Stopped-flow absorbance traces for the reductive half-reaction of pBpa-containing NicA2 mutants. (B) Stopped-flow absorbance traces for the oxidative half-reaction of pBpa-containing NicA2 mutants with CycN. Note the logarithmic time base in both panels. The kobs values from fitting reaction traces can be found in Tables S1 and S2.
We next studied the kinetics of the oxidative half reaction for the pBpa-containing variants in anaerobic stopped-flow experiments (Figure 3B). For this experiment, NicA2’s flavin was first reduced by anaerobic titration with one equivalent of nicotine in a tonometer, and the resulting reduced NicA2 was mixed with 80 μM ferric CycN in the stopped-flow and the reaction monitored by following CycN reduction at 550 nm. Here, a striking difference in kinetics was observed for some of the mutants reacting with CycN. Mutants containing pBpa at positions Phe93, Phe104, and Asp295 reduced CycN much slower than WT, with Phe104 and Asp295 taking more than 100 seconds for the reaction to complete and Phe93 not reaching completion after 400 seconds (Figure 3B). Reaction traces for all variants except for Phe93 were fit to a double exponential function as done previously for WT.11 For Phe93, the reaction was so slow that data collection was terminated before completion and the resulting partial reaction trace could only be fitted to a single exponential. The kobs values for variants containing pBpa at Lys199, Tyr370 and Arg447 were similar to WT, indicating that CycN binding and electron transfer is unaffected when pBpa is placed at these positions (Table S2). In contrast the kobs values for the reaction with CycN are 20 to 1300-fold lower for variants containing pBpa at Phe93, Phe104 and Asp295, indicating that the ability to bind and transfer electrons to CycN is severely compromised in these variants (Table S2). The kobs values for the reaction of NicA2 containing pBpa at position Arg393 are reduced only 3-4 fold relative to that of WT, in agreement with the more modest decrease in relative activity for this variant observed in Figure 2D. Importantly, the above results, combined with our analysis of the reductive half reaction, indicate that pBpa incorporation at positions Phe93, Phe104, Asp295 and Arg393 impair NicA2 binding and/or electron transfer to CycN and not the reaction with nicotine. To confirm that the observed decrease in rate constants for electron transfer to CycN in these pBpa-containing mutants is due to steric occlusion and not some other property of pBpa, we individually replaced Phe93 and Phe104, the two positions with the most dramatic decrease in electron transfer rate constants in the pBpa variants, with lysine. Our rationale for using lysine is that it is longer than phenylalanine and contains a positive charge, and we predicted that these two features would interfere with CycN binding if lysine were present at this putative binding site. We measured the kinetics of electron transfer from reduced Phe93Lys and Phe104Lys NicA2 to CycN in stopped-flow experiments, which showed that electron transfer is indeed severely compromised (kobs values ~100-fold lower than WT) in these two lysine containing variants of NicA2 (Figure S2).
Phe93, Phe104, Asp295 and Arg393 are all located in a similar region on the surface of NicA2’s structure, and this region overlaps with the CycN binding site predicted by ZDOCK and AlphaFold (Figure 2A-C).12 This, combined with the fact that electron transfer to CycN is not impaired when pBpa is present at other surface-exposed positions on NicA2 suggests that this predicted binding site is the true binding site for CycN. Phe93 and Phe104 are at the center of this putative binding site, which may explain why the pBpa-containing mutants at these positions produced the most dramatic decrease in rate constant(s) for electron transfer to CycN. Asp295 and Arg393 are located near the edge of the putative binding site, in agreement with the more modest reduction in electron transfer rate constant(s) when replaced with pBpa. Curiously, Arg96, Met275 and Phe422 are also in this same surface-exposed region on NicA2, but do not result in reduced rate constant(s) of electron transfer to CycN when replaced with pBpa. However, these three residues are on the periphery of the predicted CycN binding site and their side chains are oriented away from the space that CycN is predicted to occupy, unlike Phe93, Phe104 and Asp295 where the side chains are oriented towards this space (Figure 2B-C), and this fact may explain why pBpa incorporation at Arg96, Met275 and Phe422 does not affect electron transfer to CycN.
