Amm et al. define an amphipathic helix in Ndc1, an essential nuclear pore and spindle pole body transmembrane protein, genetically interacting with similar motifs in Nup53 and Nup59. This work suggests that the nuclear pore formation depends on a balance between amphipathic motifs in different nucleoporins.
Abstract
Nuclear pore complexes (NPCs) are embedded in the nuclear envelope and built from ∼30 different nucleoporins (Nups) in multiple copies, few are integral membrane proteins. One of these transmembrane nucleoporins, Ndc1, is thought to function in NPC assembly at the fused inner and outer nuclear membranes. Here, we show a direct interaction of Ndc1’s transmembrane domain with Nup120 and Nup133, members of the pore membrane coating Y-complex. We identify an amphipathic helix in Ndc1’s C-terminal domain binding highly curved liposomes. Upon overexpression, this amphipathic motif is toxic and dramatically alters the intracellular membrane organization in yeast. Ndc1’s amphipathic motif functionally interacts with related motifs in the C-terminus of the nucleoporins Nup53 and Nup59, important for pore membrane binding and interconnecting NPC modules. The essential function of Ndc1 can be suppressed by deleting the amphipathic helix from Nup53. Our data indicate that nuclear membrane and presumably NPC biogenesis depends on a balanced ratio between amphipathic motifs in diverse nucleoporins.
Introduction
Nuclear pore complexes (NPCs) are huge proteinaceous assemblies embedded in the double-layered nuclear envelope (NE). Each NPC is composed of multiples of 8 copies (8–64) of about 30 different nuclear pore proteins called nucleoporins (Nups) resulting in a total amount of ∼500 up to 1,000 polypeptide chains per NPC (Akey et al., 2022; Allegretti et al., 2020). The majority of the nucleoporins are organized in biochemically and structurally defined subcomplexes: the inner nuclear pore ring connecting the NPC to the nuclear membrane, the attached central channel complex, the nuclear and cytoplasmic rings with the attached peripheral elements, the nuclear basket and the cytoplasmic filaments (Hampoelz et al., 2019; Hurt and Beck, 2015; Lin and Hoelz, 2019; Schwartz, 2016). The main function of NPCs is selective macromolecular transport across the NE. A group of nucleoporins forming the central diffusion barrier contains natively unfolded, phenylalanine-glycine (FG)-rich sequences, blocking passive diffusion of molecules larger than ∼5 nm in diameter (corresponding to a molecular mass of 30–40 kD; Frey et al., 2006; Meinema et al., 2013; Mohr et al., 2009; Popken et al., 2015; Terry and Wente, 2009). Transient interactions of the FG-Nups with distinct nuclear transport receptors (NTRs) allow the active transport of large molecules and define the directionality and specificity of transport across the NE (Paci et al., 2021).
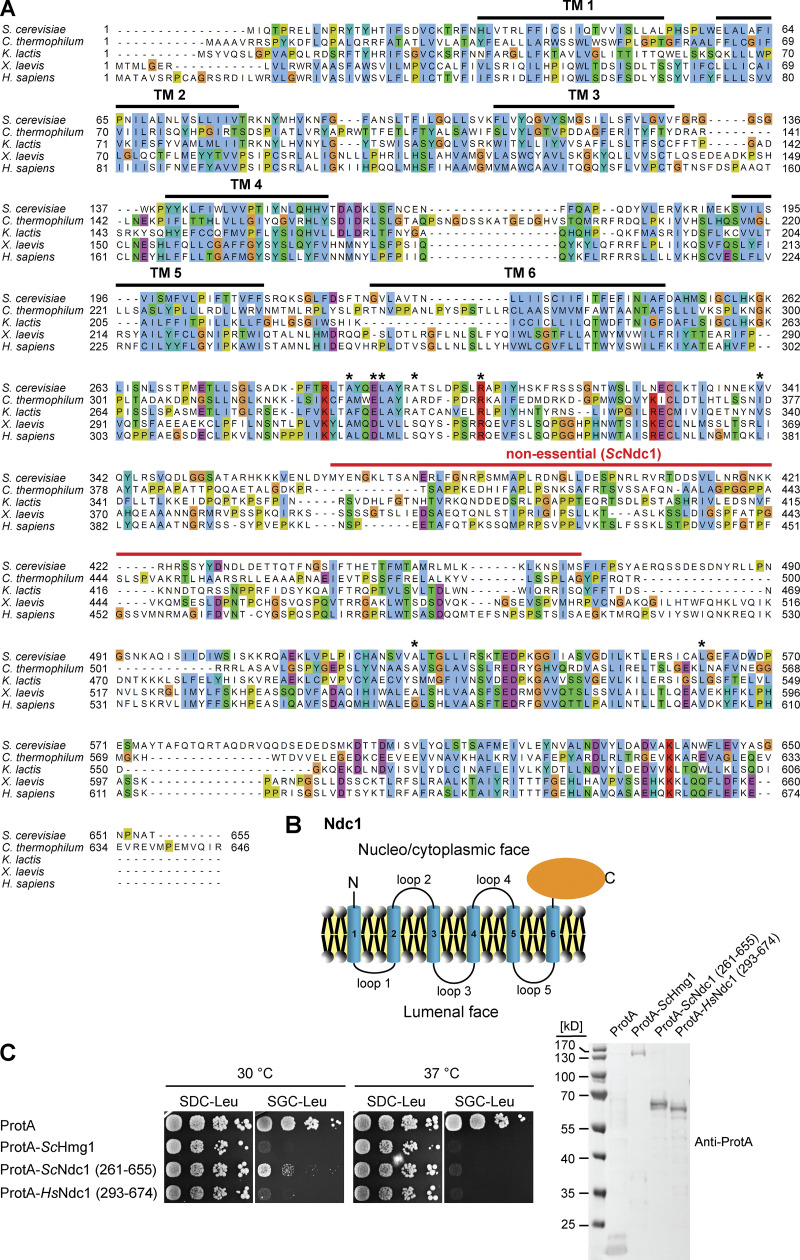
The molecular mass of the NPC up to ∼120 kD in mammals and its complex architecture raise the question of how NPCs are assembled and stabilized in the NE throughout the cell cycle. Metazoan NPC biogenesis occurs in two different cell-cycle stages. The mitotic NPC assembly mode is characteristic of organisms undergoing open mitosis (Kutay et al., 2021). At the beginning of mitosis, the NE and NPCs disassemble, and the NE, including the nuclear membrane proteins, are absorbed by the mitotic ER membrane network. In anaphase, NPC reformation is initiated by the chromatin binding of ELYS/MEL-28, which recruits the Nup107-Nup160 complex to the assembling NPC. NPC assembly continues most likely into preformed openings of the ER membranes contacting the chromatin (Otsuka et al., 2018). Whereas the mitotic assembly mode in metazoan cells establishes a transport-competent nucleus in daughter cells within minutes, NPC biogenesis occurring during interphase is much slower probably due to the requirement of NPC insertion into the intact double membrane layer of the NE (Dultz and Ellenberg, 2010). In organisms undergoing closed mitosis like Saccharomyces cerevisiae, the NE stays intact through the whole cell cycle. Thus, NPCs have to be inserted exclusively into the intact double membrane layer (Doucet and Hetzer, 2010; Otsuka and Ellenberg, 2018; Weberruss and Antonin, 2016). The major challenge is arguably bringing the inner and outer nuclear membrane (INM and ONM) close for the subsequent merger, which is an essential and rate-limiting step of interphase NPC biogenesis. In principle, membrane curvature might be generated from either side or both sides of the NE by applying forces to flat areas of the NE (D’Angelo et al., 2006; Doucet and Hetzer, 2010). Highly curved membranes represent high energy states characterized by unfavorable membrane lipid packing (Kozlov et al., 2010; Martens and McMahon, 2008). Curvature might be induced by (1) NE proteins with large luminal domains bridging between INM and ONM similar to viral fusion proteins or SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors), (2) insertion of amphipathic helices into the lipid bilayer, (3) formation of membrane-connected protein scaffolds or (4) generation of the asymmetric lipid composition of the two leaflets in the lipid bilayer (Antonin et al., 2008; Peeters et al., 2022). Membrane nucleoporins are good candidates for being involved in the assembly of the NPC into the intact NE. Yeast expresses four transmembrane nucleoporins (Ndc1, Pom34, Pom152, and Pom33) of which Ndc1, Pom34, and Pom152 form a stable transmembrane nuclear pore subcomplex and anchor the inner ring of the NPC in the equatorial plane of the nuclear pore membrane (Chial et al., 1998; Lau et al., 2004; Madrid et al., 2006; Onischenko et al., 2009; Rout et al., 2000; Upla et al., 2017; Wozniak et al., 1994). Ndc1 is the only known conserved membrane nucleoporin revealing sequence homology between lower (such as Chaetomium thermophilum) and higher eukaryotes. The C-terminal region of Ndc1 is predicted to be exposed to the cytoplasm/nucleoplasm (Lau et al., 2006; Mans et al., 2004; Mansfeld et al., 2006; Neumann et al., 2010; Stavru et al., 2006; Fig. S2, A and B). In yeasts, Ndc1 is not solely a component of NPCs but also part of spindle pole bodies (SPBs), the NE-embedded microtubule-organizing centers (Araki et al., 2006; Chial et al., 1998; Winey et al., 1993). Ndc1 is thought to be involved in the insertion of both complexes into the intact NE during interphase (Chial et al., 1998; Jaspersen and Ghosh, 2012; West et al., 1998; Winey et al., 1993). In yeast cells, Ndc1 is essential. Depletion causes cell-cycle arrest, and mislocalization of soluble nucleoporins to cytosolic foci and also affects the morphology of the NPC in the NE (Madrid et al., 2006; Onischenko et al., 2009; Winey et al., 1993).
Figure S2.
