Abstract
Grass inflorescences produce grains, which are directly connected to our food. In grass crops, yields are mainly affected by grain number and weight; thus, understanding inflorescence shape is crucially important for cereal crop breeding. In the last two decades, several key genes controlling inflorescence shape have been elucidated, thanks to the availability of rich genetic resources and powerful genomics tools. In this review, we focus on the inflorescence architecture of Triticeae species, including the major cereal crops wheat and barley. We summarize recent advances in our understanding of the genetic basis of spike branching, and spikelet and floret development in the Triticeae. Considering our changing climate and its impacts on cereal crop yields, we also discuss the future orientation of research.
Keywords: inflorescence, Triticeae, fertility, spike branching, spikelet development, dryland, high temperature
Introduction
The grass tribe Triticeae is of critical importance to the global food supply. It includes major cereal crops, such as bread wheat (Triticum aestivum), pasta wheat (T. turgidum ssp. durum), and barley (Hordeum vulgare), as well as climate-resilient crops such as rye (Secale cereale) and triticale (×Triticosecale). In addition to these cereals, the Triticeae comprises about 350 other species, including the economically significant perennial fodder grasses Agropyron, Elymus, Leymus, and Psathyrostachys (Barkworth and Bothmer 2009). The annual cereal plant species are most abundant in western Asia and around the Mediterranean region, but are also found in temperate and semi-arid regions around the world.
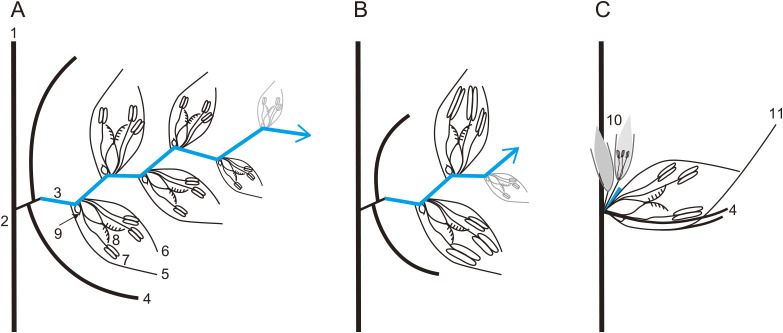
The branched compound inflorescences of grasses consist of several spikelets as fundamental units. The spikelet structure is unique to grass species which harbors one or more flowers known as florets that are subtended by a pair of bract-like structures called glumes. Typically, the floret of a Triticeae spikelet harbors reproductive organs such as two lodicules, three stamens and a pistil with two styles and stigmas all enclosed between two bract-like structures called lemma and palea (Bonnett 1966, Clayton 1990). Triticeae inflorescences, called “spikes”, differ morphologically from those of other grasses as they have an unbranched simple structure and almost sessile spikelets, meaning their spikelets attach directly to a main axis, the rachis. The Triticeae spike is basically formed by three-levels of meristematic organization, that is the inflorescence meristem producing rachis and spikelet meristems, spikelet meristem producing rachilla, and floret meristem producing florets (Figs. 1, 2). The wheat spike shape is particularly representative of the Triticeae, comprising a single spikelet per rachis node and multiple florets per spikelet (Sakuma et al. 2011). The inflorescence (spike) meristem of wheat and its relatives is determinate, with a terminal spikelet at its apex (Fig. 1). The terminal spikelet is formed at the apical end of the rachis, which determines the number of spikelets per spike. Each spikelet generates an indeterminate number of florets attached to a secondary axis, the rachilla. A hexaploid wheat spikelet can produce up to 12 floret primordia; however, generally fewer than four florets survive during development (Guo and Schnurbusch 2015) (Fig. 2). By contrast, the numbers of florets per spikelet in barley are determined to one (Fig. 2). In case of rye, though more florets are formed per spikelet (3–6), always, the first two florets become fertile and bear grains (Fig. 2). The variation in floret number per spikelet within Triticeae species is controlled by the determinate/indeterminate fates of the spikelet axis—rachilla (described in the section “regulation of spikelet determinacy”). The abortion of spikelet primordia or developing florets resulting in sterility is common in the Triticeae species. The lateral spikelets in two-rowed barley and the apical florets in wheat and rye are sterile because their development is suppressed during the growth process. Although the functional and biological significance of the sterile florets remains unknown, several genes that regulate floret development have been elucidated (Koppolu et al. 2022a, Sakuma and Schnurbusch 2020).
Fig. 1.
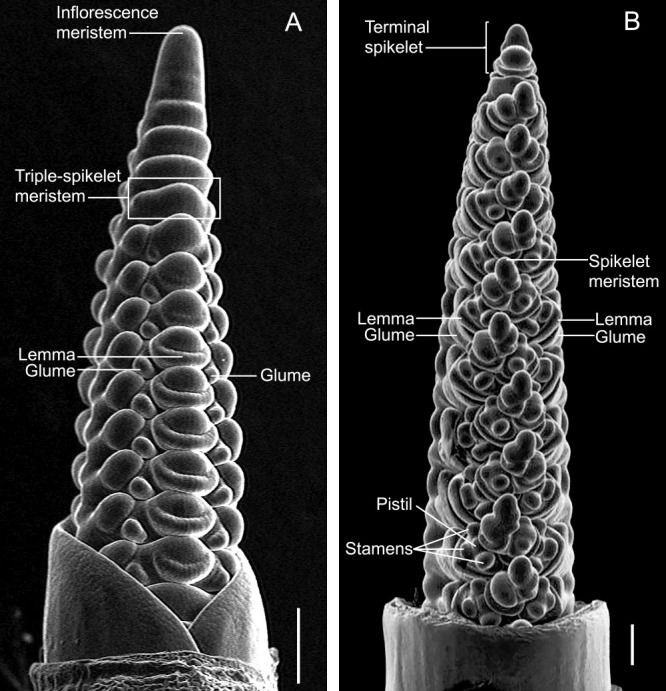
Inflorescence structure of barley and wheat. Scanning electron microscopy images of immature barley (A) and wheat (B) spikes are shown. The determinacy of the inflorescence meristem is lost in barley, whereas it is maintained to differentiate a terminal spikelet in wheat. The position of glumes is different. The barley triple-spikelet meristem is a determinate, and the wheat spikelet meristem is indeterminate. Scale bars = 200 μm.
Fig. 2.
Spikelet determinacy in Triticeae. Cartoons showing wheat (A), rye (B), and barley (C) spikelet features representative of the Triticeae spikelet structure. Spikelets in these species bear florets on the spikelet axis—rachilla (highlighted in blue). In wheat, and rye the indeterminate rachilla bears more than one floret per spikelet whereas in barley the rachilla is determinate and bears a single floret. 1 – Inflorescence meristem; 2 – Rachis; 3 – Rachilla; 4 – Glume; 5 – Lemma; 6 – Palea; 7 – Stamens; 8 – Carpel; 9 – Lodicules; 10 – Lateral spikelets; 11 – Awn.
