Abstract
Lipopolysaccharide (LPS) is a key antigen in immunity to leptospirosis. Its biosynthesis requires enzymes for the biosynthesis and polymerization of nucleotide sugars and the transport through and attachment to the bacterial membrane. The genes encoding these functions are commonly clustered into loci; for Leptospira borgpetersenii serovar Hardjo subtype Hardjobovis, this locus, named rfb, spans 36.7 kb and contains 31 open reading frames, of which 28 have been assigned putative functions on the basis of sequence similarity. Characterization of the function of these genes is hindered by the fact that it is not possible to construct isogenic mutant strains in Leptospira. We used two approaches to circumvent this problem. The first was to clone the entire locus into a heterologous host system and determine if a “recombinant” LPS or polysaccharide was synthesized in the new host. The second approach used putative functions to identify mutants in other bacterial species whose mutations might be complemented by genes on the leptospiral rfb locus. This approach was used to investigate the function of three genes in the leptospiral rfb locus and demonstrated function for orfH10, which complemented a wbpM strain of Pseudomonas aeruginosa, and orfH13, which complemented an rfbW strain of Vibrio cholerae. However, despite the similarity of OrfH11 to WecC, a wecC strain of E. coli was not complemented by orfH11. The predicted protein encoded by orfH8 is similar to GalE from a number of organisms. A Salmonella enterica serovar Typhimurium strain producing no GalE was used as a background in which orfH8 produced detectable GalE enzyme activity.
Leptospirosis is a zoonosis of worldwide importance caused by infection with spirochetes of the genus Leptospira, which is divided into 12 species based on overall genomic DNA similarity. In contrast, a serological scheme of classification divides Leptospira into more than 200 serovars (6). The predominant antigen to which agglutinating, opsonic antibodies bind is lipopolysaccharide (LPS) (8, 14). Despite the importance of this antigen in immunity to leptospirosis and in serological classification, relatively little is known about the biosynthesis of LPS in Leptospira.
Recently, the rfb locus encoding proteins involved in the biosynthesis of LPS in Leptospira borgpetersenii serovar Hardjo subtype Hardjobovis was identified and shown to span 36.7 kb and contain 31 open reading frames (ORFs). The locus was defined on the basis of the identification of putative proteins involved in the biosynthesis or polymerization of nucleotide sugars (15).
In the absence of methods that enable the production of isogenic strains in Leptospira, the approach to the characterization of the function of the genes contained on the locus has been restricted to analysis of function in heterologous host systems. In this study, we have pursued two approaches. Firstly, the locus identified by similarity analysis was cloned and introduced into Escherichia coli. The second approach utilized specific genes on the rfb locus to complement mutated genes in heterologous hosts to confirm function identified by similarity analysis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. The inserts in plasmids containing single Hardjobovis ORFs were subcloned from pLBA577 or pLBA589 (15), with the subcloned inserts oriented such that the expression of the ORF was driven by the vector-carried lac promoter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Characteristics | Reference or source |
|---|---|---|
| Plasmids | ||
| pBluescript II KS | High-copy-number cloning vector; Apr | Stratagene |
| pWSK129 | Low-copy-number cloning vector; Kmr | 29 |
| pWSK29 | Low-copy-number cloning vector; Apr | 29 |
| pUCP19 | E. coli-P. aeruginosa shuttle vector; Apr | 30 |
| pPR691 | Low-copy-number cosmid vector, pSC101 replicon, and specifies resistance to Knr, Specr, and Strepr | 12 |
| pLBA577 | 4.0-kb BamHI insert in pBluescript II KS | 15 |
| pLBA589 | 7.1-kb HindIII insert in pBluescript II KS | 15 |
