Abstract
Human infections with Escherichia coli O157:H7 cause hemorrhagic colitis that can progress to a life-threatening sequelae. The most common mode of disease transmission is ingestion of contaminated bovine food products, and it is well established that E. coli O157:H7 is a transient member of the bovine microbiota. However, the conditions that induce acquisition and subsequent clearance of this bacterium from the ruminant gastrointestinal tract (GIT) are not understood. Evidence that the rates of epithelial cell proliferation in the lower GIT of cattle are associated with the duration animals remained E. coli O157:H7 culture positive is presented. Cattle with slower rates of intestinal cell proliferation in the cecum and the distal colon were culture positive significantly longer than cohort cattle with faster cell proliferation rates. Cell death rates (apoptotic indices) between the short- and long-term culture-positive animals were not different. Typical grain-based finishing diets and forage-based growing diets did not effect GIT cell proliferation or the duration animals remained E. coli O157:H7 culture positive. To identify a dietary intervention that would effect GIT cell proliferation, we used sheep as a model ruminant. A fasting-refeeding regime that increased the rate of GIT cell proliferation was developed. The fasting-refeeding protocol was used in cattle to test the hypothesis that feeding interventions that increase the rate of GIT cell proliferation induce the clearance of E. coli O157:H7 from the bovine GIT.
Human infections with Escherichia coli O157:H7 cause hemorrhagic colitis that can progress to a life-threatening sequelae, the hemolytic-uremic syndrome (3). The major vehicle of disease transmission in ingestion of contaminated bovine food products that include undercooked contaminated ground beef and unpasteurized dairy products (29). Large-scale surveys routinely find E. coli O157:H7 culture-positive cattle (22, 23, 25, 46). Although it is well established that E. coli O157:H7 is a transient member of the bovine microbiota, the conditions that induce its acquisition, prolong or curtail its presence, and cause its clearance from the ruminant gastrointestinal tract (GIT) are not understood. The average duration an individual animal is culture positive for E. coli O157:H7 is 30 days, but the range in duration individuals shed these bacteria varies from a few days to 1 year (2, 49). Factors that may influence this variation include diet, drinking water contamination, competing microbial flora, immune response, age, breed, E. coli strain, housing conditions, and/or season.
Dietary manipulation has been suggested as a potential preharvest cattle management intervention that may reduce the prevalence of E. coli O157:H7 culture-positive cattle. Several investigations have been done on the relationship between ruminant diet and E. coli O157:H7. Under some conditions grain-fed animals shed E. coli O157:H7 for significantly shorter duration than cohort animals fed hay (27, 33, 35). Both abrupt dietary change and fasting have been shown to prolong shedding of E. coli O157:H7 by ruminants (33, 35). In addition, fasted calves are more susceptible to infection with E. coli O157:H7 than calves fed a consistent diet (6, 13). It should be noted that although dietary effects are clear in experimentally inoculated animals, there have been no epidemiological reports showing a correlation between the incidence of E. coli O157:H7 culture-positive cattle and diet (24, 25).
Abrupt changes in the dietary level of nutrients, including protein, digestible energy and fiber, and abrupt food withdrawal and refeeding are well-established dietary changes that alter the kinetics and other characteristics of the cells lining the GIT in monogastric animals. High levels of dietary fiber have been shown to enhance intestinal cell proliferation in rats (4, 17, 48) and mice (40). Growing pigs fed a high-fiber diet have higher intestinal cell proliferation indices and rates of apoptotic cell death than pigs fed a low-fiber diet (28). In rats, fasting reduces the number of colonic crypt epithelial cells undergoing cell proliferation (9). However, fasting and refeeding results in an increase in intestinal crypt epithelial cells undergoing cell proliferation compared to continuous ad libitum feeding (9, 11, 21).
To our knowledge, there are no previous reports on the effect of dietary manipulations on the cell kinetics in the GIT of ruminant animals. The goal of this study was to test the hypothesis that dietary manipulations that affect gastrointestinal E. coli O157:H7 also affect epithelial cell kinetics in the lower GIT of cattle and sheep. To this end, we (i) determined the effect of a typical grain-based finishing diet and a typical forage-based growing diet on the duration E. coli O157:H7 persisted in cattle, (ii) compared the duration E. coli O157:H7 persisted in cattle with the rates of epithelial cell proliferation and apoptosis in the intestinal tract, (iii) developed a fasting-refeeding regime to manipulate ruminant GIT cell proliferation, and (iv) tested the fasting-refeeding regime in cattle experimentally inoculated with E. coli O157:H7.
MATERIALS AND METHODS
Experimental animals.
Healthy 9- to 12-month-old Charolais × Hereford heifers and 1-year-old Holstein steers were identified by ear tags and housed without contact between animals in concrete stalls on wood-chip bedding. Healthy 1-year-old Suffolk ewes were obtained from the University of Idaho sheep farm and housed by treatment group in concrete stalls without bedding and without contact between groups.
Bacteria.
The inoculum was E. coli O157:H7 strain ATCC 43894 (American Type Culture Collection, Manassas, Va.). The bacteria were grown in Luria-Bertani broth at 37°C with aeration to a cell density of 109 CFU/ml. The number of viable bacteria was confirmed by spread plate technique. Each animal received 1010 CFU of E. coli O157:H7 via a gastric tube placed directly into the rumen.
Rations and housing.
Animals were fed daily and had water ad libitum. Cattle were fed a typical grain-based finishing diet (referred to throughout as grain), a typical forage-based growing diet (referred to throughout as forage), or alfalfa hay. The grain diet was composed of 5% grass hay, 7.29% alfalfa silage, 62% barley, and 19.33% corn. The forage diet was composed of 19.9% grass hay, 48.6% alfalfa silage, 12% barley, and 12% corn. The remaining contents of both diets were similar and contained soybean meal, ground limestone, dicalcium phosphate, and trace mineralized salt. Cattle were adapted to a diet for a minimum of 3 weeks before oral inoculation with E. coli O157:H7.
