Summary
To detect novel endometrial cancer risk variants, we leveraged information from endometrial cancer risk factors in a multi-trait GWAS analysis. We first assessed causal relationships between established and suspected endometrial cancer risk factors, and endometrial cancer using Mendelian randomization. Following multivariable analysis, five independent risk factors (waist circumference, testosterone levels, sex hormone binding globulin levels, age at menarche, and age at natural menopause) were included in a multi-trait Bayesian GWAS analysis. We identified three potentially novel loci that associate with endometrial cancer risk, one of which (7q22.1) replicated in an independent endometrial cancer GWAS dataset and was genome-wide significant in a meta-analysis. This locus may affect endometrial cancer risk through altered testosterone levels. Consistent with this, we observed colocalization between the signals for endometrial cancer risk and expression of CYP3A7, a gene involved in testosterone metabolism. Thus, our findings suggest opportunities for hormone therapy to prevent or treat endometrial cancer.
Subject areas: Bioinformatics, Systems biology, Cancer, Genomics
Graphical abstract

Highlights
-
•
5 independent endometrial cancer risk factors detected via multivariable analysis
-
•
3 potentially novel endometrial cancer risk loci revealed by multi-trait GWAS
-
•
Validation of novel 7q22.1 risk locus in an endometrial cancer GWAS replication set
-
•
GWAS variants at 7q22.1 linked to CYP3A7 expression and female testosterone levels
Bioinformatics; Systems biology; Cancer; Genomics
Introduction
Endometrial cancer is the fifth most common female cancer worldwide and the most common gynecological tumor in industrialized countries, accounting for over 380,000 new cases and nearly 90,000 deaths in 2018.1 In addition, its prevalence and mortality rate are increasing in both high- and low-income countries.2,3,4
Identification of genetic susceptibility loci lays a foundation for the understanding of cancer etiology. Using case-control genome-wide association study (GWAS) data from multiple studies in the Endometrial Cancer Association Consortium (ECAC), we have identified 16 loci significantly associated with endometrial cancer risk.5,6,7 Together, these risk loci are estimated to explain approximately a quarter of the familial relative risk attributable to common, readily imputable variants,6 indicating a large proportion of common endometrial cancer risk variants are still unidentified.
Endometrial cancer risk is likely influenced by many factors.6,8 A literature review of epidemiological evidence classified reported risk factors with strong, suggestive, and weak evidence of association with endometrial cancer.9 A series of Mendelian randomization studies, which use trait-associated genetic variants to infer causal relationships, have confirmed known, and identified new, endometrial cancer risk factors.10 These include increased body mass index (BMI),6,11,12 early onset menarche,6,13 low sex hormone binding globulin (SHBG), high testosterone,14 increased serum estradiol,15 increased fasting insulin,11 and decreased low- (LDL) and increased high-density lipoprotein (HDL) cholesterol.16
A recently developed multi-trait GWAS approach, the Bayesian GWAS (bGWAS) method,17 uses priors derived from Mendelian randomization analyses of risk factors in a Bayesian framework to identify genetic variants associated with a trait of interest. This approach not only offers an opportunity to identify novel genetic loci associated with a trait, but also highlights risk factors that may mediate their effects through specific loci.
In this study, we used GWAS summary-level data to confirm known and suspected risk factors for endometrial cancer via genetic correlation and Mendelian randomization analyses. We implemented bGWAS to identify new endometrial cancer risk loci and to explore which risk factors may mediate the effects of these loci. Additionally, we conducted subtype analysis to investigate associations with the two primary histological classifications of endometrial cancer: endometrioid and non-endometrioid. Novel loci were assessed for replication in independent endometrial cancer GWAS datasets and functional interpretation provided to elucidate the underlying genetic mechanisms.
Results
Assessment of endometrial cancer risk factors
A total of 34 known and potential risk factors were assessed for association with endometrial cancer, 19 implicated by epidemiological studies and 21 from prior Mendelian randomization publications (Table S1). We firstly assessed the degree of shared genetic architecture between each factor and endometrial cancer risk for all histologies using genetic correlation and found 19 factors that were at least nominally correlated (p < 0.05), with broadly similar results for endometrioid endometrial cancer (Table 1). However, due to the relatively small sample size and the absence of significant heritability, we could not perform correlation analysis for non-endometrioid endometrial cancer. The most significantly correlated factors included obesity-related anthropometric traits such as BMI, weight, waist circumference, and arm fat ratio (Table 1). Our inverse variance weighted (IVW) Mendelian randomization analysis findings were generally supported by the genetic correlation results, particularly for the most significantly correlated factors (Table 1). Furthermore, the IVW results were consistent with the previous Mendelian randomization analyses (Table S1) and additionally provided causal evidence for epidemiological risk factors such as waist circumference, parity and type 1 diabetes (Table 1). As bGWAS uses the IVW method to estimate causal associations, only traits that showed significant causal associations with endometrial cancer risk using this method were included in bGWAS analysis. Using an IVW p value threshold of p < 0.05, 18 risk factors for all endometrial cancer histologies, 16 risk factors for endometrioid endometrial cancer histology and 6 risk factors for non-endometrioid endometrial cancer histologies were considered for inclusion in the bGWAS analysis (Tables 1 and S2).
Table 1.
Assessment of endometrial cancer risk factors in the Endometrial Cancer Association Consortium
| Exposure | Epidemiological strengtha | Mendelian randomization study | Genetic Correlationb |
MR-IVW results |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC |
EEC |
EC |
EEC |
NEEC |
|||||||||||
| RG (SE) | P | RG (SE) | P | nsnp | OR (95% CI) | P | nsnp | OR (95% CI) | P | nsnp | OR (95% CI) | P | |||
| Anthropometric Factors | |||||||||||||||
| BMI | Strong | O'Mara et al., Nead et al., Painter et al., Masuda et al., Prescott et al.6,11,12,18,19 | 0.47 (0.05) | 2.54 × 10−23 | 0.47 (0.06) | 1.26 × 10−15 | 1091 | 1.63 (1.49–1.77) | 1.36 × 10−29 | 1092 | 1.66 (1.51–1.82) | 1.94 × 10−25 | 1085 | 1.46 (1.19–1.79) | 3.11 × 10−04 |
| Height | Highly suggestive | O'Mara et al.6c | 0.07 (0.04) | 0.13 | 0.10 (0.05) | 0.03 | 2535 | 0.99 (0.94–1.04) | 0.60 | 2535 | 0.96 (0.91–1.01) | 0.14 | 2515 | 1.06 (0.94–1.2) | 0.31 |