Overexpression of the Ndc1 C-terminus impairs cell growth. (A) Multiple sequence alignment of Ndc1 homologs from S. cerevisiae (CAA86624.1), C. thermophilum (AEL00695.1), K. lactis (QEU62154.1), X. laevis (ABA39292.1), and H. sapiens (AAZ73087.1) using the ClustalW (Larkin et al., 2007) and displayed in Jalview (Waterhouse et al., 2009). The transmembrane domains (TMs) were predicted using TMHMM (Krogh et al., 2001) and are marked with bold black lines. The red line marks the region in ScNdc1 (368-466) discovered as not being essential for ScNdc1 function (Lau et al., 2004). Asterisks mark amino acid positions in the C-terminus of ScNdc1 involved in interactions with the SPB components ScMps3 and ScNbp1, respectively (Chen et al., 2014). (B) Topology of Ndc1. Ndc1 consists of six predicted transmembrane domains connected via short loops. Both the N-terminal end and the C-terminal part of Ndc1 are exposed to the cytoplasm and nucleoplasm (Stavru et al., 2006). (C) Growth and expression were analyzed as described earlier using yeast cells expressing both ProtA-tagged Ndc1 homologs from S. cerevisiae and H. sapiens and ProtA-tagged ScHmg1 under control of the GAL1-10 promoter from LEU2 marker-containing plasmids. Source data are available for this figure: SourceData FS2.
Ndc1 interacts with the non-essential linker nucleoporins Nup53 and Nup59 thereby bridging between the core of the NPC and the NE, conserved in metazoan (Akey et al., 2022; Eisenhardt et al., 2014; Kim et al., 2018; Mansfeld et al., 2006; Onischenko et al., 2009; Petrovic et al., 2022). Both Nup53 and Nup59 additionally contain C-terminal amphipathic helices thereby exhibiting membrane binding and deformation activity (Eisenhardt et al., 2014; Lin et al., 2016; Marelli et al., 2001; Patel and Rexach, 2008; Vollmer et al., 2012). Furthermore, curvature-sensing proteins are recruited to the NE, e.g., via ArfGAP1 lipid packing sensor (ALPS) motifs (Drin et al., 2007; Vanni et al., 2013), thought to stabilize the curved membrane state. Nup120 and Nup133, conserved members of the Y-shaped Nup84 complex (Kelley et al., 2015; Lutzmann et al., 2002; Stuwe et al., 2015), contain ALPS motifs in exposed unstructured loops of their β-propeller domains capable of binding and stabilizing positive membrane curvature via membrane insertion (Doucet et al., 2015; Doucet et al., 2010; Drin and Antonny, 2010; Drin et al., 2007; Kim et al., 2014; Nordeen et al., 2020). Deletion of NUP120 or NUP133 causes NPC clustering in the NE underlying the proposed function of ALPS motives in NPC biogenesis (Li et al., 1995; Pemberton et al., 1995; Siniossoglou et al., 1996). In addition to the ALPS motif-mediated targeting to the NE, the vertebrate Nup107-Nup160 complex can directly bind the membrane nucleoporin POM121 via the N-terminal β-propeller region of Nup160 (Nup120 in lower eukaryotes; Mitchell et al., 2010). Furthermore, the Nup107-Nup160 complex can be recruited to membranes via interaction with the nuclear basket protein Nup153 which itself can bind the INM via an amphipathic helix (Vollmer et al., 2015).
In this study, we show that in C. thermophilum both Nup120 and Nup133 can be recruited to membranes independently of their ALPS domains via direct protein-protein interactions with Ndc1. In addition, we focus on a so far uncharacterized C-terminal region of the essential yeast Ndc1 protein which is dispensable for viability and therefore not further being suggested of having any crucial functions (Lau et al., 2004). We show that this region contains a previously unknown conserved amphipathic motif and elucidate its interrelationship within the nucleoporin network at the NE in the context of NPC biogenesis in S. cerevisiae.
Results
Nup120 and Nup133 can interact with Ndc1 in vitro and are recruited to Ndc1 containing GUVs
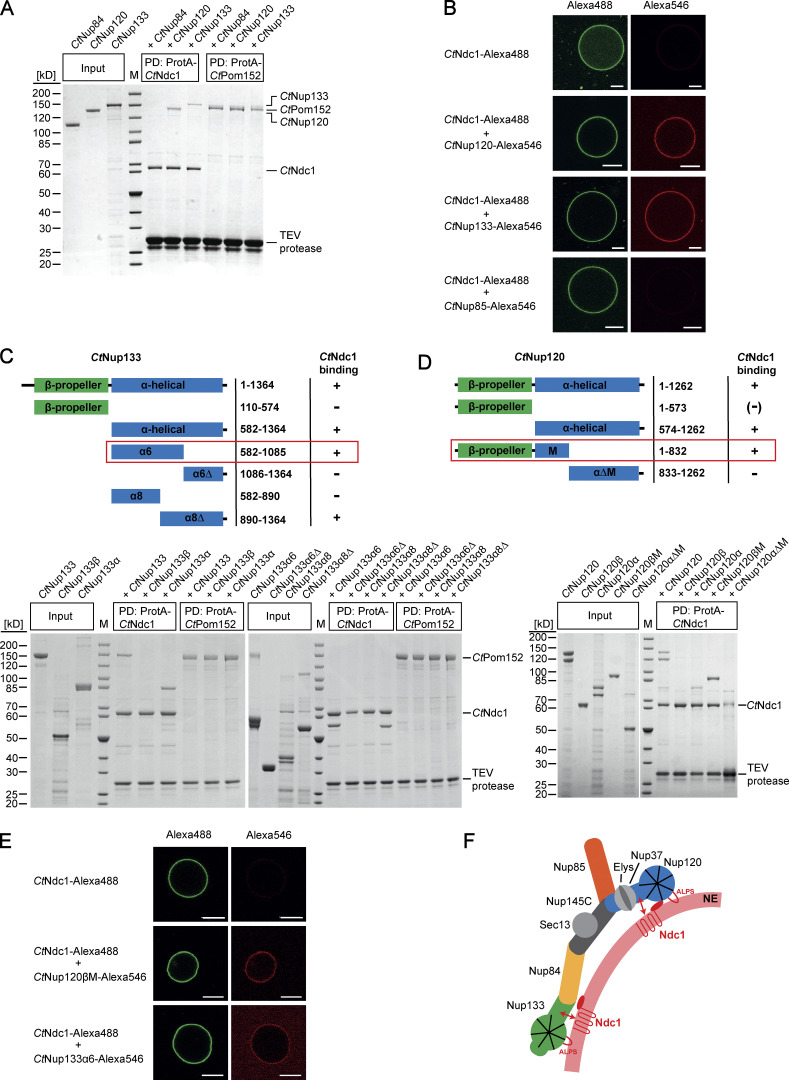
The Nup84 complex (Nup107-Nup160 complex in vertebrates) forms a scaffold coating and presumably stabilizes the high curvature of the nuclear pore membrane. To find out whether integral nuclear pore membrane proteins play a role in the recruitment and anchoring of the Nup84 complex to the NPC in fungi, we performed in vitro binding studies using nucleoporins from the thermophilic fungus C. thermophilum. For this purpose, we expressed CtNdc1 and CtPom152 in yeast, affinity-purified both membrane proteins in detergent, and tested for interaction with members of the CtNup84 complex. This analysis revealed a previously unknown direct interaction of either CtNup120 or CtNup133 with the integral membrane nucleoporin CtNdc1, whereas CtNup84 is another component of the Nup84 subcomplex (Thierbach et al., 2013) did not bind to CtNdc1 (Fig. 1 A). In a negative control, CtPom152 did not show such an interaction with CtNup120 or CtNup133 (Fig. 1 A).
Figure 1.
Chaetomium Nup120 and Nup133 interact with Ndc1 and are recruited to Ndc1-containing GUVs. (A) In vitro binding of nucleoporins of the CtNup84 complex to CtNdc1 and CtPom152. ProtA-tagged CtNdc1 and CtPom152 were expressed in S. cerevisiae and immobilized on IgG beads. The beads were incubated with affinity-purified nucleoporins of the CtNup84 complex. The purified prey Nups were added in a fivefold molar excess over the immobilized bait Nups. IgG-bound proteins were eluted with TEV protease cleaving downstream of the ProtA affinity tag of CtNdc1 and CtPom152. Input (10%) and elution fractions were analyzed via SDS-PAGE and Coomassie staining; M, marker; PD, bait protein used for the pulldown experiment. (B) Alexa Fluor 488-labeled CtNdc1 was reconstituted into giant unilamellar vesicles (GUVs). CtNdc1-GUVs were monitored by the collection of emitted light between 500 and 545 nm after argon laser-mediated excitation at 488 nm (left panels) whereas recruitment of Alexa Fluor 546-labeled CtNup120, CtNup133, or CtNup85 (red) to CtNdc1-GUVs (green) was analyzed by confocal microscopy. Scale bars, 10 µm. (C and D) In vitro binding of truncated versions of CtNup133 (C) and CtNup120 (D) purified from E. coli BL21 (DE3) to ProtA-tagged CtNdc1 immobilized on IgG beads. (E) Recruitment of either Alexa Fluor 546-labeled CtNup120βM or CtNup133α6 (red) to CtNdc1-GUVs (green) was analyzed as above (middle panels). Scale bars, 10 µm. (F) Model of recruitment and stabilization of the CtNup84 complex and CtNdc1 to NPC assembly sites in the NE. The CtNup84 complex binds directly to the NE via exposed ALPS domains in the beta-propeller domains of both CtNup120 and CtNup133 and also interacts via alpha-helical domains of CtNup120 and CtNup133 with the membrane nucleoporin CtNdc1. Source data are available for this figure: SourceData F1.