Ewes were divided into three groups and adapted to a diet of alfalfa hay for 3 weeks. One group of three ewes had feed withheld for 48 h prior to sacrifice (referred to throughout as fasted). A second group of three ewes had feed withheld for 48 h and were refed the alfalfa hay for 24 h prior to sacrifice (referred to throughout as fasted-refed). A third group of three ewes was fed consistently for all days prior to sacrifice (referred to throughout as consistently fed). Cattle were fed twice daily (referred to as continuously fed) or had feed and water withheld for 24 h starting at day 17 post-E. coli O157:H7 dose (referred to as fasted cattle only on day 18 post-E. coli O157:H7 dose) and then twice-daily feedings and ad libitum access to water was resumed (referred to as fasted-refed).
Chemical analyses of feeds.
Samples of the grain or forage feeds were dried at 60°C and ground to pass through a 1-mm screen. The samples were analyzed, using standard techniques for dry matter, crude protein, neutral detergent fiber, and acid detergent fiber (1, 30, 31, 45). The samples were also incubated in vitro as described by Terry et al. (43) to determine dry matter degradability.
Fecal culture.
Cattle were cultured as indicated in Table 3 to monitor fecal E. coli O157:H7. Fecal samples of 10 g were obtained by aseptic rectal palpation and cultured for E. coli O157:H7 by both nonenrichment and enrichment culture protocols that have been described previously (34). Colonies with typical morphologies were confirmed to be E. coli O157:H7 serologically, using a latex agglutination test (ProLab Diagnostics, Roundrock, Tex.). Heifers were considered culture negative for E. coli O157:H7 after three consecutive negative fecal samples spanning 10 days.
TABLE 3.
E. coli O157:H7 culture status in fasted-refed and continuously fed cattle
| Time (days) post-E. coli O157:H7 dosea | No. of animals positive/no of animals negativeb
|
|
|---|---|---|
| Fasted-refedc | Continuously fedd | |
| −1 | 0/8 | 0/8 |
| 1 | 8/0 | 8/0 |
| 9 | 7/1 | 7/1 |
| 17 | 2/6 | 6/2 |
| 18 | 5/3 (fasted for 24 h) | 6/2 |
| 19 | 3/5 (refed for 24 h) | 6/2 |
| 21 | 1/7 (refed for 72 h) | 5/3 |
| 24 | 2/6 | 4/4 |
| 30 | 2/6 | 4/4 |
Animals were given one oral dose of E. coli O157:H7 on day 0.
Fecal samples were cultured for E. coli O157:H7, and the results are expressed as the number of O157 culture-positive animals/the number of O157 culture-negative animals.
Animals in this group were fed twice daily and had water ad libitum except on day 17, when feed and water were withheld for 24 h.
Animals in this group were fed twice daily and had water ad libitum.
Tissue preparation.
Animals were euthanized and laid in right lateral to dorsal recumbency. The abdomen and left thoracic wall were opened. Transections (2.5 to 5 cm long) of the full circumference of the GIT were taken. In cattle, samples were taken from the ileum, 10 cm proximal to the ileocecal juncture; the cecum, 20 cm distal to the ileocecal juncture; the proximal colon, 75 cm distal to the cecocolic juncture; the central colon at the central flexure; and the distal colon 180 cm distal to the central flexure. In sheep, samples were taken from the ileum, 10 cm proximal to the ileocecal juncture; the cecum, 10 to 13 cm from the ileocecal juncture; the proximal colon, 30 cm distal to the ileocecal juncture; the central colon at the central flexure; and the distal colon, 150 cm distal to the central flexure. The transects were rinsed in ice-cold phosphate-buffered saline (PBS), cut, pinned mucosal side up on balsa wood, and submerged in 10% buffered formalin. Strips of the entire circumference of tissue were cut to approximately 0.6 cm wide, rolled Swiss roll style, and embedded in paraffin oriented to result in visualization of longitudinal crypts. Sections 5 μm thick were cut, adhered to glass slides pretreated with Histogrip (Zymed Laboratories, Inc., San Francisco, Calif.), and air dried overnight.
Cell proliferation measurements.
Proliferating cell nuclear antigen (PCNA) immunohistochemistry was used to measure cell proliferation. PCNA is a cell-cycle-associated protein that is maximally expressed during S phase (10). PCNA immunohistochemistry is considered a reliable marker of proliferation in intestinal epithelial cells (5, 47). Sections of embedded tissues were deparaffinized, rehydrated, and treated with 1.5% hydrogen peroxide to quench endogenous peroxidase activity. Sections were incubated sequentially, at room temperature for 1 h in 3% normal horse serum (Vector Laboratories, Inc., Burlingame, Calif.) and in mouse PCNA monoclonal PC10 antibody (NovaCastra Division of Vector Laboratories, Inc.) diluted 1:200. All incubations were carried out in a humidified chamber. Sections were incubated with biotinylated anti-mouse immunoglobulin G as the second antibody (Vector Laboratories, Inc.). Immunostaining was performed using the ABC method (Vector Laboratories, Inc.) and 3,3′-diaminobenzidine tetrahydrochloride-hydrogen peroxide as the chromagen. A light Mayer hematoxylin counterstain was used to visualize crypts.
Only complete crypts, defined as crypts sectioned longitudinally from top to bottom with the full length of the crypt and muscularis mucosa at the base visible, were scored. The number of epithelial cells in each crypt column (side) was defined as the crypt height. The number and position of PCNA labeled in the crypt were recorded. The proliferation index was the number of stained cells divided by total number of cells in the crypt (equal to two times the crypt height) × 100. A minimum of 20 values were obtained (10 crypts of 2 crypt columns each) for each location from each animal. The mean of these 20 values for each animal was then used in subsequent statistical analyses.
Quantification of apoptotic cells.
As the rate of cell turnover in the intestine is affected by both cell proliferation and cell death, we quantified apoptotic cells in the distal and proximal colon regions of the cattle. The 3′ labeling of apoptotic cell DNA was performed by using an ApopTag in situ apoptosis detection kit (Oncor, Gaithersburg, Md.) according to the manufacturer's instructions. Tissues were counterstained with methyl green. The number of apoptotic cells per crypt was recorded for 20 randomly selected complete crypts per animal in the proximal and distal colon segments. Sections used for measurement of apoptotic cells were from the same tissue blocks used for PCNA measurements.
Statistical methods.
