SUMMARY
The complex nature of the transcriptional networks underlying addictive behaviors suggests intricate cooperation between diverse gene regulation mechanisms that go beyond canonical activity-dependent pathways. Here we implicate in this process a nuclear receptor transcription factor, Retinoid X Receptor Alpha (RXRα), which we initially identified bioinformatically as associated with addiction-like behaviors. In the nucleus accumbens (NAc) of male and female mice, we show that, while its own expression remains unaltered after cocaine exposure, RXRα controls plasticity- and addiction-relevant transcriptional programs in both dopamine receptor D1- and D2-expressing medium spiny neurons, which in turn modulate intrinsic excitability and synaptic activity of these NAc cell types. Behaviorally, bidirectional viral and pharmacological manipulation of RXRα regulates drug reward sensitivity in both non-operant and operant paradigms. Together, this study demonstrates a key role for NAc RXRα in promoting drug addiction, and paves the way for future studies of rexinoid signaling in psychiatric disease states.
Keywords: Nucleus accumbens, Medium spiny neurons, Addiction, Motivation, Cocaine, Transcription factor, Retinoic acid, Rexinoids, Neuronal excitability, RNA-sequencing
eToc blurb
The transcriptional substrates of vulnerability to addiction are diverse and complex. Godino et al. use cell-type-specific transcriptomics, electrophysiology and behavior to single out RXRα, a putatively druggable transcription factor that governs larger gene networks to calibrate the physiology of NAc neurons and, in turn, individual sensitivity to drugs of abuse.
INTRODUCTION
Drugs of abuse such as cocaine perturb coordinated activity within the brain’s reward circuitry 1 initially in part by increasing dopamine signals to levels far exceeding those of natural reinforcers 2, thus hijacking more classic mechanisms of reward learning 3 to build pathological drug memories upon repeated exposure 4. The transition to a compulsive state of drug seeking and taking even in spite of negative consequences – a behavioral hallmark of substance use disorders 5 – however results from a composite interplay between a drug’s pharmacological properties and an individual’s sensitivity to drug reward, which depends on both innate and environmental factors 6-9.
Genetic, pharmacological and environmental effects converge onto the establishment of drug-induced transcriptional programs that underlie the several molecular-, cellular-, synaptic-, circuit- and behavioral-level alterations that define the drug-addicted phenotype 4,10. In a recent effort to better link brain-wide transcriptional patterns with individual behavioral responses to cocaine, we performed RNA-sequencing (RNAseq) on six regions of the mouse brain’s reward circuitry after cocaine self-administration, withdrawal, and relapse 11. One major finding of this study was the large differences in transcriptional landscapes across time points and brain regions 11. This indicated that the sole recruitment of canonical activity-dependent signaling pathways upstream of transcription factors such as CREB or ΔFOSB – extensively studied in addiction 12 – could not account fully for such heterogeneity in transcriptional responses. Subsequent in silico analyses 11 led us to propose that nuclear receptors – a large but understudied family of ligand-activated transcription factors that have the ability to form heterogeneous dimers and thus to coordinate gene expression across pathways yet with high specificity 13 – could be critical co-regulators of temporal- and region-specific transcriptional programs.
In the nucleus accumbens (NAc), a key striatal region of the brain’s reward circuitry that integrates midbrain dopaminergic inputs together with cortical and limbic glutamatergic signals to shape reward- and motivation-related behaviors 14, the nuclear receptor Retinoic X Receptor Alpha (RXRα) stood out. Its transcript levels were not altered by cocaine exposure but were positively correlated with addiction-relevant behavioral features 11, together making RXRα a strong mechanistic candidate to explain individual vulnerability to cocaine addiction. While burgeoning evidence implicates retinoic acid-related signaling 15-17 and other RXR family members – especially RXR Gamma (RXRγ) 18-21 – in striatal-dependent motivation-related behaviors, data on RXRα in the mature brain remain scarce. Genome-wide association studies have linked human Rxra mutations with schizophrenia 22-24, cognitive ability 25,26 and dementia 27,28, and a recent study demonstrated that RXRα can control spine and synapse formation in the adult mouse cortex 29, suggesting a role for RXRα in brain plasticity mechanisms.
Therefore, we hypothesized RXRα to be an important transcriptional regulator of NAc function in drug-related behaviors. One key question was to identify whether RXRα action in NAc is cell-type-specific, with a main focus on the two largely non-overlapping subpopulations of GABAergic medium spiny projection neurons (MSNs) that express either the dopamine D1 (Drd1) or dopamine D2 (Drd2) receptor 30, and which have been shown to play different – even antagonistic – roles in drug-related motivated and reward-learning behaviors 31-33. To that end, we here assess RXRα regulation after drug exposure, as well as the transcriptional, physiological and behavioral consequences of bidirectional viral manipulation of RXRα levels in D1- and D2-MSNs to propose a model for RXRα’s contribution to drug reward and addiction. We also provide early preclinical evidence for targeting RXRα using a small molecule inhibitor as a possible new pharmacotherapeutic avenue for patients with substance use disorders.
RESULTS
RXRα mediates addiction-like transcriptional programs in NAc
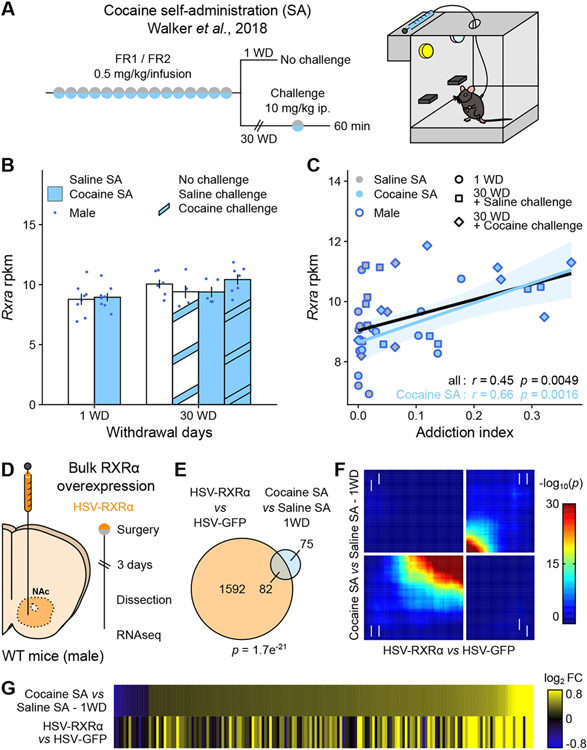
In a previous brain-wide transcriptomics study following cocaine self-administration in male mice (Figure 1A 11), Rxra transcript levels were not affected at any of the time points analyzed in the NAc (Figure 1B) or in any of the five other brain regions analyzed in that study (Supplemental Figure S1A 11). However, the correlation between individual animals’ Rxra transcript levels and their Addiction Index, a multi-factorial, bioinformatically-derived composite metric summarizing addiction-like behaviors, was the strongest in NAc (Figure 1C and Supplemental Figure S1B), even when only considering cocaine-exposed animals. This analysis suggests that pre-existing individual levels of RXRα expression in NAc might influence the severity of addiction-like behaviors upon drug exposure, presumably by contributing to cocaine-induced gene regulation in this brain region.
Figure 1. RXRα expression in NAc is linked to addiction-relevant behavior and transcription.
(A) Experimental design of the original cocaine self-administration (SA) study 11, using male mice (n = 5-8/group).
(B) RNAseq of NAc tissue showed no regulation of Rxra transcripts across experimental groups 11.
(C) Rxra levels in NAc correlated with the Addiction Index, a composite metric representative of addiction-like behavioral domains computed using exploratory factor analysis 11 (across all samples: Pearson’s r = 0.45, p = 0.0049; across cocaine SA samples only: Pearson’s r = 0.66, p = 0.0016).
(D) Experimental design of bulk RXRα overexpression transcriptomics study, using male mice (n = 4/group).
(E) RNAseq of virally-infected NAc tissue revealed a significant overlap between transcripts significantly regulated by RXRα overexpression and following 1 WD from cocaine SA 11 (Fisher’s exact test p = 1.7e−21).
(F) Comparison of transcriptome-wide expression profiles using rank-rank hypergeometric overlap (RRHO) plots confirmed overlap between the two datasets in a threshold-free manner (white arrows indicate change directionality in each dataset).
(G) Similar patterns of gene expression visualized as heatmaps of expression fold changes from respective controls.
Bar graphs represent mean ± SEM. Correlation graphs represent regression line with its 95% confidence interval.
To test this hypothesis, we compared genes regulated by RXRα in male NAc to those regulated by cocaine exposure in the original study 11. To detect direct or indirect gene targets of RXRα, we first performed bulk RNAseq on RXRα-overexpressing or control NAc tissue (Figure 1D). RXRα overexpression was achieved by infusing a Herpes Simplex Virus (HSV) encoding RXRα (or GFP as a control) and confirmed at both the RNA and protein levels (Supplemental Figure S2). Among the 1674 genes significantly regulated by RXRα overexpression (all genes and corresponding statistics available in Supplemental Table S1), 82 were also regulated in NAc one day after cocaine self-administration – more than 50% of the 157 genes affected in that condition (Figure 1E). This significant overlap strengthened the idea that RXRα contributes to NAc transcriptional programs in response to chronic self-administered cocaine. Subsequent comparisons using rank-rank hypergeometric overlap (RRHO) plots (Figure 1F) or expression-based heatmaps (Figure 1G) highlighted overlapping genes as being mostly upregulated, suggesting that RXRα acts predominantly as a permissive transcription factor 13,34,35 in that context. Collectively, these initial findings warranted further study of RXRα in the transcriptional mechanisms of cocaine reinforcement.
Acute or chronic cocaine does not affect RXRα expression in NAc
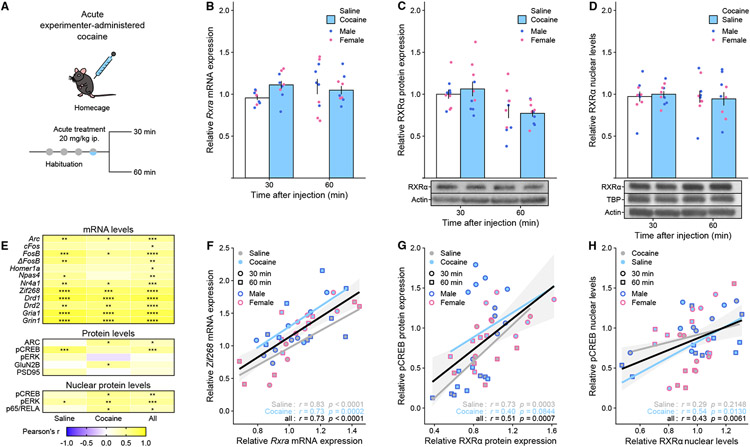
Our next goal was to examine RXRα regulation at both the RNA and protein levels after cocaine exposure in both male and female mice (Figure 2A). We first confirmed the absence of regulation of Rxra transcript levels after acute cocaine (Figure 2B). In addition, no significant changes in RXRα expression were detected at the total protein level (Figure 2C) or in nuclear-enriched fractions from NAc tissue (Figure 2D) – as another process through which RXRα could mediate its genomic effects is through regulation of its nuclear localization 29. Rxra transcript levels were also unaffected 60 min (Supplemental Figure S3A) or 30 days (Supplemental Figure S3B) after chronic experimenter-administered cocaine, and likewise RXRα nuclear levels were unaltered after 30 days of withdrawal from chronic cocaine or heroin self-administration (Supplemental Figure S3C). However, this does not exclude regulation via other mechanisms like phosphorylation or truncation 34, or regulation in other more complex experimental settings.
Figure 2. RXRα expression levels in NAc are not regulated by acute cocaine but correlate with markers of striatal activity and plasticity.
(A) Experimental design, using male and female mice (n = 4-5/group).
(B) Rxra mRNA levels were not significantly regulated after acute cocaine exposure (LMM-ANOVA: main effect of Drug: F1,31 = 0.8185, p = 0.3726; interaction Drug:Time: F1,31 = 2.5762, p = 0.1186).
(C) RXRα protein levels in whole tissue lysates were not significantly regulated after acute cocaine exposure (LMM-ANOVA: main effect of Drug: F1,32 = 0.1004, p = 0.7534; interaction Drug:Time: F1,32 = 0.4864, p = 0.4906).
(D) RXRα protein levels in nuclear fractions were not significantly regulated after acute cocaine exposure (LMM-ANOVA: main effect of Drug: F1,32 = 0.4631, p = 0.2353; interaction Drug:Time: F1,32 = 0.0070, p = 0.9340).
(E) Summary heatmap showing that Rxra/RXRα levels correlated strongly with other genes/proteins implicated in striatal function – including several IEGs – across saline only, cocaine only or all samples (Pearson’s r, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
(F) Representative example of Rxra mRNA levels correlation with the IEG Zif268 (across saline samples only: Pearson’s r = 0.83, p < 0.0001; across cocaine samples only: Pearson’s r = 0.73, p = 0.0002; across all samples: Pearson’s r = 0.73, p < 0.0001).
(G) Representative example of RXRα protein levels correlation with active transcription factor pCREB (across saline samples only: Pearson’s r = 0.73, p = 0.0003; across cocaine samples only: Pearson’s r = 0.40, p = 0.0844; across all samples: Pearson’s r = 0.51, p = 0.0007).
(H) Representative example of RXRα nuclear levels correlation with pCREB (across saline samples only: Pearson’s r = 0.29, p = 0.2148; across cocaine samples only: Pearson’s r = 0.54, p = 0.0130; across all samples: Pearson’s r = 0.43, p = 0.0061).