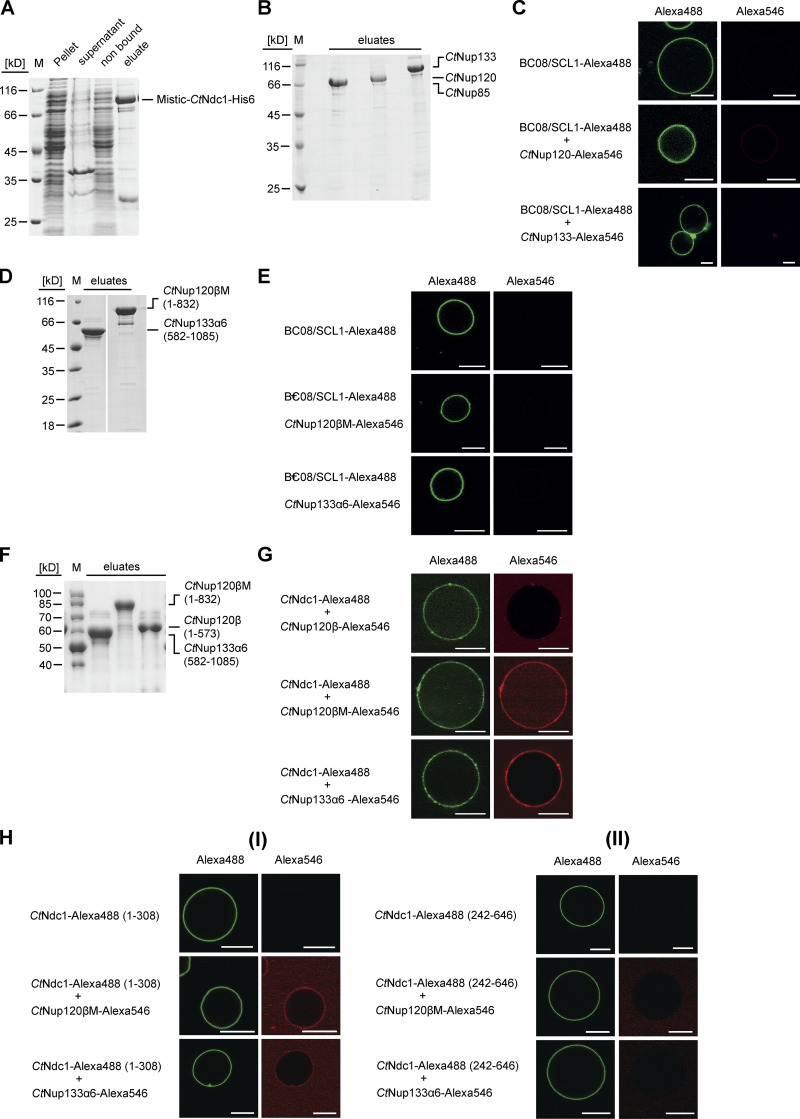
To reveal this Ndc1-Nup84 complex interaction not only in a micellar detergent system but in a real membrane environment, we reconstituted purified CtNdc1 (Fig. S1 A) into giant unilamellar vesicles (GUVs). This physiological-like reconstitution assay showed CtNup120 and CtNup133 (Fig. S1 B) recruitment to CtNdc1 embedded in GUV membranes, whereas CtNup85 was still not bound (Fig. 1 B). Moreover, CtNup120 and CtNup133 did not associate with GUVs containing the unrelated inner nuclear membrane protein BC08/SCL1 (Fig. S1 C). Although possessing ALPS domains, membrane association of both CtNup120 and CtNup133 in the absence of Ndc1 was probably hindered by the low curvature of the GUV membranes. This is similar to the vertebrate Y-complex, which does not bind to GUVs (Vollmer et al., 2015). Mapping of the protein–protein interaction sites revealed that sequences in the C-terminal α-helical domains of CtNup120 and CtNup133-mediated CtNdc1 binding, whereas the β-propeller domains did not bind to CtNdc1 (Fig. 1, C and D). Further fine-mapping indicated that the truncated α-helical domain of CtNup133 (CtNup133α6) was still able to bind to CtNdc1 (Fig. 1 C). Since the truncated α-helical domain of CtNup120 (CtNup120 [574-832]) was insoluble, we succeeded expressing it together when still attached to the β-propeller domain (CtNup120βM), which finally revealed strong binding of CtNup120βM to CtNdc1 (Fig. 1 D). Accordingly, purified CtNup133α6 and CtNup120βM (Fig. S1 D) were efficiently recruited to CtNdc1-GUVs (Fig. 1 E), but not to SCL1/BC08 containing GUVs (Fig. S1 E). CtNup120β without its α-helical extension could not be recruited to CtNdc1-GUVs (Fig. S1 G) suggesting that aa 574-832 is required for the CtNup120-CtNdc1 interaction. To find out which parts of CtNdc1 bind to CtNup120 and CtNup133, we separated CtNdc1 N-terminal transmembrane (Fig. S1 H [I]) and C-terminal soluble domains (Fig. S1 H [II]). This revealed that only the CtNdc1 N-terminal transmembrane part was sufficient to recruit CtNup120 or CtNup133 to the GUV membrane (Fig. S1 H [I]).
Figure S1.
Chaetomium Nup120 and Nup133 interact with Ndc1. (A) CtNdc1 used for reconstitution in GUVs was Ni-NTA-purified from E. coli BL21 (DE3). CtNdc1 was additionally N-terminally fused to MISTIC, a B. subtilis protein allowing high-level expression. Elution was done by imidazole treatment. Before GUV reconstitution, the MISTIC tag was cleaved off using thrombin protease. The different fractions obtained during purification were analyzed via SDS-PAGE and Coomassie staining. (B) ProtA-tagged CtNup85, CtNup120, and CtNup133 were expressed in S. cerevisiae under the control of the GAL1-10 promoter and purified using IgG beads. Purified proteins were cleaved off the beads using TEV protease. Elution fractions were subjected to SDS-PAGE and Coomassie staining. (C) SCL1/BC08 was expressed and purified from E. coli BL21 (DE3), as described for CtNdc1, Alexa Fluor 488-labeled, and reconstituted into GUVs. SCL1/BC08-GUVs were incubated with either Alexa Fluor 546-labeled CtNup120 or CtNup133. Binding was monitored as described in Fig. 1. Bars, 10 µm. (D) Expression and purification of CtNup120βM consisting of an α-helical extension of the β-propeller domain of CtNup120 and CtNup133α6, an alpha-helical stretch in the C-terminal region of CtNup133. Elution fractions were subjected to SDS-PAGE and Coomassie staining. (E) SCL1/BC08-GUVs were incubated with either Alexa 546-labeled Nup120βM or Nup133α6. Binding was monitored as described earlier. Bars, 10 µm. (F) Expression and purification of CtNup120β, CtNup120βM, and CtNup133α6. (G) CtNdc1-GUVs were incubated either with Alexa Fluor 546-labeled CtNup120β, CtNup120βM or CtNup133α6. Bars, 10 µm. (H) (I) N- (1-308) or (II) C-terminal (242-646) fragments of CtNdc1, expressed, purified and labeled as above, were reconstituted into GUVs and incubated with Alexa Fluor 546-labeled Nup120βM or Nup133α6. The CtNdc1 (242-646) fragment additionally harbored the sixth transmembrane helix of CtNdc1 to ensure proper reconstitution into GUV membranes. Source data are available for this figure: SourceData FS1.
Together, these data suggest that the CtNup84 complex, besides its ALPS-mediated interaction with the NE, has a second way to attach to the nuclear pore membrane, namely via direct interaction of the Ndc1 transmembrane domain with either CtNup120 or CtNup133. In this way, the integral part of the pore membrane protein Ndc1 could function in local Nup84 complex attachment at the fused and curved double nuclear membrane, which is the site where NPCs are inserted (Fig. 1 F).
Overexpression of Ndc1 induces ER membrane expansions with pore-like holes
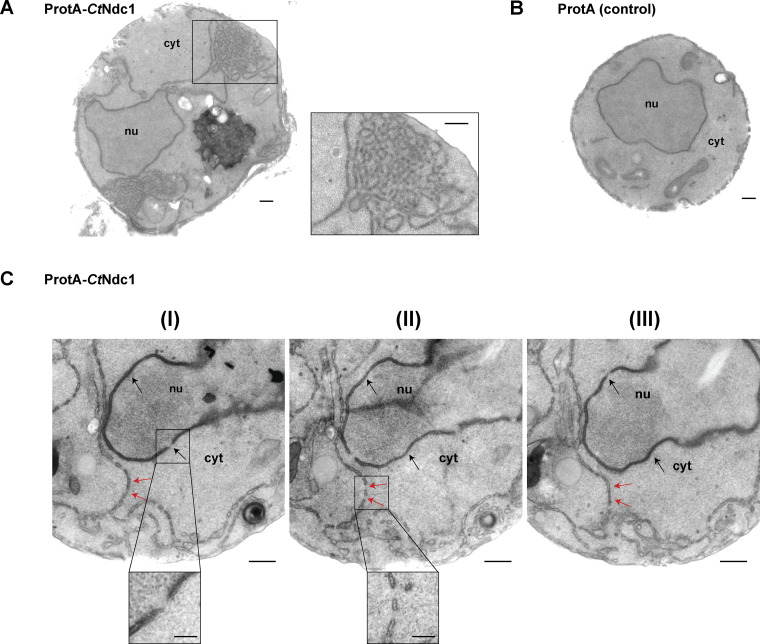
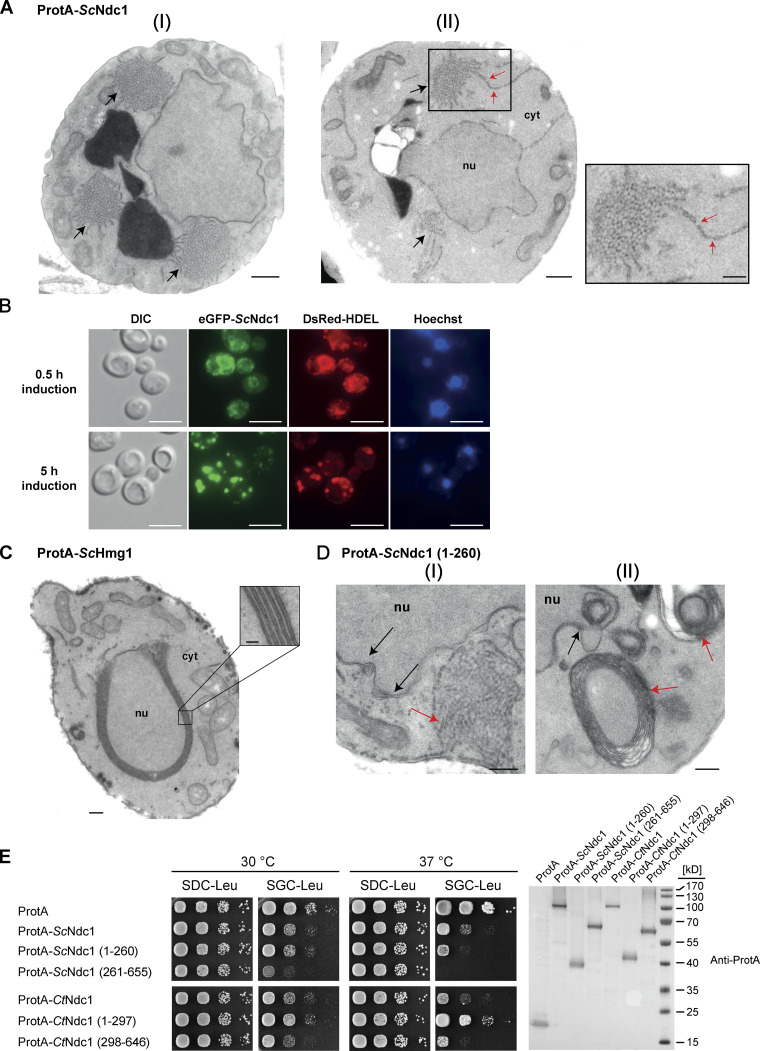
In the course of GAL1-10-promoter-induced ProtA-CtNdc1 expression in yeast, we noticed the formation of extensive extranuclear membranes, tubules, and cisternae, often organized as membrane clusters which were not observed in the control strain expressing only ProtA (Fig. 2, A and B). These abnormal membranous structures are derived from the ER, which is continuous with the ONM and cortical ER. Similar membrane proliferations were observed earlier upon overexpression of GFP-HsNDC1 or GFP-HsPOM121 in human HeLa cells (Volkova et al., 2011). Serial ultrathin sections of fixed yeast cells overexpressing ProtA-CtNdc1 revealed that these unusual extranuclear membrane proliferations exhibited pore-like structures (Fig. 2 C). These pore-like structures had a reduced diameter as compared to the diameter of NPCs within the nuclear membrane (44.8 ± 3.4 nm [mean ± SD; n = 20] versus 67.1 ± 2.3 nm [mean ± SD; n = 20]).
