Summary
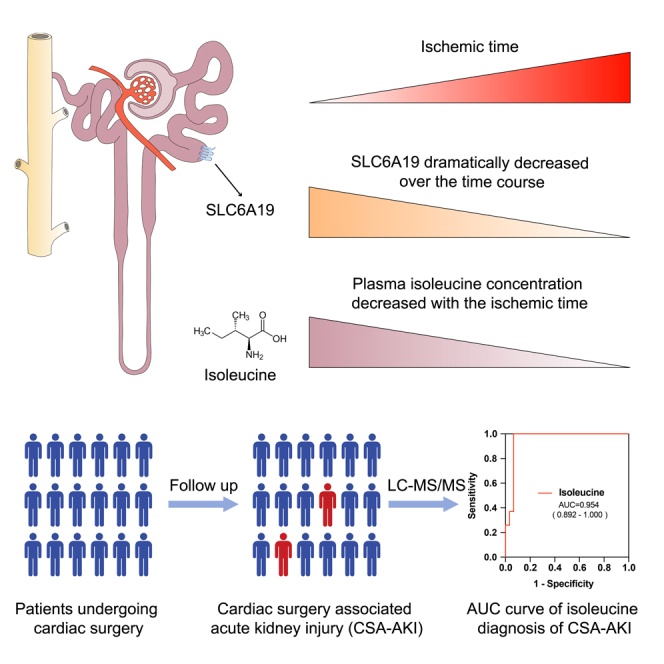
Ischemia reperfusion injury (IRI), often related to surgical procedures, is one of the important causes of acute kidney injury (AKI). To decipher the dynamic process of AKI caused by IRI (with prolonged ischemia phase), we performed single-cell RNA sequencing (scRNA-seq) of clinically relevant IRI murine model with different ischemic intervals. We discovered that Slc5a2hi proximal tubular cells were susceptible to AKI and highly expressed neutral amino acid transporter gene Slc6a19, which was dramatically decreased over the time course. With the usage of mass spectrometry–based metabolomic analysis, we detected that the level of neutral amino acid isoleucine dropped off in AKI mouse plasma metabolites. And the reduction of plasma isoleucine was also verified in patients with cardiac surgery–associated acute kidney injury (CSA-AKI). The findings advanced the understanding of dynamic process of AKI and introduced reduction of isoleucine as a potential biomarker for CSA-AKI.
Subject areas: Disease, Metabolomics, Transcriptomics, Model organism
Graphical abstract
Highlights
-
•
Injured renal cells and neutrophils increased gradually under ischemic injury
-
•
Transporter Slc6a19 was downregulated in proximal tubular cells during AKI
-
•
Decrease of plasma isoleucine might be a biomarker for acute kidney injury
Disease; Metabolomics; Transcriptomics; Model organism
Introduction
Acute kidney injury (AKI) is a clinical syndrome caused by a variety of causes and a sudden decline in renal function, with limited pharmacological treatment. Ischemia reperfusion injury (IRI), often related to surgical procedures such as cardiac surgery, is one of the important causes of AKI.1 The incidence rate in intensive care unit (ICU) is as high as 30%, which increases the postoperative mortality.2 Therefore, in-depth study of the pathogenesis and early warning markers of AKI are of great significance to improve the prognosis, reduce the burden of care in families of patients and alleviate the shortage of medical resources. Studies have shown that cardiopulmonary bypass time is an independent risk factor for postoperative AKI.3 At present, it is still unknown the dynamic cell-specific gene expression changes in AKI caused by IRI (with prolonged ischemia phase).
The inherent structural and functional complexity of the kidney makes it a great challenge to identify cell-specific regulatory pathways. The mechanisms of AKI induced by ischemia-reperfusion mainly include renal tubular injury, vascular endothelial injury, immune cell infiltration and changes in metabolic level, based on traditional methods of histopathology and cell biology.4,5,6 In recent years, the emergence of single-cell RNA sequencing (scRNA-seq) helps to delineate the transcriptional signatures of human kidney and map the immune system in kidney.7,8 Those unbiased approaches also facilitate to characterize the cell composition and cell-specific gene expression changes in AKI progression and renal fibrosis.9,10,11 However, whether kidney amino acid transport and uptake is impaired during AKI and how this might influence the progression of AKI remain unknown.
In the present study, using single-cell RNA sequencing of clinically relevant IRI murine model, we aimed to uncover the dynamic processes of AKI caused by IRI with continuous ischemia intervals. We demonstrated that AKI impaired metabolic process and promoted kidney injury and inflammation in the injured cells. The infiltration of neutrophils increased gradually. Slc5a2hi proximal tubular cell subset highly expressed neutral amino acid transporter genes Slc6a19, Slc43a2 and Slc7a8, which dramatically decreases over the time course, susceptible to IRI. And the plasma level of neutral amino acid isoleucine decreases gradually in AKI mouse model. Decreased level of isoleucine was also detected in plasma of patients with cardiac surgery–associated acute kidney injury (CSA-AKI). The findings provide new insights into the proximal tubular cells response to AKI and introduce isoleucine as a potential biomarker for AKI related to clinical surgery.
Results
scRNA-seq data reveals the major cell types and subclusters of mouse IRI kidney
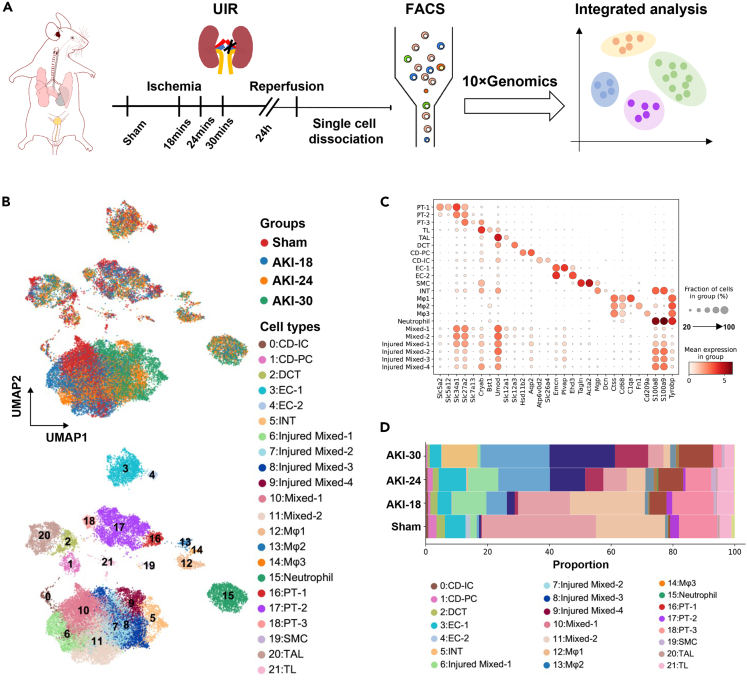
We performed single-cell RNA sequencing (scRNA-seq) with clinically relevant unilateral ischemia-reperfusion injury (IRI) model on C57BL/6N mice.12,13 To explore the early dynamic process of AKI, we set different ischemia intervals. Mice were euthanized at 24 h after IRI (Figure 1A). Acute injury was confirmed by kidney histologic changes (Figure S1A) and blood urea nitrogen (BUN) levels (Figure S1B, Table S1). We analyzed 26480 cells that remained after quality control filtering and doublets removing (Figure S2A, Tables S2 and S3). Datasets of four groups were integrated using the Harmony algorithm to reduce batch effects, and 22 separate clusters were identified using unsupervised clustering, including nephron (Cluster 2, 16, 17, 18, 20, 21), ureteric epithelium (Cluster 0, 1), endothelial cells (Cluster 3, 4), stromal cells (Cluster 5, 19), immune cells (Cluster 12, 13, 14, 15) and mixed cells (Cluster 6, 7, 8, 9, 10, 11) (Figure 1B). These cell clusters were annotated by known cell type-specific markers and cluster-specific marker genes (Figures 1C and S2B). The mixed cells highly expressed different cell type markers, including proximal tubule cells (Slc34a1, Slc27a2), loop of Henle (Cryab, Umod), principal cells (Aqp2) and injured mixed cells expressed immune and injury marker genes (S100a8, S100a9) (Figure 1C), like the mixed-identity cells as others show.10 We performed multiple labeling staining for Slc27a2, Umod and Aqp2 (Figure S3) to validate the existence of these cells. The distribution of these mixed cells was closely related to different ischemia intervals (Figure 1B). From the composition of all clusters in four groups, we found that the proportion of immune cells and injured mixed cells increased significantly, and the proportion of mixed cells and major cell clusters from nephron gradually decreased with the prolongation of ischemia time (Figure 1D and Table S4).