Representative Western Blot pictures for RXRα and control actin and TBP bands are attached to the corresponding quantifications. Bar graphs represent mean ± SEM after combining male and female data. Correlation graphs represent regression line with its 95% confidence interval.
RXRα expression levels in NAc correlate with markers of striatal function
Following up on the original conjecture that NAc RXRα levels might control drug-evoked behavioral responses, we examined whether RXRα levels would predict drug-evoked molecular responses by correlating individual RXRα transcript, total protein and nuclear levels with markers of striatal function and drug-induced plasticity. We found positive correlations with expression levels of several key players, including dopamine (Drd1, Drd2) and glutamate (Gria1, Grin1, GluN2B) receptors, signaling molecules like phosphorylated ERK and NFκB-complex transcription factor p65/RELA (Figure 2E), as well as all immediate early genes (IEGs) tested, including Zif268/Egr1 (Figure 2F). Of note, a link between RXRα action and IEG expression, which are well-established proxies of stimulus-induced neuronal activation and key intermediates in drug-induced molecular plasticity 36, has been proposed before 29. At the protein level in both total (Figure 2G) and nuclear (Figure 2H) extracts, RXRα positively correlated with levels of phosphorylated (active) CREB (pCREB), the convergence point of many drug-triggered signaling cascades 12. Although not a causal mechanistic explanation, these correlative data provide first clues as to the molecular underpinnings of RXRα striatal function: RXRα might control select target transcriptional programs that affect MSN excitability and activation of stimulus-induced signaling pathways, such as – but likely not limited to – pCREB-mediated IEG induction.
RXRα is partly enriched in NAc D1-MSNs
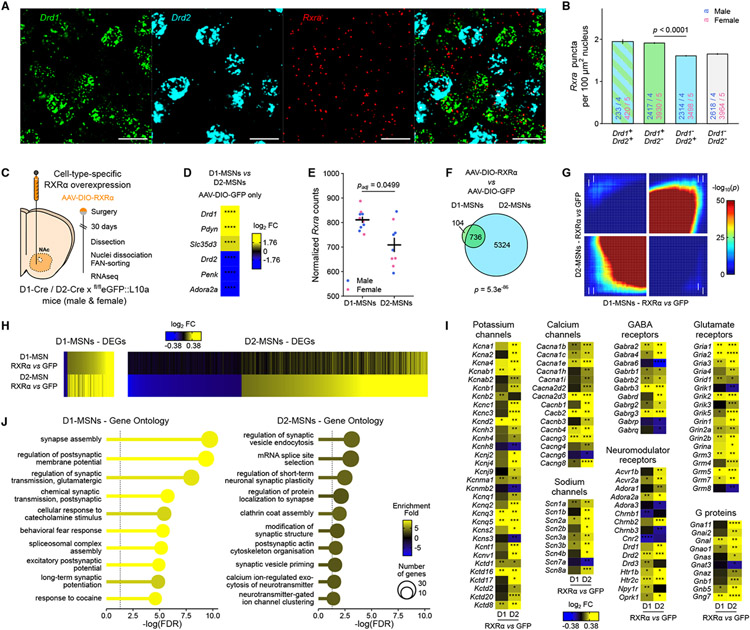
A growing literature underscores the importance of considering cell-type specificity in studies of drug-induced signaling and plasticity 12,33. Accordingly, we checked for RXRα enrichment in NAc core and medial shell cells expressing either Drd1 or Drd2 30 using RNA fluorescent in situ hybridization (FISH; Figure 3A). Quantification of Drd1 and Drd2 probes in DAPI-identified nuclei (Supplemental Figure S4) allowed for robust classification into D1-MSNs (Drd1+/Drd2−) or D2-MSNs (Drd1−/Drd2+). Quantification of Rxra puncta within cell-type-classified nuclei revealed a significant 18% higher Rxra expression level in D1-MSNs compared to D2-MSNs (Figure 3B). Rxra levels were lower in all other cell types, except in sparse microglia (data not shown), consistent with low levels of Rxra detected in NAc cholinergic and somatostatin-expressing interneurons by RNAseq of these cell types 37,38.
Figure 3. RXRα controls plasticity- and excitability-related transcriptional programs in both D1- and D2-MSNs.
(A) Representative images of RNA FISH for Drd1, Drd2 and Rxra mRNAs in NAc core. Scale bar is 20 μm.
(B) Quantification of Rxra mRNA puncta in nuclei of NAc cell types classified on Drd1 and Drd2 expression levels in both male and female mice (n = 4-5/group) showed higher Rxra expression in D1-MSNs at baseline. (LMM-ANOVA: main effect of CellType: F3,19381 = 45.1531, p < 2.2e−16, followed by Sidak’s post hoc tests).
(C) Experimental design of cell-type-specific RXRα overexpression transcriptomics study, using male and female D1-Cre xfl/fleGFP::L10a or D2-Cre x fl/fleGFP::L10a mice (n = 4-5/group).
(D) RNAseq confirmed enrichment of D1-MSNs and D2-MSNs marker genes in sorted D1 and D2 nuclei, respectively (Wald’s test D1-MSNs vs D2-MSNs, **** p < 0.0001).
(E) RNAseq also confirmed increased Rxra expression in D1-MSNs at baseline (Wald’s test with Benjamini-Hochberg correction for multiple comparisons padj = 0.0499).
(F) Significant overlap between transcripts significantly regulated by RXRα overexpression in D1-MSN and D2-MSN nuclei (Fisher’s exact test p = 5.3e−86).
(G) Comparison of D1- and D2-specific transcriptome-wide expression profiles using rank-rank hypergeometric overlap (RRHO) plots confirmed overlap (white arrows indicate change directionality in each cell type).
(H) Similar patterns of gene expression visualized as heatmaps of expression fold changes from respective controls.
(I) Selected excitability- and plasticity-related genes are regulated by RXRα overexpression in D1- or D2-MSNs (Wald’s test, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
(J) Gene ontology analyses of all genes significantly regulated by RXRα overexpression indicated enrichment of genes involved in neuronal and synaptic function in both D1- and D2-MSNs.
Bar graphs represent mean ± SEM after combining male and female data.
RXRα controls similar excitability- and plasticity-related transcriptional programs in D1- and D2-MSNs
To further investigate cell-type-specific targets downstream of RXRα signaling, we used male and female D1-Cre and D2-Cre mice – which express Cre recombinase under the promoter of the Drd1 or Drd2 gene – in combination with a Cre-dependent adeno-associated virus expressing RXRα (AAV-DIO-RXRα) infused in the NAc to achieve population-specific RXRα overexpression (Figure 3C). These mice were also crossed with a fl/fleGFP::L10a reporter line that allows for Fluorescence-Activated Nuclei Sorting (FANS) of D1- or D2-positive nuclei 39 (Supplemental Figure S5), where the vast majority (>95%) of labeled cells are respectively D1- or D2-MSNs (data not shown). RXRα overexpression was confirmed in sorted cell types at the transcript level and in whole NAc tissue at the RNA and protein levels (Supplemental Figure S2). First, we validated enrichment of canonical D1- and D2-MSNs markers 39,40 in the corresponding populations (Figure 3D). We also confirmed Rxra enrichment in D1-MSNs (Figure 3E), consistent with RNA FISH data (Figure 3B) and another D1- and D2-specific RNAseq dataset from our laboratory (Supplemental Figure S3B 41).
Next, we compared genes regulated by RXRα overexpression (AAV-DIO-RXRα vs AAV-DIO-GFP) separately in D1-MSNs or in D2-MSNs (all genes and corresponding statistics available in Supplemental Tables S2 and S3). While the total number of differentially expressed genes (DEGs) at the predefined significance threshold (±15% change, nominal p < 0.05) was much higher in D2-MSNs than D1-MSNs, DEG lists overlapped considerably, with almost 90% of the D1-MSNs DEGs also passing significance criteria in D2-MSNs (Figure 3F). Visualization in RRHO plots further confirmed strong similarities in threshold-free transcriptomic landscapes associated with RXRα overexpression in the two cell types (Figure 3G), and expression fold-change heatmaps indicated that, even for genes that do not reach significance, the change direction (up/down-regulation) was conserved across cell types (Figure 3H) although with blunted change magnitude in D1-MSNs relative to D2-MSNs. Such occluded response in D1-MSNs, both in terms of DEG number and variation amplitude, might be explained by higher levels of RXRα in D1-MSNs at baseline (Figure 3B,E). Select DEGs that encode potassium, calcium or sodium channels, as well as GABA, glutamate or neuromodulator receptors or associated G proteins (some of which have been linked to cocaine action 10,42,43) similarly showed larger fold change magnitude in D2-MSNs (Figure 3I). Unbiased gene ontology enrichment analyses further implicated RXRa-regulated genes in neuronal excitability, signaling and plasticity mechanisms (Figure 3J), with stronger enrichment for relevant gene ontology terms in D1-MSNs vs D2-MSNs, suggesting that RXRα-dependent transcriptional programs in D2-MSNs, although larger in number of DEGs, are less explicitly related to excitability or plasticity processes than in D1-MSNs. Despite these differences, this RNAseq dataset demonstrates that RXRα governs highly similar transcriptional programs in D1- and D2-MSNs, which include numerous and diverse candidate effector genes that participate in neuronal physiology.
RXRα bidirectionally and cell-autonomously controls neuronal excitability in D1- and D2-MSNs
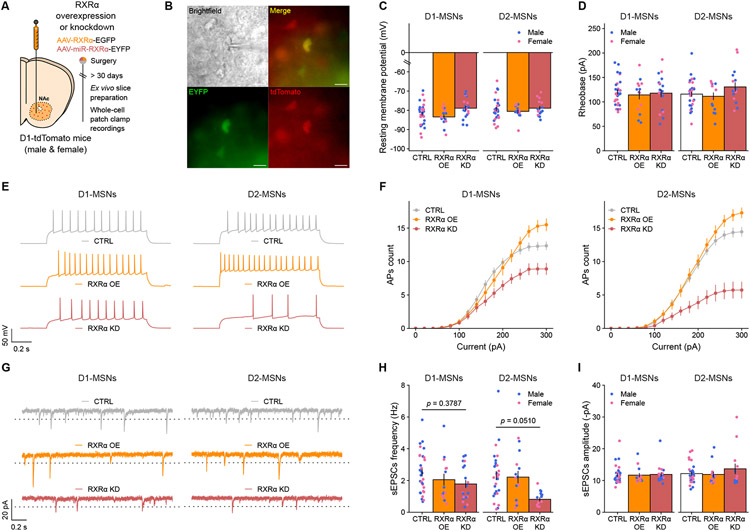
The next step was therefore to explore the cell-type-specific functional consequences of RXRα-dependent transcriptional control of excitability- and plasticity-related genes. We developed non-cell-type-specific AAV constructs for either RXRα overexpression (AAV-RXRα-GFP) or knockdown using miRNA constructs targeted against the Rxra coding sequence or a control Lacz sequence not expressed in mammals (AAV-miR-RXRα-EYFP or AAV-miR-lacZ-EYFP). Both RXRα overexpression and knockdown efficiencies were confirmed in virally-infected tissue at both the RNA and protein levels, with knockdown levels of at least 50% (Supplemental Figure S2). These constructs were then infused into the NAc of D1-tdTomato male and female mice, which express a tdTomato fluorophore under the promoter of the Drd1 gene (Figure 4A), allowing us to record from AAV-infected (GFP/EYFP+) D1-MSNs (tdTomato+) or D2-MSNs (tdTomato−) (Figure 4B). In both cell types, manipulating RXRα levels did not affect resting membrane potential (Figure 3C) or rheobase (Figure 3D), with average values for those two metrics in line with previous reports 44-46. When measuring the number of action potentials evoked in patched MSNs upon increasing steps of current injection, we found strikingly dampened responses at higher currents in both cell types with RXRα knockdown, and conversely RXRα overexpression lead to increased responses, indicating respectively blunted or enhanced intrinsic excitability (Figure 4E,F). At the synaptic level, frequency (Figure 4G,H) but not amplitude (Figure 4G,I) of spontaneous excitatory postsynaptic currents (sEPSCs) was reduced across cell types upon RXRα knockdown but unaffected by its overexpression, an effect that was stronger in D2-MSNs than in D1-MSNs. The overall blunted electrophysiological response of D1-MSNs might be related to either the fact that these cells express higher levels of RXRα at baseline (Figure 3A,B,E), or to the fact that RXRα controls many more genes with greater magnitude, including excitability- and plasticity-associated ones, in D2-MSNs than in D1-MSNs (Figure 3H,I,J). Nevertheless, these electrophysiological results substantiate that the transcriptional programs controlled by RXRα in NAc MSNs modulate intrinsic excitability and synaptic function of both MSN subtypes – likely via changes in the channel, receptor and G protein repertoires expressed at the postsynaptic membrane (Figure 3I) – and that RXRα could thus be critical in calibrating neuronal sensitivity to drug stimuli.
Figure 4. RXRα modulates intrinsic excitability of both D1- and D2-MSNs.
(A) Experimental design of cell-type-specific RXRα manipulation ex vivo electrophysiology study, using male and female Drd1-tdTomato mice (n = 4-5/group, n = 123 neurons total). Control viruses (CTRL) expressed either GFP or a miRNA-sequence targeted against Lacz sequence, and were pooled for analysis.
(B) Representative picture of a patched AAV-infected D1-tdTomato+ neuron. Scale bar is 20 μm.