| Weight (female only) | Highly suggestive | – | 0.53 (0.08) | 1.31 × 10−12 | 0.52 (0.08) | 4.90 × 10−11 | 12 | 1.48 (1.12–1.95) | 5.35 × 10−03 | 12 | 1.59 (1.15–2.19) | 4.46 × 10−03 | 12 | 1.04 (0.59–1.82) | 0.90 |
| Waist-to-hip ratio (female only) | Strong | O'Mara et al., Painter et al.6,12 | 0.19 (0.05) | 4.18 × 10−05 | 0.20 (0.06) | 3.00 × 10−04 | 331 | 1.08 (0.96–1.22) | 0.18 | 332 | 1.07 (0.94–1.21) | 0.30 | 325 | 1.40 (1.05–1.86) | 0.02 |
| Waist circumference | Highly suggestive | – | 0.48 (0.07) | 5.38 × 10−13 | 0.48 (0.07) | 1.42 × 10−11 | 44 | 2.21 (1.79–2.71) | 6.87 × 10−14 | 44 | 2.21 (1.73–2.83) | 2.39 × 10−10 | 44 | 1.59 (0.9–2.79) | 0.11 |
| Arm fat ratio | – | Freuer et al.20 | 0.40 (0.05) | 1.69 × 10−15 | 0.38 (0.06) | 6.50 × 10−10 | 153 | 1.42 (1.24–1.63) | 2.68 × 10−07 | 155 | 1.46 (1.25–1.7) | 1.03 × 10−06 | 155 | 1.01 (0.72–1.42) | 0.96 |
| Reproductive Factors | |||||||||||||||
| Age at menarche | Suggestive | O'Mara et al., Day et al.6,13 | −0.22 (0.04) | 8.98 × 10−07 | −0.21 (0.05) | 8.39 × 10−06 | 255 | 0.84 (0.78–0.9) | 2.73 × 10−06 | 256 | 0.84 (0.77–0.91) | 2.82 × 10−05 | 248 | 0.92 (0.77–1.09) | 0.31 |
| Age at natural menopause | – | O'Mara et al., Ruth et al.6,21 | 0.07 (0.05) | 0.17 | 0.04 (0.05) | 0.41 | 191 | 1.06 (1.03–1.09) | 9.31 × 10−06 | 192 | 1.06 (1.03–1.09) | 6.43 × 10−05 | 191 | 1.04 (0.98–1.1) | 0.22 |
| Number of live births | Strong (Parity) | – | −0.15 (0.07) | 0.04 | −0.15 (0.08) | 0.06 | 5 | 0.57 (0.35–0.93) | 0.02 | 5 | 0.52 (0.3–0.9) | 0.02 | 5 | 0.31 (0.08–1.21) | 0.09 |
| Circulating Factors | |||||||||||||||
| Testosterone levels (females only) | – | Ruth et al., Mullee et al.14,22 | 0.17 (0.06) | 3.20 × 10−03 | 0.19 (0.07) | 3.90 × 10−03 | 247 | 1.45 (1.31–1.61) | 3.92 × 10−13 | 248 | 1.49 (1.33–1.66) | 9.12 × 10−12 | 243 | 1.21 (0.95–1.54) | 0.12 |
| Bioavailable testosterone levels (females only) | – | Ruth et al., Mullee et al.14,22 | 0.39 (0.06) | 2.90 × 10−11 | 0.43 (0.07) | 2.49 × 10−09 | 172 | 1.74 (1.5–2.02) | 5.64 × 10−13 | 173 | 1.74 (1.48–2.05) | 2.85 × 10−11 | 167 | 1.71 (1.22–2.4) | 1.71 × 10−03 |
| Cortisol | – | Larsson et al.23 | −0.34 (0.20) | 0.08 | −0.36 (0.22) | 0.10 | 1 | 1.75 (1.13–2.71) | 0.01 | 1 | 1.96 (1.19–3.21) | 7.73 × 10−03 | 1 | 0.87 (0.26–2.92) | 0.82 |
| Estradiol (females only) | – | Thompson et al.15 | −0.15 (0.20) | 0.45 | −0.15 (0.21) | 0.46 | 2 | 2.17 (1.58–2.98) | 1.87 × 10−06 | 2 | 2.1 (1.47–3.02) | 5.59 × 10−05 | 1 | 3.08 (0.86–10.99) | 0.08 |
| HDL cholesterol | – | Kho et al.16 | −0.30 (0.05) | 4.41 × 10−11 | −0.32 (0.06) | 1.79 × 10−08 | 1189 | 0.92 (0.85–0.98) | 0.02 | 1194 | 0.9 (0.83–0.97) | 7.76 × 10−03 | 1136 | 1.00 (0.84–1.2) | 0.98 |
| LDL cholesterol | – | Kho et al.16 | 0.00 (0.04) | 0.96 | −0.02 (0.05) | 0.65 | 893 | 0.95 (0.88–1.03) | 0.21 | 894 | 0.97 (0.89–1.05) | 0.41 | 849 | 0.77 (0.63–0.94) | 9.48 × 10−03 |
| SHBG levels (females only) | – | Ruth et al.,Mullee et al.14,22 | −0.36 (0.05) | 1.85 × 10−11 | −0.37 (0.06) | 7.88 × 10−09 | 355 | 0.65 (0.56–0.75) | 5.86 × 10−09 | 356 | 0.62 (0.53–0.73) | 6.16 × 10−09 | 341 | 0.54 (0.37–0.8) | 1.97 × 10−03 |
| Tumor necrosis factor alphad | – | Yuan et al.24 | – | – | – | – | 4 | 0.27 (0.05–1.49) | 0.13 | 4 | 0.24 (0.03–1.63) | 0.14 | 4 | 0.18 (0.04–0.71) | 0.01 |
| Plasminogen activator inhibitor-1 | – | Dimou et al.25 | 0.33 (0.19) | 0.09 | 0.42 (0.22) | 0.06 | 2 | 1.01 (0.79–1.3) | 0.91 | 2 | 0.94 (0.71–1.25) | 0.69 | 2 | 1.43 (0.32–6.31) | 0.64 |
| Diabetic Factors | |||||||||||||||
| Type 2 Diabetes | Highly suggestive | Nead et al., Yuan et al.11,26 | 0.33 (0.05) | 1.85 × 10−12 | 0.33 (0.05) | 8.97 × 10−10 | 227 | 1.05 (1.00–1.10) | 0.03 | 228 | 1.05 (1.00–1.11) | 0.07 | 224 | 1.06 (0.95–1.18) | 0.32 |
| Type 1 Diabetes | Suggestive | −0.07 (0.07) | 0.30 | −0.10 (0.08) | 0.20 | 181 | 0.98 (0.96–0.99) | 7.35 × 10−03 | 184 | 0.99 (0.97–1.00) | 0.13 | 174 | 1.01 (0.98–1.04) | 0.56 | |
| Fasting insulin | – | Nead et al., Yuan et al.11,26 | 0.39 (0.11) | 4.00 × 10−04 | 0.38 (0.11) | 8.00 × 10−04 | 13 | 2.97 (1.69–5.21) | 1.50 × 10−04 | 13 | 3.15 (1.62–6.14) | 7.55 × 10−04 | 13 | 3.54 (0.83–15.14) | 0.09 |
| Early insulin secretion | – | Nead et al.11 | 0.04 (0.37) | 0.92 | −0.07 (0.41) | 0.86 | 15 | 1.07 (0.9–1.28) | 0.42 | 15 | 1.06 (0.88–1.28) | 0.55 | 15 | 0.93 (0.61–1.41) | 0.72 |
| Metformin use (ever vs. never) | Suggestive | – | 0.35 (0.07) | 1.32 × 10−07 | 0.43 (0.08) | 4.29 × 10−08 | 42 | 1.00 (0.91–1.09) | 0.96 | 42 | 1.00 (0.91–1.09) | 0.93 | 41 | 1.01 (0.87–1.16) | 0.94 |
| Other diseases | |||||||||||||||
| Hypertensione | Suggestive | – | −0.29 (0.05) | 9.82 × 10−08 | −0.27 (0.07) | 3.26 × 10−05 | 146 | 1.11 (0.87–1.4) | 0.41 | 146 | 1.12 (0.86–1.46) | 0.38 | 146 | 1.12 (0.64–1.95) | 0.70 |
| Uterine fibroids | – | Kho et al.27 | 0.25 (0.09) | 4.10 × 10−03 | 0.18 (0.10) | 0.08 | 22 | 1.18 (1.03–1.37) | 0.02 | 22 | 1.22 (1.04–1.44) | 0.02 | 22 | 1.01 (0.74–1.38) | 0.97 |
| Other Traits | |||||||||||||||
| Leukocyte telomere length | – | Telomeres Mendelian Randomization Collaboration et al.28 | 0.02 (0.06) | 0.71 | 0.01 (0.07) | 0.88 | 189 | 1.27 (1.07–1.52) | 7.01 × 10−03 | 191 | 1.30 (1.07–1.59) | 9.73 × 10−03 | 182 | 1.06 (0.73–1.56) | 0.75 |
| Physical activity (overall physical activity time) | Suggestive | – | −0.27 (0.07) | 4.33 × 10−05 | −0.29 (0.08) | 2.00 × 10−04 | 3 | 0.49 (0.20–1.20) | 0.12 | 3 | 0.58 (0.28–1.22) | 0.15 | 3 | 0.31 (0.03–3.83) | 0.36 |
| Physical activity (sedentary behavior duration) | Suggestive | – | 0.16 (0.07) | 0.01 | 0.15 (0.07) | 0.04 | 3 | 0.83 (0.34–2.05) | 0.68 | 3 | 0.78 (0.31–1.94) | 0.59 | 3 | 1.21 (0.19–7.88) | 0.84 |
| Physical activity (sleep duration) | Suggestive | – | 0.08 (0.06) | 0.17 | 0.09 (0.07) | 0.22 | 10 | 0.88 (0.63–1.22) | 0.44 | 10 | 0.76 (0.52–1.11) | 0.16 | 10 | 0.57 (0.23–1.42) | 0.23 |
| Physical activity (strenuous sports or other exercises: 2–3 vs. 0 days/week) | Suggestive | – | −0.13 (0.06) | 0.02 | 0.09 (0.07) | 0.18 | 15 | 0.70 (0.1–5.03) | 0.72 | 15 | 0.58 (0.07–4.49) | 0.60 | 15 | 15.65 (0.33–746.43) | 0.16 |
| Physical activity (vigorous physical activity: 3 vs. 0 days/week) | Suggestive | – | −0.05 (0.06) | 0.41 | 0.03 (0.07) | 0.69 | 8 | 3.46 (0.45–26.62) | 0.23 | 8 | 2.41 (0.20–28.33) | 0.48 | 8 | 3.59 (0.09–141.23) | 0.50 |
| Smoking status (ever vs. never smokers) | Suggestive | Larsson et al.29c | 0.05 (0.04) | 0.22 | 0.02 (0.05) | 0.60 | 166 | 1.17 (0.93–1.46) | 0.17 | 166 | 1.19 (0.94–1.50) | 0.15 | 166 | 1.15 (0.67–1.96) | 0.61 |
| Caffeine consumptionf | Suggestive | – | – | – | – | – | 2 | 0.62 (0.02–18.44) | 0.78 | 2 | 1.62 (0.01–212.95) | 0.85 | 2 | 0.01 (0.00–141.47) | 0.35 |
| Coffee consumption (cups/day)f | Suggestive | – | −0.06 (0.11) | 0.60 | −0.06 (0.11) | 0.63 | 16 | 0.99 (0.75–1.3) | 0.92 | 16 | 0.93 (0.71–1.22) | 0.60 | 16 | 0.82 (0.52–1.28) | 0.38 |
Findings with p < 0.05 are bolded. Abbreviations - EC: endometrial cancer (all histologies); EEC: endometrioid endometrial cancer; NEEC: non-endometrioid endometrial cancer; OR: odds ratio; CI: confidence interval; nsnp: number of SNPs included in Mendelian randomization analysis; P: p value; RG: genetic correlation; SE: standard error. See also Figures S1 and S2, Tables S1 and S2.
