Abstract
The ability to create stimuli-responsive DNA nanostructures has played a prominent role in dynamic DNA nanotechnology. Primary among these is the process of toehold-based strand displacement, where a nucleic acid molecule can act as a trigger to cause conformational changes in custom-designed DNA nanostructures. Here, we add another layer of control to strand displacement reactions through a 'toehold clipping' process. By designing DNA complexes with a photocleavable linker-containing toehold or an RNA toehold, we show that we can use light (UV) or enzyme (ribonuclease) to eliminate the toehold, thus preventing strand displacement reactions. We use molecular dynamics simulations to analyze the structural effects of incorporating a photocleavable linker in DNA complexes. Beyond simple DNA duplexes, we also demonstrate the toehold clipping process in a model DNA nanostructure, by designing a toehold containing double-bundle DNA tetrahedron that disassembles when an invading strand is added, but stays intact after the toehold clipping process even in the presence of the invading strand. This work is an example of combining multiple physical or molecular stimuli to provide additional remote control over DNA nanostructure reconfiguration, advances that hold potential use in biosensing, drug delivery or molecular computation.
INTRODUCTION
The structural features and molecular recognition properties of DNA make it a versatile building block for constructing nanoscale structures (1). The sequence programmability and reversible nature of Watson–Crick base pairing further offers dynamic control, allowing one to reconfigure the shapes of DNA nanostructures and devices, as well as to create nucleic acid based reactions and circuits. A well-known tool based on such sequence programmability is toehold-based strand displacement, a process in which two strands with full complementarity hybridize to each other, displacing one or more pre-hybridized strands in the process (2). Strand displacement reactions are often initiated at single-stranded domains (a toehold) that are complementary to part of an invading strand, and progresses through a branch migration process (Figure 1A) (3,4). The reaction proceeds because more base pairs are formed by removing the toehold-containing strand than were present when bound to its original partner. Using this approach, it has been possible to engineer a range of dynamic DNA nanostructures with applications in molecular computation [neural networks (5) and logic gates (6)], information encoding [visual tags (7) and short term memory (8)], biosensing [detecting nucleic acids (9)], drug delivery [for triggered release (10)] and materials science [controlling colloidal phase transitions (11) and size-changing hydrogels (12)]. In prior work, scientists have also developed strategies to control strand displacement reactions using photocrosslinkers (13), hidden toeholds in DNA bulges (14), additional DNA locks to regulate strand displacement rates (15) and by modulating pH to control triplex-based DNA strand displacement (16). In this work, we add to this library of tools to control toehold-based DNA strand displacement using light or enzyme to remove a toehold, a process we term ‘toehold clipping’. We demonstrate this toehold clipping mechanism in DNA duplexes using photocleavable toeholds (light-activated) and RNA toeholds (enzyme activated). Beyond control of strand displacement in duplexes, we show that this toehold clipping mechanism can be incorporated into complex DNA nanostructures, by controllably disassembling a double-bundle DNA tetrahedron.
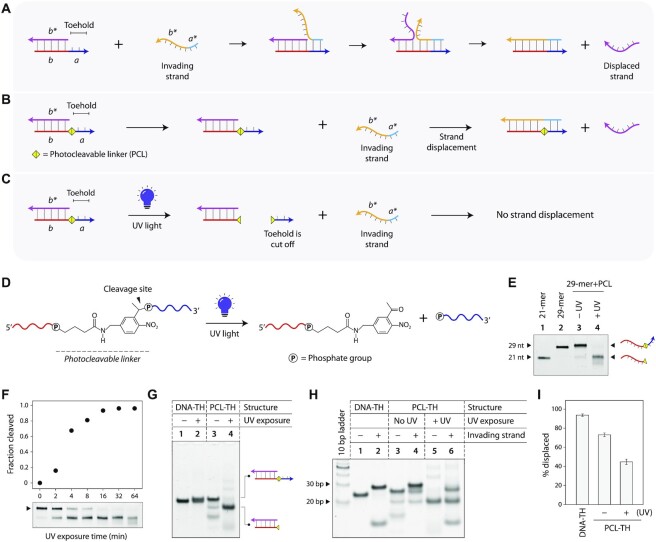
Figure 1.
Light-induced toehold clipping process. (A) Toehold-based DNA strand displacement process. Sequence identities are denoted by alphabets in italics. Arrows represent 3′ ends of strands. (B) Toehold-based strand displacement in a duplex with a photocleavable linker (PCL) at the toehold. (C) On UV exposure, the photocleavable linker is cleaved, removing the toehold and preventing strand displacement. (D) Chemical structure of the PCL incorporated in DNA strand and cleavage products after UV exposure. (E) Cleavage of ssDNA containing a PCL analyzed by denaturing PAGE. (F) Time series of photocleavage of PCL-containing DNA. (G) UV-induced toehold clipping in a DNA duplex. (H) Strand displacement in duplexes before and after toehold clipping. (I) Strand displacement efficiency in control duplex, PCL-TH duplex without UV exposure, and PCL-TH duplex on UV exposure resulting in toehold clipping.