(C) Resting membrane potential was unaffected by RXRα overexpression or knockdown in either D1- or D2- MSNs (LMM-ANOVA: main effect of Virus: F2,14.634 = 3.2852, p = 0.06630; interaction Virus:CellType: F2,102.934 = 0.8075, p = 0.44877).
(D) Rheobase, the minimal injected current required to trigger an action potential, was also unaffected by RXRα overexpression or knockdown in either D1- or D2-MSNs (LMM-ANOVA: main effect of Virus: F2,15.122 = 0.1815, p = 0.8358; interaction Virus:CellType: F2,99.563 = 1.9302, p = 0.1505).
(E) Representative membrane responses from D1- and D2-MSNs in response to a 280 pA current injection, with or without RXRα overexpression or knockdown.
(F) The number of evoked action potentials (AP) in response to increasing depolarizing current steps was increased by RXRα overexpression and blunted by RXRα knockdown in both D1- (left) and D2-MSNs (right), suggesting reduced excitability (LMM-ANOVA: interaction Virus:Current: F2,1818 = 6769.22, p < 2.2e−16; interaction Virus:Current:CellType: F2,1818 = 32.5683, p = 1.268e−14, followed by Sidak’s post hoc tests, see Supplemental Table S6 for all statistics).
(G) Representative voltage-clamp recordings of NAc D1- or D2-MSNs highlighting spontaneous Excitatory Post-Synaptic Currents (sEPSCs), with or without RXRα overexpression or knockdown. Dotted line represents the 8 pA deviation from baseline threshold for sEPSC detection.
(H) RXRα knockdown reduced the frequency of sEPSCs in both D1- and D2-MSNs, while overexpression had no effect (LMM-ANOVA: main effect of Virus: F2,14.634 = 8.0783, p = 0.004327; interaction Virus:CellType: F1,102.934 = 1.2754, p = 0.283686, followed by Sidak’s post hoc tests).
(I) RXRα overexpression or knockdown did not alter the amplitude of sEPSCs in either D1- or D2-MSNs (LMM-ANOVA: main effect of Virus: F2,15.239 = 0.6838, p = 0.1952; interaction Virus:CellType: F2,100.332 = 0.7404, p = 0.47949).
Bar graphs and line graphs represent mean ± SEM after combining male and female data.
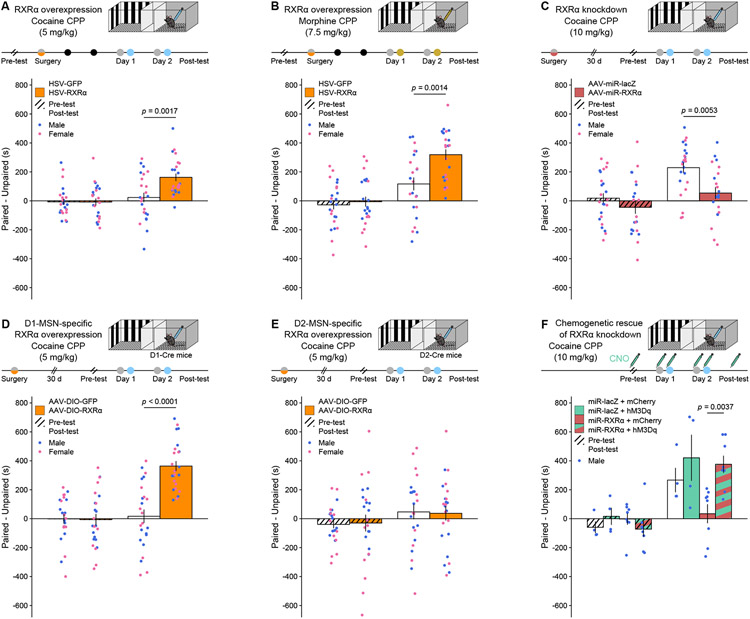
Manipulating RXRα levels in NAc influences sensitivity to drug reward contextual learning
Our final efforts were thence to characterize the behavioral consequences of RXRα-mediated transcriptional regulation of MSN excitability. To assess whether NAc RXRα levels alter the ability of an animal to associate predictive contextual cues with the rewarding properties of drugs of abuse, we used an unbiased conditioned place preference (CPP) assay in male and female mice. Bulk, HSV-mediated RXRα overexpression increased CPP for a subthreshold (5 mg/kg) dose of cocaine, which did not induce a preference in control animals (Figure 5A). This effect generalized to other drug reinforcers, as it was also observed for an intermediate (7.5 mg/kg) dose of morphine (Figure 5B), and was bidirectional, as AAV-mediated RXRα knockdown produced the opposite effect – a decrease in CPP for a higher (10 mg/kg) dose of cocaine (Figure 5C). Because D1- and D2-MSNs have been shown to play opposite roles in this specific CPP task 31,32, we hypothesized that RXRα-mediated modulation of their intrinsic excitability would exert opposite effects on cocaine CPP. We found that D1-MSN-specifc (Figure 5D) but not D2-MSN-specific (Figure 5E) RXRα overexpression increased subthreshold cocaine CPP, indicating that net bulk effects (Figure 5A) are likely mediated through RXRα action in D1-MSNs. We next assessed whether RXRα effects on neuronal excitability (Figure 4) were causal to its effects on CPP behavior by combining RXRα viral manipulations with chemogenetics. Increasing MSN excitability using an AAV expressing the activating Designer Receptor Exclusively Activated by Designer Drugs (DREADD) hM3Dq rescued the CPP deficit induced by miR-mediated RXRα knockdown (Figure 5F), indicating that RXRα effects on drug-context associative learning are mediated at least in part through regulation of MSN excitability. These experiments corroborate the correlative data shown in Figure 1, as they causally establish that differential NAc RXRα levels calibrate sensitivity to drug reward, with higher and lower RXRα levels respectively promoting and lessening drug reward learning.
Figure 5. RXRα bidirectionally and cell-type-specifically regulates dose sensitivity in drug-reward associative learning.
(A) HSV-mediated RXRα overexpression in NAc increased conditioned place preference (CPP) for a subthreshold (5 mg/kg) dose of cocaine (LMM-ANOVA: interaction Test:Virus: F1,44 = 10.1557, p = 0.002647, followed by Sidak’s post hoc tests).
(B) HSV-mediated RXRα overexpression in NAc increased CPP for a 7.5 mg/kg dose of morphine (LMM-ANOVA: interaction Test:Virus: F1,43 = 9.4374, p = 0.003681, followed by Sidak’s post hoc tests).
(C) AAV-mediated RXRα knockdown in NAc decreased CPP for a high (10 mg/kg) dose of cocaine (LMM-ANOVA: interaction Test:Virus: F1,41 = 5.1204, p = 0.02901, followed by Sidak’s post hoc tests).
(D) AAV-mediated NAc D1-MSN-specific RXRα overexpression increased CPP for a subthreshold (5 mg/kg) dose of cocaine (LMM-ANOVA: interaction Test:Virus: F1,44 = 28.4447, p < 0.0001, followed by Sidak’s post hoc tests).
(E) AAV-mediated NAc D2-MSN-specific RXRα overexpression did not increase CPP for a subthreshold (5 mg/kg) dose of cocaine (LMM-ANOVA: interaction Test:Virus: F1,44 = 0.0813, p = 0.7768).
(F) Chemogenetic activation of NAc MSNs using the hM3Dq DREADD rescued the effects of RXRα knockdown on cocaine CPP at a high (10 mg/kg) dose (LMM-ANOVA: interaction Test:DREADD:knockdown F1,20 = 5.4961, p = 0.0295, followed by Sidak’s post hoc tests).
Both male and female mice were used in all experiments except for chemogenetics (n = 11-12/group). Bar graphs represent mean ± SEM after combining male and female data.
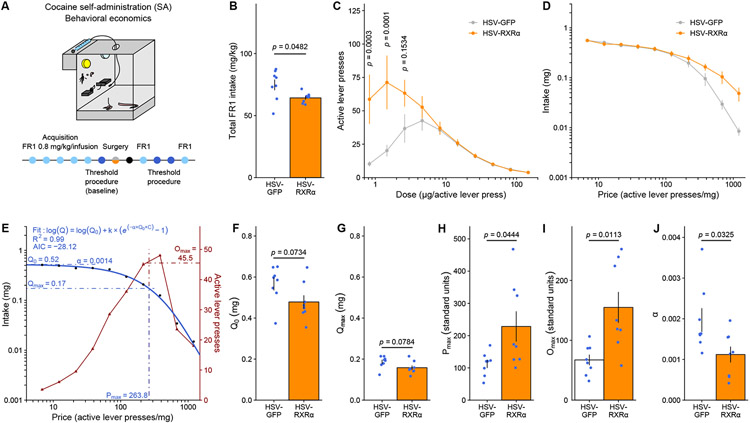
RXRα enhances cocaine reinforcing efficacy in goal-directed tasks
CPP measures have been interpreted as a direct function of a stimulus’s rewarding value, however, they also incorporate essential elements of attention, saliency and learning mechanisms 47. To better dissociate changes in dose sensitivity and reinforcing efficacy from these other factors, especially from learning components, we combined HSV-mediated RXRα overexpression with a self-administration behavioral economics threshold procedure (Figure 6A), which assesses an animal’s motivation and perseverance to self-administer a reinforcer in the face of increasing cost by generating a dose-response curve within subject within a single session 48-50. Each animal is exposed to a descending series of 11 unit doses of cocaine in an FR1 schedule: consequently, the “price” of cocaine – the number of responses required to obtain 1 mg – increases over time, and differences in consummatory and appetitive responding can be measured as a function of cocaine dose/price. We applied this procedure to male rats – where it has been best validated 51.
Figure 6. RXRα overexpression in NAc increases motivation to self-administer low cocaine doses.
(A) Experimental design for behavioral economics testing, using male rats (n = 8/group).
(B) Total cocaine intake in FR1 sessions was significantly reduced by RXRα overexpression (unpaired Welch’s t-test: t8.5723 = 2.3018, p = 0.04824).
(C) Dose response curves in the threshold procedure showed increased responding for low doses of cocaine after RXRα overexpression (LMM-ANOVA: interaction Dose:Virus: FF9,126 = 5.4513, p = 0.00000261, followed by Sidak’s post hoc tests).
(D) Averaged demand curves showed a similar trend of increased cocaine intake at higher prices (LMM-ANOVA: interaction Price:Virus: F9,126 = 1.2085, p = 0.2955).
(E) Representative example of task performance in the threshold procedure (from an HSV-GFP control), highlighting mathematical demand curve fitting and extraction of behavioral economics parameters.
(F) Consumption at low effort Q0 (unpaired Welch’s t-test: t13.987 = 1.9359, p = 0.07336) and
(G) consumption at maximum effort Qmax (unpaired Welch’s t-test: t13.977 = 1.899, p = 0.0784) both trended toward a decrease after RXRα overexpression.
(H) Motivation metrics Pmax (unpaired Welch’s t-test: t8.0394 = −2.3802, p = 0.04439) and
(I) Omax (unpaired Welch’s t-test: t8.6305 = −3.2097, p = 0.01126) were both increased by RXRα overexpression.
(J) Demand elasticity α was reduced by RXRα overexpression (unpaired Welch’s t-test: t12.208 = 2.4121, p = 0.0325).
Bar graphs and line graphs represent mean ± SEM.
After training on an FR1 schedule and HSV infusion, we first observed that HSV-RXRα rats reduced their total drug intake under FR1 responding (Figure 6B). However, during behavioral economics testing, they exhibited significantly higher responding for low (1-5 μg) doses of cocaine (Figure 6C), indicating an upward/leftward shift of the dose-response function after RXRα overexpression. Next, data were plotted as a demand curve (Figure 6D), with consumption as a function of price, and independent measures of consumption, motivation and demand elasticity were extrapolated using mathematical modeling (Figure 6E) applying basic economic principles 50,52. Consistent with FR1 consumption data (Figure 6B), both consumption at low effort Q0 (Figure 6F) and at maximum effort Qmax (Figure 6G) showed trends toward a decrease with RXRα overexpression. By contrast, motivation metrics such as the price of maximal responding after which consumption decreases Pmax (Figure 6H) and the maximal behavioral output at that price Omax (Figure 6I) were both significantly increased by RXRα overexpression. Finally, RXRα overexpression reduced demand elasticity α, which indicates how sensitive demand is to price and can be interpreted as an inverse indicator of the essential value of the reinforcer used; our findings of lower α suggesting higher cocaine essential value (Figure 6J). All of these metrics were equivalent across animals when tested before HSV surgery and group allocation (Supplemental Figure S6).
To summarize, RXRα overexpression in NAc reduced cocaine consumption but increased sensitivity to and motivation for low cocaine doses, as well as cocaine value. Decorrelation between consumption and motivation is precedented 48,53,54, and here we propose that rats with increased RXRα NAc levels consume less cocaine precisely because they are in fact more sensitive to its reinforcing properties and rewarding value, and thus consume less drug to maintain a given hedonic set point. Extending upon our CPP data (Figure 5), these operant behavioral findings demonstrate that RXRα dictates individual sensitivity to drug rewards through direct modulation of their reinforcing efficacy.