Taken from Raglan et al..9
Genetic correlation could not be assessed for NEEC because its heritability could not be estimated by LD Score Regression.
Not associated with endometrial cancer risk by Mendelian randomization in this publication.
Mendelian randomization analysis were conducted using results reported by Prins30; however, the full GWAS summary statistics was not available. Instead, we tried to estimated its genetic correlation with endometrial cancer using GWAS summary statistics of Ahola-Olli et al.31 but a value could not be assessed because the low heritability for tumor necrosis factor alpha as estimated by LD Score regression.
Genetic correlation with hypertension were calculated using GWAS summary statistics for high blood pressure from Zhu et al.32; whereas genetic variants reported by Surendran et al.33 were used as instrumental variables for hypertension in Mendelian randomization analysis.
Genetic correlation with coffee consumption (cups/day) was estimated using GWAS summary statistics from Coffee Caffeine Genetics Consortium et al.34; whereas genetic correlation with caffeine consumption could not be assessed because full summary statistics were not available for this trait. Mendelian randomization analyses were performed using trait-associated variants as reported by Li et al.35
Identification of independent endometrial cancer risk factors and multi-trait genome-wide association study
Multivariable Mendelian randomization analyses found five risk factors independently displaying a significant effect on endometrial cancer risk of all histologies: testosterone levels, SHBG levels, waist circumference, age at natural menopause, and age at menarche (Figure S1A). Multivariable Mendelian randomization analyses restricting to endometrioid endometrial cancer found bioavailable testosterone (the amount of testosterone not bound by SHBG), BMI, age at natural menopause, and age at menarche to independently affect risk (Figure S1B). This was not substantially different from the analysis of all endometrial cancer histologies as: i) BMI is genetically correlated with waist circumference (RG = 0.89, P-value <1.00 × 10−300); and ii) bioavailable testosterone is genetically correlated with both testosterone and SHBG levels (testosterone RG = 0.64, P-value = 6.86 × 10−72; SHBG RG = −0.75, P-value <1.00 × 10−300) (Figure S2). Multivariable Mendelian randomization analyses restricting to non-endometrioid endometrial cancer identified BMI and LDL cholesterol as two independent risk factors (Figure S1C).
Multi-trait GWAS analyses were conducted by bGWAS using priors constructed from the above endometrial cancer risk factors. The primary bGWAS analysis is based on Bayes factors (BFs), and identifies endometrial cancer susceptibility variants through the comparison of GWAS summary statistics for the risk factors included in the analysis and those for the largest endometrial cancer GWAS to date. We also assessed direct effects from bGWAS analysis, which identifies variants likely to affect endometrial cancer risk directly, or through risk factors not included in the multi-trait GWAS analysis. As a secondary analysis, we considered variants identified by bGWAS of posterior effects, which identifies associations displaying very large prior effects. These are associations that are largely driven by the relationship between the variant and at least one risk factor.
Our primary multi-trait BF GWAS analysis detected four loci associated with endometrial cancer risk (PBF < 5 × 10−8) (Figure 1A and Table 2), three of which are novel risk loci (7q22.1, 11p14.1 and 16q12.2), with the remaining locus (15q15.1) having been previously established through GWAS meta-analysis.10 The risk factors used in the multi-trait analysis were associated with specific loci: testosterone levels were significantly associated with the 7q22.1 and 15q15.1 risk loci; waist circumference and age at menarche were both associated with the 11p14.1 and 16q12.2 risk loci, while SHBG levels were only associated with the 16q12.2 locus (Figure 1B). Similar results were found in multi-trait analyses for endometrioid endometrial cancer, with two of the same loci (7q22.1 and 16q12.2) significantly associating with endometrial cancer risk (Figure S3A and Table 2). No genome-wide significant loci were found in multi-trait analyses for non-endometrioid endometrial cancer (Figure S3C).
Figure 1.
Multi-trait GWAS results and effects of endometrial cancer risk variants on traits included in the GWAS
(A) Manhattan plot of the -log10 p values of the Bayes Factor (x-axis; PBF) for endometrial cancer risk (all histologies) of GWAS variants (chromosmal location shown on x-axis). Novel loci are annotated in red text and known risk loci in black. The red line indicates genome-wide significance at p < 5 × 10−8.
(B) Heatmap of prior effects of endometrial cancer risk variants on risk factors included in the bGWAS analysis. Effect alleles of variants are shown. NA: variant not available for trait assessment, asterisk (∗) indicates variant is significantly associated with risk factor (p < 5 × 10−8).
Table 2.
Genome-wide significant variant associations with risk of endometrial cancer based on Bayes Factors by bGWAS
| Region | rsid | Closest/candidate genes | chr:pos (hg19) | EA/OA | All histologies |
Endometrioid histology |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Z score | μ (SE) | PBF | Z score | μ (SE) | PBF | |||||
| Novel endometrial cancer risk loci | ||||||||||
| 7q22.1 | rs117978821 | ZKSCAN5, CYP3A7 | 7:99107775 | C/T | −4.68 | −1.83 (0.38) | 1.03 × 10−10 | −4.08 | −1.34 (0.24) | 4.20 × 10−9 |
| 11p14.1 | rs962369 | BDNF | 11:27734420 | C/T | 3.94 | 1.10 (0.22) | 4.19 × 10−8 | 2.94 | 0.77 (0.13) | 2.78 × 10−6 |
| 16q12.2 | rs1421085 | FTO | 16:53800954 | C/T | 3.47 | 2.79 (0.47) | 1.54 × 10−9 | 3.42 | 1.80 (0.20) | 6.61 × 10−9 |
| Previously known endometrial cancer risk loci | ||||||||||
| 15q15.1 | rs998713 | EIF2AK4 | 15:40378467 | G/A | 5.71 | 0.99 (0.23) | 1.62 × 10−9 | 4.45 | 0.63 (0.14) | 4.11 × 10−7 |
Abbreviations – EA: effect allele; OA: other allele; Z-score: association estimate; μ: prior effect estimate; SE: standard error of μ; PBF: Bayes Factor p value. See also Figures S3 and S4, and Table S3.
Using direct effects, we detected 13 endometrial cancer risk loci (Pd < 5 × 10−8), all of which have been reported previously by endometrial cancer GWAS analyses and include the 15q15.1 locus (Figure S4 and Table S3). From, the primary multi-trait BF analysis, the 15q15.1 locus likely affects endometrial cancer risk through the effects of testosterone; whilst the other loci either affect endometrial cancer risk directly or through other risk factors not included in the analysis. An imputed singleton variant at 7p14.3 (rs9639594) associated with endometrioid endometrial cancer risk based on direct effects. This variant was identified as a potential endometrioid endometrial cancer risk variant in a previous GWAS analysis; however, as previously described, its association with endometrial cancer needs to be further investigated due to the sparse linkage disequilibrium (LD) at this region.6 In our secondary posterior effects GWAS analysis, we identified 20 potential endometrial cancer risk loci (Pp < 5 × 10−8) (Figures S5A and S5C, and Table S4). These loci had very large prior effects contributed by the risk factors included in the analysis (Figures S5B and S5D), with all of them significantly associated with at least one risk factor.
Replication of novel endometrial cancer susceptibility loci
We attempted to validate the 23 detected loci (three from the primary analysis and 20 from the secondary analysis; p < 2.17 × 10−3 for Bonferroni significance) using a replication set consisting of three publicly available GWAS datasets (UK Biobank, FinnGen, and the Japanese Biobank) and another GWAS dataset from the UK.36 All three novel loci found in our primary multi-trait BF GWAS analysis displayed a concordant direction of effect in this independent replication set, and the 7q22.1 locus replicated with a significant association with endometrial cancer risk (p = 1.33 × 10−3) (Table 3). Meta-analysis of the replication set with the larger ECAC GWAS dataset identified 7q22.1 as a genome-wide significant endometrial cancer risk locus (OR 0.80 95% CI 0.74-0.86; P-value = 3.43 × 10−8) (Figure 2A). There was little evidence for heterogeneity across studies in the meta-analysis (Table 3 and Figure S6), apart from the 5p12 locus (Phet = 0.04, I2 = 60.5%). To investigate the possibility that the inclusion of the Japanese Biobank could affect replication, we also performed a meta-analysis removing this stratum with very similar results (Table S5). From the 20 potential loci identified from the secondary posterior effects analysis, 75% of these (15/20 loci) displayed a concordant direction of effect in the independent replication set, three of which were nominally significant (p < 0.05): 2p25.3, 11q13.1 and 12p12.1.
Table 3.