Figure 2.
Overexpression of Chaetomium Ndc1 in yeast induces ER membrane expansion with pore-like structures. (A) Electron micrograph of a yeast cell overexpressing ProtA-CtNdc1 showing clusters of membrane tubules in the cytoplasm. One of these clusters is shown at a higher magnification. Bars, 250 nm. (B) As control an electron micrograph of a yeast cell expressing the ProtA tag alone is shown. Bar, 250 nm. (C) Serial sections (approximate thickness of 80 nm) of a cell overexpressing ProtA-CtNdc1. Black arrows mark nuclear pores visible in the NE. Red arrows mark local membrane openings in cytoplasmic membrane structures. Bars, 250 nm. Magnifications show a nuclear pore in the NE (I) and openings of membranes in the cytoplasm (II; lower panels; bars, 100 nm).
Overexpression of Ndc1 from S. cerevisiae (ProtA-ScNdc1) induced similar membrane morphologies (Fig. 3 A). EGFP-tagged ScNdc1 colocalized upon overexpression with the ER-marker HDEL-DsRed (Fig. 3 B). At short induction times (0.5 h) Ndc1 is mainly localized to the nuclear envelope as indicated by nuclear rim staining. Longer induction times (5 h) caused almost complete mislocalization of ScNdc1 to cytoplasmic ER-positive foci (Fig. 3 B), which likely correspond to the cytoplasmic membrane structures shown in Fig. 3 A. Overexpression of the ER membrane protein 3-hydroxy-3-methylglutaryl coenzyme A reductase (ProtA-ScHmg1), which is known to induce the formation of karmellae, i.e., membrane stacks associated with the outer nuclear membrane (Wright et al., 1988), resulted in membrane proliferations distinct from the membrane expansions with pore-like structures observed upon NDC1 overexpression (Fig. 3 C). When overexpressing the N-domain of Ndc1, which includes all six transmembrane regions (Fig. S2 A), we observed a phenotype resembling the overexpression of full-length Ndc1, i.e. extranuclear membrane proliferations with pore-like structures (Fig. 3 D [I]). Several electron micrographs showed cells containing ring-shaped, multi-layered membranes not observed upon overexpression of full-length ScNdc1 (Fig. 3 D [II]). Such karmellae-like structures, which are not attached to the NE, are described as ER whorls formed upon ER stress and further subjected to ER-phagy (Bernales et al., 2006; Schuck et al., 2014).
Figure 3.
Overexpression of budding yeast Ndc1 or its N-terminal domain induces ER membrane expansion with pore-like structures. (A) Electron micrographs of WT yeast cells overexpressing ProtA-ScNdc1 showing the membrane proliferation. Cytoplasmic tubular membrane assemblies are marked with black arrows. Bars, 500 nm. ER-expansions with local membrane discontinuities (II), marked with red arrows, are magnified. Bars, 250 nm. (B) Fluorescence microscopy of cells overexpressing eGFP-Ndc1 for 0.5 or 5 h using DsRed-HDEL as an ER marker. Chromatin is stained with Hoechst 33258. Bars, 5 µm. (C) Electron micrograph of a WT yeast cell overexpressing ProtA-tagged 3-hydroxy-3-methylglutaryl-coenzymeA reductase 1 (ProtA-ScHmg1). The observed karmellae are shown at higher magnification (bars, 250 and 50 nm for the magnified panel). (D) Electron micrographs of yeast cells overexpressing the N-terminal part of ScNdc1 (ProtA-ScNdc1 (1-260)). Black arrows mark NE/ER proliferations, red arrows cytoplasmic membrane assemblies. Tubular membrane assemblies, similar to the ones observed upon overexpression of full-length Ndc1, are shown in I, karmellae-like multi-layered membrane whorls are shown in II. Bars, 250 nm. (E) Growth tests with yeast cells overexpressing ProtA-tagged full-length Ndc1 and Ndc1 truncations from both C. thermophilum and S. cerevisiae, ProtA-ScHmg1 or ProtA alone using LEU2 as selection marker. 10-fold serial dilutions of corresponding overnight cultures were spotted onto either glucose- (expression suppressing) or galactose- (expression inducing) containing plates lacking leucine (SDC-Leu or SGC-Leu) and incubated at 30 or 37°C for up to 3 d. For expression control, whole cell lysates were prepared from overnight cultures and analyzed by Western blotting. Source data are available for this figure: SourceData F3.
Consistent with the observed dramatic membrane proliferation phenotype upon overexpression of either CtNdc1 or ScNdc1 in yeast, these cells exhibited significant growth defects both at 30 and 37°C when compared to WT yeast cells expressing ProtA alone (Fig. 3 E). To find out, which of the Ndc1 domains is responsible for the observed toxic effects, we overexpressed either the C-terminal (soluble cytoplasmic/nucleoplasmic) or N-terminal (transmembrane) part of Ndc1 from either S. cerevisiae or C. thermophilum in yeast. In both cases, the strong growth defect could be predominantly attributed to the corresponding Ndc1 C-domains, and the growth inhibition was even more severe than observed for the overexpression of the full-length Ndc1 proteins (Fig. 3 E). Overexpression of the C-domain of human Ndc1 as well as ProtA-ScHmg1 was also highly toxic to yeast cells (Fig. S2 C). Altogether, these findings showed that Ndc1 overexpression induced a strong ER membrane proliferation phenotype, yielding extranuclear membranes showing pore-like perforations.
The cytoplasmic/nucleoplasmic C-terminal domain of Ndc1 contains a conserved amphipathic motif
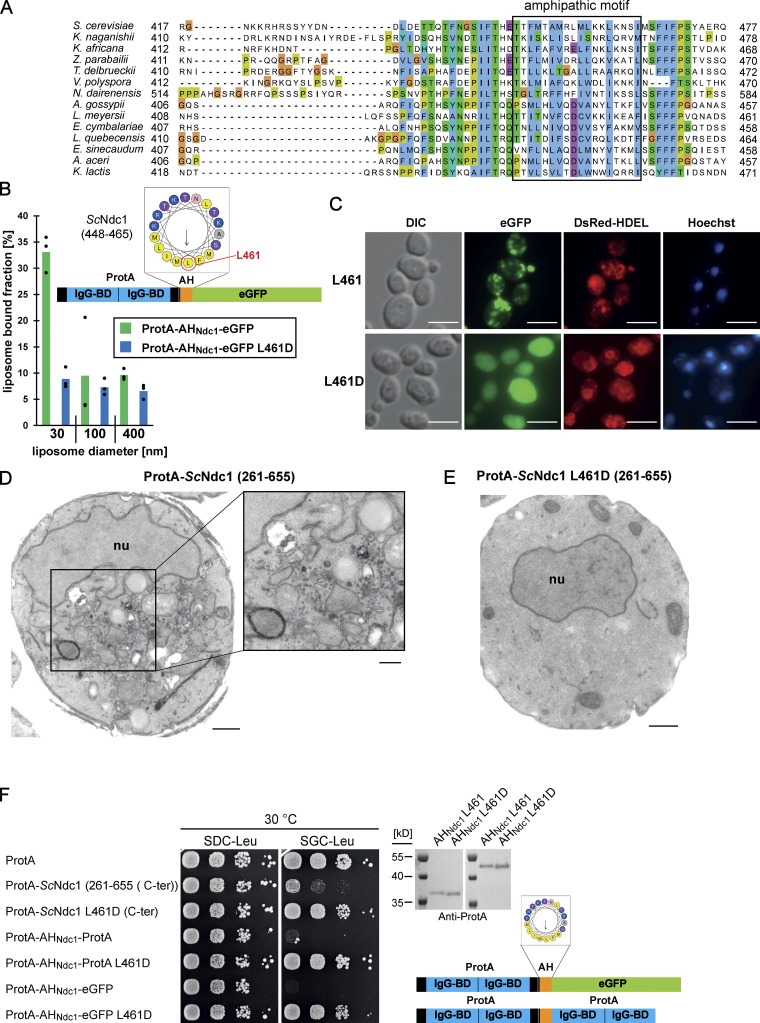
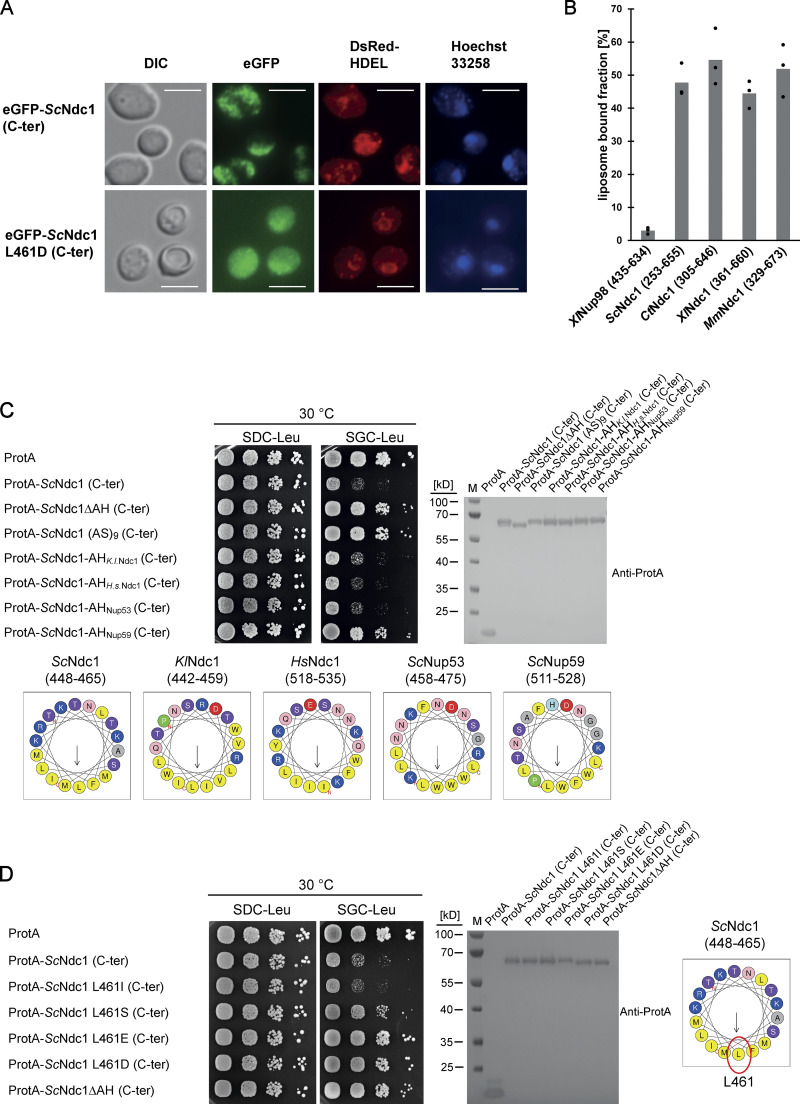
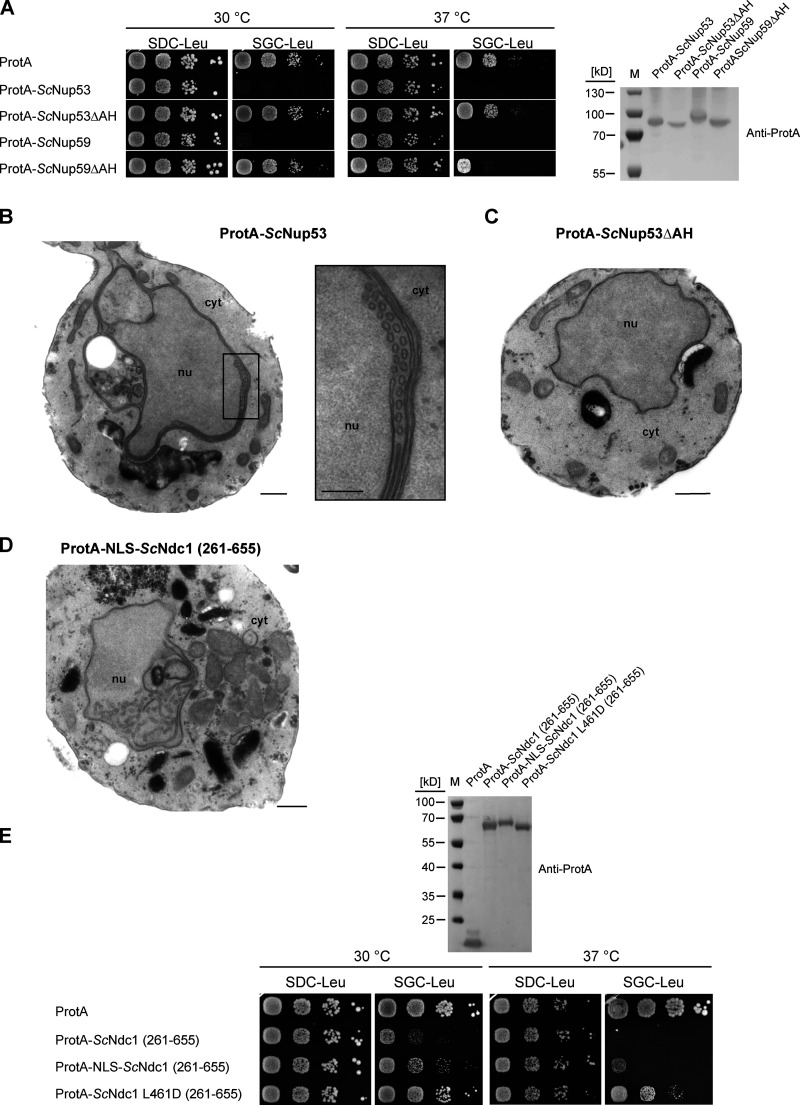
