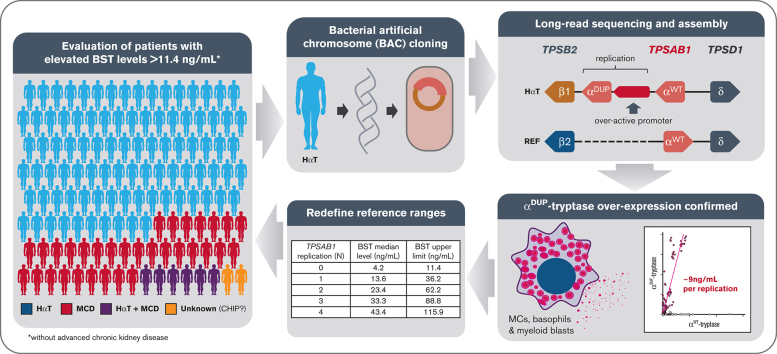
Key Points
-
•
Characterizing genetic regulation of tryptase expression redefines clinical laboratory reference ranges based on TPSAB1 replication number.
-
•
Individualized reference values for serum tryptase change and improve the utility of this biomarker in the diagnosis of myeloid neoplasms.
Visual Abstract
Abstract
Serum tryptase is a biomarker used to aid in the identification of certain myeloid neoplasms, most notably systemic mastocytosis, where basal serum tryptase (BST) levels >20 ng/mL are a minor criterion for diagnosis. Although clonal myeloid neoplasms are rare, the common cause for elevated BST levels is the genetic trait hereditary α-tryptasemia (HαT) caused by increased germline TPSAB1 copy number. To date, the precise structural variation and mechanism(s) underlying elevated BST in HαT and the general clinical utility of tryptase genotyping, remain undefined. Through cloning, long-read sequencing, and assembling of the human tryptase locus from an individual with HαT, and validating our findings in vitro and in silico, we demonstrate that BST elevations arise from overexpression of replicated TPSAB1 loci encoding canonical α-tryptase protein owing to coinheritance of a linked overactive promoter element. Modeling BST levels based on TPSAB1 replication number, we generate new individualized clinical reference values for the upper limit of normal. Using this personalized laboratory medicine approach, we demonstrate the clinical utility of tryptase genotyping, finding that in the absence of HαT, BST levels >11.4 ng/mL frequently identify indolent clonal mast cell disease. Moreover, substantial BST elevations (eg, >100 ng/mL), which would ordinarily prompt bone marrow biopsy, can result from TPSAB1 replications alone and thus be within normal limits for certain individuals with HαT.
Introduction
The tryptase locus is structurally complex in humans (Table 1). Approximately two-thirds of people have α-tryptase, encoded at TPSAB1 on 1 or both alleles, whereas everyone has β-tryptase encoded at 1 or both TPSB2 alleles, as well as at non–α-tryptase–encoding TPSAB1 loci.1, 2, 3 Copy number gain and loss of tryptase gene sequences encoding both α- and β-tryptases have also been reported.4 However, the structures of such copy number variants remain unknown. The most common copy number variants observed among Western populations are TPSAB1 gene replications encoding α-tryptase, a genetic trait known as hereditary α-tryptasemia (HαT).5 HαT is inherited in an autosomal dominant pattern and has been shown to affect nearly 6% of the general population in the United States, the United Kingdom, and Europe.6, 7, 8, 9 De novo replications have not been reported. TPSAB1 replications are associated with elevated basal serum tryptase (BST) levels of at least two- to threefold higher than median BST levels of healthy individuals without HαT.10 Although it has been hypothesized that additional TPSAB1 copies identified in HαT are present within the tryptase locus at chromosome 16p13.3, and that overexpression of α-tryptase yields the disproportionate increases in BST levels seen relative to copy number,5 this has never been demonstrated. Moreover, because only a small region of TPSAB1 has been probed to demonstrate gene replication,5,11 it remains unknown whether extra-allelic copies of TPSAB1 encode canonical or novel sequences containing homology with α-tryptase at the probe site.
Table 1.
Assemblies of the human tryptase locus at 16p13.3
|
TPSB2 |
TPSAB1REP |
TPSAB1 |
TPSD1 |
Corresponding assembly | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Orientation | Isoform | Haplotype | Orientation | Isoform | Haplotype | Orientation | Isoform | Haplotype | Orientation | |
| CACCT | REV | β3 | — | — | — | GACCC | FWD | β1 | GGTTC | FWD | GRCh37/hg19 |
| — | — | — | — | — | — | GACCC | REV | β1 | GGTTC | FWD | AC226137.3 |
| CACCT | REV | β3 | — | — | — | GACCC | FWD | β1 | CGTTC | FWD |
AC120498.2 CTD-2503P16 |
| CACCT | REV | β3 | — | — | — | GACCC | REV | β1 | CGTTC | FWD | CHM13 AL031704.24 |
| CACCT | REV | β3 | — | — | — | CACCT | REV | β3 | GGTTC | FWD | AE006466.1 |
| GACCT | REV | β2 | — | — | — | GGTTC | FWD | α | GGTTC | FWD | CHM1 AC240106.3 AC238650.2 AC213746.1 |
| GACCT | REV | β2 | — | — | — | GGTTC | REV | α | GGTTG | FWD | GM24385 |
| GACCT | REV | β2 | — | — | — | GGTTC | REV | α | GGTTC | FWD | NA12878 |
| GACCC | REV | β1 | CACCT | REV | α | GGTTC | REV | α | GGTTC | FWD | HαT BAC |
| — | — | — | — | — | — | GGTTC | — | α | — | — | AF098328.1 |
Assembly of the locus from an individual with HαT is indicated in bold font.
FWD, forward; REV, reverse.
Although HαT is the most common heritable cause for elevated BST, the clonal mast cell disorder, systemic mastocytosis (SM), is a common acquired cause,10,12 and BST levels are used routinely as a biomarker to screen for this disorder. Currently, the World Health Organization criteria define a BST level >20 ng/mL as a minor criterion for the clinical diagnosis of SM,13 and this cutoff is frequently used in clinical decision making as an indication for bone marrow (BM) biopsy in symptomatic individuals.14 However, ∼1 of every 4 individuals with HαT has a BST level >20 ng/mL, representing an estimated 7.5 million people in the United States alone.4,15 Because HαT has been shown to augment immediate hypersensitivity symptoms in a number of conditions,7,8 many of these individuals are likely to undergo unwarranted invasive workup for SM, including BM biopsy, if tryptase genotype is not taken into account. Conversely, because HαT appears to account for most elevations in BST, observed elevations in patients who do not have HαT could be considerably enriched for other pathologies, most notably myeloid neoplasms, warranting workup when such elevations in BST may be modest (ie, <20 ng/mL).
To address these questions, we cloned, long-read sequenced, and assembled the human tryptase locus containing a TPSAB1 replication, finding the sequence to encode canonical α-tryptase protein. However, a series of unique proximal noncoding variants were also identified that distinguished duplicated α (αDUP)–tryptase from nonreplicated wild-type α (αWT)–tryptase sequences. An expanded DNA motif within the 5′ untranslated region (UTR) was also linked to TPSAB1 replication–associated variants and when cloned, demonstrated increased in vitro promoter activity relative to the paralogous region in the nonreplicated promoter. Using in vitro and in silico analyses of RNA sequences, we confirmed the relative overexpression of αDUP-tryptase sequences in primary basophils, cultured mast cells, and publicly available RNA sequence data sets. Applying this knowledge, we generated a new genetic-based model for BST clinical reference ranges based on TPSAB1 replication number and examined the potential real-world impact of coupling tryptase genotyping with these newly defined values in the work up of patients with clonal mast cell disorders. The potentially practice-changing implications for identification of indolent clonal mast cell disease (MCD), the diagnosis of SM, and the elimination of unnecessary BM biopsies are discussed herein.
Methods
Study participants and samples
Patients, family members, and healthy volunteers provided informed consent on institutional review board (IRB)–approved research protocols led by investigators, with expertise in allergy/immunology at institutions that specialize in clonal and nonclonal mast cell disorders designed to study mastocytosis, or genetic diseases affecting the immune system at the National Institutes of Health Clinical Center (#NCT00852943, #NCT01164241, #NCT00044122, #NCT00001756, and #NCT007197190), Antwerp University Hospital, Belgium (B300201525454), Verona University Hospital, Italy (protocol number 39620), the University of Florida (IRB 201702274), the University of Mississippi Medical Center (IRB 2019-0082), or University Clinic Golnik, Slovenia (KME 150/09/13). All study participants had BST levels measured and tryptase genotyping performed (N = 1178). Among participants referred for evaluation or diagnosed with a mast cell–associated disorder (n = 575), complete history and physical examinations were performed.
