Abstract
The advent of mRNA vaccine technology has been vital in rapidly creating and manufacturing COVID-19 vaccines at an industrial scale. To continue to accelerate this leading vaccine technology, an accurate method is needed to quantify antigens produced by the transfection of cells with a mRNA vaccine product. This will allow monitoring of protein expression during mRNA vaccine development and provide information on how changes to vaccine components affects the expression of the desired antigen. Developing novel approaches that allow for high-throughput screening of vaccines to detect changes in antigen production in cell culture prior to in vivo studies could aid vaccine development. We have developed and optimized an isotope dilution mass spectrometry method to detect and quantify the spike protein expressed after transfection of baby hamster kidney cells with expired COVID-19 mRNA vaccines. Five peptides of the spike protein are simultaneously quantified and provide assurance that protein digestion in the region of the target peptides is complete since results between the five peptides had a relative standard deviation of less than 15 %. In addition, two housekeeping proteins, actin and GAPDH, are quantified in the same analytical run to account for any variation in cell growth within the experiment. IDMS allows a precise and accurate means to quantify protein expression by mammalian cells transfected with an mRNA vaccine.
1. Introduction
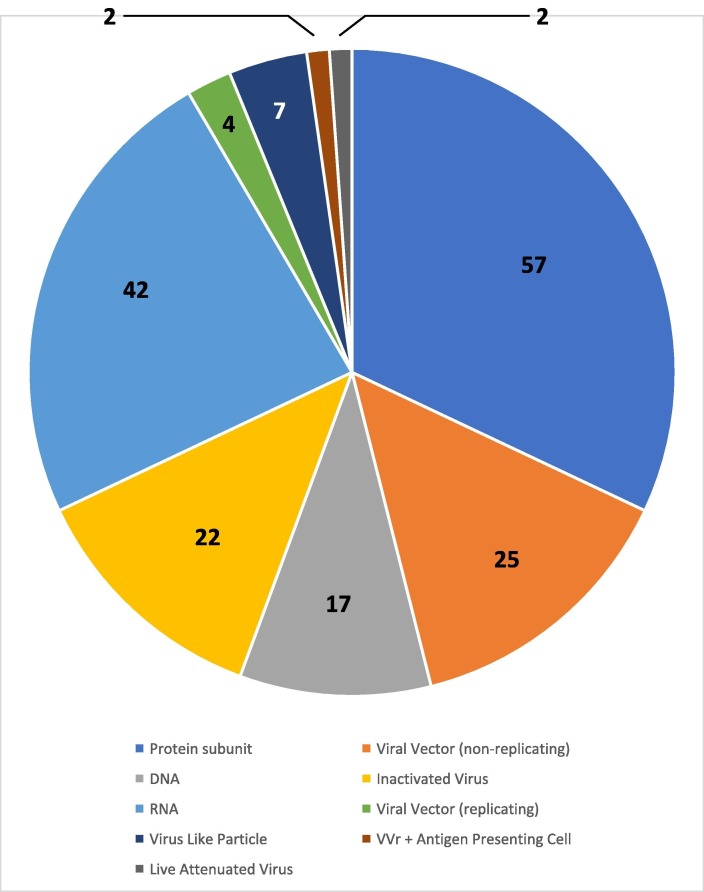
For some time now, multiple strategies for SARS-CoV-2 vaccine development have been actively pursued, including the use of inactivated virus, recombinant protein, mRNA, DNA, and viral vector vaccines [1]. Fig. 1 shows the current landscape of COVID-19 candidate vaccines according to the World Health Organization (WHO). As of March 6, 2023, there were 379 candidate vaccines with 180 having progressed to various stages of clinical trial. Forty-two off those 180 were RNA-based vaccines, 25 were viral vector non-replicating, and 4 were viral vector replicating. The spike glycoprotein is the main antigenic target for vaccine development. The nucleocapsid protein, a multifunctional protein that primarily functions for binding to the viral RNA genome and packing it into a long helical nucleocapsid structure, has also been found to be highly immunogenic and expressed abundantly during coronavirus infections [2].
Fig. 1.
Landscape of COVID-19 candidate vaccines obtained from the World Health Organization on March 6th, 2023. COVID-19 vaccine tracker and landscape (who.int). Of the vaccine candidates in clinical trials, 51 are recombinant (blue), 29 are RNA (yellow), 22 are non-replicating viral vector (green), 21 are inactivated viruses (grey), and 16 are DNA based (brown).
The mRNA vaccines possess unique advantages, including short development time, production in a cell-free environment, and their use of the translational machinery of the host [3]. Two classes of mRNAs—non-replicating and self-amplifying mRNA (SAM), are currently being developed for use as vaccines. Non-replicating mRNA encodes only the protein of interest, while SAM also encodes proteins enabling RNA replication [4]. While the technology has been in development for more than two decades [5], the use of mRNA as a vaccine had not previously been given extensive consideration because of the ease in which RNA degrades, the ubiquitous presence of ribonucleases, and the challenge of mRNA intracellular delivery [6], [7].
Since the eukaryotic initiation factor (eIF) recognizes and binds to the cap of mRNA, efforts to improve synthetic mRNA material have focused on development of new cap-analogs [8], [9] to protect the mRNA from rapid degradation by intra-cellular exonucleases and to aid in protein translation [6]. Other areas of investigation include the incorporation of 5′ and 3′ untranslated regions (UTRs) and a Poly(A) tail to also aid in stability and translation efficiency [10]. In addition, the delivery of mRNA can be mediated by both viral and non-viral vectors. Non-viral delivery vectors include lipid-based, poly-based, and hybrid lipid-polymer systems [11], while viral RNA delivery typically involves engineering of adeno-associated viruses to carry nucleic acid cargos [12].
All modifications made to the mRNA construct can affect translational efficiency, which influences the amount of protein expressed. Studies conducted at the mRNA level are largely based on a key assumption that mRNA expression is informative in predicting protein expression levels [13]. However, it is well documented that there is often poor correlation between mRNA and protein abundance [14], [15]. An accurate method is needed to monitor the vaccine during development and evaluate the effect that any changes to the construct or other vaccine components have on the expression of the desired antigen. In addition, this method should be able to accurately quantify multiple protein targets in the event that multicistronic constructs are explored. In short, an accurate method to quantify the amount of antigen produced in cell culture could be useful to evaluate potency prior to vaccine release and the ability to gauge vaccine stability over time and under various storage conditions [16], [17].
Protein quantification has traditionally been accomplished using sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), two dimensional polyacrylamide gel electrophoresis (2D-PAGE), enzyme-linked immunosorbent assay (ELISA), western blots, surface plasmon resonance, and mass spectrometry [18], [19], [20]. SDS-PAGE is typically used to check the purity of the protein, and image analysis and densitometry have been employed to quantify protein bands [21], [22]. 2D-PAGE is the standard method used in comparative proteomic studies for measuring changes in protein expression levels by comparing spot intensities, but it suffers from limited dynamic range, gel-to-gel variations, and the inability to resolve proteins of similar molecular weight [23], [24]. Antibody-based assays such as western blots and ELISA suffer from inherent problems such as limited availability of high-quality antibodies and batch-to-batch variability of said antibodies. It is also impractical to use antibody-based approaches for large-scale quantification of multiple proteins simultaneously [25], [26]. Both gel-based and antibody-based approaches generate semiquantitative measurements. Mass spectrometry (MS) offers straightforward, gel-free, and antibody-free workflows and can easily provide highly selective absolute quantification of multiple proteins in the sample. It also can be modified to quantify protein antigens of other diseases in which mRNA and viral vector platforms are being explored.
We previously reported an isotope dilution mass spectrometry (IDMS) method to quantify spike and nucleocapsid proteins of SARS-CoV-2 [27]. We have expanded the method to quantify the amount of protein expressed in baby hamster kidney cells (BHK) after transfection with an expired commercial vaccine material. We include simultaneous quantification of multiple peptides from two housekeeping proteins, actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), to serve as internal controls and allow normalization of the data to account for possible variation between the number of cells in the samples analyzed. We discuss in detail the care that must be taken to obtain precise results, between passages, of a cell-based assay. This IDMS method provides a means to accurately quantify multiple proteins, present in a wide dynamic range, in mammalian cell samples.
2. Materials and methods
2.1. Recombinant protein and vaccine material.
Recombinant SARS-CoV-2 Spike (GCN4-IZ) His Protein was purchased from R&D systems (Minneapolis, MN) and used without further purification. Commercially available COVID-19 vaccine material that was past the expiration date was received. These vials were stored per manufactuerer's instructions until use.