MATERIALS AND METHODS
Oligonucleotide synthesis
Oligonucleotides were purchased from Integrated DNA Technologies (IDT) with standard desalting. DNA oligonucleotides with photocleavable linkers (PCL) were chemically synthesized in-house at 1.0-μmol scale by solid phase synthesis using an Oligo-800 synthesizer in DMT-off mode. Native DNA and PCL phosphoramidites were purchased from ChemGenes and Glen Research, respectively. The concentration of PCL phosphoramidite was 0.1 M and all the native DNA amidites were 0.05 M, prepared in acetonitrile. All the other reagents were standard oligonucleotide synthesis solutions obtained from ChemGenes. After synthesis, oligonucleotides were cleaved from the solid support and fully deprotected with AMA (ammonium hydroxide:methylamine = 1:1) by heating the oligonucleotides at 65 °C for 50 min in screw tubes. The cleaved oligonucleotides were dried in a Speed-Vac evaporator and concentration was measured using a NanoDrop 2000 spectrophotometer. PCL-containing DNA strands were purified by preparative denaturing polyacrylamide gel electrophoresis (PAGE, 15%). The DNA strands were extracted using butanol after purification, desalted and the oligonucleotides were suspended in deionized water. The quality of the purified oligonucleotides was assessed by 15% analytical PAGE. For the 1 μmol synthesis scale described here, the cost of the PCL phosphoramidite used was ∼$130, with only a fraction of the synthesized material used in this work. This additional cost involved in the incorporation of chemical linkers is a parameter to be considered for constructing such functionalized DNA nanostructures.
Assembly of duplexes
To form the duplex, strands S and S-complement (Supplementary Table S1) were annealed in 1× TAE/Mg2+ buffer (40 mM Tris base (pH 8.0), 20 mM acetic acid and 12.5 mM magnesium acetate) at a final concentration of 500 nM. The DNA samples were annealed from 90 °C to 20 °C over 30 min in a T100 Thermal Cycler (Bio-Rad). For toehold containing duplexes, strand S-complement was hybridized with S-DNA-TH, S-PCL-TH or S-RNA-TH for duplexes with DNA toehold, PCL-containing toehold or RNA toehold, respectively.
Assembly of three-point-star motif and tetrahedron
To form the three-point-star motif, strands L, M and S-blunt were mixed in 1:3:3 ratio in 1× TAE/Mg2+ at a final concentration of 250 nM (sequences in Supplementary Table S1). The mixture was annealed from 90 °C to 20 °C over 2 h in a T100 Thermal Cycler. Strand S-blunt was used to create a motif without sticky ends so as to prevent the formation of higher order structures such as the tetrahedron. To form the DNA tetrahedron, strands L, M and S were mixed in 1:3:3 ratio at 30 nM in 1× TAE/Mg2+. For toehold containing tetrahedra, strand S was substituted with S-DNA-TH, S-PCL-TH or S-RNA-TH for DNA toehold, PCL-containing toehold or RNA toehold respectively. The DNA solution was slowly cooled down from 90 °C to 20 °C over 48 h by placing the tubes in 2 liters of hot water in a beaker placed in a Styrofoam box.
Toehold clipping
For UV-induced toehold clipping, single stranded DNA or annealed DNA complexes in 0.5 ml microtubes were placed in an ice bucket with the tubes submerged in ice. Tube lids were removed and tubes were placed ∼1 inch from the UV light source (handheld UV light Spectroline EF 240C with an output of 4 W). Samples were exposed to UV (λ = 254 nm) typically for 1 h (except for the time series experiment). For ribonuclease-induced toehold clipping, 10 μl of single stranded DNA or annealed DNA complexes were mixed with 1 μl of RNase A (20 mg/ml) and incubated at room temperature for 1 h.
Tetrahedron disassembly
For tetrahedron disassembly, the invading strand was added to DNA tetrahedra solution (typically 20 μl) in 30-fold excess and incubated at 20 °C for 1–2 h.
Electrophoretic mobility shift assay (EMSA)
Assembly of DNA structures and toehold clipping was examined by subjecting the complexes to an EMSA. Complexes were run on non-denaturing gels containing 4–18% polyacrylamide (19:1 acrylamide/bisacrylamide) at 4 °C (100 or 120 V, constant voltage) in 1× TAE/Mg2+ running buffer (duplexes were run on 18% gels and DNA tetrahedra were run on 4% gels). Samples were mixed with gel loading dye containing 50% glycerol, bromophenol blue and 1× TAE/Mg2+ before loading on gels. Gels were stained in deionized water containing 0.5× GelRed (Biotium) and destained in deionized water before imaging. Denaturing gels (15% polyacrylamide and 8.3 M urea) were run at 20 °C (500 V, constant voltage) in 1× TBE running buffer (pH 8.4) containing of 8.9 mM Tris, 8.9 mM boric acid and 0.2 mM EDTA. Denaturing gels were stained in deionized water containing ethidium bromide (Sigma-Aldrich). Imaging was done on a Bio-Rad Gel Doc XR+ imager using the default settings for GelRed or ethidium bromide with UV illumination. Gel images were exported as 12-bit images and quantified using ImageJ. Quantification was done using the highest exposure image that did not contain saturated pixels in the band of interest.
