Summary
Tissue boundaries and interfaces are engines of morphogenesis in vivo. However, despite a wealth of micropatterning approaches available to control tissue size, shape, and mechanical environment in vitro, fine-scale spatial control of cell positioning within tissue constructs remains an engineering challenge. To address this, we augment DNA “velcro” technology for selective patterning of ssDNA-labeled cells on mechanically defined photoactive polyacrylamide hydrogels. Hydrogels bearing photopatterned single-stranded DNA (ssDNA) features for cell capture are then co-functionalized with extracellular matrix (ECM) proteins to support subsequent adhesion of patterned tissues. ECM protein co-functionalization does not alter ssDNA pattern fidelity, cell capture, or hydrogel elastic stiffness. This approach enables mechanobiology studies and measurements of signaling activity at dynamic cell interfaces with precise initial patterning. Combining DNA velcro patterning and ECM functionalization provides independent control of initial cell placement, adhesion, and mechanics, constituting a new tool for studying biological interfaces and for programming multicellular interactions in engineered tissues.
Subject areas: Bioengineering, Tissue engineering, Cell biology
Graphical abstract

Highlights
-
•
DNA“velcro”enables precise cell patterning on photoactive BP-PA hydrogels
-
•
BP-PA hydrogels support stiffness tuning and ECM functionalization
-
•
Multiplexed DNA“velcro” supports long-term adhesion of patterned interfaces
-
•
Epithelial ERK signaling depends on tissue interface geometry
Bioengineering; Tissue engineering; Cell biology
Introduction
Tissues often consist of multiple cell populations with fine-scale spatial organization that is tightly linked to their biological function. These vary from specialized cellular niches that persist throughout adult life such as intestinal villi1 and hair follicles2 to transitional patterns within a single cell population, such as the differentiation of germ layers in the early embryo.3 Cell sorting and boundary formation mechanisms establish and enforce this hierarchical organization. Epithelial-mesenchymal interfaces are a form of heterotypic interaction that drive vertebrate embryonic tissue patterning, for example in tooth cusp formation,4,5 pharyngeal cartilage formation,6 and branching morphogenesis in the lung, salivary gland, and kidney.7,8 Disrupted epithelial-mesenchymal interactions can cause structural defects or organ agenesis.9 Barrier breakdown also presages dissemination of epithelial-derived tumor cells into healthy tissue.10 The spatial complexity of such interfaces in vivo demands that advanced co-culture systems created to reconstitute them in vitro have controlled cellular composition and geometry. Such systems would be particularly useful in organizing tissues for regenerative medicine or as models of cellular interactions in development and disease.
Recapitulating fine-scale tissue patterning at relevant length scales in vitro is a current challenge in tissue engineering. Micro-stencils and “wound healing” tissue culture inserts loaded with multiple cell populations produce simple and reproducible tissue interfaces,11,12 but these are low throughput and take hours for cells to close the intervening gap between cell populations. Other microstencil-based approaches can provide finer-scale hierarchical tissue patterning (∼100 μm) but involve stepwise surface passivation and de-blocking protocols between patterning different cell types.13,14 Microcontact printing or UV lithography can be used to deposit extracellular matrix (ECM) proteins into patterned features that confer cell-substrate adhesion and enforce geometric constraints down to the single cell scale (∼10 μm).15 Such approaches have been successfully implemented on glass, hydrogel, and elastomeric substrates.16,17,18 However, since the initial cell patterning and eventual adhesion footprints are one and the same, most ECM microprinting approaches are limited to assays where it is not necessary to study migration outside the patterned area. In some approaches, an initial cell population selectively adheres to a patterned ECM cue, while subsequent cells are added as backfill.19 Other photolithographic methods such as digital micromirror device projection or direct laser writing afford high spatial resolution and dynamic control over ECM patterning.20,21 These techniques permit hierarchical cell co-patterning by dynamically introducing new ECM ligand availability, for example, through biotin-avidin interactions,20 but remain limited by a lack of tools to engineer patterned cell deposition independently of ECM adhesion. Other approaches use non-uniform electric fields to selectively guide cell positioning into complex tissue patterns (such as liver lobules) onto an electrode-containing substrate.22 However, substrate requirements for these techniques limit their integration with compliant hydrogels or other favorable cell culture substrates.
DNA-programmed assembly of cells (DPAC) overcomes some of these limitations by patterning cells using printed ssDNA features as temporary adhesion ligands. Target cells are labeled with lipid-modified complementary ssDNAs and captured at the surface by Watson-Crick-Franklin base pairing.23,24 ssDNA deposits can be reliably printed at tissue-relevant spatial scales (∼10–100 μm) and cell patterning can be multiplexed, using a unique ssDNA sequence to place each cell type. DPAC approaches have previously relied on aldehyde-coated glass slides to support covalent linkage of ssDNA to the substrate. While this allows for subsequent cell adhesion to the glass or release and transfer into an encapsulating hydrogel overlay, reliance on a glass substrate limits other opportunities facilitated by compliant hydrogels, such as measurement of cell mechanical forces25 or influencing differentiation through substrate stiffness.26 We recently published a DPAC approach involving rapid photolithographic patterning of ssDNAs onto polyacrylamide hydrogels containing a benzophenone-methacrylate (BPMAC) co-monomer.27 BPMAC is photoactive, creating covalent bonds with polypeptides and ssDNA oligomers upon exposure to light wavelengths in the 250–365 nm range,28,29 permitting ssDNA photocapture.27 The photolithography process is rapid, allowing up to ∼107 ssDNA features to be simultaneously deposited across a large (square centimeters) hydrogel surface area within minutes.27 However, benzophenone-polyacrylamide (BP-PA) hydrogels are not inherently cell-adhesive, presenting an opportunity to control cell adhesion to gel-bound ECM cues by orthogonal means.
Here, we present a multi-step procedure for fabricating BP-PA hydrogel substrates, photopatterning ssDNA, and ECM functionalization. These substrates enable studies of non-autonomous cell interactions across patterned heterotypic cell interfaces in a convenient 2D format and with tunable surface chemistry and hydrogel mechanics. The ssDNA photolithography procedure is orthogonal to established polyacrylamide surface chemistry approaches that enable cell adhesion through surface-bound ECM proteins, meaning that cell patterning and cell adhesion chemistry can be independently controlled. Finally, we demonstrate that the boundary geometry alone influences cytoskeletal arrangements and extracellular signal-related kinase (ERK) activity at interfaces within micropatterned composite epithelial-mesenchymal tissues.
Results
BP-PA hydrogels support a combined cell patterning and 2D tissue culture strategy
Augmenting the already versatile polyacrylamide hydrogel system with DNA-patterned cell adhesion would enable the production of high-throughput patterned tissue interfaces, a performance benefit over existing micropatterning technology. We, therefore, established a multi-step process for photopatterning ssDNA, surface functionalization with ECM, and cell patterning onto BP-PA hydrogels. Quartz-chrome photomasks enable simultaneous deposition of up to 107 ssDNA features27 (∼10–100 μm feature size) and/or polypeptides through a UV photochemical reaction with BPMAC co-monomers in the hydrogel (Figures S1A and S1B). Photopatterned ssDNA features then define capture sites for lipid-ssDNA labeled cells, through base pairing (Figures 1A and S2). Washing the hydrogel removes excess cells, leaving cell patterns anchored to ssDNA features with high specificity (Figure 1B). In the present study, we used an “open face” format using a commercially available 8-well chambered slide that facilitates subsequent culture in a convenient format for imaging (STAR Methods and key resources table).
Figure 1.
Independent cell capture and adhesion on polyacrylamide substrates through orthogonal ssDNA and ECM patterning
(A) Schematic of top, ssDNA photolithography and BP-PA gel surface functionalization with fibronectin, and bottom, capture of lipid-ssDNA labeled cells. Cell labeling with lipid-ssDNA and sequence-matched “handle” ssDNA enables specific cell capture to hydrogel-bound ssDNA patterns. Captured cells subsequently adhere to the substrate using fibronectin ligands.
(B) Brightfield (top) and fluorescence (bottom) images of MDCK cells captured on a 500 μm diameter circular ssDNA feature.
(C) Quantification of MDCK cell capture (#) on 50, 100, 200, and 500 μm diameter ssDNA circles. Data depict n = 9 features per ssDNA diameter per three biological replicate experiments (n = 27 features total per ssDNA diameter) and means of each experiment (markers with black borders). Experiment means are shifted +30 μm along the +x axis for clarity.
(D) Top, maximum intensity projection (xy) of photopatterned ssDNA (polyT20F) and fibronectin on a BP-PA hydrogel from a 10x z stack (2.5 μm per frame, 147.5 μm total height, 59 total frames). Bottom, maximum xz intensity across the highlighted 25-pixel region.
(D’) Normalized mean fluorescence intensity sampled across the outlined portion of the xy image in panel D.
(E) Time lapse image sequences (10 h total) of cells adhering to circular, triangular, square, and star-shaped ssDNA features with fixed areas equivalent to a 200 μm diameter circle (A = 3.14x104 μm2).
(E’) Inset, detail of the star-shaped pattern. All experiments were performed on 7.5%/0.25% (Am/Bis) BP-PA hydrogels photopatterned with 200 μM polyT20G ssDNA for 90 s and functionalized with 20 μg mL−1 fibronectin. Cells were labeled with lipid anchors and “G’ handle” ssDNA. Cells in panel b are visualized using CellTracker Deep Red and ssDNA in B and E is visualized using 2x SYBR Gold nucleic acid stain. Photopatterned polyT20F ssDNA in panels D-D’ is visualized with FAM_F′ ssDNA probe and fibronectin are visualized with a rabbit anti-fibronectin primary antibody and Alexa 647 secondary antibodies. See also Figures S1 and S2 and Video S1.
To achieve specific cell capture, we used a previously described lipid-ssDNA “universal” anchor/co-anchor pair27,30 and a modular ssDNA adhesion system. Photopatterning ssDNA oligonucleotides each contain a string of 20 thymine bases followed by a 20-base pair adhesion sequence (e.g., polyT20X20). We denote the two used in the present study as “F” and “G” and confirmed that there was minimal cross-reactivity between the two sequences (Figure S2 and STAR Methods). “F’ handle” or “G’ handle” ssDNA bear the reverse complement sequence as well as an overlap sequence with the lipid anchor (Figure 1A) and are introduced to cells following membrane insertion of the lipid anchor and co-anchor. We previously determined that 60–120 s of 254 nm UV light exposure was sufficient to ligate concentrated ssDNA to the hydrogel and that these supported patterned cell capture.27 For our “open face” approach here, we first benchmarked cell capture efficiency on features of varying size. Madin-Darby canine kidney (MDCK) cells were efficiently captured on 50–500 μm diameter circular features using the G/G′ ssDNA pair, with larger features predictably capturing more cells (Figure 1C). However, we note that patterning on smaller features reduces the capture yield (only 11/27 or 41% of the 50 μm diameter ssDNA features we measured contained any captured cells). Capture efficiency can be improved to single cell resolution using a microfluidic flow cell to introduce and wash cells.27 These data show that our open-face assay design achieves similar cell capture properties to previous DPAC approaches for pattern sizes relevant to the study of tissue interfaces, but in a radically simpler format.
