Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) is a central regulator of mammalian cell growth that is dysregulated in a number of human diseases, including metabolic syndromes, aging, and cancer. Structural, biochemical, and pharmacological studies that have increased our understanding of how mTORC1 executes growth control often relied upon purified mTORC1 protein. However, current immunoaffinity-based purification methods are expensive, inefficient, and do not necessarily isolate endogenous mTORC1, hampering their overall utility in research. Here we present a simple tool to isolate endogenous mTORC1 from various cellular sources. By recombinantly expressing and isolating mTORC1-binding Rag GTPases from Escherichia coli and using them as affinity probes, we demonstrate that mTORC1 can be isolated from mouse, bovine, and human sources. Our results indicate that mTORC1 isolated by this relatively inexpensive method is catalytically active and amenable to scaling. Collectively, this tool may be utilized to isolate mTORC1 from various cellular sources, organs, and disease contexts, aiding mTORC1-related research.
Keywords: mTORC1, protein isolation, Rag-GTPases, affinity assay, cell metabolism
The mechanistic target of rapamycin complex 1 (mTORC1) integrates a multitude of environmental cues to direct mammalian cell growth (1, 2). When cells are exposed to nutrients and growth factors, mTORC1 is activated at lysosomes by the Rheb GTPase, leading to the stimulation of anabolic processes including protein and lipid synthesis, and the concomitant suppression of macroautophagy (3, 4, 5). Given its central role in the regulation of anabolic metabolism, dysregulated mTORC1 is implicated in a growing number of human pathological conditions, including cancer, autoimmune diseases, and aging, and is the focus of several therapeutic programs (6).
mTORC1 consists of three core components: the ∼250 kDa serine/threonine mTOR kinase, a 35 kDa mammalian lethal with SEC13 protein 8 (LST8, also known as GβL), and the 150 kDa scaffold protein regulatory-associated protein of mTOR (Raptor) (7). Additional splice isoforms of mTOR have been identified, including an ∼80 kDa mTORβ splice isoform which is abundant in the liver and other tissues (8). Raptor is essential for the translocation of mTORC1 to the lysosomal surface, which is mediated by binding to lysosomal-bound Rag GTPases during conditions of amino acid sufficiency (9, 10). Rag GTPases form obligate heterodimers of RagA or RagB bound to RagC or RagD (11). In their active state, RagA/B is bound to GTP and RagC/D is bound to GDP and strongly bind Raptor, thereby translocating mTORC1 to the lysosomal surface (9, 10, 12). In addition to mTORC1, mTOR is also the catalytic subunit of mTOR complex 2 (mTORC2), defined by its components Rictor and Sin1, and its unique downstream substrates including Akt (6, 13).
Despite our increased understanding of the mTORC1 pathway over the last few decades, how mTORC1 coordinates the multitude of anabolic processes in various organs or disease tissues remains poorly understood. Importantly, many of the studies that have significantly advanced our knowledge regarding how mTORC1 executes cell growth control relied upon purified protein for enzyme:function studies (14, 15, 16, 17). However, current methods for isolating mTORC1 depend entirely on ectopic expression of tagged mTORC1 components (18, 19, 20, 21). Tag-based purification strategies frequently lead to protein misfolding, incomplete expression, or cellular toxicity and are also relatively expensive given the cost of antibody-bound resins. More importantly, products of tagged recombinant proteins can introduce various confounding variables, for example, by altering the endogenous state of the protein, interfering with binding partner interactions or creating new unintended ones, or remaining unbound in its apo form (22, 23, 24). For example, N-terminal–tagged Sin1, a key component of mTORC2, cannot enter the complex (25). Also immunocomplex-based isolation methods of mTORC1 are problematic, as the immunocomplex cannot be eluted without also disrupting the mTORC1 complex, restricting downstream utility for most types of assays. Moreover, these approaches do not scale well as the source of their mTORC1 is from cell culture. Taken together, methods to isolate endogenous mTORC1 are arguably needed and would enable investigation of mTORC1 in its physiological form, at its natural equilibrium, where intraprotein interactions and activation states are conserved.
Here, we present a novel tool to isolate endogenous mTORC1, which is based on the high and specific binding affinity between the RagA/C GTPase heterodimer and the mTORC1 core-component Raptor. By using recombinant Rag GTPases expressed in Escherichia coli that are set to the correct GTP/GDP state, we show that endogenous and catalytically active mTORC1 can be eluted from various cellular origins, and even animal tissues. Hopefully, this tool will aid research that advances the knowledge of the mTORC1 pathway and support the development of novel therapeutics.
