Summary
Approaches to study therapy resistance in HCC (hepatocellular carcinoma) are limited, especially when using HCC models in vitro. Here, we present a protocol to establish an in vitro Sorafenib-resistant human HCC cell model and conduct an shRNA-mediated synthetic lethal screen in established Sorafenib-resistant HCC cell lines to identify critical regulators of Sorafenib resistance. We describe steps for RNA sequencing and functional analysis to reveal the mode of action of potential candidates in conferring therapy resistance to HCC cells.
For complete details on the use and execution of this protocol, please refer to Gao et al. (2021a)1 and Gao et al. (2021b).2
Subject areas: Cell Biology, Cancer, Sequencing, RNAseq, High Throughput Screening, Molecular Biology
Graphical abstract
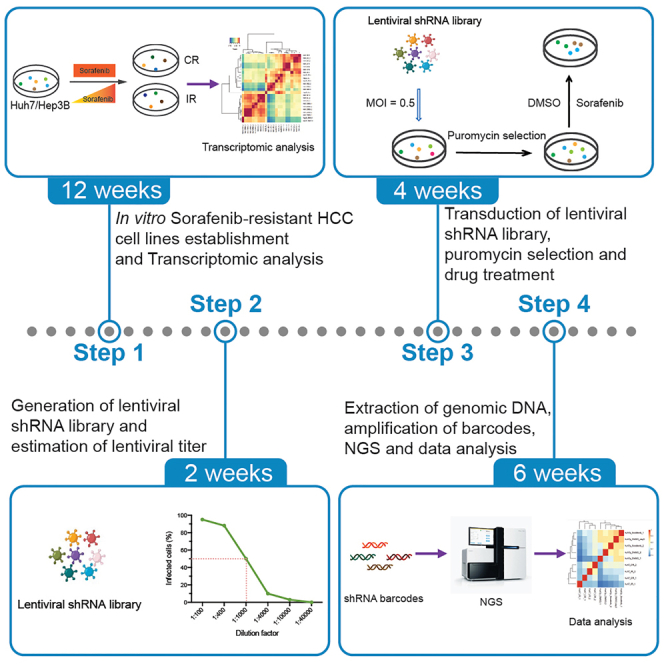
Highlights
-
•
Establishment of in vitro Sorafenib-resistant HCC cell models
-
•
Establishment of intrinsic or acquired drug-resistant cancer cell lines
-
•
Global transcriptomic analysis by RNA sequencing
-
•
Synthetic lethal screen using genome-wide pooled lentiviral shRNA libraries
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Approaches to study therapy resistance in HCC (hepatocellular carcinoma) are limited, especially when using HCC models in vitro. Here, we present a protocol to establish an in vitro Sorafenib-resistant human HCC cell model and conduct an shRNA-mediated synthetic lethal screen in established Sorafenib-resistant HCC cell lines to identify critical regulators of Sorafenib resistance. We describe steps for RNA sequencing and functional analysis to reveal the mode of action of potential candidates in conferring therapy resistance to HCC cells.
Before you begin
Sorafenib is a small compound multikinase inhibitor, which targets c-RAF, PDGFR, EGFR, VEGFR, FLT-3 and c-KIT. It is a major first-line systemic treatment for advanced HCC patients, yet drug resistance develops very fast in most cases. Due to limited methods of studying Sorafenib resistance in HCC, establishment of in vitro models is a critical step for studying therapy resistance in HCC.
This protocol describes the details of the establishment of in vitro Sorafenib-resistant HCC cell models, their global transcriptomic analysis and a synthetic lethal screen of established Sorafenib-resistant HCC cell lines by using a human genome-wide pooled lentiviral shRNA library module 1 (50467 shRNAs; Vector: PRSI16cb, Cellecta) which targets 6317 mRNAs involved in signaling pathways.
Institutional permissions
Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Establishment of Sorafenib-resistant human HCC cell lines
To delineate the molecular mechanisms underlying Sorafenib resistance in HCC, we established Sorafenib-resistant HCC cell models in vitro.
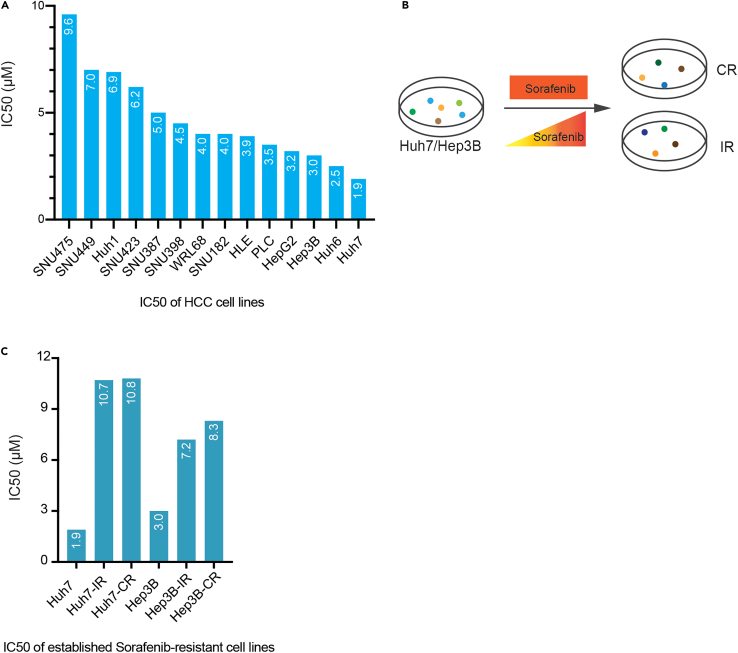
To find the most appropriate cell lines for the establishment of in vitro Sorafenib-resistant HCC cell models, we chose 14 different HCC cell lines and measured their IC50 values against the treatment with Sorafenib. The four cell lines most sensitive to Sorafenib were selected ones as candidates for the establishment of Sorafenib-resistant cell lines in vitro (Figure 1A). To closely recapitulate the features of the establishment and progression of Sorafenib resistance in patients, we conducted two different treatment protocols on the selected four parental (Sorafenib-sensitive) HCC cell lines; They were either treated with a constant high concentration of Sorafenib or with gradually increasing concentrations of Sorafenib (Figure 1B). After approximately 8 weeks of treatment, when cells stably grow in the presence of the specific concentrations of Sorafenib, four different Sorafenib-resistant HCC cell lines were established. They were named Huh7-CR and Huh7-IR and Hep3B-CR and Hep3B-IR, originating from Sorafenib-sensitive Huh7 and Hep3B cell lines, respectively, and with CR standing for a constant high concentration of Sorafenib and IR referring to the cell lines generated by gradually increasing the concentration of Sorafenib.