Circular dichroism (CD) experiments
CD spectra were collected on 5 μM annealed complexes. Experiments were performed on a Jasco-815 CD spectrometer at room temperature in a quartz cell with a 1 mm path length. CD spectra were collected from 350 to 200 nm with a scanning speed of 100 nm/min and with three accumulations. The bandwidth was 1.0 nm, and the digital integration time was 1.0 s. CD spectra were baseline-corrected for signal contributions due to the buffer.
Molecular dynamics simulations
Structural effects of PCL accommodation during strand displacement were studied by molecular dynamics (MD) simulations. AMBER (17) type force-field parameters were developed for the PCL, which was treated as a DNA modification for parameterization purposes. Geometry of the molecule was optimized using Hartree-Fock level theory and 6–31G* basis-set. Partial charges on the atoms were then obtained using the online RESP charge-fitting server R.E.D (18). AMBER99 force-field parameters with bsc1 modification (19) were used to generate bonded and non-bonded interaction parameters for the PCL. Initial structure of the DNA duplex (toehold-containing strand paired with the invading strand) was built in MOE using the same sequence as in the experiments but a shorter length (a 15-mer duplex such that the PCL insertion site was in the middle). For MD simulations, we used this duplex as control and used MOE (20) to model the PCL into the DNA duplex.
MD simulations were performed using GROMACS 2020 (21) in a solution of 100 mM excess KCl in a cubic box with an edge length of ∼13 nm containing 183 K+, 125 Cl− ions and ∼20 000 water molecules. All simulations utilized the leap-frog algorithm with a 2 fs time step to integrate the equations of motion. The system was maintained at 300 K using the velocity rescaling thermostat (22) and pressure was maintained at 1 atm using the Berendsen barostat for equilibration (23). Long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) algorithm with a real space cut-off of 1.0 nm (24) and Lennard-Jones and short-range electrostatic interactions were calculated at a 1 nm cut-off. The TIP3P model was used to represent the water molecules, and the LINCS algorithm (25) was used to constrain the motion of hydrogen atoms bonded to heavy atoms. The system was subjected to energy minimization to prevent any overlap of atoms, followed by a short equilibration (0.5 ns) and 50-ns production run. Starting with the equilibrated structure, three independent trajectories with randomized starting velocities were performed, a cumulative total of 150 ns of simulation for each construct. To prevent fraying, all heavy atom pairs involved in hydrogen bonding in the terminal base pairs were constrained to a distance of 3–4 Å with a force of 1000 kJ mol−1 nm−2 using harmonic distance restraints. Coordinates of the DNA complexes were stored every 2 ps for further analysis. Simulations were visualized using Visual Molecular Dynamics (VMD) software (26) and analyzed using gmx_distance utility included with GROMACS (21).
RESULTS AND DISCUSSION
We first designed the toehold clipping strategy in a DNA duplex containing an 8-nt toehold. By incorporating a photocleavable linker (PCL) at the junction of the duplex and the toehold, UV light can be used to cleave the toehold, thus preventing strand displacement (Figure 1B, C). To demonstrate this, we synthesized a PCL-containing DNA strand in-house (Figure 1D) and first validated that exposure to UV light can cleave the linker and thus remove the toehold portion of the ssDNA. We used 21-mer and 29-mer DNA strands as controls, corresponding to the lengths of the PCL-DNA strand without and with the toehold (Figure 1E: lanes 1 and 2, full gel in Supplementary Figure S1). We exposed the PCL-containing ssDNA to UV light (λ = 254 nm) and tested the samples before and after UV exposure using denaturing polyacrylamide gel electrophoresis (PAGE). Gel results showed that on UV exposure, the 29-mer PCL-DNA strand is cleaved to a 21-mer and an 8-mer (Figure 1E). Next, we performed a time series of UV exposure to obtain kinetics of the photocleavage of the PCL-DNA strand (Figure 1F, full gel in Supplementary Figure S2). We exposed the PCL-DNA to UV light for different time points up to 64 min, and ran the UV-exposed samples on a denaturing PAGE. We quantified the reduction in intensity of the band corresponding to the 29-mer PCL-DNA, showing 90% cleavage in ∼32 min.
After validating UV-induced cleavage of the PCL-DNA strand, we next tested toehold clipping in a duplex. We annealed a 21-mer DNA and the 29-mer PCL-DNA to form a DNA duplex containing an 8-nt toehold and characterized it using non-denaturing PAGE and circular dichroism (CD) (Figure 1G and Supplementary Figure S3). First, we confirmed that a control duplex with a DNA toehold (abbreviated DNA-TH) is not affected by UV exposure (Figure 1G: lanes 1 and 2). Next, we validated UV-induced cleavage of the toehold in the duplex with a PCL toehold (PCL-TH). On UV exposure, the toehold is cleaved from the PCL-TH duplex (25 bp complex), resulting in a smaller complex without a toehold (21 bp) (Figure 1G: lanes 3 and 4).
