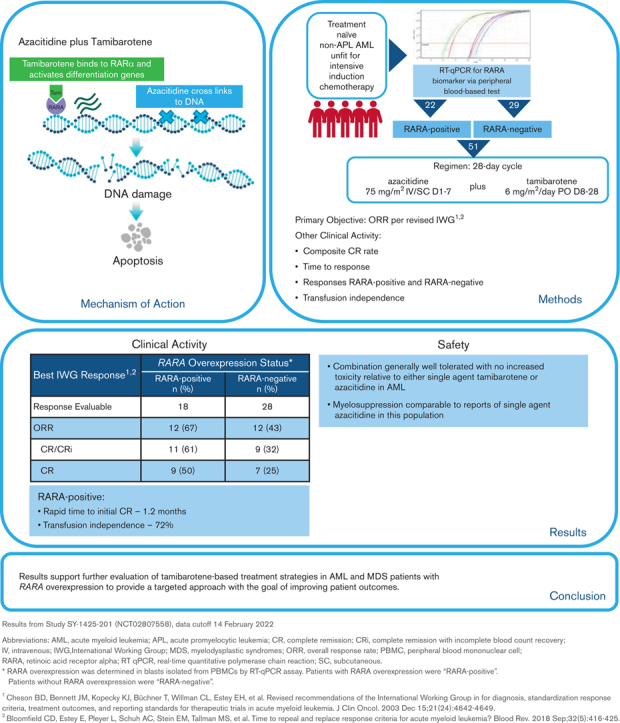
Key Points
-
•
Tamibarotene plus azacitidine was associated with a high CR rate and a rapid onset of response in RARA-positive newly diagnosed unfit AML.
-
•
Tamibarotene-based treatment in AML with RARA overexpression is a novel-targeted approach with potential to improve current therapy.
Visual Abstract
Abstract
A superenhancer at the retinoic acid receptor alpha (RARA) gene is associated with RARA mRNA overexpression in ∼30% of non-acute promyelocytic leukemia acute myeloid leukemia (AML) and in ∼50% of myelodysplastic syndromes (MDS). RARA overexpression is an actionable target for treatment with tamibarotene, an oral potent and selective RARα agonist. Sensitivity to the RARα agonist tamibarotene was demonstrated in RARA-high but not RARA-low preclinical AML models. The combination of oral tamibarotene plus azacitidine was evaluated in a phase 2 clinical study in 51 newly diagnosed unfit patients with AML identified as RARA-positive (n = 22) or RARA-negative (n = 29) for RARA mRNA overexpression in peripheral blasts using a blood-based biomarker test. In 18 response-evaluable RARA-positive patients, complete remission (CR)/CR with incomplete hematologic recovery rate was 61%, CR rate was 50%, and time to initial composite CR was rapid at 1.2 months. Transfusion independence was attained by 72% of RARA-positive patients. In contrast, 28 response-evaluable RARA-negative patients had response rates that were consistent with azacitidine monotherapy. Tamibarotene in combination with azacitidine was well tolerated. The majority of nonhematologic adverse events were low grade and hematologic adverse events were comparable to single-agent azacitidine, demonstrating that there was no additional myelosuppression when tamibarotene was combined with azacitidine. These results support further evaluation of tamibarotene-based treatment strategies in patients with AML or MDS with RARA overexpression to provide a targeted approach with the goal of improving patient outcomes. This trial was registered at www.clinicaltrials.gov as #NCT02807558.
Introduction
Acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) are biologically heterogenous diseases driven by genetic, epigenetic, and functional pathways that may serve as the target for therapeutic interventions. To date, the development of new therapies in AML and MDS has focused primarily on identification and targeting of somatic genetic mutations, cell-surface antigens, and antiapoptotic pathways.1 More recently, highly active chromatin regions, named superenhancers (SEs) have been described. SEs are large stretches of DNA that regulate the expression of genes through the binding of transcription factors. SE mapping in samples from patients with primary nonacute promyelocytic leukemia (non-APL) AML or MDS, as well as preclinical models led to the identification of a SE associated with the retinoic acid receptor alpha (RARA) gene resulting in RARA messenger RNA (mRNA) overexpression. In cells overexpressing RARA, tamibarotene, a selective RARα agonist binds to RARα, saturating unliganded RARα receptors to trigger a transcriptional activation switch to restore myeloid differentiation, inhibit blast proliferation, and promote blast cell clearance in AML and MDS.2
Tamibarotene (formerly SY-1425) is a next-generation synthetic oral retinoid with improved pharmacological properties over first-generation pan-retinoids, such as ATRA.3,4 In in vitro studies, tamibarotene as a single agent demonstrated antiproliferative effects, induction of retinoic acid response genes, and differentiation effects in non-APL AML cell line models with a RARA SE expressing relatively high levels of RARA mRNA (RARA-high). In in vivo efficacy studies, single-agent tamibarotene demonstrated antitumor activity in RARA-high but not RARA-low AML patient-derived xenograft models.2 Further preclinical work demonstrated synergy of tamibarotene with azacitidine or decitabine in RARA-high cell lines, whereas there was no additive effect when combining tamibarotene and azacitidine or decitabine in RARA-low cell lines.5 Concordantly, tamibarotene plus azacitidine produced deeper, more durable responses than either agent alone in a RARA-high AML patient-derived xenograft model.5 Using RARA mRNA as a biomarker for a RARA SE, a novel blood-based biomarker test was developed to identify patients with RARA mRNA overexpression, classified as RARA-positive, who could benefit from tamibarotene treatment.6
The clinical investigation of the relationship between the RARA biomarker and the response to tamibarotene was explored in a phase 2 study evaluating tamibarotene in AML and MDS (study SY-1425-201).7 Early cohorts of the study investigated tamibarotene monotherapy in RARA-positive low-risk MDS, relapsed/refractory (R/R) AML, and R/R higher-risk MDS (HR-MDS).8 We previously reported that, as a monotherapy, in these cohorts, tamibarotene was associated with clinical activity, as demonstrated by myeloid differentiation (supplemental Figure 1), hematologic improvement, and reduced bone marrow blasts.8 A marrow complete remission (CR) plus hematologic improvement with both a platelet and neutrophil response was observed in a patient with R/R HR-MDS.8 The evaluation of tamibarotene monotherapy was followed by evaluation of tamibarotene plus azacitidine in newly diagnosed (ND) unfit patients with AML in this same phase 2 study.9, 10, 11
Herein, we report the mature results from this cohort describing the clinical activity and tolerability of the tamibarotene/azacitidine combination in ND patients with AML with RARA overexpression deemed unfit to receive standard cytotoxic chemotherapy. We describe the development of a novel blood-based clinical assay used prospectively to identify RARA-positive patients who are most likely to respond to treatment, and report correlative analyses of patient samples from this study, identifying the association of RARA-positive AML with a monocytic gene expression phenotype.