BM biopsy and aspirate
Individuals presenting with signs or symptoms suggestive of a clonal MCD13 had additional clinical workup that included BM biopsy and aspirate. Immunohistochemistry of BM sections was performed for enumeration and characterization of mast cells (KIT and tryptase) and evaluation of CD2 and CD25 expression in aspirate and/or tissue section. Allele-specific polymerase chain reaction (PCR) for KIT p.D816V in peripheral blood and BM was also performed in these patients. In the rare case that an individual declined BM biopsy, peripheral blood KIT p.D816V was performed by allele-specific or droplet digital PCR (ddPCR). If the result was positive, these individuals were considered to have clonal MCD and included. Whereas, if KIT p.D816V screening of peripheral blood was negative in an individual with an incomplete workup for a suspected myeloid neoplasm, such individuals were excluded from the study.
Total BST quantification
Total BST levels were measured using the commercially available ImmunCAP assay (Thermo Fisher Scientific, Phadia AB, Uppsala, Sweden) or enzyme-linked immunosorbent assay as described,16 performed in Clinical Laboratory Improvement Amendments–certified laboratories (Mayo Clinic, Rochester, NY and Virginia Commonwealth University, Richmond, VA).
Bacterial artificial chromosome (BAC) library generation
Genomic DNA was isolated from peripheral blood mononuclear cells (PBMCs) of an individual homozygous for ααβ-tryptase alleles. High-molecular-weight DNA fragments were then partially digested with the restriction enzyme BamHI and size-selected before ligation of fragments into the pCC1BAC vector (Epicentre Biotechnologies, Madison, WI) and transformation of DH10B Escherichia coli cells.17,18 The clones were robotically chosen and replicated. Replicated clone copies were used as a source plate for constructing nylon filters. In-filter hybridization with digoxigenin-labeled probes was carried as previously described19 (Amplicon Express, Pullman, WA). Verification of putative clones was carried out by conventional and ddPCR.
BAC sequencing and assembly
Genomic DNA from selected clones was isolated20 and sequenced using single-molecule real-time (SMRT) sequencing technology (PacBio, Menlo Park, CA) at DNA Link (South Korea). De novo assembly was accomplished using HGAP2 and the Canu assembler for PacBio on SMRT Analysis platform version 2.2.0, SMRTPortal:SMRTAnalysis build 133377, Daemon version 2.2.0 build 132105, SMRTpipe version 2.2.0 build 132739, SMRT Portal version 2.2.0 build 133335, and SMRT View version 2.2.0 build 132578.
Bioinformatic analyses
Paired-read tryptase haplotype analysis. Genome sequence reads mapping to the tryptase locus were realigned to a 142–base pair (bp) region (supplemental Table 1). Detailed alignments for each read were parsed to extract the sequence at each of the 5 specified variable positions if covered by the read. Haplotype assignments were determined based on the collection of positional sequence assignments determined for each read pair. Generated haplotypes were then correlated to tryptase genotype and linkage to inherited alleles was confirmed when pedigrees were available.
RNA sequencing data set analyses
Using a 39-bp consensus sequence (supplemental Table 2), gene expression data sets (see “Accession codes” for list of data sets) were reanalyzed using National Center for Biotechnology Information (NCBI) Magic-BLAST (https://ncbi.github.io/magicblast/), to map reads to 3 gene isoforms: αDUP-, αWT-, and β-tryptase. Samples were designated as tryptase-positive, tryptase-negative, αDUP-, αWT-, and β-tryptase based only on the criteria of having qualified reads that mapped exactly without any mismatches to the interval between 25 and 45 bp from the 5′ end of the transcripts (see supplemental Methods).
Promoter amplification and cloning
Promoter amplicons were generated from genomic DNA from individuals with increased TPSAB1 copy number, using primers designed on conserved sequences present in all identified α- and β-tryptase sequences (forward: GGGCAAGTCCACAGGGAGCT; reverse: CTGGGGAGCAAGGAGGAGCA), to amplify all sequences between the ATG start site and a conserved region ∼1 to 2 kb 5′ of the variably expanded repeat region (Figure 1). Amplification was confirmed by gel electrophoresis, and clones of products were generated using the TOPO Cloning Kit (Thermo Fisher Scientific, Waltham, MA) and transformed into One Shot TOP10 Chemically Competent E. coli (Thermo Fisher Scientific). Single colonies were selected, confirmed to contain single intact clones by PCR and gel electrophoresis, and Sanger sequenced using the forward and reverse, as well as 2 additional internal primers (TGCAGGTGCAACCCCAGGA and TCCTGGGGTTGCACCTGCA).
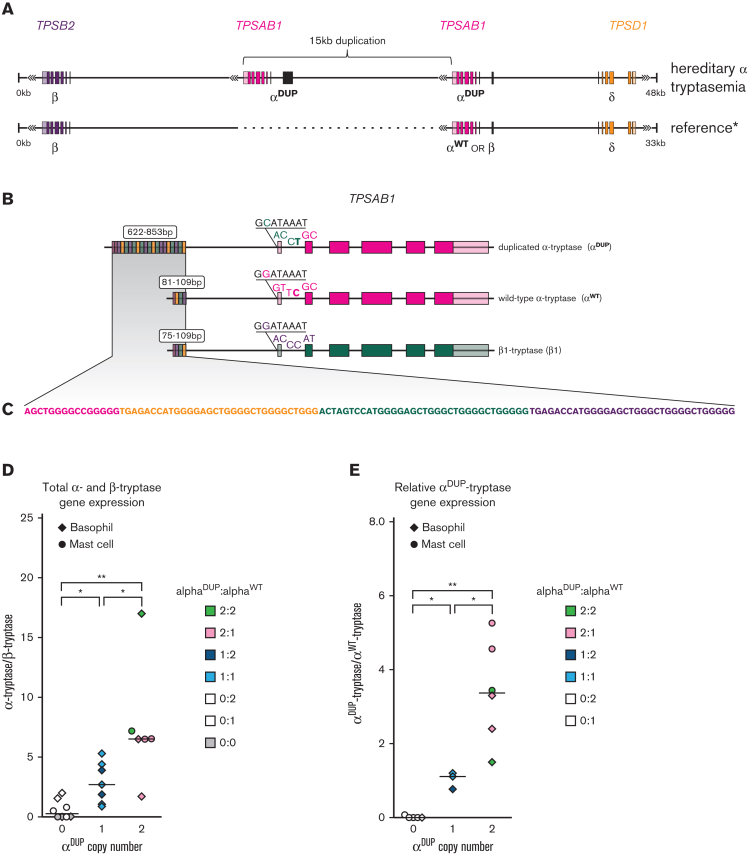
Figure 1.
TPSAB1 duplications occur at 16p13.3 and are linked to an expanded promoter associated with αDUP-tryptase overexpression. (A) Location and orientation of the 15-kb tandem duplication of TPSAB1. (B) Alignment of αDUP-, αWT-, and β-tryptases with unique 5′ variants and size of promoter repeat regions indicated; (C) repeat motif within promoter regions. Relative total α- to β-tryptase gene expression (D) and αDUP-tryptase relative to αWT-tryptase gene expression (E) in ex vivo basophils and cultured primary mast cells. ∗P < .05; ∗∗P < .005.
For αDUP- and αWT-tryptase–specific promoter cloning, the reverse primer (GACGATACCCGCTTGCTGCAG) was located across α-tryptase–specific exonic sequence used for the isoform-specific genotyping assay; for αWT-tryptase the universal forward primer was used; and for αDUP-tryptase a different sequence normally only seen linked to β-tryptase was used (GGGCAAGTCCACAGGGAGCT). Resulting amplicon size was confirmed by gel electrophoresis (supplemental Figure 2).
Reporter assay
Sequence-validated clones corresponding to αDUP- and αWT-tryptase promoters identified in the BAC assembly were subcloned into the reporter plasmid pDD-AmCyan1 (Takara Bio USA, Inc, Mountain View, CA) and verified by Sanger sequencing. MonoMac-6 cells were transfected with the reporter plasmid pDD-AmCyan1 containing αDUP- or αWT-tryptase promoter clones by electroporation using Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) using the setting U-005 according to manufacturer’s instructions. Cells were cultured in standard media in the presence of Shield1 (Takara Bio) according to manufacturer’s instructions, and basal fluorescence was measured at indicated time points and recorded using an LSR Fortessa (BD Biosciences) and analyzed using FlowJo (Treestar, Ashland, OR).
Basophil isolation
Following isolation of PBMC via density centrifugation, basophils were negatively selected using the MACS Diamond Basophil Isolation Kit (Miltenyi Biotec, San Diego, CA).