Replication of endometrial cancer risk variants identified by multi-trait GWAS analyses
| Region | Rsid | chr:pos (hg19) | EA/OA | EAF | ECAC GWAS (minus UKBB) |
Replication Set |
ECAC GWAS + Replication Set |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | Phet | I2 | OR (95% CI) | P-value | Phet | I2 | |||||
| BF analysis | ||||||||||||||
| 7q22.1 | rs117978821 | 7:99107775 | C/T | 0.03 | 0.79 (0.72–0.87) | 2.82 × 10−6 | 0.80 (0.7–0.92) | 1.33 × 10−03 | 0.65 | 0 | 0.8 (0.74–0.86) | 3.43 × 10−08 | 0.83 | 0.00 |
| 11p14.1 | rs962369 | 11:27734420 | C/T | 0.30 | 1.07 (1.03–1.11) | 8.25 × 10−5 | 1.03 (0.98–1.08) | 0.30 | 0.80 | 0 | 1.06 (1.03–1.09) | 2.23 × 10−4 | 0.61 | 0.00 |
| 16q12.2 | rs62033406 | 16:53800954 | C/T | 0.43 | 1.06 (1.03–1.09) | 3.02 × 10−4 | 1.01 (0.97–1.06) | 0.59 | 0.36 | 6.9 | 1.04 (1.01–1.07) | 2.08 × 10−3 | 0.20 | 33.70 |
| Posterior analysis | ||||||||||||||
| 1p34.3 | rs61779310 | 1:39942297 | G/C | 0.24 | 1.04 (1.01–1.08) | 0.02 | 1.00 (0.95–1.06) | 0.87 | 0.68 | 0.00 | 1.03 (1.00–1.06) | 0.06 | 0.57 | 0.00 |
| 1q32.1 | rs2820292 | 1:201784287 | C/A | 0.56 | 1.04 (1.00–1.07) | 0.03 | 1.01 (0.96–1.05) | 0.82 | 0.14 | 45.90 | 1.02 (1.00–1.05) | 0.07 | 0.15 | 40.30 |
| 2p25.3 | rs2867131 | 2:610603 | C/T | 0.83 | 1.04 (1.00–1.09) | 0.05 | 1.08 (1.01–1.14) | 0.02 | 0.74 | 0.00 | 1.05 (1.02–1.09) | 3.22 × 10−3 | 0.74 | 0.00 |
| 2q13 | rs6750599 | 2:111893869 | T/A | 0.55 | 1.04 (1.01–1.07) | 0.02 | 1.02 (0.97–1.06) | 0.44 | 0.50 | 0.00 | 1.03 (1.00–1.06) | 0.03 | 0.58 | 0.00 |
| 2q22.2 | rs6747717 | 2:144013526 | T/A | 0.17 | 1.06 (1.02–1.11) | 4.17 × 10−3 | 1.01 (0.94–1.08) | 0.84 | 0.77 | 0.00 | 1.05 (1.01–1.08) | 0.02 | 0.51 | 0.00 |
| 4p12 | rs10938397 | 4:45182527 | G/A | 0.43 | 1.03 (1.00–1.06) | 0.07 | 0.98 (0.94–1.02) | 0.34 | 0.74 | 0.00 | 1.01 (0.99–1.04) | 0.41 | 0.34 | 12.30 |
| 4p11 | rs2768950 | 4:49064487 | G/A | 0.73 | 0.96 (0.93–1.00) | 0.04 | 0.99 (0.95–1.04) | 0.80 | 0.64 | 0.00 | 1.02 (0.99–1.05) | 0.18 | 0.45 | 0.00 |
| 4q12 | rs6856974 | 4:52728324 | T/G | 0.26 | 1.04 (1.00–1.08) | 0.03 | 0.99 (0.95–1.04) | 0.82 | 0.71 | 0.00 | 1.02 (0.99–1.05) | 0.15 | 0.48 | 0.00 |
| 5p12 | rs782971 | 5:43124688 | G/A | 0.26 | 1.05 (1.01–1.09) | 0.01 | 0.97 (0.92–1.01) | 0.17 | 0.33 | 13.00 | 1.02 (0.99–1.05) | 0.31 | 0.04 | 60.50 |
| 5q21.3 | rs40071 | 5:107496102 | C/T | 0.18 | 0.96 (0.92–1.00) | 0.05 | 1.02 (0.97–1.07) | 0.51 | 0.44 | 0.00 | 0.98 (0.95–1.02) | 0.31 | 0.22 | 30.40 |
| 6p21.33 | rs1265097 | 6:31106459 | A/C | 0.09 | 1.07 (1.01–1.12) | 0.01 | 1.01 (0.94–1.09) | 0.70 | 0.74 | 0.00 | 1.05 (1.00–1.09) | 0.03 | 0.64 | 0.00 |
| 11q13.1 | rs2276014 | 11:64081445 | A/G | 0.15 | 0.94 (0.90–0.98) | 4.66 × 10−3 | 0.94 (0.88–0.99) | 0.03 | 0.39 | 0.90 | 0.94 (0.9–0.97) | 4.86 × 10−4 | 0.55 | 0.00 |
| 12p12.1 | rs2900478 | 12:21368797 | A/T | 0.17 | 1.08 (1.04–1.12) | 2.56 × 10−4 | 1.08 (1.02–1.14) | 0.01 | 0.25 | 27.60 | 1.08 (1.04–1.11) | 1.34 × 10−5 | 0.39 | 3.50 |
| 14q31.1 | rs7141420 | 14:79899454 | T/C | 0.53 | 1.05 (1.02–1.09) | 1.55 × 10−3 | 1.03 (0.98–1.07) | 0.21 | 0.17 | 40.20 | 1.04 (1.02–1.07) | 1.62 × 10−3 | 0.21 | 31.40 |
| 15q23 | rs4776970 | 15:68080886 | T/A | 0.36 | 0.94 (0.91–0.98) | 5.23 × 10−4 | 0.96 (0.91–1.00) | 0.05 | 0.12 | 48.60 | 0.95 (0.92–0.97) | 1.33 × 10−4 | 0.20 | 33.60 |
| 16p12.3 | rs4782289 | 16:19859332 | A/G | 0.14 | 0.93 (0.88–0.97) | 9.38 × 10−4 | 1.02 (0.95–1.10) | 0.52 | 0.44 | 0.00 | 0.95 (0.92–0.99) | 0.02 | 0.07 | 56.60 |
| 16p11.2 | rs12446550 | 16:28543381 | A/G | 0.43 | 1.05 (1.02–1.08) | 3.86 × 10−3 | 1.04 (1.00–1.09) | 0.08 | 0.83 | 0.00 | 1.05 (1.02–1.07) | 1.03 × 10−3 | 0.92 | 0.00 |
| 18q22.3 | rs17230390 | 18:71935195 | A/G | 0.01 | 1.22 (1.07–1.38) | 2.19 × 10−3 | 1.01 (0.82–1.24) | 0.93 | 0.51 | 0.00 | 1.15 (1.03–1.29) | 0.01 | 0.27 | 23.90 |
| 19q12.32 | rs429358 | 19:45411941 | C/T | 0.14 | 0.94 (0.89–0.98) | 4.86 × 10−3 | 0.97 (0.91–1.03) | 0.26 | 0.96 | 0.00 | 0.95 (0.91–0.98) | 5.01 × 10−3 | 0.92 | 0.00 |
| 22q13.1 | rs4820408 | 22:40604945 | G/T | 0.60 | 0.96 (0.93–0.99) | 0.01 | 0.98 (0.94–1.03) | 0.55 | 0.33 | 11.00 | 0.97 (0.94–0.99) | 0.01 | 0.38 | 1.80 |
Abbreviations – EA: effect allele; OA: other allele; OR: odds ratio; CI: confidence interval; Phet: heterogeneity p value; I2: heterogeneity estimate; BF: Bayes factor; UKBB: UK Biobank. Variants with a nominal significance (P-value <0.05) in the replication set are noted in italics; genome-wide significant variants (P-value < 5 x 10−8) are in bold. See also Figures S5 and S6, Tables S4 and S5.
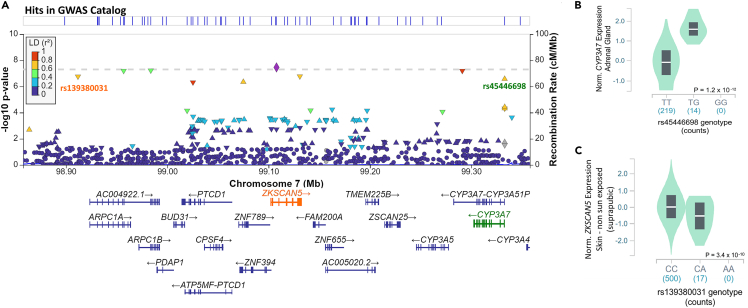
Figure 2.
Endometrial cancer risk locus at 7q22.1 identified by multi-trait BF GWAS
(A–C) Regional association plot for 7q22.1. Genetic variants at each locus are plotted by their genomic position (hg19; x-axis) and GWAS P-value -log10(P) for association with endometrial cancer risk from meta-analysis of all endometrial cancer GWAS datasets on the left y-axis. Recombination rate (cM/Mb) is on the right y axis and plotted as blue lines. The color of the circles indicates the level of linkage disequilibrium between each variant and the lead variant (purple diamond) from the 1000 Genomes 2014 EUR reference panel (see legend, inset). Expression quantitative trait loci variants (eQTLs) are labeled with associated genes highlighted in the same color. Violin plots of expression by genotype for eQTLs are provided for (B) rs45446698 and CYP3A7 (adrenal gland) and (C) rs1139380031 and ZKSCAN5 (skin not sun exposed – suprapubic). See also Tables S6–S8.