To identify the toxicity-inducing motif in the Ndc1 C-domain, we generated a series of Ndc1 deletion constructs. In the past, several ScNdc1 truncations have been constructed, e.g., by removing a large part from the non-essential C-terminus (Fig. S2 A, ScNdc1 Δ368-466), but also other parts upstream and downstream of residues 368–466, which were suggested to be crucial for ScNdc1 function and localization (e.g., for interaction with the SPB components ScNbp1 and ScMps3; Chen et al., 2014; Lau et al., 2004). Prompted by the observed severe toxicity of the Ndc1 C-domain upon overexpression even in absence of the N-terminal transmembrane domains (Fig. 3 E), we searched for a possible membrane-interacting motif in the ScNdc1 C-terminal domain by HELIQUEST (Gautier et al., 2008), which indeed detected such a sequence between residues 448–465 (Fig. 4, A and B). The primary structure of this predicted amphipathic sequence shows low conservation, even among closely related Saccharomycetales, but its amphipathic character is strongly conserved (Fig. 4 A). We cloned this predicted amphipathic helix (AH) from yeast Ndc1 and inserted it between N-terminal ProtA and C-terminal eGFP. After expression and affinity-purification of this ProtA-AHNdc1-eGFP construct, we tested whether it can bind to membranes employing a liposome flotation assay (Fig. 4 B). This revealed that ProtA-AHNdc1-eGFP can efficiently bind to liposomes, with a preference for smaller and highly curved liposomes, which is typical for the well-characterized ALPS (ArfGAP1 lipid packing sensor) motifs (Drin et al., 2007; Vanni et al., 2013). When disturbing the hydrophobic face of the putative amphipathic helix by changing Ndc1 L461>D, this mutation strongly reduced liposome binding (Fig. 4 B). To analyze whether the Ndc1-AH alone is capable of inducing membrane abnormalities upon overexpression in yeast, we analyzed the intracellular localization of ProtA-AHNdc1-eGFP constructs by fluorescence microscopy. This revealed cytoplasmic foci and patches, partially overlapping with a DsRed-HDEL ER marker and often seen close to either the plasma membrane or nuclear membrane, whereas, in the case of the mutant Ndc1-AH, only continuous cytoplasmic staining was seen (Fig. 4 C). Similar distribution patterns were also observed for cells expressing the Ndc1-AH when part of the entire ScNdc1 C-terminus (Fig. S3 A, eGFP-ScNdc1 (261-655) and eGFP-ScNdc1 L461D (261-655)). As anticipated, the C-terminal domains of the employed higher eukaryotic Ndc1 homologs from Xenopus laevis or mouse were also capable of binding to liposomes, whereas the purified C-terminal part of XlNup98 serving as a negative control was not recruited (Fig. S3 B).
Figure 4.
The C-terminus of Ndc1 contains a conserved amphipathic sequence motif. (A) Multiple sequence alignment of a C-terminal region of Ndc1 homologs from different fungal species. The alignment was done using the ClustalW (Larkin et al., 2007) and displayed in Jalview (Waterhouse et al., 2009). The putative amphipathic regions in all sequences were detected using HELIQUEST (Gautier et al., 2008). (B) In vitro liposome binding assay with the isolated amphipathic helix (ProtA-AHNdc1-eGFP) and the corresponding L461D mutation using 30, 100, and 400 nm liposomes. Columns represent the mean of three independent experiments, individual data points are indicated. (C) Fluorescence microscopy of cells overexpressing the isolated amphipathic helix (ProtA-AHNdc1-eGFP) and the corresponding L461D mutation. DsRed-HDEL serves as ER marker. Chromatin is stained with Hoechst 33258. Bars, 5 µm. (D) Electron micrograph of a yeast cell overexpressing the C-terminal part of ScNdc1 (ProtA-ScNdc1 (261-655)) showing accumulation of cytoplasmic vesicles. Bar, 500 nm. In the right panel a higher magnification is shown (bar, 250 nm). (E) Electron micrograph of a yeast cell expressing ProtA-ScNdc1 (261-655) carrying the point mutation L461D. Bar, 500 nm. (F) Growth and expression were analyzed as in Fig. 3 E using yeast cells overexpressing the artificial model constructs ProtA-AHNdc1-ProtA and ProtA-AHNdc1-eGFP. The domain structures of both artificial constructs were schematically illustrated. Source data are available for this figure: SourceData F4.
Figure S3.
The Ndc1 C-terminus contains an amphipathic membrane binding motif. (A) Fluorescence microscopy of cells expressing eGFP-tagged ScNdc1 (261-655), (upper panels) or eGFP-ScNdc1 (261-655) additionally carrying the point mutation L461D (lower panels). Hoechst 33258 served as a chromatin stain. Bars, 5 µm. (B) In vitro liposome binding assay using in E. coli expressed and purified C-termini from different Ndc1 homologs using 30 nm liposomes. Columns represent the average of three independent experiments, individual data points are indicated. A fragment of Xenopus Nup98 (435-634) served as the negative control. (C) Growth and expression were analyzed as described in Fig. 3 using yeast cells expressing chimeric Ndc1 constructs comprised of the C-terminal part of ScNdc1 (ProtA-ScNdc1 (C-ter)) and different amphipathic motifs substituted for the endogenous amphipathic motif of ScNdc1 (448-465). The corresponding sequences are illustrated as helical wheel projections. As controls ProtA-ScNdc1 (C-ter) either deleted in the amphipathic stretch (ProtA-ScNdc1ΔAH (C-ter)) or instead the AH containing an 18 aa long alanine-serine spacer (ProtA-ScNdc1 (AS)9 (C-ter)) were expressed. (D) Growth and expression were analyzed using yeast cells expressing the C-terminus of ScNdc1 (ProtA-ScNdc1 (C-ter)) either mutated in the hydrophobic face of the amphipathic motif (as illustrated) or deleted in the complete 18 aa amphipathic stretch (ProtA-ScNdc1ΔAH (C-ter)). Source data are available for this figure: SourceData FS3.