Turning to the goal of combining long-term cell adhesion with patterned cell capture, we established an orthogonal method for functionalizing the BP-PA hydrogel surface with ECM proteins. Providing ECM ligands allows captured cells to adhere to the hydrogel substrate through integrins and other cell adhesion receptors. To functionalize hydrogels with ECM, we modified a previously described photochemical reaction to derivatize acrylamide chains with N-hydroxysuccinimide (NHS) ester groups that couple proteins to the hydrogel surface through primary amines.31 We applied a mixture consisting of the UV photoinitiator compound lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), the hydrogel crosslinker N,N-methylenebisacrylamide (Bis), and an NHS ester-containing compound (acrylic acid-N-hydroxysuccinimide) to the hydrogel surface and exposed the entire hydrogel to 365 nm light for 10 minutes. After washing, we coupled ECM proteins (20 μg mL−1 fibronectin) to the hydrogel surface through overnight incubation (Figure 1D). To demonstrate the uniformity of ECM functionalization and ssDNA photopatterning, we probed the fibronectin distribution across ssDNA-patterned and unexposed hydrogel regions by immunofluorescence and confocal imaging. Fibronectin was distributed uniformly across non-exposed hydrogel regions, decreasing marginally in intensity (∼25%) across areas bearing photopatterned ssDNA (Figure 1D’). This NHS derivatization method gave the best cell adhesion results relative to protein ligation via UV-activation of BPMAC or another commonly used ECM functionalization technique (Sulfo-SANPAH). In both cases, we observed de-wetting of epithelial layers from previously UV-exposed regions (Figures S1C and S1D and STAR Methods).
To test whether BP-PA hydrogels functionalized with ECM would support long-term cell adhesion, we photopatterned ssDNA features of fixed area (A = 3.14x104 μm2, equivalent to a circle with 100 μm radius) but varying shape and introduced lipid-ssDNA-labeled MDCK epithelial cells (Figure 1E). Captured MDCK cells subsequently adhered to the ECM, formed cell-cell junctions, and began to collectively spread as epithelial colonies (Video S1). Importantly, the intervening ECM functionalization step did not prohibit the capture of lipid-ssDNA labeled cells, nor did ssDNA photopatterning prevent cell adhesion to hydrogel-bound ECM proteins. We tested the capacity of our functionalized BP-PA gels to support the adhesion of various cell types following capture on arrays of 250 μm circular ssDNA features, including epithelial cells (MDCK, LLC-PK1), fibroblasts (3T3), and embryonic tissue-derived cells (HEK 293T) (Figure S1E). After 24 hours, MDCK and LLC-PK1 cells maintained cell-cell contacts and spread as distinct colonies, while 3T3s spread and covered much of the hydrogel surface. HEK 293T cells are a weakly adherent cell line and spread poorly on fibronectin, suggesting that some cell types may require optimization of the ECM ligand or concentration to promote attachment. These data together show that augmenting our hydrogel-based DPAC approach with ECM functionalization facilitates long-term adhesion time-resolved studies of patterned tissue behavior over time.
Patterns were designed with a fixed area equivalent to a 200 μm diameter circle (A = 3.14 × 104 μm2). Images were acquired every 20 min for 10 h, starting 1 h after DNA patterning. See also Figure 1E.
BP-PA hydrogel photopatterning retains gel properties suited to mechanobiology studies
Polyacrylamide gels are desirable for mechanobiology studies because their elastic modulus (E) can be tuned by changing the relative amounts of acrylamide (Am) and Bis crosslinker in the pre-polymer solution.32 Changes in substrate elastic modulus influence mechanosensitive cell migration, proliferation, differentiation, and intracellular signaling.26,33,34,35,36,37 To validate that our co-functionalization approach is compatible with these favorable tuning properties, we cast BP-PA hydrogels with varying Am/Bis composition (3–7.5% Am, 0.035–0.25% Bis, Table S1) and indented them with a 255 μm blunt-ended cylindrical indenter attached to a force sensor (Figure 2A).38 Polyacrylamide hydrogels can be approximated as linear elastic materials,32,39 so we were able to directly measure E from the time-invariant linear part of force versus indentation depth curves.38 To test whether UV-induced ssDNA patterning influenced E, we exposed half of each hydrogel in the presence of ssDNA through a quartz microscope slide to permit 254 nm light transmission (+UV) and blocked the other half from UV exposure (ctrl). We measured similar values of E for ctrl and +UV hydrogel regions across a range of BP-PA gels cast from 3%–7.5% Am and 0.05%–0.25% Bis (Figure 2B). Hydrogels cast with 3%/0.05% Am/Bis ratio were ∼14% stiffer on the +UV side (ctrl: 2.4 ± 0.6 kPa, +UV: 2.8 ± 0.7; mean ± s.d. of n = 4 hydrogels, p = 0.016 by paired t-test), while UV exposure had an insignificant effect on E in all other cases. These data validate that BP-PA gels retain tunable elastic material properties following ssDNA photopatterning.
Figure 2.
Photopatterned BP-PA hydrogel mechanics, fibroblast spreading, and focal adhesion formation are comparable to control BP-PA gels
(A) Schematic of hydrogel UV exposure and microindentation with example force vs. indentation depth curve obtained for a 255 μm diameter cylindrical indenter on a 3%/0.05% Am/Bis ratio BP-PA hydrogel. Hydrogels were incubated in ssDNA (200 μM polyT20G) and exposed to 254 nm light for 90 s through one-half of a quartz slide (+UV), with the other side blocked from UV exposure (control).
(B) Quantification of E for BP-PA hydrogels cast with 3–7.5% Am, 0.01–0.25% Bis, and 3 mM BPMAC. See Table S1 for mean ± standard deviation (s.d.) for n = 2–4 hydrogels per Am/Bis composition. Individual data points are identified by shape, bar heights represent the overall mean.
(C) 3T3 fibroblasts spreading on +UV and control hydrogel regions functionalized with 20 μg mL−1 fibronectin. Cells adhered for 16–24 h prior to fixation and staining for F-actin and nuclei (DAPI).
(D) Quantification of 3T3 spread area on control and +UV hydrogel regions from BP-PA hydrogels cast with varying Am/Bis compositions; control, n = 108, 117, 116, 119, 110 cells and +UV, n = 92, 128, 129, 135, 110 cells. Data are pooled from two independent biological replicates (identified by marker shape), individual experiment means are overlaid onto distributions (black borders), p value comparisons between groups are from two-sided Kolmogorov-Smirnov tests.
(E) 3T3 fibroblast adhered to a 7.5%/0.25% (Am/Bis) BP-PA hydrogel containing 250 μm circular ssDNA features (labeled with FAM_F′), nuclei (DAPI), F-actin, and focal adhesions (vinculin). Insets, detail on regions marked by dashed boxes.
(F) Quantification of Ladhesion for cells on unexposed (off DNA) and UV-exposed (on DNA) hydrogel regions. Vertical dashed lines represent the mean of n = 94 (off DNA) and 87 (on DNA) focal adhesions measured from n = 29 cells pooled from two biological replicates. p value was computed using a two-sided Kolmogorov-Smirnov test.
We next asked whether photopatterning of BP-PA hydrogel surfaces influenced cell-hydrogel interactions. Fibroblast spreading increases with increasing E within a physiologically relevant range.36 Adherent 3T3 fibroblasts spread to similar extents on the control and +UV regions of BP-PA hydrogels across each Am/Bis composition that we tested (Figures 2C and 2D, Table S1). We also characterized the formation of focal adhesions, which are protein complexes that enable cell interpretation of substrate mechanical properties and ECM ligand density.40 To test whether focal adhesion formation differed between control and +UV regions, we let fibroblasts adhere for 16-24 h to 7.5%/0.25% (Am/Bis) hydrogels functionalized with 20 μg mL−1 fibronectin and bearing arrays of 250 μm circular photopatterned ssDNA features. Immunostaining for the focal adhesion protein vinculin allowed us to measure focal adhesion lengths (Ladhesion) on unexposed hydrogel surfaces and FAM_F′ probe-labeled ssDNA features (Figure 2E). We detected no difference in Ladhesion between adhesions on unexposed hydrogel (Ladhesion = 6.3 ± 3.2 μm, mean ± s.d. of n = 94 adhesions) and ones on ssDNA features (Ladhesion = 5.9 ± 2.6 μm, mean ± s.d. of n = 86 adhesions) (Figure 2F). ssDNA photopatterning on BP-PA hydrogels therefore adds a precise cell patterning capability without altering subsequent focal adhesion formation and cell spreading induced by ECM co-patterning.
Combining multiplexed ssDNA photolithography with ECM functionalization enables production and adhesion of patterned tissue interfaces
Precise spatial control over multiple cell populations would benefit studies of cell contact-related behaviors such as juxtacrine signaling or heterotypic cell adhesion. However, existing micropatterning techniques cannot reproduce complex multicellular patterns or interfaces at tissue-relevant length scale. To overcome this limitation, we took advantage of the minimal cross-reactivity between F/F′ and G/G′ ssDNA pairs during cell capture23,24,27 and used them to place multiple cell populations on photopatterned hydrogels. To align and register multiple ssDNA sequences on the same substrate, we adapted an iterative photopatterning process (Figure 3A), where each successive photomask pattern is registered to fiduciary features from previous steps using a complementary fluorescent ssDNA probe.23,27 We first used this process to create an alternating “chess board” ssDNA pattern and captured two distinct populations of MDCK cells expressing different fluorescent histone 2B (H2B) constructs to label cell nuclei (Figure 3B). We found no overall difference in mean cell capture efficiency between the two populations (Figure S3). Patterned cells adhered to surface-bound fibronectin ligands within 1 hour of patterning and spread outward to form cell-cell contacts with neighboring patterned populations, ultimately establishing a confluent monolayer after 24 hours. Within this time period, cell patterns retained aspects of their initial geometric organization, while showing some boundary evolution through movement and mixing, which typically occurs on longer timescales (days) in confluent monolayers consisting of a single epithelial cell type.13 These data show that serial ssDNA photopatterning steps can be performed without compromising cell patterning fidelity or ECM functionalization, enabling the formation of controlled tissue interfaces.
Figure 3.
ssDNA multiplexing supports production and adhesion of patterned tissues containing multiple cell populations
(A) Schematic of iterative ssDNA photopatterning.
(B) Schematic for lipid-ssDNA labeling and patterning of multiple cell populations. Note that surface fibronectin functionalization is not shown schematically but is performed before cell capture.
(C) Confocal micrograph of polyT20F and polyT20G sequences photopatterned into a chess board pattern of alternating squares (250 μm side length) across an entire culture well. ssDNAs were sequentially patterned onto a 7.5%/0.25% (Am/Bis) BP-PA hydrogel (t = 60 s exposure for each ssDNA). Hydrogels were subsequently functionalized with 20 μg mL−1 fibronectin. ssDNA patterns are visualized with 1 μM FAM_G′ and 1 μM Cy5_F′ fluorescent ssDNA probes.
(D) Two populations of MDCKs expressing fluorescent H2B constructs captured on a similar chess board pattern. Cells were imaged t = 1 h after capture and at t = 24 h. MDCK H2B-Venus cells were patterned using the G/G′ ssDNA pair and MDCK H2B-iRFP cells were patterned using the F/F′ ssDNA pair (Table S1). Inset, detailed view of interface. Data are representative of two biological replicates. See also Figure S3.