Recent advances in genome assemblies of the Triticeae species with relatively large genome sizes have accelerated genomics-based research, including the study of developmental biology (Mascher et al. 2019). Like rice (Oryza sativa), maize (Zea mays), and sorghum (Sorghum bicolor), the Triticeae are now the subject of extensive genomic analyses (Jayakodi et al. 2020, Rabanus-Wallace et al. 2021, Walkowiak et al. 2020). The genetic diversity of the entire domesticated barley collection (~20,000 accessions) maintained at IPK, the German federal ex situ genebank, has been elucidated (Milner et al. 2019). Combining colossal sample sizes and ultra-dense DNA marker data has afforded great power for genome-wide association scans studies (GWAS). Following this, several studies reported population genomic analysis of wheat and rye (Gaurav et al. 2022, Sun et al. 2022). Importantly, abundant genetic stocks are available from genebanks for research and breeding purposes upon request (Knüpffer 2009, Schulthess et al. 2022). Furthermore, several induced mutant populations have been developed, with the TILLING platform being particularly highly efficient and cost effective for generating targeted mutant lines (Jiang et al. 2022). The exomes of 2,735 mutant lines of tetraploid and hexaploid wheat have been sequenced, revealing more than 10 million mutations in the protein-coding regions (Krasileva et al. 2017). This public collection of mutant wheat stocks enables rapid gene identification and the elucidation of previously hidden variation. Along with genomic datasets, transcriptional data also have been collected for barley and wheat (Pfeifer et al. 2014, Thiel et al. 2021). These multi-omics data have revolutionized research strategies and accelerated functional genomics in Triticeae species. These resources are all freely available on web-accessible, user-friendly platforms (Ma et al. 2021, Zhang et al. 2021) listed in Table 1. In addition, advances in genetic transformation and targeted genome modification have impacted Triticeae research (detailed in a review by Hisano et al. (2021). In the present review, our current understanding of the genetic basis of Triticeae inflorescences with a focus on barley and wheat, especially the genes controlling 1) spike branching and 2) spikelet/floret development, is described (Table 2).
Table 1.
Useful databases for functional genomics in the Triticeae
| Website name | URL |
|---|---|
| GrainGenes | https://wheat.pw.usda.gov/GG3/ |
| BARLEX | https://apex.ipk-gatersleben.de/apex/f?p=284:10 |
| IPK Galaxy Blast Suite | https://galaxy-web.ipk-gatersleben.de/ |
| Barley Genes and Barley Genetic Stocks | https://www.nordgen.org/bgs/ |
| Barley spike transcriptional landscape | http://bar.utoronto.ca/eplant_barley/ |
| Wheat eFP Browser | http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi |
| Wheat Expression Browser | http://www.wheat-expression.com/ |
| WheatOmics | http://wheatomics.sdau.edu.cn/ |
| NBRP-wheat | https://shigen.nig.ac.jp/wheat/komugi/ |
| NBRP-barley | http://earth.nig.ac.jp/~dclust/cgi-bin/index.cgi |
| SeedStor | https://www.seedstor.ac.uk/ |
| Wheat training | http://www.wheat-training.com/ |
Table 2.
List of genes discussed in this review
| Phenotype | Gene/locus-barley | Gene/locus-wheat | Morex V2 gene ID | Chinese Spring v2.1 gene ID | Protein | ||
|---|---|---|---|---|---|---|---|
| Spike branching | COMPOSITUM 1 (COM1) | Unknown function | HORVU.MOREX.r2.5HG0397500.1 | TraesCS5A03G0537300.1 | TraesCS5B03G0548700.1 | TraesCS5D03G0500500.1 | class II CYC/TB1 TCP TF |
| COMPOSITUM 2 (COM2) | branched headt | HORVU.MOREX.r2.2HG0093930.1 | TraesCS2A03G0239400.1 | TraesCS2B03G0329100.1 | TraesCS2D03G0248500.1 | AP2-ERF TF | |
| Six-rowed spike 4 (vrs4) | Unknown function | HORVU.MOREX.r2.3HG0194160.1 | TraesCS3A03G0206100.1 | TraesCS3B03G0249000.1 | TraesCS3D03G0184600.1 | LOB domain TF | |
| HvMADS1 | Unknown function | HORVU.MOREX.r2.4HG0329790.1 | TraesCS4A03G0052800.1 | TraesCS4B03G0666300.1 | TraesCS4D03G0586600.1 | SEPALLATA TF | |
| Supernumerary spikelets | COMPOSITUM 2 (COM2) | MULTI ROW SPIKE (MRS) | HORVU.MOREX.r2.2HG0093930.1 | TraesCS2A03G0239400.1 | TraesCS2B03G0329100.1 | TraesCS2D03G0248500.1 | AP2-ERF TF |
| Unknown function | DUO-B1 | HORVU.MOREX.r2.1HG0055140.1 | TraesCS1A03G0780000.1 | TraesCS1B03G0895200.1 | TraesCS1D03G0747200.1 | AP2-ERF TF | |
| PHOTOPERIOD-H1 (Ppd-H1) | PHOTOPERIOD RESPONSE LOCUS1 (PPD1) | HORVU.MOREX.r2.2HG0088300.1 | TraesCS2A03G0159800.1 | TraesCS2B03G0222600.1 | TraesCS2D03G0156800.1 | PSEUDO RESPONSE REGULATOR (PRR) | |
| FLOWERING LOCUS T1 (FT1) | FLOWERING LOCUS T1 (FT1) | HORVU.MOREX.r2.7HG0542540.1 | TraesCS7A03G0272100.1 | TraesCS7B03G0031800.1 | TraesCS7D03G0250300.1 | Phosphatidylethanolamine-binding protein (PEBP) | |
| Intermedium spike-C (vrs5) | TEOSINTE BRANCHED 1 (TB1) | HORVU.MOREX.r2.4HG0280760.1 | TraesCS4A03G0702300.1 | TraesCS4B03G0092100.1 | TraesCS4D03G0066700.1 | class II TCP TF | |
| Unknown function | HOMEOBOX DOMAIN-2 (HB2) | HORVU.MOREX.r2.1HG0033880.1 | TraesCS1A03G0425300.1 | TraesCS1B03G0515400.1 | TraesCS1D03G0402600.1 | class III homeodomain-leucine zipper TF | |
| EXTRAFLORET (FLO) | Unknown | not yet cloned | — | ||||
| Spike/spikelet determinacy | HvAP2L-H5 | Q | HORVU.MOREX.r2.5HG0437030.1 | TraesCS5A03G1116700.1 | TraesCS5B03G1184600.1 | TraesCS5D03G1069300.1 | APETALA 2-like TF |
| MULTIFLORUS 2 (MUL2) | Unknown | not yet cloned | — | ||||
| Floret development | Unknown function | VEGETATIVE TO REPRODUCTIVE TRANSITION 2 (VRT2) | HORVU.MOREX.r2.7HG0551090.1 | TraesCS7A03G0411400.1 | TraesCS7B03G0215800.1 | TraesCS7D03G0398300.1 | MADS-box TF |
| third outer glume 1 (trd1) | Unknown function | HORVU.MOREX.r2.1HG0075620.1 | TraesCS1A03G1020200.1 | TraesCS1B03G1202900.1 | TraesCS1D03G0980700.1 | GATA TF | |
| Six-rowed spike 1 (vrs1) | Grain Number Increase 1 (GNI1) | HORVU.MOREX.r2.2HG0152930.1 | TraesCSU03G0009700.1 | TraesCS2B03G1031100.1 | TraesCS2D03G0878100.1 | class I homeodomain-leucine zipper TF | |
| Six-rowed spike 2 (vrs2) | Unknown function | HORVU.MOREX.r2.5HG0413830.1 | TraesCS5A03G0773600.1 | TraesCS5B03G0808200.1 | TraesCS5D03G0732800.1 | SHORT INTERNODES (SHI) TF | |
| Six-rowed spike 3 (vrs3) | Unknown function | HORVU.MOREX.r2.1HG0041980.1 | TraesCS1A03G0526300.1 | TraesCS1B03G0615000.1 | TraesCS1D03G0507600.1 | Jumonji C-type H3K9me2/me3 demethylase | |
| Unknown function | Tipped1 (B1) | HORVU.MOREX.r2.2HG0141060.1 | TraesCS5A03G1268100.1 | TraesCS4B03G0896100.1 | TraesCS4D03G0789800.1 | C2H2-type zinc finger TF | |