| pLBA591 | 1.65-kb HindIII-EcoRV fragment from pLBA589 cloned into pBluescript II KS (orfH8 complete) | This study |
| pLBA593 | 1.70-kb HindIII-BamHI fragment from pLBA589 cloned into pBluescript II KS (orfH8 complete) | This study |
| pLBA655 | Cosmid; insert containing orfH1 through to orfH31 from the Hardjobovis rfb locus | This study |
| pLBA658 | galE from the C. jejuni 81116 rfb locus in pBluescript II KS | B. Frya |
| pLBA661 | 2.04-kb BamHI-EcoRV fragment from pLBA589 cloned into pBluescript II KS (orfH10 complete) | This study |
| pLBA662 | 2.61-kb XbaI-BamHI fragment from pLBA589 cloned into pWSK129 (orfH11 complete) | This study |
| pLBA663 | 1.64-kb BamHI-HindIII fragment from pLBA577 cloned into pBluescript II SK (orfH13 complete) | This study |
| pLBA664 | 1.64-kb BamHI-HindIII fragment from pLBA577 cloned into pWSK29 (orfH13 complete) | This study |
| pLBA665 | 2.61-kb XbaI-BamHI fragment from pLBA589 cloned into pBluescript II KS (orfH11 complete) | This study |
| Bacterial strains | ||
| L. borgpertersenii | ||
| L171 | Serovar Hardjo subtype Hardjobovis | 15 |
| S. enterica serovar Typhimurium | ||
| SL696 | Wild type | 32 |
| SL761 | galE strain | 25 |
| E. coli | ||
| VCS257 | DP50 sup F[supE44 supF58 hsd53(rB mB) dapD8 lacY1 glnV44 Δ(gal-uvrB)47 tyrT58 gyrA29 tonA53 Δ(thyA57)] | Stratagene |
| DH5∝ | F−endA1 hsdR17 (mK+ rK−) supE44 thi-1 recA1 gyrA96 relA1 (φ80dlacZΔM15) | 10 |
| AB1133 | K-12 thr-1 leuB6 Δ(gpt-proA)66 hisG4 argE3 thi-1 rfbD1 lacY1 ara-14 galK2 xyl-5 mgl-51 rpsL31 supE44 | 20 |
| EC21566 | As AB1133 but rff::Tn10-66 | 20 |
| V. cholerae | ||
| O17 | Ol El Tor Ogawa | 7 |
| V1217 | Strain O17 with Knr insertion in rfbW; rough phenotype | 7 |
| P. aeruginosa | ||
| PAO1 | Band B+; serotype O5 | 4 |
| wbpM-2 | Strain PAO1 with wbpM gentamicin resistance cassette inserted at NruI; LPS B-band negative phenotype | 4 |
Department of Applied Biology and Biotechnology, RMIT, Australia.
Bacterial culture and preparation of competent cells.
Leptospira was cultured in EMJH medium (13). E. coli, Salmonella enterica serovar Typhimurium, Vibrio cholerae, and Pseudomonas aeruginosa were cultured in Luria-Bertani broth. Where necessary, ampicillin (100 μg/ml) or kanamycin (50 μg/ml) was added to media. Electrocompetent E. coli and V. cholerae cells were prepared as described previously (7, 24), as were chemically derived competent S. enterica and P. aeruginosa cells (5, 10).
DNA manipulations.
Genomic DNA was prepared from 100-ml stationary-phase leptospiral cultures using a procedure similar to the cetyltrimethylammonium bromide (CTAB) precipitation method (2), while plasmid DNA was prepared as described previously (3). Restriction endonuclease digestions, ligation reactions, and the analysis of DNA by agarose gel electrophoresis were performed using standard methods (2, 23). Southern hybridization was performed using probes labeled with digoxigenin-dUTP; the conditions used for hybridization were as specified by the manufacturer (Roche). Hybridization under high-stringency conditions was performed overnight at 68°C followed by washing at 68°C in 0.1% sodium dodecyl sulfate (SDS)–0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (2). Nucleotide sequencing was performed using the BigDye DyeDeoxy terminator cycle-sequencing kit (PE Biosystems) and an Applied Biosystems 373A automated sequencer. Sequence data were analyzed with Sequencher 3.1 (GeneCodes), while DNA and protein database comparisons were made by using the BLAST program of Altschul et al. (1).
Cosmid library.
Hardjobovis genomic DNA was partially digested with BglII. The enzyme concentration and time of digestion were varied to maximize the generation of digestion products in the range 35 to 50 kb. The partially digested genomic DNA was dephosphorylated using calf intestine alkaline phosphatase (Roche) and ligated into cosmid arms prepared by digesting pPR691 with BamHI and PvuII. DNA for transfection was prepared using in vitro packaging extracts (Gigapack III XL-4; Stratagene). Transfection of E. coli strain VCS257 resulted in a library of approximately 1,000 colonies.
Preparation of extracts. (i) LPS extract.