Figure 1.
Establishment of Sorafenib-resistant HCC cell lines
(A) IC50 values of 14 HCC cell lines: 14 HCC cell lines were treated with increasing doses of Sorafenib, and the IC50 values for cell growth inhibition by Sorafenib were determined.
(B) Schematic representation of the establishment of Sorafenib-resistant derivatives of Huh7 and Hep3B HCC cell lines. Huh7 or Hep3B cells were treated with increasing (for Huh7 is from 0 to 10 μM, for Hep3B is from 0 to 5 μM) or with consistently high (7 μM for Huh7, 5 μM for Hep3B) concentration Sorafenib to establish acquired Sorafenib-resistant cell lines.
(C) IC50 values of Huh7-IR, Huh7-CR and Hep3B-IR and Hep3B-CR. These IC50 values are on the range of Sorafenib’s clinically relevant concentration of 10 μM.
Below we provided the details of establishment of the Sorafenib-resistant HCC cell lines. This protocol could also be used as a reference for the establishment of drug resistance in other types of cancer cells.
Selection of Sorafenib-sensitive cell lines
Timing: ∼ 2 weeks
-
1.
Culture of HCC cell lines: all 14 HCC cell lines are cultured with complete high glucose DMEM at 37°C, 5% CO2;
-
2.IC50 measurements:
-
a.Seed each cell line at 5 ×104 cells into 24-well plates, 24 h later, add Sorafenib at final concentrations of 0 μM, 0.5 μM, 2 μM, 5 μM, 10 μM and 20 μM, determine cell growth in a Cell-IQ (Automated cell culture and analysis system, CM-Technologies) or a comparable device;
-
b.Refresh medium every 24 h for 3 times (at time points 48 h, 72 h and 96 h) with new medium containing the respective concentrations of Sorafenib as defined in step a.
-
c.Cell growth is recorded using a Cell-IQ, with the series of dose-response data to plot a x-y linear regression as Y = a∗X + b (Y is the cell confluence/viability in %, X is the drug concentration), when Y = 0.5, X is termed the IC50. Thus IC50 values are calculated with the formulation:
-
a.
All IC50 values of 14 HCC cell lines are shown in Figure 1A.
-
3.
Huh7, HepG2, Hep3B and Huh6 present the 4 most sensitive HCC cell lines in response to Sorafenib treatment, with the IC50 values of 1.9 μM, 3.2 μM, 3 μM and 2.5 μM, these four cell lines are used for the next steps of establishing Sorafenib-resistant HCC cell lines.
Note: Alternative option: If Cell-IQ or a comparable device is not available, cell viability could be assessed using the CellTiter-Glo 2.0 viability Assay kit (Promega: Cat# G9241): Cells are seeded and cultured refer to step 2a–b. 96 h later, measure the cell viability with the CellTiter-Glo 2.0 viability Assay kit according to the manufacturer’s instructions, calculate the IC 50 values refer to step 2c.
Establishment of Sorafenib-resistant HCC cell lines
Timing: ∼ 8 weeks
As HepG2 and Huh6 failed to survive upon long-term treatment with Sorafenib, below we provide the details of the establishment of Sorafenib-resistant cell lines derived from Huh7 and Hep3B cell lines, respectively.
-
4.Establishment of Huh7 Sorafenib-resistant cell lines:
-
a.Seed 1 × 106 Huh7 cells into three T75 flask each. Treat the first flask with DMSO, treat the second flask with 7 μM Sorafenib;
-
b.Treat the last flask of cells with increasing Sorafenib concentration at the starting concentration of 1.5 μM. Increase the concentration by 0.2 μM after each two passages, terminate the concentration increase when the cells could not survive from higher concentrations of Sorafenib treatment.
-
c.Keep the treatment until all cells remain stable, which needs approximately 8 weeks.
-
a.
-
5.Establishment of Hep3B Sorafenib-resistant cell lines:
-
a.Seed 1 × 106 Hep3B cells into three T75 flask each. Treat the first flask with DMSO, treat the second flask with 5 μM Sorafenib;
-
b.Treat the last flask of cells with increasing Sorafenib concentrations at the starting concentration of 2.5 μM. Increase the concentration by 0.2 μM after each two passages, terminate the concentration increase when the cells could not survive from higher concentrations of Sorafenib treatment.