We then demonstrated that UV-induced toehold clipping affects strand displacement in DNA duplexes. First, we tested strand displacement in a toehold-containing control duplex. We added an invading strand to the control duplex (25 bp complex), incubated the mixture at room temperature, and tested the samples using non-denaturing PAGE. Occurrence of strand displacement was confirmed by the formation of a longer (29 bp) duplex (Figure 1H, lanes 1–2, full gel in Supplementary Figure S4). Next, we confirmed that the presence of the PCL does not interfere with the strand displacement process by adding the invading strand to the PCL-TH duplex. Formation of the longer duplex indicated that strand displacement proceeded even in the presence of the PCL (Figure 1H, lanes 3 and 4). To analyze toehold clipping and its effect on strand displacement, we shined UV on the PCL-TH duplex and incubated it with the invading strand, and ran the samples on a non-denaturing gel (Figure 1H, lanes 5 and 6). We quantified the bands corresponding to the strand displacement product in each case to obtain the displacement efficiency. When the duplex was not exposed to UV, strand displacement occurred, with a displacement yield of ∼93% (Figure 1H, lanes 3 and 4 and Figure 1I). However, on exposure to UV, the strand displacement yield was only ∼44%, indicating that toehold clipping by photocleavage can be used to control strand displacement reactions (Figure 1H, lanes 5 and 6 and Figure 1I). We attribute the occurrence of strand displacement after toehold to the remnant toehold post-UV as seen in Figure 1G.
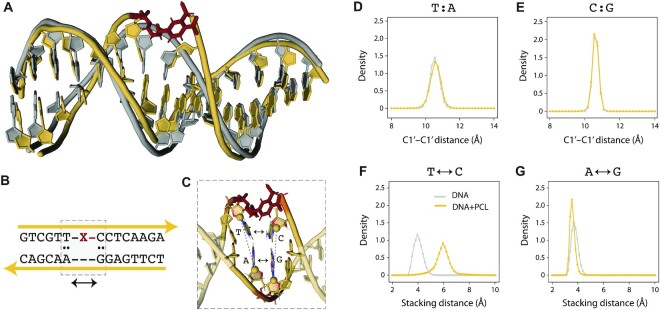
While experimental results show that the presence of the PCL does not affect strand displacement, we performed MD simulations to understand PCL accommodation and its structural effects on the duplex. We modeled and simulated a duplex of the toehold-containing strand and the invading strand with and without the PCL (Figure 2A). We found that the nucleotides adjacent to the PCL (5′ T and 3′ C) maintain canonical base pairing configurations with their partners on the invading strand (A and G respectively) throughout the simulation (Figure 2B-C). The distance between the C1′ atoms of the T:A and C:G base pairs 5′ and 3′ adjacent to the PCL is effectively the same in the constructs with and without the PCL (Figure 2D and E). The stacking distance, measured as the distance between the center of mass of the heavy atoms in the nucleobases, shows that stacking of the nucleotides (T and C) next to the PCL in the toehold-containing strand is disrupted (Figure 2F). The stacking between the corresponding paired bases on the invading strand (A and G) remained intact, as indicated by the overlapping stacking distance distribution of the constructs with and without the PCL (Figure 2G). These results suggest minimal local structural stress on the invading strand in the presence of the linker after strand displacement. In our simulations, the PCL acts as a bulge in the DNA duplex and can be accommodated with minor restructuring of the duplex, thus minimally affect the strand displacement process.
Figure 2.
Molecular dynamics simulation of a PCL-containing duplex. (A) Representative structures from MD simulation of the DNA duplex with (yellow) and without PCL (gray). PCL is shown in red. (B) Sequence identity of the DNA construct. The nucleotides adjacent to the PCL are highlighted by the dashed box. (C) Structural details of the nucleotides adjacent to the PCL. The C1′ atoms are shown as spheres. (D) Distribution of the C1′-C1′ distance for the base pair 5′ adjacent to the PCL (yellow) and the same base pair in the construct without PCL (gray). (E) Distribution of the C1′-C1′ distance for the base pair 3′ adjacent to the PCL (yellow) and the same base pair in the construct without PCL (gray). (F) Stacking distance distribution for the nucleotides 5′ and 3′ adjacent to PCL on the toehold-containing strand. (G) Stacking distance distribution for the nucleotides 5′ and 3′ adjacent to PCL on the invading strand. All distance distributions are averages from three independent simulations (50 ns each). Error bars in panels D-G represent standard deviations from these triplicates.