LPS was prepared using a method modified from that of Westphal and Jann (31). The cells from a 500-ml overnight culture were pelleted and resuspended in 10 ml of phosphate-buffered saline. An equal volume of 90% (wt/vol) phenol was added to the bacterial suspension and incubated at 68°C for 15 min with stirring. The suspension was cooled in an ice bath to approximately 10°C and centrifuged at 1,500 × g for 10 min at room temperature. The upper, aqueous phase was transferred to a fresh tube, and absolute ethanol was added to 50% (vol/vol). A few pellets of sodium acetate were added, and the solution was stirred overnight at 4°C. Precipitated material was removed by centrifuging at 15,000 × g for 15 min at 4°C; further ethanol was then added to a final concentration of 90% (vol/vol). A few pellets of sodium acetate were added, and the solution was stirred overnight at 4°C. The LPS was pelleted by centrifuging at 15,000 × g for 15 min at 4°C and resuspended in deionized water.
(ii) Cell envelope preparation.
The method for cell envelope preparation was based on previously published methods (11, 19). Bacterial cells from a 100-ml culture were pelleted at 8,000 × g for 10 min at 4°C and washed once with sterile phosphate-buffered saline. The pellet was then resuspended in 4 ml of solution I (50 mM Tris-HCl, 2 mM EDTA [pH 8.5]). The cell suspension was then subjected to sonication using a Branson B12 sonifier (four 30-s bursts at 80 W, with incubation on ice for 2 min between bursts). The remaining intact cells were removed by centrifuging at 1,200 × g for 20 min at 4°C. The supernatant was then transferred to a fresh microcentrifuge tube and centrifuged at 13,000 × g and 4°C for 1 h. The cell envelope pellet was suspended in 100 μl of solution II (2 mM Tris-HCl [pH 7.8])–100 μl of 2× sample buffer (23) and boiled for 10 min. The solution was cooled, 40 μg of proteinase K (in solution) were added, and the mixture was incubated at 56°C for 1 h. Between 10 and 20 μl of the preparation was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
SDS-PAGE.
A Bio-Rad Mini-PROTEAN II apparatus was used with 12.5% resolving gels and a 4.5% stacking gel buffered with Tricine (18). LPS was stained using the silver-staining method of Tsai and Frasch (26).
Colorimetric assay for UDP-glucose 4-epimerase activity.
The colorimetric assay was performed as described previously (21).
RESULTS
Cloning of the rfb locus.
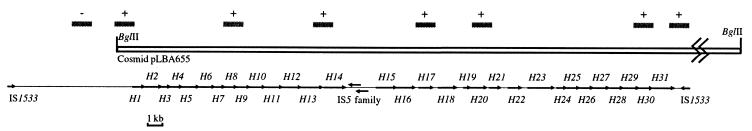
Partially BglII-digested Hardjobovis genomic DNA was mixed with cosmid arms and ligated. After packaging and transfection into E. coli strain VCS257, a library of approximately 1,000 cosmid strains was obtained. Plasmid preparations from transfected strains were screened by dot blot hybridization analysis to identify strains containing both the 5′ (probe within orfH1) and 3′ (probe in the intergenic region between orfH31 and IS1533) ends of the leptospiral rfb locus (Fig. 1). Both probes were found to hybridize to cosmid pLBA655. This cosmid was further characterized by Southern analysis using the probes indicated by bars in Fig. 1. The hybridization profile derived from the cosmid digested with a series of restriction endonuclease enzymes was consistent with patterns predicted from the nucleotide sequence of the locus (data not shown). The insert in pLBA655 was thus determined to extend from a BglII site upstream of orfH1 to a point beyond the IS1533 element downstream of orfH31 (Fig. 1). Western blot analysis of whole-cell preparations from strain VCS257 containing pLBA655 using anti-Hardjo immune sera from a series of patients revealed that no Hardjo polysaccharide was detectable (data not shown).
FIG. 1.
The insert from cosmid pLBA655 aligned with the Hardjobovis rfb locus. The BglII sites bounding the cosmid insert are shown, and the bars indicate the locations of probes used in Southern hybridization to confirm the cosmid insert (+, hybridization; −, no hybridization). Arrows indicate ORFs included in the rfb locus or transposase genes from putative IS elements.
Determination of function.