Figure 1.
scRNA-seq data reveals the major cell types and subclusters in the ischemia reperfusion–induced AKI mouse kidney
(A) Overview of experimental design. n = 3 mice at each timepoint. IRI, ischemia-reperfusion injury; UIR, unilateral ischemia/reperfusion; FACS, flow cytometry and fluorescence-activated cell sorting.
(B) UMAP plot illustrates the distribution of cell clusters at each timepoint integrated with scVI.
(C) Dot plot displaying expression pattern of genes specific to each cluster shown in (B).
(D) Stacked bar plot displaying the composition of identified clusters by group. CD-IC, intercalated cells of collecting duct; CD-PC, principal cells of collecting duct; DCT, distal convoluted tubule; EC, endothelial cells; INT, interstitial cell; Mφ, macrophages; PT, proximal tubule; SMC, smooth muscle cells; TAL, thick ascending limb of loop of Henle; TL, thin ascending limb of loop of Henle and descending limb of loop of Henle.
See also Figures S1–S3, Tables S1–S4.
Functional diversity and dynamic regulation processes of injured cells during AKI
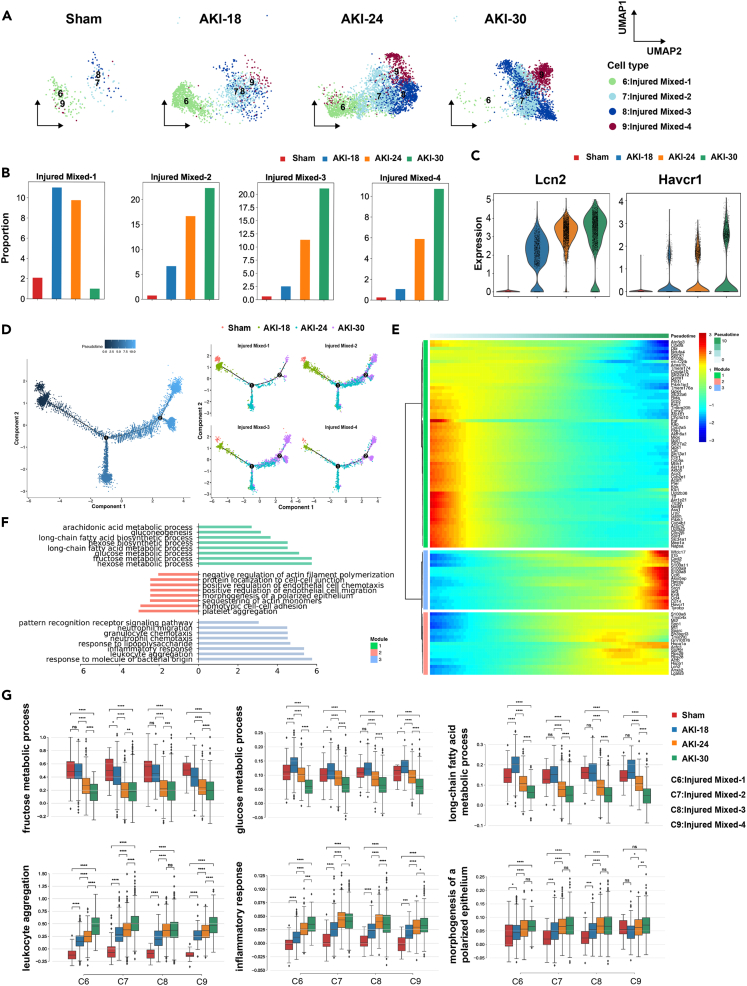
Because the injured cell subsets account for a large proportion in AKI groups compared with the sham group, we firstly studied the function and regulation mechanisms of injured mixed cells. The four clusters were merely present in the sham group. Injured Mixed-1 (Cluster 6) were enriched in AKI-18 and AKI-24 groups, whereas the proportion of Injured Mixed-2 (Cluster 7), Injured Mixed-3 (Cluster 8) and Injured Mixed-4 (Cluster 9) increased gradually in AKI groups (Figures 2A and 2B). The well-known kidney injury markers lipocalin 2 (Lcn2) and Kim1 (Havcr1)14,15 were significantly upregulated after IRI, and their expression increased in a time-dependent manner (Figure 2C), indicating that the severity of renal injury was related to the ischemia time. We reconstructed injured cells relationships by pseudotemporal order. The trajectory analysis showed the transition from normal to injured stages over the time course (Figure 2D). Injured cells from different groups were associated with distinct cell states (Figures S4A and S4B) with simulating AKI progression from mild to severe stage. To further characterize the transition process, we identified differentially expressed genes (DEGs) that significantly changed along the pseudotime in kidney injured cell clusters and defined three gene expression modules (Figure 2E and Table S5). Gene ontology (GO) analysis of the three gene modules revealed metabolic processes downregulated and inflammatory response upregulated during AKI (Figure 2F). Enriched GO biological process (GOBP) terms of Module 1 mainly included glucose metabolic and fatty acid metabolic pathway, and the representative genes in urea cycle enzyme (Ass1), mitochondrial respiration and homeostasis (Cox8a, Atp5g3), gluconeogenesis (Pck1), fatty acid catabolism (Slc27a2) were decreased gradually after AKI (Figure S4C, left panel). Modules 2 and 3 highly expressed chemokines including Ccl4, Ccl6, Cxcl2, inflammatory signals including Il1b, S100a8, S100a9 and injury marker Lcn2, indicating enhanced inflammatory cell infiltration, cell adhesion process, inflammatory response and kidney cell injury during AKI (Figures 2F, S4C, right panel). We evaluated the enrichment scores of different biological processes in the four injured clusters over the time course: fructose metabolic process, glucose metabolic process and long-chain fatty acid metabolic process were gradually downregulated; but inflammatory response, leukocyte aggregation and neutrophil chemotaxis were significantly upregulated from mild to severe stage (Figure 2G). These results indicated that the occurrence and development of acute renal injury is accompanied by the decline of metabolic level and the increase of inflammation and injury response.
Figure 2.
Functional diversity and dynamic regulation processes of injured cells during AKI
(A) UMAP plots showing the injured mixed cell cluster distribution at each group with ischemia time series.
(B) Bar plots display the percentage of cells of four subclusters in different groups.
(C) Violin plots show the expression of selected markers (Lcn2 and Havcr1) for renal injury in different groups.
(D) Pseudotime trajectory shows the transition from normal to injured stages colored by the time course. Numbers 1 and 2 represent the branch points of different cell states.
(E) Heatmap showing differentially expressed genes (DEGs) alone with the pseudotime as in (D), catalogs into three gene modules.
(F) Gene ontology (GO) terms enriched for each gene module as in (E).
(G) Boxplots of the expression levels of selected functions across four groups in each cluster. Groups are shown in different colors. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns. for not significant, Mann-Whitney-Wilcoxon test.