Patterned epithelial-mesenchymal interactions support investigation of physical and biochemical interactions between cell populations
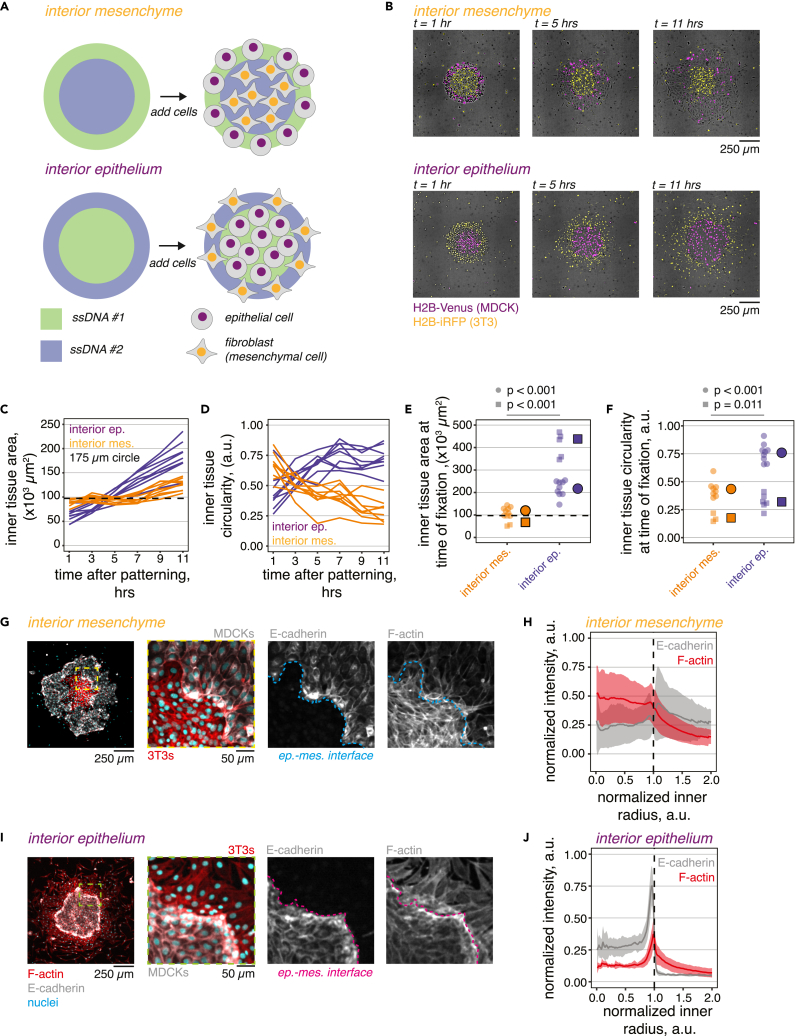
Interacting epithelial and mesenchymal cell populations are crucial to the early formation of a number of embryonic organs.4,6,9 This motivates development of culture technologies such as the present DNA patterning approach to accelerate understanding and synthetic construction of epithelial-mesenchymal interfaces and tissue boundaries. We first sought to test whether heterotypic interface configuration impacts cell behavior by designing a two-part photomask pattern that splits a 500 μm diameter circle into a concentric circle and annulus of equal area (rinner = 176 μm, router = 250 μm, Ainner = Aouter = 97264 μm2). This design creates composite tissues having one cell type enclosed within another (Figure 4A), and we refer to the two tissue configurations by the “interior” cell population (e.g., “interior mesenchyme” when 3T3s are placed within a ring of MDCKs).
Figure 4.
Epithelial-mesenchymal tissue interfaces show emergent organization that depends on initial tissue geometry
(A) Schematic of two-part ssDNA patterning design consisting of concentric circles.
(B) Example images of co-patterned H2B-iRFP 3T3s and H2B-Venus MDCKs acquired starting 1 h after patterning and every 2 h for 10 h total.
(C and D) Inner tissue area and circularity measured from n = 8 interior mesenchyme and n = 9 interior epithelium tissues from an example experiment.
(E and F) Summary plots of tissue area and circularity at time of fixation (t = 12–15 h after patterning) measured for n = 11 interior mesenchyme and n = 15 interior epithelium tissues collected from two independent replicates. Individual experiment means (markers with black borders) are offset to the right of each group and data points for each experiment are organized by shape. p values for each experiment are computed using Welch’s two-sided t-test. Dashed horizontal line in panels c and e represents the inner tissue patterning radius (r = 175 μm).
(G) Confocal micrograph of an “interior mesenchyme” microtissue stained for F-actin and E-cadherin. Inset, composite and single channel images show alignment of epithelial cells and fibroblasts at the interface (dashed lines in each panel).
(H) Radial quantification of normalized E-cadherin and F-actin fluorescence intensity relative to inner tissue radius.
(I and J) Example “interior epithelium” tissue stained for E-cadherin and F-actin and radial quantification of normalized fluorescence intensity relative to inner tissue radius. Vertical dashed line marks the tissue interface position (r = 1.0 a.u.), ribbons are mean ± s.d. of tissues analyzed in panel e. In all experiments, MDCK cells are patterned using the G/G′ ssDNA pair, while 3T3 cells are patterned using the F/F′ ssDNA pair. Cells were patterned on 7.5%/0.25% (Am/Bis) BP-PA hydrogels sequentially photopatterned with polyT20G and polyT20F ssDNA (t = 90 s exposure each) and functionalized with 20 μg mL−1 fibronectin. See also Figures S4 and S5 and Videos S2 andS3.
After producing BP-PA hydrogels bearing the two photopatterned ssDNAs and functionalizing them with ECM, we co-patterned MDCK epithelial cells and 3T3 fibroblasts and tracked their interactions using live time lapse imaging (Figure 4B). In interior mesenchyme tissues, the inner fibroblast population underwent modest expansion in area outward against the epithelial boundary (Figures 4B and 4C and Video S2). In some cases, fibroblasts exploited discontinuities in the epithelial ring to escape from the interior (see an example in Figure S4A). We also observed a progressive decrease in circularity among interior mesenchyme tissues (Figure 4D), reflecting increasingly irregular shapes of the fibroblast fields caused by tissue interface instability and motion over time. In the reverse configuration (“interior epithelium” tissues) the collective outward expansion of epithelial cells (Figures 4B and 4C) caused compression of fibroblast fields at interfaces, while more distant fibroblasts were able to move toward less densely packed areas. Compression increased their local density and alignment at the tissue edge (Video S3). As a result, we observed a progressive increase in inner tissue area (Figure 4C) and circularity (Figure 4D) across the imaging duration as the inner epithelial colony expanded. Endpoint measurements of interior tissue area and circularity in culture (two experiments fixed and stained at 12 and 15 h after patterning, respectively) revealed consistent tissue organization between the two timepoints (Figures 4E and 4F). We patterned and followed a separate batch of composite tissues in culture over a period of 5 days (Figure S5A). In these experiments, MDCK cells expanded to fill the majority of the well, forming new boundaries with adjacent tissues and compressing groups of 3T3 cells upon contact. We found similar results for tissues patterned on soft BP-PA gels (3%/0.05% Am/Bis) (Figure S5B), indicating that our approach is compatible with hydrogels of varying elastic stiffness. These data show that patterning boundary conditions can impact transient interface properties, even though the context of cell interaction at the interface is identical at the single-cell scale.
Patterns consist of an inner circle (r = 175 μm) surrounded by an annulus (r = 75 μm), each with a nominal area of A = 97264 μm2. Images were acquired every 2 h for 10 h total, starting 1 h after DNA patterning. See also Figure 4B.
Images were acquired every 2 h for 10 h total, starting 1 h after DNA patterning. See also Figure 4B.
Heterotypic tissue contacts can induce collective alignment of cells and formation of supra-cellular cytoskeletal structures at the interface.14,41,42,43 To investigate cytoskeletal organization at tissue interfaces in our system, we stained tissues for an epithelial cell marker (E-cadherin) and F-actin. Patterning tissues into a reproducible starting geometry enabled us to quantify the radial intensity of each marker.44 Fibroblasts within interior mesenchyme tissues were densely packed and showed an accumulation of actin stress fibers, leading to an increase in F-actin staining intensity within the tissue compared to the surrounding epithelium (Figures 4G and 4H). By contrast, interior epithelial tissues had a sharp accumulation of actin and E-cadherin at the interface (Figure 4J). Elongated cell morphologies and alignment perpendicular to the interface (accompanied by accumulation of actin stress fibers within both cell populations, Figure 4I) suggest elevated tension in the interface cells, which may prohibit cell movements or division across the boundary.14,41 Differences in cell density between colliding tissues may also influence the time evolution of boundary formation, with less dense tissues being displaced by denser ones at the collision site.12 In our experiments, cell density (using radially binned average signal from DAPI-labeled nuclei as an approximation) was markedly higher in interior fibroblast tissues compared to their density in the opposite patterning scheme when they bordered epithelial cells (Figure S5C). These data indicate that boundary configuration alone impacts cell behavior and cytoskeletal arrangements within cells present at the interface.
Elevated ERK signaling activity in interfacial populations of epithelial cells
We next investigated how signaling may change between the interfaces, since mesenchymal ERK signaling regulates epithelial-mesenchymal boundary integrity and sorting behaviors in cell mixing and wound healing assays.11 In epithelial tissues consisting of a single cell type, ERK activity is often spatially anisotropic across the tissue, with highest activity at sites of active remodeling, growth, mechanical stress, and the free edges of collectively migrating colonies.45,46,47 By contrast, epithelial ERK signaling activity has not been as thoroughly investigated in the context of a tissue interface with mesenchymal cells, providing an opportunity to investigate ERK signaling within the patterned epithelial-mesenchymal tissues produced by our system. We fixed and stained for active (phosphorylated) ERK (pERK, Figure 5A) and quantified the intensity within each tissue and across the interface (Figure 5B). Radial quantification of immunofluorescence signal revealed that pERK levels were highest within the epithelial population in each patterning scheme, with peak intensities in cells located immediately adjacent to the interface and decreasing farther away within the tissue. Epithelial ERK activation was higher in the “interior mesenchyme” arrangement than the opposite arrangement. Our results indicate that the tissue-level context of epithelial-mesenchymal cell contacts modulate ERK activity within the epithelial population. Interfacial effects on cell signaling may also impact tissue proliferation, motility, and morphogenesis depending on the spatial organization of cell types within the tissue, providing opportunities for future study.
Figure 5.
ERK signaling activity at tissue interfaces depends on interface boundary conditions
(A) Confocal immunofluorescence for pERK, F-actin, E-cadherin, and nuclei in “interior mesenchyme” and “interior epithelium” tissues and intensity-colored images of pERK staining.
(B) Radial quantification of normalized pERK intensity relative to inner tissue radius for each pattern type. Vertical dashed line marks the tissue interface position (r = 1.0 a.u.), ribbons are mean ± s.d. of normalized pERK intensity for n = 10 interior epithelium and n = 15 interior mesenchyme tissues pooled from two biological replicates. Cells were patterned on 7.5%/0.25% (Am/Bis) BP-PA hydrogels sequentially photopatterned with polyT20G and polyT20F ssDNA (t = 90–120 s exposure) and subsequently functionalized with 20 μg mL−1 fibronectin. All tissues were patterned as described in Figure 4 and kept in culture for 12-15 h to permit interface formation before fixation and staining.