2.2. Growth, maintenance, and transfection of BHK cells
Baby Hamster Kidney cell (BHK-21) stocks were purchased from the American Type Culture Collection (ATCC) (Manassas, VA USA). According to ATCC quality control specifications, no mycoplasma contamination was detected. The cells were removed from liquid nitrogen and thawed quickly in a 37 °C water bath. Under a biosafety cabinet, cells were transferred into a Corning (Corning, NY USA) 15 mL conical centrifuge tube along with 9 mL of complete BioWhittaker® Dulbecco’s Modified Eagle’s Medium (DMEM) (Lonza), Walkersville, MD USA), containing 5 % fetal bovine serum (FBS) (ATCC), 1 % BioWhittaker® 200 mM L-glutamine solution in 0.85 % NaCl (Lonza), and 1 % antibiotic–antimycotic solution (Corning). This suspension was centrifuged in a Beckman Coulter (Indianapolis, IN USA) Allegra X-I4R centrifuge with SX4750A rotor at 250 xg for 5 min. The supernatant was aspirated to remove any dimethyl sulfoxide (DMSO) that had been added to the cells they were frozen. The cells were resuspended in 10 mL of complete DMEM and 20 µL was removed for cell counts. Cell counts were performed by adding 20 µL of trypan blue (Invitrogen, Carlsbad, CA USA) to the 20 µL of cell suspension. Ten microliters of trypan blue/cells were added to each side of a disposable Countess™ cell counting chamber slide (Invitrogen) and the slide was placed into a Countess™ 3 automated cell counter (Invitrogen) to assess cell counts and viability using the automatic brightness and contrast setting. The 10 mL suspension was added to a 75 cm2 Corning cell culture flask and placed in a humidified VWR (Radnor, PA USA) water jacketed CO2 incubator at 5 % CO2 and 37 °C. Cells were allowed to grow until 80–90 % confluency, which was determined visually under an Olympus IX51 inverted microscope (Olympus Corporation, Tokyo, Japan), and then the cells were split or passaged.
For cell passage, media was removed from the flask and cells were washed once with 10 mL of BioWhittaker® 1X phosphate buffered saline (PBS) without calcium or magnesium (Lonza). Five milliliters of BioWhittaker® 1X trypsin-versene mixture (Lonza) was added to cover the cell monolayer and then aspirated immediately. Flasks were placed in the incubator at 5 % CO2 and 37 °C for 3 min. Cells were visually observed under a microscope to ensure dissociation. Ten milliliters of complete DMEM were added to the flask and pipetted gently up and down over the cell monolayer to detach the cells. Cell detachment was confirmed under a microscope. Once detached, the suspension was pipetted up and down multiple times to make a homogeneous suspension. The mixture was transferred to a 15 mL conical centrifuge tube and cell counts were taken as described above. For ongoing passages, 2 mL of cell suspension and 8 mL of complete DMEM were added to multiple 75 cm2 flasks. To upscale for transfection, 5 mL of cell suspension and 15 mL of complete DMEM were added to two 150 cm2 flasks. The flasks were placed in the incubator at 5 % CO2 and 37 °C and allowed to grow until 80–90 % confluency, at which point they were passaged again or upscaled for transfection, using the same procedure described here, or used for transfection as described below.
In preparing cells for transfection, cells were passaged as described above, until the step involving resuspension in 10 mL of complete DMEM. After obtaining cell counts, the suspension was centrifuged at 250 xg for 5 min, the supernatant was removed, the cells were washed by resuspending in 10 mL of Opti-MEM (Gibco, Grand Island, NY USA), and centrifuged again as described previously. Based on cell counts, cells were resuspended in enough Opti-MEM for a final concentration of 2.0x106 cells/mL. Two hundred fifty microliters of cell suspension, 5.0x105 cells, were added to 1.5 mL Eppendorf Protein LoBind® microcentrifuge tubes (Eppendorf, Hamburg, Germany). Varying amounts of expired commercial vaccine material were added to each tube. Since the vaccine material contained lipid nanoparticles, no transfection reagent was used in these experiments. The cell suspension and vaccine were mixed by pipetting up and down multiple times in the tube. The entire cell suspension/vaccine complex was added to Fisherbrand™ 6-well surface treated sterile tissue culture plates (Fisher Scientific, Waltham, MA USA) with 2 mL of outgrowth media, DMEM containing 1 % FBS, 1 % L-glutamine, and 1 % antibiotic–antimycotic solution, in each well. A control sample of 250 µL of the cell suspension, with no vaccine, was also plated. Plates were placed in the incubator at 5 % CO2 and 37 °C. After 24 h post transfection, media was removed from each well, cells were washed with 1X PBS, and plates were taken to begin IDMS. These were labeled 24-hour samples. For the 48-hour samples, media was removed after 24 h post transfection, and 2.4 mL of fresh complete DMEM was added to each well. The plates were placed back in the incubator and allowed to grow an additional 24 h, after which they were treated identically to the 24-hour samples.
2.3. Preparation of working stock, calibration, and labeled solutions
SARS-CoV-2 spike protein (S) and housekeeping (HK) peptide mixtures (both native/light and labeled/heavy) were synthesized by Vivitide (Gardner, MA USA). Vials contained 1 nmol of each target peptide for both S and HK, and separate native vials contained 5 nmol of each target HK peptide. The isotopically labeled/heavy peptides were 13C15N labeled on the N-terminal arginine or lysine. The two vials of the 1 nmol native peptide mixtures (S and HK) were reconstituted in 0.5 mL of Optima™ LC/MS grade water with 0.1 % formic acid (v/v) (0.1 % FA (aq)) (Fisher Scientific, Watham, MA USA) and allowed to sit at room temperature for 30 min with occasional vortex mixing. The contents of the two native vials were then combined yielding 1 mL of 1.0 pmol/µL native working stock standard mixtures. The process was repeated for the 2 vials of 1 nmol labeled peptide mixtures. Nine 1.0 mL stock calibration standard mixtures were prepared by adding 1, 5, 10, 30, 50, 70, 90, 180, and 250 µL of the native standard peptide mixture, 50 µL of the labeled standard peptide mixture, and enough 0.1 % FA (aq) for a final volume of 1.0 mL. This resulted in calibration standards with the following concentrations: 1, 5, 10, 30, 50, 70, 90, 180, and 250 fmol/µL. Samples prepared for protein quantification were spiked with 5 µL of the 1.0 pmol/µL labeled working stock standard mixture for quantitative analysis.
One vial of 5 nmol HK native peptide mixture was reconstituted in 0.5 mL of 0.1 % FA (aq) yielding 10.0 pmol/µL native working stock standard mixture. The vial was allowed to sit at RT for 30 min with occasional vortex mixing. Five 0.5 mL stock calibration standard mixtures were prepared by adding 10, 20, 40, 80, 120, and 180 µL of the native standard peptide mixture, 25 µL of the labeled standard peptide mixture used above, and enough 0.1 % FA (aq) for a final volume of 0.5 mL. This resulted in calibration standards with the following concentrations: 200, 400, 800, 1600, 2400, and 3600 fmol/µL.
2.4. Preparation of BHK cells and IDMS quantification
Directly to each well of the plates, 600 µL of Pierce™ RIPA lysis buffer (Thermo Scientific, Waltham, MA USA) was added and aspirated then dispensed repeatedly with a pipette over the cell monolayer to ensure complete lysis of the cells. The lysates were transferred to 2 mL Eppendorf Protein LoBind® microcentrifuge tubes (Hamburg, Germany) and 2 µL (∼678 U) of Deoxyribonuclease 1 (DNase 1) (Invitrogen) was added to the tube to degrade the DNA. The samples were then sonicated in an AQUASONIC model 150D sonicating water bath (VWR Scientific, Radnor, PA USA) for 30 min. After sonicating, 1200 µL of cold Optima™ acetone (Fisher Scientific, Waltham, MA USA) was added to each vial to precipitate the protein from the cell lysate. If a sample needed to be split, 300 µL of cell lysate was transferred to a separate 2.0 mL microcentrifuge tube after sonication, and 900 µL of cold acetone was added to each vial. The samples were then placed in a −20 °C freezer overnight.
Samples were removed from the freezer and centrifuged at 16,162 xg in an Eppendorf (Enfield, CT USA) model 5430 R centrifuge for 30 min. A protein pellet was visible on the bottom and side of the tube. The supernatant was carefully removed and 900 µL of cold absolute (200 proof) ethanol (Fisher Scientific) was added to wash the pellet to remove lipids [28]. The samples were resuspended by vortexing before being centrifuged again for 15 min at 16,162 xg and the ethanol was carefully removed from the protein pellet located at the bottom and side of the Eppendorf tube. The remaining ethanol was evaporated with a Labconco Refrigerated CentriVap Concentrator (Kansas City, MO USA). The samples were spun down using the preset speed of the CentriVap at 27 °C until the pellet was dry. Then 75 µL of a 0.05 % solution of Rapigest™ SF Surfactant (Waters Corporation, Milford, MA USA) in 50 mM ammonium bicarbonate was added to each sample, samples were heated for 5 min at 100 °C, and allowed to cool to RT. After cooling, 10 µL (∼172 pmol) of Promega Sequencing Grade Modified Trypsin (Promega) was added to each vial. The protein pellet was not immediately soluble. Samples were place on an Eppendorf ThermoMixer F 1.5 at 1500 rpm to ensure that the pellet would be constantly mixed. The samples were subject to enzymatic digestion at 37 °C for 4 hrs.