| Cleistogamy 1 (Cly1) | TaAP2 | HORVU.MOREX.r2.2HG0170380.1 | TraesCS2A03G1196100.1 | TraesCS2B03G1363800.1 | TraesCS2D03G1150300.1 | APETALA 2 TF | |
Genetic basis of spike/spikelet architecture in the Triticeae
Grass species display a vast array inflorescence architectures, ranging from branched panicle/compound spikes to inflorescences with highly reduced branching, as seen in the spike inflorescences of the Triticeae (Kellogg et al. 2013). Within the spike-type inflorescences, the inflorescence branches (as seen in panicle inflorescences) are highly reduced to single spikelets attached to the rachis (Koppolu et al. 2022a). In barley, the spikelet ridge meristem formed during the double ridge stage differentiates into three spikelet meristems, giving rise to the canonical triple spikelet meristem (TSM; Koppolu et al. 2013) that develops into the triple spikelet structure (three spikelets per rachis node; Fig. 1). By contrast, the spikelet ridge in wheat differentiates into a single spikelet meristem, limiting the number of spikelets per rachis node to one, indicating a further reduction in spike inflorescence complexity within the Triticeae (Bonnett 1935, 1936) (Fig. 1). Canonical Triticeae spikes are therefore devoid of visible long or short lateral inflorescence branches. Another important component of spike architecture within the Triticeae is spikelet determinacy, which dictates the number of florets generated per spikelet. The spikelets of barley are determinate as they bear a single floret on the spikelet axis, known as the rachilla, whereas the wheat spikelets are indeterminate, bearing up to 12 florets on elongated rachillas (Fig. 2). The genetic basis of the spike/spikelet architecture within the Triticeae has been well elucidated over the last 10 years. Various barley and wheat developmental mutants showing perturbations in canonical spike/spikelet architecture (spike branching/spikelet indeterminacy) have been characterized and the underlying genetic and molecular mechanisms were revealed. In the following sections, we elucidate these mechanisms under the subsections (i) Regulation of spike branch outgrowth, (ii) Regulation of supernumerary spikelet formation, and (iii) Regulation of spikelet determinacy.
Regulation of spike branch outgrowth
Within the Triticeae cereals, the active branch suppression and spike inflorescence shape are modulated by a class of genes called COMPOSITUMs (COM). COM1 encodes a class II TCP transcription factor (TF), whereas COM2 (an ortholog of rice FRIZZY PANICLE) encodes an AP2-ERF TF (Poursarebani et al. 2015, 2020). Mutations in barley com1 and com2 result in the loss of spikelet meristem identity, leading to the development of lateral branch-like structures (secondary spikes) in place of spikelets (Fig. 3A–3C). Intriguingly, the barley com1 com2 double mutants display an enhanced spike-branching phenotype in comparison with the single mutants, indicating the additive nature of these genes towards the branching phenotype (Poursarebani et al. 2020). Similarly in tetraploid wheat, branched headt (bht; TtBH1), the ortholog of barley COM2, functions to suppress spike branching (Poursarebani et al. 2015) (Fig. 3E, 3F). Another important regulator of the branch suppression pathway in barley is the row-type gene SIX-ROWED SPIKE 4 (VRS4; HvRAMOSA2). Like the com mutants, vrs4 mutants also display a loss of spikelet meristem identity and lateral branch outgrowth (Fig. 3A, 3D) (Poursarebani et al. 2015). VRS4 is believed to directly or indirectly regulate the transcription of both COM1 and COM2 to modulate the unbranched spike inflorescence shape of barley (Poursarebani et al. 2015, 2020). The roles of COM1 and VRS4 in wheat spike development are unknown; however, it is interesting to speculate that these genes may play similar roles in suppressing branching to maintain a spike-shaped inflorescence across the Triticeae. Two other wheat spike-branching loci causing false spike ramifications (extended rachilla elongation), sham ramification 1 and 2 (shr1, shr2), have been genetically mapped, but their underlying genes are not known (Amagai et al. 2014, 2015, Dobrovolskaya et al. 2017).
Fig. 3.
Spike branching regulation in barley and wheat. (A) Unbranched wildtype barley spike (2-rowed). (B–D) mutant spikes of compositum 1 (com1; B), compositum 2 (com2; C), and six-rowed spike 4 (vrs4; D) showing long lateral branches developing from the basal to mid region of the spike. (E) Unbranched tetraploid wheat spike. (F) Branched tetraploid wheat spike. (G) Unbranched hexaploid wheat spike. (H) Multiple-spikelet hexaploid wheat spike. White arrowheads indicate spike branches/supernumerary spikelets. Scale bars = 5 cm.
In a recent study, another barley gene, HvMADS1 (encoding the SEPALLATA TF), was shown to suppress spike branching at high temperatures to maintain a regular spike shape (Li et al. 2021a). Under high temperatures, HvMADS1 activates the transcription of genes associated with inflorescence differentiation and phytohormone signaling, especially the gene encoding the cytokinin-degrading enzyme CYTOKININ OXIDASE 3 (HvCKX3), to integrate the temperature response and cytokinin homeostasis, which is required to repress cell division in the meristems (Li et al. 2021a).
Regulation of supernumerary spikelet formation
Archetypal wheat spikes harbor distichously arranged single spikelets at each rachis node, whereas barley spikes harbor three spikelets per rachis node. In both wheat and barley, some spike forms with supernumerary/paired spikelets (SSs; > typical spikelet number per node position) that deviate from the canonical spikelet arrangement exist. These SSs generally form adaxially to the primary spikelet position (Boden et al. 2015), and are often referred to as short spike branches.
The genetic and molecular regulation of the SS phenotype has been well characterized in wheat (Fig. 3G, 3H). The first step towards understanding the mechanism of SS formation came from the cloning of MULTI ROW SPIKE (wheat FRIZZY PANICLE; WFZP; encoding an AP2-ERF TF; (Dobrovolskaya et al. 2015)). In hexaploid wheat, mutations in the WFZP-D homoeolog drive the SS phenotype while the mutations in WFZP-A along with WFZP-D mutation enhance the SS phenotype (Du et al. 2021, Li et al. 2021b) (Fig. 3G, 3H). Recently, DUO-B1, yet another AP2-ERF TF, has been implicated in the control of SS formation in wheat. Interestingly, mutants of wheat DUO-B1 showed an SS phenotype similar to the multirow spike mutants, and a further molecular analysis revealed that DUO-B1 suppresses cell division and positively regulates the expression of WFZP to control SS development (Wang et al. 2022b).