Methods
Clinical study methods
Between 6 September 2017 and 13 November 2019, 51 patients at 12 sites in the United States and France were enrolled into the ND AML cohort of this study. Male and female patients aged ≥18 years were eligible for participation if they had ND, treatment-naive, non-APL AML and, at the time of study entry, were unlikely to tolerate standard intensive chemotherapy owing to age, performance status, or comorbidities.7,12
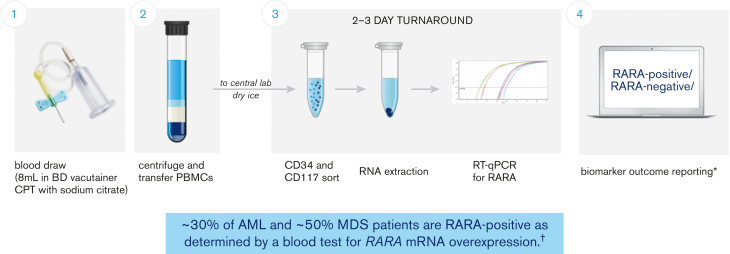
A Food and Drug Administration Investigational Device Exemption–approved blood-based RARA biomarker test was developed for patient screening before treatment with tamibarotene. Patients were evaluated for the RARA biomarker from a peripheral blood sample collected and processed into peripheral blood mononuclear cells at the clinical site and subsequently sent to a central diagnostic laboratory. The test measures, via quantitative reverse transcription polymerase chain reaction, the relative RARA mRNA expression levels against a panel of control genes with stable expression levels in CD34+ and/or CD117+ blasts isolated from peripheral blood mononuclear cells and applies a predefined cutoff to determine whether any given patient sample is RARA-positive (patients with RARA mRNA overexpression) or RARA-negative (patients without RARA mRNA overexpression) (Figure 1; refer to supplement for detailed description of the blood-based RARA biomarker test).6 Both RARA-positive and RARA-negative patients were enrolled.
Figure 1.
Patients with RARA overexpression identified with peripheral blood–based clinical trial assay. The blood-based biomarker test (clinical trial assay) was performed at a central laboratory, using frozen peripheral blood mononuclear cells (PBMCs) prepared and shipped from the clinical sites. The assay measures relative RARA mRNA expression levels against a panel of control genes via quantitative reverse transcription polymerase chain reaction (RT-qPCR) in CD34+ and/or CD117+ blasts isolated from PBMCs and applies a predefined cutoff to determine whether any given patient sample is RARA-positive or RARA-negative (patients with RARA overexpression were characterized as RARA-positive and patients without RARA overexpression as RARA-negative). ∗Details of the sample collection, sample analysis, and biomarker outcome reporting process as described in Vigil et al, 2017;6 †Syros Pharmaceuticals, Inc data on file as of 27 May 2022 from studies SY-1425-201 (all cohorts) and SELECT-MDS-1 (#NCT04797780). CPT, cell preparation tube.
Each 28-day treatment cycle included azacitidine at 75 mg/m2 intravenously or subcutaneously daily on days 1 through 7 followed by oral tamibarotene at 6 mg/m2 per day in divided doses twice daily on days 8 through 28.
The primary objective was characterization of activity by the overall response rate (ORR) per the revised International Working Group AML criteria.13,14 Additional clinical activity end points included composite CR rate (CR or CR with incomplete hematologic recovery [CRi] or CR with partial hematologic recovery), time to response, analysis of responses among RARA-positive and RARA-negative cohorts, and transfusion independence. Safety and tolerability were evaluated using Common Terminology Criteria for Adverse Events version 4.03. The mutation profiles and cytogenetic risk of patients were site reported and the association of molecular and cytogenetic characteristics with response were summarized. RNA sequencing (RNA-seq) was performed on AML blasts from enrolled patients to evaluate whether RARA-positive ND unfit patients with AML with RARA overexpression were enriched for the monocytic phenotype associated with venetoclax resistance.
Statistical methods
The safety population includes all patients who received any dose of tamibarotene or azacitidine. The safety analysis set was used to summarize demographics, baseline characteristics, disposition, and analyses of safety and efficacy data, unless otherwise specified. The response-evaluable population includes all patients in the safety population who completed at least 1 cycle of treatment with at least 1 postbaseline response evaluation or discontinued the study before completion of cycle 1 because of documented disease progression. The response-evaluable population was used to summarize selected response data, as specified in “Results.” SAS (version 9.4) was used to support the analyses. The data cutoff was 14 February 2022.
Overall survival (OS) was summarized for all patients using Kaplan-Meier estimates. CR rates, composite CR rates, and ORRs were summarized in the response-evaluable population.
Prognostic significance of RARA biomarker
RNA-seq data sets from The Cancer Genome Atlas (TCGA)34 and Beat AML17 were evaluated to describe the association of RARA expression with survival and response to azacitidine monotherapy in patients with AML (refer to supplement for detailed description).
Survival of the RARA-high and RARA-low populations was estimated using Kaplan-Meier methods and their survival was compared using a log-rank test. In addition, patient treatment data from the Beat AML data set were used to compare the CR rate with azacitidine monotherapy in RARA-high and RARA-low patients using Fisher exact test. RARA expression was determined using RNA-seq from patient samples (as described in supplement).
Monocytic gene expression signature development and analysis
To evaluate the relationships between RARA expression, monocytic AML features, and venetoclax resistance in patients in our clinical study, a monocytic gene expression signature was developed to estimate the monocytic status of patient AML blasts collected at study entry. The TCGA LAML RNA-seq data set was used to develop a signature through regularized regression of the expression of 9 well-established monocytic and primitive genes (CD14, CLEC7A, CD86, LYZ, MAFB, CD34, ITGAM, FCGR1A, and KIT) onto French-American-British classification (FAB) across 130 patients with non-APL AML with known FAB status. The resulting monocytic expression score (MES) was then applied to the RNA-seq data sets from ND unfit patients with AML treated with tamibarotene plus azacitidine in the ongoing phase 2 study (refer to supplement for detailed description).
Study oversight
Syros Pharmaceuticals, Inc provided financial support for the study and participated in the design, study conduct, analysis, and interpretation of the data. All authors had full access to the data, signed confidentiality agreements with the sponsor regarding the data, and vouch for the completeness and accuracy of the data and analyses.
The protocol was approved by the applicable institutional review boards, and all participants provided informed consent before any study procedures. This trial was registered at www.clinicaltrials.gov as #NCT02807558 and was conducted in accordance with the Declaration of Helsinki.7
Results
Patient characteristics
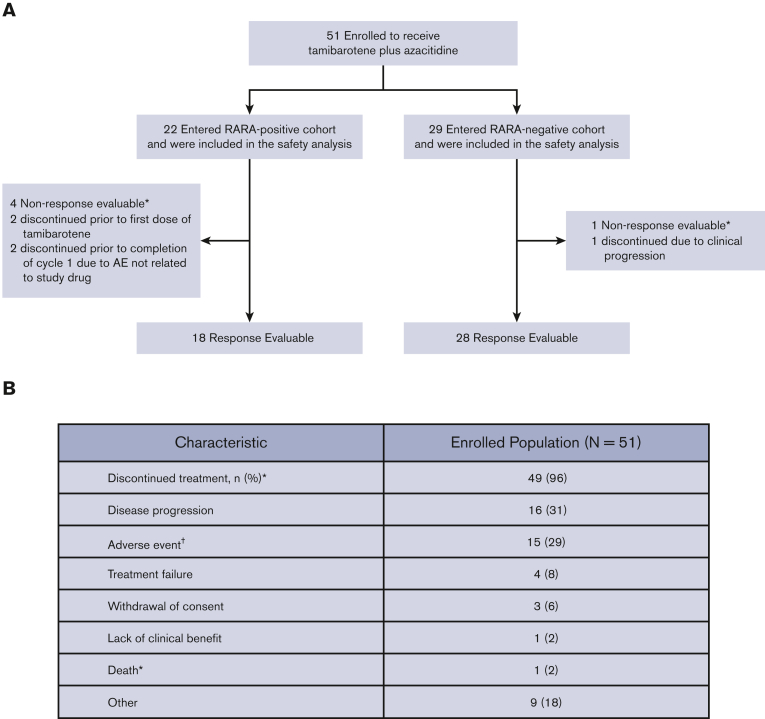
Of the 125 ND unfit patients with AML screened using the blood-based biomarker test, 37 (30%) were RARA-positive and in 88 (70%) were RARA-negative. After the completion of screening procedures, 51 patients enrolled in the study to receive tamibarotene in combination were azacitidine; 22 were RARA-positive, and 29 were RARA-negative (Figure 2). Baseline demographics and disease characteristics are presented in Table 1. The median patient age was 76 years, and 63% of patients were male. Enrolled patients had either intermediate- (67%) or poor-risk (31%) cytogenetics (Table 1) per National Comprehensive Cancer Network Clinical Practice Guidelines for AML.15 Secondary AML was reported in 43% of patients, and 35% of patients had low–blast count AML with a baseline bone marrow blast count of ≤30% (Table 1).