After incubation, 10 µL of a 0.45 M HCl solution was added to reduce the pH and samples were incubated at RT for 30 min to cleave the acid-labile surfactant [29]. To each of the samples, 5 µL of the 1.0 pmol/µL combined S and HK labeled working stock mixture was added. All samples were vortexed and centrifuged at 16,162 xg for 10 min, to ensure all hydrophobic particles, proteins, and peptides were on the side of the vial to avoid clogging the LC column. Samples were then transferred to LC autosampler vials for analysis.
2.5. LC/MS instrumentation parameters
A Thermo Scientific Vanquish Horizon Ultra High-Pressure Liquid Chromatographic (UHPLC) system was configured for separation of target peptides. The analytical column utilized was 150 mm × 2.1 mm i.d. Hypersil GOLD™ Vanquish reversed-phase C18 UHPLC column (1.9 µm particle size, Thermo Scientific). The injection volume was 5 µL. The aqueous mobile phase (A) was Optima™ LC/MS grade water with 0.1 % formic acid (v/v), while the organic mobile phase (B) was Optima™ LC/MS grade acetonitrile with 0.1 % formic acid (v/v). The flow rate was held constant at 200 µL/min. Initially, the mobile phase consisted of 98 % A and 2 % B and was held at these initial conditions for 2 min to concentrate the sample at the head of the column. At 2 min the gradient was stepped to 50 % A and 50 % B over the next 23 min. The gradient was stepped to 2 % A and 98 % B at 25 min and held at high organic for 6 min to wash the column. The mobile phase was then switched back to initial conditions (98 % A and 2 % B) at 35 min and the column was allowed to equilibrate for 22 min. The total analysis run time was 57 min.
A Thermo Scientific TSQ Altis™ triple quadrupole tandem mass spectrometer with an electrospray interface was used to analyze the compounds eluting from the LC column. The instrument was operated in positive ion mode and a selected reaction monitoring (SRM) method was used. For each peptide, a quantification transition and two additional confirmation transitions were monitored. All quantitative and confirmation transitions are listed in Table 1 . Instrument parameters were as follows: spray voltage 3500 V, sheath gas 20, auxiliary gas 0, ion transfer tube temperature 300 °C, and collision gas pressure of 1.5 mTorr. Collision energies were empirically determined using Skyline (MacCoss Lab Software, Seattle, WA USA) which uses the base collision energy (CE) linear equation that is optimized for the Altis [30]. Instrument control and data processing was performed via Thermo Scientific TraceFinder™.
Table 1.
Target peptides employed for the quantitation of SARS-CoV-2 spike (S) protein. Housekeeping proteins glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin (A) were also quantified to allow normalization for the number of mammalian cells that were processed. All peptides were 13C15N isotopically labeled on the C-terminal lysine or arginine and are indicated in bold font and underlined. M/z = mass/charge.
| Target Peptide | Protein | Precursor ion (m/z) | Quantification ion (m/z) | Confirmation ion (m/z) | Confirmation ion (m/z) | Collision energy (eV) |
|---|---|---|---|---|---|---|
| GVYYPDK | Spike(S1) | 421.208 (+2) | 685.319 (y5) | 359.193 (y3) | 522.256 (y4) | 15.5 |
| GVYYPDK | Spike (S1) | 425.215 (+2) | 693.332 (y5) | 367.207 (y3) | 530.270 (y4) | 15.5 |
| GIYQTSNFR | Spike (S1) | 543.272 (+2) | 624.310 (y5) | 752.369 (y6) | 915.432 (y7) | 19.2 |
| GIYQTSNFR | Spike (S1) | 548.276 (+2) | 634.318 (y5) | 762.377 (y6) | 925.440 (y7) | 19.2 |
| FLPFQQFGR | Spike (S1) | 570.303 (+2) | 879.447 (y7) | 635.326 (y5) | 782.394 (y6) | 20.0 |
| FLPFQQFGR | Spike (S1) | 575.307 (+2) | 889.455 (y7) | 645.334 (y5) | 792.403 (y6) | 20.0 |
| VTLADAGFIK | Spike (S2) | 517.797 (+2) | 721.388 (y7) | 650.351 (y6) | 535.324 (y5) | 18.4 |
| VTLADAGFIK | Spike (S2) | 521.804 (+2) | 729.402 (y7) | 658.365 (y6) | 543.338 (y5) | 18.4 |
| ASANLAATK | Spike (S2) | 423.737 (+2) | 688.399 (y7) | 617.362 (y6) | 503.319 (y5) | 15.6 |
| ASANLAATK | Spike (S2) | 427.744 (+2) | 696.413 (y7) | 625.376 (y6) | 511.333 (y5) | 15.6 |
| AAFTSGK | GAPDH | 341.181 (+2) | 539.282 (y5) | 392.214 (y4) | 610.320 (y6) | 13.1 |
| AAFTSGK | GAPDH | 345.189 (+2) | 547.297 (y5) | 400.228 (y4) | 618.334 (y6) | 13.1 |
| AITIFQER | GAPDH | 489.274 (+2) | 793.420 (y6) | 579.289 (y4) | 692.373 (y5) | 17.6 |
| AITIFQER | GAPDH | 494.278 (+2) | 803.429 (y6) | 589.297 (y4) | 702.381 (y5) | 17.6 |
| VIPELNGK | GAPDH | 435.258 (+2) | 657.356 (y6) | 560.303 (y5) | 770.440 (y7) | 16.0 |
| VIPELNGK | GAPDH | 439.265 (+2) | 665.370 (y6) | 568.318 (y5) | 778.454 (y7) | 16.0 |
| AGFAGDDAPR | Actin | 488.727 (+2) | 630.284 (y6) | 701.321 (y7) | 573.263 (y5) | 17.6 |
| AGFAGDDAPR | Actin | 493.731 (+2) | 640.292 (y6) | 711.330 (y7) | 583.271 (y5) | 17.6 |
| GYSFTTTAER | Actin | 566.767 (+2) | 678.342 (y6) | 577.294 (y5) | 825.410 (y7) | 19.9 |
| GYSFTTTAER | Actin | 571.771 (+2) | 688.350 (y6) | 587.302 (y5) | 835.418 (y7) | 19.9 |
| IIAPPER | Actin | 398.239 (+2) | 569.304 (y5) | 498.267 (y4) | 401.214 (y3) | 14.9 |
| IIAPPER | Actin | 403.243 (+2) | 579.312 (y5) | 508.275 (y4) | 411.223 (y3) | 14.9 |
2.6. Polyacrylamide gel electrophoresis (PAGE) and western blot analysis
Acetone-precipitated, RIPA-cell lysed proteins (non-vaccine control and vaccine-transfected BHK-21 preparations) were resuspended in 40 µL sample buffer (comprising lithium dodecyl sulfate, pH 8.4 and dithiothreitol) (Invitrogen). Samples were denatured at 70 °C for 10 min, followed by a 5–10 sec spin using a table-top centrifuge. For each sample, 18 µL was loaded in duplicates, on one-dimensional precast Novex NuPAGE™ 4 to 12 % Bis-Tris, 1.0 mm, mini gels (Invitrogen) and electrophoresed at 200 V for 45 min. Precision plus protein™ dual color standards (ranging from 10 to 250 kDa) were used as a size marker (Bio-Rad Laboratories, Hercules, CA USA). Additionally, 0.1 µg purified recombinant SARS-CoV-2 spike protein (CDC, Division of Scientific Resources, Atlanta, GA USA) and 2 µg human endothelial cell lysate (Becton, Dickinson and Company, Madison, GA USA) were used as positive protein controls.
First, separated proteins were fixed 30 min in 30 % methanol, 5 % acetic acid and visually examined using GelCode™ Blue Safe Protein Stain (Thermo Scientific, Rockford, IL USA). Second, to confirm spike protein expression, western blot analysis was performed. Proteins were transferred out of the unstained duplicate gel to a polyvinylidene fluoride (PVDF) membrane using the iBlot-2 rapid dry blotting system (Invitrogen). The PVDF blot was subsequently probed with affinity purified primary SARS-CoV-2 spike protein (receptor binding domain) monoclonal antibody (Invitrogen, 1 µg/µL; 1:100 final dilution). For immunodetection, the anti-rabbit WesternBreeze™ chromogenic kit (Invitrogen) was used. For optimal antibody reactivity, the primary antibody was incubated overnight at +4 °C and the next day, the final steps in the WesternBreeze™ chromogenic kit protocol were completed. After development and image scanning using Epson software (Epson America, Inc., Alamitos, CA USA), PVDF blots were subsequently washed twice with 10 mL phosphate buffer saline with tween™ detergent for 10 min. The PVDF blot was re-probed with mouse-β-actin monoclonal antibody (1 µg/µL, 1:1000 final) (Novus™ Biologicals, LLC, Centennial, CO USA) as a gel loading control for protein normalization.