Discussion
Despite their crucial role in morphogenesis and disease, the spatial evolution and cell-cell signaling properties of tissue interfaces are challenging to study in vivo since signaling is often non-cell-autonomous and interface responses can be transient and difficult to analyze in the full 3D tissue context. Creating precise interfaces between multiple cell types at tissue-relevant length scale (∼10–100 μm) has also been challenging to achieve with existing hydrogel micropatterning approaches. New engineering approaches to interface construction are required to increase the spatial uniformity, imaging accessibility, and throughput of such studies. Our results extend our previous work,27 establishing BP-PA hydrogels as a chassis for orthogonal control of ssDNA-directed cell patterning and cell-ECM adhesion through stepwise photopatterning. This enables both precise design of initial conditions and tracking of adhered interface dynamics for the first time. Importantly, this fabrication process does not significantly alter hydrogel mechanics, cell spreading, or focal adhesion formation, demonstrating that ssDNA patterned hydrogels are suitable for mechanobiology studies. Finally, we identified distinct changes in cellular and cytoskeletal organization and cell signaling at heterotypic cell interfaces consisting of the same two cell populations as a function of interface organization alone.
We demonstrate that cell capture can be reliably performed on ∼100 μm-scale features within commercially available clip-on slide chambers, allowing closer adherence to traditional culture protocols. Previous DPAC approaches required specialized, time-intensive, and error-prone microfluidic deposition of ssDNA and/or cells23,27 or iteratively stripping and re-applying photoresist layers onto the substrate between each ssDNA photopatterning step.24 Here, hundreds to thousands of ssDNA features can be simultaneously deposited across each BP-PA hydrogel using 1–2-min UV exposure times and ∼20 min of subsequent washing steps, allowing a trained user to complete the entire process in ∼30 min. Manually aligning and creating a second ssDNA pattern to the first can then be completed within ∼1 h. Despite vastly simplifying the cell patterning workflow, one tradeoff to our use of commercially available slide chambers is that fluidic control during washing is less precise than previous approaches and potentially exposes cells to higher shear forces during patterning. This can be mitigated by using a microfluidic flow chamber23,24,27 to reduce the shear forces from washing and direct patterning down to the single cell scale (∼10 μm feature size). Finally, some cell lines may require additional optimization to improve patterning efficiency or ECM adhesion (Figure S2).
The DNA patterning approach described here affords several experimental handles to tune properties of the engineered tissue microenvironment. Lipid-oligonucleotide labeling of cells is performed in standard culture medium and is therefore compatible with a wide variety of cell types, including stem cells and primary cells.23,24 Modifying polyacrylamide hydrogel chemistry allows us to tune the elastic hydrogel stiffness and could be extended to incorporate fluorescent fiduciary markers or beads for the measurement of cell-generated traction forces.25 Moreover, our fabrication process uses standard, commercially available reagents, and UV light sources, with both BPMAC and lipid-ssDNA available by commercial synthesis. We note that BPMAC can also be obtained through straightforward chemical synthesis.29 Recent work by Cabral and colleagues also identified commercially available cholesterol-modified ssDNA48 that is interchangeable with custom lipid-ssDNA used here.
In summary, we have established techniques to spatially control initial cell placement on mechanically defined BP-PA hydrogels using multiplexed ssDNA photolithography and lipid-ssDNA cell capture. We find that ssDNA photopatterning is compatible with existing polyacrylamide functionalization chemistry and elastic stiffness tuning (Figure S5B), allowing the future incorporation of additional spatially varying features such as stiffness gradients37 into the fabrication process. Access to patterned co-cultures would complement mechanistic in vivo studies of tissue boundary integrity41,43 or benefit in vitro studies to engineer signaling pathways that confer boundary or positional information to tissues, such as Eph/ephrin49 or planar cell polarity.50 For example, Brayford et al. reported that mesenchymal contact inhibition drives sorting of epithelial and mesenchymal cells into distinct populations in vitro, as well as the formation of stable boundaries where the two populations interact.11 They also found that boundary stability in vitro involves signaling through Eph/ephrin and mesenchymal ERK signaling. However, as their experiments used wound healing inserts to pattern a millimeters-long interface, it is unclear how epithelial-mesenchymal interfaces would evolve given different patterning configurations and length-scales. Eph/ephrin signaling establishes stable boundaries in homotypic epithelial cultures,51 suggesting that boundary properties are influenced by cell identity or mechanical state - an interesting question for continued exploration in patterned co-culture systems. DNA-patterned cell colonies on BP-PA hydrogels could also be used in conjunction with synthetic “sender/receiver” stem cell models18 to interrogate the combined roles of mechanics, geometry, and diffusible signaling relays in germ layer patterning. Advanced co-culture models of tumor-stroma interactions52 or cell competition mechanisms53 are other promising applications for this approach.
Limitations of the study
In the present work, we demonstrate co-patterning of up to two cell types using ssDNA, while other DPAC approaches have patterned up to four biological components (cells or proteins) using unique ssDNA sequences.23,24,48 However, new patterning ssDNA sequences can be readily designed to minimize cross-reactivity with other oligonucleotides in the experimental scheme. To support cell adhesion, we functionalized BP-PA hydrogel surfaces with ECM proteins by adapting established hydrogel chemistry.31 After patterning, we also observed a decrease in fibronectin staining intensity across photopatterned DNA features via immunofluorescence (Figures 1D and 1D’). Optimizing hydrogel chemistry in future studies will reveal whether other ECM functionalization schemes can provide more even protein ligation conditions. Finally, we leveraged precise multiplexed cell patterning to construct chimeric tissues consisting of MDCK cells (canine epithelial cells) and 3T3 fibroblasts (mouse mesenchymal cells). This allowed us to produce patterned heterotypic tissue interfaces and observe distinct organizational and ERK signaling phenotypes for different interface configurations (Figures 4 and 5). Interfacial interactions will likely be organism- and tissue-specific, requiring users to adapt our technology and perform additional validation using epithelial and mesenchymal cells suited to their biological question.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-E-cadherin (clone 34) | BD Biosciences | Cat#610404; RRID: AB_397787 |
| Mouse anti-vinculin (7F9) | eBioscience | Cat#14-9777-82; RRID: AB_2573028 |
| Rabbit anti-E-cadherin (24E10) | Cell Signaling Technologies | Cat#3195; RRID: AB_2291471 |
| Rabbit anti-fibronectin | Abcam | Cat#ab23750; RRID: AB_447655 |
| Rabbit anti-phospho-p44/42 MAPK (ERK1/2) | Cell Signaling Technologies | Cat#4370; RRID: AB_2315112 |
| Donkey anti-mouse Alexa Fluor™ 555 | ThermoFisher | Cat#A31572; RRID: AB_162543 |
| Donkey anti-rabbit Alexa Fluor™ 488 | ThermoFisher | Cat#A21206; RRID: AB_2535792 |
| Donkey anti-rabbit Alexa Fluor™ 555 | ThermoFisher | Cat#A31572; RRID: AB_162543 |
| Donkey anti-rabbit Alexa Fluor™ 647 | ThermoFisher | Cat#A31573; RRID: AB_2536183 |
| Chemicals, peptides, and recombinant proteins | ||
| CD-26 developer | Singh Center for Nanotechnology; Shipley | N/A |
| Remover 1165 | Singh Center for Nanotechnology; Shipley | N/A |
| Chromium etchant | Sigma | Cat#651826 |
| SU-8 2025 photoresist | Kayaku Advanced Materials Inc. | Cat#NC9981681 |
| SU-8 developer | Kayaku Advanced Materials Inc. | Cat#NC9901158 |
| 3-(trimethoxysilyl)propyl methacrylate | Sigma | Cat#440159; CAS: 2530-85-0 |
| Acrylamide (40% Am solution) | BioRad | Cat#1610140; CAS: 79-06-1 |
| N,N-methylenebisacrylamide crosslinker (2% Bis solution) | BioRad | Cat#1610142; CAS: 110-26-9 |
| N-[3-[(4-benzoylphenyl) formamido]propyl] methacrylamide | PharmAgra custom synthesis; Hughes & Herr29 | CAS: 165391-55-9 |
| Dichlorodimethylsilane | Sigma | Cat#440272; CAS: 75-78-5 |
| Sodium dodecyl sulfate | BioRad | Cat#161–0301; CAS: 151-21-3 |
| Triton X-100 | MilliporeSigma | Cat#T9284; CAS: 9036-19-5 |
| Ammonium persulfate | Sigma | Cat#A3678; CAS: 7727-54-0 |
| N,N,N′,N′-Tetramethyl ethylenediamine | Sigma | Cat#T9281; CAS: 110-18-9 |
| Bovine serum albumin | Sigma | Cat#A2153; CAS: 9048-46-8 |
| Bovine serum albumin, Alexa Fluor™ 555 conjugate | ThermoFisher | Cat#A34786 |
| HEPES free acid | BioWorld | Cat#40820004; CAS: 75277-39-3 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate | Sigma | Cat#900889; CAS: 85073-19-4 |
| Acrylic acid-N-hydroxysuccinimide ester | Sigma | Cat#A8060; CAS: 38862-24-7 |
| Sodium chloride | Sigma | Cat#S6191; CAS: 7647-14-5 |
| Fibronectin | Sigma | Cat#F1141 |
| Glycine | Sigma | Cat# G8898; CAS: 56-40-6 |
| Sulfo-SANPAH | ThermoFisher | Cat#22589 |
| Alexa Fluor™ 647 phalloidin | Sigma | Cat#A22287 |
| Critical commercial assays | ||
| SYBR™ Gold nucleic acid gel stain | ThermoFisher | Cat#S11494 |
| CellTracker™ Deep Red dye | ThermoFisher | Cat#C34565 |
| Deposited data | ||
| Photomask designs | This paper; Mendeley Data | https://data.mendeley.com/datasets/93pvd2tff2; https://doi.org/10.17632/93pvd2tff2.1 |
| MATLAB and Fiji/ImageJ analysis scripts | This paper; Mendeley Data | https://data.mendeley.com/datasets/93pvd2tff2; https://doi.org/10.17632/93pvd2tff2.1 |
| Raw and analyzed data | This paper; Mendeley Data | https://data.mendeley.com/datasets/93pvd2tff2; https://doi.org/10.17632/93pvd2tff2.1 |
| Experimental models: Cell lines | ||
| Canine: MDCK-II cell line | gift from Arjun Raj; MilliporeSigma | Cat#00062107-1VL |
| Canine: MDCK-II H2B-Venus | Viola et al.27 | N/A |
| Canine: MDCK-II H2B-iRFP670 | This paper | N/A |
| Human: LentiX™ HEK 293T cells | gift from Lukasz Bugaj; Takarabio | Cat#632180 |
| Mouse: NIH 3T3 fibroblasts | gift from Lukasz Bugaj; ATCC | Cat#CRL-1658 |
| Mouse: NIH 3T3 H2B-iRFP670 | Viola et al.27 | N/A |
| Pig: LLC-Pk1 cells | gift from David Odde; ATCC | Cat#CL-101 |
| Oligonucleotides | ||
