Abstract
In situ capturing technologies add tissue context to gene expression data, with the potential of providing a greater understanding of complex biological systems. However, splicing variants and full-length sequence heterogeneity cannot be characterized at spatial resolution with current transcriptome profiling methods. To that end, we introduce spatial isoform transcriptomics (SiT), an explorative method for characterizing spatial isoform variation and sequence heterogeneity using long-read sequencing. We show in mouse brain how SiT can be used to profile isoform expression and sequence heterogeneity in different areas of the tissue. SiT reveals regional isoform switching of Plp1 gene between different layers of the olfactory bulb, and the use of external single-cell data allows the nomination of cell types expressing each isoform. Furthermore, SiT identifies differential isoform usage for several major genes implicated in brain function (Snap25, Bin1, Gnas) that are independently validated by in situ sequencing. SiT also provides for the first time an in-depth A-to-I RNA editing map of the adult mouse brain. Data exploration can be performed through an online resource (https://www.isomics.eu), where isoform expression and RNA editing can be visualized in a spatial context.
INTRODUCTION
Post-transcriptional modification such as alternative splicing and RNA editing generates substantially more transcripts than there are genes. These modifications increase transcriptome complexity and have important implications for cellular function, as evidenced by their tight regulation and role in development and tissue homeostasis (1). Alternatively spliced transcripts are particularly important in neurogenesis and brain development, contributing to the complex architecture of the mammalian central nervous system (CNS) by regulating a vast array of neuronal functions through cell-type-specific expression patterns (2,3). Several links have been reported between defective alternative splicing and diseases including epilepsy, autism spectrum disorders, schizophrenia, or spinal muscular atrophy (4). Transcriptomic diversity can also be generated through adenosine-to-inosine (A-to-I) RNA editing, a process mediated by a specific family of enzymes called adenosine deaminases (5). This process is involved in proper neuronal function (6), and dysregulated and aberrant A-to-I RNA editing has also been reported in neurological and neurodegenerative diseases such as epilepsy, amyotrophic lateral sclerosis, and developmental disorders (7).
Information on the spatial distribution of post-transcriptional modifications is crucial for a better understanding of their roles in physiology and disease. Recent technological advances have enabled high-throughput quantification of gene expression in a spatial context (8). These methods can broadly be categorized into those that detect the presence of a predefined set of target genes and those where observations stem from sampling across the entire transcriptome. The latter is required for a priori free exploratory analysis and novel hypothesis generation. Such methods are usually based on in situ capture of poly-adenylated RNA on spatially barcoded reverse transcription primers, which allows capturing all mRNAs in the transcriptome. The captured transcripts are then sequenced ex situ, and their spatial barcodes are used to infer their spatial origins. Even though several new such in situ-capture-based methods for large-scale transcriptome profiling have recently emerged (9–12), they only assess transcripts as 3′ cDNA tags and not as complete transcripts. The fundamental reason behind this is that all those methods are based on short-read library preparation and sequencing, which inevitably implies that full-length transcript information is lost. As a large amount of diversity in the transcriptome stems from post-transcriptional modifications, a truly comprehensive description of the transcriptome can only be obtained by characterizing the full-length sequences of the transcripts.
The situation was similar in droplet-based single-cell transcriptomics where until recently just the end of the cDNA was typically sequenced. Recent bioinformatics and methodological developments have enabled detection, characterization, and quantification of full-length transcripts in single-cell experiments using either short reads (13) or long reads generated with Pacific Biosciences (PacBio) (14) or Oxford Nanopore Technology (Nanopore) (15,16). Long-read sequencing is the only option to unambiguously access the full exonic structure of captured transcripts (17). PacBio sequencing has a higher accuracy (>99%) than Nanopore sequencing (97%) (18), while the latter provides a higher sequencing throughput. A single PromethION flow cell generates more than 100 million reads, whereas only 4 million usable reads are obtained with the most recent 8M PacBio SMRTcell (19). Considering the vast amount of RNA molecules captured in current high throughput single-cell or spatial transcriptomics approaches, Nanopore sequencing is a more attractive option to generate a sufficient amount of reads to reach the sequencing saturation needed for comprehensive transcript isoform and sequence heterogeneity exploration.
We explored whether those recent developments in single-cell transcriptomics can be applied in spatial transcriptomics to obtain spatially resolved full-length sequence information.
The Visium platform by 10x Genomics appeared well suited since cDNA from tissue sections is synthesized in situ on spatially barcoded slides in this approach. Consequently, the resulting sequencing reads contain those spatial barcodes and can be assigned to coordinates on the slide. A recent study proposed a hybrid approach in which single-cell transcript isoforms were characterized by PacBio sequencing (20). The obtained isoforms were then inferred to spatial coordinates by shallow Nanopore sequencing of spatial transcriptomics (Visium) libraries. However, the low coverage and the lack of unique molecular identifiers (UMIs) did not enable broad exploration of the spatial sequence heterogeneity of full-length isoforms nor RNA editing in the studied tissue. Another study (21) recently used the Visium system to perform full-length spatial transcriptomics of the mouse heart. However, the insufficient coverage of the 5′ ends of transcripts led the authors to assemble full-length cDNA from partial transcript sequences without using UMIs for expression quantification.
Currently, no efficient approach is available to generate comprehensive spatially resolved databases of full-length RNA sequences, and define the spatial landscape of splicing and single nucleotide variations (SNVs). We introduce spatial isoform transcriptomics (SiT), which combines an existing approach that yields spatial gene expression information after short-read sequencing of cDNA reverse transcribed in situ on tissue sections with long-read sequencing to generate spatially resolved full-length mRNA sequence data. While we opted for the Oxford Nanopore Promethion long-read sequencing technology, our approach is easily transferrable to other sequencing technologies. To analyze the sequencing data, we took advantage of the fact that spatially barcoded cDNA has a similar design as single-cell cDNA with the cell barcode replaced by a spatial barcode. This similarity allowed us to adapt a strategy we recently developed for the analysis of long-read single-cell RNA sequencing data (ScNaUmi-seq (15)) to analyze spatially resolved long-read sequencing data. SiT enables spatial exploration of isoform expression and RNA sequence heterogeneity in an un-targeted manner, by interrogating all captured isoforms and SNVs rather than a single isoform or SNV at a time.
We demonstrate the workflow for two different areas of the mouse brain and show that deep long-read sequencing identifies multiple genes that display spatially distinct alternative isoform expression. Furthermore, exploration of full-length sequence heterogeneity provides for the first time a global map of A-to-I RNA editing in adult mouse brain.
MATERIALS AND METHODS
Mouse brain samples
Olfactory bulbs were isolated from C57BL/6 mice (>2 months old), snap-frozen in Isopentane (Sigma-Aldrich) and embedded in cold optimal cutting temperature (OCT, Sakura) before sectioning. A left hemisphere was isolated from a C57BL/6J (8–12 weeks old) mouse and processed similarly. Two adjacent (50 μm apart) left hemisphere sections and one olfactory bulb section was used for the Visium workflow. For in situ sequencing (ISS), left hemisphere sections were from the same individual, while for the olfactory bulb, sections from a different individual were used. Mouse procedures were performed by Adlego Biomedical AB (Uppsala, Sweden), approved by the Stockholm ethics committee (project 4570–2019 with amendment 4566–2020 Stockholms djurförsöksetiska nämnd). They followed Directive 2010/63/EU of the European Parliament and of the Council, the Swedish Animal Welfare Act (Djurskyddslagen: SFS 1988:534), the Swedish Animal Welfare Ordinance (Djurskyddsförordningen: SFS 1988:539) and the provisions regarding the use of animals for scientific purposes (DFS 2004:15 and SJVFS 2012:26).