-
c.Keep the treatment until all cells remain stable, which needs approximately 8 weeks, when cells could proliferate under the treatment of highest concentrations of Sorafenib.
-
a.
-
6.
We finally have succeeded in getting Huh7-IR/CR and Hep3B-IR/CR cell lines. Once the cells continually grow upon further culture in the presence of the highest doses of Sorafenib, freeze the cells for storage and further use (Figure 1B).
-
7.Measurement of the IC50 values:
-
a.To determine whether the established HCC cell lines are Sorafenib-resistant or not. Measure the IC50 values of the cells (Ref. Step 2) with a Cell-IQ or the CellTiter-Glo 2.0 viability Assay kit as described above. The respective IC50 values are shown in Figure 1C.
-
b.Compare the IC50 values with the clinically achievable Sorafenib drug level of 10 μM (at which concentration has no clinical effect on non-responders). The IC50 values of established Sorafenib-resistant HCC cell lines (Figure 1C) are in the range of the concentrations obtained in patients in the clinics, meaning that the established cell lines are appropriately Sorafenib-resistant.3
-
a.
Note: To define the concentrations of Sorafenib on the establishment of constant resistant cell lines, we tested gradient concentrations of 10 μM, 9 μM, 8 μM, 7 μM, 6 μM, 5 μM Sorafenib both on Huh7 and Hep3B and found that 7 μM is the maximum tolerated concentration for Huh7 to maintain basal proliferation, and 5 μM for Hep3B.
Global transcriptomic analysis of Sorafenib-resistant HCC cell lines
Timing: ∼ 2 weeks
To delimit the molecular changes upon the establishment of Sorafenib-resistance in HCC cells, global transcriptomic analysis by RNA sequencing is performed on both Huh7/Hep3B-DMSO and Huh7-IR/CR and Hep3B-IR/CR Sorafenib-resistant cells.
-
8.
Culture Huh7/Hep3B-DMSO cells and Huh7-IR/CR and Hep3B-IR/CR cells in T75 flasks:
(Huh7-IR/CR and Hep3B-IR/CR cells are always cultured at 7 μM or 5 μM Sorafenib, respectively). Once confluence reaches 80%, the cells are harvested, and RNA is extracted in biological triplicates by using miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions.
-
9.
RNA quality control is performed using a fragment analyzer and standard or high-sensitivity RNA analysis kits (Labgene; DNF-471-0500 or DNF-472-0500).
-
10.
Measure the RNA concentrations with a Quant-itTM RiboGreen RNA assay Kit (Thermo Fisher Scientific).
-
11.
Take a total of 200 ng RNA per sample for the library preparation, with a TruSeq Stranded mRNA Library Prep Kit (Illumina) to generate mRNA and transcript the mRNA into cDNA.
-
12.
Prepared cDNA library is sequenced using HiSeq SBS kit V4 (Illumina) on an Illumina HiSeq 2500 using protocols defined by the manufacturer.
-
13.Analysis of the RNA sequencing data:
-
a.Map Single-end RNA-seq reads (81-mers) to the human genome assembly, version hg19 (GRCh37.75), with RNASTAR,4 with default parameters except for allowing only unique hits to genome (outFilterMultimapNmax = 1) and filtering reads without evidence in spliced junction table (outFilterType = "BySJout").
-
b.Quantify expression levels per gene (counts over exons) for the RefSeq mRNA coordinates from UCSC using qCount function from QuasR package (version 1.12.0).
-
c.Analyze the differentially expressed genes with the edgeR package (version 3.14.0). Genes with P-values smaller than 0.05 and minimum log2-fold changes of ± 0.58 were considered as differentially regulated and were selected for functional and pathway enrichment analysis.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| DMSO | Sigma | Cat# D8418 |
| Sorafenib | Selleckchem | Cat# S7397 |
| Puromycin | Sigma | Cat# P8833 |
| Crystal violet powder | Sigma | Cat# C0075 |
| Polybrene | Sigma | Cat# 107689 |
| DMEM high glucose | Sigma | Cat# D5671 |
| L-Glutamine | Sigma | Cat# G7513 |
| Penicillin-Streptomycin | Sigma | Cat# P4333 |
| 10 × Trypsin/EDTA | Sigma | Cat# T4174 |
| FBS | Sigma | Cat# F7524 |
| PBS | Sigma | Cat# D8537 |
| Opti-MEM | Gibco | Cat# 51985034 |
| FuGENE | Promega | Cat# E2311 |
| RNase A | Qiagen | Cat# 19101 |
| Buffer P1 | Qiagen | Cat# 19051 |