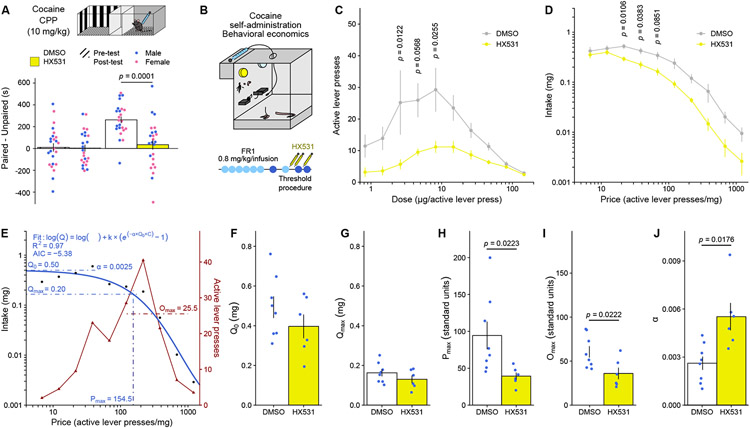
Systemic RXRα pharmacological inhibition weakens cocaine reward
To determine whether RXRα could be leveraged for addiction prevention or treatment, we evaluated the potential for systemic administration of an RXRα inhibitor in regulating drug-related behaviors. First, intraperitoneal injection of the RXR-family antagonist HX531 55, at a dose (20 mg/kg) known to interfere with striatal processes like amphetamine-induced hyperlocomotion 56 and haloperidol-induced dyskinesias 57, blocked cocaine CPP (Figure 7A). Next, we tested the effects of HX531 on operant responding for cocaine in the same rat behavioral economics task (Figure 7B) as above. Intraperitoneal injections of HX531 (12 mg/kg) blunted responding across the dose-response (Figure 7C) and demand price curves (Figure 7D). Mathematical modeling (Figure 7E) further demonstrated that, while consumption at minimal (Figure 7F) and maximal (Figure 7G) effort remained unchanged, HX531 treatment decreased motivation to self-administer cocaine – as evidenced by decreased Pmax (Figure 7H) and Omax (Figure 7I) – and decreased cocaine essential value and perseverative responding – as evidenced by increased demand elasticity α (Figure 7J). Again, these metrics were equivalent across animals when tested before HX531 treatment and group allocation (Supplemental Figure S7). Together, these experiments support systemically targeting RXRα as a therapeutically accessible approach to prevent the development of drug addiction by limiting the reinforcing efficacy of – and learning processes associated with – addictive substances.
Figure 7. Treatment with the RXR inhibitor HX531 reduces behavioral responses to cocaine.
(A) Systemic administration of HX531 (20 mg/kg) decreased CPP in male and female mice (n = 11-12/group) for a high (10 mg/kg) dose of cocaine (LMM-ANOVA: interaction Test:Virus: F1,44 = 13.4504, p = 0.0006572, followed by Sidak’s post hoc tests).
(B) Experimental design for behavioral economics testing, using male rats (n = 6-8/group) treated systemically with HX531 (12 mg/kg).
(C) Dose-response curves in the threshold procedure showed decreased responding across doses of cocaine after HX531 treatment (LMM-ANOVA: main effect of Treatment: F9,12 = 7.2432, p = 0.01962, followed by Sidak’s post hoc tests).
(D) Averaged demand curves showed decreased cocaine intake across cocaine prices (LMM-ANOVA: main effect of Treatment: F9,12 = 5.4469, p = 0.0378, followed by Sidak’s post hoc tests).
(E) Representative example of task performance in the threshold procedure (from an HSV-GFP control), highlighting mathematical demand curve fitting and extraction of behavioral economics parameters.
(F) Consumption at low effort Q0 (unpaired Welch’s t-test: t11.218 = 1.1917, p = 0.258) was not affected by HX531 treatment.
(G) Consumption at maximum effort Qmax (unpaired Welch’s t-test: t11.305 = 1.2226, p = 0.2463) was similarly not affected.
(H) Motivation metrics Pmax (unpaired Welch’s t-test: t7.953 = 2.8289, p = 0.0223) was decreased by HX531 treatment.
(I) Omax (unpaired Welch’s t-test: t11.607 = 2.6369, p = 0.0222) was similarly decreased.
(J) Demand elasticity α was increased by HX531 treatment (unpaired Welch’s t-test: t7.3526 = −3.0459, p = 0.01761).
Bar graphs and line graphs represent mean ± SEM.
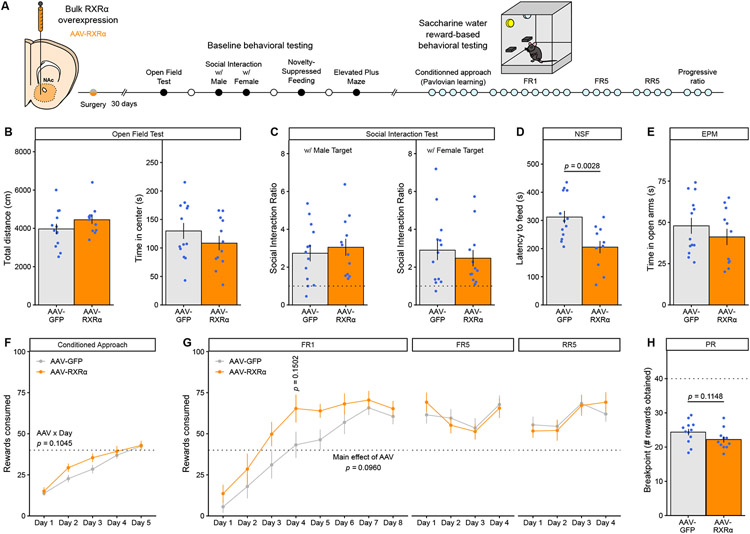
RXRα only mildly affects striatal-dependent natural behaviors
Finally, to test whether RXRα in NAc affects behaviors not involving drugs of abuse, we examined male mice with AAV-mediated RXRα overexpression in NAc neurons on a battery of striatal-dependent reward- or stress-related behaviors (Figure 8A). RXRα overexpression did not affect locomotor activity or time in the center – a proxy metric for innate anxiety-like responses – in an open-field test (Figure 8B). RXRα overexpression also did not affect social interaction towards either a male or female conspecific (Figure 8C), suggesting that RXRα signaling in NAc does not regulate baseline social reward behaviors. In a novelty-suppressed feeding test, however, food-restricted RXRα-overexpressing mice engaged in feeding behavior under environmental conflict faster than controls (Figure 8D). The observation that RXRα overexpression had no effect in either the open-field or elevated-plus maze (Figure 8E) – another anxiety-related test – indicates that the shorter latency to feed in the novelty-suppressed feeding test might represent increased goal-directed approach towards a natural food reward rather than decreased anxiety or an overall hyper-locomotor phenotype. Further investigation revealed that RXRα-overexpressing mice tended to learn both Pavlovian conditioned approach (Figure 8F) and instrumental FR1 operant responding (Figure 8G left and Supplemental Figure S8A) for natural saccharine water rewards faster than controls, although neither effect achieved statistical significance, which could be interpreted as weakly increased motivation to explore natural rewards consistent with the novelty-suppressed feeding data (Figure 8D). The effects disappeared when increasing the motor and motivation requirement via use of an FR5 schedule (Figure 8G middle), or when increasing uncertainty under an RR5 schedule (Figure 8G right), indicating that they likely do not result from changes in thirst and satiety sensitivity. Testing for perseveration under a progressive ratio (PR) schedule showed a weak trend towards decreased motivation after RXRα overexpression (Figure 8H and Supplemental Figure S8B), a moderate yet opposite effect than that for drug rewards (Figure 6). Together, these results suggest that NAc RXRα might affect other striatal behaviors selectively geared towards reward processing for natural reinforcers, yet to a considerably milder extent than for drug rewards, thus supporting a putative therapeutic approach.
Figure 8. RXRα overexpression in NAc mildly affects natural reward behaviors.
(A) Experimental design for behavioral testing, using male mice (n = 12-13/group).
(B) RXRα overexpression did not affect baseline locomotor activity (left, unpaired Welch’s t-test: t22.392 = −1.3742, p = 0.1830) or anxiety-like behaviors (right, unpaired Welch’s t-test: t22.916 = 1.1312, p = 0.2697) in the open-field test.
(C) RXRα overexpression did not alter social investigation behaviors towards either a male (left, unpaired Welch’s t-test: t22.827 = −0.5106, p = 0.6145) or female (right, unpaired Welch’s t-test: t22.475 = 0.6265, p = 0.5373) target. Dotted line represents a Social Interaction Ratio of 1.
(D) RXRα overexpression reduced the latency to feed in a novel anxiogenic context during NSF testing (unpaired Welch’s t-test: t21.688 = 3.3708, p = 0.002796).
(E) RXRα overexpression did not affect open-arm exploration in the EPM test (unpaired Welch’s t-test: t21.44 = 0.94804, p = 0.3537).
(F) RXRα overexpression induced a trend towards faster saccharine water Pavlovian learning of conditioned approach behavior (LMM-ANOVA: interaction Virus:Day: F4,91 = 1.9778, p = 0.1045).
(G) RXRα overexpression induced a trend towards faster saccharine water instrumental learning of operant responding on FR1 schedule (left, LMM-ANOVA: main effect of Virus: F1,22 = 3.0245, p = 0.09599), but had no effect on stable FR5 (middle, LMM-ANOVA: main effect of Virus: F1,22 = 0.0011, p = 0.9734) or RR5 (right, LMM-ANOVA: main effect of Virus: F1,21.98 = 0.0014, p = 0.9700) responding.
(H) RXRα overexpression induced a trend towards decreased motivation for saccharine water reward as measured by reduced PR breakpoint (unpaired Welch’s t-test: t20.978 = 1.6453, p = 0.1148).
Bar graphs and line graphs represent mean ± SEM.
DISCUSSION
In this study, we identify RXRα as a key mediator of the NAc transcriptional mechanisms underlying sensitivity to drug reinforcers in both male and female rodents. Using a combination of viral approaches, we causally demonstrate a positive association between individual RXRα levels in NAc and addiction-relevant molecular, cellular and behavioral features: the higher levels at which NAc MSNs express RXRα, the more excitable they are (Figure 4) and the more responsive to low drug doses the animal is (Figure 5, 6). Our data also support the idea that these effects are transcriptionally mediated at baseline before drug exposure (Figure 2, 3), and are also recruited as important mediators of the transcriptional plasticity mechanisms induced after acute or repeated drug exposure (Figure 1, 2).
First, this study links molecular control of NAc MSN excitability with reward-related motivational and learning processes, adding to a complex and often convoluted literature on the matter 58,59. One particular distinction may reside in differences between NAc core and shell function, where it has been proposed that these two subregions encode distinct yet related parameters for both associative learning and primary reinforcement, likely via differential input-output activity 58,60,61. Here, we primarily targeted the NAc core, but still observed behavioral consequences on both types of metrics. A more precise anatomical and circuit dissection, and especially of D1- vs D2-MSNs within these NAc subregions, would undoubtedly help clarify the respective contribution of their excitability to reward behaviors. How RXRα signaling affects inhibitory vs excitatory transmission in NAc circuits is also an interesting focus for future studies. We postulate that RXRα might act as a general master regulator of excitability transcriptional programs, and consequently hypothesize that the functional consequences of its manipulation might be mostly dictated by the recruitment (or not) of targeted circuits or cell types during execution of a specific behavior. As an example, increasing excitability of NAc core and medial shell D1-MSNs via cell-type-specific RXRα overexpression promotes the same behavioral outcome – increased cocaine reward learning – as found for optogenetic activation of those same neurons 31.
Transcriptional processes represent a fundamental building block of neuronal plasticity, including after drug exposure 4,10. In that context. RXRα is particularly well-suited to fine-tune these transcriptional programs in a very specific yet stimulus-dependent manner, because of its ability to function either as homodimers or as an obligatory heterodimerization partner for several other nuclear receptors, like brain-expressed retinoic acid receptors (RARs), peroxisome proliferator-activated receptors (PPARs), vitamin D receptors (VDRs) and thyroid hormone receptors (TRs), among many others 13,34,35,62. In addition to bridging together different signaling pathways, these heterodimers can be either permissive or repressive for transcription depending on subsequent recruitment of distinct co-regulators and epigenetic enzymes 13,34,35. This posits RXRα as a potential integrative signaling hub at key gene promoters to subtly dictate locus-specific gene expression upon activation of generic drug-evoked signaling cascades and transcription factors such as CREB 12. Consistent with this idea, RXRα motifs exist in close proximity to CREB binding sites at promoters of select cocaine-regulated genes 11. Other promising candidate RXRα binding partners include 1) RXRγ, which is known to contribute to striatal- and dopamine-dependent locomotion 18 and depressive-like 19-21 behaviors, 2) NURR77/NR4A1, an IEG and orphan receptor associated with cocaine 63 and stress 64 behaviors that also positively correlates with RXRα expression (Figure 2E), 3) RARs and PPARs because the enzymes that deliver retinoic acid to RAR/RXR or PPAR/RXR complexes – CRABP2 and FABP5 respectively – contribute to addiction- 15,16 and depression-related 17 behaviors, and 4) VDRs, as vitamin D deficiency was recently linked with opioid addiction 65. Ultimately, a finer dissection of RXRα binding partners, as well as potential coordinated shifts in RXRα binding partners and/or genomic binding sites upon drug exposure, could better explain some of the cell-type-, region-, drug- and drug-regimen-specificity of drug-induced transcriptional programs, as these likely result from the recruitment of not one but several factors and enzymes, which could be partly harmonized by RXRα.