10x genomics visium experiments
The Visium Spatial Tissue Optimization Slide & Reagent kit (10x Genomics, Pleasanton, CA, USA) was used to optimize permeabilization conditions for mouse brain tissue. Spatially barcoded full-length cDNA was then generated using Visium Spatial Gene Expression Slide & Reagent kit (10x Genomics) following the manufacturer's protocol. Two coronal sections of the left hemisphere (IDs: CBS1 and CBS2) and one section of olfactory bulb (ID: ‘MOB’) were processed. Tissue permeabilization was performed for 6 and 9 min (CBS1, CBS2) and 12 min (MOB). cDNA amplification was conducted with 12 (CBS) and 17 (MOB) cycles. A fraction of each cDNA library was used for long-read sequencing (see ‘Oxford Nanopore sequencing’), whereas 10 μl each was used in the 10x Genomics Visium library preparation protocol of fragmentation, adapter ligation, and indexing. The libraries were sequenced on a NextSeq500 (Illumina), with 28 bases from read 1 and 91 from read 2, and at a depth of 253, 217 and 210 million reads for MOB, CBS1, and CBS2 samples, respectively. The raw sequencing data was processed with a pre-launch (version 4509.7.5) of the Space Ranger pipeline (10x Genomics) and mapped to the mm10 genome assembly.
Oxford nanopore sequencing
Nanopore sequencing of full-length cDNA libraries prepared from the 10x Genomics workflow yields 20–50% reads without the 3′ adapter sequence and thus lacks the spatial barcode and UMI (15). To deplete such fragments, we initially selected for cDNA that contains a biotinylated 3′ primer. 10 ng of the 10x Genomics Visium PCR product were amplified for 5 cycles with 5′-AAGCAGTGGTATCAACGCAGAGTACAT-3′ and 5′-Biotine-AAAAACTACACGACGCTCTTCCGATCT-3′. Excess biotinylated primers were removed by 0.55x SPRIselect (Beckman Coulter) purification, and the biotinylated cDNA (in 40 μl EB, Qiagen) was bound to 15 μl 1× SSPE washed Dynabeads™ M-270 Streptavidin beads (Thermo) in 10 μl 5× SSPE for 15 min at room temperature on a shaker. Beads were washed twice with 100 μl 1× SSPE and once with 100 μl EB. The beads were suspended in 100 μl 1× PCR mix and amplified for 8 cycles with the primers NNNAAGCAGTGGTATCAACGCAGAGTACAT and NNNCTACACGACGCTCTTCCGATCT to generate enough material (1–2 μg) for Nanopore sequencing library preparation. To deplete small fragments which are typically of little interest for transcript isoform analysis (cDNA from degraded RNA, ribosomal RNAs), small cDNA (<1 kB) was depleted with a 0.5× SPRI select purification. If fragments between 0.5 and 1 kB need to be retained, SPRIselect concentration should be increased to 0.8x. Nanopore sequencing libraries were prepared with the LSK-109 or LSK-110 kit from Oxford Nanopore (1 μg cDNA) following the instructions from the manufacturer. PromethION flow cells were loaded with 200 ng libraries each. PCR amplifications for Nanopore library preparations were made with Kapa Hifi Hotstart polymerase (Roche Sequencing Solutions): initial denaturation, 3 min at 95°C; cycles: 98°C for 30 s, 64°C for 30 s, 72°C for 5 min; final elongation: 72°C for 10 min, primer concentration was 1 μM.
Oxford nanopore data processing
Nanopore reads were processed according to the scNaUmi-seq protocol (15) with slight modifications. Ligation of the amplified cDNA with the Oxford Nanopore adapters (LSK-109 or LSK-110 kit) leads to the generation of 3–20% of chimeric cDNA where two or, to a far lesser extent, multiple cDNA molecules are ligated together. cDNA from the 10xGenomics Visium system is flanked at the 5′ by a Template Switching Oligonucleotide (TSO, AAGCAGTGGTATCAACGCAGAGTACAT) and at the 3′ by a poly(A) followed by an adapter sequence (CTACACGACGCTCTTCCGATCT). Junctions of individual cDNAs in chimeric reads are characterized by the presence of those 5′ and/or 3′ terminal sequences joined together within the read sequence. To split reads that originate from chimeric cDNA, we initially scanned reads for internal (>200 nucleotides from end) TSO and 3′ adapter sequences flanked by a poly(T) (poly(T)-adapter). When two adjacent poly(T)-adapters, two TSOs or one TSO adjacent to a poly(T)-adapter and thus junctions of two cDNA molecules were found, the read was split into two separate reads. Next all reads were scanned for poly(A/T) tails and the 3′ adapter sequence to define the orientation of the read and strand-specificity. Scanned reads were then aligned to Mus musculus mm10 with minimap2 (22) v2.17 in spliced alignment mode. Spatial barcodes and UMIs were then assigned to nanopore reads using the strategy and software previously described for single-cell libraries (15). SAM records for each spatial spot and gene were grouped by UMI after removal of low-quality mapping reads (mapqv = 0) and potentially chimeric reads (terminal Soft/Hard-clipping of > 150 nt). A consensus sequence per molecule (UMI) was computed depending on the number of available reads for the UMI using the ComputeConsensus sicelore-2.0 pipeline. For molecules supported by more than two reads (RN > 2), a consensus sequence was computed with SPOA (23) using the sequence between the end of the TSO (SAM Tag: TE) and the base preceding the polyA sequence (SAM Tag: PE). Quality values for consensus nucleotides were assigned as –10*log10(n Reads not conform with consensus nucleotide/n Reads total). Consensus cDNA sequences were aligned to the Mus musculus mm10 build with minimap2 v2.17 in spliced alignment mode. SAM records matching known genes were analyzed for matching Gencode vM24 transcript isoforms (same exon makeup). To assign a UMI to a Gencode transcript, we required a full match between the UMI and the Gencode transcript exon-exon junction layout authorizing a two-base margin of added or lacking sequences at exon boundaries, to allow for indels at exon junctions and imprecise mapping by minimap2. Detailed statistics of each step of Nanopore read processing are provided in Supplementary Table S1.
Sequencing quality control metrics
The 5′ and 3′ coverage of Illumina and Nanopore sequencing reads were compared by analyzing the coverage along length-normalized transcripts using Picard CollectRnaSeqMetrics pipeline (http://broadinstitute.github.io/picard; Broad Institute, 2019). Long-read sequencing saturation curves were computed using sicelore-2.0 SaturationCurve pipeline implementing the saturation curve calculation proposed by 10x Genomics (ref.: https://kb.10xgenomics.com/hc/en-us/articles/115003646912-How-is-sequencing-saturation-calculated-).
Spatial multi-assays storage
Raw gene expression matrices generated by Space Ranger were processed using R/Bioconductor (version 4.0.2) and Seurat (24) package (version 3.9.9). We created Seurat objects for each of the three samples (MOB, CBS1 and CB2) with different assays for the analysis as follows: (i) ‘Spatial’ containing gene-level raw short-read data from the Space Ranger output, (ii) ‘ISOG’ containing the gene-level Nanopore long-read data, (iii) ‘ISO’ containing isoform-level transcript information where only the molecules where all exons are observed are kept, (iv) ‘JUNC’ containing each individual exon-exon junction observation per isoform and (v) ‘AtoI’ containing exonic editing sites from the RADAR database (mm9 UCSC liftover to mm10) and from the Licht et al., 2019, study, for which we observed at least one UMI in our dataset. The ‘AtoI’ assay stored non edited UMI count (@counts slot), edited UMI count (@data slot), and the editing ratio (@scale.data slot) per editing site.