Differences between groups for time-to-culture-negative status were analyzed using the Wilcoxon test for censored data. Intestinal cell proliferation data are presented as means with the standard errors. The Student t test was used to assess the effect of diet or culture-positive duration on cell proliferation indices at each location in the cattle experiment. One-way analysis of variance (ANOVA) was used to assess the effect of fasting and refeeding at each individual location in the intestine of the sheep. When significant effects were observed (P < 0.05), the Student-Neuman-Keuls test was used to determine specific differences among the means. The general linear model analysis was used for a split-plot ANOVA to assess whether there was an effect on cell proliferation indices across locations with a diet treatment or the culture-positive duration as a whole plot factor and location as a split-plot factor. The number of culture-positive animals following fasting-refeeding was compared to the number in the continuously fed group using the chi-square test and the more conservative Fisher exact test. All analyses were conducted using the SAS System for Windows package (release 6.11; SAS Institute, Inc., Cary, N.C.).
RESULTS
All animals remained healthy for the duration of the experiment. The cattle that were inoculated with E. coli O157:H7 were all culture positive for the bacteria 24 h after inoculation. Fecal E. coli O157:H7 titers decreased with time until all animals became culture negative.
The grain diet was higher in energy and lower in fiber than the forage diet.
The ingredients of the grain and forage diets were typical of cattle grain-based finishing and forage-based growing rations. Both diets are considered high-quality rations, and average weight gains were >0.9 kg/day with the forage diet and 1.4 kg/day with the grain diet. The chemical analyses of grain and forage, respectively, were as follows: dry matter, 74.1 and 63.5%; in vitro dry matter degradability, 83.1 and 62.7%; neutral detergent fiber, 25.1 and 41.8%; acid detergent fiber, 9.6 and 26.4%; and crude protein, 15.2 and 14.2%. As expected, the grain diet, which was higher in grain content, was higher in protein and digestible energy and lower in fiber than the forage diet. Acid and neutral detergent fiber values are negative indicators of digestible energy, and both values were lower for the grain diet than for the forage diet. In addition, fiber concentration is inversely related to in vitro dry-matter degradability values, and dietary fiber was lower for forage than for grain.
The grain and forage diet did not affect the duration or concentrations of fecal E. coli O157:H7.
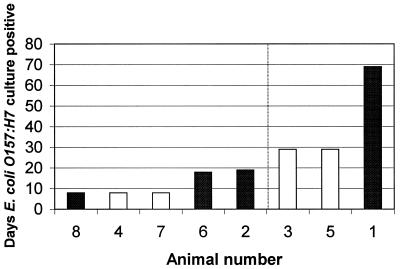
Heifers fed the forage or grain diet were inoculated with E. coli O157:H7, and fecal samples were cultured for the bacteria (Fig. 1). Animals on either diet were similarly E. coli O157:H7 positive by nonenrichment culture for an average of 5 days, after which enrichment culture was required to detect the bacterium in fecal samples. Although we observed a wide variation in the duration that individuals remained culture positive for the bacteria, there was no difference between the groups fed grain or forage (Fig. 1). The Wilcoxon test for censored data gave a P value of 0.78 for a diet effect on the time-to-culture-negative status. This test failed to detect a difference in the pattern of E. coli O157:H7 culture status in animals eating grain or forage. Also, the ANOVA of the mean CFU/gram value (data not shown), while culture positive at titers detected by nonenrichment, had a P value of 0.77.
FIG. 1.
Effect of diet on the duration cattle remained culture positive for E. coli O157:H7. Heifers were adapted to a typical forage-based growing diet (open bars) or grain-based finishing diet (shaded bars) and inoculated with a single oral dose of E. coli O157:H7. Each bar represents a single animal. The vertical line separates the animals into short-term (<19 days) (left side) and long-term (>29 days) (right side) E. coli O157:H7 culture-positive groups.
The grain and forage diet did not affect cell proliferation or apoptosis indices.
There was no difference between the grain and forage diet groups in the number of PCNA-labeled cells or the proliferation index for any segment of the intestine (all P values were >0.3) (data not shown). Split-plot ANOVA did not detect a significant diet effect for any of the cell proliferation indices (all P values were >0.4) (data not shown). There was no effect of diet on the distribution of labeled cells in the crypts or in the number of apoptotic cells per crypt in any location (all P values were >0.3) (data not shown). However, crypt height was significantly higher (P = 0.03) in the distal colon of animals fed the forage diet (65.7 ± 2.3 cells) compared with animals fed the grain diet (57.4 ± 1.7 cells).
Short-term E. coli O157 culture-positive status was associated with high indices of intestinal cell proliferation.
To evaluate GIT cell proliferation with E. coli O157:H7 culture status, we categorized the eight heifers fed the grain or forage diets according to the duration they harbored the bacteria (Table 1). The mean duration cattle were culture positive for E. coli O157:H7 was 23 days. Three animals (animals 1, 3, and 5) were culture positive for longer than 29 days postinoculation (above the mean) and were designated as the long-term culture-positive group (Fig. 1). Five animals (animals 2, 4, 6, 7, and 8) were culture positive for 8 to 19 days (below the mean) and were designated as the short-term culture-positive group (Fig. 1). The cell proliferation indices in the short-term and long-term groups at each GIT location are summarized in Table 1. The short-term culture-positive group had higher indices of intestinal cell proliferation than the long-term culture-positive group. The greatest difference in the number of PCNA-labeled cells between the long-term and short-term groups was observed in the cecum (P = 0.016) and the distal colon (P = 0.049) (Table 1). Over all GIT locations, the number of PCNA-labeled cells tended to be higher in the short-term group than in the long-term group (effect of culture-positive duration by split-plot ANOVA: P = 0.02). There was no difference (all P values were >0.31) in mean crypt heights between the short-term and long-term groups at any GIT location. The proliferation index, which takes into account the number of labeled cells, and the total crypt height was higher in the short-term group than in the long-term group (effect of culture-positive duration by split-plot ANOVA: P = 0.043). Again, the differences in proliferation index between groups were greatest in the cecum (P = 0.027). Interestingly, animal 1 (Fig. 1) was culture positive for 69 days, 39 days longer than the other animals in this study, and also had the lowest GIT cell proliferation indices. The values for number of labeled cells/crypt in this animal were all well below the mean: ileum, 21.5; cecum, 0.8; proximal colon, 5.1; central colon, 2.3; and distal colon, 7.8. Although the crypt heights at all GIT locations in animal 1 were similar to those for the other animals, the cell proliferation indices were well below the mean: ileum, 18.9%; cecum, 0.9%; proximal colon, 6.8%; central colon, 2.2%; and distal colon, 6.6%.