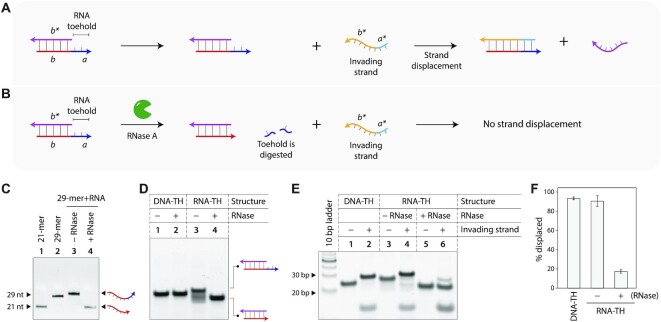
We also designed a system to demonstrate toehold clipping using enzymes (Figure 3A). We demonstrated this concept using a duplex containing an RNA toehold, and used a ribonuclease (RNase A) for toehold clipping. We first designed a chimera strand that contains a DNA sequence (forms part of the duplex) and an RNA sequence (forms the toehold). On addition of RNase A, only the RNA part is degraded, thus removing the toehold and preventing strand displacement (Figure 3B). We first tested toehold clipping using the single stranded DNA/RNA chimera strand, incubating it with RNase A and analyzing the samples on a denaturing gel (Figure 3C, full gel in Supplementary Figure S5). On addition of RNase A, the RNA portion of the 29-mer chimera strand is cleaved, resulting in a 21-nt DNA strand. Next, we created a duplex with a 21-nt DNA strand and a 29-nt RNA-toehold containing chimera strand (25 bp complex) and characterized it using non-denaturing PAGE and CD (Figure 3D and Supplementary Figure S3). RNase A addition has no effect on an all-DNA duplex (Figure 3D: lanes 1 and 2, full gel in Supplementary Figure S6), and only the duplex containing the RNA toehold is affected by RNase, as seen by the faster moving 21 bp complex (Figure 3D: lanes 3–4). Similar to the study with the PCL, we confirmed that the RNA toehold does not interfere with the strand displacement process (Figure 3E: lanes 3 and 4, full gel in Supplementary Figure S7). Next, we incubated the duplex containing the RNA toehold (RNA-TH) with RNase, followed by addition of the invading strand to the RNase-treated sample (Figure 3E: lanes 5 and 6). We analyzed the samples on a non-denaturing gel and quantified the strand displacement yields. The control duplex without RNase showed ∼93% displacement yield while the RNase-treated duplex only showed ∼17% displacement, indicating that toehold clipping affects the displacement reaction (Figure 3F).
Figure 3.
Ribonuclease-induced toehold clipping process. (A) Toehold-based strand displacement in a duplex with an RNA toehold. Sequence identities are denoted by alphabets in italics. (B) On RNase A addition, the RNA toehold is degraded, preventing strand displacement. (C) Degradation of the RNA portion of a chimera strand containing DNA and RNA analyzed by denaturing PAGE. (D) RNase-induced toehold clipping in DNA duplex. (E) Strand displacement in duplexes before and after toehold clipping. (F) Strand displacement efficiency in control duplex, RNA-TH duplex without RNase A addition, and RNA-TH duplex on RNase A addition resulting in toehold clipping.
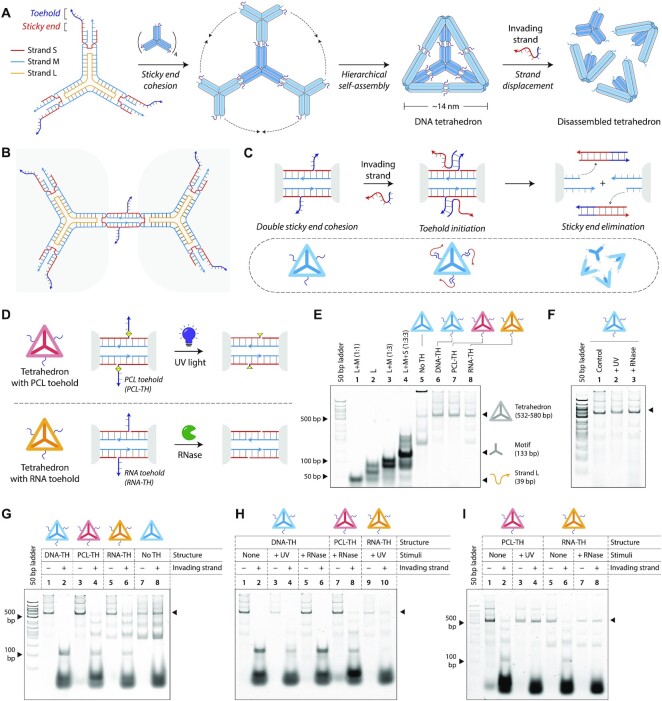
To demonstrate the broader adaptability of the toehold clipping process in DNA nanotechnology, we chose a double-bundle DNA tetrahedron as a model system. We designed a toehold-containing DNA tetrahedron that can be disassembled by the addition of an invading strand, and show that using the toehold clipping process, tetrahedron disassembly can be prevented. We assembled the DNA tetrahedron from the three-point-star motif (27), a branched DNA structure containing seven strands: a long strand (L), three medium strands (M) and three short strands (S) (Figure 4A and Supplementary Figure S8). The motif we use is three-fold symmetric, thus the three medium strands have identical sequences, so do the short strands (identical sequences are indicated by colors of the strands in Figure 4A). The sequence of the long strand is three-fold repeated within the motif, and contains 5T loops at the center of the motif that provide flexibility for the arms to associate into higher order structures. Each arm of the three-point-star motif contains two helices connected by a single crossover. Both helices are tailed with 7-nt sticky ends (one extending from S and one extending from M) which allow the motifs assemble into higher order structures through double sticky end cohesion (Figure 4B). At specific DNA concentrations (30–75 nM), a DNA tetrahedron is the predominant product, containing four units of the three-point-star motifs. The edges of the assembled tetrahedron contain four helical turns of DNA (∼14 nm).