10x Genomics visium data-driven annotation of anatomical regions
The Spatial assay was normalized with SCTransform (25) using standard parameters. The first 30 principal components of the assay were used for uniform manifold approximation and projection (UMAP) representation and clustering (resolution = 0.4). Brain regions defined by clustering were assigned to known anatomical regions based on the Allen Mouse Brain Atlas and expression of representative gene markers (Supplementary Table S2). Spot clustering was similar between short- and long-read data (Supplementary Figure S1). As short-read data contains more UMIs per spot, our different gene markers representations are based on short-read data.
Spatial spot deconvolution
For MOB, spatial spots were deconvoluted using SPOTlight (26) release 0.1.4 and signature genes identified from (27) wild type samples Plp1 expresser cell types (mean normalized expression > 1) identified using Seurat FindAllMarkers (logfc.threshold = 0.25, min.pct = 0.1). Cell types contributing to at least 8% were selected and SPOTlight deconvolution scores were used for correlation computation with Plp1 isoforms expression. The same approach was performed for the coronal brain sections (CBS1 and CBS2) using Zeisel et al. (28) external dataset (mean expression > 1 UMI/cell).
Differential splicing detection
Seurat FindMarkers function (logfc.threshold = 0.25, test.use = ‘wilcox’, min.pct = 0.1) was used to detect genes showing at least two isoforms as markers of different brain regions using the Nanopore isoform-level ‘ISO’ assay. Results were filtered for non-majority isoforms, i.e. not the isoform showing the highest bulk expression, requiring Bonferroni-adjusted P-value ≤0.05.
Coronal brain sections transcriptome correlation
Images were masked and then aligned using the functions MaskImages and AlignImages of the STUtility (29) R package (version 1.0.0). We then computed and minimized the physical distance between spots to define pair of spots showing the smallest distance between sections. We then computed the whole transcriptome correlation per pair of spots using cor.test function from Stats R package using gene-level short-read (Spatial assay) and long-read (ISOG assay) UMI count matrices.
In situ sequencing validation
Two cryosections of the olfactory bulb and two coronal sections of the left hemisphere (at 10 μm thickness) were placed on SuperFrost Plus microscope slides (ThermoFisher Scientific), stored at –80°C and shipped on dry ice to CARTANA for library preparation, probe hybridization, probe ligation, rolling circle amplification, and fluorescence labeling using the HS Library Preparation Kit (P/N 1110) and for the in-situ sequencing using the ISS kit (P/N 3110) and sequential imaging using a 20× objective. ISS probes. The specific isoform targets used in this study are listed in Supplementary Table S3. The result table of the spatial coordinates of each molecule of all targets together with the reference DAPI image per sample were provided by CARTANA. Quantification of the ISS signal was done with the use of TissUUmaps (https://tissuumaps.github.io/, version 3.0.9) (30). The ISS DAPI images (ISS-1, ISS-2) from two coronal brain sections were loaded into the software tool one at a time, along with a table of identified genes and their corresponding x,y-coordinates delivered by CARTANA. In TissUUmaps, regions were manually drawn using the ‘draw tool’ to match the gene-level data driven annotation performed on CBS1. For each region (polygon), the software matched all genes with coordinates that fall within those of the polygon. The output from the software was a table of each gene and its matched region, which was subsequently concatenated to arrive at the final count per region.
Long-read calibration for high-confidence RNA editing call
To only keep high confidence Nanopore base calls, we analyzed the agreement between the Nanopore data (consensus sequences for UMIs) and Illumina data for editing sites covered by both sequencing technologies. We first examined the agreement of long- and short-read data as a function of the number of Nanopore reads used to generate the consensus sequence for the molecule (UMI) (Supplementary Table S4). Next, we analyzed how the agreement between Illumina and Nanopore UMIs depends on the consensus quality (QV) of the Nanopore UMI at the editing site (agreement of the reads backing the UMI, see 'Oxford Nanopore data processing’ section for consensus QV calculation). We then defined the minimal number of Nanopore reads per UMI and the minimal consensus quality required for 99% agreement with Illumina data and retained Nanopore UMIs backed by at least three reads and a consensus quality at the editing position of at least 6.
RNA editing ratios
Samples CBS2 and MOB were used for calculating global editing ratios. To test the significance of our findings, resampling of capture spots across the sample was performed. Observed editing ratios per spot were kept, and each spot was randomly assigned a region label from the pool of original labels without replacement 10k times. A normal distribution was fitted to the simulated editing ratios to calculate the probability of observing a value equal to, or more extreme, than the observed value. An editing ratio was computed for each individual editing site at a region level as the total edited molecules divided by the total molecules observed at the editing site position within each region.
RESULTS
Spatial isoform detection enabled through in situ capture and long-read sequencing
We fixed fresh-frozen tissue samples on spatially barcoded glass slides using methanol. After staining and imaging, mRNA molecules were captured in situ and tagged with barcodes and UMIs (10x Genomics Visium). Full-length cDNA libraries were then split for preparation of 3′ short-read sequencing libraries as well as long-read Nanopore libraries (Figure 1A). Resulting gene expression data were clustered to define distinct anatomical regions used as landmarks for regional isoform usage analysis.
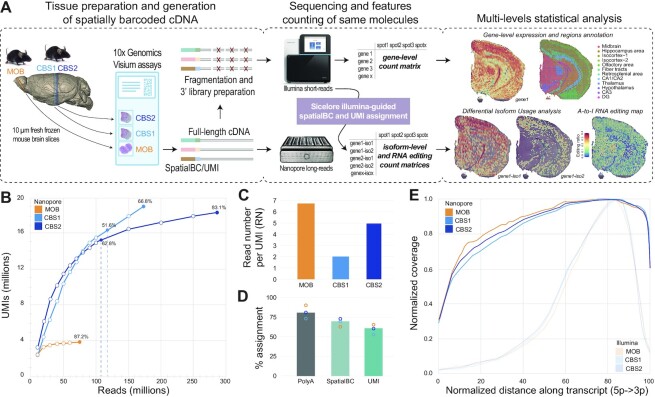
Figure 1.
SiT methodology and datasets. (A) Experimental and computational steps for SiT analysis. Right side shows unsupervised gene expression clustering and gene- (short-read) and isoform-level (SiT) expression of Snap25 in a mouse coronal brain section (CBS2). (B) Nanopore sequencing saturation curves for three Visium samples showing the number of UMIs observed as a function of the number of Nanopore reads. Labels indicate sequencing saturations obtained with all flow cells (CBS1, CBS2, MOB) and with just one latest generation Promethion flow cell per sample (vertical dotted lines, CBS1, CBS2). (C) Mean read number (RN) per molecule (UMI) observed for each of the three samples. (D) Percentage of assignment at each step of the workflow: Reads with polyA tail (PolyA) expressed as percentage of total reads; Assigned spatial barcode (SpatialBC) expressed as percentage of reads with PolyA tail found; UMI assigned reads (UMI) as percentage of reads with spatial barcode. Details about the spatial barcode/UMI assignment strategy are in (15). (E) Normalized transcript coverage plot for Nanopore and for Illumina sequencing.
We demonstrate the isoform landscape in situ in two regions of the mouse brain: the olfactory bulb (MOB) and two coronal sections of the left hemisphere (CBS1, CBS2). We provide a dataset of 13 Nanopore PromethION flow cells with a total of 535 million reads, reaching a high long-read sequencing saturation for the three samples (Figure 1B, C). The initial Nanopore PromethION flow cells (before July 2020) generated a median of 40 million reads, while the most recent flow cells (after December 2020) yielded more than 100 million reads (Figure 1B, Supplementary Table S1), a throughput that provided a high sequencing saturation of 51.6% and 62.8% for CBS1 and CBS2, respectively, with just one sequencing run. We used short-read data to assign spatial barcodes and UMIs to Nanopore reads, using the previously described scNaUMI-seq protocol (15) (Figure 1D). Our experimental approach, which included a cDNA size selection step for full-length cDNA enrichment (Supplementary Figure S2) provided a nearly uniform representation of full-length transcripts, enabling the exploration of splicing and full-length sequence heterogeneity (Figure 1E).