Figure 2.
Patient disposition, enrollment, and treatment. (A) Patient enrollment and analysis overview. Of the 125 ND unfit patients with AML screened using the blood-based biomarker test, 37 (30%) were RARA-positive and 88 (70%) were RARA-negative. The most frequent reasons for which screened ND unfit patients with AML were not enrolled included RARA-negative status before implementation of a protocol amendment allowing both RARA-positive and RARA-negative patients to enroll, and patients declined. Of the 51 patients with non-APL AML enrolled to receive tamibarotene and azacitidine, all were included in safety and efficacy analyses. The response-evaluable population comprised all patients enrolled who (1) completed 1 cycle of tamibarotene and had a follow-up assessment of disease status or (2) were withdrawn from the study before completion of cycle 1 because of documented disease progression. Figure includes patient status as of data cutoff, 14 February 2022. ∗No postbaseline response evaluation was performed for nonevaluable patients. (B) Patient disposition. ∗One patient died during treatment due to cardiac arrest that was not drug related; †Includes 2 patients who discontinued treatment before the first dose of tamibarotene. Of the 15 patients who discontinued because of AE, 3 patient discontinuations were assessed as related to study treatment; 1 was due to fatigue; 1 was due to fatigue, myalgia, arthralgia, and nausea, and 1 was due to pulmonary embolism. There were no hematologic AEs considered related to study treatment that led to treatment discontinuation.
Table 1.
Baseline demographic and clinical characteristics of patients
| Characteristic |
RARA overexpression status∗ |
||
|---|---|---|---|
| RARA-positive (n = 22) | RARA-negative (n = 29) | Total (N = 51) | |
| Age | |||
| Median (range), y | 77 (60-91) | 76 (64-86) | 76 (60-91) |
| ≥75 y, n (%) | 16 (73) | 15 (52) | 31 (61) |
| Male sex, n (%) | 13 (59) | 19 (66) | 32 (63) |
| AML type, n (%)† | |||
| De novo | 16 (73) | 13 (45) | 29 (57) |
| Secondary | 6 (27) | 16 (55) | 22 (43) |
| Evolved from antecedent hematologic malignancy | 6 (27) | 13 (45) | 19 (37) |
| Associated with treatment from prior malignancy | 0 (0) | 3 (10) | 3 (6) |
| AML cytogenetic risk category, n (%)‡ | |||
| Intermediate | 16 (73) | 18 (62) | 34 (67) |
| Poor | 6 (27) | 10 (34) | 16 (31) |
| Not determined | 0 (0) | 1 (4) | 1 (2) |
| Hypomethylating agent prior cancer treatment, n (%) | |||
| Any hypomethylating agent | 2 (9) | 6 (21) | 8 (16) |
| Azacitidine | 2 (9) | 3 (10) | 5 (10) |
| Decitabine | 0 (0) | 3 (10) | 3 (6) |
| Baseline bone marrow blasts, n (%) | |||
| ≤30% (low–blast count AML) | 7 (32) | 11 (38) | 18 (35) |
| >30% | 15 (68) | 18 (62) | 33 (65) |
| Eastern Cooperative Oncology Group performance status score, n (%)§ | |||
| 0-1 | 12 (55) | 18 (62) | 30 (59) |
| 2-3 | 8 (36) | 10 (34) | 18 (35) |
| Missing | 2 (9) | 1 (4) | 3 (6) |
Data are presented as number (%) unless indicated otherwise.
RARA overexpression was determined in blasts isolated from PBMCs by qRT-PCR assay. The presence of RARA overexpression was characterized as RARA-positive, and the absence of RARA overexpression as RARA-negative.
AML denotes non-APL AML.
Cytogenetic risk was assessed per NCCN AML guidelines 2018.15
Eastern Cooperative Oncology Group performance status scores range from 0 to 5, with 0 indicating no symptoms and higher scores indicating greater disability.
Patient disposition
All 51 enrolled patients received at least 1 dose of azacitidine, 49 received at least 1 dose of tamibarotene, and 2 discontinued before first dose of tamibarotene (Figure 2). Patients received a median of 4.0 months of study treatment (range, 0.1-36.8 months). The RARA-positive and RARA-negative cohorts included 18 and 28 response-evaluable patients, respectively. Of the 51 enrolled patients, 96% discontinued study treatment, for whom the most common reasons included progressive disease (31%) and adverse event (AE) (29%) (Figure 2). Of the 15 patients who discontinued owing to AE, 3 patient discontinuations were assessed as related to study treatment; 1 due to fatigue; 1 due to fatigue, myalgia, arthralgia, and nausea; and 1 due to pulmonary embolism (Figure 2). There were no hematologic AEs that were considered to be related to the study treatment by the investigator that led to discontinuation.
Clinical activity
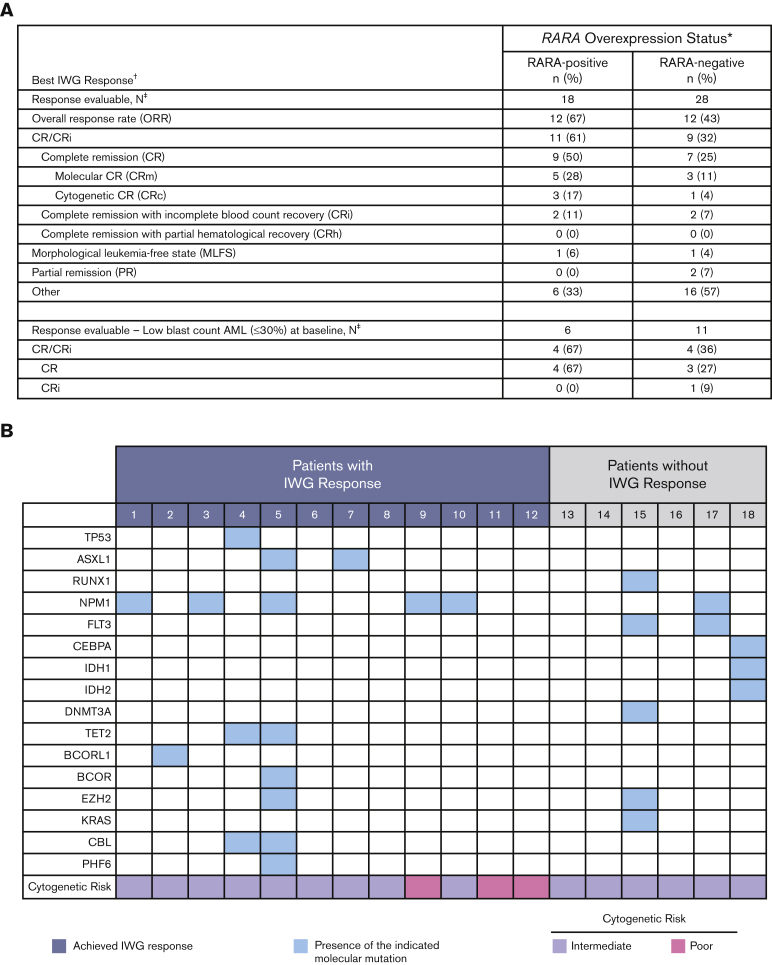
The 18 response-evaluable RARA-positive patients were evaluated for best OR (response definitions and results in Figure 3). The ORR was 67% (12/18), with a CR/CRi rate of 61% (9 CR, 2 CRi), a CR rate of 50%, and morphologic leukemia-free state achieved by 1 patient. Of the 9 patients with CR, 89% (8/9) had either a molecular CR (5/9) or cytogenetic CR (3/9). In those with low–blast count AML with a blast percentage of ≤30%, CR was observed in 67% (4/6) of RARA-positive patients.