Cell culture
Primary mast cells. PBMCs were isolated using density centrifugation and CD34+ cells were positively selected using the ferromagnetic bead-based MACS system (Miltenyi), and the number of cells was quantified. Mast cells were then differentiated under the established conditions described.21
Cell lines. MonoMac-6 cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum, 2 mM L-glutamine, nonessential amino acids, 1 mM sodium pyruvate, and 10 μg/mL human insulin.
ddPCR
Tryptase genotyping. α- and β-tryptase sequences at TPSAB1 and TPSB2 were genotyped using ddPCR as previously described5. Briefly, genomic DNA was extracted from PBMCs, cell lines, or obtained from the HapMap biorepository and restriction endonuclease–treated. Custom primer/probe sets specific for α- and β-tryptase were then employed using the PrimePCR ddPCR copy number reference AP3B1, according to the manufacturer's specifications (Bio-Rad, Hercules, CA).
Gene expression. Using Trizol reagent (Thermo Fisher Scientific) and the RNeasy Mini Kit (Qiagen) total RNA was extracted from basophils, primary cultured mast cells, or cell lines under stated conditions and reverse-transcribed using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). ddPCR was performed on a QX200 system (Bio-Rad) according to the manufacturer’s instructions using custom primer/probe sets for tryptase isoforms as indicated (supplemental Table 3), to quantify gene expression, transcripts were normalized to HPRT1 and TBP expression, which was quantified using commercially available ddPCR primer/probe sets (Bio-Rad).
Promoter detection assay. A custom primer/probe set was designed to amplify the proximal promoter linked to α-tryptase (supplemental Figure 4). The forward primer corresponded to a conserved sequence present in all identified α- and β-tryptases. The reverse primer hybridized to sequences only present in α-tryptases. The probes competed for hybridization with the C>T variant that destinguished αDUP- from αWT-tryptase–specific sequences. To successfully amplify the large 71% GC–rich amplicon, the reaction was run with 10% 1 M Betaine (Sigma, St. Louis, MO) under the following conditions: 95°C 10 minutes, 96°C 30 seconds, 66°C 1 minute (ramp 1.5°C), 50 cycles, 96°C 10 minutes.
Exome sequencing and bioinformatic analysis for clonal variants
BM aspirates were fractionated using density centrifugation into granulocyte and mononuclear fractions using a double gradient: 1.077 g/mL (Histopaque-1077) and 1.119 g/mL (Histopaque-1119) (Sigma, St. Louis, MO). Genomic DNA was extracted, and libraries were prepared with the Twist Biosciences Comprehensive Exome capture kit. Deep (>300×) exome sequencing was subsequently performed using the Illumina HiSeq 2500, on both the granulocyte and mononuclear BM fractions. Paired peripheral blood samples from the same individuals were also exome sequenced consistent with clinically recommended standards.22
Raw FastQ files were trimmed for quality and adapter contamination using Trimmomatic version 0.3923 and mapped to the human hg38 reference genome using Burrows-Wheeler Aligner-MEM version 0.7.17 (http://bio-bwa.sourceforge.net/). PCR duplicates were marked using Samlaster version 0.1.2.5,24 and Genome Analysis Toolkit (GATK) version 4.1.9.0 was used to perform base recalibration. For the peripheral blood samples, germ line variation was called using GATK version 4.1.9.0, following the recommended Best Practices (https://gatk.broadinstitute.org/hc/en-us/articles/360035535932-Germline-short-variant-discovery-SNPs-Indels-). For granulocyte and mononuclear fractions, somatic variants were called using 2 approaches. First, paired somatic calling with the PBMC sample treated as germ line was performed using 3 somatic detection tools: (1) MuTect2 from GATK version 4.1.9.0, following the recommended Best Practices, (2) Strelka version 2.9.0,25 and (3) MuTect version 1.1.7.26 In addition to paired calling, we also performed tumor-only calling with a combination of MuTect2 GATK version 4.1.9.0, MuTect version 1.1.7, and Vardict version 1.7.0.27 The latter approach of performing somatic variant detection without including the paired PBMC sample was done to account for the fact that somatic variants in the BM granulocytes and BM mononuclear cells could also be present at low fractions in the PBMC sample, and therefore, be excluded as contaminating germ line variants in the paired calling. For both approaches, variants were then merged across all callers and annotated with VEP version 10428 for downstream analysis. Reported variants were prioritized based on commonly implicated genes in the development of myeloid neoplasms.
Statistical modeling and analyses
To predict BST levels based on tryptase genotypes, specifically determined by TPSAB1 replication number (including those without replications), a log transformation was applied to the BST levels because the values were not normally distributed. Additionally, we log transformed the replication number, because transforming BST back to the normal scale resulted in a model fit that was approximately linear, with a near constant increase in BST for each increase in replication number; this supported results from early clinical data and data generated from in silico and in vitro experiments. We also added 0.5 to replication number before log transformation to include individuals without TPSAB1 replications in the model, thus, allowing us to also determine a cutoff for individuals without HαT. Thus, we used a linear regression model to predict log(BST) from log(replication number + 0.5), and from that, created an upper 1-sided 99.5% prediction interval. The 99.5% threshold was chosen as it provided high specificity, with very few individuals identified as false positives, and it correlated well with the predicted prevalence of elevated BST in the absence of HαT based on available population data.10 The model was developed using R version 3.6.3.
For clinical and experimental data, Mann-Whitney, Kruskal-Wallis, and paired and unpaired 2-tailed t tests were used where appropriate to test significance of differences, prevalence, or deviation, using Prism (GraphPad).
BST calculator code
We developed an online calculator, the Basal Serum Tryptase Clinical cutoff Assigned by Locus Copy number of UTR-Linked element and Associated TPSAB1 Encoded Replication (BST CALCULATER) with Shiny R framework, https://bst-calculater.niaid.nih.gov/. The code is available at https://github.com/niaid/BST-calculater.
Results
Increased TPSAB1 copy number occurs at the tryptase locus and encodes canonical α-tryptase protein
A BAC library was generated from an individual homozygous for TPSAB1 duplications (genotype αα/β:αα/β) and a clone containing the complete human tryptase locus was identified. Using SMRT technology, we sequenced and subsequently assembled the locus de novo (Figure 1A), allowing study of the structure and sequence of this variant-containing allele, to the best of our knowledge, for the first time. As anticipated, the assembly displayed marked structural dissimilarity with the reference human genome (GRCh37/hg19) but did not contain novel sequence (supplemental Figure 1, and online data repository). The additional TPSAB1 copy encoding α-tryptase (αDUP) was located within the tryptase locus mapping to 16p13.3, between TPSB2 and the nonreplicated TPSAB1 locus, as had been suggested based on prior in vitro experiments.5 The coding sequence was identical to that of the adjacent α-tryptase sequence present at the WT locus (αWT) (supplemental Table 4; online repository), and both sequences were in reverse orientation.
The sequence at TPSB2 in our BAC clone was identified as β1-tryptase. Although β1-tryptase at TPSB2 is uncommon, it has been reported in ∼5% of individuals screened from the HapMap cohort.29 Given that this prevalence was comparable to the prevalence of HαT reported in Western populations, we obtained the HapMap samples reported to contain β1-tryptase at TPSB2 to determine whether the donors were all individuals with HαT. However, only 4 of 15 of these individuals (27%) with β1-tryptase at TPSB2 were found to have HαT. Thus, β1-tryptase at TPSB2 is not exclusive to HαT.
TPSAB1 replications encoding αDUP-tryptase are linked to an expanded promoter
Elevated BST levels among patients with HαT have been shown to result from increased constitutive release of protryptases.5,30 To determine whether this increase results from overexpression of αDUP-tryptase transcripts, we compared noncoding sequences at the 2 TPSAB1 loci and identified an expanded (∼800 bp) repetitive DNA motif approximately 1 kb 5′ of the consensus transcription start site for αDUP-tryptase (Figure 1B). This sequence was ∼700 bp longer than the paralogous region of the αWT promoter containing 27 repeated compared to 3 repeated sequences upstream of αWT (Figure 1C). Repeat regions at the same 5′ location were also observed in publicly available assemblies of α-tryptase sequences and confirmed by gel electrophoresis in amplified αWT-tryptase and αDUP-tryptase promoters from 9 unrelated individuals with HαT (supplemental Figure 2). Subsequent cloning and Sanger sequencing of 3 additional αW- and αDUP-tryptase promoters confirmed these relative differences (supplemental Figure 3). Together these data demonstrate an expanded DNA repeat motif in the promoter of TPSAB1 replications encoding αDUP-tryptase that are consistently coinherited together.
Increased basal αDUP-tryptase expression is associated with elevated basal promoter activity
To quantify the functional outcome of the expanded promoter region linked to αDUP-tryptase sequences, we interrogated total α- and β-tryptase as well as αWT- and αDUP-tryptase transcript expression arising from TPSAB1 and TPSB2. Cultured primary human mast cells and isolated basophils from individuals with HαT expressed higher levels of total α-tryptase transcripts, and overexpressed αDUP-tryptase sequences relative to other tryptase isoforms (Figure 1D-E). Importantly, total α-tryptase gene expression levels were several orders of magnitude higher than those of β-tryptases, even when they were present in allelic balance (eg, 2α,2β vs 3α,3β).
