Abstract
The therapeutic success of an antiretroviral salvage regimen containing protease inhibitors (PI) is limited by PI-resistant viral strains exhibiting various degrees of resistance and cross-resistance. To evaluate the extent of cross-resistance to the new PI amprenavir, 155 samples from 132 human immunodeficiency virus type 1-infected patients were analyzed for viral genotype by direct sequencing of the protease gene. Concomitantly, drug sensitivity to indinavir, saquinavir, ritonavir, nelfinavir, and amprenavir was analyzed by a recombinant virus assay. A total of 111 patients had been pretreated with 1-4 PI, but all were naive to amprenavir. A total of 105 samples (67.7%) were sensitive to amprenavir; 25 samples (16.1%) were intermediately resistant, and another 25 samples were highly resistant (4- to 8-fold- and >8-fold-reduced sensitivity, respectively). The mutations 46I/L, 54L/V, 84V, and 90M showed the strongest association with amprenavir resistance (P < 0.0001). The scoring system using 84V and/or any two of a number of mutations (10I/R/V/F, 46I/L, 54L/V, and 90M) predicted amprenavir resistance with a sensitivity of 86.0% and a specificity of 81.0% within the analyzed group of samples. Of 62 samples with resistance against 4 PI, 23 (37.1%) were still sensitive to amprenavir. In comparison, only 2 of 23 samples (8.7%) from nelfinavir-naive patients with resistance against indinavir, saquinavir, and ritonavir were still sensitive to nelfinavir. Amprenavir thus appears to be an interesting alternative for PI salvage therapy.
The introduction of protease inhibitors (PI) into antiretroviral therapy leads to a profound and sustained suppression of viral load, slower disease progression, and prolonged survival in the majority of human immunodeficiency virus type 1 (HIV-1)-infected patients (12, 15). The success of antiretroviral treatment, however, is impaired by the emergence of drug-resistant viral strains (23). Due to the high structural similarity of PI, resistant viruses may exhibit various degrees of cross-resistance even to a PI to which the patient has not yet been exposed (2, 3, 4, 7, 8, 19, 25). This makes the management of second- or third-line therapies difficult.
Recently, the new PI amprenavir has been approved for antiretroviral treatment in several countries. Amprenavir proved to be highly potent in antiretroviral combination therapies, had good bioavailability, and was well tolerated, with the most frequent adverse events being nausea, mild gastrointestinal symptoms, perioral paresthesia, and rash (1, 6, 10, 13, 20, 22); A. Fetter, P. Nacci, J. Yeo, J. May, I. Vafidis, P. Tymkewycz, and L. Pedneault, 7th Eur. Conf. Clin. Aspects Treatment HIV Infect., abstr. 813, 1999). Furthermore, in vitro data suggest a unique resistance profile, with the mutation 50V developing as an initial active-site mutation (14). Clinical data, however, indicate a different resistance profile with mutations at positions 54, 82, 84, and 90, which are usually associated with broad cross-resistance (18, 21); R. Elston, R. Myers, S. Randall, B. Sadler, M. Tisdale, and W. Snowden, 7th Conf. Retrovir. Opportunistic Infect., abstr. 727, 2000). Although cross-resistance was lower for amprenavir than for the other PI in clinical samples (18); V. Calvez, C. Tamalet, J. M. Molina, C. Katlama, J. P. Mamet, Z. Antoun, A. Goetschel, and M. Ait-Khaled, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 442, 1999), these changes may result in the failure of the salvage therapy.
To characterize the cross-resistance profile of amprenavir, 155 samples from 132 HIV-1-infected patients were retrospectively analyzed for drug sensitivity against amprenavir. These samples were from patients being treated at 14 clinical centers specializing in the care of HIV-1-infected patients from January 1998 until June 1999 and had been sent for resistance testing because of treatment failure. Samples from patients with treatment interruptions were excluded. All samples had previously been characterized for viral genotype and phenotype, including indinavir, saquinavir, ritonavir, and nelfinavir resistance. Of 127 samples with detailed treatment histories, 16 (12.6%) were from PI-naive patients, and 111 (87.4%) were from patients pretreated with one (n = 24), two (n = 35), three (n = 35), or four (n = 17) PI, respectively. Indinavir, saquinavir, ritonavir, and nelfinavir had previously been used in 65, 84, 55, and 64 patients, respectively, but all patients were naive for amprenavir.