By definition, activation of specific nuclear receptor complexes is dependent on proper recognition of their respective ligands 13,34,35,62. However, the exact nature of the endogenous brain ligand(s) for RXR family members including RXRα – termed rexinoids – remains debated, partially because of technical difficulties in detecting and studying naturally-occurring retinoids and rexinoids 34,62. All-trans-retinoic acid (atRA), the undisputed physiological ligand for RARs 66, does not bind RXRs 66. 9-cis-retinoic acid (9CRA), which binds RXRs with nanomolar affinity in vitro 66, was considered as an RXR endogenous ligand until several reports failed to detect endogenous 9CRA levels in vivo 62,67. Several other molecules have since been proposed, but to date the two strongest candidates for rexinoid activity in brain are 1) free fatty acid metabolites of docosahexaenoic acid (DHA), and 2) dihydroretinoid metabolites, especially 9-cis-13,14-dihydroretinoic acid (9CDHRA). On the one hand, DHA is abundant in the mammalian brain 68 (although whether in forms and at concentrations sufficient to induce transactivation is questioned 62), DHA has been shown to control cortical excitability and synaptic architecture in an RXRα-dependent manner 29, and DHA supplementation can reverse depressive-like behavior in an RXRγ-dependent manner 20. On the other hand, 9CDRHA is detected in brain tissue at high levels sufficient for RXR binding 67, and 9CDHRA supplementation also reversed depressive-like behavior in an RXRγ-dependent manner 21. Whether DHA, 9CDRHA or other related molecules bind RXRα in NAc, as well as the exact kinetics of their action (baseline levels, but also hypothetical release temporal dynamics), and whether these are affected by drug exposure, remain to be elucidated, and would greatly benefit from improved resolution of metabolomics techniques.
One shared property of rexinoids is that they or their precursors can be nutritionally provided 19-21,62,68. This has great preclinical relevance for several other pathologies linked to RXR signaling including Alzheimer’s disease, Parkinson’s disease, multiple sclerosis and diabetes 34,35,62 and has sparked interest in testing phyto-pharmacologically derived rexinoids or de novo synthetic RXR ligands to clinically combat or prevent these conditions 34,35,62. Here we show that inhibition of RXRs using a systemically administered RXR antagonist (HX531) reduced cocaine-induced associative learning and operant responding (Figure 7), consistent with previous reports showing that HX531 blocked amphetamine-induced hyperlocomotion in a NURR77/NR4A1-dependent manner 56 – another hint at further studies of RXR/NURR77 dimers in psychostimulant action (see above). Moreover, this inhibitor has been shown to block RXR/RXR, RAR/RXR and PPARγ/RXR, but not PPARα/RXR heterodimers 55, highlighting rexinoid drugs as a potential platform from which to derive pharmacological compounds with increased specificity for certain RXR-containing nuclear receptor dimers – i.e., for some target pathways, cell type or condition. Finally, because full cortical and hippocampal RXRα knockout mice behaved normally on an extensive battery of baseline behavioral tests 29, our preliminary data illustrate that systemically or nutritionally targeting NAc RXRα using rexinoids might be leveraged as a prevention initiative for drug addiction with tolerable side effects. This work also underscores the effectiveness of bioinformatically harnessing large-scale datasets 11 as part of the effort to unravel innovative answers to highly complex brain maladaptations from both fundamental and translational standpoints.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Eric J. Nestler (eric.nestler@mssm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All RNAseq data reported in this study are deposited publicly in the Gene Expression Omnibus under accession code GSE198527. Unmodified Western blots scans and other supporting raw data are available from the lead contact upon request. Scripts and code utilized in this study, including for statistical analysis, are available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Male and female C57BL/6J mice (8-16 weeks old, 20-30 g, The Jackson Laboratory) were maintained on a 12:12 h light/dark cycle (07:00 lights on; 19:00 lights off) and were provided with food and water ad libitum. Transgenic mouse lines (D1-Cre: MGI:3836633, D2-Cre: MGI:3836635, D1-tdTomato: MGI:4360387, fl/fleGFP::L10a: IMSR_JAX:022367) were bred in-house on a C57BL/6J background. All mice were maintained according to the National Institutes of Health guidelines for Association for Assessment and Accreditation of Laboratory Animal Care accredited facilities. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Mount Sinai.
Rats
All rats were housed in the animal facilities at Mount Sinai. Male Sprague-Dawley rats (300-400g) were maintained on a 12:12 h reverse light/dark cycle (07:00 lights off; 19:00 lights on) and restricted to 95% of free-feeding body weight to improve consistency of the self-administration behavior. All rats were maintained according to the National Institutes of Health guidelines for Association for Assessment and Accreditation of Laboratory Animal Care accredited facilities. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Mount Sinai.
METHOD DETAILS
Drug treatments
Cocaine HCl (from the National Institute on Drug Abuse) was diluted in 0.9% NaCl saline solution (ICU Medical) and injected intraperitoneally at 20, 10 or 5 mg/kg. Morphine SO4 (from the National Institute on Drug Abuse) was diluted in 0.9% NaCl saline solution (ICU Medical) and injected subcutaneously at 7.5 mg/kg. HX531 (Tocris, Cat. No. 3912) was first diluted in DMSO (Sigma), then in Dulbecco’s Phosphate Buffered Saline (PBS, Gibco) to a 1% DMSO final concentration and injected intraperitoneally 20 min before testing at 20 mg/kg in mice, and to a 5% DMSO final concentration and injected intraperitoneally 30 min before testing at 12 mg/kg in rats. For chemogenetics, clozapine-N-oxide (CNO) dihydrochloride (Tocris, #6329) was diluted in PBS and injected intraperitoneally 15 min before testing at 2 mg/kg.
Viral reagents
For HSV-mediated RXRα overexpression, an RXRα insert consisting of a codon-optimized mouse sequence reverse-translated from RXRα mouse protein sequence was synthesized de novo by Genewiz into a pENTR plasmid, then sub-cloned downstream of a IE4/5 promoter into a bicistronic p1005 plasmid, which also expresses GFP under a separate CMV promoter, using Gateway LR Clonase Kit (Invitrogen). Packaging into HSVs of the overexpression (HSV-RXRα-GFP) and empty control (HSV-GFP) vectors was performed at the Gene Delivery Technology Core of Massachusetts General Hospital in Boston, Massachusetts, USA. For AAV-mediated RXRα overexpression, the RXRα insert was excised from the p1005 plasmid and sub-cloned into an AAV2 expression backbone downstream of an EGFP reporter sequence separated by a P2A cleavage sequence. For non-Cre-dependent vectors, the insert expression was controlled by a CMV promoter. For Cre-dependent expression, the insert was flanked by two inversed loxP sites to only express the transgene upon Cre-dependent recombination and expression was controlled by a CAG promoter. Cloning and then packaging into AAV9 capsids of the overexpression (AAV9-CMV-EGFP-P2A-RXRα and AAV9-CAG-DIO-EGFP-P2A-RXRα) and control (AAV9-CAG-DIO-EGFP) vectors were performed at the Duke University Viral Vector Core in Durham, North Carolina, USA. The control non-Cre-dependent AAV9-CMV-EGFP vector was obtained from Addgene (#105530-AAV9). For AAV-mediated RXRα knockdown, miRNA plasmids were prepared using the BLOCK-iT Pol II miR RNAi Expression Vector Kit (ThermoFisher Scientific) with top and bottom sequences targeting mouse Rxra cDNA (Top: 5’ - TGC TGA GAT GTT GGT AAC AGG GTC ATG TTT TGG CCA CTG ACT GAC ATG ACC CTT ACC AAC ATC T - 3’; Bottom: 5’ – CCT GAG ATG TTG GTA AGG GTC ATG TCA GTC AGT GGC CAA AAC C - 3’) or a control Lacz sequence (Top: 5’ – TGC TGA AAT CGC TGA TTT GTG TAG TCG TTT TGG CCA CTG ACT GAC GAC TAC ACA TCA GCG ATT T - 3’; Bottom: 5’ – CCT GAA ATC GCT GAT GTG TAG TCG TCA GTC AGT GGC CAA AAC GAC TAC ACA AAT CAG CGA TTT C - 3’) obtained from Invitrogen. The miRNA plasmids were then sub-cloned into an AAV2 expression backbone downstream of a CMV promoter and of an EYFP reporter sequence. Cloning and then packaging into AAV9 capsids of the knockdown (AAV9-CMV-EYFP-miR-RXRα) and control (AAV9-CMV-EYFP-miR-lacZ) vectors were performed by the Duke University Viral Vector Core in Durham, North Carolina, USA. All viral vectors were validated in vivo (Supplemental Figure S2). For chemogenetics, AAV9-hSyn-DIO-hM3D(Gq)-mCherry (#44361) and AAV9-hSyn-DIO-mCherry (#50459) were both purchased from Addgene.
Stereotaxic surgeries
Mice or rats were anesthetized with an intraperitoneal bolus of ketamine (100 mg/kg) and xylazine (10 mg/kg), then head-fixed in a stereotaxic apparatus (Kopf Instruments). Syringe needles (33G, Hamilton) were used to bilaterally infuse 1 μl of virus at a 0.1 μl/min flow rate. Needles were kept in place for 10 minutes after injection before being retracted to allow for virus diffusion. Viruses were used at the following titers: HSV-GFP and HSV-RXRα-GFP at ± 1 × 108 IU/mL, AAV9-CMV-EGFP-P2A-RXRα, AAV9-CMV-EGFP, AAV9-CAG-DIO-EGFP-P2A-RXRα, AAV9-CAG-DIO-EGFP, AAV9-hSyn-DIO-hM3D(Gq)-mCherry and AAV9-hSyn-DIO-mCherry at ± 1 × 1012 VG/mL, AAV9-CMV-EYFP-miR-RXRα and AAV9-CMV-EYFP-miR-lacZ at ± 5 × 1012 VG/mL. Coordinates for NAc were as follows, from Bregma: AP + 1.6 mm , ML + 1.6 mm, DV − 4.5 mm, 10° angle for mice, and AP + 1.9 mm , ML + 2.8 mm, DV − 7.2 mm, 10° angle for rats. These injection coordinates targeted the anterior NAc core and representative images are shown in Supplemental Figure S2. For HSV vectors, expression remained limited to NAc core. For AAV vectors, expression was strong in both NAc core and NAc medial shell.
RNA extraction and quantitative real-time PCR
Mouse brains were collected after cervical dislocation and followed by rapid bilateral punch dissections of NAc (core and shell) from 1 mm-thick coronal brain sections using a 14G needle and frozen on dry ice. RNA extraction was performed using the RNeasy Micro Kit (Qiagen) following manufacturer instructions. RNA 260/280 ratios of 2 were confirmed using spectroscopy, and reverse transcription was achieved using the iScript cDNA Synthesis Kit (BioRad). Quantitative PCR using PowerUp SYBR Green (Applied Biosystems) was used to quantify cDNA using an Applied Biosystems QuantStudio 5 system and Applied Biosystems QuantStudio Design & Analysis Software v1.4.3. Each reaction was performed in triplicate and relative expression was calculated relative to the geometric average of 3 control genes (Ppia, Tbp, Rpl38) according to published methods 69. Sequences of all used primers are available in Supplemental Table S4.
Protein extraction and Western blotting
Mouse brains were collected after cervical dislocation and followed by rapid bilateral punch dissections of NAc (core and shell) from 1 mm-thick coronal brain sections using a 14G needle and frozen on dry ice. For whole tissue extracts, frozen NAc samples were homogenized and then incubated for 30 min with agitation in 200 μL of ice-cold RIPA buffer (10 mM Trizma Base, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton-X-100, 1% sodium deoxycholate, pH 7.4, complemented with protease and serine/threonine and tyrosine phosphatase inhibitors), before 5 cycles of 20 s on/off sonication using a Bioruptor (Diagenode). Samples were centrifuged for 15 min at 14,000 g to pellet insoluble debris and lipids, and supernatant was transferred to new tubes. For nuclear fractions, frozen NAc samples were first subjected to subcellular fractionation using our established procedures 48. Briefly, frozen NAc samples were homogenized in 150 μL of HEPES-buffered sucrose (0.32 M sucrose, 4 mM HEPES, pH 7.4, complemented with protease and serine/threonine and tyrosine phosphatase inhibitors) before a 10 min centrifugation at 1000 g at 4°C to pellet nuclei, followed by 3 washes in HEPES-buffered sucrose and finally by RIPA extraction (see above). For both whole tissue and nuclear extracts, protein concentration was quantified using a Pierce BCA Protein Assay Kit (ThermoFisher Scientific). SDS-PAGE protein separation and Western blotting were performed according to manufacturer instructions and our published protocols 48. Equal amounts of proteins were mixed β-mercaptoethanol-supplemented Laemmli buffer (BioRad), heated to 95°C for 5 min before being separated by SDS-PAGE with Criterion Precast Gels (4 −15% Tris-HCl; BioRad) and transferred onto Immun-Blot PVDF 0.2 μm (BioRad) membranes. Membranes were blocked in Tris-buffered saline containing 5% bovine serum albumin (Sigma) and 0.1% Tween-20 (Fisher Bioreagents) for 1 h at room temperature. Primary antibodies (full list and concentrations used available in Supplemental Table S5) were diluted in blocking solution and incubated overnight at 4°C. After washing, membranes were incubated with anti-mouse (#PI-2000, Vector Laboratories) or antirabbit (#PI-1000, Vector Laboratories) peroxidase-conjugated secondary antibodies diluted 1:50,000 in blocking solution for 2 h, washed thoroughly, and developed using SuperSignal West Dura Substrate (ThermoFisher Scientific). Quantification was performed by densitometry using Image J v1.53c (U.S. National Institutes of Health). Protein levels were normalized to actin for whole tissue extracts or to actin and TBP for nuclear extracts. Between primary antibodies, membranes were stripped using Restore Plus Stripping Buffer (ThermoFisher Scientific). Male and female samples were run on separate gels and later normalized to their respective controls before being pooled for analysis. Unmodified Western blots scans are provided as Source Data.