Figure 3.
Summary of OR. (A) Summary of best OR in ND unfit patients with AML. Table shows a summary of the best efficacy response achieved by all response-evaluable patients. The response-evaluable population comprised all patients enrolled who (1) completed 1 cycle of tamibarotene and had a follow-up assessment of disease status or (2) were withdrawn from the study before completion of cycle 1 because of documented disease progression. Patients listed in the “other” category did not achieve an International Working Group (IWG) response. ∗RARA overexpression was determined in blasts isolated from PBMCs by qRT-PCR assay. The presence of RARA overexpression was characterized as RARA-positive, and the absence of RARA overexpression as RARA-negative; †Disease status was assessed per the revised IWG AML criteria;13,14 ‡All response-evaluable patients. (B) Association of IWG response with DNA mutations and cytogenetic risk in RARA-positive patients. Data are shown for the 18 RARA-positive response evaluable patients. Cytogenetic risk was assessed per National Comprehensive Cancer Network (NCCN) AML guidelines 2018.15 The mutation profiles and cytogenetic risk of patients were site reported. Response was assessed per the revised IWG AML criteria.13,14
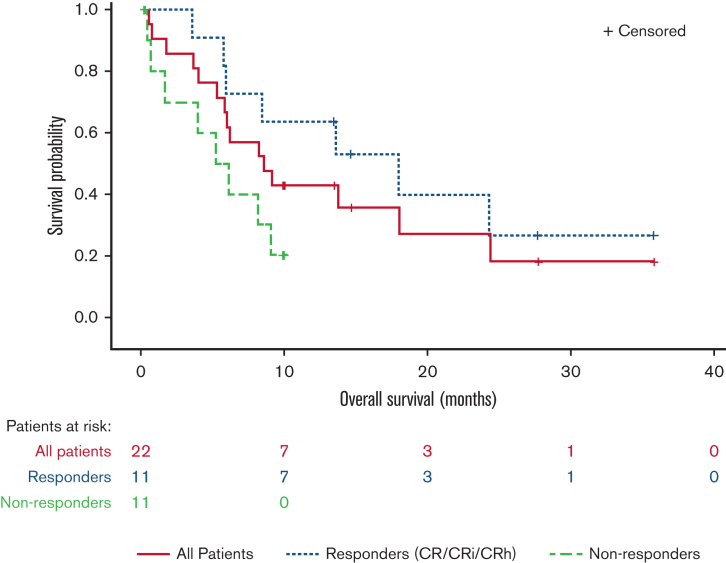
Median time to initial composite CR for RARA-positive patients was 1.2 months and the median duration of composite CR was 10.8 months (95% confidence interval [CI]: 2.9, NE). Responses were observed irrespective of site-reported mutation profile or cytogenetic risk (Figure 3). For all enrolled RARA-positive patients (n = 22), the median OS was 8.4 months (95% CI: 5.2, 15.6). For RARA-positive patients achieving CR/CRi (n = 11) the median OS was 18.0 months (95% CI: 5.7, NE) and 5.6 months (95% CI: 0.4, 9.0) for those not achieving CR/CRi (n = 11) (Figure 4).
Figure 4.
OS in RARA-positive patients summarized by response status. The OS graph includes all RARA-positive patients who enrolled in the study. Responders (CR/CRi/CRh), patients who achieved CR, CRi, or CR with partial hematologic recovery (CRh). Nonresponders, patients who did not achieve CR/CRi or CRh.
Of the 18 RARA-positive patients who were on treatment for ≥56 days and who were evaluable for transfusion independence, defined as an 8-week period on treatment without needing transfusion, red blood cell transfusion independence was achieved or maintained by 13 patients (72%), platelet transfusion independence by 16 patients (89%), and both by 13 patients (72%).
For the 28 response-evaluable RARA-negative patients, the ORR was 43% (12/28), the CR/CRi rate was 32% (7 CR, 2 CRi), the CR rate was 25%, 1 patient achieved morphologic leukemia-free state, and 2 had partial remission. In 11 RARA-negative patients and low–blast count AML the CR rate was 27%. For RARA-negative patients with CR/CRi (n = 9), the median time to initial composite CR was 3.0 months, median duration of composite CR was 10.3 months (95% CI: 3.1, 32.3), and the median OS was 15.8 months (95% CI: 6.5, NE). Of the 23 RARA-negative patients who were evaluable for transfusion independence, red blood cell transfusion independence was achieved or maintained by 13 patients (57%), platelet transfusion independence by 19 patients (83%), and both by 13 patients (57%).
Safety and AEs
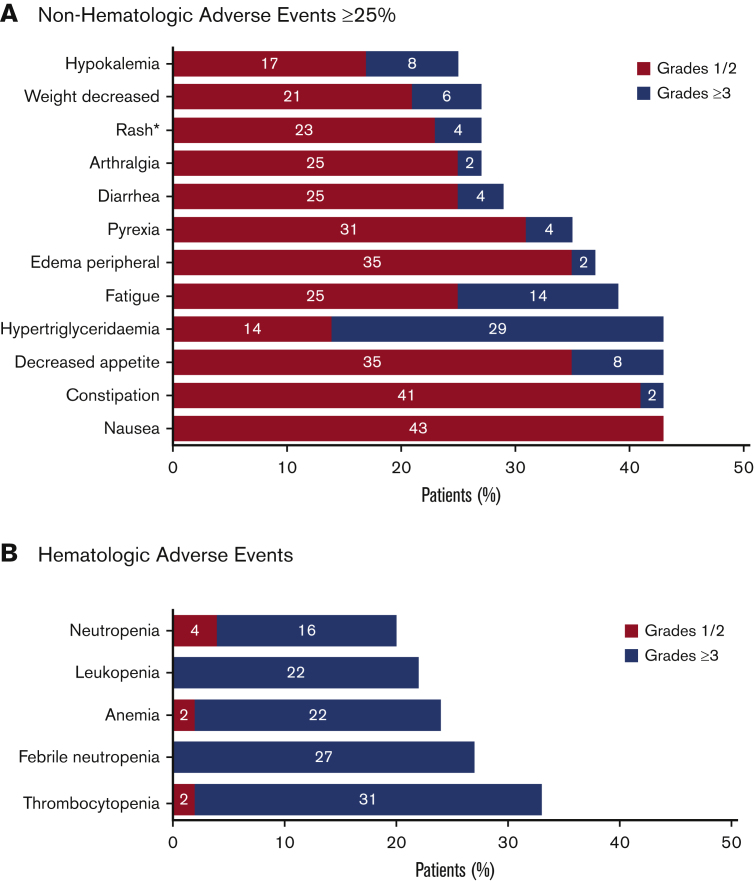
The safety analysis of the data cut for this ongoing study included all 51 enrolled patients (Figure 2). All 51 enrolled patients had at least 1 AE. Common AEs are summarized in Figure 5. The most common hematologic AEs across all grades and causalities included thrombocytopenia (33%), febrile neutropenia (27%), and anemia (24%). The most frequently reported nonhematologic AEs across all grades and causalities included hypertriglyceridemia (43%) and gastrointestinal AEs, including constipation (43%), nausea (43%), and decreased appetite (43%). The majority of nonhematologic AEs were low grade, except for hypertriglyceridemia of grade ≥3 in 29% of patients. Serious AEs of any grade were reported for 43 (84%) patients, of which the most frequent included febrile neutropenia (24%), pneumonia (14%), pyrexia (10%), and sepsis (10%).