Results
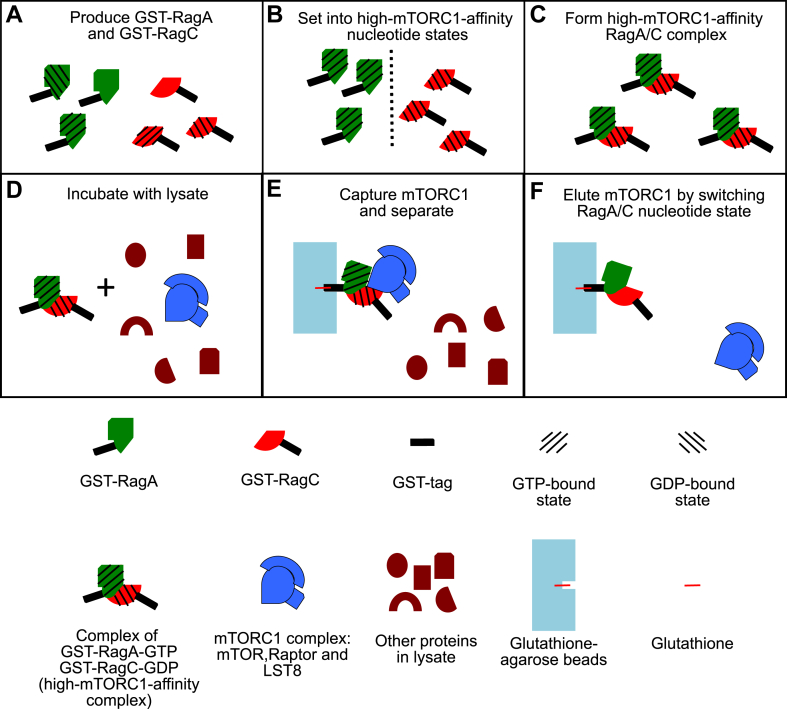
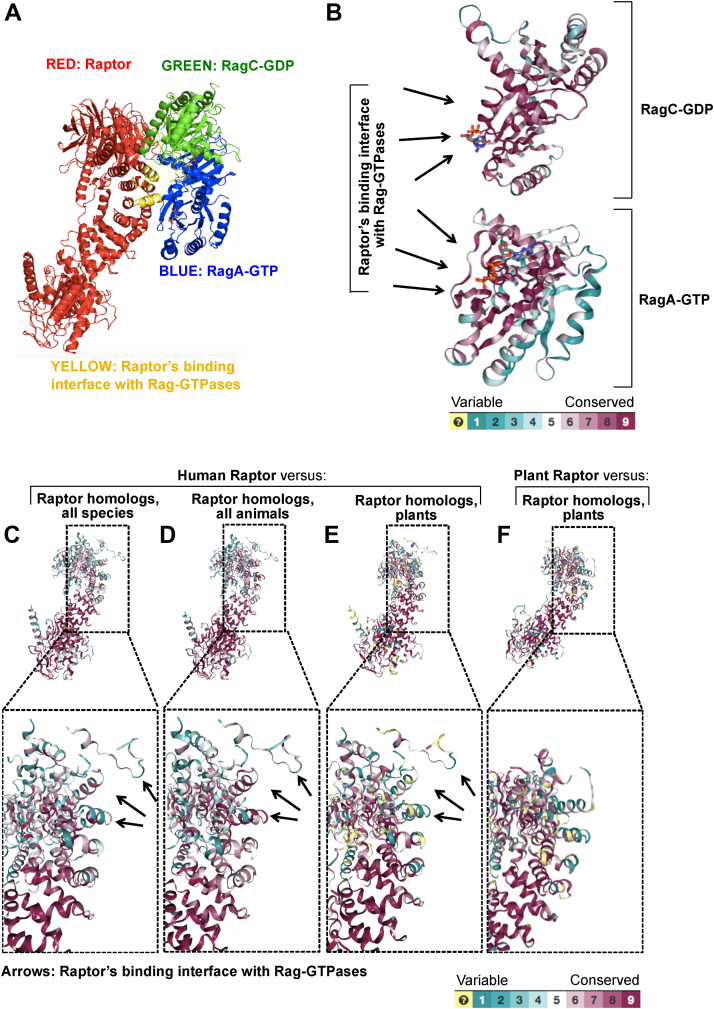
Mammalian RagA and RagC bind the mTORC1 component Raptor with high affinity when RagA is bound to GTP and RagC to GDP (9, 10, 12). Based on this, we set out to build a cost-effective tool that isolates endogenous mTORC1. The basis of this strategy is to express GST-tagged recombinant human RagA and RagC in E. coli (Fig. 1A), set these GTPases into their high-mTORC1-affinity nucleotide state: GST-RagA-GTP and GST-RagC-GDP (Fig. 1B), and utilize the GTPase heterodimer as an affinity probe to isolate mTORC1 from various cellular sources by GST affinity purification using glutathione beads (Fig. 1, C–F). By replacing GTP with the slow/nonhydrolysable GTPgammaS (GTPγS), the potential hydrolysis of GST-RagA-GTP by intrinsic GTPase activity or GTPase-activating proteins present in lysates is suppressed (Fig. 1B). At the end, mTORC1 can be eluted by changing the GST-Rag GTPase nucleotide state by simple chelation of magnesium which disrupts the nucleotide binding within the Rags (26) (Fig. 1F). Atomic-scale in silico modeling of the Rag–Raptor binding interface across the animal and plant kingdoms indicated that this strategy potentially can isolate mTORC1 from various animal cells, but not plant cells, since they lack Rag homologs (27) and the Raptor-binding interface is not conserved between humans and plants (Fig. 2).
Figure 1.
Schematic diagram of method to isolate endogenous mTORC1 using recombinant Rag GTPase proteins.A, GST-tagged RagA and RagC are expressed and purified from E. coli. B, GST-Rag GTPases are individually set into the high-mTORC1-affinity states: GST-RagA-GTPγS and GST-RagC-GDP. C, GST-RagA-GTPγS and GST-RagC-GDP are combined at a 1:1 ratio to form a high-mTORC1-affinity dimer. D, GST-Rag GTPase dimers are incubated with cell lysate containing mTORC1. E, GST-Rag GTPases are isolated using glutathione beads, which bind the GST tag. In this way, mTORC1 is captured on the beads and separated from other lysate proteins. F, mTORC1 is eluted by switching the nucleotide state of the GST-Rag GTPases to a low-mTORC1-affinity state. GTPγS, GTPgammaS; mTORC1, mechanistic target of rapamycin complex 1.
Figure 2.