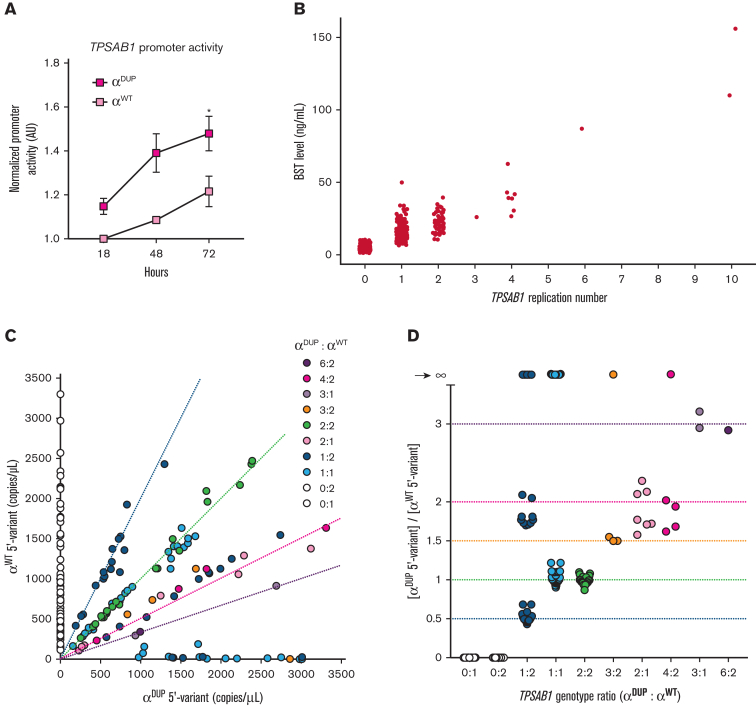
To determine whether the expanded 5′ sequences uniquely associated with αDUP-tryptase contributed to the relative overexpression of αDUP- to αWT-tryptase, we cloned the promoters containing these paralogous regions into fluorescent reporter plasmids using our BAC assembly and other SMRT-based de novo assemblies of the locus as sequence references (see Methods for referenced assemblies). These constructs were sequence verified and transfected into MonoMac-6 cells, an acute monocytic leukemia line that expresses tryptases.31,32 The expanded αDUP-tryptase promoter demonstrated significantly greater basal activity relative to the αWT-tryptase promoter (Figure 2A). This in vitro finding correlated directly with BST levels among individuals with HαT, where the absolute number of TPSAB1 replications was found to best correlate with BST levels regardless of tryptase genotype derived from TPSAB1 and TPSB2 (Figure 2B). Therefore, elevated BST in HαT results from overexpression of αDUP-tryptase at replicated TPSAB1 loci linked to a promoter with increased basal activity.
Figure 2.
The expanded promoter at duplicated TPSAB1 is conserved and has increased basal activity. (A) Normalized fluorescent reporter activity in unstimulated MonoMac-6 cells transfected with promoters cloned from endogenous (αWT) or replicated (αDUP) TPSAB1 loci. (B) BST from individuals grouped by TPSAB1 replication number. (C-D) Relative allelic frequency (C) and ratios (D) of αWT-tryptase (y-axis) and αDUP-tryptase (x-axis) associated 5′ variants determined by ddPCR. Dashed lines indicate predicted αWT:αDUP ratios by genotype. ∗P < .05.
A unique haplotype demonstrates conservation of the identified overactive promoter at replicated TPSAB1 loci
To examine the generalizability of our finding of an expanded promoter linked to αDUP-tryptase at TPSAB1, we again examined proximal noncoding sequences associated with different tryptase isoforms and identified 5 unique 5′ variants: 2 substitutions in intron 1, 2 in the 5′ UTR, and 1 in the proximal promoter (Figure 1B). Using these variants, we defined haplotypes, which, when combined with coding sequences, uniquely distinguished αDUP-tryptase (CACCT) from αWT-tryptase (GGTTC), β1-tryptase (GACCC), and δ-tryptase (GGTTC) in our BAC assembly (Figure 1B; see supplemental Table 1 for complete contextual sequences).
Next, we examined publicly available genome assemblies to determine if these haplotypes were conserved. Critically, TPSAB1 and TPSB2 are reportedly in nearly complete linkage disequilibrium and are coinherited in virtually all individuals such that major haplotypes exist and inheritance can be inferred.29,33 The major tryptase isoforms encoded at TPSAB1 are β1-tryptase followed by αWT-tryptase, and at TPSB2 are β3-tryptase and β2-tryptase, when on β/β and α/β alleles respectively. As expected, we observed GACCC linked to β1-tryptase at TPSB2 on β1/β3 (β/β) alleles, and GGTTC linked to αWT-tryptase sequences at TPSAB1 on αWT/β2 (α/β) alleles in all 13 queried assemblies as well as in the GRCh37/hg19 reference assembly (Table 1). Additional isoform-specific haplotypes at these 2 loci, GACCT linked to β2-tryptase at the TPSB2 locus only when αWT/β2 (α/β) alleles were present, and CACCT linked to β3-tryptases at the TPSB2 locus only when β1/β3 (β/β) alleles were present, were also identified. Neither the αDUP-tryptase–specific promoter expansion nor the linked CACCT haplotype were observed in association with αWT-tryptase sequences in any of the 7 assemblies containing αWT/β2 (α/β) (Table 1).
Using these haplotypes, we reexamined genome sequence reads from 183 individuals who lacked increased TPSAB1 copy number, and from 32 affected individuals from 10 families with HαT.5 Using a paired-read approach, we found that GACCC (β1-tryptase) and CACCT (β3-tryptase) were present in association with 324 of 331 individuals (98%) with β/β alleles. Moreover, the CACCT haplotype consistently segregated with the β/β haplotype in the 16 families without HαT in whom tryptase sequence inheritance could be examined. Although all α/β haplotypes (n = 6) were associated with the αWT-tryptase–linked GGTTC and β2-tryptase–linked GACCT (Table 2), the CACCT haplotype linked to αDUP-tryptase in our BAC clone, segregated universally with TPSAB1 replications in all 32 individuals with HαT. The GACCC haplotype, linked to β1-tryptase most often present at TPSAB1 but present at TPSB2 in our BAC clone, was also found to be universally present among individuals with HαT and cosegregated with αDUP-tryptase–containing alleles suggesting that β1-tryptase at TPSB2 is frequently present on alleles containing TPSAB1 replications, as seen in our BAC clone (Table 2). Therefore, these in silico findings suggest conservation of the unique promoter haplotypes we identified in our BAC assembly, and that overexpression of αDUP-tryptase could be a generalizable phenomenon.
Table 2.
Haplotypes identified in linkage with allelic tryptase genotypes
|
TPSB2 |
TPSAB1DUP |
TPSAB1 |
TPSD1 |
Allele count | Allelic genotype | Haplotype frequency (by genotype), % | |||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Isoform | Haplotype | Isoform | Haplotype | Isoform | Haplotype | |||
| CACCT | β3 | — | — | GACCC | β1 | GGTTC | 172 | β/β | 52 |
| CACCT | β3 | — | — | GACCC | β1 | CGTTC | 152 | β/β | 46 |
| GACCT | β2 | — | — | GACCC | β1 | GGTTC | 7 | β/β | 2 |
| GACCT | β2 | — | — | GGTTC | α | GGTTC | 65 | α/β | 96 |
| GACCT | β2 | — | — | GGTTC | α | CGTTC | 3 | α/β | 4 |
| GACCC | β1 | CACCT | α | GGTTC | α | GGTTC | 15 | αα/β | 63 |
| GACCC | β1 | CACCT | α | - - - - T | α | GGTTC | 5 | αα/β | 21 |
| GACCC | β1 | CACCT | α | GGTT (-6delC)C∗ | α | GGTTC | 2 | αα/β | 8 |
| GACCC | β1 | CACCT | α | GGTTT | α | GGTTC | 2 | αα/β | 8 |
A single cytosine (C) base deletion was identified 6 bases before (−6) the C terminal of the haplotype.
To confirm these findings at scale in vitro, we had to overcome the repetitive and GC-rich nature of the region. To do so, we developed a ddPCR assay capable of detecting the most proximal variant that distinguished the αDUP-tryptase promoter haplotype (CACCT) from that of αWT-tryptase (GGTTC) (see supplemental Figure 4 for description and representative data). The corresponding variant listed in the reference genome (GRCh37/hg19 16:1290818C>T) has only been reported in association with TPSB2, ostensibly linked to dominant isoform β3-tryptase at this locus. However, the reverse primer in our assay was designed to only hybridize with α-tryptase sequences.