Figure 5.
Summary of AEs. All treatment emergent AEs for all enrolled patients (N = 51) were evaluated. The safety population included all patients who received at least 1 dose of study drug (tamibarotene or azacitidine). AEs were evaluated using Common Terminology Criteria for Adverse Events version 4.03. (A) Nonhematologic AEs that were reported in at least 25% of patients. *The term “rash” included the preferred terms of rash maculo-papular, rash, drug eruption, nodular rash, rash erythematous, and rash pruritic. Rash maculopapular and rash were each reported in 5 (10%) of patients, with other terms reported in 1 patient each (2%). (B) Hematologic AEs.
Prognostic significance of RARA biomarker in AML
To evaluate whether the clinical activity in RARA-positive patients was because of an underlying association with favorable-risk features and RARA-positive status, we interrogated 2 independent public databases, TCGA database, and the Beat AML data set. These analyses identified no association between RARA-positive status and favorable risk. Analysis of the association of RARA expression with survival in the TCGA database showed survival was inferior for RARA-high patients (log-rank P = .031) (supplemental Figure 2A). Furthermore, in the Beat AML data set, RARA expression was not associated with survival (supplemental Figure 2B).
Finally, in the analysis of azacitidine monotherapy response in 54 patients in the Beat AML data set, similar rates of CR were seen in RARA-high and RARA-low patients (supplemental Figure 2C), supporting the hypothesis that RARA-high patients are not enriched for sensitivity to azacitidine monotherapy.
Gene expression features associated with RARA expression
We investigated the association of gene expression features in patients with AML with the RARA SE and with clinical response to tamibarotene. Preliminary analyses focused on expression of features initially identified as associated with the RARA SE by SE mapping, such as expression of monocytic markers.16
After transcriptome-wide normalization, an analysis of the TCGA AML data set by FAB demonstrated that RARA expression was highest in patients with monocytic AML (46% and 67% of patients with FAB M4 and FAB M5 subtypes, respectively, compared with between 0% and 21% in other non-APL AML subtypes; supplemental Figure 3). These results suggest an association between high RARA expression and monocytic features in AML.
RARA expression and MES from TCGA/beat AML data sets
To further elucidate the relationships between RARA expression and monocytic features in the phase 2 study, an MES was developed (supplemental Figure 4) as a surrogate biomarker for FAB status, to enable the prediction of a monocytic status of patient AML blasts collected at study entry (before treatment).
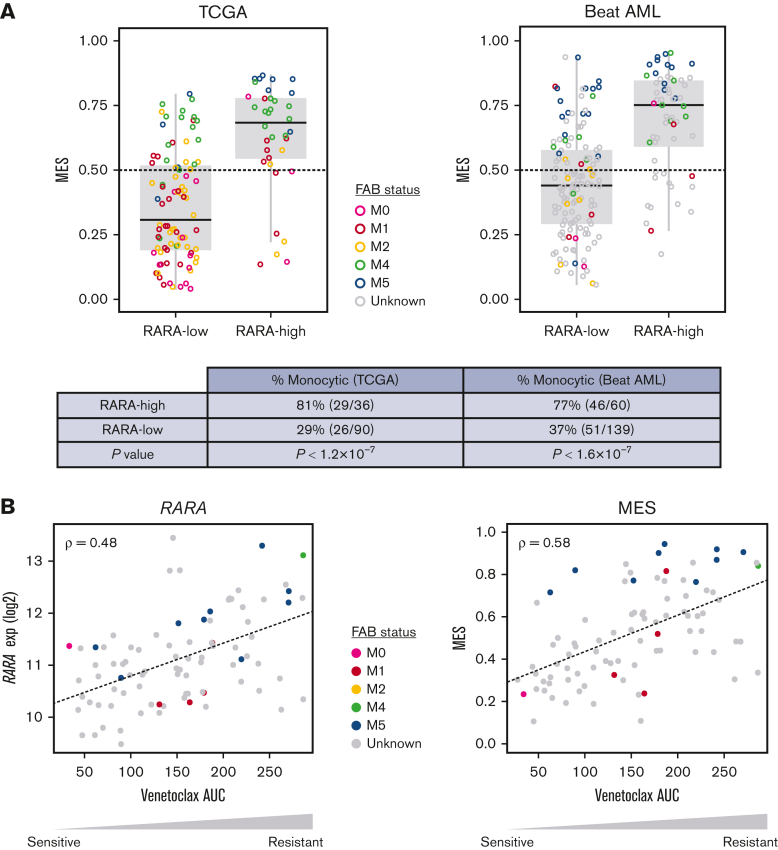
The analysis of RARA expression status in TCGA and Beat AML RNA-seq data sets was compared with monocytic status by MES and demonstrated that high RARA expression significantly enriches for a high MES, with 81% (TCGA) and 77% (Beat AML) of RARA-high patients classed as monocytic compared with 29% (TCGA) and 37% (Beat AML) of RARA-low patients (Figure 6). In summary, these analyses demonstrate that RARA-high AML is enriched for monocytic features.
Figure 6.
Association among high RARA expression, monocytic features, MES, and ex vivo venetoclax resistance. Colors denote FAB status, gray indicates unknown. (A) High RARA expression (RARA-high) identifies a population of patients with AML that is enriched for high monocytic gene expression (MES, y-axis) in the TCGA and Beat AML databases. P values by Fisher exact test. Monocytic: MES > 0.5. (B) RARA expression (left, y-axis) and MES (right, y-axis) are associated with venetoclax resistance ex vivo, quantified as area under the dose-response curve (AUC, x-axis). Plots show Spearman correlation (ρ) of normalized RARA expression or MES vs venetoclax response across 90 AML primary cultures (Beat AML).17 M0, undifferentiated acute myeloblastic leukemia; M1, acute myeloblastic leukemia with minimal maturation; M2, acute myeloblastic leukemia with maturation; M4, acute myelomonocytic leukemia; M5, acute monocytic leukemia.
Association of RARA expression with monocytic features and venetoclax resistance
Given the emerging reports of the association of monocytic AML with resistance to venetoclax ex vivo18, 19, 20 and venetoclax/azacitidine clinically,21 we evaluated the association of RARA expression, monocytic features, and venetoclax resistance.
First, we interrogated RARA expression levels in primary leukemic stem cell (LSC) samples from a cohort of 12 patients with AML treated with venetoclax ± azacitidine ex vivo, which were previously used to associate venetoclax resistance with monocytic AML features.20 RARA expression was significantly higher in venetoclax-resistant monocytic LSCs than venetoclax-sensitive primitive LSCs (P = .005) (supplemental Figure 5A). In a separate cohort of samples from patients with primary AML in the Beat AML data set, 121 inhibitors were evaluated ex vivo in primary cultures.17 Although the majority of the samples were sensitive to venetoclax, RARA-high samples were most resistant to venetoclax (supplemental Figure 5B). Furthermore, both RARA expression (ρ = .48) and MES (ρ = .58) are associated with venetoclax resistance in cultures from patients with primary AML from the Beat AML data set (Figure 6B). Together, these data suggest that high RARA expression is not only associated with monocytic features but also with venetoclax resistance in samples from patients with AML.