Conserved interacting regions of Raptor and Rag GTPases highlight the utility of the affinity assay in all mTOR homolog-expressing species except plants.A, the crystal structure of human Raptor with the dimer of human RagA-GTP with RagC-GDP at 3.18 Ångström with the interacting regions between the supercomplex highlighted in yellow. The structure was obtained from PDB file 6U62 (7). A conserved protein sequence analysis comparing human versus all homolog-containing species for RagA and RagC (B) or Raptor (C–E) is superimposed onto PDB 6U62 using ConSurf software. Conserved protein sequences for human Raptor versus Raptor homologs from all species (C), all animals (D), and plants (E) are shown. F, Raptor-conserved protein sequences for Arabidopsis thaliana (plant) versus homologs from all other plants superimposed onto Arabidopsis thaliana Raptor PDB 5WBI (14). Computed conservation scores are assigned color grades 1 to 9, going from variable to conserved, as indicated. Unreliable regions are colored in light yellow. For more info, see Experimental procedures and references therein. mTOR, mechanistic target of rapamycin.
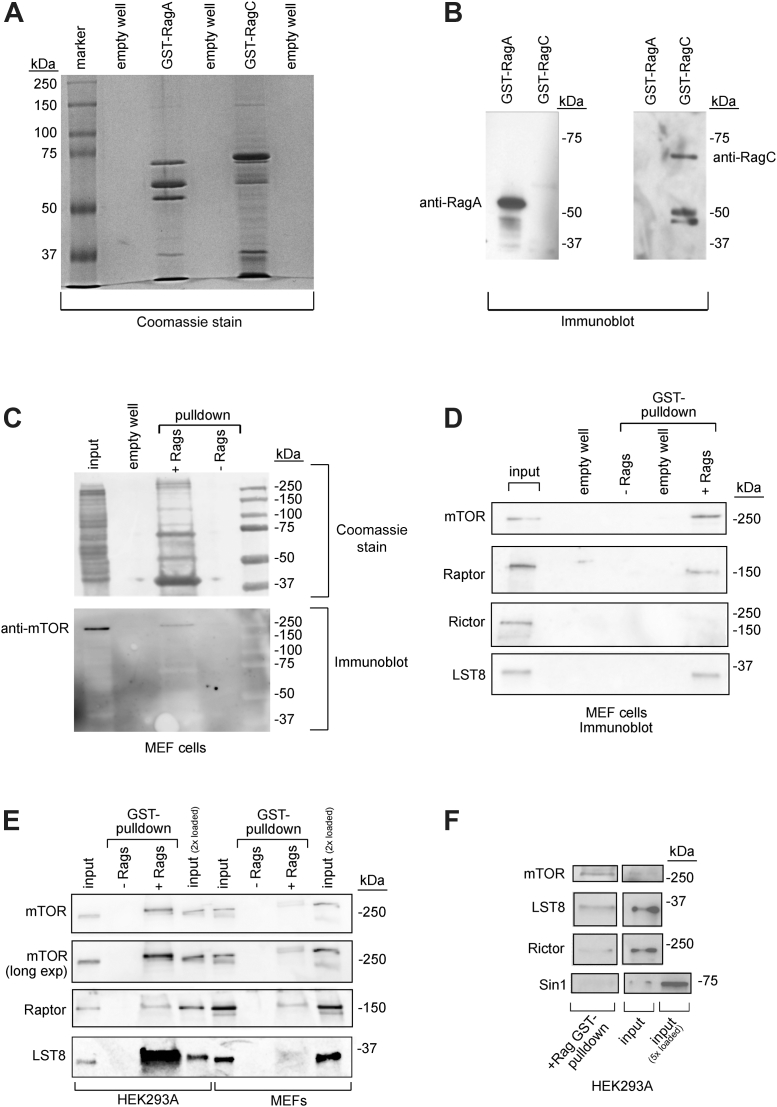
To explore whether it is possible to isolate mTORC1 using this strategy, GST-tagged human RagA and human RagC were independently expressed and purified from BL21 E. coli using glutathione beads. Coomassie staining of the purified samples demonstrated that the primary bands in isolated GST-RagA and GST-RagC corresponded to the expressed proteins, a 55- and 70-kDa protein, respectively (Fig. 3, A and B). Additional bands suggested that partial proteolysis/degradation of the Rags occurred in the bacterial lysates, which was minimal, indicating that human RagA and RagC can be readily produced in vitro. The GST-RagA isolate also contained a larger unknown protein (∼65 kDa). Next, we exchanged these recombinant proteins with GDP or GTPγS, setting their high-affinity mTORC1-binding state in vitro, whereafter they were combined at a 1:1 ratio and added to lysates of mouse embryonic fibroblasts (MEFs). After GST pull-down, we assessed whether mTORC1 components could be detected in the eluate. Indeed, Coomassie staining revealed a band at 250 kDa, which corresponded to mTOR, as assessed by immunoblotting (Fig. 3C). Furthermore, the Rag-affinity pull-down elute was enriched in mTOR, Raptor, and LST8, in contrast to the mTORC2’s Rictor, as judged by immunoblotting analysis (Fig. 3D), indicating that Rag GTPases can be utilized as affinity probes to isolate endogenous mTORC1. We next isolated mTORC1 from human HEK293A cell lysates and compared the pull-down to MEFs. By immunoblotting, we detected more mTORC1 components being isolated from HEK293A cells than MEFs (Fig. 3E) and only minor levels of mTORC2’s components Rictor and Sin1 (Fig. 3F), altogether suggesting that mTORC1 can be isolated from human HEK293A cells.