TABLE 1.
Cattle intestinal cell proliferation and persistence of E. coli O157:H7
| Parameter | Duration culture positive for E. coli O157:H7d | Mean ± SE results in:
|
|||||
|---|---|---|---|---|---|---|---|
| Ileum | Cecum | Proximal colon | Central colon | Distal colon | All locations | ||
| Labeled cellsa (no. of cells/crypt) | Long-term | 27.5 ± 3.7 | 3.1 ± 1.5e | 7.8 ± 1.5 | 6.6 ± 2.2 | 8.8 ± 1.0e | 10.8 ± 2.4 |
| Short-term | 29.1 ± 3.0 | 10.9 ± 1.6e | 11.3 ± 2.3 | 8.2 ± 1.8 | 11.4 ± 0.6e | 14.2 ± 1.7 | |
| Crypt heightb (no. of cells/crypt column) | Long-term | 68.7 ± 7.6 | 40.0 ± 2.3 | 47.6 ± 5.6 | 51.0 ± 2.5 | 61.1 ± 1.7 | 53.7 ± 3.2 |
| Short-term | 65.1 ± 2.3 | 41.8 ± 1.6 | 51.1 ± 1.5 | 45.5 ± 4.2 | 61.8 ± 3.3 | 53.1 ± 2.2 | |
| Proliferation indexc (%) | Long-term | 20.3 ± 2.9 | 3.9 ± 1.9e | 8.0 ± 0.7 | 6.5 ± 2.2 | 7.3 ± 0.9 | 9.2 ± 1.0f |
| Short-term | 22.3 ± 2.0 | 13.2 ± 2.2e | 11.0 ± 2.1 | 8.5 ± 1.2 | 9.4 ± 0.6 | 12.9 ± 1.2 | |
Labeled cells are the number of PCNA-labeled cells per crypt.
Crypt height is the total number of cells in each crypt column.
The proliferation index is the percentage of PCNA-labeled cells calculated as follows: [labeled cells/(2 × crypt height)]100.
Long-term, animals that remained E. coli O157:H7 culture positive for 29 to 69 days (n = 3); short-term, animals that remained E. coli O157:H7 culture positive for 8 to 19 days (n = 5).
Labeled cells were fewer in the long-term than in the short-term group in the cecum (P = 0.016) and distal colon (P = 0.027) (t test).
Significant effect of E. coli O157:H7 culture-positive duration over all locations (P = 0.043, split-plot ANOVA).
Therefore, short-term culture-positive animals had a significantly higher overall rate of cell proliferation in the GIT than did long-term culture-positive animals. This was most pronounced in the cecum and distal colon. The GIT location affected all indices. For example, the ileum had the highest number of labeled cells and labeling index of all GIT locations (P < 0.0001). Crypt height was higher in the ileum and distal colon than in the other locations (P < 0.0001). However, these location effects had no interaction effects with the duration animals were E. coli O157:H7 culture positive.
There were no differences (all P values were >0.16) between short- and long-term culture-positive animals in the distribution of labeled cells throughout individual crypts at any location (data not shown). In other words, the percentages of total labeled cells that were in the basal third, mid-third, or lumenal third of the crypts were similar in each location for both groups. In all locations, approximately 60 to 70% of the total number of labeled cells occurred in the lower third of the crypts. There was also no difference (all P values were >0.39) in the number of apoptotic cells in crypts in the proximal or distal colon between short-term and long-term culture-positive groups (data not shown).
Intestinal cell proliferation indices were increased in fasted-refed sheep.
To identify a dietary intervention that would effect GIT cell proliferation we used sheep as a model ruminant. Sheep that were fasted-refed had a significantly higher number of PCNA-labeled cells per crypt in the distal colon (62.9 ± 9.2) than did sheep that were either fasted (26.3 ± 8.5) or continuously fed (33.6 ± 6.1) (P = 0.0395 by one-way ANOVA) (Table 2). There was no difference (all P values were >0.28) between groups in the number of PCNA-labeled cells in the ileum, cecum, central colon, or proximal colon. However, there was a difference among groups in the proliferation indices of the distal colon. Sheep that were fasted and refed had a higher labeling index (53.9 ± 6.0) than sheep that were either fasted (23.5 ± 7.3) or continuously fed (25.6 ± 3.5) (P = 0.017). A similar trend occurred in the proximal and central colon, but differences did not reach statistical significance. Split-plot ANOVA was used to assess this dietary intervention effect over all GIT locations. The fasted-refed sheep had significantly higher proliferation indices over all GIT locations than either fasted or continuously fed sheep (P = 0.024). The proliferation index was significantly higher in the cecum and proximal colon than in the central colon (P = 0.0014). There were no significant differences among groups or locations in the crypt height (all P values were >0.24). Interactions between diet and GIT location effects were not significant.
TABLE 2.