RARA positivity and MES in clinical samples from the ND unfit AML cohort
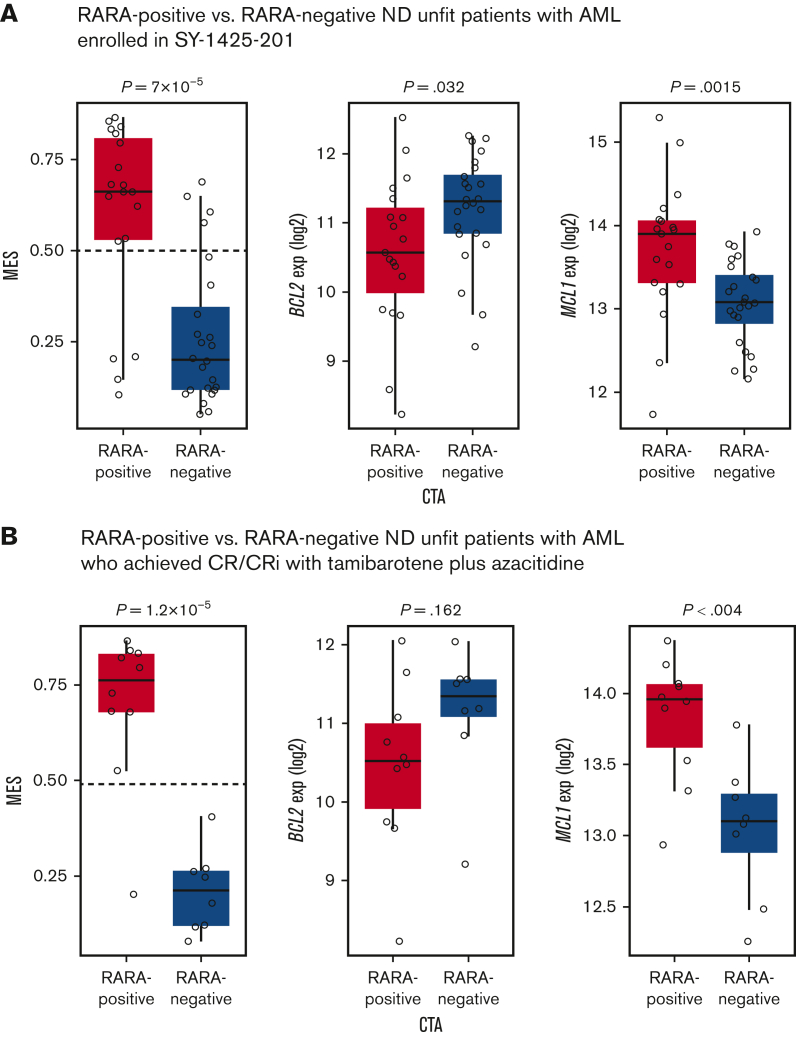
We followed these evaluations with exploratory analyses of pretreatment blood samples from patients enrolled in the ND unfit AML cohort in the SY-1425-201 study. RNA-seq was used to determine the MES and the expression of B-cell lymphoma 2 (BCL2) and myeloid-cell leukemia 1 (MCL1), 2 key genes associated with venetoclax sensitivity and resistance. Among 51 ND unfit patients with AML enrolled, 86% (19/22) of RARA-positive patients and 83% (24/29) of RARA-negative patients had blast RNA available and yielded RNA-seq results (Figure 7). Consistent with the results from the TCGA and Beat AML data sets, 80% (15/19) of RARA-positive patients were classified as monocytic by the MES (MES > 0.5) whereas only 17% (4/24) of RARA-negative patients were classified as monocytic by MES (MES > 0.5). In addition, significantly higher MCL1 expression (P = .0015) and lower BCL2 expression (P = .032) were observed in RARA-positive patients vs RARA-negative patients. Enrichment for high MES (P = 1.2 × 10−5) and high MCL1 (P < .004) was also observed in RARA-positive compared with RARA-negative patients who achieved CR/CRi (Figure 7). Collectively, these data demonstrate enrichment for a venetoclax-resistance pattern of high MES, higher MCL1, and lower BCL2 expression in RARA-positive patients.
Figure 7.
Association of RARA overexpression with monocytic features (MES) and venetoclax resistance markers in the SY-1425-201 clinical study ND unfit patients with AML. The MES and venetoclax resistance–associated features were profiled in ND unfit patients with AML. RARA-positive patients (red) were significantly enriched for features associated with venetoclax resistance including a high MES (left, y-axis), and low BCL2 (middle, y-axis) and high MCL1 expression (exp) (right, y-axis) compared with RARA-negative patients (blue). (A) Eighty percent (15/19) of RARA-positive patients and 17% (4/24) of RARA-negative patients are classified as monocytic by MES (MES > 0.5). (B) The majority of RARA-positive ND unfit patients with AML who achieved CR/CRi with tamibarotene plus azacitidine have a monocytic phenotype (high MES) associated with venetoclax resistance, which includes lower BCL2 and higher MCL1 expression.
Discussion
Evaluation of SE landscapes in samples from patients with primary non-APL AML identified a particularly strong SE associated with the RARA gene and concomitant RARA mRNA overexpression in a subset of patients with AML and MDS. This novel patient subset had a SE profile similar to mature monocytes and was not associated with mutations reported to confer responsiveness to azacitidine in AML or HR-MDS.2 The presence of the SE was associated with RARA mRNA overexpression and was predictive of response to tamibarotene, an oral potent and selective RARα agonist. A novel blood-based biomarker test was developed to identify RARA overexpression status (RARA-positive vs RARA-negative). Biomarker test results were significantly associated with differentiation of patient blood samples after ex vivo treatment with tamibarotene,6 supporting the use of the biomarker test for the identification of patients for treatment in this phase 2 study. In this study, the biomarker test successfully identified RARA-positive ND unfit patients with AML for response to the synergistic combination of tamibarotene plus azacitidine. Notably, the response rates in the RARA-negative cohort were comparable to response rates observed in historical studies evaluating azacitidine monotherapy in this patient population, demonstrating utility of the biomarker in selecting for patients with RARA overexpression who could benefit from this targeted therapy.21, 22, 23
A high CR rate, rapid onset of response, and a high rate of transfusion independence were observed in the RARA-positive ND unfit patients with AML treated with tamibarotene plus azacitidine with responses observed irrespective of cytogenetic risk groups or mutation profiles. In contrast, ORR and CR rate were lower and time to initial response was longer for RARA-negative patients relative to RARA-positive patients, consistent with azacitidine monotherapy trial outcomes and consistent with our preclinical findings demonstrating insensitivity to tamibarotene in combination with azacitidine with in vivo and in vitro RARA-low AML models.5 The duration of response of 10.8 months and median OS of 18.0 months for responders suggest that a clinically meaningful benefit was achieved for the high proportion of the RARA-positive cohort that responded. Responses were identified in RARA-positive patients across cytogenetic and mutational risk groups.24,25 This observation suggests that activity of tamibarotene is dependent on the biology of RARA overexpression and supports the potential importance of biological targeting in a complex disease, such as AML, which is associated with a multitude of mutational drivers.
We evaluated whether RARA pathway activation, as determined by increased RARA gene expression, was associated with more favorable disease (independent of tamibarotene treatment) that may explain the encouraging outcomes emerging during the course of our study. Analyses of 2 large, publicly available databases (TCGA and Beat AML) failed to demonstrate a correlation between RARA expression and an improvement in survival. In summary, these results suggest that AML with RARA overexpression is not a favorable prognostic subtype.
The tamibarotene plus azacitidine combination was generally well tolerated. The rates of myelosuppression were comparable to azacitidine monotherapy in this population suggesting no added hematologic toxicity from tamibarotene when used in combination with azacitidine.20,26 The majority of nonhematologic AEs, including rash, were low grade and generally consistent with AEs observed with other retinoids.27,28 Hypertriglyceridemia was the only frequently observed high-grade nonhematologic AE. Hypertriglyceridemia is a known side effect of retinoids such as tamibarotene and tends to be manageable and reversible with dose holds and/or reductions. Although increased white blood cells were observed during therapy in some patients, classical differentiation syndrome was not observed29,30 which is in contrast to the use of retinoids in APL31 or differentiating agents in isocitrate dehydrogenase mutant AML.32 Although limited by the small sample size and nonrandomized design, the clinical activity and generally well-tolerated AE profile of tamibarotene plus azacitidine in this study support further development of tamibarotene combinations in patients with RARA overexpression across the myeloid disease spectrum.