3. Results and discussion
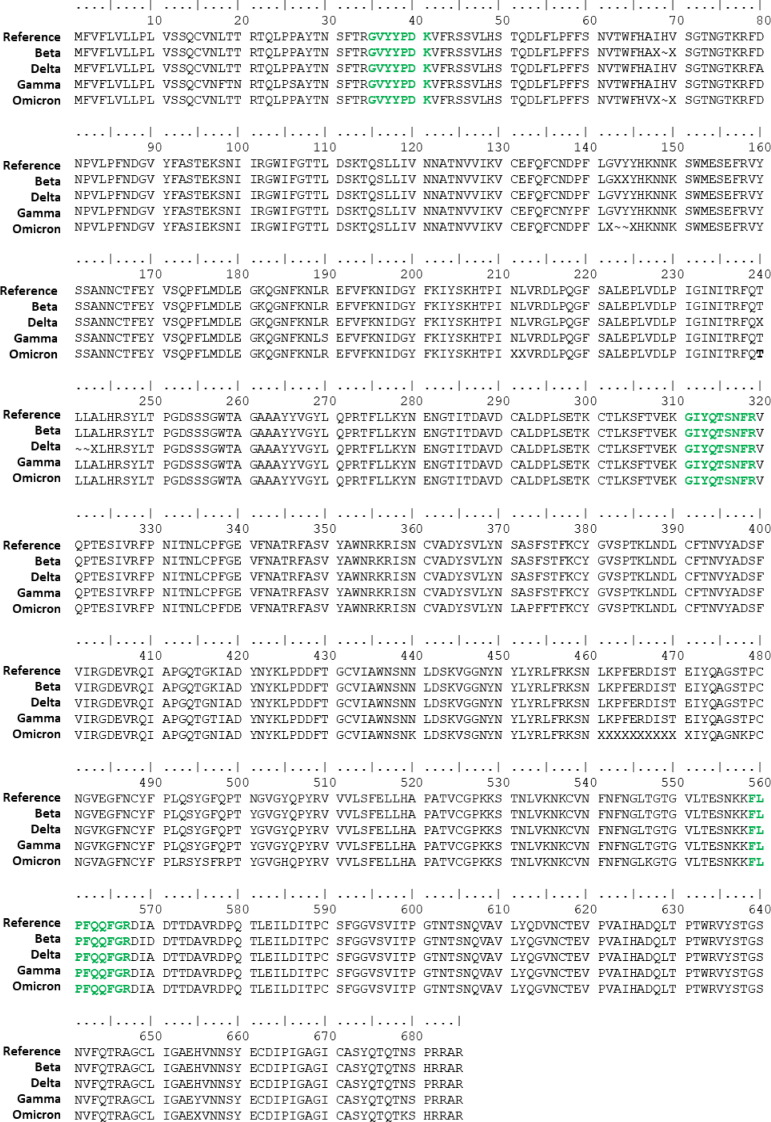
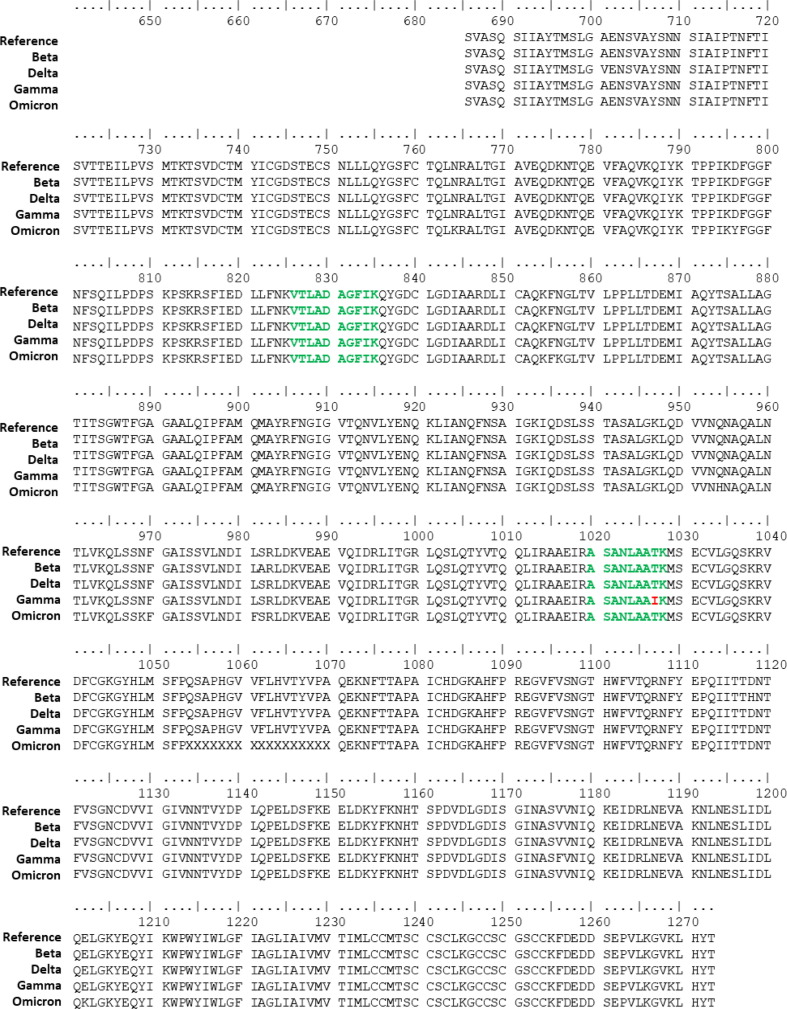
We have developed an isotope dilution mass spectrometry method to quantify SARS-CoV-2 spike protein from mammalian cells that have been transfected with a mRNA COVID-19 vaccine. In this experiment BHK-21 cells were used. In future studies, other cell lines can be tested to compare protein expression of cells transfected with mRNA vaccine. Expired vaccines were used for method development and in this study. Details of the spike peptide standard mixture required for accurate and precise quantification have been previously described [27]. Briefly, five tryptic peptides that lacked methionine, cysteine, and tryptophan that cover both the S1 (Fig. 2 A) and S2 (Fig. 2B) region of the spike protein were chosen. A critical assumption of the IDMS method is that there is one mole of peptide per mole of protein. Therefore, quantifying the amount of peptide directly correlates to the amount of protein. These peptides are conserved and can be used to quantify numerous SARS-CoV-2 lineages including the ancestral strain, Alpha, Beta, Gamma, Delta, and Omicron variants of concern. These protein sequences were obtained from Global Initiative on Sharing Avian Influenza Data (GISAID) [31], [32], [33] and the details of the strains used in the alignment are described in the acknowledgements. There is a T1027I mutation in the Gamma variant which impacts the ASANLAATK peptide, however the other four peptides are conserved and would be used for analysis should a vaccine targeting the Gamma variant be produced. Multiple target peptides for each protein provides assurance that complete digestion of the protein in the region of the target peptide is achieved as the quantification of each target peptide is an independent measurement and the expectation is that each peptide target yields the same result for the protein in that sample. Also, especially for viral proteins, the possibility of mutations in peptide sequence must be anticipated. Multiple target peptides provide redundancy in case a target peptide is rendered inadequate because of a mutation in that region of the protein. This redundancy provides assurance that the method will be applicable to multiple SARS-CoV-2 variants.
Fig. 2.
Amino acid sequences of (A) the S1 and (B) S2 regions of spike protein for variant strains of interest showing conservation of IDMS peptides. Target peptides chosen for the quantification method are indicated in bold green font. In the Gamma variant, the ASANLAATK peptide has a T1027I mutation, leaving 4 peptides for quantification of spike protein.