Nuclei purification and Fluorescence-Activated Nuclei Sorting (FANS)
Mouse brains were collected after cervical dislocation and followed by rapid bilateral punch dissections of virally-infected NAc (core and medial shell) tissue under fluorescent light from 1 mm-thick coronal brain sections using a 14G needle and frozen on dry ice. To obtain a single nuclei suspension, frozen NAc samples were homogenized in 4 mL of low-sucrose lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM Mg(Ace)2, 0.1 mM EDTA, 10 mM Tris-HCl) using a large clearance then a small clearance pestle of a glass dounce tissue grinder (Kimble Kontes). Homogenates were filtered through a 40 μm cell strainer (Pluriselect) into ultracentrifuge tubes (Beckman Coulter), underlaid with 5 mL of high-sucrose solution (1.8 M sucrose, 3 mM Mg(Ace)2, 1 mM DTT, 10 mM Tris-HCl) and centrifugated at 24,000 rpm for 1 h at 4°C in a SW41Ti Swinging-Bucket Rotor (Beckman Coulter). Supernatant was discarded and nuclei pellets were re-suspended in 800 μL of PBS. DAPI was added at a 1:5000 dilution. Nuclei were sorted on a BD FACS Aria II three-laser device with a 70 μm nozzle and using BD FACSDiva Software v8.0.2. Gating strategy from representative sorts are visualized in Supplemental Figure S5. Briefly, debris and doublets were excluded using FSC and SSC filters, nuclei were then selected as DAPI-positive (Violet1-A laser) events, and finally GFP-positive nuclei (Blue1-A laser) were sorted directly into TRIzol LS (Ambion) and flash frozen. Between 30,000 and 60,000 nuclei were recovered for each sample.
RNA-sequencing (RNAseq)
For bulk tissue RNAseq, mouse brains were collected after cervical dislocation and followed by rapid bilateral punch dissections of virally-infected NAc (core and medial shell) tissue under fluorescent light from 1 mm-thick coronal brain sections using a 14G needle and frozen on dry ice. RNA extraction was performed using the RNeasy Micro Kit (Qiagen) following manufacturer instructions. Sequencing libraries were prepared by Genewiz/Azenta with polyA selection and sequenced with Genewiz/Azenta on an Illumina HiSeq 4000 machine using a 2 × 150 bp pair-end read configuration to a minimum depth of 20 million reads per sample. For cell-type-specific RNAseq, RNA was extracted from frozen TRIzol LS (Ambion) homogenates using the Direct-zol RNA Microprep Kit (Zymo Research) following manufacturer instructions. Ribo-depleted sequencing libraries were prepared with the SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (TaKaRa Biotech) following manufacturer instructions and sequenced with Genewiz/Azenta on an Illumina NovaSeq S4 machine using a 2 × 150 bp pair-end read configuration to a minimum depth of 40 million reads per sample. Bioinformatics were run on a Java v1.8.0 environment. For both experiments, quality control was performed using FastQC v0.11.9 (www.bioinformatics.babraham.ac.uk/projects/fastqc/), adapter contaminants were trimmed using Trimmomatic v0.27 (github.com/usadellab/Trimmomatic), reads were aligned to a custom-built reference genome (RXRα and GFP plasmid sequences were added to GENCODE GRCm38 genome) using hisat2 v2.1.0 (daehwankimlab.github.io/hisat2/), duplicate reads were removed using Picard v2.18.10 MarkDuplicate tool (broadinstitute.github.io/picard/), and finally count matrices were generated using the featureCounts function of the subread v2.0.0 package (subread.sourceforge.net/featureCounts.html) and a comprehensive gene annotation file obtained from GENCODE (GRCm38 vM23). Reads mapping to the plasmid codon-optimized Rxra overexpression sequence were added to reads mapping to the endogenous Rxra sequence (ENSMUSG00000015846). The top 30% most expressed annotated genes/features (highest normalized read counts average across all samples) were kept for subsequent analysis in order to filter out poorly expressed genes. Differential expression was analyzed in R v4.0.2 using the DESeq2 package v1.34.0 70. Of note, for the cell-type-specific RNAseq both males and females were used, and biological sex was included as a factor in the model design (~ Sex + Virus). To confirm cell-type specificity, only GFP control samples were selected and differential expression was run between cell types (~ Sex + CellType model). Significance cut-offs were of at least 15% expression fold change ( ∣log2(FoldChange)∣ > log2(1.15) ) and nominal p < 0.05, except in the cases when genes were examined individually, where the Benjamini-Hochberg corrected padj was used. Gene lists and corresponding statistics are available in Supplemental Table S2 (D1-MSNs) and S3 (D2-MSNs). RRHO plots were generated using the RRHO2 package 71 (github.com/RRHO2/RRHO2). Gene ontology analyses were performed using the PANTHER knowledgebase and classification system (pantherdb.org). Analysis code is available upon request.
RNA fluorescent in situ hybridization (FISH)
Mice were transcardially perfused with a fixative solution containing 4% paraformaldehyde (PFA). Brains were post-fixed for 24 h in 4% PFA at 4°C. Sections of 20 μm thickness were cut in the coronal plane with a vibratome (Leica) and stored at −20°C in a cryoprotectant solution containing 30% ethylene glycol (v/v), 30% glycerol (v/v) and 0.1 M phosphate buffer. NAc slices were mounted on charged Superfrost Plus microscope slides (Fisher Scientific) and processed for RNA FISH using RNAscope Multiplex Fluorescent Reagent Kit v2 (ACD Bio) according to manufacturer instructions using mouse probes for Rxra (Mm-Rxra, #463121), Drd1a (Mm-Drd1a-C2, #406491-C2) and Drd2 (Mm-Drd2-C3, #406501-C3) transcripts. Sections were counterstained with DAPI and mounted using ProLong Gold Antifade Mountant (ThermoFisher Scientific). Confocal images (6-10 per animal, 1024 × 1024 pixels, 16 bits pixel depth) were acquired on a SP8 inverted confocal microscope (Leica) using a 40X objective and Leica Application Suite X v3.5.7.23225. A custom-made automated Image J v1.53c (U.S. National Institutes of Health) pipeline was used to extract channel intensity in every nucleus identified on DAPI staining (Supplemental Figure S4A). Individual Rxra puncta were detected using ComDet v0.5.4 (github.com/ekatrukha/ComDet). Analysis code is available upon request.
Ex vivo slice electrophysiology
After at least 4 weeks of recovery from AAV surgery, male and female D1-Tomato mice were anesthetized using isoflurane. Brains were rapidly extracted, and coronal sections (250 μm) were prepared using a Compresstome (Precisionary Instruments) in cold (0-4°C) sucrose-based artificial cerebrospinal fluid (SB-aCSF) containing 87 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 4 mM MgCl2, 23 mM NaHCO3, 75 mM Sucrose, 25 mM Glucose. After recovery for 60 min at 32°C in oxygenated (95% CO2 / 5% O2) aCSF containing 130 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 2.4 mM CaCl2, 1.2 mM MgCl2, 23 mM NaHCO3, 11 mM Glucose, slices were kept in the same medium at room temperature for the rest of the day and individually transferred to a recording chamber continuously perfused at 2-3 mL/min with oxygenated aCSF. Patch pipettes (4-6 MΩ) were pulled from thin wall borosilicate glass using a micropipette puller (Sutter Instruments) and filled with a K-Gluconate-based intra-pipette solution containing 116 mM KGlu, 20 mM HEPES, 0.5 mM EGTA, 6 mM KCl, 2 mM NaCl, 4 mM ATP, 0.3 mM GTP (pH 7.2). Cells were visualized using an upright microscope with an IR-DIC lens and illuminated with a white light source (Olympus for Scientifica), and fluorescence visualized through eGFP and mCherry bandpass filters upon LED illumination through the objective (p3000ULTRA, CoolLed) using μManager v2.0 (https://micro-manager.org/). All recordings were made on anterior NAc core MSNs. Excitability was measured in current-clamp mode by injecting incremental steps of current (0-300 pA, +20 pA at each step). For recording of spontaneous Excitatory Post-Synaptic Currents (sEPSCs), neurons were recorded in voltage-clamp mode at −70mV and sEPSCs detected with a 8 pA threshold. Whole-cell recordings were performed using a patch-clamp amplifier (Axoclamp 200B, Molecular Devices) connected to a Digidata 1550 LowNoise acquisition system (Molecular Devices). Signals were low pass filtered (Bessel, 2 kHz) and collected at 10 kHz using Axon pCLAMP 11 Software Suite (Molecular Devices). Electrophysiological recordings were extracted using Clampfit (Molecular Devices). All groups were counterbalanced by days of recording and all recordings were performed blind to experimental condition.
Conditioned place preference (CPP)
Unbiased CPP was carried out using three-chambered CPP Med Associates boxes and MED-PC IV v4.2 software. The two end chambers have distinct visual (gray vs striped walls) and tactile (small grid vs large grid flooring) cues to allow differentiation. On the pre-test day, animals were allowed to freely explore all three chambers for 20 min. Groups were then attributed and pairing sides were adjusted to balance out any pre-existing chamber bias. Next, drug-context learning was achieved by pairing an injection of saline with one chamber in the morning, and a second injection of cocaine or morphine with the other chamber in the afternoon for two consecutive days. CPP testing was carried out on the fourth day when each animal was again allowed to explore all chambers freely. Place preference score was taken as the difference in time spent on the cocaine-paired side time vs on the saline-paired side.
Rat intravenous self-administration
Rats were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) and implanted with chronic indwelling jugular catheters as described previously 48. Animals were singly housed, and all sessions took place during the active/dark cycle (12:00–15:00). After a 4-day recovery period, animals underwent training for self-administration where they were given access to a drug-paired lever on a fixed ratio one (FR1) schedule using a cocaine dose of 0.8 mg/kg/infusion or using a heroin dose of 0.03 mg/kg/infusion, both delivered over 5 s,. After each infusion, the lever was retracted and a stimulus light was illuminated for a 20 s timeout period. Responding on a second inactive lever was recorded but resulted in no programmed consequence. Acquisition was defined as the first session in which an animal allocated > 70% of their responses to the active lever, and when a stable pattern of inter-infusion intervals was present.
Behavioral economics testing
For behavioral economics testing, after acquisition of cocaine responding, rats then went through a within-session threshold procedure, described in detail below, before being split into two unbiased groups (Supplemental Figures S6 and S7) for HSV surgery or HX531 treatment. After surgery and one recovery day, rats were subjected to 2 FR1 consumption sessions and 2 threshold sessions, or to only 2 threshold sessions for the HX531 experiment. The threshold procedure has been used by our lab 48 and others 49,50,52-54. Briefly, a descending series of 11-unit doses of cocaine (259.9, 146.1, 82.2, 46.2, 26.0, 14.6, 8.2, 4.6, 2.6, 1.5 and 0.83 μg/infusion) are available for 10 min upon an FR1 schedule with no timeouts, and levers remain extended. Both the number of lever presses and the total cocaine intake are measured for each price bin and plotted as a function of dose/price. The resulting demand curves can be used for mathematical curve fitting using the following equation: where Q is consumption and C is the varying cost of the reinforce 50,52. Here, curve fitting was achieved using custom-made R code utilizing the stats::nls function, and goodness of fit was assessed by computing a pseudo-R2 coefficient (the square of the correlation between observed and predicted log10(Q) values) and the Akaike Information Criterion (AIC) for each individual fit. The k value was determined by fitting all individual curves with 0.1 increments of k values between 1 and 5, and the k value that maximized the overall pseudo-R2 and minimized the overall AIC was selected and was here equal to 2.7. The parameters Q0, Qmax, Pmax, Omax and α were then calculated for each animal. Q0 is a measure of the preferred level of cocaine consumption of the animals and was measured as the amount of consumption at a theoretical minimally constraining price. Pmax is the inflection point of the demand curve and was calculated as the point for which the first derivative slope of the demand function is equal to −1 in the log-log space, and corresponds to the price at which the animal no longer emits enough responses to maintain intake and consumption decreases. Qmax is the consumption at Pmax, and Omax the behavioral output (active lever presses) at Pmax. The measure of elasticity α, also termed the essential value, is a measure of how sensitive the demand is to price. With high elasticity, responding will drop off quickly as price increases, whereas with low elasticity, animals will be motivated to continue consuming drug, regardless of cost. Curve fitting and analysis code is available upon request.
Open-field test (OFT)
Mice were placed in the open field arena (44 × 44 cm) for 5 min to compare the distance traveled and time spent in the peripheral zone (7.5 cm from each border wall) compared to the center zone. Testing conditions occurred under red-light (10 lux) in a room isolated from external sound sources. The OFT apparatus was thoroughly hand cleaned between mice with an odorless 30% ethanol cleaning solution. The mouse’s activity – distance, velocity, and time spent in specific open field areas – was captured with a video tracking system (Ethovision) set to localize the mouse center point at each time of the trial.
Social interaction test
Testing for social interaction was assessed with a novel, non-littermate, aged-matched C57BL/6J male or female mouse under red-light (10 lux) in a room isolated from external sounds. In the first 2.5 min, the experimental mouse was allowed to freely explore an arena (44 × 44 cm) containing a custom-made Plexiglass circular enclosure (10 cm diameter) centered against one wall of the arena. In the next 2.5 min, the target mouse was placed in the enclosure. The apparatus was thoroughly hand cleaned between mice with an odorless 30% ethanol cleaning solution. Time spent in a defined ‘interaction zone’ (a 7.5 cm-wide circular band surrounding the enclosure) was measured using video tracking (Ethovision). A “Social Interaction Ratio” was calculated as time spent exploring the interaction zone with the target mouse present, divided by time spent exploring the interaction zone with the target mouse absent.