Intestinal cell proliferation in fasted, fasted-refed, or continuously fed sheep
| Parametera | Feeding groupb | Mean ± SE results in:
|
|||||
|---|---|---|---|---|---|---|---|
| Ileum | Cecum | Proximal | Central | Distal | All locations | ||
| Labeled cells (no. of cells/crypt) | Fasted | 40.1 ± 11.8 | 67.6 ± 7.2 | 45.8 ± 10.9 | 22.9 ± 5.0 | 26.3 ± 8.5 | 40.5 ± 5.5 |
| Fasted-refed | 51.2 ± 13.8 | 55.2 ± 3.0 | 63.2 ± 12.3 | 38.7 ± 9.1 | 62.9 ± 9.2c | 54.2 ± 4.5 | |
| Continuously fed | 66.4 ± 2.2 | 65.9 ± 16.1 | 52.3 ± 4.5e | 40.3 ± 14.4 | 33.6 ± 6.1 | 51.7 ± 5.6 | |
| Crypt height (no. of cells/crypt column) | Fasted | 79.6 ± 10.0 | 73.9 ± 6.0 | 64.8 ± 10.9 | 70.2 ± 6.8 | 55.5 ± 2.7 | 68.8 ± 3.7 |
| Fasted-refed | 70.2 ± 1.2 | 61.8 ± 2.3 | 61.4 ± 8.1 | 68.2 ± 4.5 | 58.0 ± 3.5 | 63.9 ± 2.1 | |
| Continuously fed | 88.7 ± 20.6 | 77.2 ± 13.7 | 70.4 ± 8.4 | 84.1 ± 7.4 | 65.1 ± 4.0 | 77.1 ± 5.2 | |
| Proliferation index (%) | Fasted | 26.5 ± 8.3 | 45.5 ± 1.2 | 34.3 ± 3.0 | 16.0 ± 2.6 | 23.5 ± 7.3 | 29.2 ± 3.4 |
| Fasted-refed | 36.8 ± 10.2 | 44.7 ± 1.8 | 50.7 ± 5.5 | 27.9 ± 5.6 | 53.9 ± 6.0c | 42.8 ± 3.5d | |
| Continuously fed | 40.9 ± 7.5 | 42.0 ± 3.9 | 41.7 ± 0.4e | 24.8 ± 8.9 | 25.6 ± 3.5 | 34.5 ± 3.2 | |
Parameters are as defined in Table 1.
Fasted animals had feed and water withheld for 48 h, refed animals were fasted and had alfalfa hay for 24 h, and fed animals had continuous daily feeding.
Refed values were higher than fasted or fed values (P = 0.04 for labeled cells and P = 0.017 for proliferation index as determined by one-way ANOVA).
Significant effect of refeeding over all locations (P = 0.024; split-plot ANOVA).
n = 2; in all other cases, n = 3.
Fasted-refed cattle were E. coli O157:H7 culture positive for a shorter duration than continuously fed cattle.
The effect of fasting and fasting-refeeding dietary manipulation on the duration that cattle remained E. coli O157:H7 culture positive was tested (Table 3). All animals were fecal O157 culture negative before receiving an oral dose of E. coli O157:H7, and all animals were fecal O157 culture positive 24 h after the dose. The number of O157 culture-positive animals in the continuously fed group declined steadily with time. This steady decline was not observed in the fasting-refeeding treatment group. Prior to the fast, six of the eight animals in this treatment group were culture negative. After a 24-h fast, three animals that were O157 culture negative prior to fasting (Table 3, day 17 post-E. coli O157:H7 dose) tested O157 culture positive (Table 3, day 18). Also, 24 and 72 h after feeding was resumed, the number of culture-positive animals declined rapidly so that only one of the eight steers in that group remained O157 culture positive on day 21. This finding is in contrast to the animals in the continuously fed group, where five of eight animals remained O157 culture positive at the same time (Table 3, day 21: chi-square test, P = 0.04; Fisher exact test, P = 0.059).
DISCUSSION
The most significant finding of the present study was that the rates of cell proliferation in the GIT of cattle were associated with the duration animals remained E. coli O157:H7 culture positive. Cattle with slower rates of intestinal cell proliferation in the cecum and the distal colon were culture positive significantly longer than cohort cattle with faster cell proliferation rates. Also, we identified a fasting-refeeding regime that increased the rate of GIT cell proliferation in sheep, and we tested this dietary intervention in O157 culture-positive cattle.
Little is known about the relationship between E. coli O157:H7 and the ruminant intestinal mucosal surface. A perplexing issue has been the inability to identify a GIT site of bacterial colonization. Although experimentally induced enterocolitis in neonatal calves results in intimately adhered E. coli O157:H7 in the ileum, cecum, colon, and rectum (14, 16), these findings do not reflect E. coli O157:H7 colonization patterns seen in healthy cattle populations. In calves at 1 to 2 weeks after oral inoculation, E. coli O157:H7 has been isolated from tissue and digesta of the rumen, reticulum, abomasum, jejunum, ileum, cecum, and colon (12). Similar analyses still later (13 to 28 days) after oral inoculation isolated E. coli O157:H7 only from the rumen, omasum, or colon (6), suggesting that the O157:H7 serotype, as with other E. coli, is best adapted to the lower GIT. No study in healthy adult cattle or sheep has found histologic or immunohistochemical evidence that E. coli O157:H7 adheres intimately to the mucosa (15).
It has been suggested that E. coli O157:H7 persists in the lumen as the source of fecal shedding (6). However, it is likely that the bacteria restricted to the lumen would be flushed from the system by digesta passage in a few days. The fact that E. coli O157:H7 can persist in the colonic digesta of some animals for many months suggests that the bacteria associate with the mucosa in some way. The lack of evidence for this association may be a reflection of the extremely low numbers of E. coli O157:H7 in the ruminant GIT. The anatomy of the lower GIT mucosa is comprised of invaginated surfaces covered with crypts. We measured the lower GIT crypts of sheep and cattle to average between 40 and 89 epithelial cells long and to create cavities of approximately 400 to 900 μm deep (data not shown). The crypts contain microbial flora and may provide a physical niche that sequesters replicating E. coli O157:H7 and prevents complete clearance of the bacteria by digesta movement. Alternatively, E. coli O157:H7 may be on the mucosa surface, in association with cells at the top of the crypts. In either scenario, persistence of the bacteria in the GIT may be dependent on the balance between bacterial replication and the rate of epithelial cell proliferation and thus migration and sloughing of epithelial cells from the top of the crypts.
Among the inoculated heifers in this study, a widely varying persistence of E. coli O157:H7 occurred, and animals were grouped as short-term (≤19 days) and long-term (≥29 days) culture-positive animals. An association was found between increased epithelial cell proliferation in the lower GIT and the more rapid clearance of E. coli O157:H7. Increased cell division in the absence of increased crypt height (total number of cells) suggests that rates of cell death or cell sloughing into the lumen may be increased. Differences in the cell death rates (apoptotic indices) between the short- and long-term culture-positive animals were not detected, but the method used only detects apoptotic cells that have not been sloughed into the lumen. The mechanism by which increased intestinal cell proliferation is associated with the clearance of E. coli O157:H7 is beyond the scope of this investigation. However, it may be that this bacterium is associated with the colonic crypts so that increased proliferation and sloughing of crypt epithelial cells may physically remove the niche more rapidly than bacterial replication takes place.