Functional analyses of novel endometrial cancer susceptibility loci
We assessed quantitative trait loci (QTL) data from the Genotype-Tissue Expression (GTEx v8) project to determine whether candidate causal risk variants at 7q22.1 (determined by fine-mapping analysis, see STAR Methods; Table S6) also associated with gene expression or splicing. We found two candidate casual risk variants to be lead expression quantitative trait loci (eQTLs), associated with expression of ZKSCAN5 in suprapublic skin not sun exposed (rs139380031) and CYP3A7 in the adrenal gland and visceral adipose (rs44546698) (Figures 2A–2C and Table S7). Analysis of the 7q22.1 locus provided evidence for the colocalization of endometrial cancer risk and the eQTL signals: ZKSCAN5 (suprapubic skin not sun-exposed, posterior probability for colocalization PPH4 = 0.81) and CYP3A7 (adrenal gland PPH4 = 0.8 and visceral adipose PPH4 = 0.82) (Table S7). Relevant to the association with testosterone levels at this locus, CYP3A7 encodes an enzyme (cytochrome P450 family 3, subfamily A, polypeptide 7) that metabolizes testosterone37 and we observed that the risk alleles associated with lower CYP3A7 expression in the GTEx tissues. The lead expression QTL for CYP3A7 in adrenal gland and visceral adipose (candidate causal variant rs45446698), is located only 128 bp from the summit of a CYP3A7 promoter that has activity in adipocytes38 (promoter p1@CYP3A7; http://slidebase.binf.ku.dk/human_promoters/). In silico analysis predicted that this variant modifies several transcription factor binding motifs (Table S8). These include motifs created by the risk allele that are bound by transcriptional repressors (MEIS1, FOXP1, and FOXD3) and whose expression is positively correlated with CYP3A7 expression, in the adrenal gland or visceral adipose, providing a potential cis-regulatory mechanism for the QTL (Table S8).
Discussion
Using multivariable Mendelian randomization and multi-trait GWAS approaches we have identified five independent risk factors and three potentially novel risk loci for endometrial cancer. In an independent dataset, one of these loci, 7q22.1 replicated its association with endometrial cancer risk and demonstrated genome-wide significance in a subsequent meta-analysis, establishing it as a new risk locus. Secondary analysis using multi-trait posterior effects GWAS analysis identified another 20 potential endometrial cancer risk loci, three of which had nominally significant associations in the independent dataset, highlighting the need for further investigation in larger endometrial cancer datasets.
We used IVW Mendelian randomization and multivariable Mendelian randomization analyses to screen suggested endometrial cancer risk factors for inclusion in a multi-trait GWAS analysis. For endometrial cancer all histologies, these analyses ultimately resulted in the selection of five independent risk factors (testosterone levels, SHBG levels, waist circumference, age at natural menopause, and age at menarche). All these risk factors, except for age at natural menopause, demonstrated some degree of genetic correlation with endometrial cancer risk. However, it should be noted that the genetic correlation was measured as an average across the entire genome, whereas the Mendelian randomization analysis assessed a minor component of genetic variation. Thus, a null finding for genetic correlation between traits does not preclude the presence of local genetic correlation or a causal effect of one trait on another. Similar risk factors were selected for multi-trait GWAS analysis of endometrioid endometrial cancer. For non-endometrioid endometrial cancer analysis, BMI and LDL-cholesterol were selected. All selected risk factors had previously been reported as affecting endometrial cancer risk by Mendelian randomization approaches, apart from waist circumference, which had been found to be highly suggestive for endometrial cancer risk by epidemiological studies.9
The novel 7q22.1 risk locus harbors a gene (CYP3A7) encoding an enzyme that metabolizes testosterone. Estrogen, which plays a crucial role in endometrial carcinogenesis,39 is synthesized from testosterone; thus, the perturbation of testosterone metabolic pathways may affect endometrial cancer risk. In this study, we show that risk alleles at the 7q22.1 endometrial cancer risk signal are associated with decreased expression of CYP3A7 in biologically relevant tissues. Furthermore, this same signal has been associated with increased total and bioavailable testosterone in women14 and increased estradiol levels in men.14 Hence, we propose that CYP3A7 is the likely causal gene at 7q22.1 and the risk signal mediates its effect by reducing CYP3A7 expression, resulting in increased sex-hormone levels. Analyses at 7q22.1 highlighted candidate causal variant rs45446698 as a likely functional variant whose risk allele may reduce CYP3A7 expression by the generation of motifs bound by transcriptional repressors. Consistent with this effect, two repressors (MEIS1 and FOXP1) also inhibit testosterone signaling through effects on the androgen receptor.40,41
The findings from the Mendelian randomization and GWAS analyses support therapeutic strategies to prevent or treat endometrial cancer (e.g. weight loss or inhibition of testosterone signaling). Weight loss, in particular that induced by bariatric surgery, has been shown to reduce endometrial cancer risk by up to 80% (reviewed by Njoku et al.42). However, there have been limited studies of the effect of weight loss on endometrial cancer survival and the few studies to date have been small and inconclusive.43 Although there are a number of medications available to inhibit testosterone signaling, through a variety of mechanisms (reviewed in Crawford et al.44), there have been very few studies that have intentionally targeted testosterone in endometrial cancer. However, an androgen receptor inhibitor that blocks testosterone signaling (enzalutamide) has been reported to slow the proliferation of primary endometrial tumor cells45 and is currently being studied in combination with chemotherapy in a phase II trial of endometrial cancer (ClinicalTrials.gov Identifier: NCT02684227). Despite testosterone having been identified as a therapeutic target in endometrial cancer, it is not clear if testosterone exerts its effects simply as a precursor to estrogen or through other pathways such as androgen receptor signaling (reviewed by Gibson et al.46) Further studies are needed in this area to determine the effects of testosterone on endometrial cancer development and its potential for therapeutic targeting.
In conclusion, we have used genetic approaches to comprehensively assess reported risk factors for endometrial cancer. We were then able to leverage risk factor genetic information in a multi-trait GWAS analysis to identify novel endometrial cancer risk loci. Multivariable Mendelian randomization analysis highlighted the importance of androgens in endometrial cancer development, with all the identified independent risk factors having some relationship with testosterone. Indeed, the novel locus identified by multi-trait GWAS analysis is strongly associated with testosterone levels in women. The findings of this study suggest opportunities for androgen suppression therapy to reduce the risk of endometrial cancer development.
Limitations of the study
This study demonstrates the strength of using the largest endometrial cancer GWAS dataset available and leveraging genetic information from risk factors to identify new susceptibility loci for endometrial cancer. To maximize statistical power, where available, we preferentially selected sex-combined GWAS summary statistics for inclusion, which could mask sex-specific effects. We attempted to overcome this by using female-specific summary statistics for traits that have been reported to have strong sex heterogeneity in GWAS effects. However, we acknowledge that results could be confounded by subtle or unreported sex-specific effects. Only one of the potentially novel loci was statistically validated in an independent endometrial cancer GWAS dataset, indicating the need for larger replication datasets to verify findings. All studies included in the discovery ECAC GWAS and the datasets included in the multi-trait bGWAS analysis were of European ancestry. Thus, it is not known if these findings are applicable to other ethnic groups. Furthermore, as samples from several of the studies in the replication dataset did not include the histology information on endometrial cancer cases (i.e. UK Biobank, FinnGen, and Japanese Biobank), we were unable to validate the identified risk loci associated with endometrioid endometrial cancer. We were also unable to fully assess non-endometrioid endometrial cancer in our study. Due to the small number of cases in this subset (n = 1,230),6 we could not quantify the heritability of non-endometrioid endometrial cancer and therefore could not run genetic correlation analyses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| GWAS summary statistics for enodmetrial cancer and risk factors | GWAS Catlog | https://www.ebi.ac.uk/gwas/; accession numbers: GCST006464 (endometrial cancer), GCST007293 (arm fat ratio), GCST006047 (no. live births), GCST90012112 (testosterone), GCST90012102 (bioavailable testosterone), GCST90020092 (estradiol), GCST90012107 (SHBG), GCST004426 (TNFa), GCST90010422 (PAI1), GCST90014023 (T1D), GCST007610 (hypertension), GCST009158 (uterine fibroids), GCST006912, GCST006913, GCST006914, GCST006100, GCST006098 (physical activity), GCST007327 (smoking status) |
| FinnGen endometrial cancer GWAS summary statistics (C3_CORPUS_UTERI) | FinnGen Data freeze 6 | http://r6.finngen.fi |
| Japanese Biobank endometrial cancer GWAS summary statistics | Sakaue et al.47 | https://pheweb.jp |
| UK Biobank case and control genotypes; approved application no: 25331 | UK Biobank | https://www.ukbiobank.ac.uk/ |
| GWAS summary statistics for endometrial cancer risk factors: BMI and WHC | Pulit et al.48 | https://zenodo.org/record/1251813/#.Ys4dcnZBw2w |
| GWAS summary statistics for endometrial cancer risk factors: height, weight | GIANT consortium | https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files |
| GWAS summary statistics for endometrial cancer risk factors: age at menarche, age at menopause | Reprogen consortium | https://www.reprogen.org/data_download.html |
| GWAS summary statistics for endometrial cancer risk factors: fasting insulin, early insulin secretion | MAGIC consortium | https://magicinvestigators.org/downloads/ |
| GWAS summary statistics for endometrial cancer risk factors: HDL and LDL cholesterol | GLGC consortium | http://csg.sph.umich.edu/willer/public/glgc-lipids2021/results/ancestry_specific/ |
| GWAS summary statistics for endometrial cancer risk factors: T2D | DIAGRAM consortium | http://diagram-consortium.org/downloads.html |
| Multi-trait GWAS of endometrial cancer (bGWAS: Bayes Factor, posterior and direct summary statistics) | This paper | Zenodo: https://doi.org/10.5281/zenodo.7545994 |
| eQTL and sQTL data | GTEx consortium v849 | GTExportal.org |
| Software and algorithms | ||
| LDSC | https://github.com/bulik/ldsc | |
| PLINK v1.90 | http://pngu.mgh.harvard.edu/purcell/plink/ | |
| TwoSampleMR | https://mrcieu.github.io/TwoSampleMR/ | |
| bGWAS | https://github.com/n-mounier/bGWAS | |
| METAL release 2020-05-05 | http://csg.sph.umich.edu/abecasis/Metal/ | |
| R meta package | https://cran.r-project.org/web/packages/meta/index.html | |
| SuSiE R package | https://stephenslab.github.io/susieR/index.html | |
| LocusFocus | https://locusfocus.research.sickkids.ca/ | |
| TOMTOM | https://meme-suite.org/meme/tools/tomtom | |
| GEPIA | http://gepia2.cancer-pku.cn/#index | |
| FANTOM5 promoters | https://slidebase.binf.ku.dk/human_promoters/ | |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Tracy O’Mara (Tracy.OMara@qimrberghofer.edu.au).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
GWAS summary statistics
GWAS summary statistics of endometrial cancer risk were from the latest ECAC GWAS analysis (12,906 cases and 108,979 controls).6 To avoid bias due to overlapping sample sets, UK Biobank samples were removed from the ECAC summary statistics for Mendelian randomization and bGWAS analyses, resulting in 12,270 endometrial cancer cases and 46,126 controls. Subtype-specific analyses were also performed using GWAS summary statistics restricted to endometrioid endometrial cancer histology only (8,758 cases and 46,126 controls).