The CACCT variant was confirmed to be universally present in linkage with αDUP-tryptase in 120 affected individuals with increased TPSAB1 copy number from 101 families. Conversely, CACCT was never found in linkage with αWT-tryptase in 81 unaffected individuals from 69 families who did not have increased TPSAB1 copy number but carried 1 or 2 αWT-tryptase–encoding copies (Figure 2C). Furthermore, the ratios of identified promoter copy number confirmed that all 166 HαT-associated αDUP-tryptase sequences on 156 alleles were linked to the C>T variant (Figure 2D) and by inference, the expanded promoter.
Interestingly, in 30 of 120 individuals with HαT, αWT-tryptase was not linked to GGTTC; 10 of these αWT-tryptase promoters appeared to contain the C>T missense, and the remaining 19 alleles may have had this or another unknown missense variant or indel, the latter of which may have resulted in failed probe or primer hybridization. Importantly, our in silico analysis identified a GGTTT haplotype linked to αWT-tryptase in 1 family with HαT. In a second family, a single base pair deletion in the −6 position from C>T was observed, which we have labeled GGTT(−6delC)C (Table 2). This deletion interfered with probe hybridization in our assay, potentially accounting for some of the 19 αWT-tryptase–containing alleles with ambiguous promoter linkage. The αWT-tryptase promoter from these samples also failed to amplify and was not successfully cloned, suggesting yet another possible αWT-tryptase promoter may exist. Indeed, public database interrogation yielded at least 6 different α-tryptase sequences exhibiting a high degree of homology (supplemental Table 4). Regardless, αWT-tryptase–containing alleles with undefined promoter haplotypes, or even those linked to the C>T intronic variant, were not associated BST differences in vivo (supplemental Figure 5), indicating that these undefined promoters are not associated with differences in gene expression at linked αWT-tryptase–encoding TPSAB1 loci. Collectively, these data support conservation of a unique haplotype linked to an overactive promoter element at replicated TPSAB1 loci that is associated with overexpression of α-tryptase by mast cells and basophils.
αDUP-tryptase transcripts are overexpressed in public data sets
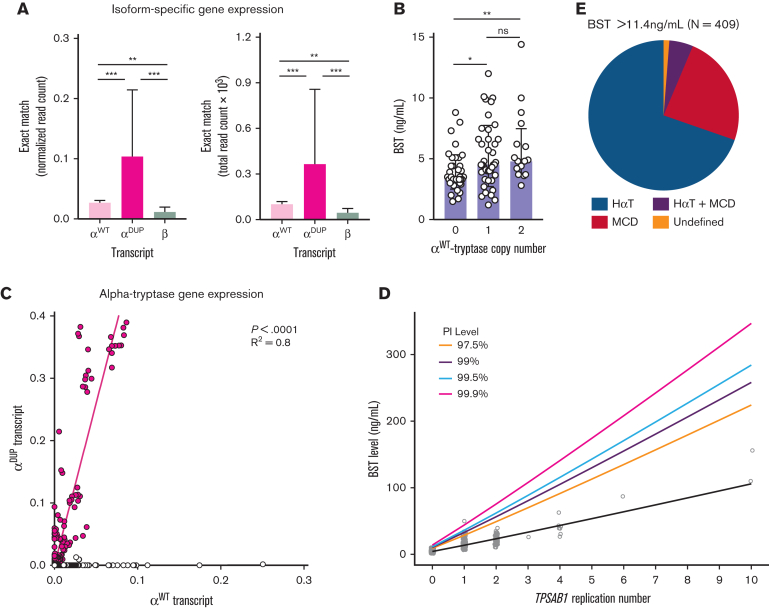
Using α- and β-tryptase isoform–specific coding variants and the uniquely linked proximal noncoding haplotypes, a 39-bp consensus sequence was defined, which contained 4 variants that could distinguish αWT-, αDUP-, and β-tryptase transcripts; given the proximity of the first variant to the transcriptional start site, only the distal 3 variants were used for read alignment (Figure 1B; supplemental Table 2). Querying 58 non–disease-associated transcriptome data sets, 863 of 4160 samples were identified with exact transcript matches to ≥1 of the 3 defined 39-bp consensus sequences. αDUP-tryptase was observed in 25 of 863 individuals (3%), and αWT-tryptase transcripts were identified in 289 of 863 individuals (34%) (supplemental Table 5). Because linked metadata were limited, the race and ethnicities of these samples are unknown. Although White individuals are likely to be overrepresented in these samples, other ethnicities and racial groups are likely included, which may explain the lower-than-expected prevalence of HαT (3% vs 5.6%).
Using the same method, we examined the expression levels of αWT-, αDUP-, and β-tryptase transcripts in these and other public data sets. This analysis confirmed overexpression of both αWT- and αDUP-tryptase transcripts relative to β-tryptase (Figure 3A). Although overexpression of αWT-tryptase was also observed in our primary gene expression data, it is possible that some δ-tryptase reads could align to the αWT-tryptase consensus sequence owing to haplotype sequence homology but not to αDUP-tryptase, where the linked haplotype is not observed in linkage with δ-tryptase. Therefore, misalignment of δ-tryptase reads could lead to a potential overestimation of αWT-tryptase expression levels. To exclude this possibility, we examined BST levels, which represent only total secreted α- and β-tryptases, among individuals without HαT. As reported,34 we saw a modest but significant positive correlation between BST levels and increasing αWT-tryptase copy number at TPSAB1 (Figure 3B). Thus, consistent with the RNA sequence read alignment results, αWT does appear to be modestly overexpressed relative to β-tryptases. Because of the variable levels of total tryptase transcript in any given sample, we employed regression analysis to quantify more precisely the relative expression of αDUP-tryptase to αWT-tryptase. Consistent with all our prior data, αDUP-tryptase was found to be overexpressed relative to αWT-tryptase by a factor of 5.2 fold (95% confidence interval, 4.864-5.499) in these data sets (R2 = 0.8, P < .0001) (Figure 3C). Thus, in silico, in vitro, and in vivo findings collectively indicate that elevated BST in HαT results from basal overexpression of canonical α-tryptase sequence at replicated TPSAB1 loci linked to coinherited overactive promoter elements.
Figure 3.
Modeling serum tryptase levels based on genotype improves clinical utility. (A) Normalized (left) and total (right) read counts for reads aligning exactly to the 39-bp consensus sequences that identify β-, αWT-, and αDUP-tryptase. (B) BST levels among individuals with conserved 4n tryptase copy number (combined from TPSAB1 and TPSB2). (C) Regression analysis of relative expression levels of αDUP-tryptase (y-axis) and αWT-tryptase (x-axis) transcripts. (D) Prediction intervals for BST levels based on TPSAB1 replication number. (E) Prevalence of HαT, clonal MCD, and those without either among individuals referred with BST levels above the predicted upper limit of normal (>11.4 ng/mL). ∗P < .01; ∗∗P < .005; ∗∗∗P < .0001.
Modeling BST levels based on TPSAB1 replication number redefines clinical reference ranges
Using this knowledge, we created a data-driven model from 204 individuals with normal TPSAB1 copy number and 309 individuals with HαT in whom there was no clinical evidence of clonal MCD, to generate prediction intervals with upper limits for predicted BST levels based on TPSAB1 replication number (Figure 3D), and created an online application for clinical use, the BST CALCULATER. These thresholds can be used to determine where a BM or further clinical evaluation may be needed for individuals with or without HαT when another clinical indication for such evaluation may be lacking.
This model not only redefined clinically meaningful upper limits for BST levels among individuals with HαT but it also, for the first time, defines a clinically actionable upper limit among individuals without HαT as 11.4 ng/mL (Table 3). Remarkably, this has been the upper limit of normal for BST commonly used in most clinical laboratories. Moreover, based on the data, this cutoff of 11.4 ng/mL would appear to be valid in defining a clinically abnormal elevated BST when considering the diagnosis of clonal MCD, as opposed to the currently accepted 20 ng/mL, provided that HαT has been ruled out by tryptase genotyping.
Table 3.