Various other genes have recently been reported to regulate the SS phenotype, including the PHOTOPERIOD RESPONSE LOCUS1 (PPD-1), FLOWERING LOCUS T 1 (FT1), and the major domestication gene TEOSINTE BRANCHED 1 (TB1) (Boden et al. 2015, Dixon et al. 2018). It has been shown that TB1 and PPD1 regulate FT1—the central regulator of the floral meristem identity gene cascade to control SS formation in wheat. In the photoperiod-insensitive mutant Ppd-D1, FT1 expression is attenuated, promoting SS formation by delaying spikelet meristem maturation (Boden et al. 2015). Intriguingly, TB1 also promotes SS formation by attenuating FT1. However, TB1 mediates FT1 attenuation in a manner distinct to Ppd-1, where TB1 forms protein complex with FT1. In the gain-of-function wheat TB1 alleles, the TB1 protein competitively binds to FT1, making it less available to promote meristem maturation (Dixon et al. 2018). In another study, Dixon et al. (2022) showed that semidominant alleles of the wheat A and D homoeologous genes encoding the class III homeodomain-leucine zipper TF HOMEOBOX DOMAIN-2 (HB-2) promote SS formation. In contrast to the previous mechanisms, the regulation of SS formation by HB-2 is modulated through microRNA-based regulation; in the semi-dominant alleles of HB-2, the complementary microRNA165/166 (miR165/166) binding site is disrupted, leading to elevated levels of HB-2 transcripts known to promote leaf and vascular development and increase the amino acid supply required for grain development in the SS (Dixon et al. 2022).
Various uncharacterized genomic regions controlling SS formation have also been identified through quantitative trait loci (QTL) studies, indicating that SS is a highly quantitative phenotype (Boden et al. 2015, Echeverry-Solarte et al. 2014, Wolde et al. 2021). Despite the wealth of genetic evidence available for the regulation of SS formation in wheat, our genetic knowledge of this phenotype in barley is completely lacking. Although a class of barley mutants called extrafloret (flo.a, -.b, and -.c) and another mutant multiflorus 2.b display the SS phenotype (Koppolu et al. 2022b). However, the genes underlying these mutant phenotypes are not yet known.
Regulation of spikelet determinacy
Spikelet determinacy in wheat and barley is largely determined by the elongation or suppression of the spikelet axis, known as the rachilla. In the determinate barley spikelet, the rachilla degenerates after producing one floret, whereas in the indeterminate wheat spikelet the rachilla continues to elongate, producing up to 12 florets before being degenerated (Fig. 2). In the majority of grasses, orthologous APETALA 2 (AP2) genes, maize INDETERMINATE SPIKELET1 (IDS1)/TASSELSEED 6 (Chuck et al. 1998, 2007, 2008), rice IDS1 (Lee and An 2012), and wheat Q (AP2L5) (Debernardi et al. 2017, Greenwood et al. 2017) regulate rachilla elongation, thereby controlling the floret number per spikelet. In a recent discovery, barley researchers showed that HvAP2L-H5 (an ortholog of Q) regulates the determinate fate of barley spikelets, with ap2l5 mutants losing the determinate nature of spikelets and producing more than one floret on elongated rachilla (Zhong et al. 2021). Another barley mutant, multiflorus 2, produces indeterminate spikelets bearing up to three florets on its elongated rachillas (Koppolu et al. 2022b); however, the gene(s) regulating this phenotype are yet to be identified.
Interestingly, in the lateral spikelets of the barley spike-branching mutants com1, com2, and vrs4, the rachilla elongates to produce more than one floret/spikelet, indicating a loss of spikelet determinacy in these lines (Koppolu and Schnurbusch 2019). From these studies, it is evident that the elongation or suppression of the rachilla (the spikelet axis) can dictate the floret number per spikelet, making it an important yield-determining organ in these grass crops.
Genes regulating spikelet/floret development
The structure and development of the spikelet are key determinants of the grass reproductive organ (Kellogg 2022, Wang et al. 2022a). The number of spikelets per rachis node and florets within a spikelet are diagnostic characters of the Triticeae (Sakuma et al. 2011). The most common Triticeae spikelet form is a single spikelet per rachis node, as seen in wheat and rye (Bonnett 1966). Unlike the single spikelet type, barley produces a triple-spikelet meristem, resulting in one central spikelet and two lateral spikelets per rachis node. This character is unique to barley and wild Hordeum species (Bothmer et al. 1995).
The spikelet is distinguished by two glumes surrounding one or more florets; thus, the differentiation of the glume primordia is also a key determinant of spikelet identity. The function of the glumes is not yet well understood; however, their toughness is very important for grain retention or easy threshability in domesticated wheat. During wheat domestication, a dominant mutation at the Q locus was fixed during the artificial selection of spikes that were easier to thresh (Simons et al. 2006). The expression of Q, encoding an AP2-like TF, is negatively regulated by miR172; a reduction in miR172 expression led to higher levels of Q expression and greater similarity between glumes and lemmas. Conversely, high levels of miR172 and the loss of function of Q (three homoeoalleles) leads to sterile lemmas and the indeterminacy of the spikelet meristem (Debernardi et al. 2017). In the lowermost spikelets, the transition between glumes and lemmas appeared particularly malleable, such that more miR172 and less AP2L5 could lead to glume-like organs in the position of lemmas (i.e., sterile lemmas).
Variation in the size and position of the glumes is also evident in Triticeae species, with enlarged distichous glumes in wheat and smaller and more pointed parallel glumes in barley (Fig. 1). Longer glume is particularly evident in the tetraploid wheat Triticum turgidum ssp. polonicum (also termed T. polonicum). Recent studies revealed that the ectopic expression of VEGETATIVE TO REPRODUCTIVE TRANSITION 2 (VRT2, P1 locus, chromosome 7A) in the spikelet organs underlies the elongated glume phenotype of T. polonicum (Adamski et al. 2021). The gene encodes a MADS-box TF belonging to the SHORT VEGETATIVE PHASE family. In addition, the paralog of VRT2 (SVP-A1) is proposed to be a candidate gene of the P2 locus on chromosome 6A (Chen et al. 2022). In Triticum isphanicum, which develops enlarged glumes, a 482-bp deletion in the SVP-A1 promoter was found to be associated with the ectopic and higher expression of this gene in the elongated glumes. In barley, the third outer glume 1 (trd1) mutant, is characterized by the outgrowth of leaf-like structures (bracts) in between the two glumes of the central spikelets. The gene underlying trd1 has been shown to encode the GATA TF, and is orthologous to rice NECKLEAF 1 (NL1) and maize TASSELSHEATH 1 (TSH1) (Houston et al. 2012). Interestingly, rice spikelets show rudimentary glumes and empty glumes (called sterile lemmas). The mutation of rice long sterile lemma1 (g1), encoding an ALOG protein, appears to promote the homeotic transformation of the sterile lemma into a lemma-like structure (Yoshida et al. 2009). Several studies suggest that the putative Oryza ancestor had three florets, with the two lateral florets degenerated during evolution (Ren et al. 2013, Yoshida et al. 2009).