Regional isoform switching in the olfactory bulb
To investigate the spatial isoform landscape in MOB, we followed the SiT workflow depicted in Figure 1A. We generated 253 million Illumina short reads and 74 million long reads from two PromethION flow cells reaching a sequencing saturation of 87.2% (93.1% for short reads). For the long-read data, we applied a stringent filter to only retain molecules (UMIs) that contain all exon-exon junctions (mean exon number 6.7) of the reference isoform (Mouse Gencode vM24). Following this strategy, we unambiguously defined the full transcript structure for 2.19 million UMIs, sequenced with 59.9% of the 25.5 million spatialBC-UMI associated reads (mean 6.8 reads per UMI).
Across the tissue section, we observed 23560 different Gencode reference isoforms of 13291 distinct genes. We computed a Pearson correlation of the UMIs number per capture-spot equal to 0.95 between short and long reads (Supplementary Figure S3). Per spatially barcoded spot (55 μm diameter), we observed a median of 1917 UMIs corresponding to a median of 974 distinct isoforms (Supplementary Figure S4). Standard clustering of the short-read data defined five anatomic regions, as previously demonstrated (31) (Figure 2A). Based on this unsupervised clustering, we mined for genes showing a differential isoform usage between regions and identified 36 such genes, out of which Myl6 and Plp1 showed the most prominent patterns (Materials and Methods, Supplementary Table S5).
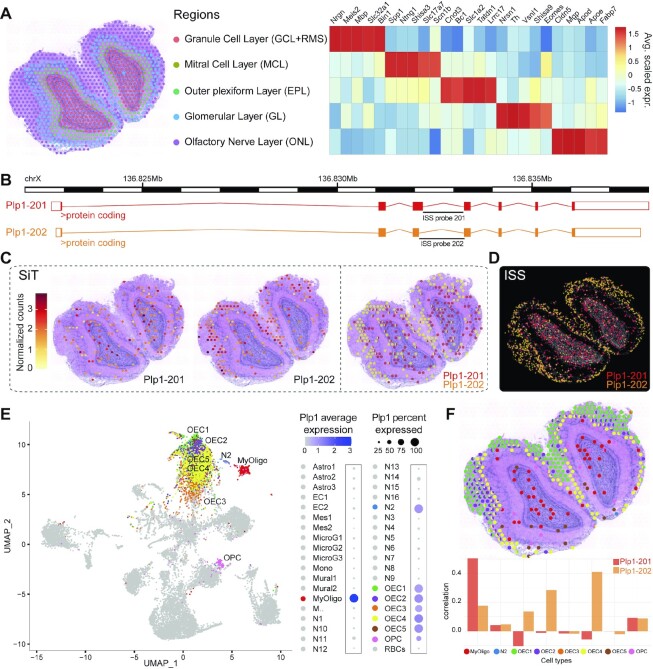
Figure 2.
SiT reveals isoform switches in the mouse olfactory bulb. (A) Data-driven annotation (left panel) of mouse olfactory bulb spatial regions through transcriptome-clustering of short-read data. Heatmap shows the expression of prominent marker genes for each region. (B) Exonic structure of the different Plp1 isoforms (mm10 genome build) detected by SiT. (C, D) Expression of Plp1 isoforms detected by SiT (C) and ISS (D). (E) Uniform Manifold Approximation and Projection (UMAP) representation of an external MOB single cell dataset (27). The dot plot on the right indicates Plp1 expression per cell type in the single cell dataset. (f) Spatial spot deconvolution of cell types with high/prominent Plp1 expression. Each dot corresponds to a pie graph indicating cell type composition in this spot (upper panel). Per spatial spot correlation observed between deconvolution score and Plp1 isoform expression (lower panel). Results show that Plp1 is predominantly expressed by olfactory ensheathing cells (OEC) in the olfactory nerve layer (ONL) and by myelinating-oligodendrocytes (MyOligo) in the granule cell layer (GCL).
Myosin Light Chain 6 (Myl6), codes for the non-phosphorylatable alkali light chain component of the hexameric Myosin motor protein, which has been shown to be involved in neuronal migration and synaptic remodeling in immature and mature neurons (32,33). Myl6 produces two main polypeptides of same size, that just differ in five of the last nine carboxy terminal amino acids: the non-muscle isoform Myl6-206 (Lc17a) and the smooth-muscle isoform Myl6-201 (Lc17b) (34). Our data revealed a high expression of Myl6-201 in the granule cell layer while Myl6-206 is preferentially expressed in the olfactory nerve and mitral cell layer (Supplementary Figure S5).
Proteolipid protein 1 (Plp1), a gene involved in severe pathologies associated with CNS dysmyelination (35), demonstrated a clear regional difference in isoform expression between the inner granule cell layer, expressing full Plp1-201 (PLP) isoform and the outer regions of the olfactory nerve layer, expressing preferentially the truncated Plp1-202 (DM20) isoform (Figure 2B, C, Supplementary Figure S6). SiT allows to clearly identify 35 codons that are exclusively present in PLP and to quantify the PLP/DM20 splicing balance in a spatial context, a balance that has been shown to be implicated in Pelizaeus-Merzbacher disease (36).
Interestingly, while the Myl6 SiT isoform switch could be confirmed by short reads due to its proximity to the 3′ end of the transcript, short reads were unable to confirm the alternate splicing of Plp1 detected by SiT since the alternate exon is located far from the 3′ end. This illustrates the huge advantage of SiT for comprehensive differential isoform usage analysis (Supplementary Figure S7).
We validated the differential regional isoform expression of Myl6 and Plp1 using an independent hybridization-based technology, in situ sequencing (ISS), on a tissue section from another individual (Figure 2d, Supplementary Figure S5).
Cell type inference reveals origin of plp1 isoforms
Each spatially barcoded spot typically captures transcripts from multiple cells. Single-cell RNA-seq data allow to deconvolute the transcriptional signal into the likely constituent cell types of the spot, and to associate specific cell type(s) to spatial isoform expression data. We used a previously published mouse olfactory bulb single-cell RNA-seq dataset (27) to perform a deconvolution strategy based on identifying pairwise cell correspondence using SPOTlight. This approach nominated the myelinating-oligodendrocyte (MyOligo) cell type within the granule cell layer as the predominant origin of the Plp1 standard isoform and the olfactory ensheathing cell (OEC) within the olfactory nerve layer as the predominant origin of the truncated Plp1 isoform DM20 (Figure 2E, F).
Deep sequencing of coronal brain sections
SiT was then applied to two 50 μm spaced adjacent coronal brain sections (CBS1, CBS2). The higher complexity of the spatial coronal brain section libraries motivated a deeper sequencing than for the MOB (Figure 1B). We generated a total of 174 and 287 million long reads with three and eight Oxford Nanopore flow cells, for CBS1 and CBS2 respectively (Supplementary Table S1). Clustering based on short-read gene expression data defined 12 anatomical regions (Figure 3a, Supplementary Figure S8) that broadly corresponded to regions in the Allen mouse brain reference atlas (37) (Figure 3B). To assess the robustness of our method, we computed the transcriptome expression correlation between the two sections after image alignment and minimization of the spot-to-spot distance between sections (Supplementary Figure S9). We observed a Pearson correlation of 0.98 and 0.93, respectively for short-read and long-read gene-level profiles of corresponding pairs of spatial spots (Figure 3C).