To reveal the specific membrane alterations induced by the toxic Ndc1 amphipathic motif in more detail, we performed transmission electron microscopy of cells overexpressing ProtA-ScNdc1 (261–655). Surprisingly, many cells accumulated extra cytoplasmic membrane structures, in particular clusters of small vesicles (Fig. 4 D), distinct from what was observed upon overexpression of full-length Ndc1 or its N-domain (Fig. 3, A and D). However, similar results were previously seen in yeast overexpressing the ALPS-domain-containing α-synuclein, which induced curved membrane structures including a large number of small vesicles (Outeiro and Lindquist, 2003; Pranke et al., 2011). Cells expressing the Ndc1 C-terminus carrying the mutated AH at L461>D (ProtA-ScNdc1 L461D [261-655]), however, exhibited a normal membrane organization (Fig. 4 E) like cells expressing ProtA alone (Fig. 2 B).
To prove that the Ndc1-AH is responsible for the toxicity initially observed when overexpressing the complete C-terminus of Ndc1 in yeast cells (Fig. 3 E), we generated minimal constructs, ProtA-AHNdc1-eGFP and ProtA-AHNdc1-ProtA, either intact or carrying the L461D mutation, and induced expression in vivo via the strong GAL1-10 promoter, which yielded a similar growth pattern like observed for the longer constructs (Fig. 4 F). Furthermore, we generated chimeric ScNdc1 constructs composed of ProtA-ScNdc1 (261-655), but carrying instead of its own amphipathic helix (residues 448-465) the amphipathic motifs from other Ndc1 orthologs including human, or the well-characterized amphipathic helices of ScNup53 and ScNup59. Except for the AH from ScNup59, all the other AHs induced a toxic phenotype in yeast (Fig. S3 C). Replacing the Ndc1-AH with an 18 amino acid long stretch of alternating alanine-serine residues, called (AS)9, or deleting the complete AH (ΔAH) rescued the growth defect (Fig. S3 C). Moreover, only L461D or L461E point mutations, destroying the hydrophobic character of the Ndc1-AH, but not a conserved L461I mutation, could inactivate the toxicity upon overproduction (Fig. S3 D).
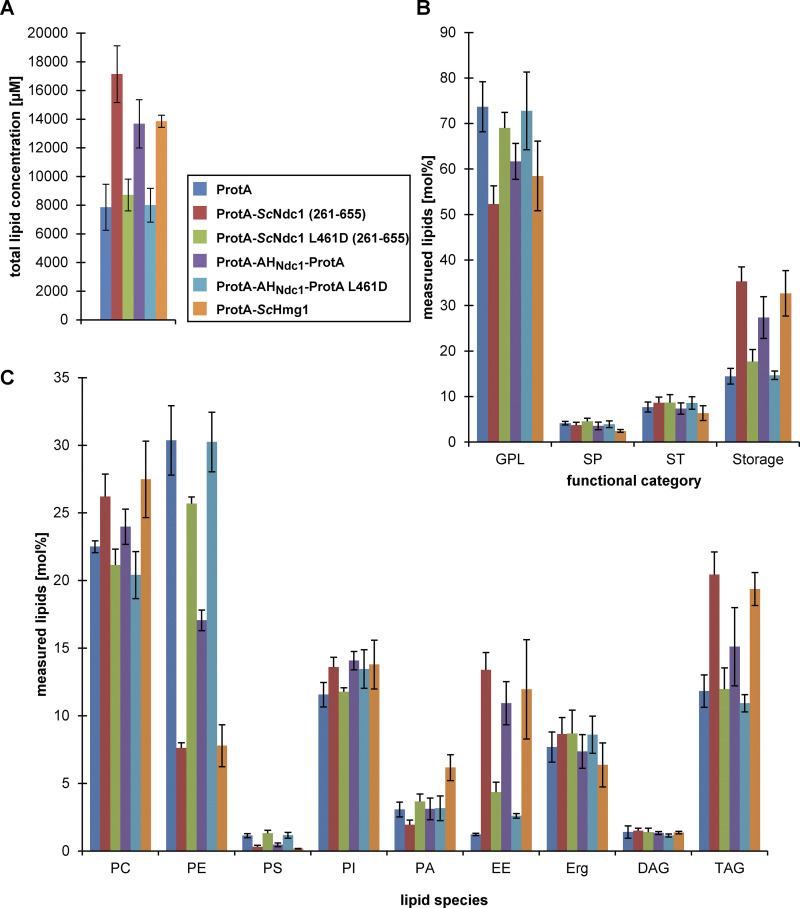
Overexpression of ProtA-ScNdc1 (261-655) or solely ProtA-AHNdc1-ProtA was accompanied by altered cellular lipid profiles (Fig. 5). The total lipid content was drastically increased in cells expressing ProtA-ScNdc1 (261-655), ProtA-AHNdc1-ProtA but also ProtA-ScHmg1 (Fig. 5 A). This is consistent with the massive membrane proliferation observed. As the ER and NE are the main sites of lipid synthesis, it is conceivable that the increase in these membrane areas also increases the cell's lipid synthesis capabilities. The Ndc1 L461D mutation in the ProtA-ScNdc1 (261-655) construct and the isolated amphipathic helix (ProtA-AHNdc1-ProtA L461D) alleviated this effect.
Figure 5.
Overexpression of the C-terminus of Ndc1 or its amphipathic motif alters cellular lipid profiles. Lipid profiles of cells overexpressing the indicated proteins under the control of the GAL1-10 promoter. Lipids were extracted from cells grown overnight to an OD600 of ≈ 1.0 and analyzed by nano-ESI-MS/MS. (A) Total lipid concentrations of lipid extracts prepared from equal cell numbers. (B) Plots of the functional categories of lipids glycerophospholipids (GPL), sphingolipids (SP), sterols (ST), and storage lipids (Storage). (C) Amount of the lipid species phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid (PA), ergosteryl ester (EE), ergosterol (Erg), diacylglycerol (DAG), and triacylglycerol (TAG) were plotted as mol% of measured lipids. Error bars represent the standard deviation of the mean of four independent experiments.
When analyzing the lipid classes, we found that the proportion of storage lipids, especially triacylglycerol (TAG) and ergosteryl ester (EE) increased upon overproduction of an intact Ndc1-AH and ScHmg1 in comparison with control cells (Fig. 5, B and C). This was mirrored by a reduction of the proportion of the membrane phospholipids phosphatidylethanolamine (PE) and phosphatidylserine (PS). A similar increase in storage lipids was previously observed upon overexpression of the amphipathic helix-containing α-synuclein in S. cerevisiae (Fanning et al., 2019), suggesting that excess of a membrane interacting amphipathic helix can massively disturb lipid homeostasis. Accordingly, for the L461D mutation, both in the C-domain and in the isolated AH this effect was lost. The increased concentrations of EE and TAG might point toward an enhanced biosynthesis/stability of lipid droplets (LDs) triggered by the amphipathic motif of ScNdc1 or the ScHmg1-mediated ER-expansion. This could be similar to what was shown for the α-synuclein AH, which can bind LDs and prevent their turnover (Cole et al., 2002; Girard et al., 2021).
Since also overexpression of ProtA-ScNup53 or ProtA-ScNup59 was toxic to cells, which, like in the case of the Ndc1-AH, was caused by the corresponding C-terminal amphipathic motifs (Fig. S4 A), we wondered why ProtA-ScNup53 overproduction triggered the formation of inner nuclear membrane abnormalities (Fig. S4 B; see also Marelli et al., 2001) whereas Ndc1-AH caused cytoplasmic membrane abnormalities (see Fig. 4 D). We suspected that ScNup53 may exert its toxic effect in the nucleus due to NLS-mediated nuclear import (Lusk et al., 2002). Hence, we fused the ScNdc1 C-domain to the NLS of SV40 large T antigen (Kalderon et al., 1984). As anticipated, this construct, which was still toxic for the cells, induced extensive intranuclear membrane proliferations not observed upon overexpression of ScNdc1 261-655 alone (Fig. S4, D and E).
Figure S4.
Nup53 or Nup59 overexpression-induced growth defects depend on their amphipathic helices. (A) Growth tests with cells overexpressing the linker nucleoporins Nup53 and Nup59 either non-mutated or mutated in their C-terminal amphipathic motifs. Growth and expression were analyzed as described earlier. (B) Electron micrograph of a yeast cell overexpressing ProtA-ScNup53; bar, 500 nm. Membrane sheets and tubules close to the INM are magnified (bar, 250 nm). (C) Electron micrograph of a yeast cell overexpressing ProtA-ScNup53ΔAH. Bar, 500 nm. (D) Electron micrograph of a yeast cell overexpressing ProtA-NLS-ScNdc1 (261-655) showing intranuclear vesicular membrane structures. Bar, 500 nm. (E) Growth tests with cells overexpressing the C-terminus of ScNdc1 N-terminally fused to the NLS sequence of the SV40 large T-antigen. Growth and expression were analyzed as described earlier. Source data are available for this figure: SourceData FS4.
Taken together, a transferable and conserved amphipathic motif in the C-terminus of ScNdc1, similar to related motifs in functionally interacting nucleoporins such as ScNup53, exerts a dominant-negative effect on intracellular membrane organization, which may have its primary cause in affecting membrane curvatures (see Discussion).