Ionic interactions between complementary charged amino acid side chains have been shown to be important for the interaction between other cytochrome c proteins and their binding partners, such as the flavocytochrome b2-cytochrome c complex and the interaction between cytochrome c and cytochrome c oxidase.21-23 However, we previously reported that CycN lacks most of the lysine residues in the region near the exposed heme that are important for binding in these other cytochrome c proteins.11 Accordingly, the heme displaying surface of CycN is less charged and is more enriched in nonpolar amino acids (Figure S3). The CycN binding surface of NicA2 is similarly lacking in charged amino acids and has a nonpolar center that includes Phe93 and Phe104, making it suitable for binding to CycN (Figure S3). Notably, the NicA2-CycN interface predicted by AlphaFold Multimer is strikingly devoid of specific contacts between the two proteins apart from a salt bridge between Asp294 of NicA2 and Arg108 of CycN on the edge of the binding interface. This lack of specific contacts in the structural model is consistent with the weak affinity observed previously for CycN binding to NicA2 and would allow for the required rapid exchange of CycN at the CycN binding site since two CycN molecules are necessary to oxidize NicA2’s reduced flavin during each turnover of the enzyme.11
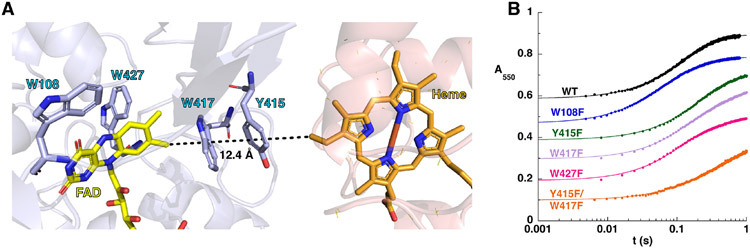
The CycN binding surface on NicA2 identified in this study is >12 Å away from the C7-C8 edge of the isoalloxazine of NicA2’s flavin, which is buried in the core of the protein at the active site (Figure 4A). Trp417 in NicA2 is adjacent to the C7-C8 edge of the isoalloxazine and Trp417 is stacked against Tyr415 at the CycN binding site. Since Trp and Tyr residues have the potential to be redox active,24 we wondered if these two residues constitute a protein-derived wire to transport electrons from NicA2’s flavin to CycN’s heme. To evaluate this, we individually mutated Trp417 and Tyr415 to redox inactive phenylalanine in addition to making a Tyr415Phe/Trp417Phe double mutant and measured the kinetics of electron transfer to CycN for the mutant enzymes in anaerobic stopped-flow experiments. We also performed similar experiments with Trp108Phe and Trp427Phe mutants of NicA2 since these two Trp residues are near the isoalloxazine, but not in the direction of the CycN binding site. All of the phenylalanine-substituted mutants transferred electrons from their reduced flavin to CycN with kinetics comparable to WT (Figure 4B and Table S3), indicating that none of these tryptophan or tyrosine residues are critical for electron transfer to CycN in WT NicA2. These results suggest that electron transfer between NicA2 and CycN can occur without the assistance of amino acid residues in NicA2. Notably, the ~12 Å distance between the isoalloxazine and heme in our structural model is consistent with that of redox cofactor pairs known to undergo unassisted long range electron transfer in other protein systems (Figure 4A).24-27
Figure 4.
(A) Residues in NicA2 (light blue) adjacent to the FAD that are potentially redox active and could facilitate electron transfer between NicA2’s FAD and CycN’s heme in the NicA2-CycN complex predicted by AlphaFold2 Multimer. (B) Stopped-flow absorbance traces for the reaction of reduced NicA2 variants with CycN. Traces have been manually offset to facilitate comparison. Note the logarithmic time base. The solid line for each trace is the fit to a two-exponential function, and the kobs values from these fits can be found in Table S3.
In summary, we have identified the CycN binding site on NicA2 and have proposed that electron transfer between these two redox proteins does not utilize a protein-derived wire in NicA2. Several of the surface-exposed residues in this CycN binding site are strictly conserved between NicA2 and Pnad, including Phe93 and Asp295, but are not conserved in other oxidase homologs (Figure S4),12 and the evolution of this binding site presumably allowed these two dehydrogenase “outliers” to use CycN as an oxidant instead of the O2 more widely used by most members of the FAO enzyme family.
Supplementary Material
Funding Sources
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM139069 (to F. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. E. J. M. and J. A. W. Jr. were each supported by an undergraduate Research and Creative Scholarship from the Lee Honors College at Western Michigan University.
Footnotes
Notes
The model of the NicA2-CycN complex is available in ModelArchive at https://modelarchive.org/doi/10.5452/ma-9e5s7 using the access code T3F8XEo2a5.
Supporting Information
Supporting figures, tables and experimental procedures (PDF).