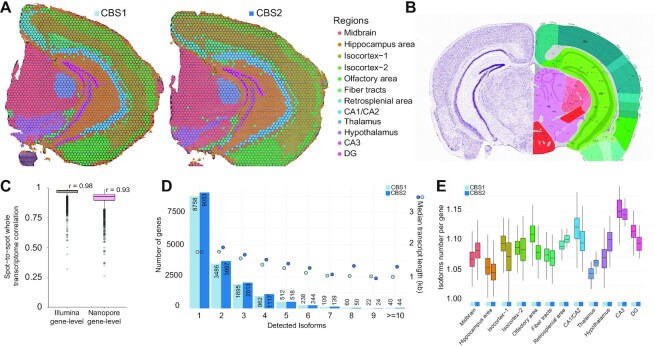
Figure 3.
SiT robustness assessment using two coronal brain sections. (A) Data-driven annotation of two mouse coronal brain regions through transcriptome clustering of short-read data. (B) Allen Mouse Brain Atlas reference map for the coronal brain section region annotation. (C) Gene-level short-read and long-read transcriptome correlation between corresponding spatial spots of the CBS1 and CBS2 section after image alignment and distance minimization. (D) Histogram showing the frequency distribution of the number of isoforms per gene for CBS1 and CBS2. The median length of transcripts is indicated for each category. (E) Average number of isoforms per gene detected for each spatial region.
Regional isoform switching in coronal brain sections
Next, we sought to identify genes with differential isoform usage across the 12 anatomical regions. Using the UMI filtering described for MOB, we successfully assigned 10 million molecules (UMIs) to a precise isoform in CBS2, corresponding to 33097 distinct isoforms encoded by 16899 genes. Among those genes, we observed 9053 (53.6%) that expressed a single isoform and 7846 (46.4%) that expressed multiple isoforms across the tissue section (Figure 3D). We obtained a median of 3644 UMIs for each spatially barcoded spot, corresponding to a median of 1524 unique isoforms (Supplementary Figure S4, cf. Supplementary Table S1 for CBS1 statistics). We noticed small variations in the number of isoforms per gene across the different brain regions and a slightly higher isoform complexity in the CA3 region of the hippocampus (Figure 3E, Supplementary Table S6). Among the multi-isoform genes, we mined those showing a splicing pattern change across regions to decipher differences in spatial cell organization. We identified 126 and 166 significant (Bonferroni-adjusted P-value < 0.05) regional isoform switching genes in CBS1 and CBS2 respectively, out of which 61 were identified in both sections (Materials and Methods, Supplementary Tables S5–S7).
Hypothalamus expresses high levels of a snap25a isoform
For both sections, our data revealed a pronounced regional isoform switching for several genes involved in brain function (Figure 4A, B). A first example is Snap25 which was expressed as the Snap25-202 (Snap25a) isoform in the hypothalamus, in contrast to the midbrain where Snap25-201 (Snap25b) was the predominant isoform (Figure 4C, D). Both isoforms result from the inclusion/exclusion of two closely spaced sequences that encode distinct fifth exons. This results in 9 out of 206 amino acid changes between the two polypeptides, a difference that has been shown to play a role in plasticity at central synapses (38). As previously described, Snap25a is the dominant transcript during embryonic and early postnatal development in mouse brain, while in adulthood, Snap25b becomes the dominant mRNA. Snap25a remains the dominant isoform in endocrine and neuroendocrine cells throughout life (39). The observed spatial isoform expression pattern was confirmed by ISS (Figure 4E, F).
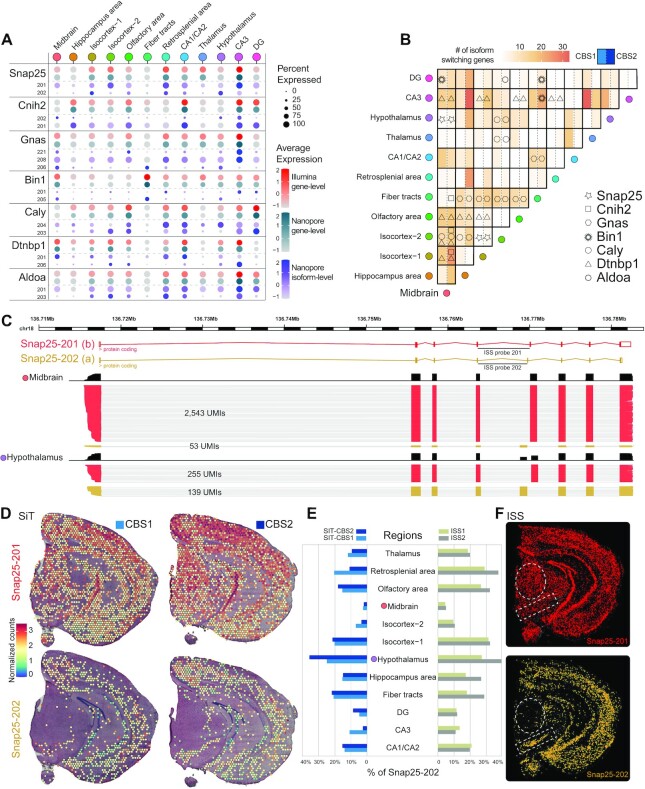
Figure 4.
SiT reveals Snap25 isoform-switch between mouse brain regions. (A) Dotplot showing the seven most significant (adjusted P-value < 0.05) regional differences in isoform usage detected in CBS1 and CBS2 sections (CBS2 expression value is shown). (B) Heatmap illustrating the number of isoform-switching genes identified for each pair of brain regions (same order for the horizontal and vertical axes). Genes discussed in this study are highlighted with symbols when differential transcript usage was called for CBS1 (left part) and/or CBS2 (right part) for each pair of regions. (C) Snap25 isoforms (alternative exon 5) tracks and number of molecules (UMI) observed in Midbrain and Hypothalamus (CBS2). (D) Snap25 isoform expression in coronal brain sections revealed by SiT. (E) Contribution of Snap25-202 to the total expression of Snap25 in different brain regions for two independent replicates of SiT (left) and ISS (right). (F) Snap25 isoform expression validation by in situ-sequencing (ISS).
Dense neuronal regions and white matter express different bin1 isoforms
A second example of regional isoform switching is Bin1, which belongs to the Bin-Amphiphysin-Rvs167 (BAR) domain superfamily proteins. Bin1 is involved in the regulation of membrane curvature, particularly in clathrin-coated synaptic vesicles (40). The Bin1 locus has been identified as a leading modulator of genetic risk in Alzheimer's disease (41). Our data revealed a clear regional differential Bin1 isoform usage. Midbrain and fiber tracts expressed Bin1-205 (human Bin1iso9), while the Bin1-201 isoform (human Bin1iso1) was expressed in a sparse pattern including in the isocortex, and the hippocampal formation, with enrichment in the Dentate Gyrus (DG) and the CA3 region. Spatial deconvolution using the single-cell Mouse Brain Atlas dataset (28) revealed a high correlation between Bin1-205 isoform expression and Oligodendrocytes especially the MOL1 and MOL3 subtypes, which delineates more precisely the expression of Bin1-205 (Bin1iso9) in these two subtypes of mature oligodendrocytes (42) (Supplementary Figure S10).
The Gnas locus shows complex isoform expression pattern across brain regions
A third example of regional isoform switching is Gnas, encoded by a complex imprinted locus (43) as the alpha-subunit of the stimulatory G protein (Gsα), an important component of the cyclic AMP signaling pathway (44). In both coronal brain sections, we observed a high expression of Gnas α-L (Gnas-208) across all regions making it the most abundant Gnas isoform. SiT identified multiple Gnas isoforms such as the lowly expressed splice variant Gnas α-S (Gnas-206) present mainly in fiber tracts as well as Gnas-221, a paternally imprinted allele-specific isoform, expressed in isocortex and in the CA3 region of the hippocampus, including a restricted expression in or adjacent to the posterior amygdalar nucleus (PA) region according to the Allen Mouse Brain reference atlas (Supplementary Figure S11).