Genotyping of the original plasma samples was performed by direct sequencing of the protease gene using the primers H2720a (5′-TATTGTATGGATTTTCAGGCC-3′) and H2254s (5′-TCAGGTCACTTTTGCAAC-3′). The sequences were analyzed for the mutations described by Schinazi et al. (21). If both wild-type and mutant forms were present at a position described for PI resistance, the mutation was counted. The detection limit for mixed populations was about 30%. For phenotyping, a recombinant virus assay was performed as previously described (11, 24). For statistical evaluation, the chi-square test and Fisher's exact test were used, as appropriate.
The 50% inhibitory concentration (IC50) of amprenavir for the nonresistant reference virus NL4-3 was determined to be 0.021 ± 0.007 μM in 14 independent assays, a finding which is similar to previously published data (22). The coefficient of variation of 0.36 was similar to those measured for other PI (24). A total of 105 samples (67.7%) were sensitive to amprenavir (1- to 3-fold-reduced sensitivity); 25 samples (16.1%) were intermediately resistant (4- to 8-fold-reduced sensitivity), and another 25 samples were highly resistant (>8-fold-reduced sensitivity; range, 9- to 66-fold). The extent of cross-resistance between amprenavir and previously approved PI is shown in Table 1. Only 1 of 58 samples which were sensitive to the other PI showed intermediate resistance against amprenavir. A total of 25 of 35 samples with resistance against 1-3 PI (71.4%) and 23 of 62 samples with resistance against 4 PI (37.1%) were still sensitive to amprenavir. For comparison, a similar analysis was performed for the nelfinavir-naive patients in this study (n = 63). The percentage of nelfinavir-sensitive samples with resistance against indinavir, saquinavir, and ritonavir (2 of 23, 8.7%) was significantly lower than the percentage of amprenavir-sensitive samples with resistance against all other PI (P = 0.01, chi-square test).
TABLE 1.
Cross-resistance to amprenavir in samples resistant to none, one, two, three, or four of the previously approved PI (n = 155)
| No. of PI with >3-fold-reduced sensitivity (no. of samples) | No. of samples (%) with the following fold reduction in sensitivity to amprenavir:
|
||
|---|---|---|---|
| 1–3 | 4–8 | >8 | |
| None (58) | 57 (98.3) | 1 (1.7)a | 0 |
| 1 (17) | 15 (88.2) | 1 (5.9) | 1 (5.9) |
| 2 (5) | 4 (80.0) | 1 (20.0) | 0 |
| 3 (13) | 6 (46.2) | 6 (46.2) | 1 (7.7) |
| 4 (62) | 23 (37.1) | 16 (25.8) | 23 (37.1) |
| Total (155) | 105 (67.7) | 25 (16.1) | 25 (16.1) |
This sample, which contained the mutations 10I and 71V, exhibited fivefold-reduced sensitivity against amprenavir.
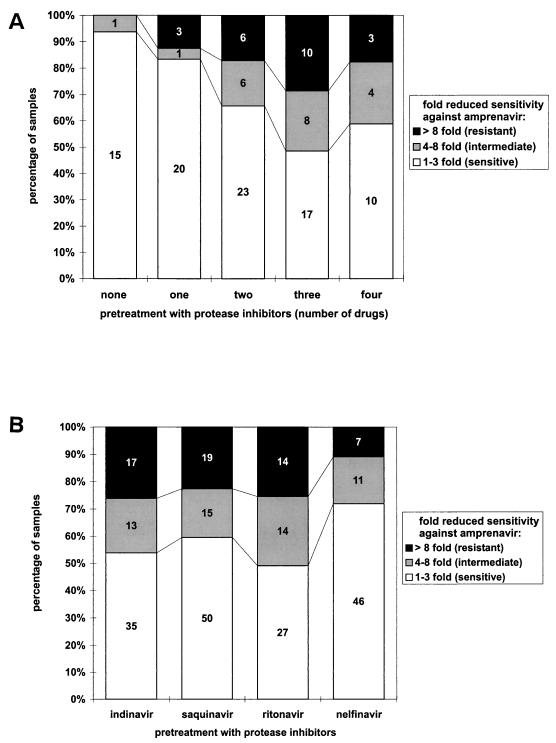
The number of PI in the pretreatment was also significantly correlated with amprenavir resistance (chi-square test for trend, 13.97 [P = 0.007]; Fig. 1A). With respect to individual PI in the treatment history, samples from patients with indinavir or ritonavir pretreatment were more likely to be resistant against amprenavir than samples from patients with previous nelfinavir therapy (Fig. 1B).