Novelty-suppressed feeding (NSF) test
Mice were food-restricted for 24 h before testing. They were then placed in the corner of a novel, black open-field arena (44 × 44 cm) covered in a different type of saw-dust bedding and where a single pellet of chow food was placed in the center of the arena. Testing conditions occurred under dim white-light (6 lux) in a room isolated from external sounds. The NSF boxes were thoroughly hand cleaned between mice with an odorless 30% ethanol cleaning solution, bedding replaced and a new food pellet was used for each mouse. Latency to feed was hand-scored by an experimenter blind to the experimental groups as the start of the first bout of uninterrupted feeding that was longer than 5 s.
Elevated-Plus maze (EPM) test
EPM apparatuses were designed in black Plexiglass (arm length 75 cm, arm width 8 cm, height 80 cm) and fitted with white surfaces to provide contrast. Testing conditions occurred under red light (10 lux) in a room isolated from external sounds. The apparatuses were thoroughly hand cleaned between mice with an odorless 30% ethanol cleaning solution. Mice were positioned in a closed arm, and behavior was video tracked for a 5 min period. Time in EPM compartments, locomotion and velocity were measured by video tracking (Ethovision).
Saccharine water self-administration
Mice were restricted to 15 min of water access per day for the entire duration of the experiment, given after behavioral testing. Natural reward-related behaviors were tested in 5 consecutive phases, all carried out using the same MedAssociates operant boxes and associated MedPC IV software. First, Pavlovian conditioned approach behavior learning was assessed using a dipper training procedure: for 30 min, a dipper exposing 20 μL of 0.2% saccharine water raised for 5 s every 30 s inside a head port equipped with laser beams to detect magazine head entries. A reward was considered ‘consumed’ when a head entry was detected while the dipper was raised. Conditioned approach behavior was considered ‘acquired’ when mice on average consumed 40 rewards, i.e., two thirds of the total 60 rewards offered, which was observed after 5 days. Second, water-restricted mice learned to press an active lever for a 5 s access to the same saccharine water reward under a fixed ratio 1 (FR1) schedule of reinforcement. Responding on a second inactive lever was recorded but resulted in no programmed consequence. FR1 testing was continued for a total of 8 days to allow all mice to reach a responding plateau of stable reward consumption. One of 25 mice failed to acquire operant responding and was removed from further testing. Third, the responding requirement was increased to an FR5 schedule for 4 days, and then to a random ratio 5 (RR5) schedule for 4 days, where each lever press had a 20% chance of resulting in reward presentation. Finally, mice were tested on a progressive ratio (PR) schedule for 3 consecutive days, where, for each session, the FR requirements to trigger the next reward delivery increased following an exponential suite defined by the following equation: where n is the number of rewards previously obtained and FRreq was rounded to the nearest integer. Breakpoints were defined as the total number of rewards obtained and averaged for each mouse across the 3 testing sessions.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical power estimation analyses were used to predetermine sample sizes, which instead were chosen to match previous publications 11,31,39,48. All statistics were performed in R v4.0.2 mostly relying on stats v4.0.2, tidyverse v1.3.1 and lmerTest v3.1-3 packages. Detailed statistics, including the exact functions and arguments used, are provided for each figure panel in Supplemental Table S6. In summary, pairwise comparisons were performed with Welch’s t-tests (stats::t.test function), enrichment tests using Fisher’s LSD (stats::fisher.test function), correlations using Pearson’s r (stats::cor.test function) and more complex multifactorial designs were analyzed using linear models computed with the stats::lm function for fixed effects-only models or lmerTest::lmer function for mixed effects models. Random effects (repeated measures and/or nested observations) were modeled as random intercept factors. Subsequent analysis of variance was performed using type III sums of squares with Kenward-Roger’s approximation of degrees of freedom. Of note, biological sex was always included as a fixed effect factor according to best practice guidelines in considering sex as a biological variable 72. Pooled data are represented in figures, but individual data points are color-coded by sex. Post hoc testing (on pooled data when relevant) was performed using the emmeans package and significance was adjusted using Sidak’s correction, except for RNAseq data where Benjamini-Hochberg correction was used as part of the DESeq2 pipeline. Bar and line graphs represent mean ± SEM. Correlation graphs represent regression line with its 95% confidence interval. Significance was set at p < 0.05.
Supplementary Material
Table S1. Full DEG list after HSV-mediated bulk RXRα overexpression. (related to Figure 1)
Table S2. Full DEG list after AAV-mediated RXRα overexpression in D1-MSNs. (related to Figure 3
Table S3. Full DEG list after AAV-mediated RXRα overexpression in D2-MSNs. (related to Figure 3)
Table S6. Full statistics. (related to all Figures)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| see Supplemental Table S4 | ||
| anti-mouse peroxidase-conjugated secondary antibody | Vector Laboratories | Cat#PI-2000 |
| anti-rabbit peroxidase-conjugated secondary antibody | Vector Laboratories | Cat#PI-1000 |
| Oligonucleotides | ||
| see Supplemental Table S5 for qPCR primers | ||
| see Method Details for miRNA sequences | ||
| FISH probe: Mm-Rxra | ACD Bio | Cat#63121 |
| FISH probe: Mm-Drd1a-C2 | ACD Bio | Cat#406491-C2 |
| FISH probe: Mm-Drd2-C3 | ACD Bio | Cat#406501-C3 |
| Chemicals | ||
| Cocaine HCl | NIDA | N/A |
| Morphine SO4 | NIDA | N/A |
| HX531 | Tocris | Cat#3912 |
| PBS | Thermo Fisher Scientific | 14-190-144 |
| DMSO | Sigma | D4540 |
| CNO dihydrochloride | Tocris | Cat#6329 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
| Mouse: Tg(Drd1-cre)FK150Gsat | GENSAT | MGI:3836633 |
| Mouse: Tg(Drd2-cre)ER44Gsat | GENSAT | MGI:3836635 |
| Mouse: Tg(Drd1-tdTomato)5Calak | Shuen et al., 2008 | MGI:4360387 |
| Mouse: Rosa26fsTRAP | The Jackson Laboratory | IMSR_JAX:022367 |
| Rat: SAS Sprague Dawley | Charles River | Cat#400 |
| Viruses | ||
| HSV-GFP | Gene Delivery Core, MGH | N/A |
| HSV-RXRa-GFP | Gene Delivery Core, MGH | N/A |
| AAV9-CAG-DIO-EGFP | Viral Vector Core, Duke | BK277 |
| AAV9-CAG-DIO-EGFP-P2A-RXRa | Viral Vector Core, Duke | BK1432 |
| AAV9-CMV-EYFP-miR-lacZ | Viral Vector Core, Duke | BK1202 |
| AAV9-CMV-EYFP-miR-RXRa | Viral Vector Core, Duke | BK1149 |
| AAV9-CMV-EGFP | Wilson lab | Addgene #105530 |
| AAV9-CMV-EGFP-P2A-RXRa | Viral Vector Core, Duke | BK1418 |
| AAV9-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene #44361 |
| AAV9-hSyn-DIO-mCherry | Roth lab | Addgene #50459 |
| Software and algorithms | ||
| FastQC v0.11.9 | Babraham Institute | RRID:SCR_014583 |
| Trimmomatic v0.27 | Usadel lab | RRID:SCR_011848 |
| hisat2 v2.1.0 | Kim lab | RRID:SCR_015530 |
| Picard v2.18.10 | Broad Institute | RRID:SCR_006525 |
| subread v2.0.0 | RRID:SCR_009803 | |
| DESeq2 v1.34.0 | Love et al., 2014 | RRID:SCR_015687 |
| RRHO2 | Cahill et al., 2015 | github.com/RRHO2/RRHO2 |
| PANTHER | Thomas et al., 2003, 2006 | pantherdb.org |
| ComDet v0.5.4 | github.com/ekatrukha/ComDet | |
| Axon pCLAMP 11 Software Suite | Molecular Devices | RRID:SCR_011323 |
| R v4.0.2 | R foundation | RRID:SCR_001905 |
| Image J v1.53c | NIH | RRID:SCR_003070 |
Highlights.
NAc RXRα expression correlates with addiction-relevant behavioral and molecular features
RXRα transcriptionally controls neuronal excitability of NAc D1- and D2-MSNs
Manipulating NAc RXRα levels modulates cocaine reward learning and reinforcing efficacy
Systemic, pharmacological inhibition of RXRα weakens cocaine-related behaviors
ACKNOWLEDGEMENTS
This work was supported by funding from the Boehringer Ingelheim Fonds (PhD Fellowship to A.G.) and the National Institute of Health (P01DA047233 to E.J.N.). The authors would like to thank Katherine Beach, Catherine McManus, Kyra Schmidt and Ezekiell Mouzon for transgenics breeding and genotyping, Dr. Logan Brown and Dr. Boris Kantor from the Duke University Viral Vector Core for cloning and packaging AAV vectors, Dr. Guillermo Villegas and Dr. Edgardo Aritzia from the Dean’s Flow Cytometry CoRE at the Icahn School of Medicine at Mount Sinai for assistance in nuclei sorting, Drs. Yasmin Hurd and Paul Kenny for access to their rat self-administration equipment, and Dr. Mark Baxter for statistical advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Koob GF, and Volkow ND (2016). Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 3, 760–773. 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Chiara G, and Imperato A (1988). Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. PNAS 85, 5274–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelley AE, and Berridge KC (2002). The Neuroscience of Natural Rewards: Relevance to Addictive Drugs. J. Neurosci 22, 3306–3311. 10.1523/JNEUROSCI.22-09-03306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nestler EJ (2013). Cellular basis of memory for addiction. Dial. Clin. Neurosci 15, 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lüscher C, Robbins TW, and Everitt BJ (2020). The transition to compulsion in addiction. Nat Rev Neurosci 21, 247–263. 10.1038/s41583-020-0289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piazza PV, and Le Moal ML (1996). Pathophysiological basis of vulnerability to drug abuse: role of an interaction between stress, glucocorticoids, and dopaminergic neurons. Annu Rev Pharmacol Toxicol 36, 359–378. 10.1146/annurev.pa.36.040196.002043. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Prescott CA, Myers J, and Neale MC (2003). The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry 60, 929–937. 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 8.Volkow ND, and Muenke M (2012). The genetics of addiction. Hum Genet 131, 773–777. 10.1007/S00439-012-1173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitesell M, Bachand A, Peel J, and Brown M (2013). Familial, Social, and Individual Factors Contributing to Risk for Adolescent Substance Use. J Addict 2013, 579310. 10.1155/2013/579310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nestler EJ, and Lüscher C (2019). The Molecular Basis of Drug Addiction: Linking Epigenetic to Synaptic and Circuit Mechanisms. Neuron 102, 48–59. 10.1016/j.neuron.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, et al. (2018). Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biological Psychiatry 84, 867–880. 10.1016/j.biopsych.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teague CD, and Nestler EJ (2021). Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol Psychiatry. 10.1038/s41380-021-01163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivares AM, Moreno-Ramos OA, and Haider NB (2015). Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases. J Exp Neurosci 9, 93–121. 10.4137/JEN.S25480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox J, and Witten IB (2019). Striatal circuits for reward learning and decision-making. Nat Rev Neurosci 20, 482–494. 10.1038/s41583-019-0189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Kong F, Crofton EJ, Dragosljvich SN, Sinha M, Li D, Fan X, Koshy S, Hommel JD, Spratt HM, et al. (2016). Transcriptomics of Environmental Enrichment Reveals a Role for Retinoic Acid Signaling in Addiction. Front. Mol. Neurosci 9. 10.3389/fnmol.2016.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crofton EJ, Nenov MN, Zhang Y, Tapia CM, Donnelly J, Koshy S, Laezza F, and Green TA (2021). Topographic transcriptomics of the nucleus accumbens shell: Identification and validation of fatty acid binding protein 5 as target for cocaine addiction. Neuropharmacology 183, 108398. 10.1016/j.neuropharm.2020.108398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Crofton EJ, Smith TES, Koshy S, Li D, and Green TA (2019). Manipulation of retinoic acid signaling in the nucleus accumbens shell alters rat emotional behavior. Behav Brain Res 376, 112177. 10.1016/j.bbr.2019.112177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krężel W, Ghyselinck N, Samad TA, Dupé V, Kastner P, Borrelli E, and Chambon P (1998). Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science 279, 863–867. 10.1126/science.279.5352.863. [DOI] [PubMed] [Google Scholar]
- 19.Krzyżosiak A, Szyszka-Niagolov M, Wietrzych M, Gobaille S, Muramatsu S, and Krezel W (2010). Retinoid x receptor gamma control of affective behaviors involves dopaminergic signaling in mice. Neuron 66, 908–920. 10.1016/j.neuron.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Wietrzych-Schindler M, Szyszka-Niagolov M, Ohta K, Endo Y, Pérez E, de Lera AR, Chambon P, and Krezel W (2011). Retinoid x receptor gamma is implicated in docosahexaenoic acid modulation of despair behaviors and working memory in mice. Biol Psychiatry 69, 788–794. 10.1016/j.biopsych.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Krzyżosiak A, Podleśny-Drabiniok A, Vaz B, Alvarez R, Rühl R, de Lera AR, and Krężel W (2021). Vitamin A5/X controls stress-adaptation and prevents depressive-like behaviors in a mouse model of chronic stress. Neurobiol Stress 15, 100375. 10.1016/j.ynstr.2021.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J, Chen J, Yan J, Jones IR, Craddock N, Cook EH Jr., Goldman D, Heston LL, and Sommer SS (2005). Structural variants in the retinoid receptor genes in patients with schizophrenia and other psychiatric diseases. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 133B, 50–53. 10.1002/ajmg.b.30113. [DOI] [PubMed] [Google Scholar]