Inflammatory cells response to acute kidney injury
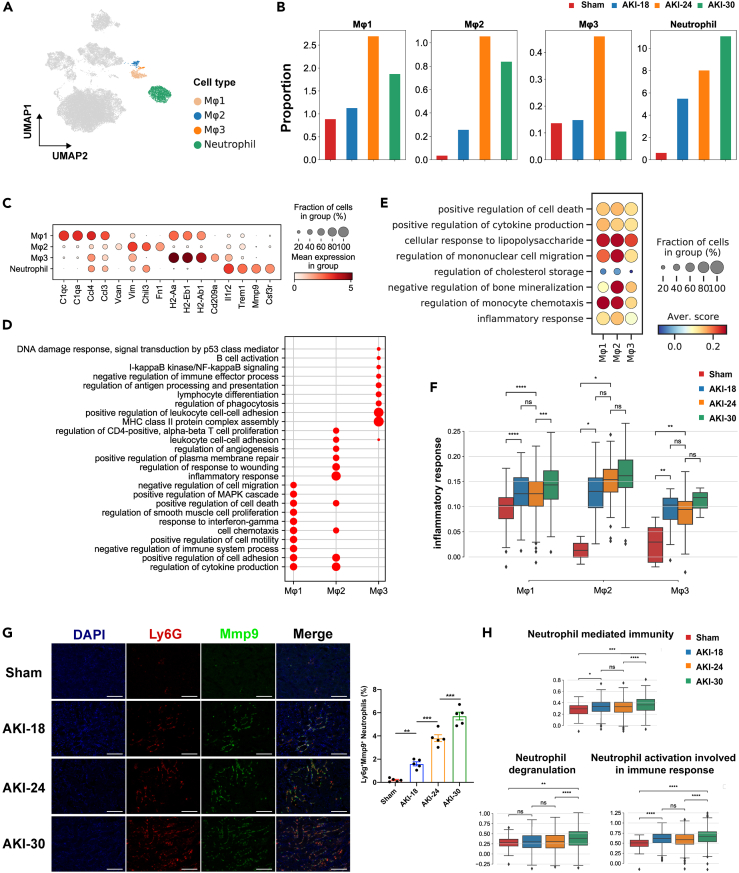
In view of the markedly increased inflammatory response during AKI, we sought to explore the immune microenvironment. We observed three clusters of macrophages and one cluster of neutrophils (Figure 3A). We next compared the proportion of these clusters in the different timepoints (Figure 3B). Of note, the abundance of macrophages was significantly increased in the AKI-24 group, which was then decreased in the group with the longest ischemia time. Of interest, the percentage of neutrophils was increased gradually along the ischemia time series. The representative genes for each cluster were C1qa, C1qc, and chemokines Ccl4, Ccl3 in Macrophage 1, Vim and Fn1 in Macrophage 2, MHC-II genes (H2-Aa, H2-Eb1, H2-Ab1) in Macrophage 3, Il1r2 and Csf3r in Neutrophil (Figure 3C). Gene ontology (GO) terms enriched in macrophages were with some differences among three clusters. Macrophage 1 represented the tissue-resident macrophages and mainly involved in GO terms inflammatory, anti-inflammatory, cell proliferation and migration. Macrophage 2 played more roles in inflammatory response and wound healing. Macrophage 3 was more focused on antigen presentation and regulation of leukocyte cell-cell adhesion (Figure 3D). After injury, of the three macrophage clusters, Macrophage 2 had the highest inflammation score such as regulation of mononuclear cell migration and inflammatory response scores (Figure 3E and Table S6), and three macrophage clusters showed the early response to acute injury (Figure 3F). As shown in Figure 3G, Ly6g+Mmp9+ neutrophils infiltrated heavily after kidney injury. GO terms for neutrophils were more enriched in neutrophil degranulation, neutrophil activation and immune response (Figure S5A). Neutrophil degranulation was not obvious until the later stage of injury, whereas neutrophil activation and neutrophil mediated immunity occurred early in response to acute injury, with highly expressed Cxcl2 and Icam1 representing enhanced chemotactic and adhesion of neutrophil (Figures 3H and S5B). These results suggest that macrophages and neutrophils played complementary roles in the early phase of AKI and promoting inflammatory injury.
Figure 3.
Characterization of inflammatory cells response to acute kidney injury
(A) UMAP plot of clusters of macrophages and neutrophils colored as in Figure 1.
(B) Bar plots showing the distribution of each cluster in different groups.
(C) Dot plot displaying expression patterns of marker genes in each cluster.
(D) The enriched GO terms of each macrophage subcluster.
(E) Dot plot showing the scores of GO biological process (GOBP) terms compared between AKI-24 with Sham group among three subclusters of macrophages.
(F) Boxplots of the expression levels of inflammatory response across four groups in each macrophage cluster. Groups are shown in different colors. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns. for not significant, Mann-Whitney-Wilcoxon test.
(G) Representative images of Ly6g and Mmp9 co-staining in mouse kidney sections from sham and AKI subgroups. n = 5 in each group. ∗∗p < 0.01, ∗∗∗p < 0.001, One-way ANOVA. Scale bar, 100 μm. Data are presented as mean ± SEM.
(H) Boxplots of expression levels of three GOBP terms in neutrophils across four groups. Groups are shown in different colors. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns. for not significant, Mann-Whitney-Wilcoxon test.
Downregulation of neutral amino acid transporters in proximal tubular cells due to AKI
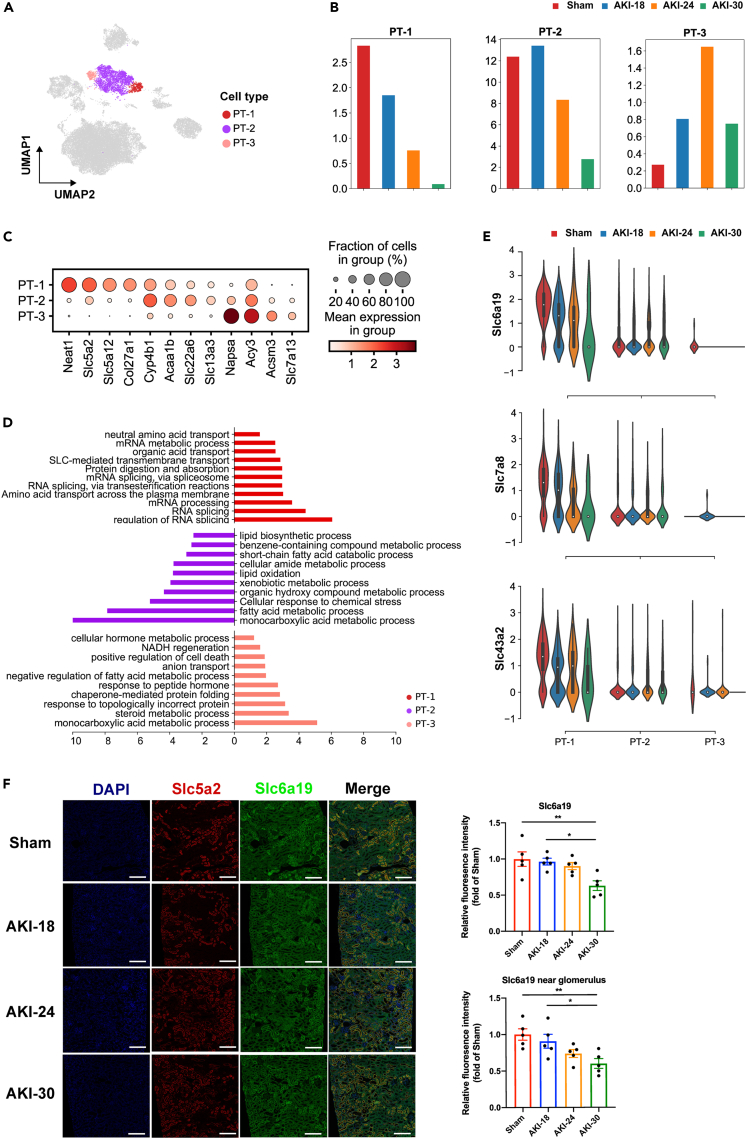
Because the development of acute renal injury is accompanied by a significant decline in metabolic levels, we next focused our analysis on proximal tubular cells, which were very vulnerable to injury because of high metabolic activity. The proximal tubular cells were subdivided into three clusters: PT-1, PT-2 and PT-3 (Figure 4A). These subpopulations had different distribution characteristics along the ischemia time series (Figure 4B). The percentage of PT-1 was gradually decreased markedly, whereas the abundance of PT-2 started to decrease in AKI-24 group. The proportion of PT-3 increased with the elongation of ischemia time, and reached the highest at 24 min, and then decreased. The representative genes for PT-1 were Slc5a2 and Slc5a12. PT-2 highly expressed solute linked carriers Slc22a6 and Slc13a3.16 The representative genes for PT-3 were enzyme-encoding genes (Napsa, Acy3, Acsm3) and Slc7a13 (Figure 4C). GO terms enriched in PT-1 were regulation of RNA splicing and SLC-mediated transmembrane transport. PT-2 is mainly involved in GO terms monocarboxylic acid metabolic process and lipid oxidation. The genes in PT-3 were significantly enriched in monocarboxylic acid metabolic process and response to protein misfolding (Figure 4D). Of interest, PT-1 highly expressed neutral amino acid transporters Slc6a19, Slc43a2 and Slc7a8,17,18,19 and the expression levels of Slc6a19, Slc43a2 and Slc7a8 were downregulated in PT-1 during AKI progression (Figure 4E). As shown in Figure 4F, Slc6a19 was colocalized with Slc5a2 on the luminal membrane of proximal tubule. With the prolongation of ischemia time, the expression of Slc6a19 was downregulated in PT-1 cells, and loss of brush border, vacuolization or dilated proximal tubules increased. Surprisingly, the cells expressing Slc6a19 in close approximation to glomerulus (macula densa) were greatly decreased (Figure 4F), which may also affect the reabsorption of sodium ions and homeostasis. These results suggested that AKI might lead to downregulation of Slc6a19 and dysregulation of neutral amino acid transport and reabsorption in proximal tubules.