FIG. 1.
(A) Percentage of amprenavir sensitive, intermediately resistant, or highly resistant isolates with respect to the number of PI in the treatment history (indinavir, saquinavir, ritonavir, and nelfinavir). (B) Percentage of amprenavir sensitive, intermediately resistant, or highly resistant isolates with respect to individual PI in the treatment history.
To determine the genotypes relevant for cross-resistance, the frequency of individual mutations in amprenavir-sensitive samples was compared to their frequency in both intermediately and highly resistant isolates. The mutations 10I/R/V/F and 46I/L, 54L/V, 84V, 90M were found to be significantly associated with reduced sensitivity to amprenavir (P < 0.001 and P < 0.0001, respectively [chi-square test]), whereas 20R/M, 24I, 30N, 32I, 33F, 36I, 48V, 63P, 71V/T, 73S, 77I, and 82A/F/T/S had P values of >0.003 and thus did not achieve significance at the 95% level after adjustment for multiple comparisons (n = 17). Of 16 samples with 84V, 14 were highly resistant to amprenavir; the remaining two samples exhibited five- and eight-fold-reduced sensitivity. More will undoubtedly be learned from the isolates of patients treated with amprenavir.
Two scoring systems describing genotypes associated with amprenavir resistance were established. 84V and/or any two of a number of mutations (10I/R/V/F, 46I/L, 54L/V, and 90M) predicted amprenavir drug resistance (intermediate or high level) with a sensitivity of 86% (i.e., the percentage of phenotypically resistant viruses that were also scored as genotypically resistant) and a specificity of 81% (i.e., the percentage of phenotypically sensitive viruses that were also scored as genotypically sensitive). 84V and/or any two of several mutations (46I/L, 54L/V, and 90M) predicted high-level amprenavir resistance with a sensitivity of 88.0% and a specificity of 79.2%.
This study revealed a relatively low prevalence of cross-resistance to amprenavir in samples from patients pretreated with other PI (18; Calvez et al., 39th ICAAC). The extent of cross-resistance was significantly lower for amprenavir than for nelfinavir, which was introduced as a fourth PI, even though the resistance against three PI (nelfinavir) was compared with the resistance against four PI (amprenavir).
It has been controversially discussed how resistance against new antiretroviral drugs can be predicted reliably. Since resistance mutations selected in vitro for a particular compound may be different from those developing in vivo, the activity of new antiretroviral compounds should be assessed against currently circulating highly resistant clinical strains (16). This study was performed according to this recommendation. Since the predictive value of phenotypic resistance testing was recently demonstrated in three retrospective studies (5, 9, 17), our data on the cross-resistance profile of amprenavir should reliably reflect the in vivo situation. However, it is still unclear which level of resistance will ultimately be predictive for therapy failure. Recent clinical studies suggest an increased risk of therapy failure if PI resistance above interassay variability was observed at baseline (5, 9). Nevertheless, it may be possible that patients with low-level amprenavir resistance still profit from a combination of two PI with synergistic effects. Therefore, the data of this study were analyzed both for low- and high-level amprenavir resistance.
Two prospective clinical studies recently showed that salvage regimens with amprenavir were less successful in PI-treated than in PI-naive children (S. Blanche, A. Fetter, H. Cox, J. Yeo, S. Randall, and W. Snowden, 7th Conf. Retrovir. Opportunistic Infect., abstr. 695, 2000; J. Church, M. Rathore, T. Rubio, P. Flynn, J. Ramos, L. Rosado, L. Kahl, J. Yeo, A. Fetter, S. Hetherington, D. Brown, T. Kelley, and N. Mustafa, 7th Conf. Retrovir. Opportunistic Infect., abstr. 693, 2000). However, in both studies, baseline characteristics for PI-treated children indicated more advanced disease and more extensive prior antiretroviral therapy. Therefore, multiresistant viral strains may have already been present. If only one or two drugs of a rescue regimen are still active, the virus will soon acquire resistance against these drugs as well. Furthermore, it also has to be considered that viruses which are still sensitive in the phenotypic assay will develop clinical resistance against amprenavir more rapidly if various resistance-related mutations are already present in the protease gene.