Protein expression is typically estimated by comparison against the levels of signal observed for proteins inherent in the cells, commonly called “housekeeping” (HK) proteins [34], [35]. These proteins are universally expressed in every cell type in the organism. Some common housekeeping proteins are structural proteins such as actin and tubulin while others, like glyceraldehyde-3-phosphate dehydrogenase (GAPDH), are enzymes involved in multiple complex cellular functions [36]. These housekeeping proteins are often used to check for equal loading or to compensate for potential loading differences in western blot analysis. IDMS quantification of these housekeeping proteins does not require proper choice and testing of antibodies and is more accurate than measuring stain density. By quantifying these housekeeping proteins by IDMS simultaneously in the same analytical run as that being used to quantify the viral antigen, an alternative to cell-counting is provided that is more accurate and can be evaluated without any loss of sample. It is the expectation that each well of the 6-well plate contained the same number of cells after growth under consistent conditions and time. However, the housekeeping proteins are used to verify this assumption, and to compensate for technical discrepancies or unknown cell growth variation which may cause cell counts to fluctuate between wells. By quantifying the housekeeping proteins, a means to ensure consistency of the experiment in terms of the number of cells analyzed is provided. Since the housekeeping proteins provide the means to correct for the variable number of cells in a well, more reproducible results are achieved. This is reflected in a lower percent relative standard deviation ( % RSD).
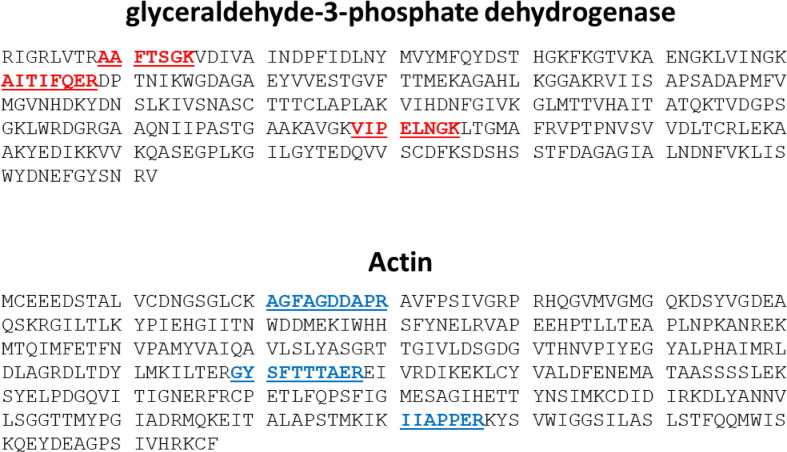
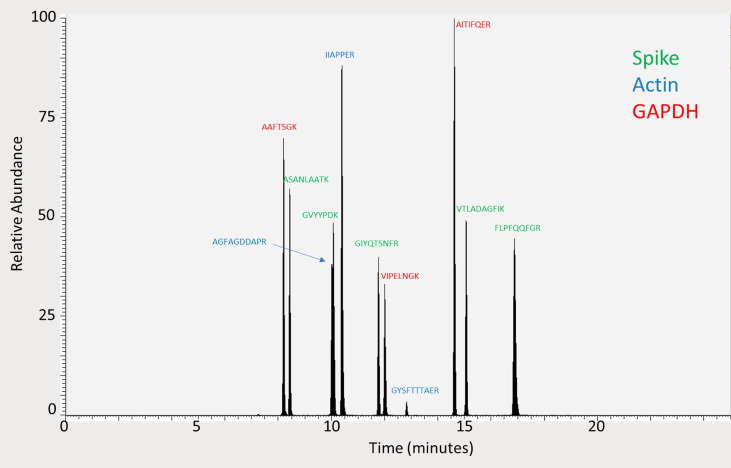
Because the HK proteins are often far more abundant than the protein whose expression is being measured, a calibration curve with an extended dynamic range was used for actin and GAPDH. A calibration curve that contained both spike and HK proteins was made that spanned the range from 1 to 250 fmol/µL. A second calibration curve was made for the HK proteins that spanned the range from 200 to 3600 fmol/µL. In all samples analyzed, spike protein was quantified using the 1–250 fmol/µL calibration curve, while GAPDH often could be quantified using either calibration curve. Actin required the 200–3600 fmol/µL calibration curve. All standards and samples were spiked with the same internal standard so that the target peptides could be analyzed with their respective calibration curves in one analytical run. The peptides chosen to quantify these two HK proteins are shown in Fig. 3 and the mass spectrometry transitions are listed in Table 1. The LC run time was 57 min and peptides from each protein are color-indicated in the following manner in Fig. 4 : spike (green), GAPDH (red), and actin (blue).
Fig. 3.
Amino acid sequences of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin from Mesocricetus auratus (Golden hamster) were obtained from The Universal Protein Resource (UniProt). Target peptides for quantification of GAPDH and actin are underlined and indicated in red bold font and blue bold font respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Total ion chromatogram illustrating that all proteins are eluted from the column within 20 min. Peptides of spike protein are in green font, GAPDH in red font, and actin in blue font.
Table 2 shows an example of the average protein amounts, standard deviations, and % RSDs, as well as the agreement between target peptides, for spike, actin, and GAPDH in a sample harvested 48-hours post transfection. The average amount of spike, GAPDH, and actin was 5.2 pmol/cell pellet, 45.1 pmol/cell pellet, and 294.5 pmol/cell pellet, with a standard deviation of 0.8 pmol/cell pellet, 5.5 pmol/cell pellet, and 25.4 pmol/cell pellet, and a % RSD of 14.5, 12.2, and 8.6, respectively.
Table 2.
Cells were transfected in triplicate with expired commercial vaccine material, lysed, and analyzed by IDMS. Target peptides were were used to quantify spike, GAPDH, and actin. The average and % RSD were calculated by considering each peptide result of each replicate as an independent data point. The % RSD for each protein was less than 15.
| Spike (fmol/cell pellet) | ASANLAATK | GVYYPDK | GIYQTSNFR | FLPFQQFGR | VTLADAGFIK | Average | Std. Dev. | % RSD |
|---|---|---|---|---|---|---|---|---|
| 50 µg vaccine prep 1 | 5.7 | 5.2 | 4.7 | 6.5 | 5.6 | 5.2 | 0.8 | 14.5 |
| 50 µg vaccine prep 2 | 4.7 | 4.3 | 4 | 5.2 | 4.4 | |||
| 50 µg vaccine prep 3 | 5.6 | 5.4 | 5.1 | 6.7 | 5.5 | |||
| GAPDH (pmol/cell pellet) | AAFTSGK | VIPELNGK | AITIFQER | |||||
| 50 µg vaccine prep 1 | 39.7 | 54.1 | 43.6 | 45.1 | 5.5 | 12.2 | ||
| 50 µg vaccine prep 2 | 37.5 | 48.9 | 40.7 | |||||
| 50 µg vaccine prep 3 | 44.8 | 51.2 | 45.4 | |||||
| Actin (pmol/cell pellet) | AGFAGDDAPR | IIAPPER | GYSFTTTAER | |||||
| 50 µg vaccine prep 1 | 297.8 | 255.6 | 300.5 | 294.8 | 25.4 | 8.6 | ||
| 50 µg vaccine prep 2 | 322 | 281.8 | 310.5 | |||||
| 50 µg vaccine prep 3 | 328 | 258.3 | 298.4 |
The effect of the cell matrix on the accuracy of IDMS quantification was tested by the addition of a recombinant spike protein (R&D Systems, Minneapolis, MN USA) to cells immediately prior to cell lysis. A recombinant spike protein solution (20 µL) (R&D Systems, Minneapolis, MN USA) was analyzed by IDMS in triplicate. The concentration of the solution was determined to be 221.18 ± 1.87 fmol/µL. The effect of the cell matrix on the accuracy of IDMS quantification was tested by the addition of this recombinant spike protein to cells immediately prior to cell lysis. Before the cells were lysed using RIPA buffer, 20 µL of the recombinant spike protein solution was added to three individual wells of the plate. The cells with the addition of the solution of recombinant spike protein were then lysed, the proteins precipitated, and subsequently subjected to enzymatic digestion. The quantitative result of adding the same recombinant spike protein to the plates containing BHK cells was 213.73 ± 9.06 fmol/µL with a recovery of 96.6 %. This demonstrates that the cell matrix has no detrimental effects on the quantification of targeted proteins.
When developing a quantitative assay, it is important to design the experiment in such a way as to minimize sample handling, and thus, unintended sample loss. This is critical when the peptide internal standards are not introduced until after all the sample preparation steps are complete. To ensure complete lysis of all cells in the wells and to minimize sample loss, trypsin was not used to harvest and pellet cells. Instead, cell lysis was performed directly in the sample plate. To ensure efficient lysis of the cells and protein extraction, Pierce™ RIPA lysis buffer was used. This cell lysis reagent from Thermo Scientific has a fully disclosed formulation, contains three non-ionic and ionic detergents, and enables the simultaneous extraction of membrane, nuclear, and cytoplasmic proteins. No protease inhibitors were added to allow DNase 1 mediated degradation of the DNA and the downstream trypsin digestion of the proteins.
Since detergents can cause degradation of chromatographic resolution and severely suppress ionization in mass spectrometry analyses, detergent removal is required prior to LC/MS analysis. Molecular weight cut-off spin filters and dialysis membranes can result in sample loss and reduced recovery of some proteins [37], [38]. Therefore, it was concluded that protein precipitation would be used to remove the detergents from the samples. Protein precipitation is simple and has been demonstrated to be a robust and reproducible means to recover intact proteins for quantitative assays [39], [40].