Ndc1’s essential function can be suppressed by deleting the Nup53 amphipathic helix
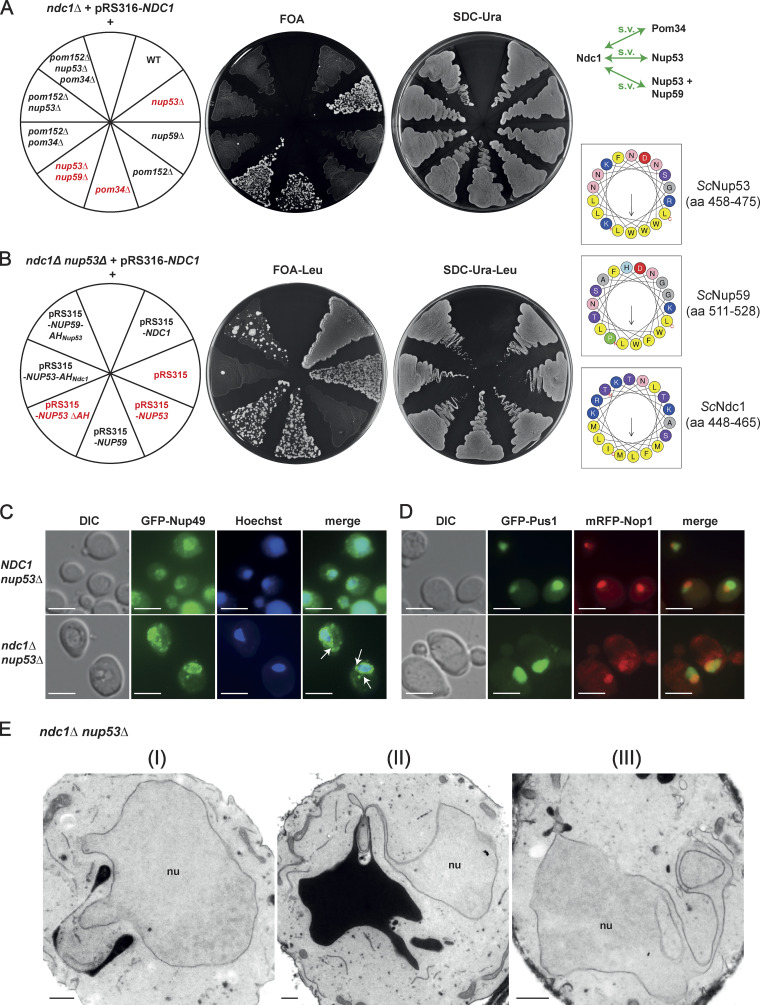
Prompted by the finding that Ndc1 and Nup53/Nup59 amphipathic motifs may affect the nuclear membrane curvature at the NPC insertion sites in a coordinated fashion, we wondered whether this is also manifested in a genetic relationship, for instance in form of a synthetic lethal interaction. However, we discovered the opposite, namely that the lethal phenotype of a complete NDC1 gene disruption was rescued by further deleting the non-essential NUP53 gene, which quite unexpectedly allowed the ndc1Δ nup53Δ double disruption strain to regain growth (Fig. 6 A). In contrast, the chromosomal nup59Δ deletion did not rescue the ndc1Δ cells (Fig. 6A), whereas an ndc1Δ nup53Δ nup59Δ triple knock-out strain was still viable. Notably, also a pom34Δ deletion could restore growth of the lethal ndc1Δ strain (Fig. 6 A) consistent with the observed viability of GAL1-NDC1 pom34Δ cells when shifted from galactose to glucose media (Madrid et al., 2006). Thus, the absence of the non-essential integral pore membrane protein Pom34 or lack of the linker nucleoporin Nup53, which via its C-terminal amphipathic helix interacts with the nuclear membrane (Eisenhardt et al., 2014; Marelli et al., 1998; Patel and Rexach, 2008; Vollmer et al., 2012), rescues the lethal phenotype of the ndc1Δ null yeast strain.
Figure 6.
ScNdc1 genetically interacts with the amphipathic helix of ScNup53. (A) Nup53 and Pom34 interact genetically with Ndc1. The used yeast strains were all endogenously deleted in NDC1 and expressed ScNdc1 from a centromeric URA3 marker-containing plasmid under the control of its native promoter (pRS316-ScNDC1). Loss of pRS316-ScNDC1 was forced by spreading out on plates containing 5-fluoroorotic acid (FOA). Growth on SDC-Ura plates served as control. Green arrows in the illustration mark the observed positive genetic interactions resulting in synthetic viability (s.v.) of corresponding double/triple deletion strains. (B) Ndc1 shuffle strains deleted in NUP53 and expressing different proteins, as indicated, from pRS315, a centromeric LEU2 marker-containing plasmid, were spread on FOA-Leu plates forcing the loss of pRS316-ScNDC1. Growth on SDC-Ura-Leu plates served as control selecting for the presence of both plasmids. The amphipathic motifs present in the expressed proteins were illustrated as helical wheel projections. (C) Fluorescence microscopy of yeast strains, as indicated, expressing the GFP-tagged nucleoporin ScNup49 as NPC marker showing interphase cells. White arrows mark GFP-ScNup49 as part of NE/ER expansions. Hoechst 33258 served as a chromatin stain. Bars, 5 µm. (D) Fluorescence microscopy of interphase yeast cells, as indicated, coexpressing GFP-tagged ScPus1 as nucleoplasmic and the mRFP-tagged ScNop1 as a nucleolar marker. Hoechst 33258 served as a chromatin stain. Bars, 5 µm. (E) Electron micrographs of yeast cells deleted in both NDC1 and NUP53 (I–III). Bars, 500 nm.
To find out whether the nup53Δ-dependent suppression of ndc1Δ is dependent on the Nup53 C-terminal amphipathic helix (AH), we combined nup53 ΔAH and ndc1Δ alleles, which however still allowed the cells to grow (Fig. 6 B). In contrast, replacing Nup53’s C-terminal amphipathic helix by the AHNdc1 abolished the suppression phenotype and ndc1Δ cells were no longer viable (Fig. 6 B, Nup53-AHNdc1). On the other hand, a Nup59-AHNup53 construct enabled the ndc1Δ suppression phenotype (Fig. 6 B) indicating that Nup59 performs (an) unique role(s) at the NE independent of its C-terminal AH.
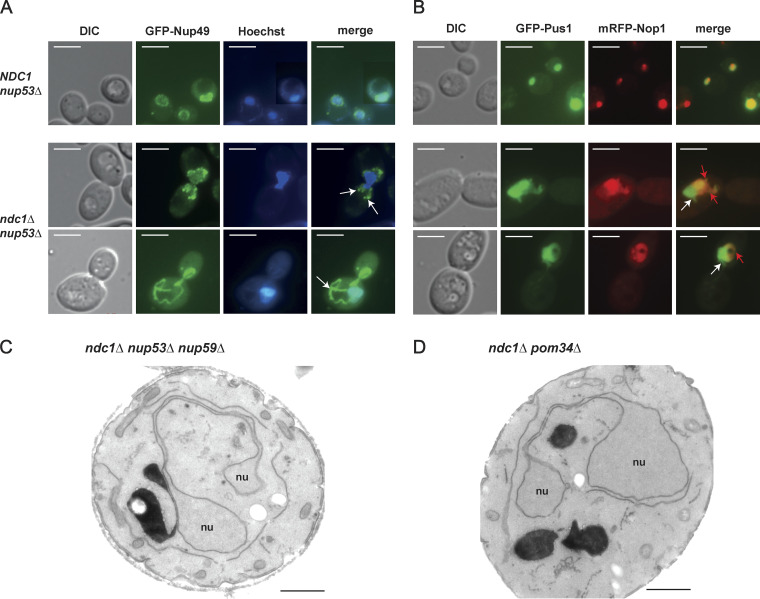
ndc1Δ nup53Δ cells grown in an asynchronous culture, showed enlarged nuclei in comparison with NDC1 nup53Δ cells (data not shown, but see Fig. 6 C). To further characterize the ndc1Δ nup53Δ strain, we analyzed the subcellular distribution of the nucleoporin marker GFP-Nup49 in interphase (Fig. 6 C) and mitotic cells (Fig. S5 A). Whereas WT and nup53Δ cells exhibited predominantly round-shaped nuclei, the GFP-Nup49 staining observed in ndc1Δ nup53Δ cells often showed nuclei with irregular shapes, frequently with an isthmus-like restriction or with extensive nuclear membrane protrusions, which could reach deeply into the cytoplasm and were devoid of chromatin (Fig. 6 C and Fig. S5 A). Electron microscopic analysis of ndc1Δ nup53Δ cells revealed these NE abnormalities and extensions in greater detail (Fig. 6 E [I–III]), which were also observed in ndc1Δ nup53Δ nup59Δ cells (Fig. S5 C) or ndc1Δ pom34Δ cells (Fig. S5 D). Interestingly, related NE abnormalities called “flares” were reported in earlier studies upon deletion of NEM1 or SPO7, which encode a phosphatase complex controlling phospholipid biosynthesis and nuclear membrane growth (Campbell et al., 2006; Santos-Rosa et al., 2005; Siniossoglou et al., 1998) but also upon deletions of genes important for spindle functions or chromosome segregation as they cause a cell cycle delay (Witkin et al., 2012). Here, the expanded NE forms large nuclear extensions adjacent to the nucleolus (Campbell et al., 2006; Walters et al., 2014; Witkin et al., 2012). In an asynchronously grown ndc1Δ nup53Δ cell culture expressing GFP-Pus1 as nucleoplasmic (Campbell et al., 2006; Hellmuth et al., 1998) and mRFP-Nop1 as nucleolar marker (Henriquez et al., 1990) mitotic but not interphase cells showed nuclear expansions not observable in the control strain NDC1 nup53Δ (Fig. 6 D and Fig. S5 B). This might be due to the involvement of Ndc1 in the late steps of SPB duplication, particularly in anchoring the duplicated SPB into the NE. Reportedly, deletion of NDC1 causes a monopolar spindle phenotype and subsequent mitotic arrest (Chial et al., 1999; Lau et al., 2004; Madrid et al., 2006; Winey et al., 1993).
Figure S5.
Deletion of NDC1 causes alterations in NE morphology. (A) Fluorescence microscopy of mitotic yeast cells, as indicated, expressing the GFP-tagged nucleoporin ScNup49 as NPC marker. White arrows mark GFP-ScNup49 as part of cytoplasmic NE/ER expansions. Hoechst 33258 serves as a chromatin stain. Bars, 5 µm. (B) Fluorescence microscopy of mitotic yeast cells, as indicated, coexpressing GFP-tagged ScPus1 as nucleoplasm marker protein and the mRFP-tagged nucleolar marker protein ScNop1. White arrows mark nuclear expansions not overlapping with nucleoli. Red arrows mark Nop1-positive nuclear flares. Hoechst 33258 serves as a chromatin stain. Bars, 5 µm. (C and D) Electron micrographs of ndc1Δ nup53Δ nup59Δ (C) and ndc1Δ pom34Δ (D) cells both showing abnormalities of the nuclear morphology. Bars, 1 µm.