Floret abortion/fertility
Floret fertility is the most important trait determining the final grain number of each inflorescence. A single floret is composed of, from the outside, a lemma with or without an awn, a palea, three stamens, two lodicules, and a pistil (Fig. 2). The lemma and palea are considered to be leaf-like structures containing chlorophyll. In Triticeae crops, several key genes regulating floret development have been identified in the last two decades. A six-rowed spike phenotype is a major target for barley researchers, with the row-type determining gene Vrs1 first cloned as key suppressor of floret fertility at the lateral spikelets (Komatsuda et al. 2007). The gene encodes a homeodomain leucine zipper class I TF, which is unique to the plant kingdom. The wheat Vrs1 ortholog (Grain Number Increase 1; GNI1) was also found to be a suppressor of apical florets within the spikelets (Sakuma et al. 2019). The loss of Vrs1/GNI1 function results in more grains formed per spike. Interestingly, the wheat spikes appear to be evolved to produce more fertile florets per spikelet with the increase in ploidy level and associated heterochrony in floret development (Shitsukawa et al. 2009). The diploid einkorn wheats usually set one grain, while the tetraploid wheats set two or three, and the hexaploid wheats set more than three grains per spikelet (Sakuma et al. 2019, Shitsukawa et al. 2009). It is interesting to speculate that the diploidy in barley and rye could be one of the reasons for lower number of florets/grains formed per spikelet, compared to hexaploid wheat, however research-based evidence is necessary to back this hypothesis. A special allele of barley Vrs1, originally endemic to Ethiopia, was also identified (Sakuma et al. 2017). The causal mutation of this mutant allele, known as deficiens, is a single amino acid substitution located at an unknown C-terminal domain. The deficiens spike shows rudimentary lateral spikelets and enlarged grains in the central spikelet. In barley, the induced six-rowed spike mutants vrs2, vrs3, vrs4, and vrs5 have also been cloned, although these mutant alleles are not yet utilized in breeding programs (Bull et al. 2017, Koppolu et al. 2013, Ramsay et al. 2011, van Esse et al. 2017, Youssef et al. 2017). The function of the wheat orthologs of vrs2, vrs3, and vrs4 remain unknown; however, the wheat vrs5 ortholog TB1 was found to be a regulator of paired spikelet formation (Dixon et al. 2018). Wheat TB1 also regulates plant height and the length of the stem internode (Dixon et al. 2020).
Awn development
Awns are needle-like structures, elongated from apex of the lemma in grasses (Fig. 2). Triticeae species present diverse awn morphologies, ranging from long to short awns. Spring-type bread wheat cultivars tend to have long awns, while winter-types have short/tipped awns. Awns in Triticeae crops contribute to photosynthesis and grain yields in warmer and drier rainfed environments (Guo and Schnurbusch 2016, Rebetzke et al. 2016). Several studies have attempted to understand awn function, with some researchers hypothesizing that awns use C4 photosynthesis rather than the C3 photosynthesis typically used by the Triticeae (AuBuchon-Elder et al. 2020, Tambussi et al. 2021). Awns are also important from the point of view of domestication and adaptation of wild species. Wild emmer wheat characteristically has two long awns per spikelet. The arrangement of cellulose fibrils causes the awns to bend with changes in humidity, helping them to play a role in seed dispersal by either fixing into the soil or attaching to wild animals (Elbaum et al. 2007). The genetic basis of awn bending is currently unknown. In wheat, awn elongation is suppressed by three dominant genes, Tipped1 (B1) on chromosome 5A, Tipped2 (B2) on chromosome 6B, and Hooded (Hd) on chromosome 4A (Yoshioka et al. 2017). B1 was identified as a gene encoding C2H2 zinc finger with ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs (Huang et al. 2020). Constitutive overexpression of B1 is responsible for awn inhibition together with pleiotropic effects on plant height and fertility. Haplotype analysis revealed that four SNPs located in B1 promoter region are associated with awn length, grain length and thousand-grain weight (Wang et al. 2020). The investigation of 231 wheat lines from the NIAB MAGIC population found that the presence of awns increased the grain calcium content without decreasing the flour extraction rate, despite the negative correlation between these traits (Fradgley et al. 2022).
Floret opening and its importance towards hybrid breeding
Hybrid breeding has the potential to boost Triticeae crop yields. Indeed, breeding programs for allogamous rye have succeeded in enhancing its inflorescence structure, large anther extrusion, pollen production, and the development of efficient cytoplasmic male sterility (CMS) coupled with nuclear Restorer-of-fertility (Rf) genes (Miedaner et al. 2022, Vendelbo et al. 2020). Hybrid seed production in autogamous plants such as wheat and barley remains challenging however, because they require a self-pollination block. Although the molecular mechanisms underlying Rf genes have been elucidated in wheat (Melonek et al. 2021), the hybrid seed production system is currently insufficient. Understanding floret structure, including the development of the anthers and lodicule, is therefore important. Three anthers are produced in Triticeae florets, there is a correlation between anther length and number of pollen grains. Although the size and number of pollen grains is crucial for the success of hybrid breeding, its genetic basis remains unknown. The Rht-D1a allele results in a tall stature in bread wheat, but is also associated with large anthers and a high anther extrusion, despite not enhancing the anther filament length (Okada et al. 2021). The lodicule functions to open the lemma and palea during anthesis. Lodicules in cleistogamous barley remain small due to the lack of vascular tissue, a phenotype which is under the control of the Cleistogamy 1 (Cly1) locus encoding an AP2 TF (Nair et al. 2009). The cleistogamous barley cultivars possess the cly1.b allele, which is distinguished by a synonymous mutation at the miR172 binding site. This change results in the reduced abundance of the CLY1 protein, but not of its transcript (Anwar et al. 2018).
Grain shattering
Grain shattering has long been recognized to cause yield losses in cereal crops (Bolland 1984, Clarke 1981, Sugimoto et al. 2010). Grain shattering is distinguished from the brittle rachis trait shown in wild barley and wild emmer wheat (Pourkheirandish and Komatsuda 2022), with the causal loss-of-function mutations located at Non-brittle rachis 1 (btr1) and btr2 (Avni et al. 2017, Pourkheirandish et al. 2015). In grain shattering, spikelet disarticulation from rachis occurs above the glume whereas in brittle rachis phenotype, disarticulation occurs below the glume (Sakuma et al. 2011). Some wheat varieties growing in heat-prone and drylands such as Sudan show grain-shattering habits, in which grains fall to the ground when harvesting is delayed. This results in a yield loss of up to 30%; thus, the development of cultivars with reduced shattering while maintaining threshability is an important breeding goal. Recent study by Bokore et al. (2022) revealed that a major QTL on chromosome 4BS is associated with reduced grain shattering, and a second QTL was detected on chromosome 5AL. These works shed light on grain shattering resistance and provide DNA markers for developing new cultivars.
The causes and impact of grain shattering (syn. head shattering, head loss) have been reported in barley too (Kandemir et al. 2000). A major QTL for head shattering (designated Hst-3) has been mapped onto the centromeric region of chromosome 3H, which is different from the Btr1/Btr2 loci. Some barley cultivars drop their spikes onto the ground during a heatwave (Curry and Paynter 2019). In barley, a spontaneous mutant in a cultivar named Kamairazu, which means “sickle not needed to harvest”, has leaves and stems that are easily broken when physically bent (Takahashi et al. 1953). This extraordinary fragility is exhibited even after maturity. The locus controlling this fragile phenotype is located on chromosome 5HL, and has been renamed fragile stem 1 (fst1). The identification of the gene underlying the fst1 and a deeper understanding of spike shattering would be valuable for future cereal breeding under ongoing climate change.
Future perspectives
As discussed above, inflorescence shape fundamentally contributes to final grain number and size, which is critical to final grain yield. The branched inflorescences in general tend to produce more grains per inflorescence. For example, the panicle inflorescences of rice and sorghum produce more grains per inflorescence compared to species possessing reduced or unbranched inflorescences as in barley and wheat. However, tillering and the grain size trade-offs are evident between branched and unbranched inflorescences. Comparative yield studies across species bearing different inflorescence types can give an understanding about the influence of inflorescence forms on final grain yield in these species. Also, a thorough understanding of the genetic basis underlying these inflorescence shapes helps boosting the yield potential and improve yield stability.