A requirement for accurate quantification of proteins by IDMS is that complete digestion of the protein in the region of the target proteins is achieved. Parameters that can affect the digestion efficiency are the addition of denaturing detergents, the amount of trypsin used for enzymatic digestion, and the time in which the sample incubates in the presence of trypsin [41]. In-plate lysis was chosen as an efficient means to lyse the cells without significant sample loss. The traditional use of trypsin to harvest and pellet cells resulted in leftover media being present in the samples. Additionally, the media contains fetal bovine serum, which is rich with proteins, that would also be precipitated after adding acetone. The presence of these undesirable FBS proteins increases the total amount of protein in the sample so that the ratio of trypsin to protein would be lower than desired, and thus, the digestion efficiency of target proteins would be adversely impacted. Since multiple peptides are used to quantify a protein, incomplete digestion is inferred when multiple target peptides do not yield the same quantitative results. Lack of peptide agreement was more frequently observed in the 48-hour samples due to the extra growth time, which resulted in more cells than 24-hour samples. Therefore, for 48-hour samples, the cell pellet was split prior to tryptic digestion to achieve peptide agreement for these samples. While the amount of trypsin added could have been increased and the time of incubation could have been extended, splitting the cell pellet was simple and did not significantly add to the time or cost of the method.
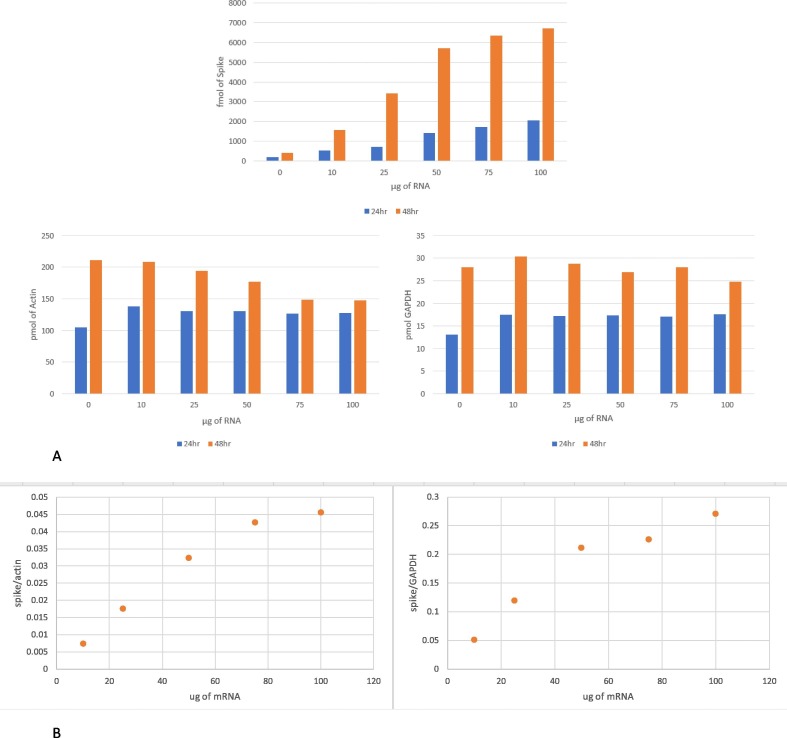
Fig. 5A shows the results for quantification of spike, actin, and GAPDH in passage 18 cells transfected for 24 and 48 h with 10, 25, 50, 75, or 100 µg of expired commercial vaccine material. A control sample for each time point, where no expired commercial vaccine was added, resulted in no detection of spike protein, as expected. The 24-hour samples transfected with 10 and 25 µg of expired commercial vaccine material resulted in less than 1 pmol/cell pellet of spike. The 24-hour samples transfected with 50, 75, and 100 µg of expired commercial vaccine material resulted in 1.43, 1.72, and 2.04 pmol/cell pellet of spike, while the 48-hour samples transfected with 10, 25, 50, 75, and 100 µg of expired commercial vaccine material resulted in 1.55, 3.43, 5.71, 6.33, and 6.71 pmol/cell pellet of spike, respectively. Based on these data, it can be concluded that samples within the same passage can be compared since housekeeping protein quantitation remained consistent from sample to sample. This indicates that cells seeded in separate wells are growing at the same rate. Since the media, and therefore the vaccine, was aspirated from the 48-hour samples after the first 24 h, the increased spike expression was not the result of cells growing and then up taking more vaccine. This means that any increased spike expression in the 48-hour samples only came from cells transfected in the first 24 h, and therefore, harvesting cells 48 h post transfection was necessary to ensure the cells had enough time to express sufficient spike for quantitation. It is possible that harvesting cells 24 h post transfection would be sufficient with a vaccine that had not reached its expiration date, however, access to unexpired vaccine was not possible at the time of this investigation. Harvesting cells 72 h post transfection was attempted. While the amount of actin and GAPDH increased, the amount of spike decreased. When the cells were observed microscopically before harvesting, dead cells were observed floating in the media along with a high confluency of 95–100 %. Confluency refers to the percentage of the surface area of the well that is covered by the adherent cells. As the cells grow and multiply, there eventually reaches a point where there is no more room for the cells to grow, stated as 100 % confluent. Once this happens, the cells compete for the limited number of resources available to survive. As the resources dwindle, cells will begin to die and lyse, resulting in their contents spilling into the media. This happens indiscriminately. When the media was aspirated, any spike that had been produced by these dead cells was also aspirated. The result is less spike protein quantified in the cells that were harvested, demonstrating that a longer post transfection incubation time was not beneficial.
Fig. 5.
Bar graphs of (A) protein quantitation in pmol/cell pellet for spike, GAPDH, and actin vs amount of mRNA vaccine material added in µg and (B) the ratio between actin and spike, and GAPDH and spike vs amount of mRNA vaccine material added in µg.
For both the 24- and 48-hour samples, increasing the amount of expired commercial vaccine transfected also resulted in an increase in spike expression. When comparing samples from the same timepoint, it is important to use the housekeeping proteins to normalize for the number of cells in the sample. To minimize sample loss, cell lysis was performed directly in the well, so cell counts are not measured by other means. Fig. 5B shows the ratio of actin to spike and GAPDH for samples harvested 48 h post transfection. Each well in the 6-well plate was seeded with 5.0×105 cells.
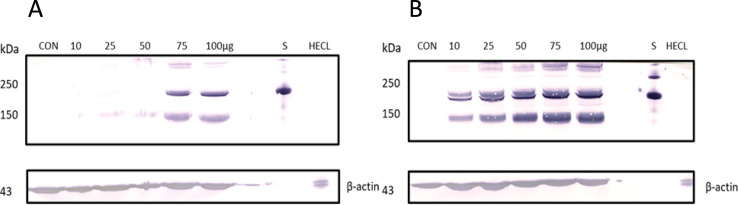
We also analyzed the spike protein expression of 24- and 48-hours by western blot analysis (Fig. 6 ) as an orthogonal method to confirm expression of spike protein. The antibody used for western blot targeted the receptor binding domain portion of the spike protein. Antibody reactive bands demonstrate that full length spike was expressed from the BHK cells. Additionally, immune reactive band intensity of spike protein increases with vaccine concentration. This linearity correlates with IDMS spike peptide quantitative values. Like the use of the housekeeping proteins in IDMS, mouse-β-actin was used to verify equal loading of the gel, ensuring that the band intensities between samples were comparable.
Fig. 6.
Western blot analysis confirming the expression of spike protein in BHK for both (A) 24- and (B) 48-hour samples. M = Precision plus protein™ dual color standards; CON = control; S = recombinant SARS-CoV-2 spike protein; HECL = human endothelial cell lysate.
To determine the effect of the cell culture on protein expression, the same experiment was repeated across five subcultures of the reference cell line obtained from ATCC. These subcultures are known as “passages” [42]. It is important to allow the cells to fully recover after being thawed from liquid nitrogen storage. This means it is necessary to passage the cells multiple times before performing transfection experiments, but after too many passages, the genetic characteristics of the cells may differ from the original culture which could impact protein expression [43], [44], [45]. Thus, all experiments were performed after 10 passages and concluded by 22 passages. Transfection using the same levels of vaccine were performed at passages 10, 12, 14, 16, 18, and 20. Since two cell passages were performed a week, this translates to one transfection experiment performed each week.