SiT identified several additional differences in the regional isoform usage for an additional set of genes including Cnih2, Caly, Dtnpb1 and Aldoa, which were confirmed by ISS (Supplementary Figure S12).
SiT reveals A-to-I RNA editing mouse brain map
We next examined whether SiT enables exploration of Single Nucleotide Variation (SNV). We investigated RNA adenosine-to-inosine (A-to-I) editing events, the principal source of transcript sequence heterogeneity in the mammalian transcriptome. RNA editing has been shown to be essential for neurotransmission and other neuronal functions (45). While other studies examined editing events on bulk samples from mouse brain (46), or spatially resolved by ISS for a limited number of targeted editing sites (47), none has yet provided a large-scale spatially resolved RNA editing map of the mouse brain.
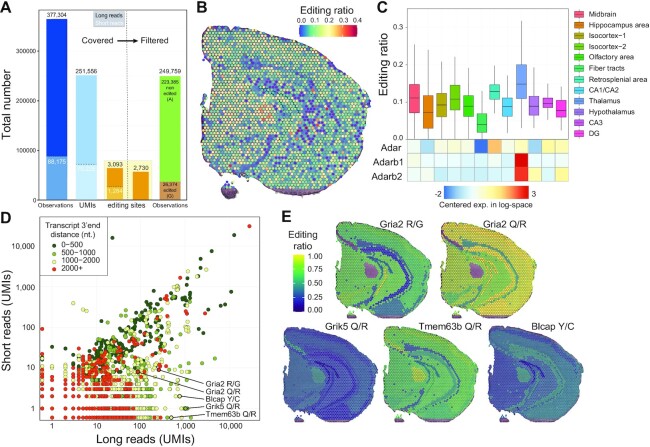
Robust SNV calling requires substantial sequencing depth, and we therefore focused on CBS2 for this purpose (Supplementary Table S1). We explored a total of 5817 exonic A-to-I RNA editing sites described in the literature (46,48). To ensure high-confidence calls with Nanopore long reads, we defined an ad hoc UMI sequencing depth and a consensus call accuracy threshold for Nanopore editing site calls. To that end, we examined the agreement between long- and short-read editing site data for the same UMI (RNA molecule) for 70225 UMIs where at least one known editing site was observed in both Illumina and Nanopore molecules (total 88175 editing site observations). Based on the analysis shown in Supplementary Figure S13, we retained UMIs backed by at least 3 reads with a consensus quality at the editing site greater than 6 for further analysis. This resulted in a 99% agreement between Illumina and Nanopore editing site calls. Out of the 377304 Nanopore editing site observations, 249 759 (66.2%) passed those filters and were used for downstream analysis (Figure 5A, Materials and Methods). Globally, we observed an A-to-I RNA editing ratio of 10.56% for 2730 distinct editing sites covered by at least one UMI (46.9% of the 5817 known editing sites explored, Supplementary Table S8). Interestingly, editing ratios displayed a non-uniform spatial distribution (Figure 5b). Consistent with a previous report (47), we observed a significantly higher editing ratio in Thalamus (mean 17.6%; 749/4247 UMIs edited) than in Fiber tracts (mean 5.4%; 657/12063 UMIs edited). We also noticed a positive correlation between the expression levels of the A-to-I editing enzymes (adenosine deaminases, ADARs) and the editing ratios for the brain regions (Figure 5C, Supplementary Table S9). The same variation between distinct brain areas was independently noticed in both coronal brain sections (Supplementary Figure S14; cf. Supplementary Figure S15 for MOB analysis), which display similar profiles. We observed a Pearson correlation score of 0.96 between CBS1 and CBS2 editing ratio for the 483 editing sites showing at least 20 UMIs in CBS1 and CBS2 long reads profiles (Supplementary Figure S16).
Figure 5.
SiT reveals spatial variations in A-to-I RNA editing in the mouse brain. (A) Histogram showing the sum of known exonic editing site observations (Observations, blue bars), the total number of unique UMIs covering at least one editing site (UMIs, light blue bars), the number of unique editing sites covered by at least one UMI (orange bars) by long-read (black numbers) and short-read (white numbers) data before (left part) and after (right part) quality filtration of Nanopore UMIs (consensus sequence quality ≥6 and UMI sequencing depth ≥3, see Supplementary Figure S13). (B) Spatial map of A-I editing ratios. (C) Editing ratio and gene-level expression of ADAR enzymes per region. (D) Editing site short- and long-read coverage colored by distance to transcripts 3′-end. (E) Spatial map of the A-I editing ratio for a selection of editing sites. Editing ratios were computed as the average editing ratio of molecules observed per region.
We next sought to compare the potential of short- and long-read sequencing to identify RNA editing events. As expected, we observed that long reads yield more information than short reads (Figure 5D) since they allowed investigation of sequence heterogeneity beyond the 3′-end editing sites. This resulted in the identification of editing variation across brain regions in several key genes of neuronal function such as Gria2 (49), Grik5 (50), Tmem63b (51) or Blcap (52) (Figure 5E).
AMPA receptors (AMPARs) mediate most of the fast excitatory neurotransmission in the brain and are composed of four different subunits. The transcript for the Gria2 (GluA2) subunit is known to be edited at two positions: the R/G site (mm10, chr3:80692286), which is in the ligand-binding domain where editing causes faster desensitization and recovery from desensitization, and the Q/R site (mm10, chr3:80706912) located within the channel pore, which editing renders AMPARs virtually Ca2+-impermeable and thereby affects a key aspect of neurotransmission (53). Consistent with previous observations (54), we observed less editing at the R/G site (mean = 55.5%, 142/256 UMIs edited) than at the Q/R site (mean = 94.2%, 81/86 UMIs edited). We also observed regional differences in R/G site editing with low editing levels in subregions of the hippocampus (DG granule cell layer: mean = 36.4%, 4/11 edited UMIs; CA1/CA2: mean = 20.5%, 8/39 UMIs edited) and high editing levels in the isocortex region (mean = 67.5%, 79/117 UMIs edited) (Figure 5E).
DISCUSSION
Here we present SiT, the first transcriptome-wide approach to explore isoform expression and sequence heterogeneity in a tissue context. We demonstrated the ability of SiT to identify differential isoform usage across brain regions. We further validated the ability of SiT to detect A-to-I RNA editing events in full-length transcript sequences. In the mouse olfactory bulb, we demonstrated a clear isoform switch of Plp1, a gene coding for the major myelin protein in the nervous system, between the outer regions of the olfactory nerve layer and the inner granule cell layer. In coronal brain sections we showed differential isoform usage for several other key genes, including Snap25, Bin1 or Gnas. We further established the spatial pattern of A-to-I RNA editing events to provide for the first time a map of global editing ratios in the adult mouse brain and show that SiT enables investigation of individual editing sites in a spatial context.