| Photopatterning oligo: polyT20F: 5′-TTTTTTTTTTTTTTTTTTTTA GAAGAAGAACGAAGAAGAA-3′ |
IDT | N/A |
| Photopatterning oligo: polyT20G: 5′-TTTTTTTTTTTTTTTTTTTTA GCCAGAGAGAGAGAGAGAG-3′ |
IDT | N/A |
| Lipid ssDNA: Universal Anchor: 5′-TGGAATTCTCGGGTGCCAA GGGTAACGATCCAGCTGTCACT-lignoceric-amide-3′ |
Oligo Factory custom synthesis; Viola et al.27 | N/A |
| Lipid ssDNA: Universal Co-Anchor: 5′-palmitic-amide-AGTGAC AGCTGGATCGTTAC-3′ |
Oligo Factory custom synthesis; Viola et al.27 | N/A |
| Cell patterning oligo: F′ handle: 5′-CCTTGGCACCCGAGAATT CCATTTTTTTTTTTTTTTTTTTTTTCTTCTTCGTTCTTCTTCT-3′ |
IDT | N/A |
| Cell patterning oligo: G′ handle: 5′-CCTTGGCACCCGAGAAT TCCATTTTTTTTTTTTTTTTTTTCTCTCTCTCTCTCTCTGGCT-3′ |
IDT | N/A |
| Fluorescent probe oligo: FAM_F’: 5′-5(6)-carboxyfluorescein-TT TCTTCTTCGTTCTTCTTCT-3′ |
IDT | N/A |
| Fluorescent probe oligo: FAM_G’: 5′-5(6)-carboxyfluorescein-CT CTCTCTCTCTCTCTGGCT-3′ |
IDT | N/A |
| Fluorescent probe oligo: 5′-Cy5-TTTCTTCTTCGTTCTTCTTCT-3′ | IDT | N/A |
| Recombinant DNA | ||
| Plasmid: pCMV delta R8.91 | gift from Didier Trono; Addgene | Cat#12263; RRID: Addgene_12263 |
| Plasmid: pLentiPGK DEST H2B-iRFP | gift from Markus Covert; Addgene | Cat#90237; RRID: Addgene_90237 |
| Plasmid: pMD2.G | gift from Didier Trono; Addgene | Cat#112854; RRID: Addgene_112854 |
| Software and algorithms | ||
| LayoutEditor | juspertor GmBH | https://layouteditor.com/ |
| Fiji/ImageJ | Schindelin et al.54 | https://imagej.nih.gov/ij/ |
| Clock Scan ImageJ plugin | Dobretsov et al.55 | https://sourceforge.net/projects/clockscan/ |
| MATLAB_R2022a | TheMathWorks Inc. | https://www.mathworks.com/products/matlab.html |
| MATLAB Curve Fitting Toolbox | TheMathWorks Inc. | https://www.mathworks.com/products/curvefitting.html |
| MATLAB Optimization Toolbox | TheMathWorks Inc. | https://www.mathworks.com/products/optimization.html |
| R v 4.2.0 | R Core Team | https://www.r-project.org/ |
| Other | ||
| DLW-66+ direct laser writer | Singh Center for Nanotechnology; Heidelberg Instruments | N/A |
| 5”x5”x0.90″ chrome-on-quartz photomask, IP3500 photoresist | Singh Center for Nanotechnology | N/A |
| 4″ mechanical grade silicon wafer | Singh Center for Nanotechnology; University Wafer, Inc. | N/A |
| Spin coater with vacuum chuck | Instras | SCK-300P |
| 365 nm mounted LED | ThorLabs | M365LP1 |
| Ultrasonic cleaning bath | ThermoFisher | Cat#15-337-411 |
| UV crosslinker oven | Spectro-UV | XL-1000 |
| Portable glove box | SP Bel-Art | H50028-2001 |
| 8-well cell culture slide | Mattek | Cat#CCS-8 |
| FluoSpheres™ carboxylate-modified microspheres, Dark Red, 0.2 μm diameter | ThermoFisher | Cat#F8807 |
| 1″ x 3″ quartz slide, 1 mm thick | Ted Pella | Cat#26011 |
| Stepper motor | Levental et al.38; Nanotec | L4018S1204-M6 |
| Langmuir monolayer trough tensiometer | Levental et al.38; Kibron Inc. | MicroTrough X |
| SAE 316L stainless steel wire, 255 μm cylindrical diameter | Levental et al.38 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alex Hughes (ajhughes@seas.upenn.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines
MDCK-II epithelial cells (female, 00062107-1VL, Millipore Sigma) were cultured in minimum essential medium (MEM, Earle’s salts and L-glutamine, MT10-010-CV, Corning) supplemented with 10% fetal bovine serum (FBS, MT35-010-CV, Corning) and 1x pen/strep (100 IU mL−1 penicillin, and 100 μg mL−1 streptomycin, 100x stock, 15140122, Invitrogen) and passaged with 0.25% trypsin-EDTA (2530056, Corning). 3T3 mouse embryonic fibroblasts (CRL-1658, ATCC) were maintained in Dulbecco’s minimum essential medium (DMEM, 4.5 g L−1 glucose, L-glutamine, and sodium pyruvate, MT10-013-CV, Corning) supplemented with 10% calf serum (SH3008703, HyClone) and 1x pen/strep. LLC-Pk1 porcine kidney epithelial cells (male, CL-101, ATCC) were cultured in MEM supplemented with 10% FBS, 1 mM sodium pyruvate (100 mM stock, 11360070, Invitrogen), and 1x pen/strep and passaged with 0.25% trypsin-EDTA. Human embryonic kidney (HEK 293T) cells (Lenti-X 293, 632180, Takarabio) were cultured in DMEM supplemented with 10% FBS and 1x pen/strep and passaged with 0.05% trypsin-EDTA. MDCK H2B-Venus and 3T3 H2B-iRFP cell lines were generated by viral transduction in a previous publication.27 All cells were cultured at 37°C and 5% CO2 in polystyrene 75 cm2 or 182 cm2 flasks kept in a humidified incubator. Cell lines used in this study have not been authenticated.
Method details
Benzophenone-polyacrylamide gels
1″ x 3″ glass microscope slides (12-550-A3, ThermoFisher) were cleaned with 0.1% Triton X-100 (T9284, Sigma-Aldrich), rinsed in water, etched in 1 N sodium hydroxide (NaOH, S8045, Sigma) for 10 min, and dried under compressed air before coating one slide surface with a layer of 3-(trimethoxysilyl)propyl methacrylate (440159, Sigma) and acetic acid in DI water for 30 min to create a surface layer of methacrylate groups that facilitates hydrogel attachment to the slide.29 Methacrylate-coated slides were immersed in methanol, washed in DI water, dried under compressed air, and stored for up to 2 weeks before use.
Uniform 30 μm thick hydrogel sheets were cast between the methacrylate-coated glass slide surface and a mechanical grade 4″ diameter silicon wafer (University Wafer) containing guide rail shims made from cured SU-8 2025 photoresist (NC9981681, Kayaku Advanced Materials) and rendered hydrophobic by vapor deposition of ∼1 mL dichlorodimethylsilane (DCDMS, 440272, Sigma-Aldrich) in vacuo for ∼10 min. To make wafers, SU-8 was spin coated to 30 μm thickness (SCK-300P, Instras Scientific) onto the wafer surface and exposed to 365 nm UV light (M365LP1, ThorLabs) for 30 s through a custom Mylar mask printed at 20,000 d.p.i. (CAD/Art Services). Excess photoresist was removed using SU-8 developer (NC9901158, Kayaku) followed by alternating washes in isopropanol and acetone. All spin coating, exposure, and heating steps were performed according to manufacturer guidelines. Wafers were washed with alternating 0.1% Triton X-100 and water and dried under compressed air between uses.
BP-PA hydrogel precursor solution contained 3–7.5% acrylamide (40% w/v Am stock, 1610142, BioRad), 0.035–0.25% N,N-methylenebisacrylamide crosslinker (2% w/v Bis stock, 1610140, BioRad), 0.06% SDS (5% w/v stock in DI water; 161–0301, Bio-Rad), 0.06% Triton X-100 (5% w/v stock in DI water; T9284, Sigma), and 3 mM N-[3-[(4-benzoylphenyl) formamido]propyl] methacrylamide (BPMAC, custom synthesis, PharmAgra) and 10x Dulbecco’s phosphate buffered saline (DPBS; 14200075, ThermoFisher), diluted to a 1x final DPBS concentration in DI water. First, a partial precursor solution containing Am, Bis, DPBS, and DI water was degassed under vacuum in an ultrasonic cleaning bath (15-337-411, ThermoFisher). SDS, Triton X-100, and BPMAC were then successively added to the precursor with brief vortexing steps between each addition. To initiate polymerization, we successively added 0.05% (w/v) ammonium persulfate (APS, A3678, Sigma) and 0.05% (v/v) N,N,N′,N′-tetramethylenediamine (TEMED, T9281, Sigma) and briefly mixed by vortexing. Precursor was injected into the gap between the methacrylate-coated glass slide surface and hydrophobic silicon wafer using a P200 pipette tip and allowed to polymerize for ∼30 min. Polymerized hydrogels were rehydrated in 1x DPBS (MT21-031-CV, Corning), carefully lifted off the wafer using a razor blade, and either stored in 1x DPBS at 4°C overnight or used immediately for photolithography. BPMAC was diluted to 100 mM in DMSO and stored in working aliquots at −20°C. APS was diluted to a 10% w/v stock solution in DI water and stored in working aliquots at −80°C and TEMED was freshly diluted to a 10% v/v stock solution in DI water. Component volumes and Am/Bis ratios are reported in Table S1.
Oligonucleotides
Photopatterning ssDNA consists of 3′-T20X20-5′ sequences, where T20 is a 20-thymine base sequence and X20 is a unique 20 base sequence. We used two previously described sequences that exhibit minimal cross-reactivity during cell patterning,24,27 which we refer to as polyT20F and polyT20G. For the “universal” anchor and co-anchor lipid ssDNAs, lignoceric acid (C23H47COOH, lipid numbers: C24:0) was covalently attached to the anchor 3′ end and palmitic acid (CH3(CH2)14COOH, lipid numbers: C16:0) was covalently attached to the co-anchor 5′ end through amide linkages.30 All ssDNA oligos were ordered as custom syntheses (Integrative DNA Technologies for standard ssDNA and fluorescent probes; OligoFactory for lipid-ssDNA oligos), resuspended to either 5 mM (photopatterning ssDNA) or 100 μM (lipid-ssDNA, handles, and fluorescent probes) in deionized (DI) water (Direct-Q 3 UV water purification system, MilliporeSigma), and stored at −20°C. All ssDNA sequences are reported in Key Resources.
Photomasks
Photomask designs were drawn to scale in LayoutEditor (juspertor) and fabricated in cleanroom facilities by direct writing on 5”x5”x0.90″ chrome-on-quartz photomasks coated with a layer of IP3500 photoresist (Shipley) using a DWL 66+ laser lithography system (Heidelberg Instruments) with a 10 mm write head. After writing, exposed photoresist was removed from features with CD-26 developer (Shipley), masks were washed in DI water, and dried under compressed nitrogen gas. Chromium was removed from exposed patches using chromium etchant (Sigma), the surface was washed again with DI water and dried with compressed nitrogen, and excess photoresist was removed by submersion in resist stripper 1165 (Shipley) for 3 min at 60°C. Photomasks were cleaned with alternating acetone and isopropyl alcohol washes before their first use, then rinsed with 0.1% Triton X-100 and water and dried under compressed air between uses.
ssDNA photolithography
Photopatterning ssDNA oligos (polyT20F or polyT20G, Key Resources) were diluted to 200–250 μM in DI water and were degassed under vacuum. For simultaneous ssDNA-protein patterning, 200 μM polyT20G and either 10 mg mL−1 bovine serum albumin (BSA, A2153, Sigma-Aldrich) or 1 μM Alexa Fluor 555-conjugated BSA (A34786, Fisher) were mixed in 1x DPBS and not degassed. To photopattern ssDNA, BP-PA hydrogels were first completely dried under compressed air, moved to a nitrogen-filled glove box (H50028-2001, Bel-Art), rehydrated with 200–300 μL ssDNA solution, and sandwiched against the photomask region containing the desired pattern. Excess ssDNA solution was removed from the sides by gently wicking with a disposable wipe and the BP-PA gel was exposed through the photomask (I254 nm = 7 mW cm−2, t = 60–120 s) in a UV crosslinker oven (SpectroLinker XL-1000, Spectronics Corporation). Photopatterned BP-PA hydrogels were rehydrated in Tris-acetate-EDTA buffer (1x TAE: 40 mM Tris, 40 mM acetate, 1 mM EDTA, pH 8.0), gently lifted off the photomask using a razor blade, and then washed in 1x TAE +0.1% SDS followed by 1x TAE and stored in 1x DPBS.