Despite several recent drug approvals for AML, the clinical need remains for more effective treatment options for patients with AML who cannot receive intensive chemotherapy. Venetoclax in combination with hypomethylating agents has emerged as a standard-of-care for treatment of this patient population, although challenges remain.21 Despite the high response rate and prolonged OS associated with the venetoclax/azacitidine combination, approximately one-third of patients do not respond, nearly all patients eventually relapse, and prolonged suppression of normal marrow function frequently occurs in vulnerable patients.16,33 Thus, understanding mechanisms of underlying resistance to venetoclax is critical. Several reports have recently shown that primary venetoclax resistance and inferior OS are associated with monocytic features in AML, which may be associated with reduced expression of the venetoclax target, BCL2, in monocytic AML.18, 19, 20 Given that RARA expression is highest in monocytic AML (M4, M5), we hypothesized that RARA overexpression may also be associated with venetoclax resistance. In patient samples from this phase 2 study, RARA-positive patients were significantly more likely to be classified as monocytic using a MES than RARA-negative patients, and with lower BCL2 and higher MCL1 expression than RARA-negative patients. These data suggest that RARA-positive patients with AML have monocytic gene expression features that have been associated with resistance to venetoclax-based therapies.20
The discovery of a SE that drives RARA pathway activation has identified a biologically defined subset of patients with AML and MDS with RARA mRNA overexpression who might be responsive to treatment with tamibarotene. Given that tamibarotene is oral and generally well tolerated, it is anticipated that tamibarotene may be combined with other therapies used to treat myeloid malignancies to provide a targeted foundation of treatment for RARA-positive patients. Tamibarotene-based therapy for RARA-positive malignancies is a novel approach that targets RARA pathway activation and is not limited to targeting a specific cytogenetic or molecular mutation.
The clinical development of tamibarotene is being advanced in combination with venetoclax/azacitidine in patients with AML with RARA overexpression and in a phase 3 study of tamibarotene/azacitidine in patients with HR-MDS with RARA overexpression. The targeting of RARα with tamibarotene is a novel approach in HR-MDS and is supported by the activity and tolerability of tamibarotene plus azacitidine in RARA-positive AML, particularly in low–blast count AML. Combining tamibarotene with venetoclax/azacitidine in AML may also provide benefit by targeting both BCL2 and RARA pathway activation in RARA-positive patients.
In summary, progress in myeloid malignancies will require novel targeted therapies in addition to broadly applicable treatments. Further development of tamibarotene may provide a new foundation for the treatment of patients with RARA overexpression across the spectrum of myeloid diseases.
Conflict-of-interest disclosure: S.d.B. received honoraria from AbbVie, Astellas, Bristol Myers Squibb (BMS), Jazz Pharmaceuticals, and Servier; had a consulting or advisory role at BMS, GlaxoSmithKline, Servier, and Syndak; is a member of the speakers bureau at AbbVie, Astellas, BMS, Jazz Pharmaceuticals, and Servier; received research funding from Auron and Forma Therapeutics; and received travel, accommodations, and expenses from AbbVie and Servier. T.C. is a member on the advisory boards at AbbVie, Agios, Celgene, Jazz Pharmaceuticals, Novartis, and Roche; performed clinical research for Alexion, Amgen, Arog, Celgene/BMS, Janssen, Kartos, Novartis, Servier, Syros Pharmaceuticals, and Takeda; received training from Amgen, Astellas, Celgene/BMS, Novartis, and Stemline/Menarini; and attended international congresses for Celgene/BMS, Novartis, and Pfizer. J.L.L. is a member on the advisory boards at BMS, Blueprint Sciences, and Pharmacosmos. J.G.J. provided consultancy for Celgene/BMS, Jazz Pharmaceuticals, Novartis, and Syros Pharmaceuticals and received research funding from AbbVie, Arog, Astellas, Celgene/BMS, Celularity, Forma Therapeutics, Gilead/Forty Seven, GlycoMimetics, PTC Therapeutics, and Syros Pharmaceuticals. M.A.S. is a member on the advisory boards at BMS, Kurome, Novartis, and Syros Pharmaceuticals. M.R.S. had an advisory and consulting role at AbbVie, BMS, CTI, Geron, Karyopharm, Novartis, Ryvu, Sierra Oncology, Taiho, Takeda, and TG Therapeutics; received research support from ALX Oncology, Astex, Incyte, Takeda, and TG Therapeutics; holds equity at Karyopharm and Ryvu; and is on the data safety monitoring boards at Celgene, Sierra Oncology, and TG Therapeutics. G.J.R. provided consultancy or is a member on the advisory boards or data and safety monitoring committee at AbbVie, Actinium, Agios, Amgen, Astellas, AstraZeneca, Blueprint Medicines, bluebird bio, BMS, Celgene, GlaxoSmithKline, Janssen, Jasper Therapeutics, Jazz Pharmaceuticals, MEI Pharma (IDMC Chair), Mesoblast, Novartis, Pfizer, Syndax, and Takeda (IRC Chair) and received research support from Janssen. E.M.S. is a member on the advisory boards at AbbVie, Agios, Aptose, Astellas, Blueprint, BMS, Calithera, CTI Biopharma, Daiichi, Foghorn, Genentech, Genesis, Gilead, Janssen, Jazz Pharmaceuticals, Neoleukin, Novartis, Menarini, OnCusp, Ono Pharma, PinotBio, Servier, Syros Pharmaceuticals, and Syndax; received honoraria from Kura; provided safety monitoring for Cellectis and Epizyme; received research funding from BMS and Eisai; and holds equity at Auron. A.V., K.S., K.B., S.P., J.C., G.H., M.K., and D.A. Roth are employees of Syros Pharmaceuticals, Inc. K.M., Q.K.-F., M.E., M.M., and C.F. are formed employees of Syros Pharmaceuticals, Inc. A.V., K.S., Q.K.-F., K.B., S.P., J.C., M.E., G.H., M.K., C.F., and D.A. Roth are shareholders at Syros Pharmaceuticals, Inc. The remaining authors declare no competing financial interests.
Acknowledgments
The authors acknowledge and thank all investigators, coinvestigators, study staff, patients, and families who contributed to this clinical study. The authors thank Calogera L. McCormick for providing medical writing support, funded by Syros Pharmaceuticals, Inc.
This study was supported by research funding from Syros Pharmaceuticals, Inc.
Authorship
Contribution: S.d.B., E.M.S., G.H., D.A. Roth, and K.S. designed the research; S.d.B., T.C., C.V., R.J.C., P.R., D. A. Rizzieri, J.L.L., P.F., T.B., A.B., J.G.J., M.A.S., M.R.S., G.J.R., D.B., Q.K.-F., M.M., J.C., M.E., C.F., and E.M.S. performed the research; S.d.B., K.M., A.V., Q.K.-F., K.B., S.P., M.M., J.C., M.E., G.H., C.F., M.J.K., D.A. Roth, and E.M.S. analyzed and interpreted data; S.P. performed statistical analysis; and M.J.K., A.V., Q.K.-F., M.E., and K.B. wrote the manuscript.
Footnotes
The Cancer Genome Atlas data are available through the The Cancer Genome Atlas data portal at https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga and Beat AML data can be accessed at www.vizome.org.