The effects of climate change are already felt around the world, and are expected to worsen significantly over time. Ongoing increases in temperature and rainfall variability in Triticeae crop–growing areas, including temperate, semi-arid, and dryland regions, limit yield improvements, with large reductions and problematic variability in production predicted to occur as a result of climate change. Drylands are a frontier of global warming; hence, understanding crop growth phenomena in drylands is crucial for predicting future patterns of agriculture. Record high temperatures and droughts occur almost every year in relatively stable regions such as Europe, with regular flooding across Japan. To adapt to these climate shifts, the development of new crop varieties using novel strategies and genomic diversity is urgently required. However, our understanding of inflorescence shape and development against such kinds of stress and response is still limited. The first step towards understanding the influence of temperature on spike branch outgrowth regulation has been laid out by characterization of HvMADS1 and its role in maintaining spike shape under ambient temperatures (Li et al. 2021a). However, there is still a strong demand for research and investigation for precise understanding of the influence of temperature and water stress towards inflorescence morphogenesis. The outputs from such research activities can ultimately pave the way for the development of climate-resilient cultivars adapted to various environments by leveraging genetic resources and genomic tools.
Author Contribution Statement
SS conceptualized the review and participated in drafting the initial version, preparation of illustrations and revision of the review. RK participated in drafting the initial version, preparation of illustrations and revision of the review. Both authors have read and agreed upon the final content of the review.
Acknowledgments
This work was supported in part by JSPS KAKENHI grant number 22H02312, the Science and Technology Research Partnership for Sustainable Development (SATREPS) grant JPMJSA1805 by Japan Science and Technology Agency, and Tottori University Research Support Project for the Next Generation.
Literature Cited
- Adamski, N.M., Simmonds J., Brinton J.F., Backhaus A.E., Chen Y., Smedley M., Hayta S., Florio T., Crane P., Scott P.et al. (2021) Ectopic expression of Triticum polonicum VRT-A2 underlies elongated glumes and grains in hexaploid wheat in a dosage-dependent manner. Plant Cell 33: 2296–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai, Y., Aliyeva A.J., Aminov N.K., Martinek P., Watanabe N. and Kuboyama T. (2014) Microsatellite mapping of the genes for sham ramification and extra glume in spikelets of tetraploid wheat. Genet Resour Crop Evol 61: 491–498. [Google Scholar]
- Amagai, Y., Aliyeva A.J., Aminov N.K., Martinek P., Watanabe N. and Kuboyama T. (2015) Microsatellite mapping of the gene for sham ramification in spikelets derived from a hexaploid wheat (Triticum spp.) accession 171ACS. Genet Resour Crop Evol 62: 1079–1084. [Google Scholar]
- Anwar, N., Ohta M., Yazawa T., Sato Y., Li C., Tagiri A., Sakuma M., Nussbaumer T., Bregitzer P., Pourkheirandish M.et al. (2018) miR172 downregulates the translation of cleistogamy 1 in barley. Ann Bot 122: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AuBuchon-Elder, T., Coneva V., Goad D.M., Jenkins L.M., Yu Y., Allen D.K. and Kellogg E.A. (2020) Sterile spikelets contribute to yield in sorghum and related grasses. Plant Cell 32: 3500–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni, R., Nave M., Barad O., Baruch K., Twardziok S.O., Gundlach H., Hale I., Mascher M., Spannagl M., Wiebe K.et al. (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Barkworth, E.M. and R. Bothmer (2009) Scientific Names in the Triticeae. In: Muehlbauer, G.J. and C. Feuillet (eds.) Genetics and Genomics of the Triticeae, Springer, New York, pp. 3–10. [Google Scholar]
- Boden, S.A., Cavanagh C., Cullis B.R., Ramm K., Greenwood J., Jean Finnegan E., Trevaskis B. and Swain S.M. (2015) Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants 1: 14016. [DOI] [PubMed] [Google Scholar]
- Bokore, F.E., Cuthbert R.D., Knox R.E., Campbell H.L., Meyer B., N’Diaye A., Pozniak C.J. and DePauw R. (2022) Main effect and epistatic QTL affecting spike shattering and association with plant height revealed in two spring wheat (Triticum aestivum L.) populations. Theor Appl Genet 135: 1143–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland, M.D.A. (1984) Grain losses due to delayed harvesting of barley and wheat. Aust J Exp Agric 24: 391–395. [Google Scholar]
- Bonnett, O.T. (1935) The development of the barley spike. J Agric Res 51: 451–460. [Google Scholar]
- Bonnett, O.T. (1936) The development of the wheat spike. J Agric Res 53: 445–451. [Google Scholar]
- Bonnett, O.T. (1966) Inflorescences of maize, wheat, rye, barley and oats: their initiation and development. University of Illinois College of Agriculture, Agricultural Experimental Station, Illinois, p. 105. [Google Scholar]
- Bothmer, R., N. Jacobsen, C. Baden, R. Jorgensen and I. Linde-Laursen (1995) An ecogeographical study of the genus Hordeum, 2nd edn. IBPGR, Rome, p. 127. [Google Scholar]
- Bull, H., Casao M.C., Zwirek M., Flavell A.J., Thomas W.T.B., Guo W., Zhang R., Rapazote-Flores P., Kyriakidis S., Russell J.et al. (2017) Barley SIX-ROWED SPIKE3 encodes a putative Jumonji C-type H3K9me2/me3 demethylase that represses lateral spikelet fertility. Nat Commun 8: 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Liu Y., Zhang J., Torrance A., Watanabe N., Adamski N.M. and Uauy C. (2022) The Triticum ispahanicum elongated glume locus P2 maps to chromosome 6A and is associated with the ectopic expression of SVP-A1. Theor Appl Genet 135: 2313–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Meeley R.B. and Hake S. (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev 12: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Meeley R., Irish E., Sakai H. and Hake S. (2007) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517–1521. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Meeley R. and Hake S. (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019. [DOI] [PubMed] [Google Scholar]
- Clarke, J.M. (1981) Effect of delayed harvest on shattering losses in oats, barley and wheat. Can J Plant Sci 61: 25–28. [Google Scholar]
- Clayton, W. (1990) The spikelet. In: Chapman, G.P. (ed.) Reproductive versatility in the grasses, Cambridge University Press, Cambridge, pp. 32–51. [Google Scholar]
- Curry, J. and B. Paynter (2019) Causes and impact of barley head loss in the Western Region. GRDC progress report, Western Australia, p. 68. [Google Scholar]
- Debernardi, J.M., Lin H., Chuck G., Faris J.D. and Dubcovsky J. (2017) microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, L.E., Greenwood J.R., Bencivenga S., Zhang P., Cockram J., Mellers G., Ramm K., Cavanagh C., Swain S.M. and Boden S.A. (2018) TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell 30: 563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, L.E., Pasquariello M. and Boden S.A. (2020) TEOSINTE BRANCHED1 regulates height and stem internode length in bread wheat. J Exp Bot 71: 4742–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, L.E., Pasquariello M., Badgami R., Levin K.A., Poschet G., Ng P.Q., Orford S., Chayut N., Adamski N.M., Brinton J.et al. (2022) MicroRNA-resistant alleles of HOMEOBOX DOMAIN-2 modify inflorescence branching and increase grain protein content of wheat. Sci Adv 8: ebn5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya, O., Pont C., Sibout R., Martinek P., Badaeva E., Murat F., Chosson A., Watanabe N., Prat E., Gautier N.et al. (2015) FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol 167: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya, O.B., Amagai Y., Popova K.I., Dresvyannikova A.E., Martinek P., Krasnikov A.A. and Watanabe N. (2017) Genes WHEAT FRIZZY PANICLE and SHAM RAMIFICATION 2 independently regulate differentiation of floral meristems in wheat. BMC Plant Biol 17: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, D., Zhang D., Yuan J., Feng M., Li Z., Wang Z., Zhang Z., Li X., Ke W., Li R.et al. (2021) FRIZZY PANICLE defines a regulatory hub for simultaneously controlling spikelet formation and awn elongation in bread wheat. New Phytol 231: 814–833. [DOI] [PubMed] [Google Scholar]