To register and align multiple ssDNA patterns on the same gel, we incorporated a previously described “cross and window” alignment scheme into the photomask design.27 Briefly, the hydrogels were dried under compressed air and “cross” features were labeled using a drop of 1 μM of the appropriate 5′-6-fluorescein amidite (FAM) probe (e.g., FAM_G′) for ∼5 min, followed by washing in 1x TAE +0.1% SDS and 1x TAE. Hydrogels were then incubated in the second ssDNA sequence, assembled against the photomask in a nitrogen glove box, and then manually aligned to the photomask “window” regions using the GFP filter set on a benchtop epifluorescence microscope. The second ssDNA patterns were UV-exposed, washed, and stored as described above.
Fibronectin functionalization of BP-PA hydrogels
To derivatize acrylamide chains with bioreactive NHS esters, a solution of 0.01% (w/v) N,N-methylenebisacrylamide crosslinker buffered with 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 40820004, bioWORLD) from a 0.5 M stock (pH 6.0) and was degassed under vacuum in an ultrasonic bath. Next, 0.09% (w/v) lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP photoinitiator, 900889, Sigma) and 0.1% (w/v) acrylic acid-N-hydroxysuccinimide ester (N2 acrylic acid, A8060, Sigma) were successively added and mixed by vortexing. Bis solution was stored as a 0.2% (w/v) stock in DI water at 4°C. LAP stock solutions were 0.9% (w/v) in DI water and were stored at 4°C for up to 2 weeks. N2 was stored at −20°C and freshly prepared to 0.3% (w/v) in 50% ethanol/50% DI water.
Hydrogels were dried and rehydrated in the NHS mixture, drawing the liquid evenly across the hydrogel surface using a DCDMS-treated glass slide. Hydrogels were sandwiched between the supporting slide and the DCDMS-treated slide and UV-exposed (I365 nm = 15 mW cm−2) for 20 min, then rehydrated in ice-cold DI water. We gently lifted off the DCDMS-treated slide and washed twice in ice-cold DI water containing 100 μM sodium chloride (NaCl, S6191, Sigma). Hydrogels were then partially dried and a plastic clip-on 8-well slide with a silicone gasket (CCS-8, MatTek) was assembled around the region containing photopatterned ssDNA features and wells were incubated in 10–20 μg mL−1 bovine plasma fibronectin (F1141, Sigma) diluted from a 1 mg mL−1 stock in sterile 50 mM HEPES (pH 8.5) overnight at 4°C. Next day, fibronectin solution was removed, and wells were washed in 50 mM HEPES (pH 8.5) containing 100 mM glycine (G8898, Sigma) for 1 h at room temperature, followed by 3–4 washes in sterile 1x DPBS.
Sulfosuccinimidyl 6-(4′-azido-2′-nitrophenylamino)hexanoate (Sulfo-SANPAH; 22589, ThermoFisher) was used as an alternative approach to derivatize hydrogels with NHS esters (Figure S2). After drying the photopatterned hydrogel and assembling the clip-on slide, hydrogels were rehydrated with 1 mg mL−1 Sulfo-SANPAH in DI water and irradiated with collimated UV light (I365 nm = 15 mW cm−2) for 10 min. Exposed gel surfaces were washed in 1x DPBS followed by two washes in 50 mM HEPES (pH 8.5) and incubated overnight in 10–20 μg mL−1 fibronectin in 50 mM HEPES (pH 8.5) at 4°C before washing and adding cells. Sulfo-SANPAH aliquots (1 mg in 20 μL DMSO) were flash frozen in liquid nitrogen and stored at −80°C. However, when we used Sulfo-SANPAH as our NHS source, we found that MDCK cells could adhere to the unexposed hydrogel surface but cleared from ssDNA patterned features (Figure S2B). This occurred even in cases where ssDNA had not been present in the photopatterning solution (data not shown) but could be overcome by adding bovine serum albumin (BSA) or fibronectin to the ssDNA patterning mixture (Figure S2C). This dramatically improved the adhesion of MDCK cells, yet other cell types typically remained poorly adherent to UV-exposed hydrogel regions when compared side-by-side with the previously described method.
Lentiviral transduction
MDCK cells expressing H2B fused to near infrared fluorescent protein 670 (H2B-iRFP) were established by lentiviral transduction. To generate H2B-iRFP lentiviral particles, 7x105 Lenti-X 293T cells were plated in each well of a 6-well plate and were transiently co-transfected 24 h later with 1.5 μg of pLentiPGK DEST H2B-iRFP transfer vector (#90237, Addgene, RRID: Addgene_90237) and viral packaging plasmids: 1.33 μg pCMV delta R8.91 (#12263, Addgene, RRID: Addgene_12263) and 0.17 μg pMD2.G (#12259, Addgene, RRID: Addgene_112854). Transfection of HEK 293T cells were performed using the calcium phosphate method as described in.56 Approximately 48 h after transfection, media containing lentiviral particles was collected and centrifuged for 3 min at 800 xg to remove cell debris, then passed through a sterile 0.45 μm syringe filter (723–2545, Thermo Scientific). Filtered viral particles were introduced to MDCK cells (105 cells in each well of a 6-well plate) in antibiotic-free culture medium supplemented with 8 μg mL−1 polybrene (TR-1003-G, EMD Millipore) or frozen in single use aliquots and stored at −80°C until use. After ∼48 h, transduced cell lines were expanded and enriched for fluorescent expression using a BD Influx cell sorter (BD Biosciences).
Lipid-ssDNA cell labeling and patterning
For a typical patterning experiment (1–2 BP-PA gels) we prepared a 70–80% confluent T182 flask (∼1-2.5x107 cells) for each cell line. Cells were dissociated from flasks with trypsin and (if appropriate for the experiment) labeled with 1 μm CellTracker Deep Red (C34565, Fisher) cytoplasmic dye in serum-free DMEM for 15–30 min at 37°C prior to lipid-ssDNA labeling. Suspended cells were pelleted and washed twice in 1x DPBS (without Ca2+ and Mg2+), transferred to a 2 mL Eppendorf tube, and resuspended in 100 μL DPBS. Lipid anchor, lipid co-anchor, and handle ssDNA (5 μL each) were successively added with 10-min incubations between each. Labeled cells were washed twice and resuspended in 0.5–1 mL DPBS.
To introduce cells, we first aspirated the liquid from each well and added ∼100 μL of labeled cell suspension. Cells were allowed to settle for ∼5 min before excess cells were removed by gently washing with 1x DPBS using a P200 pipette tip. Excess cells could be returned to the Eppendorf tube and stored on ice to be reintroduced to a new substrate. Next, the hydrogel surface was gently washed in DPBS using a P200 tip or vacuum aspirator to remove liquid. Alternatively, the 8-well slide could be disassembled and labeled cells introduced to the ssDNA deposits across the entire hydrogel surface. After allowing cells to settle on the ssDNA patterns, we iteratively washed the hydrogel surface by immersion in 1x DPBS to remove non-adherent cells. After the last wash, the remaining DPBS was gently removed from wells and replaced with a complete imaging medium. Imaging medium consisted of phenol-free DMEM (4.5 g L−1 glucose, L-glutamine, and 25 mM HEPES, 21063-029, Invitrogen) supplemented with 10% FBS, 1 mM sodium pyruvate, and 1x pen/strep). To visualize ssDNA patches during time lapse imaging (Figure 1E) we stained ssDNA with 1-5x SYBR Gold nucleic acid gel stain (S11494, ThermoFisher) in 1x DPBS for 5–10 min and washed twice in 1x DPBS before introducing culture medium. SYBR Gold was resuspended to a 10,000x stock in DMSO and stored in working aliquots at −20°C.
Microindentation
We adapted a previously described micro-indenter device and approach to measure E of BP-PA hydrogels.38 Hydrogels were cast on methacrylate-coated glass slides at various Am/Bis ratios (Table S1) containing 0.1% (v/v) 0.2 μm diameter far red fluorescent carboxylate microspheres (FluoSpheres™, F8807, ThermoFisher) in the pre-polymer mixture. To cast hydrogels, ∼250 μL of pre-polymer mixture was introduced into a gap between the methacrylate-treated glass slide and a DCDMS-treated hydrophobic 2″ x 3″ slide (2947, Corning) created using double layered laboratory tape shims as guides. Following polymerization and rehydration in 1x DPBS, we uniformly photopatterned half of each hydrogel with 200 μM polyT20G ssDNA (I254 nm = 7 mW cm−2, t = 90 s) through a 1 mm thick quartz microscope slide (26011, Ted Pella) using aluminum foil to mask the other side from UV exposure.
The microindentation apparatus consisted of a 255 μm diameter blunt end indenter fabricated from cylindrical 30 gauge (AWG) SAE 316L stainless steel wire attached to a tensiometric sensor and stepper motor (L4018S1204-M6, Nanotec) to control indenter z position. After calibrating the spring constant of the sensor on a rigid (glass) surface, we indented BP-PA hydrogels using an approach rate of 12.5 μm s−1 while simultaneously recording time, force, and indenter z-position. E was calculated from the linear part of force versus indentation depth curves by the following relationship (Equation 1), assuming a soft homogeneous material of finite thickness and a rigid, blunt-ended cylindrical indenter:
| (Equation 1) |
In Equation 1 is the slope of the F-d curve, ν is the Poisson ratio (0.457 for polyacrylamide hydrogels57), κ is the Hayes correction factor for finite sample thickness,58 and 2r is the probe diameter (255 μm). Values for κ were estimated using hydrogel thickness (t) measurements obtained from confocal z-stacks (10 μm interval) of fluorescent microspheres embedded within the gel. F-d curves were fit to a linear model in MATLAB (TheMathWorks Inc.) using the Optimization and Curve Fitting toolboxes.
Imaging
In experiments quantifying cell capture (Figure 1C) and fibroblast spreading experiments (Figures 2C and 2D), patterned hydrogels were imaged using a Nikon Ti2 epifluorescence microscope (Nikon Instruments) equipped with a motorized stage, CMOS camera (DS-Qi2, Nikon), LED transmitted and epifluorescence illumination (Sola II light engine, Lumencor), single-pass DAPI, FITC, TRITC, and Cy5 filter sets (Chroma), 4x/0.25 numerical aperture (NA), 10x/0.45 NA, and 20x/0.45 NA lenses. For other live and immunofluorescence imaging, we used a Ti2 microscope equipped with a CSU-W1 spinning disk (Yokugawa), solid state laser launch (100 mW 405, 488, and 561 nm lasers and a 75 mW 640 nm laser), a white light LED for transmitted illumination, a motorized stage, a Prime 95B back-thinned CMOS camera (Teledyne Photometrics), and 4x/0.25 NA, 10x/0.45 NA, and 20x/0.45 NA lenses. A sealed enclosure built around the microscope stage (OkoLab) provided stable environmental conditions at 37°C and 5% CO2 for the duration of the experiment. Microscopes were under control of NIS Elements AR software (version AR 5.11.00).