Figure 4.
Downregulation of neutral amino acid transporters in Slc5a2hi proximal tubular cells because of AKI
(A) UMAP plot of clusters of proximal tubular cells colored as in Figure 1.
(B) Bar plots showing the distribution of each cluster of proximal tubular cells in different groups. Groups are shown in different colors.
(C) Dot plot displaying expression patterns of marker genes in subclusters of proximal tubular cells.
(D) Bar diagram showing the enriched GO biological process (GOBP) terms of each subcluster of proximal tubular cells.
(E) Violin plots showing the downregulation of Slc6a19, Slc43a2 and Slc7a8 specifically in PT-1 during AKI. Groups are shown in different colors.
(F) Multiple labeling staining for Slc5a2 (red) and Slc6a19 (green) in kidney tissues from four groups.n = 5, ∗p < 0.05, ∗∗p < 0.01, One-way ANOVA. Scale bar, 200 μm. Data are presented as mean ± SEM.
Reduction of neutral amino acid isoleucine in plasma from mice of AKI and from human with cardiac surgery–associated acute kidney injury (CSA-AKI)
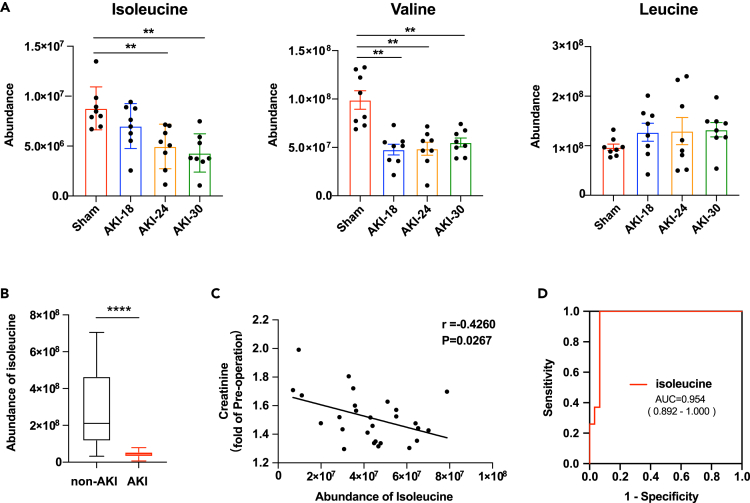
Because Slc6a19 is neutral amino acid uniporter,19 with a preference for branched-chain amino acids (BCAA),20,21 we analyzed the levels of branched-chain amino acids in AKI mouse plasma by liquid chromatography-tandem mass spectrometry using targeted metabolomic approaches. And we observed gradually decreasing plasma levels of isoleucine, and plasma levels of valine were also decreased in all AKI groups, but without a gradient change in our AKI model (Figure 5A). Besides, we have also detected the plasma levels of phenylalanine, methionine and L-alanine, which were transported by Slc43a2 and Slc7a8. We observed that the levels of phenylalanine were decreased in AKI-30 group compared with AKI-18 group, but the levels of methionine and L-alanine were not changed significantly (Figure S6). We next explored whether the level of isoleucine reduced in plasma of patients with CSA-AKI. We have collected plasma samples from patients undergoing cardiac surgery in ICU within 48 h after operation (Tables S7 and S8). We found that the plasma level of isoleucine was significantly downregulated in patients with AKI (27 patients with AKI and 30 patients without AKI) (Figure 5B). And the isoleucine abundance had a negative correlation with the serum creatinine (fold of pre-operation) of patients with AKI, as shown in Figure 5C. And we identified isoleucine as a potential biomarker with the area under the receiver operating characteristics curve (AUC-ROC) value 0.954 with a 95% confidence interval of 0.892–1.000 (Figure 5D).
Figure 5.
Reduction of neutral amino acid isoleucine in plasma of AKI mouse and human with CSA-AKI
(A) Boxplots showing the abundance of isoleucine, valine and leucine in plasma of sham or AKI mice. n = 8 for each group. ∗∗p < 0.01, compared to Sham group, One-way ANOVA. Data are presented as mean ± SEM.
(B) Boxplots showing the concentration of isoleucine in plasma of patients with or without CSA-AKI. n = 30 for non-AKI group, n = 27 for AKI group. ∗∗∗∗p < 0.0001, Student’s t test. Data are presented as mean ± SEM.
(C) The correlation analysis between the abundance of isoleucine and the serum creatinine (fold of pre-operation) of patients with CSA-AKI. n = 27, Pearson correlation analysis.
(D) ROC curve for diagnostic value of isoleucine to differentiate patients with AKI from patients without AKI after cardiac surgery. CSA-AKI indicates cardiac surgery–associated AKI; ROC, receiver operating characteristic; AUC, area under the curve.
The findings suggested that Slc5a2hi proximal tubular cells could be the early responder to AKI with downregulation of neutral amino acid transporter Slc6a19. And decreased neutral amino acid isoleucine in plasma could be early signals of kidney injury because of dysregulation of isoleucine transport and absorption in proximal tubular cells after AKI.
Discussion
Single-cell RNA sequencing (scRNA-seq) is helpful to find specific cell subsets and deep into understanding the regulatory mechanisms in human kidney diseases. Rodent models are often used to study the mechanisms of AKI owing to easy handling and breeding, whereas translational value of therapeutics from rodent models is questionable. More medical research uses porcine models to investigate clinically important issues, such as pig-to-human kidney xenotransplantation.22 There are also porcine models of AKI because pig kidneys are anatomically and physiologically similar to human kidneys. Nevertheless, we chose murine models of AKI rather than porcine models in this study because there are few studies on scRNA-seq because of lack of full gene sequence, transcriptomics, and proteomics information in pigs.23,24 Although we would consider using porcine models of AKI for future target validation and clinical transformation research.
Previous studies using scRNA-seq with murine IRI models to study the mechanism of AKI mostly set a fixed ischemia time with different duration for reperfusion, a few hours to a few days, to explore the mechanisms of AKI from occurrence to recovery and fibrosis.10,11 Because ischemia for 18 min of IR could lead to AKI,25 and ischemia for 30 min before reperfusion could lead to severe renal injury,12,13 we for the first time performed scRNA-seq with IRI model by tightening the mouse renal pedicle for 18 min, 24 min, 30 min separately, then loosening them (reperfusion) for 24 h to simulate different stages of renal injury from mild to moderate to severe and explore the dynamic changes from AKI onset. We performed contralateral nephrectomy after clamping the left renal pedicle to prevent the compensatory effect of the contralateral kidney, because nephrectomy may have effects on kidney recovery after AKI.26 There was no death of mice in the study and the model was successfully. We found a group of injured cell clusters, and the expression of kidney injury markers Lcn2 and Havcr1 were significantly upregulated after IRI in a time-dependent manner. Meanwhile, we identified DEGs that significantly changed mainly enriched on upregulated inflammatory response and downregulated metabolic processes over the time-course. Rudman-Melnick et al.10 also reported that crucial injury response factors increased at day 1 after unilateral ischemia/reperfusion. And many genes were also changed consistently with that reported in the mentioned study,10 such Slc34a1, Aqp1, Lrp2, Slc7a13, Slc22a6, Slc37a4, Aco2, Cyp2e1, Miox, Slc27a2, Tnfrsf12a, Nfkbia, Lgals1, Ctsd S100a11, Lcn2, Cstb, Anxa2, Mgst1, Gpx3. Although the genes in cell cycle regulation were not significant changed of in our study (Figure 2E, Table S5). There are differences in the mice strains, method of AKI model establishment and scRNA-seq analysis methods in both of our study. But we both found a group of mixed identity cells, expressing various renal cell type markers, such as Umod, Slc34a1, Aqp2. They performed RNAScope to validate these cells, and we verified the mixed-identity cells using multiple labeling staining at protein levels (Figure S3). In the study of Suszták K’s lab,27 they reported another type of mixed cell subgroup of intercalated cells (ICs) and principal cells (PCs). These studies above showed that there are mixed identity cells in normal and disease kidneys, but studies using single-nucleus RNA sequencing (snRNA-seq) did not find similar clusters,16,28 and this difference may be caused by the bias between scRNA-seq and snRNA-seq.29,30 It may be more likely to detect mixed identity cells by single-cell RNA sequencing than single-nucleus RNA sequencing.