Previous studies comparing grossly different grain and forage diets show significant dietary effect on the duration cattle were E. coli O157:H7 culture positive (27). However, the duration cattle were E. coli O157:H7 culture positive was not affected by the nutritious forage and grain diets used in this study. In addition, the grain and forage diets did not affect the cattle GIT cell proliferation. Although the concentrations and proportions of acetate and propionate or the pH of ruminal and colonic digesta were not measured, the grain and forage diets are different enough to have affected these parameters. We must conclude, however, that the differences were not great enough to affect GIT cell proliferation or the duration of culture-positive status. In monogastric animals, changes in the short-chain fatty acid concentrations have been suggested as a mechanism by which fermentable fibers in the diet lead to an increase in intestinal cell proliferation (41). Unfortunately, a consistent correlation between diet quality (fiber) and intestinal cell proliferation has not been observed in studies of monogastrics. For example, rats fed cellulose, a poorly fermentable fiber, had higher luminal pH and lower short-chain fatty acids concentrations in the cecum and proximal and distal colon than rats fed fermentable fiber sources (pectin or oat bran) (48). A positive correlation between short-chain fatty acids and proliferation index was observed in the cecum but not in the distal colon (indices for the proximal colon were not reported) (48). In another study, the addition of wheat bran, but not pectin, to the diet of rats increased the short-chain fatty acid concentration in the cecum, whereas both fibers increased short-chain fatty acid concentrations in the proximal colon but had no effect in the distal colon (36). Increased crypt height occurred in the cecum of pectin-fed rats and in the distal colon of both pectin- and wheat bran-supplemented rats, illustrating the lack of a consistent correlation between short-chain fatty acids and cell proliferation (36). Malville-Shipan and Fleming (38) reported that neither cecal short-chain fatty acid concentrations nor the proliferation index in the cecum and proximal or distal colon were altered by the addition of wheat bran to the diets of rats when the energy intake was equivalent. The high-fiber diet did significantly lower pH of the luminal contents of the cecum and distal colon. Various fibers fed to miniature pigs significantly altered cecal short-chain fatty acid concentrations and pH, but these did not correlate with changes in cell proliferation in either the cecum or distal colon (18). The importance of other dietary variables, including total energy and nutrient intake on cell proliferation, has been emphasized (38).
To further investigate the relationship between intestinal cell proliferation and clearance of E. coli O157:H7, intestinal cell proliferation will need to be manipulated in a predictable manner. We have established the use of sheep previously as a model animal to enhance investigations of the relationship between cattle and E. coli O157:H7 (32–34). Here we used sheep as a model ruminant to test whether lower gastrointestinal cell proliferation could be manipulated using fasting-refeeding. Increased intestinal cell proliferation following fasting-refeeding in rats and mice has been reported by numerous investigators (8, 9, 11, 21, 26, 37, 39). Peak intestinal cell proliferation in these animals occurs at between 16 and 24 h and slowly returns to normal with refeeding (8, 11, 21). The timing of GIT cell proliferation response after fasting-refeeding is unknown in ruminants. Here we show in sheep a decrease with fasting and an increase 24 h after refeeding in GIT cell proliferation. We predict that the return to baseline GIT cell proliferation in ruminants would be similar or longer than in monogastrics due to the retention of digesta in the rumen. Fleming et al. (18) suggest that effects of short-chain fatty acids on intestinal proliferation may be observed only when baseline concentrations of short-chain fatty acids are very low, such as following prolonged fasting. Sakata and Tamate (42) reported that rapid intraruminal infusion of sodium n-butyrate to adult male sheep resulted in an increase in ruminal cell proliferation, while a slow administration of butyrate had no effect on ruminal cell proliferation. Galfi et al. (19) reported similar results. Only cell proliferation in the ovine rumen was evaluated in these studies. Our observation that cell proliferation in the ovine lower GIT is responsive to dietary manipulation complements this earlier work, and the fact that the greatest cell proliferation difference occurred in the distal colon is in agreement with observations in monogastric animals. For example, Butler et al. (9) showed that changes in cell proliferation with fasting-refeeding were greater in the large intestine than in the small intestine in rats.
We tested our hypothesis that a fasting-refeeding dietary manipulation would affect the duration animals were O157 culture positive in a predicted manner in Holstein steers. Feed and water were withheld for 24 h, conditions similar to those cattle may experience before processing. We began the fasting-refeeding regime when at least half of the cattle had become culture negative for O157 so that we could detect increases in the culture-positive status if they occurred with fasting. By this time postinoculation, animals were positive only by selective enrichment culture so that enumeration of O157 organisms/g of feces was not possible. We cannot explain the marked difference between the number of O157 culture-positive cattle in the two groups just before fasting (Table 3, 17 days after the E. coli O157:H7 dose); however, the influence of dietary intervention is clear. The number of culture-positive animals, among eight total, increased from two to five after this fast (Table 3, days 17 and 18). The finding that withholding feed increases the number of O157 culture-positive animals has been observed by many other investigators (6, 7, 12, 20). This phenomenon has been noted both in animals experimentally dosed with E. coli O157:H7 and in the prevalence of naturally O157 culture-positive sheep and cattle that have traveled the farthest to the market or feedlot (25, 44). As in most previous studies, whether or not E. coli O157:H7 present in the GIT is induced to proliferate to detectable numbers or if animals become more susceptible to reinfections with the bacteria from the environment was not determined. The decrease in the number of O157 culture-positive animals we observed 72 h after feeding was resumed coincides with predicted increases in lower GIT cell proliferation. We did not measure a time course of GIT cell proliferation in these steers; to do so would have required that animals were sacrificed at each time point. We caution against adoption of a fasting-refeeding dietary intervention until this hypothesis is tested further in larger numbers of cattle and with a variety of O157 strains. Elucidation of the mechanism(s) that clear E. coli O157:H7 from the ruminant GIT may lead to the development of preharvest interventions that reduce culture-positive animals from entering our food chain.