Immunofluorescence
To visualize fibronectin coupled to BP-PA gels alongside photopatterned polyT20F ssDNA (Figure 1E) we first incubated the gels in a rabbit anti-fibronectin antibody (1:100, ab23750, Abcam, AB_447655) diluted in 1x DPBS for 45 min followed by donkey anti-rabbit IgG-Alexa Fluor 647 (1:200, A31573, ThermoFisher, RRID: AB_2536183) and 1 μM FAM_F′ probe diluted in 1x DPBS for 30 min. Stained gels were washed 1x DPBS before imaging.
Cells were fixed in 4% paraformaldehyde (PFA, 16% stock; 15710, Electron Microscopy Sciences) diluted in 1x DPBS for 10 min, permeabilized in 0.5% v/v Triton X-100 (10 min) in 1x DPBS and blocked with 1% BSA in PBS-T (1x DPBS +0.1% v/v Tween 20; 9480, EMD Millipore) for 30–60 min. All fixation, permeabilization, and blocking steps were performed at room temperature. Fixed and blocked cells were incubated in primary antibodies for 2 h at room temperature or 4°C overnight and secondary antibodies for 1 h at room temperature with three PBS-T washes following each incubation. Primary antibodies were diluted in blocking buffer: mouse anti-vinculin (7F9) (1:100, 14-9777-82, eBiosceince, RRID: AB_2573028), mouse anti-E-cadherin (clone 34) (1:200, 610404, BD Biosciences, RRID: AB_397787), rabbit anti-phospho-p44/42 MAPK (ERK1/2) (1:200, 4370, Cell Signaling Technologies, RRID: AB_2315112), and rabbit anti-E-cadherin (24E10) (1:200, 3195, Cell Signaling Technologies, RRID: AB_2291471). Secondary antibodies (all raised in donkey) were used at 1:1000 dilution: anti-rabbit Alexa Fluor 488 (A21206, ThermoFisher, RRID: AB_2535792), anti-mouse Alexa Fluor 555 (A31572, ThermoFisher, RRID: AB_162543), anti-rabbit Alexa Fluor 555 (A31572, ThermoFisher, RRID: AB_162543), and anti-rabbit Alexa Fluor 647 (A31573, ThermoFisher, RRID: AB_2536183). Nuclei and F-actin were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, 300 nM; D1306, ThermoFisher) and phalloidin-Alexa Fluor 647 (1:100, A22287, Invitrogen) added along with the secondary antibodies. For fibroblast spreading assays (Figures 2C and 2D), we fixed and permeabilized as described and then directly stained cells for 1 h with phalloidin-Alexa Fluor 647, DAPI, and 1 μM FAM_G′ probe diluted in PBS-T without blocking.
Image analysis
All image analysis was performed using ImageJ/Fiji.54 Cells captured on ssDNA patches in Figure 1 were manually counted from fluorescence signal (CellTracker dye) or brightfield images using the multi point selection tool in ImageJ/Fiji. Patch boundaries were determined using the SYBR Gold fluorescence signal. To count multiple cell populations on ssDNA patterns that were not labeled (chess board design in Figures 3 and S3) we drew a 250 μm square region of interest (ROI) centered around the middle of each captured cell cluster and counted cells within the bounding region. Patch coverage (Figure S2) was quantified by performing a rolling ball background subtraction (subtract background function with a 150-pixel radius), manually thresholding the SYBR Gold-labeled ssDNA (FITC channel) and Cell Tracker Deep Red (Cy5 channel) signals and using analyze particles to calculate the area fraction of Cy5 to FITC object area.
To measure fibroblast spread areas (Figures 2C and 2D) we acquired image stacks at 20× magnification on the Ti2 widefield system and manually thresholded the phalloidin-AlexaFluor 647 channel in ImageJ/Fiji to obtain a binarized image containing cell outlines. Next, we used the erode, fill holes, and analyze particles functions with a minimum area (excluding objects <500 μm2) to obtain ROIs for individual cells. We excluded cells on the edge of the image and limited our analysis to single cells by using the nucleus (DAPI) channel to exclude ROIs that contained more than one nucleus. To obtain images for focal adhesion measurements (Figures 2E and 2F) we acquired z-stacks (13 frames, 2.5 μm z-step size, 30 μm total) with the 20x lens using a 1.5x intermediate zoom lens (30× objective magnification). We measured focal adhesion lengths from the vinculin immunofluorescence channel manually using the line segment tool in Fiji/ImageJ. In both sets of experiments, we used FAM ssDNA probe signal to distinguish between UV-exposed and unexposed control hydrogel regions.
We employed a custom Fiji/ImageJ radial analysis plugin44,55 to quantify fluorescence intensity in composite epithelial-mesenchymal tissues (Figures 4, 5, and S4). We used the draw polygon function to trace the boundary between the inner and outer tissues and radially transformed the ROI to obtain a 200-pixel radius circular ROI with the interface located at r = 100 pixels (normalized to r = 1.0 a.u.). The use of a radial transformation enables an “apples-to-apples” comparison of irregularly shaped tissues or interfaces. After removing background by subtracting a fixed value, fluorescence intensities of F-actin, E-cadherin, or pERK were calculated at each evenly spaced radial bin and normalized to the maximum pixel intensity across all measurements. We excluded tissues with an incomplete border (e.g., where <60% of the circumference was surrounded by the second cell population) from our analysis.
Quantification and statistical analysis
Statistics were calculated using R (version 4.2.0, R Core Team) running on RStudio (version 2022.02.0) and data were visualized using ggplot2 in tidyR.59 Exact numbers of measurements (n), numbers and types of independent replicates, and statistical tests used to compare results and compute p values are indicated in the figure legends. Two-tailed p values were computed unless otherwise noted. We used Welch’s t-test to facilitate comparisons between two groups where data were normally distributed (as determined by the Shapiro-Wilk non-normality test with a p = 0.05 cutoff) and non-parametric tests otherwise. For measurements of cell capture using different ssDNA labeling schemes (Figure S2B) we report a single value from a non-parametric Kruskal-Wallis test, since all pairwise comparisons using the matched handle ssDNA were significantly different from the “mis-matched” handle, “anchors only”, and “none” (unlabeled) groups. For microindentation measurements of E (Figure 2B) we used paired t-tests to compare mean elastic modulus of exposed (Euv) and unexposed (Ectrl) regions of the same hydrogel. E for each hydrogel is the mean of 3 measurements per side for a total of 6 measurements per hydrogel; overall mean ± s.d. of E for n hydrogels of each composition is summarized in Table S1. We used a two-tailed Kolmogorov-Smirnov test to compare cumulative distributions of fibroblast spreading (Figure 2D) or focal adhesion lengths (Ladhesion) on unexposed (ctrl) and photopatterned (+UV) hydrogel regions (Figure 2F). All summarized biological data are representative of at least two independent biological replicates and are pooled where appropriate. Technical replicates of hydrogel characterization experiments are defined in figure legends.
Acknowledgments
We thank members of the Hughes lab for helpful discussions. We also thank David Li, Jeff Byfield, and Paul Janmey for micro-indenter instrument access and training; the Singh Center for Nanotechnology staff, particularly David Jones and Eric Johnston, for cleanroom training and assistance with photomask preparation; Arjun Raj for the gift of MDCK II cells; David Odde for LLC-Pk1 cells; and Lukasz Bugaj for 3T3 and Lenti-X 293T cells, lentiviral vectors, and assistance with virus production and establishment of stable cell lines. This work was carried out in part at the Singh Center for Nanotechnology at the University of Pennsylvania, which is supported by the NSF National Nanotechnology Coordinated Infrastructure Program under grant NNCI-2025608 and at the Penn Cytomics and Cell Sorting Resource Laboratory. Funding support was provided by NIH grants R35 GM133380 and R01 DK132296 to AJH, T32 HD083185 to JMV, and F32 DK126385 to LSP; an NSF Graduate Research Fellowship to CMP; and the NSF Faculty Early Career Development Program (CAREER grant # 2047271) to AJH.
Author contributions
Conceptualization, L.S.P and A.J.H.; Methodology, L.S.P., C.M.P., J.L., J.M.V., and A.J.H.; Software, L.S.P. and A.J.H.; Formal Analysis, L.S.P.; Investigation, L.S.P. and A.J.H.; Resources, A.J.H.; Writing - Original Draft, L.S.P. and A.J.H.; Writing - Review & Editing, L.S.P., C.M.P., J.L., J.M.V., and A.J.H.; Visualization, L.S.P. and A.J.H.; Supervision, A.J.H.; Funding Acquisition, L.S.P. and A.J.H.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106657.
Supplemental information
Data and code availability
-
•
Photomask design files and microindentation data files have been deposited to Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead author upon request.