Macrophages and neutrophils in innate immune response are involved in the process of AKI.31 Recently, it was reported that the cluster of S100a9hiLy6chi macrophages is an early responder to AKI.11 Renal neutrophil recruitment is more obvious in AKI caused by IR than that caused by sepsis.32 In our experimental model, macrophages reacted most significantly in moderate AKI, the proportion of three clusters of macrophages increased and activated the immune response. Although Ly6g+Mmp9+ neutrophils increased in a time-dependent manner, running through mild to moderate to severe stages, promoting inflammation and tissue infiltration (Figure 3G). Inhibition of neutrophil infiltration by antibodies or small molecules might contribute to reduce AKI.11,33
The kidney is one of the most energy demanding organs in the human body. It consumes a lot of ATPs to remove waste from the blood, reabsorb nutrients, and maintain the electrolyte and acid-base balance of the body. Kidney injuries mainly target metabolically active proximal tubular cells.34,35,36 We found metabolic reprogramming in proximal tubular cells, especially lipid metabolism disorder in our data. Surprisingly, we found a cluster of proximal tubular cells mainly functioning in regulation of RNA splicing and SLC-mediated transmembrane transport, decreased gradually along the ischemia time series. These cells highly expressed Slc6a19, Slc7a8 and Slc43a2, whereas the expression of these genes decreased during AKI progression. The cells expressing Slc6a19 near glomerulus were greatly decreased in Group AKI-30 (Figure 4F), which may also affect the reabsorption of sodium ions and homeostasis. Slc6a19, Slc7a8 and Slc43a2 are neutral amino acid transporters,17,18,19,37 regulating the transport and reabsorption of amino acids such as leucine, isoleucine, and valine, as well as methionine and phenylalanine.38 Mutations in the gene encoding Slc6a19 were reported to cause human Hartnup disorder, an autosomal recessive defect resulting from impaired of renal and gastrointestinal neutral amino acid transport.39,40 Mice lacking Slc6a19 showed impaired nutrient signaling and reduced body weight41 and reduced plasma levels of essential amino acids after high-protein diets because of amino acid malabsorption.19 However, whether Slc6a19 participates in AKI progression is unknown. We detected the gradual decrease of isoleucine in the plasma of AKI mouse. Valine was also decreased in all AKI groups without a gradient change. Although leucine, methionine and phenylalanine did not change significantly in AKI mice compared with the Sham group. Furthermore, our mass spectrometry metabolomic data showed that isoleucine abundance decreased in the plasma of patients with CSA-AKI, and negatively correlated with the serum creatinine. Besides, isoleucine could be used to differentiate patients with AKI from patients without AKI after cardiac surgery, suggesting that dysfunction of isoleucine transport and uptake might play an important role in the progress of clinical surgery related AKI. In previous studies, branched chain amino acids (BCAA), especially valine and leucine decreased in plasma of CKD patients, but the levels of isoleucine seem to be unchanged.42 Although in another study, valine, isoleucine, and leucine were lower in the patients with end-stage renal disease (ESRD) than in control group.43 The level of plasma amino acids may be affected by different disease stages, individual differences, and different detection methods adopted by different research institutions. We defined AKI by the criteria that the serum creatinine rise was greater than 26.5 μmol/L within 48 h according to the Kidney Disease Improving Global Outcomes (KDIGO) definition.44 The cardiac surgery-associated acute renal injury (CSA-AKI) patients we included belong to stage 1 of AKI (Table S8). Decrease of isoleucine might be more sensitive to early AKI.
Overall, scRNA-seq analysis uncovered the dynamic processes of AKI caused by IRI and identified neutral amino acid transporter Slc6a19 downregulated in Slc5a2hi proximal tubular cells during AKI, which might result in dysregulation of isoleucine transport and absorption. The plasma level of neutral amino acid isoleucine decreased in AKI mouse model and CSA-AKI patients. The present data illustrate that plasma isoleucine is a potential biomarker and Slc6a19 might be a potential intervention target for AKI.
Limitations of the study
We identified four subpopulations of injured cells based on the clustering of 26480 cells. We do not find specific marker genes for these clusters, because these injured cells highly expressed injury markers, Havcr1 and Lcn2, and they also expressed markers for other cell types, such as proximal tubules, loop of Henle, collecting duct. We demonstrated that downregulated expression of Slc6a19 and reduction of the Slc5a2hi proximal tubular cells might result in dysregulation of isoleucine transport and absorption during AKI. How the gene Slc6a19 is regulated in AKI remains to be investigated in the future. Also, it is better to test the putative biomarker isoleucine in a separate clinical study involving patients undergoing cardiac surgery with high-risk of stage 2 or 3 AKI in the future and verify whether isoleucine in the urine of AKI patients increases, which will make the clinical research more complete.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SLC6A19 Polyclonal Antibody | Invitrogen | Cat# PA5-60276; RRID: AB_2647551 |
| Rabbit polyclonal anti-SLC5A2 | Origene | Cat# TA381682 |
| Rabbit monoclonal [EP1254] to MMP9 | Abcam | Cat# ab76003; RRID: AB_1310463 |
| Rabbit monoclonal [EPR22909-135] to Ly6g | Abcam | Cat# ab238132; RRID: AB_2923218 |
| Rabbit monoclonal [EPR20071] to UMOD | Abcam | Cat# ab207170; RRID: AB_2889163 |
| Rabbit monoclonal [EPR21080] to Aquaporin 2 | Abcam | Cat# ab199975; RRID: AB_2820249 |
| FATP2 Polyclonal antibody | Proteintech | Cat# 14048-1-AP; RRID: AB_2239416 |
| Biological samples | ||
| Human plasma | Fuwai Hospital, Chinese Academy of Medical Sciences | N/A |
| Critical commercial assays | ||
| OPALTM 7-COLOR MANUAL IHC KIT | Akoya Biosciences | Cat# NEL811001KT |
| Deposited data | ||
| scRNA-seq processed data | This paper | OMIX, https://ngdc.cncb.ac.cn/omix: accession no. OMIX001933. Accession numbers: OMIX001933 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6N | Vital River Laboratories | https://www.vitalriver.com/#/animalModel/detailedReading?id=20&idd=9&namecode=productserve |
| Software and algorithms | ||
| GraphPad Prism 8.3.1 | GraphPad Software | RRID: SCR_002798 |
| Image-ProPlus software | MEDIACYBERNETICS | RRID: SCR_007369 |
| SPSS Statistics 26 | SPSS, IBM | RRID: SCR_016479 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Jiangping Song (fwsongjiangping@126.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal model
Animal experiments were approved by the Animal Ethics Committee of Fuwai Hospital. C57BL/6N male mice (8 to 10 weeks, 20-25 g weight) were purchased from Vital River Laboratories (Beijing, China). Unilateral IRI plus contralateral nephrectomy were performed. 1% sodium pentobarbital was used to anesthetize mice (50 mg/kg, i.p.injection). Body temperature was maintained by a heating blanket. After incision of abdominal cavity along the midline, the left renal pedicle was clamped with a microaneurysm clip for 18 mins, 24 mins, 30 mins, separately. And the right kidney was removed. The microaneurysm clip was released, and the kidneys and plasma samples were harvested after 24 h.
For paraffin sections, mouse kidneys were collected after perfused with cold PBS, and then fixed with 10% (v/v) formalin for 48 h. Paraffin-embedded tissues were cut into 4 μm sections for histologic assessment and immunostaining.