| Recombinant Proteinase K solution (20 mg/mL) | Thermo Fisher Scientific | Cat# AM2546 |
| Critical commercial assays | ||
| miRNeasy Mini kit | Qiagen | Cat# 217084 |
| RNA analysis kit | Labgene | Cat# DNF4710500/4720500 |
| Quant-itTM RiboGreen RNA assay Kit | Thermo Fisher Scientific | Cat# R11490 |
| TruSeq stranded mRNA Library Prep Kit | Illumina | Cat# 20020594 |
| HiSeq SBS kit V4 | Illumina | Cat# FC-401-4002 |
| Hiseq TruSeq SR Cluster Kit v3-cbot-HS | Illumina | Cat# GD-401-3001 |
| QIAquick gel extraction kit | Qiagen | Cat# 28706 |
| QIAquick PCR purification kit | Qiagen | Cat# 28104 |
| NGS Prep Kit | CELLECTA | Cat# LNGS-120 |
| Deposited data | ||
| RNA-sequencing data on Sorafenib-resistant cell lines | This paper | GEO: GSE117116 |
| Synthetic lethal barcode sequencing data | This paper | GEO: GSE158458 |
| Experimental models: Cell lines | ||
| Huh7 | University Hospital Basel | N/A |
| HepG2 | ATCC | Cat# HB-8065 |
| Hep3B | ATCC | Cat# HB-8064 |
| HLE | University Hospital Basel | N/A |
| SNU398 | ATCC | Cat# CRL-2233 |
| SNU449 | ATCC | Cat# CRL-2234 |
| SNU182 | ATCC | Cat# CRL-2235 |
| SNU475 | ATCC | Cat# CRL-2236 |
| SNU387 | ATCC | Cat# CRL-2237 |
| SNU423 | ATCC | Cat# CRL-2238 |
| PLC/PRF/75 | ATCC | Cat# CRL-8024 |
| Huh1 | University Hospital Basel | N/A |
| WRL68 | ATCC | Cat# CL-48 |
| Huh6 | University Hospital Basel | N/A |
| HEK-293T | ATCC | Cat# CRL-3216 |
| Recombinant DNA | ||
| pMD2.G | Didier Trono Lab | Addgene Cat# 12259 |
| pMDLg/pRRE | Didier Trono Lab | Addgene Cat# 12251 |
| pRSV-Rev | Didier Trono Lab | Addgene Cat# 12253 |
| Pooled lentiviral shRNA library | CELLECTA | Cat# HGW-M1-P2 |
| Software and algorithms | ||
| FastQC | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| R Development Core | N/A | https://www.r-project.org/ |
| Adobe Illustrator | Adobe | N/A |
| Prism 9 | GraphPad | N/A |
| Other | ||
| 12-well plate | Milian | Cat# 353043 |
| 24-well plate | Milian | Cat# 353047 |
| T75 flask | Milian | Cat# 353136 |
| T175 flask | Milian | Cat# 353112 |
| 0.45 μm syringe filter | Sigma | Cat# SLHV033R |
| 15 mL tube | Sarstedt | Cat# 62.554.502 |
| 50 mL tube | Sarstedt | Cat# 62.547.254 |
| Cryotube | Sarstedt | Cat# 72.380.007 |
| Centrifuge | Eppendorf | 5702 |
| Cell-IQ | CM Technologies | N/A |
Materials and equipment
Complete high glucose DMEM medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM high glucose | N/A | 500 mL |
| L-Glutamine | 2 mM | 5 mL |
| Penicillin-Streptomycin | 100 U/mL | 5 mL |
| FBS | 10% | 50 mL |
| Total | 560 mL |
Note: Prepare in a sterile hood, store at 4°C for a maximum of 3 weeks.
0.5% Crystal Violet
| Reagent | Final concentration | Amount |
|---|---|---|
| Crystal violet powder | 0.5% | 0.5 g |
| Distilled H2O | N/A | 80 mL |
| Methanol | N/A | 20 mL |
| Total | 100 mL |
Note: Dissolve crystal violet powder in distilled H2O and then add methanol. Store in the dark at around 23°C for a maximum of 2 months.
Step-by-step method details
Generation of a pooled lentiviral shRNA library
Timing: ∼ 1 week
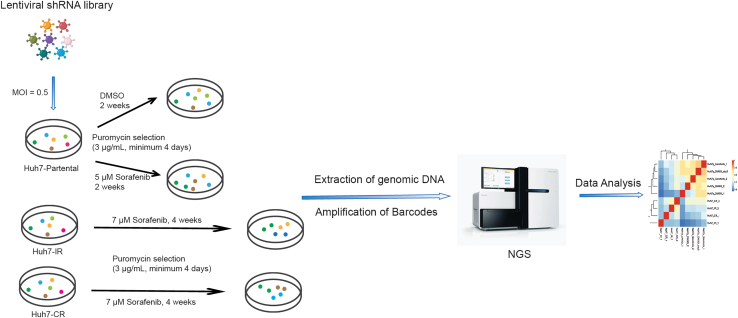
This section details the step-by-step protocol for the generation of a pooled lentiviral shRNA library. The shRNA pools are purchased from Cellecta. A scheme of the synthetic lethal screen is shown in Figure 2.
CRITICAL: The library contains 50467 pooled lentiviral shRNAs targeting 6317 genes. To generate a sufficient virus titer, a batch of 100 μg plasmid is recommended for the production.
-
1.
Seed 1 × 107 HEK-293T cells into seven T175 flasks each, culture cells with high glucose DMEM contains 10% FBS and 2 mM L-Glutamine at 37°C, 5% CO2.
Note: Don’t add antibiotics (Penicillin/streptomycin) to DMEM when producing lentivirus.
-
2.Transfection of pooled lentiviral shRNA library.
-
a.Once the cell confluence reaches approximately 50%, replace medium with 30 mL fresh medium.
-
b.Prepare two sterile 50 mL Falcon tubes and prepare the transfection system as follows: Tube A: 12 mL Opti-MEM, 37.5 μg pMD2.G, 75 μg pMDLg/pRRE, 25 μg pRSV-Rev, 100 μg library plasmid, mix well with pipette; Tube B: 12 mL Opti-MEM, 720 μL FuGENE, mix well with pipette.