Figure 3.
Expression of GST-RagA and GST-RagC and isolation of mouse mTORC1. Coomassie stain (A) and SDS-page Western immunoblotting (B) of 8% acrylamide gel of recombinant human GST-RagA and GST-RagC expressed in Escherichia coli and purified on glutathione beads, as indicated. C, Coomassie stain (upper panel) and immunoblotting (lower panel) of Rag-isolated mTORC1 from MEF lysates. D–F, Rag-isolated mTORC1 from MEFs (D and E) or HEK293A (E and F) were immunoblotted against the indicated proteins. Experiments were repeated at least three times. In C–F, mTORC1 was isolated by Rag GST pull-down, lysed, and boiled. Lysates were prepared from six confluent 15-cm plates (C, D, and F) and six confluent 15-cm plates (∼14 mg of protein) (E). MEFs, mouse embryonic fibroblasts; mTORC1, mechanistic target of rapamycin complex 1.
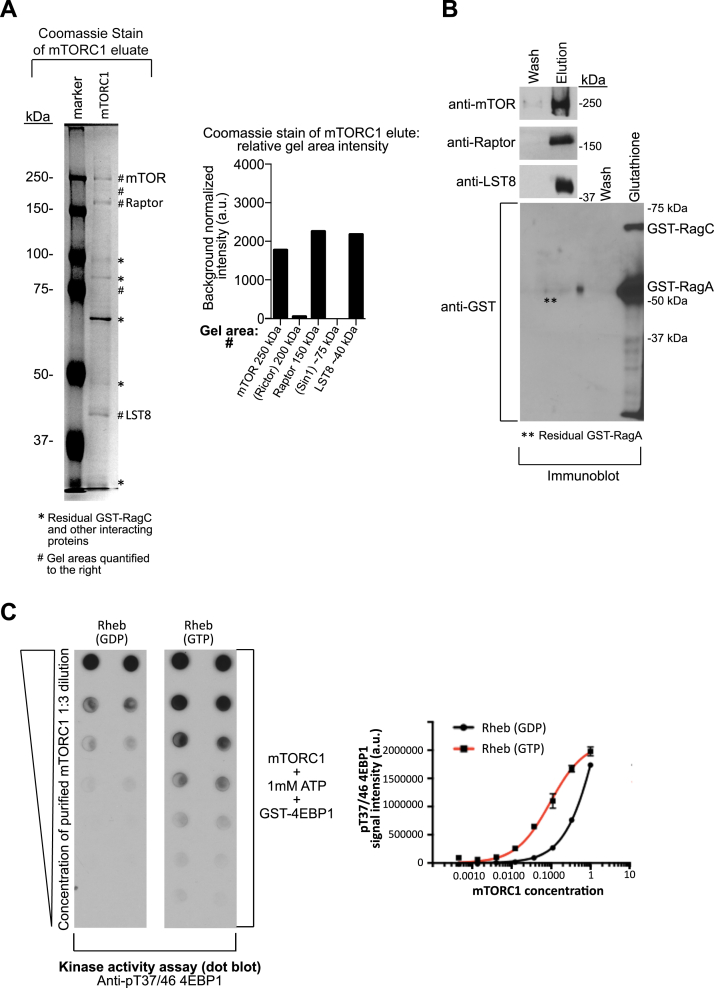
We next tested whether the assay could be scaled-up to isolate and elute mTORC1 by recombinant high affinity-state Rag binding and release (Fig. 1). To this end, we focused on HEK293A where we had found mTORC1 components to be abundant (Fig. 3E). Affinity pull-down and elution readily isolated mTORC1-sized component proteins, which were confirmed to be Raptor, mTOR, and LST8 by immunoblotting (Fig. 4, A and B). Moreover, the affinity pull-down “wash” condition lacked mTORC1, suggesting specificity to the chelation-mediated elution step (Fig. 4B). To test whether isolated mTORC1 was catalytically active, we performed an in vitro kinase assay with the mTORC1 substrate 4E-BP1 and its activator Rheb. In the experiment, purified Rheb had either been loaded with GDP or GTPγS since GTP-bound Rheb is known to activate mTORC1 (28). A significant increase in the phosphorylation of 4E-BP1 T37/46 was observed when Rheb-GTPγS was added to the reaction mixtures containing increasing amounts of isolated mTORC1, compared to the addition of Rheb-GDP (Fig. 4C). 4E-BP1 was phosphorylated by both conditions at the highest mTORC1 amount, suggesting that the eluted kinase is constitutively active, as observed previously (14, 21, 29). Taken together, these data suggest that mTORC1 could be eluted from the Rag GTPases ex vivo and that eluted mTORC1 was catalytically functional, thus retaining the structural features necessary for its further activation by Rheb.
Figure 4.
Rag isolation of human mTORC1 is catalytically active.A and B, Coomassie stain (A) and SDS-page Western immunoblot (B) of 6% acrylamide gel of Rag-purified mTORC1 from HEK293A cells. In (A), symbol ∗ denotes residual GST-RagC and other interacting proteins. Symbol # denotes areas in the gel quantified to the right, including gel areas corresponding to mTORC1 components validated by immunoblotting in (B): mTOR (250 kDa), Raptor (150 kDa), and LST (40 kDa). Gel areas corresponding to the mTORC2 complex components were extrapolated based on known molecular weights: Rictor (200 kDa) and Sin1 (75 kDa). As further described in Experimental procedures, in (B), the wash condition was collected, and mTORC1 was eluted from glutathione beads with high NaCl+EDTA. Beads were washed with Hepes buffer and GST-bound proteins were eluted with glutathione. mTORC1 components mTOR, Raptor, and mLST8 were detected in first eluate; GST-tagged RagA and RagC were detected in the second eluate. C, isolated mTORC1 was incubated with Rheb-GDP or Rheb-GTP, followed by incubation with ATP and 4E-BP1. The ability of eluted human mTORC1 to phosphorylate its downstream target 4E-BP1 at T37/46 was measured by dot blot analysis, with signal intensity quantified on the right. All experiments were repeated at least three times. In (A and B), lysates were prepared from 40 confluent 15-cm plates of cells. mTORC1, mechanistic target of rapamycin complex 1.