Measured and predicted BST levels based on TPSAB1 replication number encoding α-tryptase
| Primary data |
Modeled reference values |
||||
|---|---|---|---|---|---|
| TPSAB1 replication number | N | Median BST (ng/mL) | Standard deviation BST | Predicted BST (ng/mL) | Upper 99.5% prediction interval |
| 0 | 204 | 4.15 | 2.12 | 4.2831 | 11.3879 |
| 1 | 247 | 13.60 | 5.55 | 13.6363 | 36.2216 |
| 2 | 52 | 22.45 | 6.40 | 23.3642 | 62.1517 |
| 3 | 1 | 26.00 | Not applicable | 33.3110 | 88.7541 |
| 4 | 6 | 40.50 | 12.61 | 43.4148 | 115.8535 |
| 5 | — | — | — | 53.6420 | 143.3508 |
| 6 | 1 | 87.00 | Not applicable | 63.9708 | 171.1816 |
| 7 | — | — | — | 74.3862 | 199.3007 |
| 8 | — | — | — | 84.8774 | 227.6741 |
| 9 | — | — | — | 95.4357 | 256.2756 |
| 10 | 2 | 133.00 | 32.53 | 106.0544 | 285.0841 |
This model fits the primary data well (R2 = 0.76, P < .001), with only 1 individual being identified in the entire primary data set as outside the 99.5% threshold (Figure 3D; Table 3; supplemental Figure 6), and consistent with in vitro and in vivo data, demonstrated linearity on the log-transformed scale (supplemental Figure 6). Interestingly, this individual who did not conform to the model, had a provocative clinical phenotype, which included chronic spontaneous urticaria and angioedema as well as syncopal episodes with alcohol consumption that had prompted a BM biopsy before genotyping. Although BM sections demonstrated left-shifted megaloblastoid erythroid hyperplasia, focal areas with increased pronormoblasts, and occasional dyserythropoietic forms, allele-specific PCR for KIT p.D816V was negative, no mast cell aggregates or aberrant markers (ie, CD2 and CD25) were identified, and no definitive diagnosis was made.
Finally, to prospectively validate the modeled cutoff clinically, we measured BST levels in an additional 130 individuals with HαT caused by TPSAB1 duplications in whom BST levels were unknown and no other clinical indication for BM biopsy was present. In all 130 prospectively examined individuals, BST levels fell below the 99.5% cutoff (median, 15.1 ng/mL; range, 8.0-35.0 ng/mL).
Application of tryptase genotyping and modeled BST levels improves biomarker utility
To further examine the real-world implications of applying this new model clinically, we analyzed all samples sent or referred to the National Institutes of Health Clinical Center for tryptase genotyping from 2016 to 2021; 377 of these samples had not been used to construct the model. None of the participants had kidney disease or other clinical presentations consistent with genetic disorders known to be associated with elevated BST.13 Of these, 409 samples were identified with BST >11.4 ng/mL; 285 of these had HαT, 98 had MCD, 21 had both HαT and MCD, 2 individuals had hypereosinophilic disorders (one of whom also had HαT), and 4 individuals did not have a clear diagnosis following genotyping. Thus, in individuals with BST > 11.4 ng/mL, elevated BST levels could be attributed to HαT and/or MCD in 404 of 409 (98.8%) individuals (Figure 3E). Moreover, 255 of 262 of samples (97.3%) with BST levels between 8 and 11.4 ng/mL were associated with HαT and/or MCD. Based on the prevalence of HαT and MCD, data from previous studies35,36 have also suggested that BST > 8 ng/mL is relatively uncommon among individuals without these conditions, occurring in an estimated 5% of the populations tested compared to ∼3% of our cohort. Consistent with these data is the modeled 95% threshold of 7.99 ng/mL among individuals without HαT or MCD, indicating that <5% of the total population would be predicted to have BST > 8 ng/mL.
The 5 individuals with BST > 11.4 ng/mL who did not have HαT or evidence of MCD, underwent paired peripheral blood exome and deep exome sequencing (300×) of granulocyte and mononuclear BM aspirate fractions. In all 5 individuals, variants in genes commonly mutated in hematological malignancies (eg, TET2, RUNX1, CEBPA, MAP2K1, NOTCH2, KMT2C) were identified (supplemental Table 6); in 4, the variants were somatic, whereas in the remaining individual, RUNX1 and CEBPA variants were determined to be germline. Although pathogenicity of the variants was not experimentally determined, several including the TET2 and NOTCH2 variants were nonsense variants and predicted to be damaging. In 1 individual, the identified variant was associated with hypereosinophilic syndrome. However, BM evaluation and screening for JAK2 p.V617F as well as FIP1L1-PDGFRA fusion was unrevealing, thus the patient's provisional diagnosis remained as idiopathic hypereosinophilic syndrome. Together with those individuals meeting criteria for MCD, these data demonstrate that BST > 11.4 ng/mL in the absence of HαT frequently identifies indolent clonal MCD (98 of 103 of our referral cohort [95%]) or, less frequently, identifies patients with somatic genetic variants suggestive of occult or evolving myeloid neoplasms such as clonal hematopoiesis of indeterminant potential.
Of note, 55 of 144 samples (38.2%) with BST > 20 ng/mL were a result of HαT alone. Of these 55 participants, 54 were less than the threshold based on replication number. Thus if tryptase genotyping was not considered, and instead the current minor criterion for SM of BST > 20 ng/mL was used as a sole indication for BM evaluation, 54 of these individuals could have been subjected to unnecessary invasive BM examination. This is particularly relevant among individuals with 2 or more TPSAB1 replications, where median BST levels were >20 ng/mL and could be >100 ng/mL, as seen in 2 related individuals with 10 TPSAB1 replications in cis (Figures 2B and 3D; Table 3).
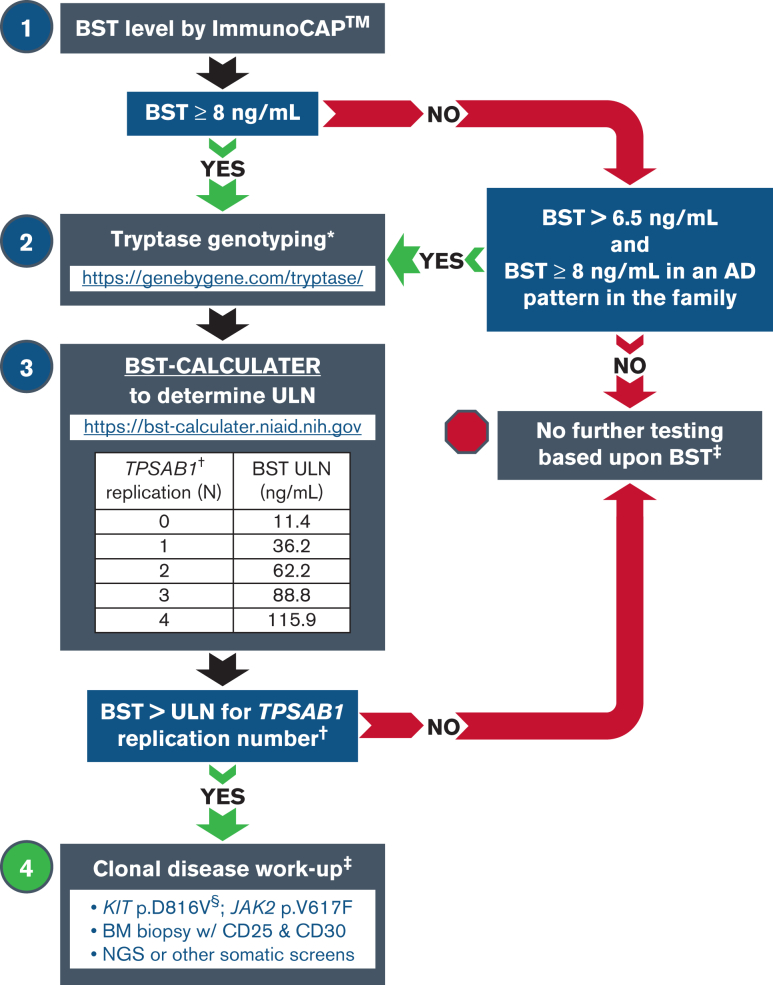
Finally, of the 21 individuals identified with both HαT and MCD (Table 4), 10 individuals had indolent SM (ISM), 8 of whom had BST levels above the 99.5% upper prediction limit dictated by TPSAB1 replication number. Ten individuals had BST levels below the 99.5% upper prediction limit based on TPSAB1 replication number in our model; 6 with monoclonal MMAS, 2 with ISM, 1 with diffuse cutaneous mastocytosis, and 1 with well-differentiated ISM. All these individuals had clinical implications for a BM beyond the BST level and would have received a BM regardless. Only one of these individuals, with the rare phenotype of well-differentiated ISM, would no longer have met clinical criteria for their diagnosis, if the upper prediction limit was used as a minor criterion for the diagnosis of SM (rather than the current level of >20 ng/mL). Together, these data demonstrate that where tryptase genotyping is clinically available, these new reference ranges based on TPSAB1 replication number can be applied to the workup of patients with elevated BST, establishing robust thresholds for which patients should undergo more extensive workup including BM aspiration and biopsy regardless of clinical presentation or symptomatology (see Figure 4 for a stepwise algorithm), and as an individualized minor criterion for the clinical diagnosis of SM.
Table 4.