-
c.Transfer tube B into tube A, mix well with pipette or vortex, then incubate for 15 min at around 23°C.
-
d.Transfer 3 mL mixture to each T175 flask and crossly rock the flasks to mix well with medium, then, put the cells to the incubator.
-
e.18 h post transfection, change medium with 30 mL fresh high glucose DMEM.
-
a.
-
3.Harvest the shRNA library virus.
-
a.48 h post-transfection, collect all the culture medium in 50 mL falcon tubes.
-
b.Centrifuge the tubes at 500 g for 5 min, pass the supernatant through a 0.45 μm syringe filter.
-
c.Freeze the viral supernatant in aliquots of 5 mL in 15 mL Falcon tubes at −80°C for further use.
-
a.
Figure 2.
Schematic of the pooled lentiviral shRNA library screen (Created with BioRender.com)
Generated pooled lentiviral library was transduced into both Huh7-parental and Sorafenib-resistant cells. With 3 μg/mL puromycin treatment for at least 4 days to confirm the library to be transduced into the cells successfully. Either DMSO or Sorafenib was added into DMEM, culture Huh7-parental for another 2 weeks, culture Huh7-IR and Huh7-CR cells for another 4 weeks; Following with the genomic DNA extraction, shRNA barcodes amplification and the NGS sequencing with Data analysis.
Lentiviral virus titration
Timing: ∼ 1 week
To determinate the virus titer, thaw one aliquot of the frozen viral supernatant and dilute it at different dilution factors and measure the transfection efficiency.
-
4.Seed cells and infect cells with diluted virus supernatant:
-
a.Seed 1 × 105 Huh7 cells per well in two 12-well plates.
-
b.Once the cell confluence reaches 50%, thaw one frozen viral supernatant on ice.
-
c.Dilute the virus supernatant with complete high glucose DMEM at a ratio of 1:100, 1:400, 1:1000, 1:4000, 1:10000, 1:40000 to a final volume of 10 mL virus solution (100 μL, 25 μL, 10 μL, 2.5 μL, 1 μL, 0.25 μL of virus respectively).
-
d.Take 3 mL diluted virus solution and add polybrene to 5 μg/mL.
-
e.Transfer 1 mL diluted solution to each well (each ratio with duplicate wells).
-
f.Keep four wells without virus infection as control.
-
a.
Note: Polybrene is used to enhance the infection efficiency.
-
5.
12 h later, change medium with 1 mL fresh complete high glucose DMEM.
-
6.
24 h post the infection, change with 1 mL fresh complete high glucose DMEM contains 3 μg/mL puromycin (for the four control wells, two with puromycin, two without puromycin to confirm the efficacy of puromycin).
-
7.
Culture cells for another 72 h, stain the cells with 0.5% Crystal Violet and calculate the transduction efficiency of different dilution factors.
-
8.
Repeat the virus transduction two more times, plot the crystal violet staining ratio versus dilution factors into a chart, as shown in Figure 3.
-
9.
Calculate the viral concentration as the formula: Titer equals cell numbers × estimated MOI/dilution factor, then calculate the virus volume required for the transduction to obtain a transduction efficiency of around 40%, which is considered a MOI of 0.5 for the next steps.
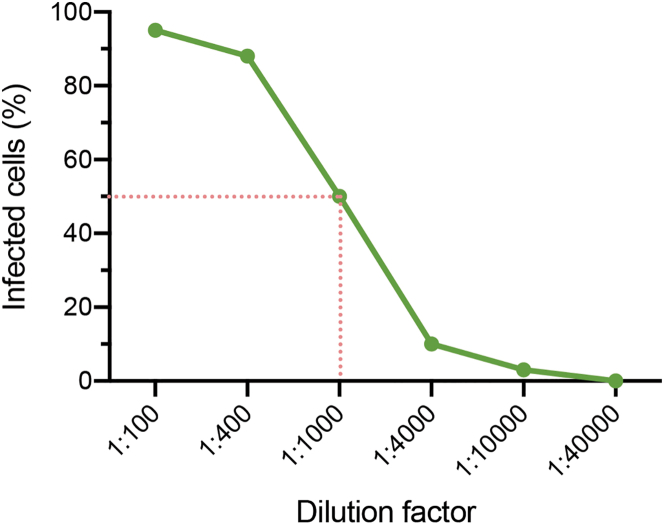
Figure 3.
Estimation of the virus titer
A plot of the infected cells (%) versus dilution factors: when the virus is diluted with 1:1000, around 50% cells are infected, the estimated MOI is around 0.7. With the estimated MOI, the virus titer is with the formula: Virus titer = Cell numbers × MO / Dilution factor.
Transduction of library
Timing: ∼ 4 weeks
The pooled lentiviral shRNA library will be transduced into Huh7-parental, Huh7-IR and Huh7-CR cell lines, a MOI of 0.5 will be used. After the transduction, cells will be treated with 3 μg/mL puromycin for at least 4 days, afterward, DMSO or Sorafenib will be added to the cells until harvest.
-
10.
Set up four experimental groups as Huh7-parental + DMSO, Huh7-parental + 5 μM Sorafenib, Huh7-IR + 7 μM Sorafenib, Huh7-CR + 7 μM Sorafenib: take eight T175 flasks for each group, seed 1 × 107 cells per flask.
-
11.Infect cells and puromycin selection:
-
a.Take seven T175 flasks of cells per group when the cell confluence reaches 50%.