Similarity analysis has been used as the basis for assigning putative functions to several genes in the rfb locus (15). Based on their sequence similarity to genes of known function, genes from the leptospiral rfb locus were used to complement specific mutations in hosts other than Leptospira. In addition, an enzyme assay was used to investigate the function of OrfH8.
OrfH8 functional analysis.
OrfH8 was proposed to be a galactose epimerase; this activity can be detected specifically by a sensitive enzyme assay (21). S. enterica strain SL761 (25), which lacks galactose epimerase activity, was chosen as a host for plasmids pLBA591 and pLBA593, which contained orfH8 such that its expression was driven by the lac promoter. Extracts from strain SL761 containing pLBA591 or pLBA593 were shown to have 52 and 133 U of galactose epimerase activity per g of protein, respectively. These activities were lower than those detected in wild-type S. enterica (strain SL696) and strain SL761 transformed with the Campylobacter jejuni galE gene (Table 2).
TABLE 2.
Determination of GalE enzyme activity
| Strain | Feature | Enzyme activity (mU/mg of protein)a |
|---|---|---|
| SL696 | S. enterica wild type | 318 |
| SL761 | galE strain | 0 |
| SL761(pBluescript II KS) | pBluescript II KS control | 0 |
| SL761(pLBA591) | orfH8 | 52 |
| SL761(pLBA593) | orfH8 | 133 |
| SL761(pLBA658) | galE from C. jejuni | 367 |
Unit definition: the amount of enzyme which produced 1 μmol of UDP-glucose in 1 min at 37°C.
Functional analysis of putative nucleotide-sugar epimerases encoded by orfH10 and orfH11.
Based on similarity analysis, orfH10 and orfH11 were proposed to encode epimerases that convert UDP-N-acetylglucosamine (UDP-GlcNAc) to unknown epimers. These epimerases may be involved in the biosynthesis of UDP-N-acetylgalactosamine (GalNAc), UDP-N-acetylmannosamine (UDP-ManNAc), UDP-N-acetylfucosamine (UDP-FucNAc), or other UDP-hexoseNAcs. OrfH10 is most closely related to Staphylococcus aureus Cap5E (80% similarity, 65% identity), while OrfH11 is most closely related to Cap5G (72% similarity, 53% identity) as well as to Cap5P (52% similarity, 27% identity) (15).
Cap5P and Cap5O are involved in UDP-ManNAcA biosynthesis; Cap5P is a 2-epimerase which converts UDP-GlcNAc to UDP-ManNAc, and Cap5O is a dehydrogenase which converts UDP-ManNAc to UDP-ManNAcA. Encoded on the same locus, Cap5G is also proposed to be a 2-epimerase, possibly with a role in the biosynthesis of UDP-FucNAc (16). Kiser and Lee (16) used E. coli strain EC21566 containing a mutation in wecC (known to encode a protein which converts UDP-GlcNAc to UDP-ManNAc) to demonstrate that cap5P complemented the mutation while cap5G did not. We have attempted to complement the wecC mutation in E. coli strain EC21566 with orfH11.
A 2.6-kb XbaI-BamHI fragment containing orfH11 was subcloned into pWSK129 and pBluescript II SK (pLBA662 and pLBA665, respectively) such that expression of orfH11 could be driven by the lac promoter. Whole-cell lysates from E. coli strain DH5α containing pLBA665, prepared after induction overnight with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), were shown to contain an additional band at 42 kDa compared to lysates from a control strain containing pBluescript II SK (data not shown). This corresponded to the predicted molecular mass of 41.5 kDa for OrfH11. No similar band was observed for strain DH5α containing pLBA665. pLBA662 and pLBA665 were transformed into EC21566. The phenotypes of the parental strain of EC21566 (AB1133), EC21566, and the two strains containing orfH11 were assessed by Western blotting using monoclonal antibody 898 (20, 22). This monoclonal antibody reacted with the parental strain but not with the control or either of the strains containing orfH11, indicating that this ORF was not able to complement the wecC mutant.