Individuals with MCD and HαT
| Diagnosis | Age, y | Gender | Tryptase genotype |
Current World Health Organization–defined criteria for SM diagnosis |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | β | MC aggregates∗ | Spindle† | KIT p.D816V† | CD2 or CD25† | BST | >20 ng/ mL† | >99.5% prediction interval | New diagnosis‡ | |||
| ISM | 66-70 | M | 3 | 2 | Yes | Yes | Positive | Positive | 236 | Yes | Yes | No |
| ISM | 20-25 | M | 4 | 2 | Yes | Yes | Positive | Positive | 107 | Yes | Yes | No |
| ISM | 66-70 | F | 3 | 2 | Yes | Yes | Positive | Positive | 88.7 | Yes | Yes | No |
| ISM | 66-70 | F | 3 | 2 | Yes | Yes | Negative | Positive | 70 | Yes | Yes | No |
| ISM | 10-15 | F | 2 | 3 | Yes | Yes | Positive | Positive | 60.1 | Yes | Yes | No |
| MMAS | 70-75 | F | 3 | 2 | No | No | Negative | Positive | 52.2 | Yes | Yes | No |
| ISM | 50-55 | M | 3 | 2 | No | Yes | Positive | Positive | 51 | Yes | Yes | No |
| ISM | 70-75 | F | 3 | 2 | No | Yes | Positive | Negative | 42 | Yes | Yes | No |
| MMAS | 66-70 | F | 2 | 3 | No | No | Positive | Negative | 38.8 | Yes | Yes | No |
| ISM (well-differentiated) | 30-35 | F | 3 | 2 | Yes | No | Negative | Negative | 37.9 | Yes | Yes | No |
| ISM | 10-15 | M | 3 | 2 | No | Yes | Positive | Positive | 37.6 | Yes | Yes | No |
| MMAS | 20-25 | M | 3 | 2 | No | No | Positive | Negative | 37 | Yes | Yes | No |
| Diffuse cutaneous mastocytosis | 6-10 | M | 3 | 2 | Na | Na | Negative§ | Na | 32.6 | Yes | No | No |
| ISM | 56-60 | F | 2 | 3 | No | Yes | Positive | Positive | 32.2 | Yes | No | No |
| MMAS | 50-55 | F | 3 | 2 | No | Yes | Negative | Negative | 27.3 | Yes | No | No |
| MMAS | 60-65 | M | 3 | 2 | No | No | Positive | Negative | 26.8 | Yes | No | No |
| ISM (well-differentiated) | 26-30 | F | 2 | 3 | Yes | No | Negative | Negative | 26.8 | Yes | No | Yes |
| ISM | 30-35 | F | 3 | 2 | No | Yes | Positive | Positive | 23.7 | Yes | No | No |
| MMAS | 60-65 | M | 2 | 3 | No | No | Positive | Negative | 23.1 | Yes | No | No |
| MMAS | 56-60 | M | 3 | 2 | No | No | Positive | Negative | 20.3 | Yes | No | No |
| MMAS | 66-70 | F | 2 | 3 | No | No | Positive | Negative | 18.8 | No | No | No |
The individual with a change in diagnosis based on genotype-defined BST reference values is indicated in bold italic font.
MMAS, monoclonal mast cell activation syndrome.
Major criteria for the diagnosis of SM.
Minor criteria for the diagnosis of SM.
A new diagnosis being assigned based on application of the 99.5% threshold for BST, rather than >20 ng/mL.
In peripheral blood only.
Figure 4.
Tryptase genotyping in the evaluation of patients with elevated BST. Stepwise approach to a patient workup based on BST level and tryptase genotype using the BST CALCULATER. Myeloid neoplasms often exist in the absence of elevated BST; this algorithm is intended only to aid in the correct interpretation of elevated BST when other indications for workup are absent/nonspecific. AD, autosomal dominant; NGS, next-generation sequencing; ULN, upper limit of normal. ∗At the time of this publication, this is the only Clinical Laboratory Improvement Amendments/College of American Pathologists–certified laboratory performing tryptase genotyping. †Only α-tryptase–encoding TPSAB1 replications are associated with elevated BST and require correction. ‡BST elevation is not a requirement for any clonal neoplasm, and evaluation should be guided by clinical presentation and findings. §Allele-specific PCR or ddPCR should be used because of low allelic frequency.
Discussion
Based on our findings, HαT is a genetic trait best described as a naturally occurring overexpression model, whereby a conserved overactive promoter is coinherited with α-tryptase–encoding TPSAB1 gene replications driving basal overexpression of αDUP-tryptase that, when translated, is indistinguishable from other α-tryptase proteins (ie, αWT-tryptase). Using this knowledge and clinical data, we have generated a model that redefines clinically meaningful cutoffs for the upper limit of normal for serum tryptase levels based on tryptase genotype. Moreover, we have demonstrated the clinical utility of using these individualized values in the evaluation of patients with elevated serum tryptase levels and in the diagnosis of SM.
We acknowledge that MCD is not the only clonal myeloid neoplasm associated with elevated BST, rather it is the predominant one referred to our centers. Moreover, several studies have now demonstrated an increased prevalence of HαT among individuals with SM who are symptomatic,6, 7, 8 likely owing to potentiation of mast cell–mediated symptoms and reactions. Additional prospective studies will be helpful in validating the use of these new reference values in additional patients with HαT, MCD, and other myeloid neoplasms. Severe renal dysfunction can also affect BST levels, as can other parasitic diseases, which are rare in most Western countries.15 These factors were not represented or evaluated in this study and should be considered when applying this model to patients. However, the model was generated based on individuals without clinical signs or symptoms of these disorders. Thus, we expect these new reference values to remain robust in their predictive value, as has been suggested by a recent retrospective study of a regional health system where 54 of 58 of individuals (93%) with BST > 11.4 ng/mL had HαT, a myeloid neoplasm, or chronic kidney disease.37
The data used to generate our model are largely from individuals with ≤2 replications. Thus, although available data for higher order copy number are consistent with our model, and indeed were incorporated into the model, the generated cutoffs at high TPSAB1 replication number (eg, 3) are largely extrapolations. The a priori evidence demonstrated a linear relationship between BST and replication number, provide support for this extrapolation. However, there is less certainty on the upper limits of BST levels beyond 2 TPSAB1 replications, and cutoffs for individuals with higher order copy number may require refinement as additional data become available.
To our knowledge, the application of genotypic information to determine clinical reference ranges in a personalized manner has not previously been used in laboratory medicine. However, many common clinical laboratory measurements (eg, immunoglobulin levels) are not normally distributed, and in some cases, outliers may be similarly determined by heritable traits. Thus, we anticipate this kind of precision approach to clinical laboratory medicine will expand in the future as next-generation sequencing becomes increasingly utilized in standard clinical practice.
Conflict-of-interest disclosure: Virginia Commonwealth University receives royalties from Thermo Fisher Scientific for their tryptase test that are shared with L.B.S. as its inventor. The remaining authors declare no competing financial interests.
Acknowledgments
The investigators thank the patients, their families, and the healthy volunteers who contributed to this research. The authors also thank Adam Phillippy for sharing his expertise on complex genetic structural rearrangement.
This project was funded in whole or in part with federal funds from the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) and with federal funds from the National Cancer Institute, NIH, under contract numbers: HHSN261201500003I and 75N910D00024. This project was also funded in part with federal funds from the Division of Intramural Research of the National Heart, Lung, and Blood Institute, NIH. This work was supported by a grant from US Public Health Service (grant NIH R21DE028378) (S.C.G.). The involvement of B.B. and J. Halstead was supported by the Intramural Research Program of the National Library of Medicine. V.S. and D.G.E. are senior clinical researchers of the Research Foundation Flanders/Fonds Wetenschappelijk Onderzoek (FWO: 1804518N and FWO: 1800614N, respectively).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Authorship
Contribution: J.J.L. designed the study; J.J.L., S.L., C.N., J.B., T.D., M.C.C., H.D.K., J.S., K.M.H., E.F., P.K., J.D.M., V.S., D.G.E., P.B., R.Z., P.K., and D.D.M., recruited and enrolled study participants; J.J.L., I.T., J. Hughes, J. Halstead, J.M., C.S.H., B.B., M.P., N.N.T., G.H.C., and J.L. performed and/or supervised the bioinformatics analyses; J.J.L. designed and J.C., J.K., and Y.L. performed droplet digital polymerase chain reaction assays; I.M. performed hematopathologic analyses; J.J.L., J.C., J.K., D.D.M., L.B.S., Y.B., Y.Y., Y.H.P., Y.L., M.P.O., Q.L., and A.P.H. designed, supervised, and/or performed the functional and genetic studies and sequencing; A.M. and E.H.B. developed the prediction interval model; J.J.L. prepared the draft manuscript; and all authors contributed to discussion of the results and to manuscript preparation.