Method details
Preparation of single cell samples
The mice were anesthetized in an isoflurane chamber, and the kidneys were perfused with ice-cold PBS via the aorta before harvesting. The left kidneys were collected (3 kidneys per group) and stored in cold DMEM, cut into small pieces, and then digested with 1 mg/ml CollagenaseType 2 (LS004176, Worthington) in a 37°C water bath with shaking for 40 min. 10% FBS/DMEM was added to stop digestion. The digested mix was passed through a 70 μm cell strainer (352350, Falcon). The cells were pelleted by centrifugation at 400g for 5min at 4°C. The cells were resuspended with 1 ml of red blood cell (RBC) lysis buffer (R1010, Solarbio) and incubated for 5min at room temperature. 9 ml cold PBS was added to dilute the RBC lysis buffer and the cells were passed through a 40 μm filter (352340, Falcon). The cells were centrifuged at 400 g for 5minat 4°C and resuspended in 1 ml pre-chilled PBS with 1% BSA. Single cell suspensions were stained with a 1:20 dilution of 7-AAD (559925, BD Biosciences) and viable cells (7-AAD-) were sorted with a FACS Aria II cell sorter (BD Biosciences).
Single-cell RNA sequencing
Cells were counted and diluted to 700-1200 cells/ul. Cell suspensions were loaded on a Chromium Single-Cell Controller (10x Genomics). 9000-12000 FACS-sorted kidney cells (more than 85% viability) were encapsulated in droplets. The scRNA-seq libraries were prepared using 10× Genomics Chromium Single Cell 3′Reagent Kits following the user guide and sequenced on an Illumina HiSeq Xten instrument.
scRNA-seq preprocessing and data analysis
Preliminary sequencing results (.bcl files) were converted to FASTQ files with CellRanger (version 3.0, https://support.10xgenomics.com/single-cell-gene-expression/soſtware/pipelines/latest/what-is-cell-ranger). We followed the 10x Genomics standard seq protocol by trimming the barcode and unique molecular identifier (UMI) end to 26 bp, and the mRNA end to 98 bp. The FASTQ files were aligned to the genome reference sequence mm10. Subsequently, we applied CellRanger version 6.0 for preliminary data analysis and generated a h5 file that contained barcode, gene and expression matrix. We further carried out preliminary quality control (QC) on the FASTQ files to ensure high quality scRNA-seq data. The preliminary count matrix was then used for the further analysis.
For all samples, the subsequent analysis was performed by Scanpy packages in Python. We firstly combined the count matrix of each sample into a merged AnnData object and the advanced quality control including total UMI counts, number of genes and proportion of mitochondrial genes were further applied. Specifically, low-quality cells with less than 200 genes deleted or the mitochondrial gene ratio more than 30% were excluded. To further remove the potential doublets, cells with counts above 25000 and gene number above 5000 were filtered out. Additionally, only genes detected in at least 10 cells were kept. As a result, a merged matrix with 26480 cells and 32285 genes were subsequently normalized and scaled for the downstream analysis.
To better integrate cells from different samples into a shared space, a deep learning algorithm namely scVI was used to correct the batch effect. The top 2000 highly variable genes were identified for the input of the scVI model and the low-dimensional latent representation matrix was generated to build nearest neighbor graph. Such neighbor graph was then used to find clusters by leiden algorithm and the 2D cell embeddings was performed by using UMAP function for visualization. Marker genes for major clusters was identified using rank_genes_groups function. To identity sub type within each major cell type,same procedure starting from the batch correction was performed as described above. All cell types were defined manually according to the maker genes. In total, we identify 22 cell types. The differentially expressed genes were further detected by “rank_genes_groups” function in Scanpy by using Wilcoxon method.
Pseudotemporal analysis
Pseudotemporal analysis was performed using Monocle2 R package. The count matrix was used as input and variable genes used for cell ordering were identified based on q-value with cutoff <0.01. The structure of the trajectory was plotted in two-dimensional space using DDRTree algorithm and the cells were ordered in pseudotime.
GO enrichment scoring
Enrichment scores were calculated using Scanpy function “score_genes” which calculated the average expression of a given gene set subtracing the aggregated expression of control gene sets.
Serum BUN measurement
Mouse blood samples were collected and placed at room temperature for 40 min. The whole blood samples were centrifuged at 4000 rpm for 10min at 4°C. Serum was collected and stored at 4°C. Serum BUN levels were detected using QuantiChrom™ Urea Assay Kit (DIUR-100, BioAssay Systems) according to the manufacturer’s protocol.
Patient cohort
This study was approved by the Human Ethics Committee of Fuwai Hospital, Chinese Academy of Medical Sciences. Written informed consent was obtained from each patient. Our research group has collected plasma samples from patients undergoing cardiac surgery with cardiopulmonary bypass (CPB) in intensive care unit (ICU) before and after operation (with 48 hours) in Fuwai Hospital. Clinical characteristics of the participants are summarized in Tables S7 and S8. According to the diagnostic criteria of Kidney Disease Improving Global Outcomes (KDIGO) definition,45 27 patients with cardiac surgery-associated acute renal injury (CSA-AKI) and 30 patients without CSA-AKI were included in this study. The CSA-AKI patients we included belong to stage 1 AKI.
Metabolite extraction from plasma samples
Plasma samples (50 ul) were extracted in a mixture of acetonitrile: methanol: H2O (v/v/v, 4:4:2, 450 ul) by vortexing for 5 min. Keep the mixtures at -20°C for 2 h. Then the mixture was centrifuged at 15000 gat 4°C for 30 min to precipitate proteins, and the supernatants were transferred to a new tube and stored at -80°C until used for LC-MS/MS analysis. Blending equal volume of each sample makes the quality control (QC) sample. Plasma samples collected from mice 24 hours post-operation were used for targeted LC-MS/MS based metabolomic analysis. Plasma samples collected from patients undergoing cardiac surgery in intensive care unit (ICU) within 48 hours after operation were used for untargeted LC-MS/MS based metabolomic analysis.
Targeted LC-MS/MS-based metabolomic analysis
With a Vanquish ultraperformance liquid chromatography system coupled to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific), an ACQUITY UPLC BEH Amide column (50 x 2.1 mm, 1.7 μm, Waters) was used for metabolite separation. Metabolites were separated with a 15 min gradient at a flow rate of 0.4 mL/min. Mobile phase A was H2O with 10 mM NH4COOH and mobile phase B was ACN. The gradient was set as follows: 0-1 min, 95% B; 1-7 min, 70% B; 7-10 min, 30% B, 10-12.5 min, 30% B, 12.5-13.0, 95% B, 13-15 min, 95% B. The above Chemicals and reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
The mass spectrometer was operated in electrospray ionization (ESI) positive ion mode. Analysis was performed in parallel reaction monitoring (PRM) scan mode. Relative quantification of metabolites was calculated by peak area. The instrument settings for the MS/MS mode were: 15,000 resolution, 1 × 106 AGC, 100 ms maximum injection time, and dynamic collision energy of 20, 40, 60 ev. Source ionization parameters were: spray oltage set at 3.5 kV for positive ion mode, capillary temperature set at 320°C, sheath gas set at 25, and aux gas set at 5.
Quantitative analysis of targeted metabolites
Relative quantification of peak area was performed for targeted metabolites in this study. For leucine, isoleucine and valine, the precursor ions with am/z of 132.102 and 118.0869 (M+H+), respectively, which yielded product ions with am/z of 86.0962, 69.0545 and 72.0728, respectively; For phenylalanine, methionine and L-alanine, the precursor ions with am/z of 166.1, 150.08 and 90.0 (M+H+), respectively, which yielded product ions with am/z of 120.08, 104.05 and 44.0, respectively, were used for quantification.
Untargeted LC-MS/MS based metabolomic analysis
Metabolomic profiling was performed as previously described.46 Using a Vanquish ultraperformance liquid chromatography system coupled to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific), samples were injected onto a Hypersil GOLD C18 column (Thermo Fisher Scientific), and metabolite separation was performed with a 15 min gradient program process at a flow rate of 0.25 mL/min. The mass spectrometer was operated in electrospray ionization (ESI) positive ion mode. Analysis was performed in the full scan [mass-to-charge ratio (m/z) = 67∼1000] and data-dependent scan (dd-MS2) modes. Peak area was used to calculate the relative quantification of isoleucine.