-
•
All original code has been deposited to Mendeley Data and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Noah T.K., Donahue B., Shroyer N.F. Intestinal development and differentiation. Exp. Cell Res. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millar S.E. Molecular mechanisms regulating hair follicle development. J. Invest. Dermatol. 2002;118:216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- 3.Solnica-Krezel L., Sepich D.S. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- 4.Mammoto T., Mammoto A., Torisawa Y.-S., Tat T., Gibbs A., Derda R., Mannix R., de Bruijn M., Yung C.W., Huh D., Ingber D.E. Mechanochemical control of mesenchymal condensation and embryonic tooth organ formation. Dev. Cell. 2011;21:758–769. doi: 10.1016/j.devcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dassule H.R., McMahon A.P. Analysis of epithelial–mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 6.Talbot J.C., Nichols J.T., Yan Y.-L., Leonard I.F., BreMiller R.A., Amacher S.L., Postlethwait J.H., Kimmel C.B. Pharyngeal morphogenesis requires fras1-itga8-dependent epithelial-mesenchymal interaction. Dev. Biol. 2016;416:136–148. doi: 10.1016/j.ydbio.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang C., Conrad L., Iber D. Organ-specific branching morphogenesis. Front. Cell Dev. Biol. 2021;9:671402. doi: 10.3389/fcell.2021.671402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varner V.D., Nelson C.M. Cellular and physical mechanisms of branching morphogenesis. Development. 2014;141:2750–2759. doi: 10.1242/dev.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller U., Wang D., Denda S., Meneses J.J., Pedersen R.A., Reichardt L.F. Integrin α8β1 is critically important for epithelial–mesenchymal interactions during kidney morphogenesis. Cell. 1997;88:603–613. doi: 10.1016/S0092-8674(00)81903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Provenzano P.P., Eliceiri K.W., Campbell J.M., Inman D.R., White J.G., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brayford S., Kenny F.N., Hiratsuka T., Serna-Morales E., Yolland L., Luchici A., Stramer B.M. Heterotypic contact inhibition of locomotion can drive cell sorting between epithelial and mesenchymal cell populations. J. Cell Sci. 2019;132:jcs223974. doi: 10.1242/jcs.223974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinrich M.A., Alert R., Wolf A.E., Košmrlj A., Cohen D.J. Self-assembly of tessellated tissue sheets by expansion and collision. Nat. Commun. 2022;13:4026. doi: 10.1038/s41467-022-31459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javaherian S., Paz A.C., McGuigan A.P. Micropatterning cells on permeable membrane filters. Methods Cell Biol. 2014;121:171–189. doi: 10.1016/B978-0-12-800281-0.00012-9. [DOI] [PubMed] [Google Scholar]
- 14.Javaherian S., D’Arcangelo E., Slater B., Zulueta-Coarasa T., Fernandez-Gonzalez R., McGuigan A.P. An in vitro model of tissue boundary formation for dissecting the contribution of different boundary forming mechanisms. Integr. Biol. 2015;7:298–312. doi: 10.1039/c4ib00272e. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez A.J., Schaeffer A., De Pascalis C., Letort G., Vianay B., Bornens M., Piel M., Blanchoin L., Théry M. Acto-myosin network geometry defines centrosome position. Curr. Biol. 2021;31:1206–1220.e5. doi: 10.1016/j.cub.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Smith Q., Rochman N., Carmo A.M., Vig D., Chan X.Y., Sun S., Gerecht S. Cytoskeletal tension regulates mesodermal spatial organization and subsequent vascular fate. Proc. Natl. Acad. Sci. USA. 2018;115:8167–8172. doi: 10.1073/pnas.1808021115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang X., Ali M.Y., Saif M.T.A. A novel technique for micro-patterning proteins and cells on polyacrylamide gels. Soft Matter. 2012;8:7197–7206. doi: 10.1039/C2SM25533B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Nemashkalo A., Rezende L., Jung J.Y., Chhabra S., Guerra M.C., Heemskerk I., Warmflash A. Nodal is a short-range morphogen with activity that spreads through a relay mechanism in human gastruloids. Nat. Commun. 2022;13:497. doi: 10.1038/s41467-022-28149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 20.Strale P.-O., Azioune A., Bugnicourt G., Lecomte Y., Chahid M., Studer V. Multiprotein printing by light-induced molecular adsorption. Adv. Mater. 2016;28:2024–2029. doi: 10.1002/adma.201504154. [DOI] [PubMed] [Google Scholar]
- 21.Missirlis D., Baños M., Lussier F., Spatz J.P. Facile and versatile method for micropatterning poly(acrylamide) hydrogels using photocleavable comonomers. ACS Appl. Mater. Interfaces. 2022;14:3643–3652. doi: 10.1021/acsami.1c17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y.-S., Tung C.-K., Dai T.-H., Wang X., Yeh C.-T., Fan S.-K., Liu C.-H. Liver-lobule-mimicking patterning via dielectrophoresis and hydrogel photopolymerization. Sensor. Actuator. B Chem. 2021;343:130159. doi: 10.1016/j.snb.2021.130159. [DOI] [Google Scholar]
- 23.Todhunter M.E., Jee N.Y., Hughes A.J., Coyle M.C., Cerchiari A., Farlow J., Garbe J.C., LaBarge M.A., Desai T.A., Gartner Z.J. Programmed synthesis of three-dimensional tissues. Nat. Methods. 2015;12:975–981. doi: 10.1038/nmeth.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheideler O.J., Yang C., Kozminsky M., Mosher K.I., Falcón-Banchs R., Ciminelli E.C., Bremer A.W., Chern S.A., Schaffer D.V., Sohn L.L. Recapitulating complex biological signaling environments using a multiplexed, DNA-patterning approach. Sci. Adv. 2020;6:eaay5696. doi: 10.1126/sciadv.aay5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelham R.J., Wang Y.-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 27.Viola J.M., Porter C.M., Gupta A., Alibekova M., Prahl L.S., Hughes A.J. Guiding cell network assembly using shape-morphing hydrogels. Adv. Mater. 2020;32:e2002195. doi: 10.1002/adma.202002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dormán G., Prestwich G.D. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 29.Hughes A.J., Herr A.E. Microfluidic western blotting. Proc. Natl. Acad. Sci. USA. 2012;109:21450–21455. doi: 10.1073/pnas.1207754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber R.J., Liang S.I., Selden N.S., Desai T.A., Gartner Z.J. Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules. 2014;15:4621–4626. doi: 10.1021/bm501467h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakins J.N., Chin A.R., Weaver V.M. Exploring the link between human embryonic stem cell organization and fate using tension-calibrated extracellular matrix functionalized polyacrylamide gels. Methods Mol. Biol. 2012;916:317–350. doi: 10.1007/978-1-61779-980-8_24. [DOI] [PubMed] [Google Scholar]
- 32.Denisin A.K., Pruitt B.L. Tuning the range of polyacrylamide gel stiffness for mechanobiology applications. ACS Appl. Mater. Interfaces. 2016;8:21893–21902. doi: 10.1021/acsami.5b09344. [DOI] [PubMed] [Google Scholar]
- 33.Farahani P.E., Lemke S.B., Dine E., Uribe G., Toettcher J.E., Nelson C.M. Substratum stiffness regulates Erk signaling dynamics through receptor-level control. Cell Rep. 2021;37:110181. doi: 10.1016/j.celrep.2021.110181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bangasser B.L., Shamsan G.A., Chan C.E., Opoku K.N., Tüzel E., Schlichtmann B.W., Kasim J.A., Fuller B.J., McCullough B.R., Rosenfeld S.S., Odde D.J. Shifting the optimal stiffness for cell migration. Nat. Commun. 2017;8:15313. doi: 10.1038/ncomms15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulrich T.A., de Juan Pardo E.M., Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solon J., Levental I., Sengupta K., Georges P.C., Janmey P.A. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys. J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo C.M., Wang H.B., Dembo M., Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levental I., Levental K.R., Klein E.A., Assoian R., Miller R.T., Wells R.G., Janmey P.A. A simple indentation device for measuring micrometer-scale tissue stiffness. J. Phys. Condens. Matter. 2010;22:194120. doi: 10.1088/0953-8984/22/19/194120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anseth K.S., Bowman C.N., Brannon-Peppas L. Mechanical properties of hydrogels and their experimental determination. Biomaterials. 1996;17:1647–1657. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- 40.Peyton S.R., Putnam A.J. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 41.Monier B., Pélissier-Monier A., Brand A.H., Sanson B. An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat. Cell Biol. 2010;12:60–69. doi: 10.1038/ncb2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucia S.E., Jeong H., Shin J.H. Cell segregation via differential collision modes between heterotypic cell populations. Mol. Biol. Cell. 2022;33:ar129. doi: 10.1091/mbc.E22-03-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canty L., Zarour E., Kashkooli L., François P., Fagotto F. Sorting at embryonic boundaries requires high heterotypic interfacial tension. Nat. Commun. 2017;8:157. doi: 10.1038/s41467-017-00146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobretsov M., Romanovsky D. “Clock-scan” protocol for image analysis. Am. J. Physiol. Cell Physiol. 2006;291:C869–C879. doi: 10.1152/ajpcell.00182.2006. [DOI] [PubMed] [Google Scholar]
- 45.Ihermann-Hella A., Hirashima T., Kupari J., Kurtzeborn K., Li H., Kwon H.N., Cebrian C., Soofi A., Dapkunas A., Miinalainen I., et al. Dynamic MAPK/ERK activity sustains nephron progenitors through niche regulation and primes precursors for differentiation. Stem Cell Rep. 2018;11:912–928. doi: 10.1016/j.stemcr.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishii M., Tateya T., Matsuda M., Hirashima T. Retrograde ERK activation waves drive base-to-apex multicellular flow in murine cochlear duct morphogenesis. Elife. 2021;10:e61092. doi: 10.7554/eLife.61092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hino N., Rossetti L., Marín-Llauradó A., Aoki K., Trepat X., Matsuda M., Hirashima T. ERK-mediated mechanochemical waves direct collective cell polarization. Dev. Cell. 2020;53:646–660.e8. doi: 10.1016/j.devcel.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Cabral K.A., Patterson D.M., Scheideler O.J., Cole R., Abate A.R., Schaffer D.V., Sohn L.L., Gartner Z.J. Simple, affordable, and modular patterning of cells using DNA. J. Vis. Exp. 2021 doi: 10.3791/61937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holland S.J., Gale N.W., Mbamalu G., Yancopoulos G.D., Henkemeyer M., Pawson T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722–725. doi: 10.1038/383722a0. [DOI] [PubMed] [Google Scholar]
- 50.Strutt H., Strutt D. Differential stability of flamingo protein complexes underlies the establishment of planar polarity. Curr. Biol. 2008;18:1555–1564. doi: 10.1016/j.cub.2008.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodríguez-Franco P., Brugués A., Marín-Llauradó A., Conte V., Solanas G., Batlle E., Fredberg J.J., Roca-Cusachs P., Sunyer R., Trepat X. Long-lived force patterns and deformation waves at repulsive epithelial boundaries. Nat. Mater. 2017;16:1029–1037. doi: 10.1038/nmat4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen K., Luk S., Hicks D.F., Elman J.S., Bohr S., Iwamoto Y., Murray R., Pena K., Wang F., Seker E., et al. Resolving cancer–stroma interfacial signalling and interventions with micropatterned tumour–stromal assays. Nat. Commun. 2014;5:5662–5711. doi: 10.1038/ncomms6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagstaff L., Goschorska M., Kozyrska K., Duclos G., Kucinski I., Chessel A., Hampton-O’Neil L., Bradshaw C.R., Allen G.E., Rawlins E.L., et al. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 2016;7:11373. doi: 10.1038/ncomms11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobretsov M., Petkau G., Hayar A., Petkau E. Clock scan protocol for image analysis: ImageJ plugins. J. Vis. Exp. 2017 doi: 10.3791/55819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benman W., Berlew E.E., Deng H., Parker C., Kuznetsov I.A., Lim B., Siekmann A.F., Chow B.Y., Bugaj L.J. Temperature-responsive optogenetic probes of cell signaling. Nat. Chem. Biol. 2022;18:152–160. doi: 10.1038/s41589-021-00917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takigawa T., Morino Y., Urayama K., Masuda T. Poisson’s ratio of polyacrylamide (PAAm) gels. Polym. Gels Netw. 1996;4:1–5. doi: 10.1016/0966-7822(95)00013-5. [DOI] [Google Scholar]
- 58.Hayes W.C., Keer L.M., Herrmann G., Mockros L.F. A mathematical analysis for indentation tests of articular cartilage. J. Biomech. 1972;5:541–551. doi: 10.1016/0021-9290(72)90010-3. [DOI] [PubMed] [Google Scholar]
- 59.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patterns were designed with a fixed area equivalent to a 200 μm diameter circle (A = 3.14 × 104 μm2). Images were acquired every 20 min for 10 h, starting 1 h after DNA patterning. See also Figure 1E.
Patterns consist of an inner circle (r = 175 μm) surrounded by an annulus (r = 75 μm), each with a nominal area of A = 97264 μm2. Images were acquired every 2 h for 10 h total, starting 1 h after DNA patterning. See also Figure 4B.
Images were acquired every 2 h for 10 h total, starting 1 h after DNA patterning. See also Figure 4B.
Data Availability Statement
-
•
Photomask design files and microindentation data files have been deposited to Mendeley Data and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead author upon request.
-
•
All original code has been deposited to Mendeley Data and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.