ACKNOWLEDGMENTS
This work was supported in part by the Idaho Agriculture Experiment Station (publication 00511), the Public Health Service grant AI33981 from the National Institutes of Health, U.S. Department of Agriculture NRICGP grants 95-37201-1979 and 99-35201-8539, and grants from the United Dairymen of Idaho and the Idaho Beef Council.
We thank Steven Parish for expert veterinary assistance and dissections, Kathleen Hendrix for technical assistance in tissue histology, and Sherilyn Haenny for animal handling and bacterial culture. We also gratefully acknowledge the use of the Washington Animal Disease Diagnostic Laboratory facilities and thank John Hobbs and the UI Farm Operations Personnel for technical assistance.
REFERENCES
- 1.Association of Official Analytical Chemists. Offical methods of analysis of the Association of Official Analytical Chemists. 15th ed. Washington, D.C.: Association of Official Analytical Chemists; 1990. [Google Scholar]
- 2.Besser T E, Hancock D D, Pritchett L C, McRae E M, Rice D H, Tarr P I. Duration of Detection of Fecal Excretion of Escherichia coli O157:H7 in Cattle. J Infect Dis. 1997;175:726–729. doi: 10.1093/infdis/175.3.726. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M F, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. [Google Scholar]
- 4.Boffa L C, Lupton J R, Mariani M R, Ceppi M, Newmark H L, Scalmati A, Lipkin M. Modulation of colonic epithelial cell proliferation, histone acetylation, and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer Res. 1992;52:5906–5912. [PubMed] [Google Scholar]
- 5.Bostick R M, Fosdick L, Lillemoe T J, Overn P, Wood J R, Grambsch P, Elmer P, Potter J D. Methodological findings and considerations in measuring colorectal epithelial cell proliferation in humans. Cancer Epidemiol Biomarkers Prevention. 1997;6:931–942. [PubMed] [Google Scholar]
- 6.Brown C A, Zhao T, Doyle M P. Experimental Escherichia coli O157:H7 carriage in calves. Appl Environ Microbiol. 1997;63:27–32. doi: 10.1128/aem.63.1.27-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownlie L E, Grau F H. Effect of food intake on growth and survival of Salmonella and Escherichia coli in the bovine rumen. J Gen Microbiol. 1967;46:125–134. doi: 10.1099/00221287-46-1-125. [DOI] [PubMed] [Google Scholar]
- 8.Burholt D R, Etzel S L, Schenken L L, Kovacs C J. Digestive tract cell proliferation and food consumption patterns of Ha/ICR mice. Cell Tissue Kinet. 1985;18:369–386. doi: 10.1111/j.1365-2184.1985.tb00668.x. [DOI] [PubMed] [Google Scholar]
- 9.Butler R N, Bruhn B, Pascoe V, Fettman M J, Roberts-Thomson I C. Regional factors affecting proliferation in the large intestine of the rat. Proc Soc Exp Biol Med. 1992;200:133–137. doi: 10.3181/00379727-200-43405. [DOI] [PubMed] [Google Scholar]
- 10.Chang C D, Ottavio L, Travali S, Lipson K E, Baserga R. Transcriptional and posttranscriptional regulation of the proliferating cell nuclear antigen gene. Mol Cell Biol. 1990;10:3289–3296. doi: 10.1128/mcb.10.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven P A, DeRubertis F R. Alterations in protein kinase C system of colonic epithelium during fasting-refeeding. Evidence for protein kinase C independent pathway of enhanced proliferative activity. Dig Dis Sci. 1992;37:1162–1169. doi: 10.1007/BF01296555. [DOI] [PubMed] [Google Scholar]
- 12.Cray W C, Jr, Moon H W. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cray W C, Jr, Bosworth B T, Rasmussen M A. Effect of dietary stress on fecal shedding of Escherichia coli O157:H7 in calves. Appl Environ Microbiol. 1998;64:1975–1979. doi: 10.1128/aem.64.5.1975-1979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean-Nystrom E A, Bosworth B T, Cray W C, Moon H W. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect Immun. 1997;65:1842–1848. doi: 10.1128/iai.65.5.1842-1848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean-Nystrom E, Bosworth B T, Moon H W, O'Brien A D. Bovine infection with Shiga toxin-producing Escherichia coli. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: ASM Press; 1998. pp. 261–267. [Google Scholar]
- 16.Dean-Nystrom E A, Bosworth B, Moon H W, O'Brien A D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards C A, Wilson R G, Hanlon L, Eastwood M A. Effect of the dietary fiber content of lifelong diet on colonic cellular proliferation in the rat. Gut. 1992;33:1076–1079. doi: 10.1136/gut.33.8.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming S E, Fitch M D, De Vries S. The influence of dietary fiber on proliferation of intestinal mucosal cells in miniature swine may not be mediated primarily by fermentation. J Nutr. 1992;122:906–916. doi: 10.1093/jn/122.4.906. [DOI] [PubMed] [Google Scholar]
- 19.Galfi P, Neogrady S, Kutas F. Dissimilar ruminal epithelial response to short-term and continuous intraruminal infusion of sodium n-butyrate. J Vet Med Ser A. 1986;33:47–52. doi: 10.1111/j.1439-0442.1986.tb00504.x. [DOI] [PubMed] [Google Scholar]
- 20.Grau F H, Brownlie L E, Smith M G. Effects of food intake on numbers of Salmonella and Escherichia coli in rumen and faeces of sheep. J Appl Bacteriol. 1969;32:112–117. doi: 10.1111/j.1365-2672.1969.tb02195.x. [DOI] [PubMed] [Google Scholar]
- 21.Hagemann R F, Stragand J J. Fasting and refeeding: cell kinetic response of jejunum, ileum and colon. Cell Tissue Kinet. 1977;10:3–14. doi: 10.1111/j.1365-2184.1977.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 22.Hancock D D, Besser T E, Kinsel M L, Tarr P I, Rice D H, Paros M G. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. J Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock D D, Besser T E, Rice D H, Herriott D E, Tarr P I. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol Infect. 1997;118:193–195. doi: 10.1017/s0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock D D, Rice D H, Herriott D E, Besser T E, Ebel E. Efects of farm manure handling practices on Escherichia coli O157 prevalence in cattle. J Food Prot. 1997;60:363–366. doi: 10.4315/0362-028X-60.4.363. [DOI] [PubMed] [Google Scholar]