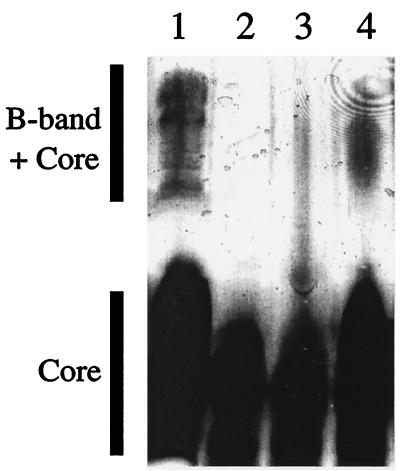
Similarity analysis revealed that OrfH10 is also related to a number of GlcNAc epimerases including Cap8E from S. aureus (65% identity and 80% similarity) and WbpM from P. aeruginosa (32% identity and 58% similarity) (15). WbpM converts UDP-GlcNAc to UDP-GalNAc, the first step in the biosynthesis of UDP-FucNAc. This function is proposed to reside in the N-terminal half of the protein. Strain wbpM-2 contains a wbpM gene which has been insertionally inactivated at the 5′ end. Phenotypically, this strain has no detectable B-band LPS and the wild-type phenotype can be restored by providing wbpM in trans (4). Accordingly, we used this mutant to determine if its mutation could be complemented by orfH10. Plasmid pLBA661 was derived by cloning a 2-kb BamHI-EcoRV fragment containing the entire orfH10 into BamHI-SmaI-digested pUCP19. This plasmid was transformed into strain wbpM-2; the B-band was partially restored in strain LBA736 compared to the situation for strain PAO1 (Fig. 2).
FIG. 2.
Silver-stained Tricine-SDS-PAGE (12.5% polyacrylamide) analysis of LPS extracts. Lanes: 1, P. aeruginosa strain PAO1, wild type, serotype O5; 2, strain wbpM-2, B-band negative; 3, strain wbpM-2 (pUCP19); 4, strain wbpM-2 (pLBA661). Stained polysaccharide features are indicated.
orfH13 functional analysis.
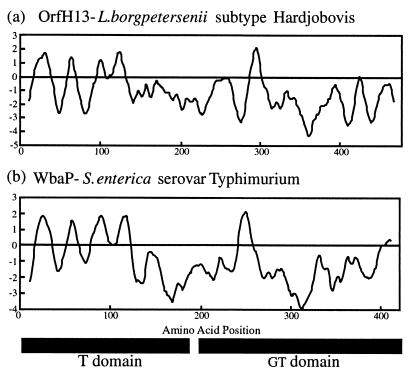
Based on similarity analysis, OrfH13 was proposed to be an undecaprenyl-glycosyl-1-phosphate transferase (und-pp-glycosyltransferase) (15). Among the proteins identified as similar to OrfH13 was WbaP from S. enterica (55% similarity, 25% identity). Importantly, similarity extends across the entire length of the compared proteins, and hence OrfH13 may contain both the T and GT domains (N- and C-terminal halves, respectively) identified previously in WbaP (28). Furthermore, the hydropathy profiles of OrfH13 and WbaP are remarkably similar; in particular, the T domain of both OrfH13 and WbaP is predicted to contain four transmembrane domains (Fig. 3).
FIG. 3.
Hydropathy profile of Hardjobovis OrfH13 (a) and WbaP from S. enterica serovar Typhimurium (b). The T and GT domains indicated are as identified by Wang et al. (28).
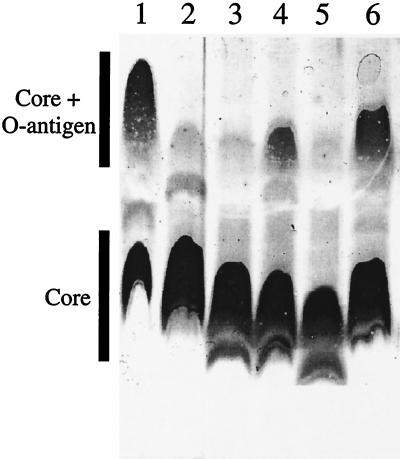
We attempted to examine the sugar specificity of OrfH13, the putative und-pp-glycosyltransferase of Hardjobovis. Based on the assumption that the two domains present in OrfH13 represent a fusion of two proteins (28) and that the original, independent functions of these proteins have not been destroyed by the fusion, it should be possible to identify the glycosyltransferase activity of the GT domain of OrfH13. RfbW (63% similarity and 43% identity to OrfH13) from V. cholerae is a galactosyltransferase. The rfbW gene in strain V1217 has been insertionally inactivated using a kanamycin resistance cassette, resulting in a rough phenotype (7). When rfbW was supplied in trans, the smooth phenotype was restored, as indicated by the presence of O-antigen and core bands in SDS-PAGE analysis (7). A comparison of the LPS produced by this strain with the LPS produced by strain V1217 transformed with either pLBA663 or pLBA664 (containing the entire orfH13 coding region) is shown in Fig. 4. The restoration of the O-antigen and core bands in the complemented strains indicated that OrfH13 has galactosyltransferase activity.