Endometrial cancer known and suspected risk factors were identified from an umbrella review of the literature.9 Endometrial cancer risk factors that were classified as “strong”, “highly suggestive” or “suggestive” and had a published GWAS available were included in our analysis. We also included factors which were previously identified as associated with endometrial cancer risk by Mendelian randomization analysis (until October 2021; reviewed in Wang et al.10). GWAS summary statistics for these risk factors were sought from publicly available resources, including the GWAS Catalog (https://www.ebi.ac.uk/gwas/), MAGIC (https://magicinvestigators.org/), GLGC (http://lipidgenetics.org/), and the ReproGen Consortium (https://www.reprogen.org). GWAS summary statistics of sex-combined cohorts were preferentially selected for inclusion, unless it was a female-specific trait or strong evidence of sex-heterogeneity in genetic effects reported. Female-specific traits are: age at menarche, age at natural menopause, number of live births and uterine fibroids. Traits reported to have strong sex-heterogeneity are: waist-hip ratio, arm fat ratio, and sex-steroid hormones (testosterone, bioavailable testosterone, SHBG and estradiol).14,48,50 Although weight has not been reported to have significant sex-heterogeneity in genetic effects,51 only sex-specific GWAS summary statistics were available for this trait. Further information for trait GWAS summary statistics used in this study is provided in Table S1.
To construct a replication set and validate our findings from the bGWAS analysis, we downloaded publicly available endometrial cancer GWAS summary statistics which were not included in the ECAC GWAS data.6 These were from the Finnish biobank study, FinnGen (data freeze 6, 1,430 cases and 116,981 controls; http://r6.finngen.fi/), and from the Japanese Biobank study (1,200 cases and 60,614 controls; https://pheweb.jp/).47 Endometrial cancer GWAS summary statistics from the Manchester cohort (UK) were provided by collaboration (560 cases and 1,202 controls).36 Since the UK Biobank endometrial cancer case-control samples were excluded from the bGWAS analysis, we additionally included GWAS summary statistics from an analysis of UK Biobank samples in our replication set (1,866 cases and 18,660 controls).27
Quantification and statistical analysis
All statistical analyses using R packages were performed using R 4.2.1, unless otherwise stated. Information on specific statistical analyses are described below.
Genetic correlation
Genetic correlation between endometrial cancer risk and available traits were estimated using publicly available GWAS summary statistics and LD score regression (LDSC version 1.0.1).52 Nominally significant correlations (p < 0.05) were identified (Figure S2).
Mendelian randomization analysis
Traits were assessed for endometrial cancer causality using two-sample Mendelian randomization analyses. Independent genetic variants robustly associated with individual traits (p < 5 × 10−8) were selected as instrumental variables using the clump method implemented in PLINK v1.9053 with arguments --clump-r2 0.01 and --clump-kb 500. Instrument variables for early insulin secretion used the same variants as selected by Nead et al.11; briefly, 17 genetic variants associated with insulin levels at 30 min during oral glucose tolerance test that showed the expected directions of effect were selected from Scott et al.54 whereas variant effects used that reported by Prokopenko et al.55 For hypertension, as variant effect and the associated standard error were not provided by Surendran et al.,33 standard error was calculated as 1/sqrt(N ∗ 2 ∗ maf ∗ (1-maf)), where N is the sample size and maf is the effect allele frequency; variant effect was calculated as standard error ∗ z score. A/T or C/G allelic variants with minor allele frequency larger than 0.42 were excluded from analyses due to the ambiguity into the identity of the effect allele in the exposure and outcome.56 Mendelian randomization analyses were performed using the “TwoSampleMR” R package (version 0.5.5).56
Bayesian genome-wide association study (bGWAS)
Multi-trait GWAS analysis of endometrial cancer was conducted using the bGWAS framework implemented in R17. Factors associated with endometrial cancer risk by univariate Mendelian randomization (p < 0.05) were included in the bGWAS analysis.
The bGWAS method involves two main steps: (1) identification of independent risk factors that jointly affect endometrial cancer risk; and (2) determination of variant-trait associations by multi-trait GWAS using risk factors identified in step 1. Specifically, in the first step, univariate regression analyses were conducted to identify endometrial cancer risk factors using instrumental variables associated with traits (Bonferroni-adjusted p < 0.05). Instrumental variables were considered independent if their physical distances were over 500 kb. Risk factors identified by univariate regression analyses were subsequently included in a multivariable Mendelian randomization analysis to determine independent risk factors of endometrial cancer via a stepwise selection procedure. In the second step, prior effects of individual variants were estimated as the sum of the products of the variant effects on individual risk factors and the causal effects of individual risk factors on endometrial cancer risk. Bayes factors (BFs) and the corresponding p value (PBF) were calculated. Significant associations identified based on BFs represent variants exerting their effects on endometrial cancer risk through risk factors included in the analysis. Direct and posterior effects and their corresponding p values were also estimated. Variants showing a significant association by direct effects are likely to affect endometrial cancer risk directly or through other risk factors not included in the analysis; whereas variants displaying large prior effects, driven by one or more risk factors, could show significant posterior effects. A genome-wide significance threshold (p < 5 × 10−8; Figures 1; S3 and S4) was used to identify significant variant-trait associations. We highlighted potential novel endometrial cancer risk loci if a genome-wide significant signal was observed more than 500 kb from a known endometrial cancer risk locus.
To identify potential subtype-specific loci, we performed an additional bGWAS analysis using the endometrioid endometrial cancer or non-endometrioid endometrial cancer risk GWAS summary statistics.
To minimize false positives in the detection of loci based on posterior effects, following bGWAS analysis, the following criteria were applied to restrict output variants: (1) concordant direction of effect on endometrial cancer risk in this study and in the most recent ECAC GWAS,6 and (2) a nominally significant association (p < 0.05) with endometrial cancer risk in the most recent ECAC GWAS.6
Endometrial cancer GWAS replication
Summary statistics for each replication study set (i.e. FinnGen, Japanese Biobank, Manchester and UK Biobank) were harmonized to the same genomic build (hg19) and variants with low minor allele frequency (<1%) or low imputation quality (R2 < 0.3) were removed. The summary statistics from the four sets were combined in a fixed-effects, inverse-variance weighted meta-analysis by METAL57 (version release 2020-05-05), adjusting each set for genomic control. The I2 and Cochran’s Q statistics as output by METAL were used to assess for heterogeneity in associations across the study sets. We additionally generated forest plots using the ‘meta’ package58 in R 3.4.1 to visually assess heterogeneity.