Figure 4.
Toehold clipping to control reconfiguration of a DNA tetrahedron. (A) Design and assembly of the 3-point-star motif into a DNA tetrahedron. The motif is constructed from three strands, a short (S), medium (M) and a long (L) strand (strand identities are indicated by different colors). An invading strand can bind to the toeholds on the DNA tetrahedron causing disassembly of the structure. (B) The 3-point-star motifs connect to each other via double cohesion. Beyond the sticky end region, the motif also contains a toehold. (C) An invading strand can bind to the toehold on strand S and remove it from the motif, this eliminating the sticky end cohesion and causing the tetrahedron to disassemble. (D) Schematic showing toehold clipping in the DNA tetrahedron using light or enzyme. (E) Non-denaturing gel showing stepwise assembly of the DNA tetrahedra. (F) Gel image showing DNA tetrahedra are unaffected on exposure to UV or on RNase A addition. (G) Tetrahedron disassembly by addition of invading strand. (H) Demonstration of stimuli specificity in DNA tetrahedron disassembly. (I) Toehold clipping by UV on PCL-TH tetrahedron and RNase A on RNA-TH tetrahedron prevents tetrahedron disassembly.
To control DNA tetrahedron disassembly by strand displacement, we included single stranded overhangs beyond the sticky end region on strand S (Figure 4A and Supplementary Figure S8). This 8-nt overhang acts as the toehold to initiate strand displacement based disassembly of the DNA tetrahedron on the addition of an invading strand. Our earlier work with DNA tetrahedron assembly and disassembly showed that an 8-nt toehold is optimal for DNA strand displacement process with minimal excess of the invading strand (28). We designed the invading strand to be complementary to the entire sequence of strand S, including the toehold region. Addition of the invading strand results in complete displacement of strand S from the tetrahedron, thus eliminating the sticky end interactions that hold the tetrahedron together, leading to disassembly of the DNA tetrahedron into its component motifs (Figure 4C). To control strand displacement by toehold clipping, we designed a DNA tetrahedron with strand S containing a PCL toehold (for toehold clipping using UV light) and a tetrahedron containing strand S with an RNA toehold (for toehold clipping using RNase A) (Figure 4D). The incorporation of a PCL-containing toehold and RNA toehold allows the removal of the toeholds by UV exposure or RNase addition respectively, thereby preventing the disassembly of the DNA tetrahedron by strand displacement.
We assembled the DNA tetrahedron by mixing strands L, M, and S in a ratio of 1:3:3 in 1× TAE/Mg2+ buffer and slowly annealing the solution from 90 °C to 20 °C over 48 h. This one-step annealing forms the individual three-point-star motifs which further self-assemble to form the DNA tetrahedron. We validated proper self-assembly of the DNA tetrahedra on non-denaturing gels, a technique well-established for characterizing this particular DNA nanostructure in prior work by us as well as others in the field (Figure 4E). We assembled four versions of the DNA tetrahedron: a structure without any toeholds (Figure 4E: lane 5), with regular toeholds (DNA-TH, lane 6), with a PCL-containing toehold (PCL-TH, lane 7) and a tetrahedron with an RNA toehold (RNA-TH, lane 8). PAGE results showed that incorporation of toeholds did not affect the self-assembly of the DNA tetrahedron. Further, results also showed that the incorporation of a PCL in the strand or the RNA toehold did not adversely affect tetrahedron assembly. Since the toehold clipping process requires the DNA tetrahedron to be exposed to UV and RNase, we first confirmed that these conditions do not affect the assembled DNA tetrahedron (Figure 4F). We exposed the DNA tetrahedron to UV for 1 h and show that the tetrahedron was intact without any visible effect. Similarly, we incubated the DNA tetrahedron with RNase A and validated the intactness of the structure using nondenaturing PAGE.
We then demonstrated disassembly of the DNA tetrahedron using an invading strand that is complementary to the toehold and remaining part of the short strand (Figure 4G). We observed that the DNA tetrahedra containing the regular toehold was completely disassembled on the addition of the invading strand, as seen by the disappearance of the band corresponding to the tetrahedron (Figure 4G, lanes 1 and 2). In the DNA tetrahedra with the PCL-toehold and the RNA toehold, the structures were 90% disassembled on the addition of the invading strand (Figure 4G, lanes 3–4 and 5–6, respectively). This difference in the disassembly efficiency could be due to the differences in displacement yields in modified toeholds as seen in the duplexes. Addition of the invading strand did not have any effect on a tetrahedron without a toehold, confirming that disassembly proceeds by toehold-mediated strand displacement (Figure 4G, lanes 7 and 8). Reconfiguration of the structure is also sequence specific, with our prior work showing that even a single nucleotide mismatch in the toehold region can prevent structural reconfiguration (28).