Genotypic assays will soon be available on a routine basis. However, no consensus algorithm is available for the interpretation of genotypic data. Therefore, two scoring systems were developed to predict low- and high-level cross-resistance to amprenavir. These may only be applied to samples before amprenavir treatment, since all samples were from amprenavir-naive patients. Even though these scoring systems have sensitivities and specificities of >80%, about one-third of all samples may be classified as either falsely resistant or falsely sensitive to amprenavir.
Phenotypic tests are time-consuming and labor-intensive and are thus restricted to specialized research laboratories. Despite these limitations, phenotypic assays appear to be the most reliable method to evaluate the cross-resistance profile of a new antiretroviral compound before the drug is broadly used in clinical practice.
Acknowledgments
This work was supported by the Bayerische Staatsministerium für Kultus, Erziehung und Wissenschaft.
We thank B. Fleckenstein for helpful discussions and continuous support. The indicator cell line was kindly provided by R. E. Means and R. C. Desrosiers. We are also indebted to T. Harrer and M. Schmitt (Erlangen, Germany), M. Helm, G. Abelein, and W. Brockhaus (Nürnberg, Germany), L. Schneider (Fürth, Germany), S. Mauss, G. Schmutz, and T. Niehues (Düsseldorf, Germany), H. Knechten and L. Habets (Aachen, Germany), J. A. Rump (Freiburg, Germany), R. Billhardt (Offenburg, Germany), F. Schlote, E. Lauenroth-Mai, C. Schauenburg, and F. Krauthausen (Berlin, Germany), A. Groh and M. Scholz (Jena, Germany), C.-J. Reiss (Halle, Germany), and U. Bohr (Magdeburg, Germany) for providing clinical samples and drug histories. We also thank U. Marcus, Robert-Koch-Institut, Berlin, Germany, for his help in obtaining amprenavir. Indinavir was provided by Merck Sharp & Dohme, saquinavir was provided by Hoffmann-La Roche, ritonavir was provided by Abbott, nelfinavir was provided by Agouron Pharmaceuticals, and amprenavir was provided by Glaxo Wellcome.
REFERENCES
- 1.Adkins J C, Faulds D. Amprenavir. Drugs. 1998;55:837–842. doi: 10.2165/00003495-199855060-00015. [DOI] [PubMed] [Google Scholar]
- 2.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 3.Craig C, Race E, Sheldon J, Wittaker L, Gilbert S, Moffatt A, Rose J, Dissanayeke S, Chirn G W, Duncan I B, Cammack N. HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS. 1998;12:1611–1618. doi: 10.1097/00002030-199813000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Deeks S G, Grant R M, Beatty G W, Horton C, Detmer J, Eastman S. Activity of a ritonavir plus saquinavir-containing regimen in patients with virologic evidence of indinavir or ritonavir failure. AIDS. 1998;12:F97–F102. doi: 10.1097/00002030-199810000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Deeks S G, Hellmann N S, Grant R M, Parkin N T, Petropoulos C J, Becker M, Symonds W, Chesney M, Volberding P A. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J Infect Dis. 1999;179:1375–1381. doi: 10.1086/314775. [DOI] [PubMed] [Google Scholar]
- 6.Drusano G L, D'Argenio D Z, Symonds W, Bilello P A, McDowell J, Sadler B, Bye A, Bilello J A. Nucleoside analog 1592U89 and human immunodeficiency virus protease inhibitor 141W94 are synergistic in vitro. Antimicrob Agents Chemother. 1998;42:2153–2159. doi: 10.1128/aac.42.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulioust A, Paulous S, Guillemot L, Delavalle A M, Boue F, Clavel F. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J Virol. 1999;73:850–854. doi: 10.1128/jvi.73.1.850-854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatkenheuer G, Hoetelmans R M, Hunn N, Schwenk A, Franzen C, Reiser M, Jutte A, Rockstroh J, Diehl V, Salzberger B. Salvage therapy with regimens containing ritonavir and saquinavir in extensively pretreated HIV-infected patients. AIDS. 1999;13:1485–1489. doi: 10.1097/00002030-199908200-00007. [DOI] [PubMed] [Google Scholar]