- 23.Lee C-H, Liu C-M, Wen C-C, Chang S-M, and Hwu H-G (2010). Genetic copy number variants in sib pairs both affected with schizophrenia. J Biomed Sci 17, 2. 10.1186/1423-0127-17-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang AW, Avramopoulos D, Lori A, Mulle J, Conneely K, Powers A, Duncan E, Almli L, Massa N, McGrath J, et al. (2019). Genome-wide association study in two populations to determine genetic variants associated with Toxoplasma gondii infection and relationship to schizophrenia risk. Prog Neuropsychopharmacol Biol Psychiatry 92, 133–147. 10.1016/j.pnpbp.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Davis OSP, Butcher LM, Docherty SJ, Meaburn EL, Curtis CJC, Simpson MA, Schalkwyk LC, and Plomin R (2010). A three-stage genome-wide association study of general cognitive ability: hunting the small effects. Behav Genet 40, 759–767. 10.1007/s10519-010-9350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo SK, Shtir C, Doyle AE, Mick E, McGough JJ, McCracken J, Biederman J, Smalley SL, Cantor RM, Faraone SV, et al. (2012). Genome-wide association study of intelligence: additive effects of novel brain expressed genes. J Am Acad Child Adolesc Psychiatry 51, 432–440.e2. 10.1016/j.jaac.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Kölsch H, Lütjohann D, Jessen F, Popp J, Hentschel F, Kelemen P, Friedrichs S, Maier TAW, and Heun R (2009). RXRA gene variations influence Alzheimer’s disease risk and cholesterol metabolism. J Cell Mol Med 13, 589–598. 10.1111/j.1582-4934.2009.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akram A, Schmeidler J, Katsel P, Hof PR, and Haroutunian V (2010). Increased expression of RXRα in dementia: an early harbinger for the cholesterol dyshomeostasis? Mol Neurodegener 5, 36. 10.1186/1750-1326-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, Li M-Y, Li G, Li S-J, Wen B, Lu Y, and Yu X (2020). Retinoid X Receptor α Regulates DHA-Dependent Spinogenesis and Functional Synapse Formation In Vivo. Cell Rep 31, 107649. 10.1016/j.celrep.2020.107649. [DOI] [PubMed] [Google Scholar]
- 30.Le Moine C, and Bloch B (1995). D1 and D2 dopamine receptor gene expression in the rat striatum: sensitive cRNA probes demonstrate prominent segregation of D1 and D2 mRNAs in distinct neuronal populations of the dorsal and ventral striatum. J Comp Neurol 355, 418–426. 10.1002/cne.903550308. [DOI] [PubMed] [Google Scholar]
- 31.Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, Dietz DM, Zaman S, Koo JW, Kennedy PJ, et al. (2010). Cell Type-Specific Loss of BDNF Signaling Mimics Optogenetic Control of Cocaine Reward. Science 330, 385–390. 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calipari ES, Bagot RC, Purushothaman I, Davidson TJ, Yorgason JT, Peña CJ, Walker DM, Pirpinias ST, Guise KG, Ramakrishnan C, et al. (2016). In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. PNAS 113, 2726–2731. 10.1073/pnas.1521238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salery M, Trifilieff P, Caboche J, and Vanhoutte P (2020). From Signaling Molecules to Circuits and Behaviors: Cell-Type-Specific Adaptations to Psychostimulant Exposure in the Striatum. Biological Psychiatry 87, 944–953. 10.1016/j.biopsych.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Dawson MI, and Xia Z (2012). The retinoid X receptors and their ligands. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1821, 21–56. 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S, Shen T, Chitranshi N, Gupta V, Basavarajappa D, Sarkar S, Mirzaei M, You Y, Krezel W, Graham SL, et al. (2022). Retinoid X Receptor: Cellular and Biochemical Roles of Nuclear Receptor with a Focus on Neuropathological Involvement. Mol Neurobiol. 10.1007/s12035-021-02709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salery M, Godino A, and Nestler EJ (2021). Drug-activated cells: From immediate early genes to neuronal ensembles in addiction. Adv Pharmacol 90, 173–216. 10.1016/bs.apha.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeiro EA, Salery M, Scarpa JR, Calipari ES, Hamilton PJ, Ku SM, Kronman H, Purushothaman I, Juarez B, Heshmati M, et al. (2018). Transcriptional and physiological adaptations in nucleus accumbens somatostatin interneurons that regulate behavioral responses to cocaine. Nat Commun 9, 3149. 10.1038/s41467-018-05657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JH, Ribeiro EA, Kim J, Ko B, Kronman H, Jeong YH, Kim JK, Janak PH, Nestler EJ, Koo JW, et al. (2020). Dopaminergic Regulation of Nucleus Accumbens Cholinergic Interneurons Demarcates Susceptibility to Cocaine Addiction. Biol Psychiatry 88, 746–757. 10.1016/j.biopsych.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kronman H, Richter F, Labonté B, Chandra R, Zhao S, Hoffman G, Lobo MK, Schadt EE, and Nestler EJ (2019). Biology and Bias in Cell Type-Specific RNAseq of Nucleus Accumbens Medium Spiny Neurons. Sci Rep 9, 8350. 10.1038/s41598-019-44798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobo MK, Karsten SL, Gray M, Geschwind DH, and Yang XW (2006). FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci 9, 443–452. 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 41.Mews P, der Zee YV, Kronman H, Gurung A, Ramakrishnan A, Browne C, Futamura R, Estill M, Ryan M, Reyes AA, et al. (2022). Cell-Type-Specific Epigenetic Priming of Gene Expression in Nucleus Accumbens by Cocaine. 2022.06.24.497533. 10.1101/2022.06.24.497533. [DOI] [Google Scholar]
- 42.Wolf ME (2016). Synaptic mechanisms underlying persistent cocaine craving. Nat Rev Neurosci 17, 351–365. 10.1038/nrn.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong S, Fayette N, Heinsbroek JA, and Ford CP (2021). Cocaine shifts dopamine D2 receptor sensitivity to gate conditioned behaviors. Neuron 109, 3421–3435.e5. 10.1016/j.neuron.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gertler TS, Chan CS, and Surmeier DJ (2008). Dichotomous anatomical properties of adult striatal medium spiny neurons. J Neurosci 28, 10814–10824. 10.1523/JNEUROSCI.2660-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Muhtasib N, Forcelli PA, and Vicini S (2018). Differential electrophysiological properties of D1 and D2 spiny projection neurons in the mouse nucleus accumbens core. Physiological Reports 6, e13784. 10.14814/phy2.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deroche MA, Lassalle O, Castell L, Valjent E, and Manzoni OJ (2020). Cell-Type- and Endocannabinoid-Specific Synapse Connectivity in the Adult Nucleus Accumbens Core. J Neurosci 40, 1028–1041. 10.1523/JNEUROSCI.1100-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huston JP, de S Silva MA, Topic B, and Müller CP (2013). What’s conditioned in conditioned place preference? Trends in Pharmacological Sciences 34, 162–166. 10.1016/j.tips.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Calipari ES, Godino A, Salery M, Damez-Werno DM, Cahill ME, Werner CT, Gancarz AM, Peck EG, Jlayer Z, Rabkin J, et al. (2019). Synaptic Microtubule-Associated Protein EB3 and SRC Phosphorylation Mediate Structural and Behavioral Adaptations During Withdrawal From Cocaine Self-Administration. J. Neurosci 39, 5634–5646. 10.1523/JNEUROSCI.0024-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oleson EB, and Roberts DCS (2012). Cocaine Self-Administration in Rats: Threshold Procedures. In Psychiatric Disorders: Methods and Protocols Methods in Molecular Biology., Kobeissy FH, ed. (Humana Press; ), pp. 303–319. 10.1007/978-1-61779-458-2_20. [DOI] [PubMed] [Google Scholar]
- 50.Bentzley BS, Fender KM, and Aston-Jones G (2013). The behavioral economics of drug self-administration: a review and new analytical approach for within-session procedures. Psychopharmacology (Berl) 226, 113–125. 10.1007/s00213-012-2899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson AR, Thibeault KC, Lopez AJ, Peck EG, Sands LP, Sanders CM, Kutlu MG, and Calipari ES (2019). Cues play a critical role in estrous cycle-dependent enhancement of cocaine reinforcement. Neuropsychopharmacology 44, 1189–1197. 10.1038/s41386-019-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christensen CJ, Silberberg A, Hursh SR, Roma PG, and Riley AL (2008). Demand for cocaine and food over time. Pharmacology Biochemistry and Behavior 91, 209–216. 10.1016/j.pbb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentzley BS, Jhou TC, and Aston-Jones G (2014). Economic demand predicts addiction-like behavior and therapeutic efficacy of oxytocin in the rat. PNAS 111, 11822–11827. 10.1073/pnas.1406324111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oleson EB, and Roberts DCS (2009). Behavioral Economic Assessment of Price and Cocaine Consumption Following Self-Administration Histories which Produce Escalation of Either Final Ratios or Intake. Neuropsychopharmacology 34, 796–804. 10.1038/npp.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebisawa M, Umemiya H, Ohta K, Fukasawa H, Kawachi E, Christoffel G, Gronemeyer H, Tsuji M, Hashimoto Y, Shudo K, et al. (1999). Retinoid X receptor-antagonistic diazepinylbenzoic acids. Chem Pharm Bull (Tokyo) 47, 1778–1786. 10.1248/cpb.47.1778. [DOI] [PubMed] [Google Scholar]
- 56.Bourhis E, Maheux J, Paquet B, Kagechika H, Shudo K, Rompré P-P, Rouillard C, and Lévesque D (2009). The transcription factors Nur77 and retinoid X receptors participate in amphetamine-induced locomotor activities. Psychopharmacology 202, 635–648. 10.1007/s00213-008-1343-0. [DOI] [PubMed] [Google Scholar]
- 57.Éthier I, Kagechika H, Shudo K, Rouillard C, and Lévesque D (2004). Docosahexaenoic acid reduces haloperidol-induced dyskinesias in mice: Involvement of Nur77 and retinoid receptors. Biol. Psychiatry 56, 522–526. 10.1016/j.biopsych.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 58.Day JJ, and Carelli RM (2007). The Nucleus Accumbens and Pavlovian Reward Learning. Neuroscientist 13, 148–159. 10.1177/1073858406295854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlezon WA, and Thomas MJ (2009). Biological substrates of reward and aversion: a nucleus accumbens activity hypothesis. Neuropharmacology 56, 122–132. 10.1016/j.neuropharm.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Chiara G (2002). Nucleus accumbens shell and core dopamine: differential role in behavior and addiction. Behav Brain Res 137, 75–114. 10.1016/s0166-4328(02)00286-3. [DOI] [PubMed] [Google Scholar]
- 61.West EA, and Carelli RM (2016). Nucleus Accumbens Core and Shell Differentially Encode Reward-Associated Cues after Reinforcer Devaluation. J Neurosci 36, 1128–1139. 10.1523/JNEUROSCI.2976-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krężel W, Rühl R and de Lera AR (2019). Alternative retinoid X receptor (RXR) ligands. Mol Cell Endocrinol 491, 110436. 10.1016/j.mce.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 63.Carpenter MD, Hu Q, Bond AM, Lombroso SI, Czarnecki KS, Lim CJ, Song H, Wimmer ME, Pierce RC, and Heller EA (2020). Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat Commun 11, 504. 10.1038/s41467-020-14331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campos-Melo D, Galleguillos D, Sánchez N, Gysling K, and Andrés ME (2013). Nur transcription factors in stress and addiction. Front Mol Neurosci 6. 10.3389/fnmol.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kemény LV, Robinson KC, Hermann AL, Walker DM, Regan S, Yew YW, Lai YC, Theodosakis N, Rivera PD, Ding W, et al. (2021). Vitamin D deficiency exacerbates UV/endorphin and opioid addiction. Sci Adv 7, eabe4577. 10.1126/sciadv.abe4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, and Chambon P (1993). Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. PNAS 90, 30–34. 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rühl R, Krzyżosiak A, Niewiadomska-Cimicka A, Rochel N, Szeles L, Vaz B, Wietrzych-Schindler M, Álvarez S, Szklenar M, Nagy L, et al. (2015). 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice. PLoS Genet 11, e1005213. 10.1371/journal.pgen.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Urquiza AM, Liu S, Sjöberg M, Zetterström RH, Griffiths W, Sjövall J, and Perlmann T (2000). Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science 290, 2140–2144. 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 69.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cahill KM, Huo Z, Tseng GC, Logan RW, and Seney ML (2018). Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep 8, 9588. 10.1038/s41598-018-27903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tannenbaum C, Ellis RP, Eyssel F, Zou J, and Schiebinger L (2019). Sex and gender analysis improves science and engineering. Nature 575, 137–146. 10.1038/s41586-019-1657-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Full DEG list after HSV-mediated bulk RXRα overexpression. (related to Figure 1)
Table S2. Full DEG list after AAV-mediated RXRα overexpression in D1-MSNs. (related to Figure 3
Table S3. Full DEG list after AAV-mediated RXRα overexpression in D2-MSNs. (related to Figure 3)
Table S6. Full statistics. (related to all Figures)
Data Availability Statement
All RNAseq data reported in this study are deposited publicly in the Gene Expression Omnibus under accession code GSE198527. Unmodified Western blots scans and other supporting raw data are available from the lead contact upon request. Scripts and code utilized in this study, including for statistical analysis, are available from the lead contact upon request.