- 25.Hancock D D, Rice D H, Thomas L A, Dargatz D A, Besser T E. Epidemiology of Escherichia coli O157 in feedlot cattle. J Food Prot. 1997;60:462–465. doi: 10.4315/0362-028X-60.5.462. [DOI] [PubMed] [Google Scholar]
- 26.Hoff M B, Chang W W, Mak K M. Effect of estrogen on cell proliferation in colonic mucosa of the mouse. Virchows Arch B Cell Pathol Mol Pathol. 1981;35:263–273. doi: 10.1007/BF02889166. [DOI] [PubMed] [Google Scholar]
- 27.Hovde C J, Austin P R, Cloud K A, Williams C J, Hunt C W. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl Environ Microbiol. 1999;65:3233–3235. doi: 10.1128/aem.65.7.3233-3235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L, Reynolds L P, Redmer D A, Caton J S, Crenshaw J D. Effects of dietary fiber on intestinal growth, cell proliferation, and morphology in growing pigs. J Anim Sci. 1994;72:2270–2278. doi: 10.2527/1994.7292270x. [DOI] [PubMed] [Google Scholar]
- 29.Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing Escherichia coli strains. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 30.Komarek A R. A filter bag procedure for improved efficiency of fiber analysis. J Dairy Sci. 1993;76:250. [Google Scholar]
- 31.Komarek A R. Improved efficiency of ADF analysis using a filter bag procedure. J Anim Sci. 1993;71:284. [Google Scholar]
- 32.Kudva I T, Hatfield P G, Hovde C J. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli isolated from sheep. J Clin Microbiol. 1996;35:892–899. doi: 10.1128/jcm.35.4.892-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudva I T, Hatfield P G, Hovde C J. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl Environ Microbiol. 1995;61:1363–1370. doi: 10.1128/aem.61.4.1363-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudva I T, Hatfield P G, Hovde C J. Escherichia coli O157:H7 in microbial flora of sheep. J Clin Microbiol. 1996;34:431–433. doi: 10.1128/jcm.34.2.431-433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudva I T, Hunt C W, Williams C J, Nance U M, Hovde C J. Evaluation of dietary influences on Escherichia coli O157:H7 shedding by sheep. Appl Environ Microbiol. 1997;63:3878–3886. doi: 10.1128/aem.63.10.3878-3886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupton J R, Kurtz P P. Relationship of colonic luminal short-chain fatty acids and pH to in vivo cell proliferation in rats. J Nutr. 1993;123:1522–1530. doi: 10.1093/jn/123.9.1522. [DOI] [PubMed] [Google Scholar]
- 37.Majumdar A P. Effects of fasting and refeeding on antral, duodenal, and serum gastrin levels and on colonic thymidine kinase activity in rats. Hormone Res. 1984;19:127–134. doi: 10.1159/000179877. [DOI] [PubMed] [Google Scholar]
- 38.Malville-Shipan K, Fleming S E. Wheat bran and corn oil do not influence proliferation in the colon of healthy rats when energy intakes are equivalent. J Nutr. 1992;122:37–45. doi: 10.1093/jn/122.1.37. [DOI] [PubMed] [Google Scholar]
- 39.Premoselli F, Sesca E, Chiara M, Binasco V, Tessitore L. Fasting/refeeding enhances the crypt multiplicity in rat colon carcinogenesis induced by azoxymethane. Boll-Soc Ital Biol Speriment. 1996;72:239–245. [PubMed] [Google Scholar]
- 40.Robblee N M, McLellan E A, Bird R P. Measurement of the proliferative status of colonic epithelium as a risk marker for colon carcinogenesis: effect of bile acid and dietary fiber. Nutr Cancer. 1989;12:301–310. doi: 10.1080/01635588909514030. [DOI] [PubMed] [Google Scholar]
- 41.Sakata T. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br J Nutr. 1987;58:95–103. doi: 10.1079/bjn19870073. [DOI] [PubMed] [Google Scholar]
- 42.Sakata T, Tamate H. Rumen epithelial cell proliferation accelerated by rapid increase in intraruminal butyrate. J Dairy Sci. 1978;61:1109–1113. doi: 10.3168/jds.S0022-0302(78)83694-7. [DOI] [PubMed] [Google Scholar]
- 43.Terry R A, Tilley J M A, Outen G E. Effect of pH on cellulose digestion under in vitro conditions. J Sci Food Agric. 1969;20:317. doi: 10.1002/jsfa.2740200514. [DOI] [PubMed] [Google Scholar]
- 44.U.S. Department of Agriculture/Animal and Plant Health Inspection Service/VS. An update: Escherichia coli O157:H7 in humans and cattle. 1997. pp. 1–28. . Report from the Centers for Epidemiology and Animal Health. U.S. Government Printing Office, Washington, D.C. [Google Scholar]
- 45.Van Soest P J, Robertson J B, Lews B A. Methods of dietary fiber, neutral detergent fiber and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 46.Wells J G, Shipman L D, Greene K D, Sowers E G, Green J H, Cameron D N, Downes F P, Martin M L, Griffin P M, Ostroff S M, Potter M E, Tauxe R V, Wachsmuth I K. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada K, Yoshitake K, Sato M, Ahnen D J. Proliferating cell nuclear antigen expression in normal, preneoplastic, and neoplastic colonic epithelium. Gastroenterology. 1992;103:160–197. doi: 10.1016/0016-5085(92)91109-h. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Lupton J R. Dietary fibers stimulate colonic cell proliferation by different mechanisms at different sites. Nutr Cancer. 1994;22:267–276. doi: 10.1080/01635589409514352. [DOI] [PubMed] [Google Scholar]
- 49.Zhao T, Doyle M P, Shere J, Garber L. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl Environ Microbiol. 1995;61:1290–1293. doi: 10.1128/aem.61.4.1290-1293.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]