Quantitative trait loci analysis
We further investigated loci which replicated in the independent GWAS dataset. Candidate causal endometrial cancer risk variants were identified from fine-mapping of the 7q22.1 locus using the meta-analysis of the ECAC and independent GWAS dataset. Fine-mapping was performed using an iterative Bayesian stepwise procedure using the summary statistics version of the Sum of Single Effects (SuSiE) v0.11.42 R package59,60 with default settings. Variants located within ±1 Mb of the lead variant were used in the analysis and LD matrices were calculated based on the UKB10K reference panel (a random subset of 10,000 unrelated participants from the UK Biobank cohort) from Kho et al.27 To assess candidate causal risk variants for effects on splicing, the GTEx database (v8)49 was accessed to determine if any variants were lead splicing quantitative trait loci (sQTLs) in available tissues. For eQTL analysis, candidate causal risk variants were interrogated with the Qtlizer tool61 by selecting the variants that were lead (i.e. ‘best’ according to Qtlizer) expression QTLs in GTEx (v8) tissues.49 Signals for identified lead eQTLs were then assessed for colocalization with the endometrial cancer GWAS risk signal using COLOC2 by LocusFocus.62 A H4 posterior probability (PPH4) > 0.8 was considered evidence that the endometrial cancer GWAS risk and relevant gene-tissue eQTL signals are explained by the same genetic variant (i.e. colocalized). To identify transcription factor binding sites, 10 bases flanking the variants of interest (effect allele or other allele) were taken to predict allele specific changes to transcription factor binding. Sequence based predictions were performed by TOMTOM63 (MEMESuitev5.3.3, mapped with the HOCOMOCOv11_full_HUMAN database). Spearman correlations between transcription factor gene expression and CYP3A7 were performed in GTEx tissues using GEPIA2.64
Acknowledgments
This work was supported by a National Health and Medical Research Council (NHMRC) of Australia Investigator Grant (APP1173170), a Cancer Australia PdCCRS Project Grant, funded by Cure Cancer Australia and the Compton Foundation (#1138084) awarded to T.A.O’M, an NHMRC Project Grant (APP1158083) awarded to T.A.O’M and D.M.G, and a project grant awarded to T.A.O’M co-funded by Worldwide Cancer Research and Cancer Australia (grant number 22-0253). T.A.O’M and A.B.S. are supported by NHMRC Investigator Fellowships (APP1173170 and APP1177524). P.F.K. was supported by an Australian Government Research Training Program PhD Scholarship and QIMR Berghofer Postgraduate Top-Up Scholarship. E.J.C. is supported by a National Institute for Health Research (NIHR) Advanced Fellowship (NIHR300650). E.J.C., C.B., and D.G.E. were supported by the NIHR Manchester Biomedical Research Centre (IS-BRC-1215-20007). D.R. and T.D. were supported by the Wilhelm Sander Foundation (2021.142.1)
This work was conducted using the UK Biobank Resource (application number 25331). We thank the participants and investigators of the FinnGen study and the Biobank Japan Project. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the U.S. National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this article were obtained from the GTEx Portal in February 2021.
We thank the many individuals who participated in the Endometrial Cancer Association Consortium and the numerous institutions and their staff who supported recruitment. We particularly thank the efforts of Deborah Thompson. The endometrial cancer genome-wide association analyses were supported by the National Health and Medical Research Council of Australia (APP552402, APP1031333, APP1109286, APP1111246, and APP1061779), the U.S. National Institutes of Health (R01-CA134958), European Research Council (EU FP7 Grant), Wellcome Trust Centre for Human Genetics (090532/Z/09Z) and Cancer Research UK. OncoArray genotyping of ECAC cases was performed with the generous assistance of the Ovarian Cancer Association Consortium (OCAC), which was funded through grants from the U.S. National Institutes of Health (CA1X01HG007491-01 (C.I. Amos), U19-CA148112 (T.A. Sellers), R01-CA149429 (C.M. Phelan) and R01-CA058598 (M.T. Goodman); Canadian Institutes of Health Research (MOP-86727 (L.E. Kelemen)) and the Ovarian Cancer Research Fund (A. Berchuck). We particularly thank the efforts of Cathy Phelan. OncoArray genotyping of the BCAC controls was funded by Genome Canada Grant GPH-129344, U.S. National Institutes of Health Grant U19 CA148065, and Cancer UK Grant C1287/A16563. All studies and funders are listed in O’Mara et al. (2018). Full acknowledgments and funding for ECAC can be found in the Supplementary Note.
Author contributions
Conceptualization, T.A.O, X.W., and D.M.G. and.; methodology, X.W., D.M.G., and T.A.O.; investigation, X.W., D.M.G., T.A.O., P.F.K., D.R., and T.D.; data curation, X.W., D.G.M., T.A.O; writing – original draft, X.W., DMG, and TAO; writing – review & editing, all authors; funding acquisition, T.A.O.; resources, T.A.O., D.M.G., T.D., A.B.S, C.B., F.A., E.L.G., R.J.S., I.T., D.G.E., and E.J.C.; supervision, T.O.M.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106590.
Supplemental information
Data and code availability
-
•
Summary statistics from the bGWAS multi-trait GWAS analysis have been deposited at Zenodo, to be made publicly available by publication (https://doi.org/10.5281/zenodo.7545994). This paper analyzes existing, publicly available data. Accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyse data reported in this paper is available from the lead author upon request.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lortet-Tieulent J., Ferlay J., Bray F., Jemal A. International patterns and trends in endometrial cancer incidence, 1978-2013. J. Natl. Cancer Inst. 2018;110:354–361. doi: 10.1093/jnci/djx214. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA. Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S., Sun K., Zheng R., Zeng H., Wang S., Chen R., Wei W., He J. Cancer incidence and mortality in China, 2015. Journal of the National Cancer Center. 2021;1:2–11. doi: 10.1016/j.jncc.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng T.H., Thompson D.J., O'Mara T.A., Painter J.N., Glubb D.M., Flach S., Lewis A., French J.D., Freeman-Mills L., Church D., et al. Five endometrial cancer risk loci identified through genome-wide association analysis. Nat. Genet. 2016;48:667–674. doi: 10.1038/ng.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Mara T.A., Glubb D.M., Amant F., Annibali D., Ashton K., Attia J., Auer P.L., Beckmann M.W., Black A., Bolla M.K., et al. Identification of nine new susceptibility loci for endometrial cancer. Nat. Commun. 2018;9:3166. doi: 10.1038/s41467-018-05427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spurdle A.B., Thompson D.J., Ahmed S., Ferguson K., Healey C.S., O'Mara T., Walker L.C., Montgomery S.B., Dermitzakis E.T., Australian National Endometrial Cancer Study Group, et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat. Genet. 2011;43:451–454. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setiawan V.W., Yang H.P., Pike M.C., McCann S.E., Yu H., Xiang Y.B., Wolk A., Wentzensen N., Weiss N.S., Webb P.M., et al. Type I and II endometrial cancers: have they different risk factors? J. Clin. Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raglan O., Kalliala I., Markozannes G., Cividini S., Gunter M.J., Nautiyal J., Gabra H., Paraskevaidis E., Martin-Hirsch P., Tsilidis K.K., Kyrgiou M. Risk factors for endometrial cancer: an umbrella review of the literature. Int. J. Cancer. 2019;145:1719–1730. doi: 10.1002/ijc.31961. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Glubb D.M., O'Mara T.A. 10 Years of GWAS discovery in endometrial cancer: aetiology, function and translation. EBioMedicine. 2022;77 doi: 10.1016/j.ebiom.2022.103895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nead K.T., Sharp S.J., Thompson D.J., Painter J.N., Savage D.B., Semple R.K., Barker A., Attia J., et al. Australian National Endometrial Cancer Study Group ANECS. Perry J.R.B. Evidence of a causal association between insulinemia and endometrial cancer: a mendelian randomization analysis. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Painter J.N., O'Mara T.A., Marquart L., Webb P.M., Attia J., Medland S.E., Cheng T., Dennis J., Holliday E.G., McEvoy M., et al. Genetic risk score mendelian randomization shows that obesity measured as body mass index, but not waist:hip ratio, is causal for endometrial cancer. Cancer Epidemiol. Biomarkers Prev. 2016;25:1503–1510. doi: 10.1158/1055-9965.EPI-16-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day F.R., Thompson D.J., Helgason H., Chasman D.I., Finucane H., Sulem P., Ruth K.S., Whalen S., Sarkar A.K., Albrecht E., et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruth K.S., Day F.R., Tyrrell J., Thompson D.J., Wood A.R., Mahajan A., Beaumont R.N., Wittemans L., Martin S., Busch A.S., et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson D.J., O'Mara T.A., Glubb D.M., Painter J.N., Cheng T., Folkerd E., Doody D., Dennis J., Webb P.M., Australian National Endometrial Cancer Study Group ANECS, et al. CYP19A1 fine-mapping and Mendelian randomization: estradiol is causal for endometrial cancer. Endocr. Relat. Cancer. 2016;23:77–91. doi: 10.1530/ERC-15-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kho P.F., Amant F., Annibali D., Ashton K., Attia J., Auer P.L., Beckmann M.W., Black A., Brinton L., Buchanan D.D., et al. Mendelian randomization analyses suggest a role for cholesterol in the development of endometrial cancer. Int. J. Cancer. 2021;148:307–319. doi: 10.1002/ijc.33206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mounier N., Kutalik Z. bGWAS: an R package to perform Bayesian genome wide association studies. Bioinformatics. 2020;36:4374–4376. doi: 10.1093/bioinformatics/btaa549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda T., Ogawa K., Kamatani Y., Murakami Y., Kimura T., Okada Y. A Mendelian randomization study identified obesity as a causal risk factor of uterine endometrial cancer in Japanese. Cancer Sci. 2020;111:4646–4651. doi: 10.1111/cas.14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prescott J., Setiawan V.W., Wentzensen N., Schumacher F., Yu H., Delahanty R., Bernstein L., Chanock S.J., Chen C., Cook L.S., et al. Body mass index genetic risk score and endometrial cancer risk. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freuer D., Linseisen J., O'Mara T.A., Leitzmann M., Baurecht H., Baumeister S.E., Meisinger C. Body fat distribution and risk of breast, endometrial, and ovarian cancer: a two-sample mendelian randomization study. Cancers. 2021;13:5053. doi: 10.3390/cancers13205053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruth K.S., Day F.R., Hussain J., Martínez-Marchal A., Aiken C.E., Azad A., Thompson D.J., Knoblochova L., Abe H., Tarry-Adkins J.L., et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature. 2021;596:393–397. doi: 10.1038/s41586-021-03779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullee A., Dimou N., Allen N., O'Mara T., Gunter M.J., Murphy N. Testosterone, sex hormone-binding globulin, insulin-like growth factor-1 and endometrial cancer risk: observational and Mendelian randomization analyses. Br. J. Cancer. 2021;125:1308–1317. doi: 10.1038/s41416-021-01518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson S.C., Lee W.H., Kar S., Burgess S., Allara E. Assessing the role of cortisol in cancer: a wide-ranged Mendelian randomisation study. Br. J. Cancer. 2021;125:1025–1029. doi: 10.1038/s41416-021-01505-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan S., Carter P., Bruzelius M., Vithayathil M., Kar S., Mason A.M., Lin A., Burgess S., Larsson S.C. Effects of tumour necrosis factor on cardiovascular disease and cancer: a two-sample Mendelian randomization study. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimou N.L., Papadimitriou N., Mariosa D., Johansson M., Brennan P., Peters U., Chanock S.J., Purdue M., Bishop D.T., Gago-Dominquez M., et al. Circulating adipokine concentrations and risk of five obesity-related cancers: a Mendelian randomization study. Int. J. Cancer. 2021;148:1625–1636. doi: 10.1002/ijc.33338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan S., Kar S., Carter P., Vithayathil M., Mason A.M., Burgess S., Larsson S.C. Is type 2 diabetes causally associated with cancer risk? Evidence from a two-sample mendelian randomization study. Diabetes. 2020;69:1588–1596. doi: 10.2337/db20-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kho P.F., Mortlock S., Endometrial Cancer Association Consortium. International Endometriosis Genetics Consortium. Rogers P.A.W., Nyholt D.R., Montgomery G.W., Spurdle A.B., Glubb D.M., O'Mara T.A. Genetic analyses of gynecological disease identify genetic relationships between uterine fibroids and endometrial cancer, and a novel endometrial cancer genetic risk region at the WNT4 1p36.12 locus. Hum. Genet. 2021;140:1353–1365. doi: 10.1007/s00439-021-02312-0. [DOI] [PubMed] [Google Scholar]
- 28.Telomeres Mendelian Randomization Collaboration, Haycock P.C., Burgess S., Nounu A., Zheng J., Okoli G.N., Bowden J., Wade K.H., Timpson N.J., Evans D.M., et al. Association between telomere length and risk of cancer and non-neoplastic diseases: a mendelian randomization study. JAMA Oncol. 2017;3:636–651. doi: 10.1001/jamaoncol.2016.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson S.C., Carter P., Kar S., Vithayathil M., Mason A.M., Michaëlsson K., Burgess S. Smoking, alcohol consumption, and cancer: a mendelian randomisation study in UK Biobank and international genetic consortia participants. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prins B.P. Doctor of Philosophy Thesis (Rijksuniversiteit Groningen; 2016. Inflammatory Biomarker Genomics: From Discovery to Causality. [Google Scholar]
- 31.Ahola-Olli A.V., Würtz P., Havulinna A.S., Aalto K., Pitkänen N., Lehtimäki T., Kähönen M., Lyytikäinen L.P., Raitoharju E., Seppälä I., et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z., Wang X., Li X., Lin Y., Shen S., Liu C.L., Hobbs B.D., Hasegawa K., Liang L., International COPD Genetics Consortium, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir. Res. 2019;20:64. doi: 10.1186/s12931-019-1036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surendran P., Feofanova E.V., Lahrouchi N., Ntalla I., Karthikeyan S., Cook J., Chen L., Mifsud B., Yao C., Kraja A.T., et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat. Genet. 2020;52:1314–1332. doi: 10.1038/s41588-020-00713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coffee and Caffeine Genetics Consortium, Cornelis M.C., Byrne E.M., Esko T., Nalls M.A., Ganna A., Paynter N., Monda K.L., Amin N., Fischer K., et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol. Psychiatry. 2015;20:647–656. doi: 10.1038/mp.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Choudhury T., Zhang M., Chen L., Wen J., Liu W., Chen P. Habitual coffee consumption increases risks for metabolic diseases: genome-wide association studies and a phenotype-wide two sample mendelian randomization analysis. medRxiv. 2021 doi: 10.1101/2021.03.08.21253114. Preprint at. [DOI] [Google Scholar]
- 36.Bafligil C., Thompson D.J., Lophatananon A., Ryan N.A.J., Smith M.J., Dennis J., Mekli K., O'Mara T.A., Evans D.G., Crosbie E.J. Development and evaluation of polygenic risk scores for prediction of endometrial cancer risk in European women. Genet. Med. 2022;24:1847–1856. doi: 10.1016/j.gim.2022.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Kandel S.E., Han L.W., Mao Q., Lampe J.N. Digging deeper into CYP3A testosterone metabolism: kinetic, regioselectivity, and stereoselectivity differences between CYP3A4/5 and CYP3A7. Drug Metab. Dispos. 2017;45:1266–1275. doi: 10.1124/dmd.117.078055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.FANTOM Consortium and the RIKEN PMI and CLST DGT. Forrest A.R.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez A.C., Blanchard Z., Maurer K.A., Gertz J. Estrogen signaling in endometrial cancer: a key oncogenic pathway with several open questions. Horm. Cancer. 2019;10:51–63. doi: 10.1007/s12672-019-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama K., Horie-Inoue K., Ikeda K., Urano T., Murakami K., Hayashizaki Y., Ouchi Y., Inoue S. FOXP1 is an androgen-responsive transcription factor that negatively regulates androgen receptor signaling in prostate cancer cells. Biochem. Biophys. Res. Commun. 2008;374:388–393. doi: 10.1016/j.bbrc.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 41.Cui L., Li M., Feng F., Yang Y., Hang X., Cui J., Gao J. MEIS1 functions as a potential AR negative regulator. Exp. Cell Res. 2014;328:58–68. doi: 10.1016/j.yexcr.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Njoku K., Abiola J., Russell J., Crosbie E.J. Endometrial cancer prevention in high-risk women. Best Pract. Res. Clin. Obstet. Gynaecol. 2020;65:66–78. doi: 10.1016/j.bpobgyn.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Kitson S., Ryan N., MacKintosh M.L., Edmondson R., Duffy J.M., Crosbie E.J. Interventions for weight reduction in obesity to improve survival in women with endometrial cancer. Cochrane Database Syst. Rev. 2018;2:CD012513. doi: 10.1002/14651858.CD012513.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crawford E.D., Heidenreich A., Lawrentschuk N., Tombal B., Pompeo A.C.L., Mendoza-Valdes A., Miller K., Debruyne F.M.J., Klotz L. Androgen-targeted therapy in men with prostate cancer: evolving practice and future considerations. Prostate Cancer Prostatic Dis. 2019;22:24–38. doi: 10.1038/s41391-018-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tangen I.L., Onyango T.B., Kopperud R., Berg A., Halle M.K., Øyan A.M., Werner H.M.J., Trovik J., Kalland K.H., Salvesen H.B., Krakstad C. Androgen receptor as potential therapeutic target in metastatic endometrial cancer. Oncotarget. 2016;7:49289–49298. doi: 10.18632/oncotarget.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson D.A., Simitsidellis I., Collins F., Saunders P.T.K. Evidence of androgen action in endometrial and ovarian cancers. Endocr. Relat. Cancer. 2014;21:T203–T218. doi: 10.1530/ERC-13-0551. [DOI] [PubMed] [Google Scholar]
- 47.Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Yamamoto K., Akiyama M., et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PubMed] [Google Scholar]
- 48.Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., Yengo L., Ferreira T., Marouli E., Ji Y., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rask-Andersen M., Karlsson T., Ek W.E., Johansson Å. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 2019;10:339. doi: 10.1038/s41467-018-08000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S., et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N., Daly M.J., Price A.L., Neale B.M., et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott R.A., Fall T., Pasko D., Barker A., Sharp S.J., Arriola L., Balkau B., Barricarte A., Barroso I., Boeing H., et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes. 2014;63:4378–4387. doi: 10.2337/db14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prokopenko I., Poon W., Mägi R., Prasad B R., Salehi S.A., Almgren P., Osmark P., Bouatia-Naji N., Wierup N., Fall T., et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarzer G. meta: an R Package for meta-analysis. R. News. 2007;7:40–45. [Google Scholar]
- 59.Zou Y., Carbonetto P., Wang G., Stephens M. Fine-mapping from summary data with the "sum of Single effects" model. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G., Sarkar A., Carbonetto P., Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. Roy. Stat. Soc. B. 2020;82:1273–1300. doi: 10.1111/rssb.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munz M., Wohlers I., Simon E., Reinberger T., Busch H., Schaefer A.S., Erdmann J. Qtlizer: comprehensive QTL annotation of GWAS results. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-75770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panjwani N., Wang F., Mastromatteo S., Bao A., Wang C., He G., Gong J., Rommens J.M., Sun L., Strug L.J. LocusFocus: web-based colocalization for the annotation and functional follow-up of GWAS. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Summary statistics from the bGWAS multi-trait GWAS analysis have been deposited at Zenodo, to be made publicly available by publication (https://doi.org/10.5281/zenodo.7545994). This paper analyzes existing, publicly available data. Accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyse data reported in this paper is available from the lead author upon request.