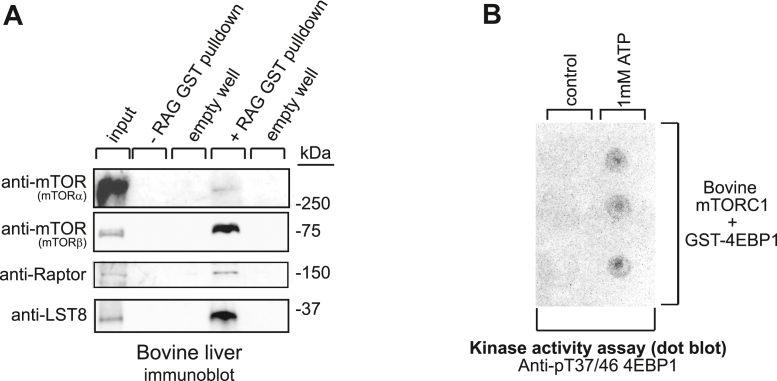
It was still unclear whether our assay could be scaled beyond cultured cells and applied to animal tissues, an abundant source of mTORC1. To test this, we applied our assay on the lysates of bovine liver. In the murine and human liver, two isoforms of mTOR exist: the full-length form designated as mTORα and a splice-isoform known as mTORβ (8). Tagged-protein overexpression studies using HEK293 cells have demonstrated that both mTORα and mTORβ can form mTORC1 in cells (8); however, this has never been demonstrated in vivo nor has mTORC1-β’s presence been demonstrated physiologically. By immunoblotting for mTORC1 components in the purified mTORC1 from bovine liver, we confirmed that the affinity pull-down isolated Raptor at 150 kDa, LST8 at 35 kDa, mTORα at ∼280 kDa, as well as a ∼80 kDa mTOR-sized protein (Fig. 5A), which was strongly immunoreactive to anti-mTOR antibody and arguably corresponds to the mTORβ isoform (8). Interestingly, an in vitro kinase assay of the isolate indicated that bovine mTORC1 was catalytically active as it phosphorylated 4E-BP1 at T37/46 (Fig. 5B). Collectively, these data suggest that the Rag affinity assay can be used to scale up mTORC1 isolation and that mTORα and mTORβ may form distinct mTOR complexes that coexist in the same tissue.
Figure 5.
Rag isolation of mTORC1 from bovine liver tissue.A and B, lysates of affinity-isolated mTORC1 were either subjected to immunoblotting against the endogenous indicated proteins (A) or incubated with ATP and 4E-BP1 in an in vitro kinase assay (B). The ability of eluted human mTORC1 to phosphorylate its downstream target 4E-BP1 at T37/46 was measured by dot blot analysis (B). As further described in Experimental procedures, mTORα was detected with polyclonal mTOR antibody ab2732, Abcam, and mTORβ with rabbit monoclonal antibody 2983, Cell Signaling Technologies. In (B), GST pull-down samples were eluted from the glutathione beads as described in Experimental procedures. In (A), GST pull-down samples were lysed and boiled in sample buffer. In (A), lysates were prepared from 1.5 g of tissue. Experiments were carried out on three individual liver samples. mTORC1, mechanistic target of rapamycin complex 1.
Discussion
Our study describes a unique approach to isolate large quantities of endogenous mTORC1 without the use of immunoaffinity reagents. We have demonstrated that the Rag GTPases, in agreement with their established role as Raptor-binding proteins (10, 12), can be used as affinity probes to isolate and elute mTORC1. Furthermore, our data suggest that purified mTORC1 is isolated in its native confirmation as it is catalytically active (Fig. 4). Importantly, our method can be scaled up and applied to a variety of cellular sources, opening up the possibility to isolate mTORC1 from abundant sources like animal tissues. Although this strategy may also isolate additional Rag-interacting proteins, as is likely the case even from bacterial lysates (Fig. 3A, RagA band ∼65 kDa), the predominant chelation-eluted proteins purified through this method correspond to known mTORC1 components (Figs. 3D and 4A), suggesting that this strategy on the whole is rather selective for mTORC1. Moreover, our results indicate that both mTORα and mTORβ isoforms form complexes with Raptor and LST8 and that both can be readily isolated from tissue samples (Fig. 5). Secondary purification steps could be applied to achieve further purity of mTORC1, such as size-exclusion chromatography and ion-exchange methods.
Given that Rag GTPase binding to Raptor is nucleotide dependent, the elution of mTORC1 from recombinant Rags is readily achieved by nucleotide switching. Thus far, we achieved this by simple chelation of the Mg2+ divalent ion that is required for the stabilization of both the RagA-GTP and RagC-GDP nucleotide binding and their interaction with Raptor, mediated by its switch-I domain (26, 30, 31, 32). An alternative elution approach could potentially be to add recombinant GATOR1, a known GTPase activating protein for RagA/B (33), but this would come at the cost of introducing more proteins to the elution that subsequently must be separated.