Data are available on request from the corresponding author, Stéphane de Botton (stephane.debotton@gustaveroussy.fr).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Kantarjian H, Kadia T, DiNardo C, et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11(2):41–66. doi: 10.1038/s41408-021-00425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKeown MR, Corces MR, Eaton ML, et al. Superenhancer analysis defines novel epigenomic subtypes of non-APL AML including an RARα dependency targetable by SY-1425, a potent and selective RARα agonist. Cancer Discov. 2017;7(10):1136–1153. doi: 10.1158/2159-8290.CD-17-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muindi J, Frankel SR, Miller WH, Jr., et al. Continuous treatment with all-trans retinoic acid causes a progressive reduction in plasma drug concentrations: implications for relapse and retinoid “resistance” in patients with acute promyelocytic leukemia. Blood. 1992;79(2):299–303. [PubMed] [Google Scholar]
- 4.Muindi J, Frankel SR, Huselton C, et al. Clinical pharmacology of oral all-trans retinoic acid in patients with acute promyelocytic leukemia. Cancer Res. 1992;52(8):2138–2142. [PubMed] [Google Scholar]
- 5.McKeown MR, Johannessen L, Lee E, Fiore C, di Tomaso E. Antitumor synergy with SY-1425, a selective RARα agonist, and hypomethylating agents in retinoic acid receptor pathway activated models of acute myeloid leukemia. Haematologica. 2019;104(4):e138–e142. doi: 10.3324/haematol.2018.192807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vigil CE, Jurcic J, Raza A, et al. 5 October 2017. RARA pathway activation biomarkers in Study SY-1425-201 define a new subset of AML and MDS patients and correlate with myeloid differentiation following ex vivo SY-1425 treatment. Abstract presented at: European School of Haematology 4th International Conference on Acute Myeloid Leukemia “Molecular and Translational”: Advances in Biology and Treatment. [Google Scholar]
- 7.A biomarker-directed phase 2 trial of SY-1425 in patients with acute myeloid leukemia or myelodysplastic syndrome. https://clinicaltrials.gov/ct2/show/NCT02807558 ClinicalTrials.gov identifier: NCT02807558. Updated 6 May 2022.
- 8.Jurcic JG, Raza A, Vlad G, et al. Early results from a biomarker-directed Phase 2 trial of SY-1425 in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) demonstrate DHRS3 induction and myeloid differentiation following SY-1425 treatment [abstract] Blood. 2017;130(suppl 1):2633. Abstract 2633. [Google Scholar]
- 9.de Botton S, Vigil CE, Cluzeau T, et al. 24 October 2019. SY-1425, a potent and selective RARα agonist, in combination with azacitidine demonstrates high response rates and a rapid onset of clinical responses in RARA-positive newly diagnosed unfit AML. Abstract presented at: European School of Haematology 5th International Conference Acute Myeloid Leukemia “Molecular and Translational”: Advances in Biology and Treatment. [Google Scholar]
- 10.Cook RJ, Moyo TK, Liesveld JL, et al. Early results from a biomarker-directed Phase 2 trial of SY-1425 in combination with azacitidine or daratumumab in non-APL acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) [abstract] Blood. 2018;132(suppl 1):2735. Abstract 2735. [Google Scholar]
- 11.de Botton S, Cluzeau T, Vigil CE, et al. SY-1425, a potent and selective RARα agonist, in combination with azacitidine demonstrates a high complete response rate and a rapid onset of response in RARA-positive newly diagnosed unfit acute myeloid leukemia [abstract] Blood. 2020;136(suppl 1):4–5. Oral presentation 134600. Abstract 112. [Google Scholar]
- 12.Ferrara F, Barosi G, Venditti A, et al. Consensus-based definition of unfitness to intensive and non-intensive chemotherapy in acute myeloid leukemia: a project of SIE, SIES and GITMO group on a new tool for therapy decision making. Leukemia. 2013;27(5):997–999. doi: 10.1038/leu.2012.303. [DOI] [PubMed] [Google Scholar]
- 13.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group in for diagnosis, standardization response criteria, treatment outcomes, and reporting standards for therapeutic trials in acute myeloid leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 14.Bloomfield CD, Estey E, Pleyer L, et al. Time to repeal and replace response criteria for acute myeloid leukemia? Blood Rev. 2018;32(5):416–425. doi: 10.1016/j.blre.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 15.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology™: Acute myeloid leukemia. v1.2018. https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf
- 16.Fiore C, Kelly M, Volkert A, et al. Selection of RARA-positive newly diagnosed unfit AML patients with elevated RARA gene expression enriches for features associated with primary resistance to venetoclax and clinical response to SY-1425, a potent and selective RARα agonist, plus azacitidine [abstract] Blood. 2020;136(suppl 1):15–16. Oral presentation 137323. Abstract 137323. [Google Scholar]
- 17.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Nakauchi Y, Köhnke T, et al. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer. 2020;1(8):826–839. doi: 10.1038/s43018-020-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuusanmäki H, Leppä AM, Pölönen P, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica. 2020;105(3):708–720. doi: 10.3324/haematol.2018.214882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pei S, Pollyea DA, Gustafson A, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov. 2020;10(4):536–551. doi: 10.1158/2159-8290.CD-19-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383(7):617–629. doi: 10.1056/NEJMoa2012971. [DOI] [PubMed] [Google Scholar]
- 22.Dombret H, Seymour JF, Butrym A, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood. 2015;126(3):291–299. doi: 10.1182/blood-2015-01-621664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenaux P, Mufti GJ, Hellström-Lindberg E, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol. 2010;28(4):562–569. doi: 10.1200/JCO.2009.23.8329. [DOI] [PubMed] [Google Scholar]
- 24.Traina F, Visconte V, Elson P, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28(1):78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 25.Itzykson R, Kosmider O, Cluzeau T, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25(7):1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 26.Bowen D, Groves MJ, Burnett AK, et al. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia. 2009;23(1):203–206. doi: 10.1038/leu.2008.173. [DOI] [PubMed] [Google Scholar]
- 27.Lübbert M, Grishina O, Schmoor C, et al. Valproate and retinoic acid in combination with decitabine in elderly nonfit patients with acute myeloid leukemia: results of a multicenter, randomized, 2 × 2, phase II trial. J Clin Oncol. 2020;38(3):257–270. doi: 10.1200/JCO.19.01053. [DOI] [PubMed] [Google Scholar]
- 28.Vesanoid. Prescribing Information. Roche Laboratories Inc. 2004 www.accessdata.fda.gov/drugsatfda_docs/label/2004/20438s004lbl.pdf [Google Scholar]
- 29.Montesinos P, Bergua JM, Vellenga E, et al. Differentiation syndrome in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline chemotherapy: characteristics, outcome, and prognostic factors. Blood. 2009;113(4):775–783. doi: 10.1182/blood-2008-07-168617. [DOI] [PubMed] [Google Scholar]
- 30.Norsworthy KJ, Mulkey F, Scott EC, et al. Differentiation syndrome with ivosidenib and enasidenib treatment in patients with relapsed or refractory IDH-mutated AML: a U.S. Food and Drug Administration systematic analysis. Clin Cancer Res. 2020;26(16):4280–4288. doi: 10.1158/1078-0432.CCR-20-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl M, Tallman MS. Differentiation syndrome in acute promyelocytic leukaemia. Br J Haematol. 2019;187(2):157–162. doi: 10.1111/bjh.16151. [DOI] [PubMed] [Google Scholar]
- 32.Fathi AT, DiNardo CD, Kline I, et al. Differentiation syndrome associated with enasidenib, a selective inhibitor of mutant isocitrate dehydrogenase 2: analysis of a phase 1/2 study. JAMA Oncol. 2018;4(8):1106–1110. doi: 10.1001/jamaoncol.2017.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byrne M, Danielson N, Sengsayadeth S, et al. The use of venetoclax-based salvage therapy for post-hematopoietic cell transplantation relapse of acute myeloid leukemia. Am J Hematol. 2020;95(9):1006–1014. doi: 10.1002/ajh.25859. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.