Histological examinations and staining
For histological examinations, paraffin-embedded sections were stained with hematoxylin-eosin (HE).
Opal multicolor IHC staining was performed according to the manufacturer’s protocol: 1) slide preparation-deparaffinization and rehydration of the tissue sections; 2) AR6 buffer was used for antigen retrieval in a microwave oven; 3) blocking with blocking/antibody diluent; 4) primary antibody incubation, anti-Slc6a19 (Invitrogen Cat# PA5-60276, RRID: AB_2647551), 1 h, RT; 5) secondary antibody incubation; 6) signal amplification, Opal 520 (1:100), 10 min, RT. The protocol was restarted at step 2 for detection of the other target (anti-Slc5a2, Origene Cat# TA381682; Opal 650 (1:100) for signal amplification); 7) counterstaining and mounting were done with Antifade Mounting Medium with DAPI. The slides were scanned by Vectra Quantitative Pathology Imaging Systems (PerkinElmer), and scanning protocols were created by optimizing the exposure time for each fluorescence channel. The intensity of Slc6a19 expression was measured with Image-ProPlus software (RRID: SCR_007369).
Mixed identity cells were detected by anti-FATP2 (Proteintech Cat# 14048-1-AP, RRID: AB_2239416), anti-Umod (Abcam Cat# ab207170, RRID: AB_2889163), anti- Aquaporin 2 (Abcam Cat# ab199975, RRID: AB_2820249). Anti-Mmp9 (Abcam Cat# ab76003, RRID: AB_1310463) and anti-Ly6g (Abcam Cat# ab238132, RRID: AB_2923218) were used to detect neutrophils. The Ly6g+Mmp9+ cells were quantified with Image-ProPlus software (RRID: SCR_007369).
Quantification and statistical analysis
Data groups (two groups) with normal distributions were compared using the two-sided unpaired Student’s t-test. Comparisons between multiple groups were assessed by a one-way ANOVA with Turkey’s multiple comparisons. Mann-Whitney-Wilcoxon test was used for comparison of different groups in analysis results from scRNA-seq data. The association between the abundance of isoleucine and the serum creatinine (fold of pre-operation) was determined by Pearson correlation analysis. ∗P<0.05; ∗∗P<0.01; ∗∗∗P<0.001; ∗∗∗∗P<0.0001; ns, not significant. Data are expressed as mean ± SEM. The receiver operating characteristic (ROC) curve analysis were conducted for isoleucine with the area under mass spectrum peak by SPSS Statistics 26, and their predictive abilities were assessed using the AUC. Statistical analysis was performed with GraphPad Prism 8.3.1 (Graphpad Software, USA, RRID: SCR_002798) and SPSS Statistics 26 (IBM, USA, RRID: SCR_016479).
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars of China (82125004; to JPS) and the National Natural Science Foundation of China (81670376; to JPS).
Author contributions
J.S. and P.J. designed and supervised the study; D.S. performed the experiments and wrote the manuscript with input from all authors; Y.W., D.S., Y.C., and H. K. analyzed and interpreted the scRNA-seq data; H.C. and Y.S. helped with mass spectrometry of plasma metabolites; M.T. performed animal experiments. J.S., P.J., and Y.W. revised the manuscript. All authors read and approved the manuscript. Y.W., D.S., and Y.C. contributed equally to this work.
Declaration of interests
The authors have nothing to disclose.
Published: April 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106646.
Contributor Information
Peilin Jia, Email: pjia@big.ac.cn.
Jiangping Song, Email: fwsongjiangping@126.com.
Supplemental information
Data and code availability
-
•
The scRNA-seq processed data reported in this paper have been deposited in the Database: OMIX (https://ngdc.cncb.ac.cn/omix: accession no. OMIX001933), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences https://ngdc.cncb.ac.cn/omix: accession no. OMIX001933and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Go A.S., Hsu C.Y., Yang J., Tan T.C., Zheng S., Ordonez J.D., Liu K.D. Acute kidney injury and risk of heart failure and atherosclerotic events. Clin. J. Am. Soc. Nephrol. 2018;13:833–841. doi: 10.2215/CJN.12591117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronco C., Bellomo R., Kellum J.A. Acute kidney injury. Lancet. 2019;394:1949–1964. doi: 10.1016/S0140-6736(19)32563-2. [DOI] [PubMed] [Google Scholar]
- 3.Yamauchi T., Miyagawa S., Yoshikawa Y., Toda K., Sawa Y., Osaka Cardiovascular Surgery Research OSCAR Group Risk index for postoperative acute kidney injury after valvular surgery using cardiopulmonary bypass. Ann. Thorac. Surg. 2017;104:868–875. doi: 10.1016/j.athoracsur.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Pefanis A., Ierino F.L., Murphy J.M., Cowan P.J. Regulated necrosis in kidney ischemia-reperfusion injury. Kidney Int. 2019;96:291–301. doi: 10.1016/j.kint.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Zuk A., Bonventre J.V. Acute kidney injury. Annu. Rev. Med. 2016;67:293–307. doi: 10.1146/annurev-med-050214-013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabelink T.J., Carmeliet P. Renal metabolism in 2017: glycolytic adaptation and progression of kidney disease. Nat. Rev. Nephrol. 2018;14:75–76. doi: 10.1038/nrneph.2017.173. [DOI] [PubMed] [Google Scholar]
- 7.Wang P., Chen Y., Yong J., Cui Y., Wang R., Wen L., Qiao J., Tang F. Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Rep. 2018;24:3554–3567.e3. doi: 10.1016/j.celrep.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 8.Stewart B.J., Ferdinand J.R., Clatworthy M.R. Using single-cell technologies to map the human immune system - implications for nephrology. Nat. Rev. Nephrol. 2020;16:112–128. doi: 10.1038/s41581-019-0227-3. [DOI] [PubMed] [Google Scholar]
- 9.Chung J.J., Goldstein L., Chen Y.J.J., Lee J., Webster J.D., Roose-Girma M., Paudyal S.C., Modrusan Z., Dey A., Shaw A.S. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J. Am. Soc. Nephrol. 2020;31:2341–2354. doi: 10.1681/ASN.2020020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudman-Melnick V., Adam M., Potter A., Chokshi S.M., Ma Q., Drake K.A., Schuh M.P., Kofron J.M., Devarajan P., Potter S.S. Single-cell profiling of AKI in a murine model reveals novel transcriptional signatures, profibrotic phenotype, and epithelial-to-stromal crosstalk. J. Am. Soc. Nephrol. 2020;31:2793–2814. doi: 10.1681/ASN.2020010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao W., Chen Y., Li Z., Ji J., You A., Jin S., Ma Y., Zhao Y., Wang J., Qu L., et al. Single cell RNA sequencing identifies a unique inflammatory macrophage subset as a druggable target for alleviating acute kidney injury. Adv. Sci. 2022;9:e2103675. doi: 10.1002/advs.202103675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B., Lan S., Dieudé M., Sabo-Vatasescu J.P., Karakeussian-Rimbaud A., Turgeon J., Qi S., Gunaratnam L., Patey N., Hébert M.J. Caspase-3 is a pivotal regulator of microvascular rarefaction and renal fibrosis after ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2018;29:1900–1916. doi: 10.1681/ASN.2017050581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tod P., Bukosza E.N., Róka B., Kaucsár T., Fintha A., Krenács T., Szénási G., Hamar P. Post-ischemic renal fibrosis progression is halted by delayed contralateral nephrectomy: the involvement of macrophage activation. Int. J. Mol. Sci. 2020;21:3825. doi: 10.3390/ijms21113825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichimura T., Bonventre J.V., Bailly V., Wei H., Hession C.A., Cate R.L., Sanicola M. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 1998;273:4135–4142. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 15.Paragas N., Qiu A., Zhang Q., Samstein B., Deng S.X., Schmidt-Ott K.M., Viltard M., Yu W., Forster C.S., Gong G., et al. The Ngal reporter mouse detects the response of the kidney to injury in real time. Nat. Med. 2011;17:216–222. doi: 10.1038/nm.