- 9.Harrigan P R, Hertogs K, Verbiest W, Pauwels R, Larder B, Kemp S, Bloor S, Yip B, Hogg R, Alexander C, Montaner J S. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS. 1999;13:1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 10.Haubrich R, Thompson M, Schooley R, Lang W, Stein A, Sereni D, van der Ende M E, Antunes F, Richman D, Pagano G, Kahl L, Fetter A, Brown D J, Clumeck N. A phase II safety and efficacy study of amprenavir in combination with zidovudine and lamivudine in HIV-infected patients with limited antiretroviral experience. Amprenavir PROAB2002 Study Team. AIDS. 1999;13:2411–2420. doi: 10.1097/00002030-199912030-00013. [DOI] [PubMed] [Google Scholar]
- 11.Means R E, Greenough T, Desrosiers R C. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J Virol. 1997;69:5431–5436. doi: 10.1128/jvi.71.10.7895-7902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mocroft A, Vella S, Benfield T L, Chiesi A, Miller V, Gargalianos P, d'Arminio Monforte A, Yust I, Bruun J N, Phillips A N, Lundgren J D. Changing patterns of mortality across Europe in patients infected with HIV-1. Lancet. 1998;352:1725–1730. doi: 10.1016/s0140-6736(98)03201-2. [DOI] [PubMed] [Google Scholar]
- 13.Murphy R L, Gulick R M, DeGruttola V, D'Aquila R T, Eron J J, Sommadossi J P, Currier J S, Smeaton L, Frank I, Caliendo A M, Gerber J G, Tung R, Kuritzkes D R. Treatment with amprenavir alone or amprenavir with zidovudine and lamivudine in adults with human immunodeficiency virus infection. AIDS Clinical Trials Group 347 Study Team. J Infect Dis. 1999;179:808–816. doi: 10.1086/314668. [DOI] [PubMed] [Google Scholar]
- 14.Myers R E, Snowden W, Randall S, Tisdale M. Unique resistance profile of the protease inhibitor amprenavir (141W94) observed in vitro and in the clinic. Antivir Ther. 1998;3(Suppl. 1):59–60. [Google Scholar]
- 15.Palella F J, Jr, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 16.Palmer S, Shafer R W, Merigan T C. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13:661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piketty C, Race E, Castiel P, Belec L, Peytavin G, Si-Mohamed A, Gonzalez-Canali G, Weiss L, Clavel F, Kazatchkine M D. Efficacy of a five-drug combination including ritonavir, saquinavir and efavirenz in patients who failed on a conventional triple-drug regimen: phenotypic resistance to protease inhibitors predicts outcome of therapy. AIDS. 1999;13:F71–F77. doi: 10.1097/00002030-199907300-00001. [DOI] [PubMed] [Google Scholar]
- 18.Race R, Dam E, Obry V, Paulous S, Clavel F. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS. 1999;13:2061–2068. doi: 10.1097/00002030-199910220-00008. [DOI] [PubMed] [Google Scholar]
- 19.Roberts N A, Craig J C, Sheldon J. Resistance and cross-resistance with saquinavir and other HIV protease inhibitors: theory and practice. AIDS. 1998;12:453–460. doi: 10.1097/00002030-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Sadler B M, Hanson C D, Chittick G E, Symonds W T, Roskell N S. Safety and pharmacokinetics of amprenavir (141W94), a human immunodeficiency virus (HIV) type 1 protease inhibitor, following oral administration of single doses to HIV-infected adults. Antimicrob Agents Chemother. 1999;43:1686–1692. doi: 10.1128/aac.43.7.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance: 1999–2000 update. Int Antivir News. 1999;7:46–69. [Google Scholar]
- 22.St. Clair M H, Millard J, Rooney J, Tisdale M, Parry N, Sadler B M, Blum M R, Painter G. In vitro antiviral activity of 141W94 (VX-478) in combination with other antiretroviral agents. Antivir Res. 1996;29:53–56. doi: 10.1016/0166-3542(95)00916-7. [DOI] [PubMed] [Google Scholar]
- 23.Vandamme A M, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antivir Chem Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]
- 24.Walter H, Schmidt B, Korn K, Vandamme A-M, Harrer T, Überla K. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. J Clin Virol. 1999;13:71–80. doi: 10.1016/s1386-6532(99)00010-4. [DOI] [PubMed] [Google Scholar]
- 25.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]