-
b.Thaw the frozen viral aliquots, dilute virus with complete high glucose DMEM at a MOI of 0.5.
-
c.Add polybrene to a final concentration of 5 μg/mL to the diluted virus and then infect the cells.
-
d.24 h later, change medium with fresh complete high glucose DMEM containing 3 μg/mL puromycin.
-
e.Take the residual one T175 flask from each group (four flasks in total) as the control.
-
f.Treat the control cells with 3 μg/mL puromycin for at least 4 days to confirm the successful transduction of the pooled lentiviral shRNA library.
-
a.
-
12.DMSO or Sorafenib treatment and cell harvest:
-
a.Treat the cells with either DMSO or Sorafenib as mentioned in step 9, culture Huh7-parental + DMSO / 5 μM Sorafenib for around 2 weeks.
-
b.Culture Huh7-IR/CR cells for over 10 passages to generate sufficient cell numbers.
-
c.Trypsinize the cells and combine all the cells from the same treatment group (6 × 107 cells are needed, which is 1000 times of the library size).
-
d.Centrifuge at 300 g for 5 min, remove the supernatant and collect the cell pellets for the next step.
-
a.
Note: Upon treatment with Sorafenib, Huh7-parental cells will only survive for approximately 2 weeks. Hence, these cells will be harvested for analysis after 2 weeks, as the shRNAs which had dropped out in Huh7-parental cells upon acute treatment with 5 μM Sorafenib will be subtracted from the synthetic lethal gene list, to specifically enrich for the genes critical for the maintenance of Sorafenib resistance.
Extraction of genomic DNA for samples
Timing: ∼ 1 week
To isolate whole genomic DNA from the cells of genetic screen, a conventional genomic DNA extraction protocol is used.
-
13.
Resuspend cell pellets in 5 mL QIAGEN Buffer P1 (Cat# 19051) with 5 μL RNase A (final concentration is 100 μg/mL, Cat# 19101) with 15 mL Falcon tubes, add 250 μL 10% SDS, mix well and incubate at around 23°C for 5 min.
-
14.
Sonicate to shear DNA into 10–100 kb sized fragments with an ultrasonic homogenizer at 4°C.
-
15.
Add 10 μL Proteinase K (final concentration is 40 μg/mL), mix well and incubate at around 23°C for 15 min.
-
16.
Add 5 mL Phenol: Chloroform: Isoamyl Alcohol solution (25:24:1), vortex and centrifuge with 6000 g for 60 min at 23°C.
-
17.
Take 4 mL of upper phase, and transfer to new 15 mL Falcon tubes, add 500 μL 3 M Sodium Acetate and 4 mL Isopropanol, mix well and centrifuge with 6000 g for 30 min at 23°C.
-
18.
Discard the supernatant and wash with 10 mL 70% Ethanol, centrifuge with 6000 g for 5 min at 23°C, discard the supernatant and dry the pellet.
-
19.
Dissolve the pellet into ddH2O, check the extraction quality by electrophoresis in 3.5% agarose gels and measure the concentrations of genomic DNA.
Amplification of Barcode Sequences from genomic DNA
Timing: ∼ 1 week
The shRNA barcodes are amplified using NGS Prep Kit (CELLECTA, Cat# LNGS-120), the protocol is based on two rounds of PCR to amplify the shRNA barcode sequences. PCR products are purified using QIAquick PCR purification kit and separated by electrophoresis in 3.5% agarose gels, bands were excised and purified using QIAquick gel extraction kit.
-
20.
First Round PCR.
The goal of the first round PCR is to amplify barcodes out of total genomic DNA. When amplifying the barcodes from samples which are generated by the positive selection screen, adequate amounts of genomic DNA are needed (up to 400 μg).-
a.Prepare the master mix according to the following table. A maximum of 50 μg genomic DNA is used in 100 μL volume per reaction. With 400 μg genomic DNA as template, eight reactions are set up at 100 μL each;
Component Volume per reaction Genomic DNA x μL Forward 1st Round PCR Primer 3 μL Reverse 1st Round PCR Primer 3 μL dNTP Mix 2 μL Taq Polymerase Buffer 10× 10 μL PCR-Grade H2O 80 – x μL Taq Polymerase 2 μL Total volume 100 μL -
b.Mix well, centrifuge briefly, and start the PCR program with the following cycling conditions;
Steps Temperature Time Cycles Initial denaturation 95°C 2 min 1 Denaturation 95°C 30 s 16–18 Annealing 65°C 30 s Extension 68°C 2 min Final extension 68°C 2 min 1 Hold 4°C ∞  CRITICAL: The numbers of cycles depend on how many genomic DNA is used. If less than 50 μg are added in a 100 μL PCR reaction, one or two more cycles are recommended.
CRITICAL: The numbers of cycles depend on how many genomic DNA is used. If less than 50 μg are added in a 100 μL PCR reaction, one or two more cycles are recommended. -
c.Combine all the individual reactions into one tube for each group sample, the PCR products will be used for the second round PCR.
-
a.
Pause point: The PCR product can be stored at −20°C for 6 months, if not perform the second round PCR immediately.
-
21.
Second Round PCR.