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirita Y., Wu H., Uchimura K., Wilson P.C., Humphreys B.D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. USA. 2020;117:15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilches C., Boiadjieva-Knöpfel E., Bodoy S., Camargo S., López de Heredia M., Prat E., Ormazabal A., Artuch R., Zorzano A., Verrey F., et al. Cooperation of antiporter LAT2/CD98hc with uniporter TAT1 for renal reabsorption of neutral amino acids. J. Am. Soc. Nephrol. 2018;29:1624–1635. doi: 10.1681/ASN.2017111205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajendran A., Poncet N., Oparija-Rogenmozere L., Herzog B., Verrey F. Tissue-specific deletion of mouse basolateral uniporter LAT4 (Slc43a2) reveals its crucial role in small intestine and kidney amino acid transport. J. Physiol. (Oxford, U. K.) 2020;598:5109–5132. doi: 10.1113/JP280234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javed K., Bröer S. Mice lacking the intestinal and renal neutral amino acid transporter SLC6A19 demonstrate the relationship between dietary protein intake and amino acid malabsorption. Nutrients. 2019;11:2024. doi: 10.3390/nu11092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böhmer C., Bröer A., Munzinger M., Kowalczuk S., Rasko J.E.J., Lang F., Bröer S. Characterization of mouse amino acid transporter B0AT1 (slc6a19) Biochem. J. 2005;389:745–751. doi: 10.1042/BJ20050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jando J., Camargo S.M.R., Herzog B., Verrey F. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLoS One. 2017;12:e0184845. doi: 10.1371/journal.pone.0184845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery R.A., Stern J.M., Lonze B.E., Tatapudi V.S., Mangiola M., Wu M., Weldon E., Lawson N., Deterville C., Dieter R.A., et al. Results of two cases of pig-to-human kidney xenotransplantation. N. Engl. J. Med. 2022;386:1889–1898. doi: 10.1056/NEJMoa2120238. [DOI] [PubMed] [Google Scholar]
- 23.Huang J., Bayliss G., Zhuang S. Porcine models of acute kidney injury. Am. J. Physiol. Ren. Physiol. 2021;320:F1030–F1044. doi: 10.1152/ajprenal.00022.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Packialakshmi B., Stewart I.J., Burmeister D.M., Chung K.K., Zhou X. Large animal models for translational research in acute kidney injury. Ren. Fail. 2020;42:1042–1058. doi: 10.1080/0886022X.2020.1830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei J., Song J., Jiang S., Zhang G., Wheeler D., Zhang J., Wang S., Lai E.Y., Wang L., Buggs J., Liu R. Role of intratubular pressure during the ischemic phase in acute kidney injury. Am. J. Physiol. Ren. Physiol. 2017;312:F1158–F1165. doi: 10.1152/ajprenal.00527.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawla L.S. Permissive azotemia during acute kidney injury enables more rapid renal recovery and less renal fibrosis: a hypothesis and clinical development plan. Crit. Care. 2022;26:116. doi: 10.1186/s13054-022-03988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake B.B., Chen S., Hoshi M., Plongthongkum N., Salamon D., Knoten A., Vijayan A., Venkatesh R., Kim E.H., Gao D., et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 2019;10:2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Sullivan E.D., Mylonas K.J., Hughes J., Ferenbach D.A. Complementary roles for single-nucleus and single-cell RNA sequencing in kidney disease research. J. Am. Soc. Nephrol. 2019;30:712–713. doi: 10.1681/ASN.2019020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang H.R., Rabb H. Immune cells in experimental acute kidney injury. Nat. Rev. Nephrol. 2015;11:88–101. doi: 10.1038/nrneph.2014.180. [DOI] [PubMed] [Google Scholar]
- 32.Li Z., Ludwig N., Thomas K., Mersmann S., Lehmann M., Vestweber D., Pittet J.F., Gomez H., Kellum J.A., Rossaint J., Zarbock A. The pathogenesis of ischemia-reperfusion induced acute kidney injury depends on renal neutrophil recruitment whereas sepsis-induced AKI does not. Front. Immunol. 2022;13:843782. doi: 10.3389/fimmu.2022.843782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouschop K.M.A., Roelofs J.J.T.H., Claessen N., da Costa Martins P., Zwaginga J.J., Pals S.T., Weening J.J., Florquin S. Protection against renal ischemia reperfusion injury by CD44 disruption. J. Am. Soc. Nephrol. 2005;16:2034–2043. doi: 10.1681/ASN.2005010054. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Lu S., Li X. The role of metabolic reprogramming in tubular epithelial cells during the progression of acute kidney injury. Cell. Mol. Life Sci. 2021;78:5731–5741. doi: 10.1007/s00018-021-03892-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legouis D., Ricksten S.E., Faivre A., Verissimo T., Gariani K., Verney C., Galichon P., Berchtold L., Feraille E., Fernandez M., et al. Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat. Metab. 2020;2:732–743. doi: 10.1038/s42255-020-0238-1. [DOI] [PubMed] [Google Scholar]
- 36.Basile D.P., Anderson M.D., Sutton T.A. Pathophysiology of acute kidney injury. Compr. Physiol. 2012;2:1303–1353. doi: 10.1002/cphy.c110041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oparija L., Rajendran A., Poncet N., Verrey F. Anticipation of food intake induces phosphorylation switch to regulate basolateral amino acid transporter LAT4 (SLC43A2) function. J. Physiol. 2019;597:521–542. doi: 10.1113/JP276714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makrides V., Camargo S.M.R., Verrey F. Transport of amino acids in the kidney. Compr. Physiol. 2014;4:367–403. doi: 10.1002/cphy.c130028. [DOI] [PubMed] [Google Scholar]
- 39.Seow H.F., Bröer S., Bröer A., Bailey C.G., Potter S.J., Cavanaugh J.A., Rasko J.E.J. Hartnup disorder is caused by mutations in the gene encoding the neutral amino acid transporter SLC6A19. Nat. Genet. 2004;36:1003–1007. doi: 10.1038/ng1406. [DOI] [PubMed] [Google Scholar]
- 40.Kleta R., Romeo E., Ristic Z., Ohura T., Stuart C., Arcos-Burgos M., Dave M.H., Wagner C.A., Camargo S.R.M., Inoue S., et al. Mutations in SLC6A19, encoding B0AT1, cause Hartnup disorder. Nat. Genet. 2004;36:999–1002. doi: 10.1038/ng1405. [DOI] [PubMed] [Google Scholar]
- 41.Bröer A., Juelich T., Vanslambrouck J.M., Tietze N., Solomon P.S., Holst J., Bailey C.G., Rasko J.E.J., Bröer S. Impaired nutrient signaling and body weight control in a Na+ neutral amino acid cotransporter (Slc6a19)-deficient mouse. J. Biol. Chem. 2011;286:26638–26651. doi: 10.1074/jbc.M111.241323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar M.A., Bitla A.R.R., Raju K.V.N., Manohar S.M., Kumar V.S., Narasimha S.R.P.V.L. Branched chain amino acid profile in early chronic kidney disease. Saudi J. Kidney Dis. Transpl. 2012;23:1202–1207. doi: 10.4103/1319-2442.103560. [DOI] [PubMed] [Google Scholar]
- 43.Hong S.Y., Yang D.H., Chang S.K. The relationship between plasma homocysteine and amino acid concentrations in patients with end-stage renal disease. J. Ren. Nutr. 1998;8:34–39. doi: 10.1016/s1051-2276(98)90035-8. [DOI] [PubMed] [Google Scholar]
- 44.Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin. Pract. 2012;120:c179–c184. doi: 10.1159/000339789. [DOI] [PubMed] [Google Scholar]
- 45.Su T., Stanley G., Sinha R., D'Amato G., Das S., Rhee S., Chang A.H., Poduri A., Raftrey B., Dinh T.T., et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018;559:356–362. doi: 10.1038/s41586-018-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui H., Shu S., Li Y., Yan X., Chen X., Chen Z., Hu Y., Chang Y., Hu Z., Wang X., Song J. Plasma metabolites-based prediction in cardiac surgery-associated acute kidney injury. J. Am. Heart Assoc. 2021;10:e021825. doi: 10.1161/JAHA.121.021825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The scRNA-seq processed data reported in this paper have been deposited in the Database: OMIX (https://ngdc.cncb.ac.cn/omix: accession no. OMIX001933), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences https://ngdc.cncb.ac.cn/omix: accession no. OMIX001933and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.