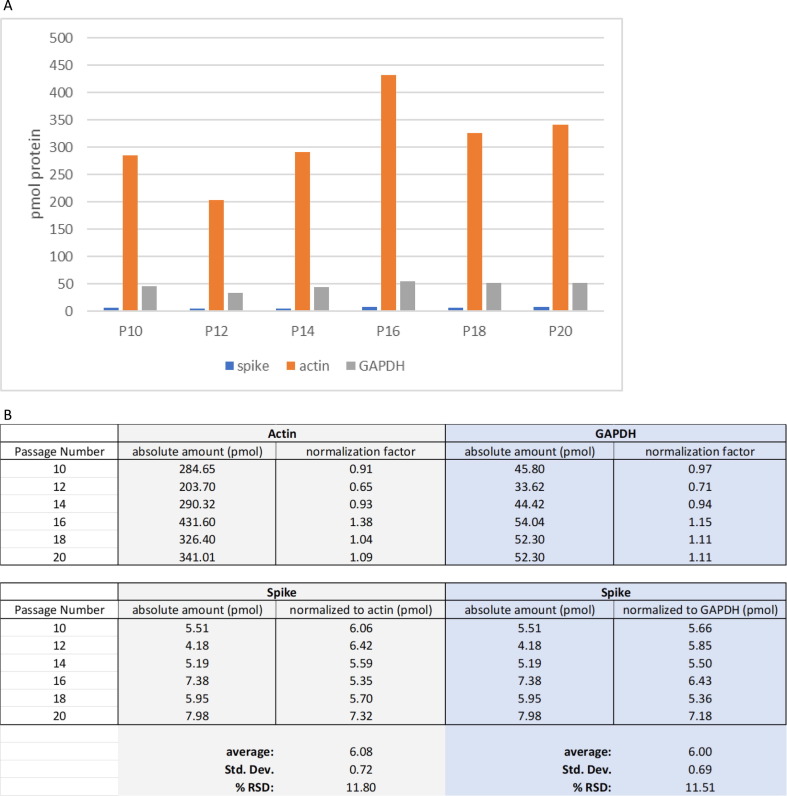
For direct comparison between the amount of mRNA used to transfect the cells and the amount of spike protein expressed, the amount of mRNA was converted from µg to pmol. To make this conversion, the molecular weight of the mRNA construct must be known. We estimated that the molecular weight of an mRNA construct that contains the instructions only to make spike protein has a molecular weight of approximately 1.4MDa. Thus, the amount of mRNA that was used to transfect the cells in these experiments ranged from 1 to 70 pmol. When 50 µg, or 36.3 pmol, of mRNA was transfected in passages 10, 12, 14, 16, 18, and 20, spike expression was quantified as 5.5, 4.2, 5.2, 7.4, 6.0, 8.0 pmol respectively. Therefore, across 11 passages, of which only 6 were sampled, the absolute amount of spike expressed was 6.1 +/− 1.4 pmol ( % RSD of 23.5). The absolute values of spike protein and the HK proteins are shown in Fig. 7 A.
Fig. 7.
(A) Bar graph of absolute protein quantitation in pmol/cell pellet for all three proteins quantified in 48-hour, 50 µg samples across the six passages used for experiments. (B) Normalization factors were calculated for actin and GAPDH and applied to spike quantitation. The amount of actin quantified in each passage was averaged and that average was used to calculate the normalization factor. The amount of actin quantified in each passage was then divided by the average to provide the normalization factor for that passage. Each passage had its own normalization factor. That amount of spike quantified in that passage was divided by the normalization factor of that passage. The same process was done for GAPDH to account for varying number of cells per well. Both housekeeping proteins provided equivalent correction indicating that either protein could be used as a housekeeping protein to quantify protein expression from BHK cells.
To consider the varying amounts of cells that might be present in the sample well, a normalization factor was calculated for both actin and GAPDH and applied to spike protein. These normalization factors and its application is shown in Fig. 7B. For these experiments, both actin and GAPDH provided equivalent corrections and the average amount of spike protein expressed with 50 µg of expired vaccine was 6.08 +/− 0.72 and 6.00 +/− 0.69 pmol, respectively. With the inclusion of the normalization factor, the precision of the measurement improved to a % RSD of less than 12. From these experiments with expired vaccine, the amount of mRNA produced approximately 1 pmol of spike protein per 6 pmol of vaccine. It is likely that the precision of the experiment would improve if sampling was conducted over a narrower range of cell passage and that the amount of spike protein expressed would increase if the cells were transfected with unexpired vaccine.
4. Conclusions
Isotope dilution mass spectrometry (IDMS) methods have been used to quantify the primary antigens of influenza vaccines, both seasonal and those of pandemic potential. These IDMS methods have been demonstrated on whole virus, split and subunit vaccines, and recombinant proteins. A similar method was developed to quantify spike and nucleocapsid proteins of SARS-CoV-2 [27]. While these methods are useful to quantify vaccine materials and reference materials for COVID-19 antigen diagnostic tests that contain these protein antigens, the emergence of mRNA and viral vector vaccines necessitated expansion of our protocol to include transfection of mammalian cells with such vaccine materials and subsequently use IDMS to quantify antigen expression. We have developed an accurate and precise IDMS method, confirmed by western blot analysis, for the quantification of spike protein expressed by transfecting BHK cells with an expired commercial COVID-19 mRNA vaccine. The IDMS method has advantages over western blot analysis because it directly quantifies the amount of spike protein based on reference peptides. Further evaluation of this method would include analysis of fresh, unexpired vaccine material. It would also be useful in developing prototype vaccines in which parameters can be changed and the effect of those changes observed in antigen expression. The amount of spike produced compared to the amount of vaccine added for transfection could be dependent on the composition of the vaccine and suitable antigen expression levels may need to be determined for each manufacturer. This method could be used to evaluate vaccine stability under various storage conditions and to help establish vaccine expiration dates. The inclusion of five target peptides for spike provides redundancy in the case an amino acid mutation occurs and renders a target peptide unusable with the emergence of a new variant of concern. Thus, IDMS is well-equipped to analyze the current, and likely any future, variants should the need arise to make a strain-specific vaccine.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to acknowledge Dr. Yuhong Xie of Seqirus, Inc. for helpful discussions regarding transfection of BHK cells.
We gratefully acknowledge the following Authors from the Originating laboratories responsible for obtaining specimens and the Submitting laboratories where genetic sequence data were generated and shared via GISAID Initiative, on which this research is based.
1. hCoV-19/Wuhan/IVDC-HB-01/2019 (reference); Accession No. EPI_ISL_402119; Collected 2019-12-30; Originating laboratory National Institute for Viral Disease Control and Prevention; Submitting laboratory Chinese Center for Disease Control and Prevention; Authors Wenjie Tan, Xiang Zhao, Wenling Wang, Xuejun Ma, Yongzhong Jiang, Roujian Lu, Ji Wang, Weimin Zhou, Peihua Niu, Peipei Liu, Faxian Zhan, Weifeng Shi, Baoying Huang, Jun Liu, Li Zhao, Yao Meng, Xiaozhou He, Fei Ye, Na Zhu Yang Li, Jing Chen, Wenbo Xu, George F. Gao, Guizhen Wu.
2. hCov-19/South Africa/KRISP-K006851/2020; (Beta); Accession No. EPI_ISL_825115; Collected 2020-11-24; Originating laboratory NHLS-IALCH; Submitting laboratory KRISP, KZN Research Innovation and Sequencing Platform; Authors Giandhari J, Pillay, S Lessels R, Mdlalose K, York D, Khan S, Tegally H, Wilkinson E, de Oliveira T.
3. hCoV-19/Brazil/SP-999/2021 (Gamma); Accession No. EPI_ISL_875688; Collected 2021-01-04; Originating laboratory Nation Influenza Center-Instituto Adolfo Lutz; Submitting laboratory Instituto Adolfo Lutz, Interdisciplinary Procedures Center, Strategic Laboratory; Authors Ana Lucia de Carvalho Avelino, Claudia Regina Goncalves, Claudio Tavares Sacchi, Erica Valessa Ramos Gomes, Karoline Rodrigues Campos, Katia Correa de Oliveira Santos.
4. hCoV-19/India/TG-CDFD-OMC11/2021 (Delta); Accession No. EPI_ISL_2924251; Collected 2021-05-24; Originating laboratory Osmania Medical College; Submitting laboratory CDFD; Authors Bashyam M, Gupta A, Basu R, Donipadi V, Kammili N, Vashisht D, Dalal A.
5. hCoV-19/Botswana/R99B82_BHP_AAC74225/2022 (Omicron); Accession No. EPI_ISL_10981810; Collected 2022-01-28; Originating laboratory Kasane Primary Hospital Laboratory; Submitting Laboratory Botswana Harvard HIV Reference Laboratory; Authors Sikhulile Moyo, Wonderful T. Choga, Dorcas Maruapula, Sefetogi Ramaologa, Thongbotho Mphoyakgosi, Boitumelo Zuze, Legodile Kooepile, Ontlametse T. Bareng, Pamela Smith-Lawrence, Kgomotso Moruisi, Shirley Johane, Roger Shapiro, Shahin Lockman, Joseph Makhema, Mphaphi B. Mbulawa, Mosepele Mosepele, Simani Gaseitsiwe.