Footnotes
Sequencing data were obtained from the NCBI Sequence Read Archive for the following BioProjects; HαT: PRJNA342304, and individuals not selected by tryptase genotype: PRJNA208369, PRJNA219425, PRJNA232669, PRJNA252605, PRJNA253059, PRJNA254943, PRJNA257389, PRJNA258216, PRJNA261011, PRJNA261251, PRJNA263242, PRJNA266512, PRJNA266572, PRJNA270371, PRJNA271942, PRJNA274028, PRJNA274360, PRJNA275801, PRJNA279249, PRJNA280990, PRJNA283839, PRJNA289905, PRJNA291619, PRJNA292690, PRJNA293555, PRJNA296379, PRJNA301173, PRJNA301364, PRJNA310988, PRJNA315611, PRJNA317535, PRJNA318253, PRJNA319220, PRJNA326113, PRJNA327986, PRJNA330840, PRJNA340161, PRJNA342177, PRJNA343985, PRJNA354367, PRJNA358081, PRJNA369563, PRJNA369684, PRJNA373887, PRJNA376200, PRJNA377555, PRJNA378385, PRJNA384963, PRJNA388978, PRJNA389466, PRJNA392116, PRJNA395367, PRJNA395589, PRJNA399103, PRJNA400331, PRJNA407731, PRJNA415746, PRJNA428940, PRJNA214592, PRJNA229548, PRJNA278364, PRJNA278767, and PRJNA263397. Referenced GenBank accession codes include GRCh37/hg19 NCBI assembly: GCF_000001405.13; tryptase locus assemblies: AC226137.3, AC120498.2, CTD-2503P16, CHM13, AL031704.24, AE006466.1, CHM1, AC240106.3, AC238650.2, AC213746.1, GM24385, NA12878, and AF098328.1.
Renewable materials, data sets, and protocols can be made available on request from the corresponding author, Jonathan J. Lyons (jonathan.lyons@nih.gov).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Caughey GH. Tryptase genetics and anaphylaxis. J Allergy Clin Immunol. 2006;117(6):1411–1414. doi: 10.1016/j.jaci.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JS, Westin EH, Schwartz LB. Cloning and characterization of complementary DNA for human tryptase. J Clin Invest. 1989;84(4):1188–1195. doi: 10.1172/JCI114284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JS, Moxley G, Schwartz LB. Cloning and characterization of a second complementary DNA for human tryptase. J Clin Invest. 1990;86(3):864–870. doi: 10.1172/JCI114786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glover SC, Carter MC, Korosec P, et al. Clinical relevance of inherited genetic differences in human tryptases: hereditary alpha-tryptasemia and beyond. Ann Allergy Asthma Immunol. 2021;127(6):638–647. doi: 10.1016/j.anai.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons JJ, Yu X, Hughes JD, et al. Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat Genet. 2016;48(12):1564–1569. doi: 10.1038/ng.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chollet MB, Akin C. Hereditary alpha tryptasemia is not associated with specific clinical phenotypes. J Allergy Clin Immunol. 2021;149(2):728–735.e2. doi: 10.1016/j.jaci.2021.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Greiner G, Sprinzl B, Gorska A, et al. Hereditary alpha tryptasemia is a valid genetic biomarker for severe mediator-related symptoms in mastocytosis. Blood. 2021;137(2):238–247. doi: 10.1182/blood.2020006157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyons JJ, Chovanec J, O’Connell MP, et al. Heritable risk for severe anaphylaxis associated with increased alpha-tryptase-encoding germline copy number at TPSAB1. J Allergy Clin Immunol. 2021;147(2):622–632. doi: 10.1016/j.jaci.2020.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Robey RC, Wilcock A, Bonin H, et al. Hereditary alpha-tryptasemia: UK prevalence and variability in disease expression. J Allergy Clin Immunol Pract. 2020;8(10):3549–3556. doi: 10.1016/j.jaip.2020.05.057. [DOI] [PubMed] [Google Scholar]
- 10.Lyons JJ. Inherited and acquired determinants of serum tryptase levels in humans. Ann Allergy Asthma Immunol. 2021;127(4):420–426. doi: 10.1016/j.anai.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons JJ. Hereditary alpha tryptasemia: genotyping and associated clinical features. Immunol Allergy Clin North Am. 2018;38(3):483–495. doi: 10.1016/j.iac.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz LB, Metcalfe DD, Miller JS, Earl H, Sullivan T. Tryptase levels as an indicator of mast-cell activation in systemic anaphylaxis and mastocytosis. N Engl J Med. 1987;316(26):1622–1626. doi: 10.1056/NEJM198706253162603. [DOI] [PubMed] [Google Scholar]
- 13.Lyons JJ, Schwartz LB. In: Mastocytosis. 1st ed. Akin C, editor. Springer; 2020. Clinical approach to a patient with elevated serum tryptase: implications of acute versus basally elevated levels; pp. 35–54. [Google Scholar]
- 14.Valent P, Horny HP, Escribano L, et al. Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res. 2001;25(7):603–625. doi: 10.1016/s0145-2126(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 15.Luskin KT, White AA, Lyons JJ. The genetic basis and clinical impact of hereditary alpha-tryptasemia. J Allergy Clin Immunol Pract. 2021;9(6):2235–2242. doi: 10.1016/j.jaip.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz LB, Bradford TR, Rouse C, et al. Development of a new, more sensitive immunoassay for human tryptase: use in systemic anaphylaxis. J Clin Immunol. 1994;14(3):190–204. doi: 10.1007/BF01533368. [DOI] [PubMed] [Google Scholar]
- 17.Tao Q, Zhao H, Qiu L, Hong G. Construction of a full bacterial artificial chromosome (BAC) library of Oryza sativa genome. Cell Res. 1994;4(2):127–133. [Google Scholar]
- 18.Tao Q, Zhang HB. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 1998;26(21):4901–4909. doi: 10.1093/nar/26.21.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtke HJ, Ankenbauer W, Muhlegger K, et al. The digoxigenin (DIG) system for non-radioactive labelling and detection of nucleic acids--an overview. Cell Mol Biol (Noisy-le-grand) 1995;41(7):883–905. [PubMed] [Google Scholar]
- 20.Reid GA, Sambroo kJ, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13) Blood. 1999;94(7):2333–2342. [PubMed] [Google Scholar]
- 22.Rehder C, Bean LJH, Bick D, et al. Next-generation sequencing for constitutional variants in the clinical laboratory, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23(8):1399–1415. doi: 10.1038/s41436-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30(17):2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15(8):591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 26.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trivedi NN, Tamraz B, Chu C, Kwok PY, Caughey H. Human subjects are protected from mast cell tryptase deficiency despite frequent inheritance of loss-of-function mutations. J Allergy Clin Immunol. 2009;124(5):1099–10105.e1-4. doi: 10.1016/j.jaci.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateja A, Wang Q, Chovanec J, et al. Defining baseline variability of serum tryptase levels improves accuracy in identifying anaphylaxis. J Allergy Clin Immunol. 2022;149(3):1010–1017.e10. doi: 10.1016/j.jaci.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang R, Abrink M, Gobl AE, et al. Expression of a mast cell tryptase in the human monocytic cell lines U-937 and Mono Mac 6. Scand J Immunol. 1993;38(4):359–367. doi: 10.1111/j.1365-3083.1993.tb01738.x. [DOI] [PubMed] [Google Scholar]
- 32.Soto D, Malmsten C, Blount JL, Muilenburg DJ, Caughey GH. Genetic deficiency of human mast cell alpha-tryptase. Clin Exp Allergy. 2002;32(7):1000–1006. doi: 10.1046/j.1365-2222.2002.01416.x. [DOI] [PubMed] [Google Scholar]
- 33.Lyons JJ, Stotz SC, Chovanec J, et al. A common haplotype containing functional CACNA1H variants is frequently coinherited with increased TPSAB1 copy number. Genet Med. 2018;20(5):503–512. doi: 10.1038/gim.2017.136. [DOI] [PubMed] [Google Scholar]
- 34.Min HK, Moxley G, Neale MC, Schwartz LB. Effect of sex and haplotype on plasma tryptase levels in healthy adults. J Allergy Clin Immunol. 2004;114(1):48–51. doi: 10.1016/j.jaci.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Fellinger C, Hemmer W, Wohrl S, Sesztak-Greinecker G, Jarisch R, Wantke F. Clinical characteristics and risk profile of patients with elevated baseline serum tryptase. Allergol Immunopathol. 2014;42(6):544–552. doi: 10.1016/j.aller.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Quintela A, Vizcaino L, Gude F, et al. Factors influencing serum total tryptase concentrations in a general adult population. Clin Chem Lab Med. 2010;48(5):701–706. doi: 10.1515/CCLM.2010.124. [DOI] [PubMed] [Google Scholar]
- 37.Waters AM, Park HJ, Weskamp AL, et al. Elevated basal serum tryptase: disease distribution and variability in a regional health system. J Allergy Clin Immunol Pract. 2022;10(9):2424–2435.e5. doi: 10.1016/j.jaip.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.