FIG. 4.
Silver-stained Tricine-SDS-PAGE (12.5% polyacrylamide) analysis of proteinase K-treated cell envelope extracts. Lanes: 1, V. cholerae strain O17, wild type; 2, strain 1217, rfbW strain; 3, strain 1217 (pBluescript KS+); 4, strain 1217(pLBA663); 5, strain 1217(pWSK129); 6, strain 1217(pLBA664). Stained polysaccharide features are indicated.
DISCUSSION
The initial objective of this work was to investigate the possibility of using the Hardjobovis rfb locus to synthesize “recombinant” leptospiral O antigen in a heterologous host such as E. coli. The cosmid (pLBA655) containing orfH1 through to orfH31 was unable to direct synthesis of Hardjobovis O antigen. Reverse transcription-PCR analysis of E. coli strains containing the cosmid revealed that orfH10 through orfH16 were not transcribed (unpublished data). A plausible explanation for the failure of the cosmid strain to produce Hardjobovis O antigen is that there was no O-antigen subunit assembly because the und-pp-glycosyltransferase (orfH13) was not expressed. This enzyme carries out a critical early function in O-antigen subunit assembly.
In the context of the development of a rational strategy for the expression of Hardjobovis O antigen in a heterologous host, the identification of the activity of the GT domain of OrfH13 is significant. Previously, 14C-labeled nucleotide sugars have been used to detect the incorporation of sugars onto the lipid carrier, and in combination with thin-layer chromatography, this approach has been used to determine the specificities of glycosyltransferases (17, 27). Significantly, the use of this approach to determine the specificities of the glycosyltransferases in the Hardjobovis rfb locus is only now possible because of the identification of galactose as the first sugar transferred to the lipid carrier.
Demonstrating that the putative und-pp-galactosyltransferase, OrfH13, can complement a galactosyltransferase mutant is not consistent with the model suggested by Wang et al. (28), which proposed that the specificity for the lipid carrier was contained in the GT domain. However, a functional dependence for glycosyltransferases on the lipid carrier (perhaps by binding to the lipid carrier) may be a means by which the function of glycosyltransferases is coordinated in the cytoplasm. It is also possible to speculate that und-pp-galactosyltransferases may have arisen through a fusion of a ‘T’ protein and a glycosyltransferase. Gene fusions involving glycosyltransferases appear to be common (e.g., OrfH23 in the Hardjobovis locus [15] and Y4gI in Rhizobium [9]) and may provide an advantage by intimately linking the functions of the fused proteins, as in the case of OrfH13, ensuring that galactose is the first sugar attached to the lipid carrier.
OrfH8, a galactose epimerase, converts UDP-glucose to UDP-galactose, thereby providing the substrate for OrfH13. Notably, a homolog of OrfH13 is present in the rfb locus of L. interrogans serovars Copenhageni and Pomona, although no OrfH8 homolog is present in either of these loci (unpublished data). The high level of similarity (86% similarity and 74% identity) between the OrfH13 homologs indicates that UDP-galactose is probably the substrate for each of the homologs and therefore suggests that the orfH8 homolog is located elsewhere on the Copenhageni and Pomona genomes.
Based on similarity analysis, we have previously proposed that the OrfH10 and OrfH11 are epimerases that have a common substrate (15). In this study, OrfH10 has been shown to convert UDP-GlcNAc to UDP-GalNAc. While it has not yet been possible to identify the function of OrfH11, it is feasible that OrfH11 converts UDP-GlcNAc to an epimer other than UDP-ManNAc. The WbpM protein is another protein that may have arisen as a result of a gene fusion. The N-terminal half of this protein is similar to OrfH10 and other UDP-GlcNAc epimerases, while the C-terminal half is similar to a number of GalE protein homologs. Notably, the C-terminal half does not play an essential role in B-band synthesis, as indicated by the unchanged phenotype in the wbpM-1 strain, in which the 3′ end of wbpM was insertionally interrupted (4).