Accession Codes
NicA2: NCBI accession ID WP_013973880
CycN: NCBI accession ID WP_080563818
Pnad: NCBI accession ID WP_013973879
REFERENCES
- (1).Tang H, Wang L, Wang W, Yu H, Zhang K, Yao Y, and Xu P (2013) Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in Pseudomonas. PLoS Genet. (Copenhaver GP, Ed.) 9, e1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wang SN, Liu Z, Tang HZ, Meng J, and Xu P (2007) Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153, 1556–1565. [DOI] [PubMed] [Google Scholar]
- (3).Xue S, Schlosburg JE, and Janda KD (2015) A New Strategy for Smoking Cessation: Characterization of a Bacterial Enzyme for the Degradation of Nicotine. J. Am. Chem. Soc 137, 10136–10139. [DOI] [PubMed] [Google Scholar]
- (4).Kallupi M, Xue S, Zhou B, Janda KD, and George O (2018) An enzymatic approach reverses nicotine dependence, decreases compulsive-like intake, and prevents relapse. Sci. Adv 4, eaat4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pentel PR, Raleigh MD, LeSage MG, Thisted T, Horrigan S, Biesova Z, and Kalnik MW (2018) The nicotine-degrading enzyme NicA2 reduces nicotine levels in blood, nicotine distribution to brain, and nicotine discrimination and reinforcement in rats. BMC Biotechnol. 18, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mattevi A (2006) To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci 31, 276–283. [DOI] [PubMed] [Google Scholar]
- (7).Tararina MA, Xue S, Smith LC, Muellers SN, Miranda PO, Janda KD, and Allen KN (2018) Crystallography Coupled with Kinetic Analysis Provides Mechanistic Underpinnings of a Nicotine-Degrading Enzyme. Biochemistry 57, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tararina MA, Dam KK, Dhingra M, Janda KD, Palfey BA, and Allen KN (2021) Fast Kinetics Reveals Rate-Limiting Oxidation and the Role of the Aromatic Cage in the Mechanism of the Nicotine-Degrading Enzyme NicA2. Biochemistry 60, 259–273. [DOI] [PubMed] [Google Scholar]
- (9).Fitzpatrick PF (2010) Oxidation of amines by flavoproteins. Arch. Biochem. Biophys 493, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Romero E, Gómez Castellanos JR, Gadda G, Fraaije MW, and Mattevi A (2018) Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. Chem. Rev 118, 1742–1769. [DOI] [PubMed] [Google Scholar]
- (11).Dulchavsky M, Clark CT, Bardwell JCA, and Stull F (2021) A cytochrome c is the natural electron acceptor for nicotine oxidoreductase. Nat. Chem. Biol 17, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Choudhary V, Wu K, Zhang Z, Dulchavsky M, Barkman T, Bardwell JCA, and Stull F (2022) The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase. J. Biol. Chem 298, 102251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, and Weng Z (2014) ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S, Zielinski M, Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Kohli P, Jumper J, and Hassabis D (2022) Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021.10.04.463034. [Google Scholar]
- (15).Cianfrocco MA, Wong-Barnum M, Youn C, Wagner R, and Leschziner A (2017) COSMIC2: A science gateway for cryo-electron microscopy structure determination. ACM Int. Conf. Proceeding Ser. Part F1287, 13–17. [Google Scholar]
- (16).Farrell IS, Toroney R, Hazen JL, Mehl RA, and Chin JW (2005) Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods 2, 377–384. [DOI] [PubMed] [Google Scholar]
- (17).Young TS, Ahmad I, Yin JA, and Schultz PG (2010) An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli. J. Mol. Biol 395, 361–374. [DOI] [PubMed] [Google Scholar]
- (18).Feissner R, Xiang Y, and Kranz RG (2003) Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal. Biochem 315, 90–94. [DOI] [PubMed] [Google Scholar]
- (19).Butt WD, and Keilin D (1962) Absorption spectra and some other properties of cytochrome c and of its compounds with ligands. Proc. R. Soc. London. Ser. B. Biol. Sci 156, 429–458. [DOI] [PubMed] [Google Scholar]
- (20).Moran GR (2019) Anaerobic methods for the transient-state study of flavoproteins: The use of specialized glassware to define the concentration of dioxygen, in Methods in Enzymology 1st ed., pp 27–49. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- (21).Capaldi RA (1990) STRUCTURE AND FUNCTION OF CYTOCHROME c OXIDASE. Annu. Rev. Biochem 59, 569–596. [DOI] [PubMed] [Google Scholar]
- (22).Daff S, Sharp RE, Short DM, Bell C, White P, Manson FDC, Reid GA, and Chapman SK (1996) Interaction of Cytochrome c with Flavocytochrome b 2. Biochemistry 35, 6351–6357. [DOI] [PubMed] [Google Scholar]
- (23).Diêp Lê KH, Lederer F, and Golinelli-Pimpaneau B (2010) Structural Evidence for the Functional Importance of the Heme Domain Mobility in Flavocytochrome b2. J. Mol. Biol 400, 518–530. [DOI] [PubMed] [Google Scholar]
- (24).Dempsey JL, Winkler JR, and Gray HB (2010) Proton-coupled electron flow in protein redox machines. Chem. Rev 110, 7024–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gray HB, and Winkler JR (1996) Electron transfer in proteins. Annu. Rev. Biochem 65, 537–561. [DOI] [PubMed] [Google Scholar]
- (26).Reece SY, and Nocera DG (2009) Proton-coupled electron transfer in biology: Results from synergistic studies in natural and model systems. Annu. Rev. Biochem 78, 673–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kang G, Taguchi AT, Stubbe JA, and Drennan CL (2020) Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex. Science (80-.). 368, 424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.