Primers for the second round PCR contain the P5 and P7 sequences which are complementary to the immobilized primers in the NGS illumine flow cells. The NGS Prep Kits provide the index primer sets which contain different index sequences and can be used to deconvolute the sequencing results for each sample mixed together in the same flow cell or sequencing lane. Details are found in the User Manual of the NGS Prep kit.-
a.Use 5 μL of PCR product from the first round PCR as the template, prepare the mix according to the following table (2 reactions with 100 μL per reaction);
Component Volume per reaction First Round PCR Product 5 μL NFwd Primer 5 μL NRev(Index) Primer 5 μL dNTP Mix 2 μL Taq Polymerase Buffer 10× 10 μL PCR-Grade H2O 71 μL Taq Polymerase 2 μL Total volume 100 μL  CRITICAL: Each sample to be sequenced together in a single lane needs to use different NRev Index primers from the NGS Prep kit.
CRITICAL: Each sample to be sequenced together in a single lane needs to use different NRev Index primers from the NGS Prep kit. -
b.Mix well, centrifuge briefly, and start the PCR program with the following cycling conditions;
Steps Temperature Time Cycles Initial denaturation 95°C 2 min 1 Denaturation 95°C 30 s 12–14 Annealing 65°C 30 s Extension 68°C 2 min Final extension 68°C 2 min 1 Hold 4°C ∞
-
a.
-
22.
PCR purification and extraction.
PCR products from the second round PCR are analyzed by gel electrophoresis to ensure equal amounts of amplified barcodes for all samples. Purify the PCR products by the following steps.-
a.Use the QIAquick PCR purification kit to purify the PCR products following the manufacturer’s protocol. For the last centrifugation step, take the maximum speed for 5 min to completely dry the membrane;
-
b.Load the purified PCR products on a 3.5% agarose gel to separate the bands by electrophoresis;
-
c.Excise the bands and extract DNA with QIAquick gel extraction kit;
-
d.Quantitate the DNA concentration at A260nm using NanoDrop, and store for next generation sequencing with an Illumina HiSeq (Illumina).
-
a.
Pause point: The PCR product can be stored at −20°C for 6 months, if not perform the NGS sequencing immediately.
NGS (next generation sequencing) deep sequencing and data analysis
Timing: ∼ 4 weeks
-
23.
NGS deep sequencing.
Pooled amplified barcodes will be sequenced by NGS on an Illumina Hiseq (80–100 million reads per sample) using the GexSeq sequencing primer. Take 2 μL of 10 nM PCR product from the purified DNA band for the cluster generation step. The number of cycles for hGW library is 44, for the other libraries, the cycle numbers required depend on the length of barcode.-
a.Adjust the purified PCR samples to a final concentration of 10 nM.
-
b.To each lane, add 2 μL sample and Phix174 control template based on the standard Illumina protocol for cluster generation using the Illumina single-read flow cell.
-
c.Add GexSeq primer to the PhiX174 primer to get a final concentration of 0.5 μM for the NGS step.
-
d.Run the NGS reaction for 44 cycles with a Hiseq TruSeq SR Cluster Kit v3-cbot-HS.
-
a.
-
24.
Data analysis.
The open-source processing pipeline in edgeR5 provides a complete analysis solution for the synthetic lethal screen data. It begins with the raw sequence reads and ends with a ranked list of candidate genes for downstream biological validation.-
a.First summarize the raw data contained in a fastq file into a matrix of counts (samples in the columns, genes in the rows) with options for allowing mismatches and small shifts in sequence position;
-
b.Process Amplicons: process raw data from pooled genetic sequencing screens (https://rdrr.io/bioc/edgeR/man/processAmplicons.html);
-
c.Multidimensional scaling plot of distances between digital gene expression profiles (https://rdrr.io/bioc/edgeR/src/R/plotMDS.DGEList.R);
-
d.Calculate Normalization Factors to Align Columns of a Count Matrix (https://rdrr.io/bioc/edgeR/man/calcNormFactors.html);
-
e.Compute GeneWise exact tests for differences in the means between two groups of negative-binomially distributed counts (exactTest: https://rdrr.io/bioc/edgeR/src/R/exactTest.R);
-
f.Camera and Roast for GeneSet enrichment analysis.
-
a.
Expected outcomes
The combination of transcriptomic analysis and pooled lentiviral shRNA library screen on Sorafenib-resistant HCC cell lines allows the identification of pivotal regulators of Sorafenib resistance in HCC. One recent example is the identification and functional validation of the YAP/TAZ-ATF4-SLC7A11 axis as a critical player in establishing and maintaining Sorafenib resistance by in vitro/vivo experiments and on HCC patients’ samples, as reported in the original article by Gao et al. (2021b).2 The raw RNA-sequencing data and pooled lentiviral shRNA library screen data has been deposited at Gene Expression Omnibus (GEO): GSE117116 (RNA-sequencing Sorafenib-resistant cell lines), GSE158458 (Synthetic lethal barcode sequencing).
Limitations
This protocol provides details of the in vitro establishment of Sorafenib-resistant HCC cell lines, a procedure which can also be used to establish other in vitro drug resistance models in different cancer cell types. Nevertheless, the establishment of drug-resistant cell lines may not be applicable for all types of cancer cell lines due to various response to different drugs, a batch of different cell lines needs to be tested to select the proper ones. The synthetic lethal pooled lentiviral shRNA library screen can also be modified for application with any other type of cell line. While such in vitro drug-resistant cell models may recapitulate the situation in drug-resistant HCC patients, they may be limited in fully mimicking the complex organismic interactions as observed in patients with drug-resistant HCC. Also, while the pooled lentiviral shRNA library screen has identified several pivotal regulators of drug-resistance, some critical players may be missed due to the complexity of a full genome-wide screening approach or due to the differences in the specific genetic and molecular make-up of the cell lines selected. Moreover, some of the newly identified players escape a functional validation in experimental animal models of HCC or in HCC patients.