Before we tested toehold clipping in the DNA tetrahedron, we first verified that an incorrect stimulus (UV exposure to an RNA toehold structure or RNase A addition to a PCL toehold structure) does not cause toehold clipping and prevent disassembly of the DNA tetrahedron. As a control, we exposed a DNA tetrahedron with regular toehold to UV light or incubated it with RNase A, followed by incubation with the invading strand. The tetrahedron was disassembled in all three cases: (i) the control tetrahedron without any stimulus (Figure 4H, lanes 1 and 2), (ii) tetrahedron exposed to UV (lanes 3 and 4) and (iii) the tetrahedron incubated with RNase A (lanes 5 and 6). The PCL-toehold containing DNA tetrahedron incubated with RNase A was disassembled on the addition of the invading strand (Figure 4H, lanes 7 and 8), showing that RNase A did not cause toehold clipping in the PCL system. Similarly, the RNA toehold containing tetrahedron was not affected by the UV exposure, and was disassembled on the addition of the invading strand (Figure 4H, lanes 9 and 10).
Finally, we tested the toehold clipping strategy in the DNA tetrahedron model system. We exposed the PCL-TH tetrahedron to UV, followed by the addition of the invading strand. Non-denaturing gel results showed that the DNA tetrahedron was intact after the addition of the invading strand, indicating that toehold clipping was successful (Figure 4I, lanes 3 and 4). In contrast, the DNA tetrahedron without UV exposure showed ∼88% disassembly on addition of the invading strand (lanes 1–2). Similarly, we tested two samples of the RNA-TH tetrahedron, one incubated with RNase A and one without. On addition of the invading strand, the DNA tetrahedron without the RNase A showed ∼84% disassembly (Figure 4I, lanes 5 and 6) while the sample incubated with RNase A showed intact DNA tetrahedron (lanes 7 and 8), indicating that toehold clipping worked.
CONCLUSION
In this work, we show that toehold-based strand displacement reactions can be controlled by physical (UV light) or molecular (RNase) cues, adding another layer of control for creating programmable, stimuli-responsive DNA nanostructures. Our demonstration of the toehold clipping process in the DNA tetrahedron shows that the strategy is scalable for multiple toeholds in a system (the DNA tetrahedron contains 12 toeholds). It is possible that such a strategy can be incorporated into a biological system, where biological RNAs (microRNAs, for example) could cause the strand displacement reaction, and externally applied light could be used for toehold clipping. For biological applications, the light-based toehold clipping strategy might require wavelengths that are less harmful than UV, and can be accomplished by the synthesis and integration of near infrared (NIR)-responsive linkers into DNA nanostructures. Our strategy is agnostic to the toehold sequence in this work, but one could tailor the sequences of the toehold to be responsive to specific enzymes or incorporate modified nucleotides at specific locations on the toehold to reduce strand displacement rates by truncating the toehold instead of fully eliminating it. Overall, our work adds to the library of molecular tools that allow control of programmable DNA nanostructures with potential use in a variety of applications.
DATA AVAILABILITY
All data supporting the findings of this study are available within the article and its supplementary information.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: H.F. and H.T. performed non-denaturing PAGE experiments. J.M. and H.T. performed denaturing PAGE experiments. J.M. synthesized and purified DNA strand containing photocleavable linker. H.F., H.Z. and S.V. performed MD simulations. H.F. and A.R.C performed experimental data analysis. J.S., A.A.C. and A.R.C acquired funding. S.V. and A.A.C. supervised the MD simulations. S.V. wrote the MD content of the manuscript. A.R.C. conceived and supervised the project, designed experiments, visualized data, and wrote the original draft of the manuscript. All authors provided comments on the manuscript.
Contributor Information
Hiba Faheem, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA.
Johnsi Mathivanan, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA; Department of Chemistry, University at Albany, State University of New York, Albany, NY, USA.
Hannah Talbot, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA.
Hana Zeghal, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA.
Sweta Vangaveti, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA.
Jia Sheng, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA; Department of Chemistry, University at Albany, State University of New York, Albany, NY, USA.
Alan A Chen, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA; Department of Chemistry, University at Albany, State University of New York, Albany, NY, USA.
Arun Richard Chandrasekaran, The RNA Institute, University at Albany, State University of New York, Albany, NY, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University at Albany, State University of New York and the RNA Institute start-up funds (to A.R.C.); National Institutes of Health (NIH) through National Institute on Aging (NIA) [R03AG076599 to A.R.C.]; National Institute of General Medical Sciences (NIGMS) [R35GM133469 to A.A.C., R01GM143749 to J.S.]; National Science Foundation (NSF) [MCB1651877 to A.A.C., CHE1845486 to J.S.]. The RNA Institute Summer Fellowship provided partial salary support for H.F. and H.T. The Center for Undergraduate Research and Creative Engagement (CURCE) at the University at Albany, State University of New York provided partial support for lab supplies to A.R.C. Funding for open access charge: University at Albany, State University of New York start-up funds to A.R.C.
Conflict of interest statement. None declared.