The data presented here provide an important insight into the biosynthesis of LPS in L. borgpetersenii subtype Hardjobovis. The identification of OrfH13 as an und-pp-galactosyltransferase indicates that the process by which leptospiral LPS is synthesized is the same as in other bacteria (i.e., the O-antigen subunit is synthesized on a lipid carrier). This knowledge is essential for the design of effective strategies for the eventual synthesis of leptospiral LPS in heterologous host systems. In addition, the identification of the nucleotide sugars synthesized by proteins encoded on the leptospiral rfb locus will assist in the eventual determination of the structure of leptospiral LPS.
ACKNOWLEDGMENTS
This work was supported by a research grant from the National Health and Medical Research Council, Canberra, Australia.
We thank Vicki Vallance and Ian McPherson for their skilled technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D M, Seldman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 3.Birnboim M J, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrows L L, Charter D F, Lam J S. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol Microbiol. 1996;22:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 5.Elleman T C, Peterson J E. Expression of multiple types of N-methyl Phe pili in Pseudomonas aeruginosa. Mol Microbiol. 1987;1:377–80. doi: 10.1111/j.1365-2958.1987.tb01945.x. [DOI] [PubMed] [Google Scholar]
- 6.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 2nd ed. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 7.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrelly H E, Adler B, Faine S. Opsonic monoclonal antibodies against lipopolysaccharide antigens of L. interrogans serovar hardjo. J Med Microbiol. 1987;23:1–7. doi: 10.1099/00222615-23-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on the transformation of E. coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X-M, Brahmbhatt H N, Quigley N B, Reeves P R. A low copy number cosmid. Plasmid. 1987;18:170–172. doi: 10.1016/0147-619x(87)90045-x. [DOI] [PubMed] [Google Scholar]
- 13.Johnson R C, Walby J, Henry R A, Auran N E. Cultivation of parasitic leptospires: effect of pyruvate. Appl Microbiol. 1973;26:118–119. doi: 10.1128/am.26.1.118-119.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jost B, Adler B, Vinh T, Faine S. A monoclonal antibody reacting with a determinant on leptospiral lipopolysaccharide protects guinea pigs against leptospirosis. J Med Microbiol. 1986;22:269–275. doi: 10.1099/00222615-22-3-269. [DOI] [PubMed] [Google Scholar]
- 15.Kalmbaheti T, Bulach D M, Rajakumar K, Adler B. Genetic organization of the lipopolysaccharide O-antigen biosynthetic locus of Leptospira borgpetersenii serovar Hardjobovis. Microb Pathog. 1999;27:105–117. doi: 10.1006/mpat.1999.0285. [DOI] [PubMed] [Google Scholar]
- 16.Kiser K B, Lee J C. Staphylococcus aureus cap5O and cap5P genes functionally complement mutations affecting enterobacterial common-antigen biosynthesis in Escherichia coli. J Bacteriol. 1998;180:403–406. doi: 10.1128/jb.180.2.403-406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the ‘major outer membrane protein’ of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 20.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 21.Moreno F, Rodicio R, Herrero P. A new colorimetric assay for UDP-glucose 4-epimerase activity. Cell Mol Biol. 1981;27:589–592. [PubMed] [Google Scholar]
- 22.Rick P D, Mayer H, Neumeyer B A, Wolski S, Bitter-Suermann D. Biosynthesis of enterobacterial common antigen. J Bacteriol. 1985;162:494–503. doi: 10.1128/jb.162.2.494-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Smith M, Jessee J, Landers T, Jordan J. High efficiency bacterial electroporation: 1 × 1010E. coli transformants/μg. Focus. 1990;12:38–40. [Google Scholar]
- 25.Sugiyama T, Kido N, Arakawa Y, Mori M, Naito S, Ohta M, Kato N. Rapid small-scale preparation method of cell surface polysaccharides. Microbiol Immunol. 1990;34:635–641. doi: 10.1111/j.1348-0421.1990.tb01039.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsai C-M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 27.van Kranenburg R, van Swam I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol. 1996;178:2598–604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 30.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky A L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 31.Westphal O, Jann K. Bacterial lipopolysaccharides; extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 32.Wilkinson R G, Gemski P, Stocker B A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972;70:527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]