While mTOR was discovered nearly 30 years ago, to this day, discoveries into its mechanisms and function continue to be made. Recent studies indicate that numerous unknown mTORC1 substrates may exist (34) and these substrates require further validation with purified mTORC1. Additionally, mTORC1 is the subject of many therapeutic development programs, including the identification of inhibitors and activators, which also require large amounts of purified mTORC1. As shown in Figure 5A, our tool may not only be used for large prep isolation but also for the isolation of varied isoforms of mTORC1 from animal tissues, opening up the possibility of studying variations of mTORC1 and its binding partners.
In sum, we have developed a novel strategy to isolate mTORC1 from various cellular sources. We hope that this tool will aid research investigating the physiological role of the mTORC1 pathway in different organs, cell types, or disease contexts, thus supporting the development of mTORC1-directed therapeutics.
Experimental procedures
Cloning
Full-length human RagA, RagC, Rheb, and 4E-BP1 were cloned in-frame with N-terminally–tagged GST in pGEX4t.1 vector system, where translation is controlled by the Lac operon.
Bacterial protein expression and purification
pGEX plasmids were transformed in BL21 DE3 competent E. coli cells, and positive clones were grown under ampicillin selection. BL21 competent pGEX-RagA, pGEX-RagC, pGEX-Rheb, and pGEX-4E-BP1 were grown in 250 l LB media each at 37 °C degrees overnight. Cultures were then inoculated into 0.5-1L LB for protein expression. Upon reaching 0.6 absorbance, IPTG was added to the cultures for a final concentration of 400 μM and incubated at 37 °C for 4 to 5 h prior to bacteria pelleting by centrifugation. Cell pellets were resuspended in Hepes buffer (40 mM Hepes, 150 mM NaCl, 5 mM MgCl2 and cOmplete protease inhibitor cocktail from Roche) and sonicated with a probe sonicator. Protein lysates were incubated with glutathione beads (Pierce Glutathione Agarose, Thermo Fisher Scientific) at 4 °C for 1 to 2 h, followed by bead isolation by centrifugation and ten times wash with Hepes buffer.
Nucleotide loading of RagA and RagC
To set RagA and RagC in their mTORC1 high-affinity states, two different methods were utilized. In Figure 4, cleared bacterial lysate was incubated with glutathione-conjugated agarose beads and washed ten times with Hepes buffer. GST-RagA and GST-RagC were isolated from glutathione-conjugated beads with 10 mM reduced glutathione. Rag proteins were processed independently unless otherwise indicated. To release the nucleotides from the Rag proteins, purified RagA or RagC was placed in 10 kDa dialysis tubing and incubated with 10 mM EDTA for 1 h at room temperature. RagA or RagC dialysis tubing was placed in Hepes dialysis buffer without MgCl2 for 24 h at 4 °C, renewing buffer 2 to 3 times. While still in the dialysis tubing, purified RagA was incubated with 1 mM GTPγS and mixed, whereas purified RagC was incubated with 1 mM GDP and mixed. Purified RagA or RagC was incubated with 10 mM MgCl2 for 30 min at room temperature. Dialysis tubing was then placed in dialysis buffer Hepes with MgCl2 for 24 to 48 h at 4 °C, renewing dialysis buffer 2 to 3 times. Purified RagA or RagC was concentrated using a 10 kDa Centricon filter, and protein concentration was quantified by nano-drop 280 nm wavelength measurement. Equal molar concentrations of purified, nucleotide-switched RagA and RagC were combined to make RagA-GTPγS/RagC-GDP dimers.
In Figures 3 and 5, glutathione beads containing GST-RagA, GST-RagC, and GST-Rheb were switched to a no-MgCl2 wash buffer (40 mM Hepes, 150 mM NaCl) and incubated at room temperature with 10 mM EDTA for 1 h on rotation to remove magnesium from the catalytic core of the GTPases and thereby dissociate bound nucleotides. EDTA was then removed by washing the samples ten times with MgCl2 wash buffer (40 mM Hepes, 150 mM NaCl, 5 mM MgCl2), whereafter samples were incubated with the specified nucleotides for 1 h at room temperature, on rotation, and washed 5 to 10 times with MgCl2 wash buffer.
Preparation of lysates
HEK293A cells were purchased from Thermo Fisher Scientific. MEFs were a kind gift from Dr John Blenis (Weill Cornell Medicine). Both cell lines were grown in 2D culture with DMEM high glucose (Thermo Fisher Scientific) and 10% fetal bovine serum (Thermo Fisher Scientific). Fresh bovine livers were kindly provided by Lövsta Kött. 0.5 to 2 g of each liver was homogenized by mincing the tissue with razor blades in the appropriate amount of ice-cold lysis buffer (40 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 0.2% CHAPS with cOmplete protease inhibitor cocktail from Roche). Mammalian cells were lysed in appropriate amount of ice-cold lysis buffer and passed through a 23G needle ten times. Samples were then centrifuged at the maximum speed for >30 min, 4 °C. Lysates were either directly incubated with nucleotide-switched GST-Rag GTPases, as described below, or processed by a dialysis step for 24 h, using a 100 kDa dialysis tubing (Spectrum Labs) and changing lysis buffer 3 to 5 times.