- Echeverry-Solarte, M., Kumar A., Kianian S., Mantovani E.E., Simsek S., Alamri M.S. and Mergoum M. (2014) Genome-wide genetic dissection of supernumerary spikelet and related traits in common wheat. Plant Genome 7: 1–16. [DOI] [PubMed] [Google Scholar]
- Elbaum, R., Zaltzman L., Burgert I. and Fratzl P. (2007) The role of wheat awns in the seed dispersal unit. Science 316: 884–886. [DOI] [PubMed] [Google Scholar]
- Fradgley, N.S., Gardner K., Kerton M., Swarbreck S.M. and Bentley A.R. (2022) Trade-offs in the genetic control of functional and nutritional quality traits in UK winter wheat. Heredity (Edinb) 128: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav, K., Arora S., Silva P., Sanchez-Martin J., Horsnell R., Gao L., Brar G.S., Widrig V., Raupp W.J., Singh N.et al. (2022) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat Biotechnol 40: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, J.R., Finnegan E.J., Watanabe N., Trevaskis B. and Swain S.M. (2017) New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development 144: 1959–1965. [DOI] [PubMed] [Google Scholar]
- Guo, Z. and Schnurbusch T. (2015) Variation of floret fertility in hexaploid wheat revealed by tiller removal. J Exp Bot 66: 5945–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. and Schnurbusch T. (2016) Costs and benefits of awns. J Exp Bot 67: 2533–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano, H., Abe F., Hoffie R.E. and Kumlehn J. (2021) Targeted genome modifications in cereal crops. Breed Sci 71: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston, K., Druka A., Bonar N., Macaulay M., Lundqvist U., Franckowiak J., Morgante M., Stein N. and Waugh R. (2012) Analysis of the barley bract suppression gene Trd1. Theor Appl Genet 125: 33–45. [DOI] [PubMed] [Google Scholar]
- Huang, D., Zheng Q., Melchkart T., Bekkaoui Y., Konkin D.J.F., Kagale S., Martucci M., You F.M., Clarke M., Adamski N.M.et al. (2020) Dominant inhibition of awn development by a putative zinc-finger transcriptional repressor expressed at the B1 locus in wheat. New Phytol 225: 340–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakodi, M., Padmarasu S., Haberer G., Bonthala V.S., Gundlach H., Monat C., Lux T., Kamal N., Lang D., Himmelbach A.et al. (2020) The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588: 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Lei M., Guo Y., Gao G., Shi L., Jin Y., Cai Y., Himmelbach A., Zhou S., He Q.et al. (2022) A reference-guided TILLING by amplicon-sequencing platform supports forward and reverse genetics in barley. Plant Commun 3: 100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandemir, N., Kudrna D.A., Ullrich S.E. and Kleinhofs A. (2000) Molecular marker assisted genetic analysis of head shattering in six-rowed barley. Theor Appl Genet 101: 203–210. [Google Scholar]
- Kellogg, E.A. (2022) Genetic control of branching patterns in grass inflorescences. Plant Cell 34: 2518–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A., Camara P.E.A.S., Rudall P.J., Ladd P., Malcomber S.T., Whipple C.J. and Doust A.N. (2013) Early inflorescence development in the grasses (Poaceae). Front Plant Sci 4: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüpffer, H. (2009) Triticeae Genetic Resources in ex situ Genebank Collections. In: Muehlbauer, G.J. and C. Feuillet (eds.) Genetics and Genomics of the Triticeae, Springer, New York, pp. 31–79. [Google Scholar]
- Komatsuda, T., Pourkheirandish M., He C., Azhaguvel P., Kanamori H., Perovic D., Stein N., Graner A., Wicker T., Tagiri A.et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu, R., Anwar N., Sakuma S., Tagiri A., Lundqvist U., Pourkheirandish M., Rutten T., Seiler C., Himmelbach A., Ariyadasa R.et al. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110: 13198–13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppolu, R. and Schnurbusch T. (2019) Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J Integr Plant Biol 61: 278–295. [DOI] [PubMed] [Google Scholar]
- Koppolu, R., Chen S. and Schnurbusch T. (2022a) Evolution of inflorescence branch modifications in cereal crops. Curr Opin Plant Biol 65: 102168. [DOI] [PubMed] [Google Scholar]
- Koppolu, R., Jiang G., Milner S.G., Muqaddasi Q.H., Rutten T., Himmelbach A., Guo Y., Stein N., Mascher M. and Schnurbusch T. (2022b) The barley mutant multiflorus2.b reveals quantitative genetic variation for new spikelet architecture. Theor Appl Genet 135: 571–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva, K.V., Vasquez-Gross H.A., Howell T., Bailey P., Paraiso F., Clissold L., Simmonds J., Ramirez-Gonzalez R.H., Wang X., Borrill P.et al. (2017) Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.Y. and An G. (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461. [DOI] [PubMed] [Google Scholar]
- Li, G., Kuijer H.N.J., Yang X., Liu H., Shen C., Shi J., Betts N., Tucker M.R., Liang W., Waugh R.et al. (2021a) MADS1 maintains barley spike morphology at high ambient temperatures. Nat Plants 7: 1093–1107. [DOI] [PubMed] [Google Scholar]
- Li, Y., Li L., Zhao M., Guo L., Guo X., Zhao D., Batool A., Dong B., Xu H., Cui S.et al. (2021b) Wheat FRIZZY PANICLE activates VERNALIZATION1-A and HOMEOBOX4-A to regulate spike development in wheat. Plant Biotechnol J 19: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S., Wang M., Wu J., Guo W., Chen Y., Li G., Wang Y., Shi W., Xia G., Fu D.et al. (2021) WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant 14: 1965–1968. [DOI] [PubMed] [Google Scholar]
- Mascher, M., Schreiber M., Scholz U., Graner A., Reif J.C. and Stein N. (2019) Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat Genet 51: 1076–1081. [DOI] [PubMed] [Google Scholar]
- Melonek, J., Duarte J., Martin J., Beuf L., Murigneux A., Varenne P., Comadran J., Specel S., Levadoux S., Bernath-Levin K.et al. (2021) The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat Commun 12: 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedaner, T., Korzun V. and Wilde P. (2022) Effective pollen-fertility restoration is the basis of hybrid rye production and ergot mitigation. Plants (Basel) 11: 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, S.G., Jost M., Taketa S., Mazon E.R., Himmelbach A., Oppermann M., Weise S., Knupffer H., Basterrechea M., Konig P.et al. (2019) Genebank genomics highlights the diversity of a global barley collection. Nat Genet 51: 319–326. [DOI] [PubMed] [Google Scholar]
- Nair, S.K., Wang N., Turuspekov Y., Pourkheirandish M., Sinsuwongwat S., Chen G., Sameri M., Tagiri A., Honda I., Watanabe Y.et al. (2009) Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 107: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T., Jayasinghe J.E.A.R.M., Eckermann P., Watson-Haigh N.S., Warner P., Williams M.E., Albertsen M.C., Baumann U. and Whitford R. (2021) Genetic factors associated with favourable pollinator traits in the wheat cultivar Piko. Funct Plant Biol 48: 434–447. [DOI] [PubMed] [Google Scholar]