REFERENCES
- 1. Seeman N.C. DNA in a material world. Nature. 2003; 421:427–431. [DOI] [PubMed] [Google Scholar]
- 2. Yurke B., Turberfield A.J., Mills A.P. Jr, Simmel F.C., Neumann J.L. A DNA-fuelled molecular machine made of DNA. Nature. 2000; 406:605–608. [DOI] [PubMed] [Google Scholar]
- 3. Simmel F.C., Yurke B., Singh H.R.. Principles and applications of nucleic acid strand displacement reactions. Chem. Rev. 2019; 119:6326–6369. [DOI] [PubMed] [Google Scholar]
- 4. Zhang D.Y., Seelig G.. Dynamic DNA nanotechnology using strand-displacement reactions. Nature Chem. 2011; 3:103–113. [DOI] [PubMed] [Google Scholar]
- 5. Qian L., Winfree E., Bruck J.. Neural network computation with DNA strand displacement cascades. Nature. 2011; 475:368–372. [DOI] [PubMed] [Google Scholar]
- 6. Zhang C., Yang J., Xu J.. Circular DNA logic gates with strand displacement. Langmuir. 2010; 26:1416–1419. [DOI] [PubMed] [Google Scholar]
- 7. Berk K.L., Blum S.M., Funk V.L., Sun Y., Yang I.-Y., Gostomski M.V., Roth P.A., Liem A.T., Emanuel P.A., Hogan M.E.et al.. Rapid visual authentication based on DNA strand displacement. ACS Appl. Mater. Interfaces. 2021; 13:19476–19486. [DOI] [PubMed] [Google Scholar]
- 8. Chandrasekaran A.R., Levchenko O., Patel D.S., MacIsaac M., Halvorsen K.. Addressable configurations of DNA nanostructures for rewritable memory. Nucleic. Acids. Res. 2017; 45:11459–11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z., Zeng D., Ma H., Feng G., Hu J., He L., Li C., Fan C.. A DNA-Origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement. Small. 2010; 6:1854–1858. [DOI] [PubMed] [Google Scholar]
- 10. Bujold K.E., Hsu J.C.C., Sleiman H.F.. Optimized DNA “Nanosuitcases” for encapsulation and conditional release of siRNA. J. Am. Chem. Soc. 2016; 138:14030–14038. [DOI] [PubMed] [Google Scholar]
- 11. Rogers W.B., Manoharan V.N.. Programming colloidal phase transitions with DNA strand displacement. Science. 2015; 347:639–642. [DOI] [PubMed] [Google Scholar]
- 12. Fern J., Schulman R.. Modular DNA strand-displacement controllers for directing material expansion. Nat. Commun. 2018; 9:3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura S., Hashimoto H., Kobayashi S., Fujimoto K.. Photochemical acceleration of DNA strand displacement by using ultrafast DNA Photo-crosslinking. ChemBioChem. 2017; 18:1984–1989. [DOI] [PubMed] [Google Scholar]
- 14. Xing Y., Yang Z., Liu D.. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. Int. Ed. 2011; 50:11934–11936. [DOI] [PubMed] [Google Scholar]
- 15. Liu L., Hu Q., Zhang W., Li W., Zhang W., Ming Z., Li L., Chen N., Wang H., Xiao X.. Multifunctional clip strand for the regulation of DNA strand displacement and construction of complex DNA nanodevices. ACS Nano. 2021; 15:11573–11584. [DOI] [PubMed] [Google Scholar]
- 16. Amodio A., Zhao B., Porchetta A., Idili A., Castronovo M., Fan C., Ricci F.. Rational design of pH-Controlled DNA strand displacement. J. Am. Chem. Soc. 2014; 136:16469–16472. [DOI] [PubMed] [Google Scholar]
- 17. Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995; 117:5179–5197. [Google Scholar]
- 18. Dupradeau F.-Y., Pigache A., Zaffran T., Savineau C., Lelong R., Grivel N., Lelong D., Rosanski W., Cieplak P.. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010; 12:7821–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivani I., Dans P.D., Noy A., Pérez A., Faustino I., Hospital A., Walther J., Andrio P., Goñi R., Balaceanu A.et al.. Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2016; 13:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Molecular Operating Environment (MOE) Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2019. 2019; [Google Scholar]
- 21. Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E.. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015; 1–2:19–25. [Google Scholar]
- 22. Bussi G., Donadio D., Parrinello M.. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007; 126:014101. [DOI] [PubMed] [Google Scholar]
- 23. Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R.. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984; 81:3684–3690. [Google Scholar]
- 24. Darden T., York D., Pedersen L.. Particle mesh ewald: an N⋅log(N) method for ewald sums in large systems. J. Chem. Phys. 1993; 98:10089–10092. [Google Scholar]
- 25. Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M.. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997; 18:1463–1472. [Google Scholar]
- 26. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graphics. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 27. He Y., Ye T., Su M., Zhang C., Ribbe A.E., Jiang W., Mao C.. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008; 452:198–201. [DOI] [PubMed] [Google Scholar]
- 28. Chandrasekaran A.R., Halvorsen K.. Controlled disassembly of a DNA tetrahedron using strand displacement. Nanoscale Adv. 2019; 1:969–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplementary information.