One limitation of SiT is that only 26–40% of the Nanopore reads both matched the reference genome and were assigned to a valid spatial barcode and a UMI for the three sections while 63–90% of the Illumina reads met those criteria. The principal reasons for the high fraction of unassigned Nanopore reads are: (i) elimination of low-quality Nanopore reads where no spatial barcode and UMI can be identified. Further improvements in Nanopore sequencing accuracy will increase assignment efficiency significantly. (ii) About 20% of the sequences do not contain the RT primer with the spatial barcode and the UMI and are discarded. Those are mainly low-quality reads and PCR artifacts lacking the 3′ end of the cDNA. Optimization of the library preparation should significantly increase assignment efficiency, for instance by further depletion of unwanted cDNA sequences that do not contain a polyA tail. (iii) While our Illumina-guided cell (or spatial) barcode and UMI assignment strategy is highly accurate (15), the efficiency of UMI assignment to Nanopore reads depends on the sequencing saturation of the short-read dataset which was 71%, 73% and 93% for the CBS1, CBS2 and MOB section respectively. Nanopore reads with UMIs that were not found in the Illumina dataset cannot be assigned and are lost. The development of an Illumina-free assignment strategy will help recover molecules missed during Illumina sequencing and increase the efficiency of UMI assignment. Another limitation of SiT is that the resolution is limited to that of the Visium technology. Several alternatives were recently described that increase the resolution of spatial transcriptomics (DBiT-seq (11), Slide-SeqV2 (55)). SiT can be readily adapted to those approaches which generate full-length cDNA. A last point, also shared with all single-cell approaches, regards the limitations on the number and integrity of captured mRNA molecules. Only half of the captured molecules contain all exons of a Gencode isoform and are unambiguously attributed to a full-length isoform. The rest corresponds to truncated cDNA, likely derived from degraded RNA. Another limitation of both single-cell and sequencing based spatial transcriptomics approaches is that only a fraction of the mRNA molecules is captured and sequenced. This leads to drop-outs and noisy expression patterns for low expressed transcripts. Further optimizations of the Visium workflow aiming at reduced RNA degradation prior to cDNA synthesis and increased capture of mRNA molecules should address those limitations.
Our results show that a throughput of 100 million long reads, now obtained routinely with one PromethION flow cell, is sufficient to explore the spatial landscape of transcript isoform expression in a typical Visium experiment. Increased sequence accuracy and throughput of the Nanopore technology will further benefit the SiT approach. We anticipate that such progresses should make SiT more and more applicable in many different environments, including clinical contexts. A straightforward application would certainly be in oncology to resolve spatially the expression of pathological isoforms (e.g. fusion transcripts) and cancer mutations in order to better characterize the heterogeneity of tumor biopsies. SiT could thus contribute to the advent of more efficient therapeutic avenues.
SiT expands the spatial transcriptomics toolbox to long-read-based exploration of transcript isoforms and SNVs, such as RNA editing or somatic mutations. These observations have thus far escaped detection due to the limitations of conventional short-read sequencing approaches. In combination with whole-brain molecular maps (56), we show here how this approach offers a new opportunity to understand the spatial and molecular organization of complex organs. While Nanopore sequencing was chosen due to a far lower cost per read over PacBio sequencing (Supplementary Table S10), SiT can easily be adapted for PacBio sequencing.
The SiT methodology is based on commercially available reagents and enables a deepened investigation of the isoform landscape, including studies of imprinting, fusion transcripts, and SNV expression in a spatial context. SiT will enable a better description of complex transcriptomes. As such, it provides an important additional resource to enrich existing Cell Atlas initiatives, as illustrated here through the online resource of mouse brain that we provide (https://www.isomics.eu/).
DATA AVAILABILITY
All relevant data have been deposited in Gene Expression Omnibus under accession number GSE153859. CARTANA ISS data and all custom software used have been deposited in Zenodo (https://doi.org/10.5281/zenodo.7664367). R figures and analysis scripts are also accessible at the same address. Seurat object .rds files for the three samples are available as supplementary files within the GEO serie and interactively browsable in the dedicated SiTx R Shiny application accessible at https://www.isomics.eu.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ludvig Bergenstråhle and Alma Andersson for advice and helpful discussions. We would like to thank the National Genomics Infrastructure (NGI) Sweden for providing infrastructure support.
Contributor Information
Kevin Lebrigand, Université Côte d’Azur, CNRS, Institut de Pharmacologie Moléculaire et Cellulaire, F06560 Sophia Antipolis, France.
Joseph Bergenstråhle, Department of Gene Technology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Science for Life Laboratory, Solna, Sweden.
Kim Thrane, Department of Gene Technology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Science for Life Laboratory, Solna, Sweden.
Annelie Mollbrink, Department of Gene Technology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Science for Life Laboratory, Solna, Sweden.
Konstantinos Meletis, Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden.
Pascal Barbry, Université Côte d’Azur, CNRS, Institut de Pharmacologie Moléculaire et Cellulaire, F06560 Sophia Antipolis, France.
Rainer Waldmann, Université Côte d’Azur, CNRS, Institut de Pharmacologie Moléculaire et Cellulaire, F06560 Sophia Antipolis, France.
Joakim Lundeberg, Department of Gene Technology, School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Science for Life Laboratory, Solna, Sweden.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut National contre le Cancer [PLBIO2018-156]; FRM [DEQ20180339158]; Inserm Cross-cutting Scientific Program HuDeCA 2018; National Infrastructure France Génomique (Commissariat aux Grands Investissements) [ANR-10-INBS-09-03, ANR-10-INBS-09-02]; 3IA@cote d’azur [ANR-19-P3IA-0002]; Swedish Research Council; Swedish Foundation for Strategic Research; European Union's H2020 Research and Innovation Program [874656 (discovAIR)]; Conseil départemental 06; Knut and Alice Wallenberg Foundation [2018.0172]; Erling-Persson Family Foundation (HDCA); Science for Life Laboratory; PPIA 4D-OMICS [21-ESRE-0052]. Funding for open access charge: France Génomique (ANR).
Conflict of interest statement. J.L. and K.T. are scientific consultants to 10xGenomics Inc.
REFERENCES
- 1. Baralle F.E., Giudice J.. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017; 18:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Su C.H., Dhananjaya D., Tarn W.Y.. Alternative splicing in neurogenesis and brain development. Front. Mol. Biosci. 2018; 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herbrechter R., Hube N., Buchholz R., Reiner A.. Splicing and editing of ionotropic glutamate receptors: a comprehensive analysis based on human RNA-seq data. Cell. Mol. Life Sci. 2021; 78:5605–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lipscombe D., Lopez Soto E.J.. Alternative splicing of neuronal genes: new mechanisms and new therapies. Curr. Opin. Neurobiol. 2019; 57:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Y., Okada S., Sakurai M.. Adenosine-to-inosine RNA editing in neurological development and disease. RNA Biol. 2021; 18:999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sapiro A.L., Shmueli A., Henry G.L., Li Q., Shalit T., Yaron O., Paas Y., Li J.B., Shohat-Ophir G.. Illuminating spatial A-to-I RNA editing signatures within the Drosophila brain. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:2318–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Costa Cruz P.H., Kawahara Y.. Rna editing in neurological and neurodegenerative disorders. Methods Mol. Biol. 2021; 2181:309–330. [DOI] [PubMed] [Google Scholar]
- 8. Asp M., Bergenstråhle J., Lundeberg J.. Spatially resolved transcriptomes—Next generation tools for tissue exploration. Bioessays. 2020; 42:e1900221. [DOI] [PubMed] [Google Scholar]
- 9. Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z.. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019; 363:1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D., Arlotta P., Macosko E.Z., Chen F.. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol. 2021; 39:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y., Yang M., Deng Y., Su G., Enninful A., Guo C.C., Tebaldi T., Zhang D., Kim D., Bai Z.et al.. High-spatial-resolution multi-omics sequencing via deterministic barcoding in. Tissue. Cell. 2020; 183:1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen A., Liao S., Cheng M., Ma K., Wu L., Lai Y., Qiu X., Yang J., Xu J., Hao S.et al.. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022; 185:1777–1792. [DOI] [PubMed] [Google Scholar]