- Pfeifer, M., Kugler K.G., Sandve S.R., Zhan B., Rudi H., Hvidsten T.R., International Wheat Genome Sequencing Consortium, Mayer K.F.X. and Olsen O.A. (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Pourkheirandish, M., Hensel G., Kilian B., Senthil N., Chen G., Sameri M., Azhaguvel P., Sakuma S., Dhanagond S., Sharma R.et al. (2015) Evolution of the grain dispersal system in barley. Cell 162: 527–539. [DOI] [PubMed] [Google Scholar]
- Pourkheirandish, M. and Komatsuda T. (2022) Grain disarticulation in wild wheat and barley. Plant Cell Physiol 63: 1584–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursarebani, N., Seidensticker T., Koppolu R., Trautewig C., Gawronski P., Bini F., Govind G., Rutten T., Sakuma S., Tagiri A.et al. (2015) The genetic basis of composite spike form in barley and ‘Miracle-wheat’. Genetics 201: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursarebani, N., Trautewig C., Melzer M., Nussbaumer T., Lundqvist U., Rutten T., Schmutzer T., Brandt R., Himmelbach A., Altschmied L.et al. (2020) COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat Commun 11: 5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanus-Wallace, M.T., Hackauf B., Mascher M., Lux T., Wicker T., Gundlach H., Baez M., Houben A., Mayer K.F.X., Guo L.et al. (2021) Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat Genet 53: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, L., Comadran J., Druka A., Marshall D.F., Thomas W.T.B., Macaulay M., MacKenzie K., Simpson C., Fuller J., Bonar N.et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172. [DOI] [PubMed] [Google Scholar]
- Rebetzke, G.J., Bonnett D.G. and Reynolds M.P. (2016) Awns reduce grain number to increase grain size and harvestable yield in irrigated and rainfed spring wheat. J Exp Bot 67: 2573–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D., Li Y., Zhao F., Sang X., Shi J., Wang N., Guo S., Ling Y., Zhang C., Yang Z.et al. (2013) MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, S., Salomon B. and Komatsuda T. (2011) The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol 52: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, S., Lundqvist U., Kakei Y., Thirulogachandar V., Suzuki T., Hori K., Wu J., Tagiri A., Rutten T., Koppolu R.et al. (2017) Extreme suppression of lateral floret development by a single amino acid change in the VRS1 transcription factor. Plant Physiol 175: 1720–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, S., Golan G., Guo Z., Ogawa T., Tagiri A., Sugimoto K., Bernhardt N., Brassac J., Mascher M., Hensel G.et al. (2019) Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc Natl Acad Sci USA 116: 5182–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, S. and Schnurbusch T. (2020) Of floral fortune: tinkering with the grain yield potential of cereal crops. New Phytol 225: 1873–1882. [DOI] [PubMed] [Google Scholar]
- Schulthess, A.W., Kale S.M., Liu F., Zhao Y., Philipp N., Rembe M., Jiang Y., Beukert U., Serfling A., Himmelbach A.et al. (2022) Genomics-informed prebreeding unlocks the diversity in genebanks for wheat improvement. Nat Genet 54: 1544–1552. [DOI] [PubMed] [Google Scholar]
- Shitsukawa, N., Kinjo H., Takumi S. and Murai K. (2009) Heterochronic development of the floret meristem determines grain number per spikelet in diploid, tetraploid and hexaploid wheats. Ann Bot 104: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K.J., Fellers J.P., Trick H.N., Zhang Z., Tai Y.S., Gill B.S. and Faris J.D. (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T.et al. (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107: 5792–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Shen E., Hu Y., Wu D., Feng Y., Lao S., Dong C., Du T., Hua W., Ye C.Y.et al. (2022) Population genomic analysis reveals domestication of cultivated rye from weedy rye. Mol Plant 15: 552–561. [DOI] [PubMed] [Google Scholar]
- Takahashi, R., Yamamoto J., Yasuda S. and Itano Y. (1953) Inheritance and linkage studies in barley. Berichte des Ohara Instituts für landwirtschaftliche Forschungen 10: 29–52. [Google Scholar]
- Tambussi, E.A., Maydup M.L., Carrion C.A., Guiamet J.J. and Araus J.L. (2021) Ear photosynthesis in C3 cereals and its contribution to grain yield: methodologies, controversies, and perspectives. J Exp Bot 72: 3956–3970. [DOI] [PubMed] [Google Scholar]
- Thiel, J., Koppolu R., Trautewig C., Hertig C., Kale S.M., Erbe S., Mascher M., Himmelbach A., Rutten T., Esteban E.et al. (2021) Transcriptional landscapes of floral meristems in barley. Sci Adv 7: abf0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, G.W., Walla A., Finke A., Koornneef M., Pecinka A. and von Korff M. (2017) Six-rowed spike3 (VRS3) is a histone demethylase that controls lateral spikelet development in barley. Plant Physiol 174: 2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelbo, N.M., Sarup P., Orabi J., Kristensen P.S. and Jahoor A. (2020) Genetic structure of a germplasm for hybrid breeding in rye (Secale cereale L.). PLoS One 15: e0239541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowiak, S., Gao L., Monat C., Haberer G., Kassa M.T., Brinton J., Ramirez-Gonzalez R.H., Kolodziej M.C., Delorean E., Thambugala D.et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Yu K., Jin D., Sun L., Chu J., Wu W., Xin P., Gregova E., Li X., Sun J.et al. (2020) Natural variations in the promoter of Awn Length Inhibitor 1 (ALI-1) are associated with awn elongation and grain length in common wheat. Plant J 101: 1075–1090. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Bi X. and Zhong J. (2022a) Revisiting the origin and identity specification of the spikelet: A structural innovation in grasses (Poaceae). Plant Physiol 190: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Du F., Wang J., Wang K., Tian C., Qi X., Lu F., Liu X., Ye X. and Jiao Y. (2022b) Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat Plants 8: 930–939. [DOI] [PubMed] [Google Scholar]
- Wolde, G.M., Schreiber M., Trautewig C., Himmelbach A., Sakuma S., Mascher M. and Schnurbusch T. (2021) Genome-wide identification of loci modifying spike-branching in tetraploid wheat. Theor Appl Genet 134: 1925–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A., Suzaki T., Tanaka W. and Hirano H.Y. (2009) The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA 106: 20103–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, M., Iehisa J.C.M., Ohno R., Kimura T., Enoki H., Nishimura S., Nasuda S. and Takumi S. (2017) Three dominant awnless genes in common wheat: Fine mapping, interaction and contribution to diversity in awn shape and length. PLoS One 12: e0176148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef, H.M., Eggert K., Koppolu R., Alqudah A.M., Poursarebani N., Fazeli A., Sakuma S., Tagiri A., Rutten T., Govind G.et al. (2017) VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat Genet 49: 157–161. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Dong C., Chen Z., Gui L., Chen C., Li D., Xie Z., Zhang Q., Zhang X., Xia C.et al. (2021) WheatGmap: a comprehensive platform for wheat gene mapping and genomic studies. Mol Plant 14: 187–190. [DOI] [PubMed] [Google Scholar]
- Zhong, J., van Esse G.W., Bi X., Lan T., Walla A., Sang Q., Franzen R. and von Korff M. (2021) INTERMEDIUM-M encodes an HvAP2L-H5 ortholog and is required for inflorescence indeterminacy and spikelet determinacy in barley. Proc Natl Acad Sci USA 118: e2011779118. [DOI] [PMC free article] [PubMed] [Google Scholar]