Troubleshooting
Problem 1
Failed to establish Sorafenib-resistant cell lines on Huh6 and HepG2 cells.
Potential solution
High doses of Sorafenib will lead to severe cell death with Huh6 and HepG2 cells and only few cells did survive. One possible solution to generate the IR (Increasing resistant) cell lines is slowly increasing the concentration of Sorafenib, prolong the culture time with each concentration to generate sufficient cell numbers before the next stage of higher concentration treatment. For the constant high concentration of Sorafenib treatment, try to seed more cell numbers and culture for long enough time in the presence of the highest concentration of Sorafenib, which a few Huh6 and HepG2 cells may survive. These may take much longer times than was used with Huh7 and Hep3B cells.
Problem 2
Sorafenib-resistant cells are not growing well (Before you begin, step 6).
Potential solution
Sorafenib has to be added to the medium when the Sorafenib-resistant cells are in culture to maintain the resistance. For Huh7-IR/CR use 7 μM Sorafenib, for Hep3B-IR/CR use 5 μM Sorafenib. Culture medium has to be appropriately changed depending on the cells’ growth conditions.
Problem 3
Low lentiviral titer (Generation of a pooled lentiviral shRNA library).
Potential solution
Low titer may be caused by: a. too high or too low cell density of HEK-293T, 50%–80% confluence is recommended; b. Low transfection efficiency: we tested 4 different transfection reagents: FuGENE, JetPEI, Lipo2000 and Lipo3000 to assess which one had the best efficiency of lentivirus production. CV staining (% to Control) of post-puromycin selected Huh7 or Hep3B was used as the criteria for the evaluation, FuGENE exhibited the highest efficiency of virus production in our system; c. Storage of aliquoted packaged library should be at −80°C. Don’t freeze-thaw the virus more than one time.
Problem 4
Concentration of puromycin used for the selection (step 10).
Potential solution
The sensitivity to puromycin treatment varies between different cell lines. It is critical to generate a killing curve for determining the optimal concentration of puromycin.
Problem 5
No PCR product (Amplification of Barcode Sequences from Genomic DNA).
Potential solution
Ensure that the correct primers need to be used for each round of PCR, 10 ng plasmid library DNA maybe used as a positive control. Low numbers of barcodes in the genomic DNA could also block the amplification, the transduction efficiency of pooled lentiviral shRNA library has to be checked.
Problem 6
No barcodes present in the NGS sequencing (step 22).
Potential solution
Ensure that the correct sequencing primers are used when performing NGS. Use the Gexseq Sequencing primers from the NGS Prep kit.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Ruize Gao (ruize.gao@lih.lu).
Materials availability
Plasmids and cell lines reported in this study will be available upon request.
Acknowledgments
We thank A. Kapus, B. Gan, and A. Hergovich for providing plasmid constructs. We thank C. Beisel and the Genomics Facility Basel for RNA and barcode sequencing. We are grateful to Luca Quagliata for sharing the HCC cell lines, David Büchel for preparing the Sorafenib-resistant cell lines, and Ravi Kiran Reddy Kalathur for the data analysis. This works was supported by the European Research Council (ERC) Synergy Project MERiC and the Swiss National Science Foundation Sinergia Project MERiC.
Author contributions
Conceptualization, F.T., R.G., G.C.; Methodology, F.T., R.G.; Investigation, R.G.; Writing – Original Draft, R.G.; Writing – Review & Editing, G.C., F.T., R.G.; Supervision, G.C.; Project Administration, G.C.; Funding Acquisition, G.C.
Declaration of interests
The authors declare no competing interests.
Data availability
The sequencing files are deposited on GEO database under the accession numbers: GSE117116 (RNA-sequencing Sorafenib-resistant cell lines) and GSE158458 (Synthetic lethal barcode sequencing).
References
- 1.Gao R., Buechel D., Kalathur R.K., Morini M.F., Coto-Llerena M., Ercan C., Piscuoglio S., Chen Q., Blumer T., Wang X. USP29-mediated HIF1α stabilization is associated with Sorafenib resistance of hepatocellular carcinoma cells by upregulating glycolysis. Oncogenesis. 2021;10:1–13. doi: 10.1038/s41389-021-00338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R., Kalathur R.K.R., Coto-Llerena M., Ercan C., Buechel D., Shuang S., Piscuoglio S., Dill M.T., Camargo F.D., Christofori G., Tang F. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol. Med. 2021;13:e14351. doi: 10.15252/emmm.202114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith M.A., Houghton P. A proposal regarding reporting of in vitro testing results. Clin. Cancer Res. 2013;19:2828–2833. doi: 10.1158/1078-0432.CCR-13-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Z., Sheridan J.M., Gearing L.J., Moore D.L., Su S., Wormald S., Wilcox S., O'Connor L., Dickins R.A., Blewitt M.E. edgeR: a versatile tool for the analysis of shRNA-Seq and CRISPR-Cas9 genetic screens. F1000Res. 2014;3:95. doi: 10.12688/f1000research.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing files are deposited on GEO database under the accession numbers: GSE117116 (RNA-sequencing Sorafenib-resistant cell lines) and GSE158458 (Synthetic lethal barcode sequencing).