mTORC1 affinity isolation and elution
Glutathione beads containing nucleotide-switched GST-Rag GTPases were combined at a 1:1 ratio and incubated with lysates for 2 to 4 h on rotation at 4 °C. Samples were then washed ten times with lysis buffer without protease inhibitors, whereafter protein was either recovered by boiling the samples 5 to 10 min in Laemmli buffer with 5% 2-mercaptoethanol, or eluted, as specified in the figure legends. For elusion, protein was washed 5× with target-no-MgCl2 wash buffer (40 mM Hepes, 150 mM NaCl, 0.2% CHAPS), then incubated with target-no-MgCl2-EDTA wash buffer (40 mM Hepes, 500 mM NaCl, 0.2% CHAPS, 5 mM EDTA) for 3 h at room temperature. The supernatant/eluate was collected, and 10 mM MgCl2 was added and incubated at 4 °C for 1 h. High NaCl-containing eluate was diluted with target-No-NaCl wash buffer (40 mM Hepes, 0.2% CHAPS, 5 mM MgCl2) to reach a final NaCl concentration of 150 mM. The target-No-MgCl2 wash buffer and target-No-MgCl2-EDTA wash buffer eluates were each concentrated using a 100 kDa cutoff Amicon concentrator.
GST-Rags, GST-Rheb, and GST-4E-BP1 elution
Glutathione beads containing GST-Rag GTPases, GST-4E-BP1, or GST-Rheb were incubated with 10 mM reduced glutathione (Thermo Fisher Scientific) in MgCl2 wash buffer (40 mM Hepes, 150 mM NaCl, 5 mM MgCl2). Eluate was concentrated using a 10 kDa cutoff Amicon concentrator.
mTORC1 kinase assay
Increasing concentrations of HEK293A-isolated mTORC1 was incubated with a set amount of GST-4E-BP1 in 40 mM Hepes buffer with 5 mM MgCl2 and GST-Rheb in either the GTP-bound or GDP-bound nucleotide state for 30 min at room temperature. One millimolar ATP was added to each mixture and incubated for 1 h at 37 °C. Similarly, bovine isolate mTORC1 was incubated with a set amount of GST-4E-BP1 in 40 mM Hepes buffer with 5 mM MgCl2. Control samples were incubated with water, and treated samples were incubated with 1 mM ATP and incubated for 1 h at 37 °C. Samples were dotted onto nitrocellulose membranes and immunoblotting techniques were applied.
Coomassie imaging, dot blot, and Western immunoblot
Samples were boiled for 5 to 10 min in Laemmli sample buffer with 5 to 10% 2-mercaptoethanol. Five to fifty micrograms of lysate were resolved by SDS-PAGE and transferred to 0.2 μm nitrocellulose membranes (Bio-Rad Laboratories), after which membranes were blocked with EveryBlot blocking buffer (Bio-Rad Laboratories) for 1 h. For dot blots, 2.5 μl of kinase reaction assays were dotted onto nitrocellulose membranes (Bio-Rad Laboratories), dried, and then blocked with 2.5% nonfat milk. Primary antibodies were diluted in blocking buffer and incubated overnight at 4 °C. Secondary antibody (horseradish peroxidase conjugated from GE healthcare) incubations were performed in blocking buffer at room temperature for 1 h. Membranes were captured with an Amersham Imager 680 (GE HealthCare), using SuperSignal West Pico PLUS Chemiluminescent Substrate or SuperSignal West Femto Maximum Sensitivity Substrate from Thermo Fisher Scientific.
The following primary antibodies were purchased from Cell Signaling Technology: mTOR (#2983), Raptor (#2280), Rictor (#2114), GbetaL/LST8 (#3274), phospho-4E-BP1 Thr37/46 (#2855), RagA (#4357), RagC (#9480). Anti-GST was purchased from Santa Cruz Biotechnology (#sc-138) and anti-Sin1 from Sigma-Aldrich (#07-2276-I). In bovine liver samples, rabbit monoclonal mTOR Cell Signaling Technology #2983 strongly detected mTORβ and for the detection of full-length mTORα in these samples, a polyclonal antibody against mTOR (ab2732, Abcam) was utilized in line with a previous study (35). In several of the experiments, proteins of similar sizes (such as Rictor and Raptor or Raptor and mTOR) had to be analyzed on separate gels due to the interference of secondary antibodies.
For Coomassie blue stainings, gels were incubated with Coomassie brilliant blue G-250 (Bio-Rad) for 1 to 12 h and rinsed/washed in deionized water until protein band resolution was attained.
In Figure 3C, the SDS-page gel that had been stained with Coomassie blue was transferred to a nitrocellulose membrane so that the Coomassie stain was detectible on the membrane. After immunoblotting against mTOR as described above, both the anti-mTOR horseradish peroxidase signal and the Coomassie staining could be detected simultaneously on the Amersham Imager 680 (GE HealthCare).
Consurf structure assessment
Consurf analysis on 6U62.pdb (7) and 5WBI.pdb (14) was performed as per website instructions (https://consurf.tau.ac.il/) (36, 37, 38, 39, 40).
Data availability
All data supporting the findings of this study are available from the corresponding authors upon request.
Conflict of interest
Y. H. I. is the founder of Araucaria Laboratories Inc.
Acknowledgments
We thank Dr Pau Castel for his critical reading of our manuscript. We thank all members of the Mutvei lab, especially Huy Dang, for valuable input and assistance. We also thank members of the Theresa Vincent lab and Michael Andäng lab for important feedback. Bovine liver samples were kindly provided by Filip Lundgren, Lövsta Kött, Uppsala, Sweden.
Author contributions
Y. H. I. conceptualization; Y. H. I. methodology; Y. H. I. and A. P. M. validation; Y. H. I., S. P., and A. P. M. investigation; Y. H. I. and A. P. M. formal analysis; Y. H. I., S. P., and A. P. M. writing–original draft; Y. H. I. and A. P. M. writing–review and editing; Y. H. I. and A. P. M. visualization; Y. H. I. and A. P. M. supervision; Y. H. I. and A. P. M. funding acquisition; Y. H. I. and A. P. M. project administration.