- 13. Hagemann-Jensen M., Ziegenhain C., Chen P., Ramsköld D., Hendriks G.J., Larsson A.J.M., Faridani O.R., Sandberg R.. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020; 38:708–714. [DOI] [PubMed] [Google Scholar]
- 14. Gupta I., Collier P.G., Haase B., Mahfouz A., Joglekar A., Floyd T., Koopmans F., Barres B., Smit A.B., Sloan S.A.et al.. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 2018; 36:1197–1202. [DOI] [PubMed] [Google Scholar]
- 15. Lebrigand K., Magnone V., Barbry P., Waldmann R.. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun. 2020; 11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Volden R., Palmer T., Byrne A., Cole C., Schmitz R.J., Green R.E., Vollmers C.. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9726–9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakamoto Y., Sereewattanawoot S., Suzuki A.. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2020; 65:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q.. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020; 21:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mincarelli L., Uzun V., Rushworth S.A., Haerty W., Macaulay I.C.. Combined single-cell gene and isoform expression analysis in haematopoietic stem and progenitor cells. 2020; bioRxiv doi:06 April 2020, preprint: not peer-reviewed 10.1101/2020.04.06.027474. [DOI] [PMC free article] [PubMed]
- 20. Joglekar A., Prjibelski A., Mahfouz A., Collier P., Lin S., Schlusche A.K., Marrocco J., Williams S.R., Haase B., Hayes A.et al.. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 2021; 12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boileau E., Li X., Naarmann-de Vries I.S., Becker C., Casper R., Altmüller J., Leuschner F., Dieterich C.. Full-length spatial transcriptomics reveals the unexplored isoform diversity of the myocardium Post-MI. Front. Genet. 2022; 13:912572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vaser R., Sović I., Nagarajan N., Šikić M.. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017; 27:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choudhary S., Satija R.. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 2022; 23:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elosua-Bayes M., Nieto P., Mereu E., Gut I., Heyn H.. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021; 49:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tepe B., Hill M.C., Pekarek B.T., Hunt P.J., Martin T.J., Martin J.F., Arenkiel B.R.. Single-cell RNA-seq of mouse olfactory bulb reveals cellular heterogeneity and activity-dependent molecular census of adult-born neurons. Cell Rep. 2018; 25:2689–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G.et al.. Molecular architecture of the mouse nervous system. Cell. 2018; 174:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergenstråhle J., Larsson L., Lundeberg J.. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics. 2020; 21:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pielawski N., Andersson A., Avenel C., Behanova A., Chelebian E., Klemm A., Nysjö F., Solorzano L., Wählby C.. TissUUmaps 3: interactive visualization and quality assessment of large-scale spatial omics data. 2022; bioRxiv doi:16 January 2023, preprint: not peer-reviewed 10.1101/2022.01.28.478131. [DOI] [PMC free article] [PubMed]
- 31. Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M.et al.. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016; 353:78–82. [DOI] [PubMed] [Google Scholar]
- 32. Kneussel M., Wagner W.. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat. Rev. Neurosci. 2013; 14:233–247. [DOI] [PubMed] [Google Scholar]
- 33. Vallee R.B., Seale G.E., Tsai J.-W.. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009; 19:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen P., Yin J., Guo Y., Xiao H., Wang X., DiSanto M.E., Zhang X.. The expression and functional activities of smooth muscle myosin and non-muscle myosin isoforms in rat prostate. J. Cell. Mol. Med. 2018; 22:576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nave K.A. Myelination and support of axonal integrity by glia. Nature. 2010; 468:244–252. [DOI] [PubMed] [Google Scholar]
- 36. Regis S., Grossi S., Corsolini F., Biancheri R., Filocamo M.. PLP1 gene duplication causes overexpression and alteration of the PLP/DM20 splicing balance in fibroblasts from Pelizaeus-Merzbacher disease patients. Biochim. Biophys. Acta - Mol. Basis Dis. 2009; 1792:548–554. [DOI] [PubMed] [Google Scholar]
- 37. Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., Hawrylycz M., Dang C.. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013; 41:D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Irfan M., Gopaul K.R., Miry O., Hökfelt T., Stanton P.K., Bark C.. SNAP-25 isoforms differentially regulate synaptic transmission and long-term synaptic plasticity at central synapses. Sci. Reports. 2019; 9:6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bark C.I., Hahn K.M., Ryabinin A.E., Wilson M.C.. Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang B., Zelhof A.C.. Amphiphysins: raising the BAR for synaptic vesicle recycling and membrane dynamics. Traffic. 2002; 3:452–460. [DOI] [PubMed] [Google Scholar]
- 41. Chapuis J., Hansmannel F., Gistelinck M., Mounier A., Van Cauwenberghe C., Kolen K.V., Geller F., Sottejeau Y., Harold D., Dourlen P.et al.. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry. 2013; 18:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Rossi P., Buggia-Prévot V., Clayton B.L.L., Vasquez J.B., Van Sanford C., Andrew R.J., Lesnick R., Botté A., Deyts C., Salem S.et al.. Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol. Neurodegener. 2016; 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bastepe M. The GNAS Locus: quintessential complex gene encoding gsalpha, xlalphas, and other imprinted transcripts. Curr. Genomics. 2008; 8:398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Turan S., Bastepe M.. GNAS spectrum of disorders. Curr. Osteoporos. Rep. 2015; 13:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Behm M., Öhman M.. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 2016; 32:165–175. [DOI] [PubMed] [Google Scholar]
- 46. Licht K., Kapoor U., Amman F., Picardi E., Martin D., Bajad P., Jantsch M.F.. A high resolution A-to-I editing map in the mouse identifies editing events controlled by pre-mRNA splicing. Genome Res. 2019; 29:1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lundin E., Wu C., Widmark A., Behm M., Hjerling-Leffler J., Daniel C., Öhman M., Nilsson M.. Spatiotemporal mapping of RNA editing in the developing mouse brain using in situ sequencing reveals regional and cell-type-specific regulation. BMC Biol. 2020; 18:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salpietro V., Dixon C.L., Guo H., Bello O.D., Vandrovcova J., Efthymiou S., Maroofian R., Heimer G., Burglen L., Valence S.et al.. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun. 2019; 10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chew L.J., Yuan X., Scherer S.E., Qie L., Huang F., Hayes W.P., Gallo V.. Characterization of the rat GRIK5 Kainate receptor subunit gene promoter and its intragenic regions involved in neural cell specificity. J. Biol. Chem. 2001; 276:42162–42171. [DOI] [PubMed] [Google Scholar]
- 51. Wu D., Zang Y.-Y., Shi Y.-Y., Ye C., Cai W.-M., Tang X.-H., Zhao L., Liu Y., Gan Z., Chen G.et al.. Distant coupling between RNA editing and alternative splicing of the osmosensitive cation channel Tmem63b. J. Biol. Chem. 2020; 295:18199–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schulz R., McCole R.B., Woodfine K., Wood A.J., Chahal M., Monk D., Moore G.E., Oakey R.J.. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum. Mol. Genet. 2009; 18:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pachernegg S., Münster Y., Muth-Köhne E., Fuhrmann G., Hollmann M.. GluA2 is rapidly edited at the Q/R site during neural differentiation in vitro. Front. Cell. Neurosci. 2015; 9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wen W., Lin C.-Y., Niu L.. R/G editing in GluA2Rflop modulates the functional difference between GluA1 flip and flop variants in GluA1/2R heteromeric channels. Sci. Rep. 2017; 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stickels R.R., Murray E., Kumar P., Li J., Marshall J.L., Di Bella D.J., Arlotta P., Macosko E.Z., Chen F.. Highly sensitive spatial transcriptomics at near-cellular resolution with slide-seqV2. Nat. Biotechnol. 2021; 39:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ortiz C., Navarro J.F., Jurek A., Märtin A., Lundeberg J., Meletis K.. Molecular atlas of the adult mouse brain. Sci. Adv. 2020; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data have been deposited in Gene Expression Omnibus under accession number GSE153859. CARTANA ISS data and all custom software used have been deposited in Zenodo (https://doi.org/10.5281/zenodo.7664367). R figures and analysis scripts are also accessible at the same address. Seurat object .rds files for the three samples are available as supplementary files within the GEO serie and interactively browsable in the dedicated SiTx R Shiny application accessible at https://www.isomics.eu.