References in this paper to any specific commercial products, process, service, manufacturer, or company do not constitute an endorsement or a recommendation by the U.S. Government or the Centers for Disease Control and Prevention. The findings and conclusions reported in this article are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
This project was supported in part by an appointment to the Research Participation Program at the Centers for Disease Control and Prevention, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC.
Data availability
Data will be made available on request.
References
- 1.Kyriakidis N.C., Lopez-Cortes A., Gonzalez E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. npj Vaccines. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4) doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J.W., Lagniton P.N.P., Liu Y.u., Xu R.-H. mRNA vaccines for COVID-19: what, why and how. Int J Biol Sci. 2021;17(6):1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., et al. Self-amplifying rna vaccines give equivalent protection against influenza to mrna vaccines but at much lower doses. Mol Ther. 2018;26(2):446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. [Google Scholar]
- 6.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mrna-based vaccines. Pharmaceutics. 2020;12(2):102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tavernier G., Andries O., Demeester J.o., Sanders N.N., De Smedt S.C., Rejman J. mRNA as gene therapeutic: how to control protein expression. J Control Release. 2011;150(3):238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Kowalska J., Wypijewska del Nogal A., Darzynkiewicz Z.M., Buck J., Nicola C., Kuhn A.N., et al. Synthesis, properties, and biological activity of boranophosphate analogs of the mRNA cap: versatile tools for manipulation of therapeutically relevant cap-dependent processes. Nucleic Acids Res. 2014;42(16):10245–10264. doi: 10.1093/nar/gku757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17(8):961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 10.Rahman M.M., Zhou N., Huang J. An overview on the development of mrna-based vaccines and their formulation strategies for improved antigen expression in vivo. Vaccines (Basel) 2021;9(3):244. doi: 10.3390/vaccines9030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trepotec Z., Lichtenegger E., Plank C., Aneja M.K., Rudolph C. Delivery of mrna therapeutics for the treatment of hepatic diseases. Mol Ther. 2019;27(4):794–802. doi: 10.1016/j.ymthe.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., et al. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4(2):43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Xiao P., Lei S., Deng F., Xiao G.G., Liu Y., et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin (Shanghai) 2008;40(5):426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- 14.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson L., Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18(3-4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 16.Sanyal G., Sarnefalt A., Kumar A. Considerations for bioanalytical characterization and batch release of COVID-19 vaccines. npj Vaccines. 2021;6:53. doi: 10.1038/s41541-021-00317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mrna vaccine stability. J Pharm Sci. 2021;110(3):997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389(4):1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 19.Kolker E., Higdon R., Hogan J.M. Protein identification and expression analysis using mass spectrometry. Trends Microbiol. 2006;14(5):229–235. doi: 10.1016/j.tim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Williams T.L., Callahan J.H., Monday S.R., Feng P.C.H., Musser S.M. Relative quantitation of intact proteins of bacterial cell extracts using coextracted proteins as internal standards. Anal Chem. 2004;76(4):1002–1007. doi: 10.1021/ac034820g. [DOI] [PubMed] [Google Scholar]
- 21.Alonso Villela S.M., Kraïem H., Bouhaouala‐Zahar B., Bideaux C., Aceves Lara C.A., Fillaudeau L. A protocol for recombinant protein quantification by densitometry. Microbiologyopen. 2020;9(6):1175–1182. doi: 10.1002/mbo3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight M.I., Chambers P.J. Problems associated with determining protein concentration: a comparison of techniques for protein estimations. Mol Biotechnol. 2003;23:19–28. doi: 10.1385/MB:23:1:19. [DOI] [PubMed] [Google Scholar]
- 23.Taylor S.C., Posch A. The design of a quantitative western blot experiment. Biomed Res Int. 2014;2014 doi: 10.1155/2014/361590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai-Kastoori L., Schutz-Geschwender A.R., Harford J.A. A systematic approach to quantitative Western blot analysis. Anal Biochem. 2020;593:113608. doi: 10.1016/j.ab.2020.113608. [DOI] [PubMed] [Google Scholar]
- 25.Mann M. Can proteomics retire the western blot? J Proteome Res. 2008;7(8):3065. doi: 10.1021/pr800463v. [DOI] [PubMed] [Google Scholar]
- 26.Tighe P.J., Ryder R.R., Todd I., Fairclough L.C. ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 2015;9(3-4):406–422. doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce-Ruiz C., Santana W.I., Sutton W.J.H., Fischler D.A., Cooper H.C., Marc L.R., et al. Quantification of SARS-CoV-2 spike and nucleocapsid proteins using isotope dilution tandem mass spectrometry. Vaccine. 2021;39(36):5106–5115. doi: 10.1016/j.vaccine.2021.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini R.K., Prasad P., Shang X., Keum Y.-S. Advances in lipid extraction methods-a review. Int J Mol Sci. 2021;22(24):13643. doi: 10.3390/ijms222413643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y.-Q., Gilar M., Lee P.J., Bouvier E.S.P., Gebler J.C. Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem. 2003;75(21):6023–6028. doi: 10.1021/ac0346196. [DOI] [PubMed] [Google Scholar]
- 30.MacLean B., Tomazela D.M., Abbatiello S.E., Zhang S., Whiteaker J.R., Paulovich A.G., et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 2010;82(24):10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khare S., et al. Gisaid's role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson R.E., Carroll H.P., Harris A., Maher E.R., Selby P.J., Banks R.E. Housekeeping proteins: A preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics. 2005;5(2):566–571. doi: 10.1002/pmic.200400941. [DOI] [PubMed] [Google Scholar]
- 35.Li R., Shen Y. An old method facing a new challenge: re-visiting housekeeping proteins as internal reference control for neuroscience research. Life Sci. 2013;92(13):747–751. doi: 10.1016/j.lfs.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y., Wu M., He G., Zhang X., Li W., Gao Y., et al. Glyceraldehyde-3-phosphate dehydrogenase: a universal internal control for Western blots in prokaryotic and eukaryotic cells. Anal Biochem. 2012;423(1):15–22. doi: 10.1016/j.ab.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Tubaon R.M., Haddad P.R., Quirino J.P. Sample clean-up strategies for esi mass spectrometry applications in bottom-up proteomics: trends from 2012 to 2016. Proteomics. 2017;17(20):1700011. doi: 10.1002/pmic.201700011. [DOI] [PubMed] [Google Scholar]
- 38.Erde J., Loo R.R.O., Loo J.A. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13(4):1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern K., Xie Y., Palladino G., Barr J.R., Settembre E.C., Williams T.L., et al. Reference antigen-free and antibody-free LTD-IDMS assay for influenza H7N9 vaccine in vitro potency determination. Vaccine. 2018;36(41):6144–6151. doi: 10.1016/j.vaccine.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper H.C., Xie Y., Palladino G., Barr J.R., Settembre E.C., Wen Y., et al. Limited tryptic digestion-isotope dilution mass spectrometry (ltd-idms): a reagent-free analytical assay to quantify hemagglutinin of a(H5N1) vaccine material. Anal Chem. 2020;92(17):11879–11887. doi: 10.1021/acs.analchem.0c02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norrgran J., Williams T.L., Woolfitt A.R., Solano M.I., Pirkle J.L., Barr J.R. Optimization of digestion parameters for protein quantification. Anal Biochem. 2009;393(1):48–55. doi: 10.1016/j.ab.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 42.Rajaratinam H., et al. Passage number of 4t1 cells influences the development of tumour and the progression of metastasis in 4t1 orthotopic mice. Malays J Med Sci. 2022;29:30–42. doi: 10.21315/mjms2022.29.3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao H., He H., Chen Y., Zeng F., Huang J., Wu L.i., et al. Effects of long-term serial cell passaging on cell spreading, migration, and cell-surface ultrastructures of cultured vascular endothelial cells. Cytotechnology. 2014;66(2):229–238. doi: 10.1007/s10616-013-9560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park M.T., Lee M.S., Kim S.H., Jo E.C., Lee G.M. Influence of culture passages on growth kinetics and adenovirus vector production for gene therapy in monolayer and suspension cultures of HEK 293 cells. Appl Microbiol Biotechnol. 2004;65:553–558. doi: 10.1007/s00253-004-1617-3. [DOI] [PubMed] [Google Scholar]
- 45.Peiser C., Riebe-Imre M., Emura M., Mohr U. Influence of culture passages on growth kinetics, xenobiotic metabolism, chromosomal stability and transformation in a clonal fetal hamster lung epithelial cell line. Mutat Res. 1993;289(2):281–290. doi: 10.1016/0027-5107(93)90079-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.