Funding and additional information
This work was funded by Araucaria Laboratories Inc and supported by grants from The Swedish Research Council (2020-01126), The Swedish Cancer Society (21 0437 JIA, 20 1188 Pj), Åke Wibergs Foundation, Lars Hierta's Memory Foundation, and Magnus Bergvall Foundation to A. P. M.
Reviewed by members of the JBC Editorial Board. Edited by Alex Toker
Contributor Information
Yasir H. Ibrahim, Email: Ibrahim.y.h@araucarialabs.com.
Anders P. Mutvei, Email: anders.mutvei@igp.uu.se, anders.mutvei@ki.se.
References
- 1.González A., Hall M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Sahra I., Manning B.D. mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melick C.H., Jewell J.L. Regulation of mTORC1 by upstream stimuli. Genes (Basel) 2020;11:989. doi: 10.3390/genes11090989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mutvei A.P., Nagiec M.J., Hamann J.C., Kim S.G., Vincent C.T., Blenis J. Rap1-GTPases control mTORC1 activity by coordinating lysosome organization with amino acid availability. Nat. Commun. 2020;11:1416. doi: 10.1038/s41467-020-15156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buel G.R., Dang H., Asara J.M., Blenis J., Mutvei A.P. Prolonged deprivation of arginine or leucine induces PI3K/Akt-dependent reactivation of mTORC1. J. Biol. Chem. 2022;298:102030. doi: 10.1016/j.jbc.2022.102030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogala K.B., Gu X., Kedir J.F., Abu-Remaileh M., Bianchi L.F., Bottino A.M.S., et al. Structural basis for the docking of mTORC1 on the lysosomal surface. Science. 2019;366:468–475. doi: 10.1126/science.aay0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panasyuk G., Nemazanyy I., Zhyvoloup A., Filonenko V., Davies D., Robson M., et al. mTORbeta splicing isoform promotes cell proliferation and tumorigenesis. J. Biol. Chem. 2009;284:30807–30814. doi: 10.1074/jbc.M109.056085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., Guan K.-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiguchi T., Hirose E., Nakashima N., Ii M., Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 12.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash V., Carson B.B., Feenstra J.M., Dass R.A., Sekyrova P., Hoshino A., et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019;10:2110. doi: 10.1038/s41467-019-10100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Jiang X., Li B., Yang H.J., Miller M., Yang A., et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aylett C.H.S., Sauer E., Imseng S., Boehringer D., Hall M.N., Ban N., et al. Architecture of human mTOR complex 1. Science. 2016;351:48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 16.Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Xu C., Kirubakaran S., Zhang X., Hur W., Liu Y., et al. Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 2013;73:2574–2586. doi: 10.1158/0008-5472.CAN-12-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarbassov D.D., Bulgakova O., Bersimbaev R.I., Shaiken T. Isolation of the mTOR complexes by affinity purification. Methods Mol. Biol. 2012;821:59–74. doi: 10.1007/978-1-61779-430-8_5. [DOI] [PubMed] [Google Scholar]
- 19.Kim D.-H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 20.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 21.Tao Z., Barker J., Shi S.D.-H., Gehring M., Sun S. Steady-state kinetic and inhibition studies of the mammalian target of rapamycin (mTOR) kinase domain and mTOR complexes. Biochemistry. 2010;49:8488–8498. doi: 10.1021/bi100673c. [DOI] [PubMed] [Google Scholar]
- 22.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 23.Pina A.S., Lowe C.R., Roque A.C.A. Challenges and opportunities in the purification of recombinant tagged proteins. Biotechnol. Adv. 2014;32:366–381. doi: 10.1016/j.biotechadv.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell M.R., Engleka M.J., Malik A., Strickler J.E. To fuse or not to fuse: what is your purpose? Protein Sci. 2013;22:1466–1477. doi: 10.1002/pro.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frias M.A., Thoreen C.C., Jaffe J.D., Schroder W., Sculley T., Carr S.A., et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Wittinghofer A., Vetter I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 27.Tatebe H., Shiozaki K. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules. 2017;7:77. doi: 10.3390/biom7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garami A., Zwartkruis F.J.T., Nobukuni T., Joaquin M., Roccio M., Stocker H., et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 29.Dennis P.B., Jaeschke A., Saitoh M., Fowler B., Kozma S.C., Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B., Zhang Y., Wang Z., Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of rho family GTP-binding proteins. J. Biol. Chem. 2000;275:25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 31.Rudack T., Xia F., Schlitter J., Kötting C., Gerwert K. The role of magnesium for geometry and charge in GTP hydrolysis, revealed by quantum mechanics/molecular mechanics simulations. Biophys. J. 2012;103:293–302. doi: 10.1016/j.bpj.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Sacher M., Ferro-Novick S. Trapp stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battaglioni S., Benjamin D., Wälchli M., Maier T., Hall M.N. mTOR substrate phosphorylation in growth control. Cell. 2022;185:1814–1836. doi: 10.1016/j.cell.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Dong J., Yue K., Loor J.J., Aboragah A., Li G., Chen L., et al. Increased adipose tissue lipolysis in dairy cows with fatty liver is associated with enhanced autophagy activity. J. Dairy Sci. 2022;105:1731–1742. doi: 10.3168/jds.2021-20445. [DOI] [PubMed] [Google Scholar]
- 36.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., et al. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013;53:199–206. [Google Scholar]
- 38.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are available from the corresponding authors upon request.