Transformed or untransformed ndc1Δ nup53Δ, ndc1Δ nup53Δ nup59Δ, or ndc1Δ pom34Δ cells could be maintained by continual re-streaking on YPD plates or FOA media after forcing plasmid loss of pRS316-ScNDC1. This indicates that the strains are stable, and the survival is not due to a temporary stabilization of the Ndc1 protein in the absence of Nup53, Nup53 and Nup59, or Pom34. However, the cells show a shortened life span (data not shown). In addition, ndc1Δ nup53Δ cells show a decreased growth rate and often possess morphological abnormalities compared to the NDC1 nup53Δ strain such as increased cell size and large buds (Fig. S5, A and B; and Fig. 7 B).
Figure 7.
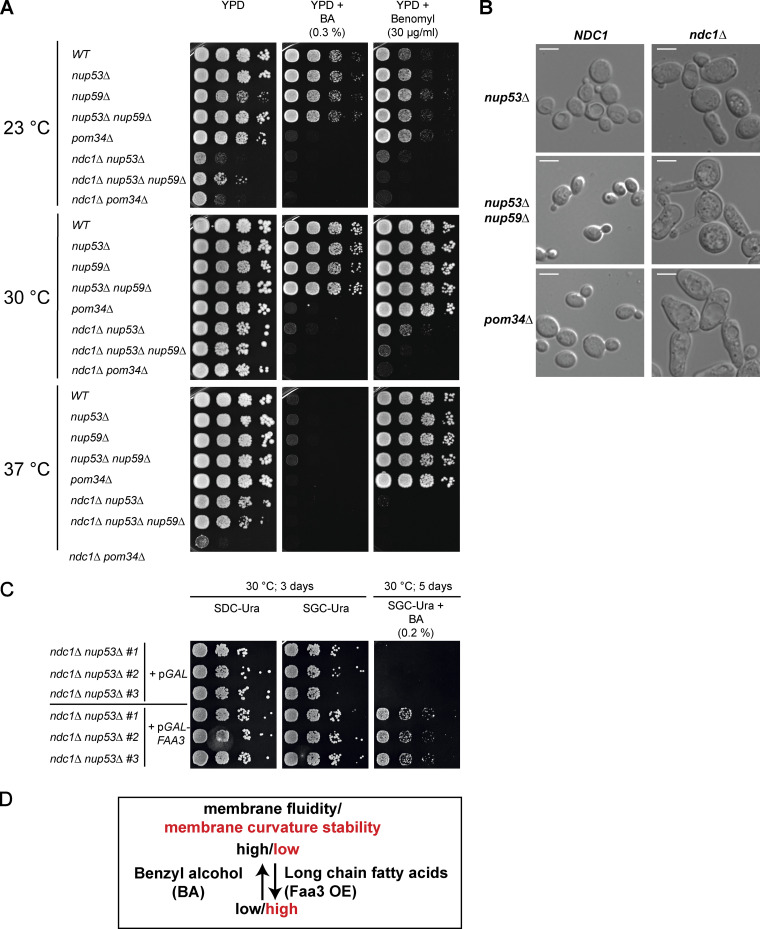
ndc1Δ nup53Δ, ndc1Δ nup53Δ nup59Δ, and ndc1Δ pom34Δ strains are sensitive to the membrane-fluidizing drug benzyl alcohol. (A) Growth tests of indicated yeast deletion strains. Growth was monitored on YPD plates using drugs either affecting membrane fluidity (benzyl alcohol, BA) or integrity of microtubules (Benomyl) at 23, 30, and 37°C. (B) Morphology analysis of yeast cells, as indicated, using differential interference contrast microscopy. Bars, 5 µm. (C) Growth tests of ndc1Δ nup53Δ cells expressing the long chain fatty acyl-CoA synthetase ScFaa3 from pYES2, an URA3-marker-containing 2 µ plasmid, under control of the GAL1 promoter. The ndc1Δ nup53Δ strain transformed with the empty pYES2 plasmid served as control. The growth tests were performed using galactose-containing agar plates (SGC-Ura) in the absence or presence of the membrane-fluidizing drug benzyl alcohol. Three different transformants of each strain were used. The plates were incubated for up to 5 d at 30°C. (D) The impact of BA and ScFaa3 expression on the membrane fluidity and topology are summarized indicating that BA increases membrane fluidity and destabilizes membrane curvature, an effect counteracted by high amounts of long-chain fatty acids induced by Faa3 overexpression.
Consistent with the observed changes in nuclear membrane morphology, the deletion strains ndc1Δ nup53Δ, ndc1Δ nup53Δ nup59Δ, and ndc1Δ pom34Δ were found to be sensitive to the membrane-fluidizing drug benzyl alcohol (BA; Fig. 7 A). This is probably due to lack of Ndc1 as ndc1-39 cells also exhibit BA sensitivity which is suggested to be caused by an hyperfluid NE (Friederichs et al., 2011; Lau et al., 2004). A recent study revealed that BA treatment of WT yeast cells causes partial Nup49-GFP mislocalization and an irregular-shaped nucleus (Erguden, 2022), similar to what we often observed for the ndc1Δ nup53Δ strain (Fig. 6). These results support the idea that the BA sensitivity of the NDC1-deleted cells is attributed to the involvement of Ndc1 in NE remodeling events during interphase needed for efficient biogenesis of NPC/SPB in the intact NE. The observed sensitivity to the microtubule-depolymerizing drug benomyl (Fig. 7 A) is in line with the proposed functions of Ndc1 in late SPB biogenesis steps (Chial et al., 1999; Lau et al., 2004; Winey et al., 1993). Surprisingly, the three NDC1-deleted strains were also cold sensitive which could hint toward an imbalanced response to changes in physical properties of the NE if Ndc1 is absent. Cold sensitivity was also previously observed for several mutant strains affected in the integral nuclear membrane Apq12-Brl1-Brr6 complex, which was shown to promote NPC biogenesis and additionally interact with Ndc1 (Hodge et al., 2010; Lone et al., 2015; Scarcelli et al., 2007; Zhang et al., 2018). The three strains deleted in NDC1 showed similar morphology. Cells were enlarged compared to the NDC1 cells and often showed large and elongated buds. These morphological abnormalities point toward an impact of the NDC1 deletion on mitotic progression (Fig. 7 B).
Finally, overexpression of Faa3, a long-chain fatty acyl-CoA synthetase, rescued the growth defect of ndc1Δ nup53Δ cells upon exposure to BA (Fig. 7 C). A similar effect of Faa3-induced recovery was observed for the BA-sensitive mps3Δ pom152Δ strain, which also showed abnormalities in NE morphology (Friederichs et al., 2011). Long fatty acids produced by Faa3 might stabilize the curved membrane state by turning the NE more rigid and by this, counteracting the increased fluidity caused by BA (Fig. 7 D).
Thus, the essential Ndc1 becomes dispensable for cell growth in yeast if the amphipathic helix of Nup53 is deleted, but such cells exhibit an abnormal NE morphology, which may be attributed to an altered lipid composition with a nuclear membrane biogenesis (see Discussion).
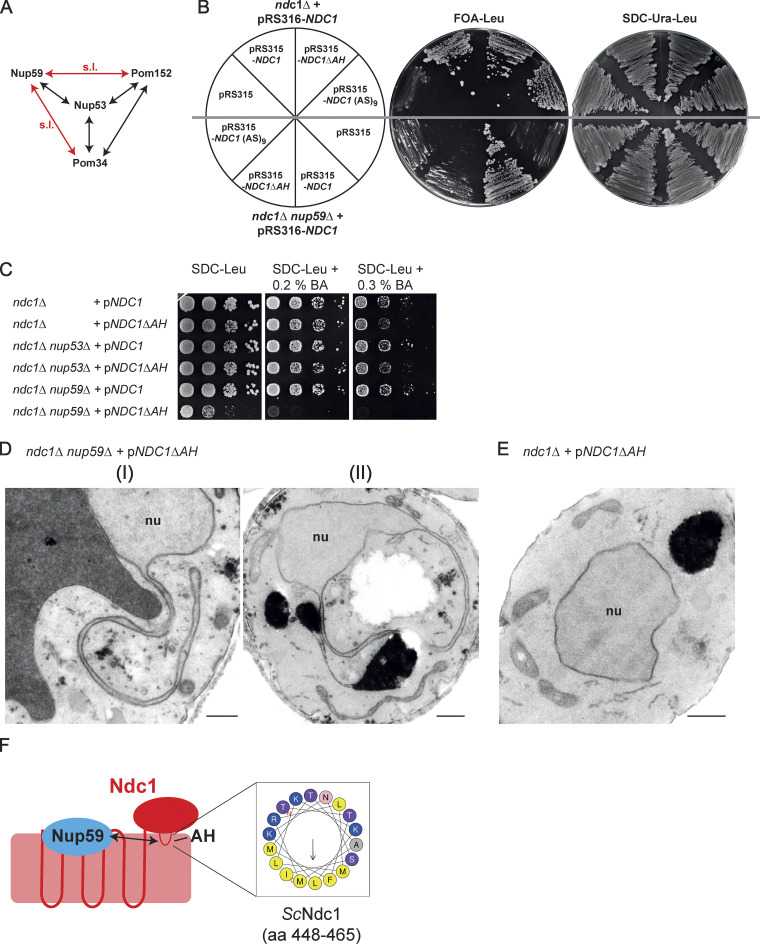
The amphipathic helix of Ndc1 becomes crucial in a nup59Δ strain
Next, we studied the function of the Ndc1-AH within the nucleoporin network, which is part of the NPC inner ring scaffold (Fig. 8 A). Since both Nup53 and Nup59 insert into the nuclear pore membrane via their C-terminal amphipathic motifs and both interact with Ndc1, an overlapping function of these highly homologous linker Nups has been suggested (Marelli et al., 2001; Onischenko et al., 2009; Patel and Rexach, 2008; Vollmer et al., 2012). On the other hand, Nup53 and Nup59 also fulfill unique roles, which among other things is indicated by the observation that the nup59Δ, but not nup53Δ deletion strain, is lethal when combined with pom34Δ or pom152Δ knock-out alleles (Marelli et al., 1998; Miao et al., 2006; Onischenko et al., 2009).
Figure 8.
The Ndc1-AH becomes crucial if Nup59 is lacking. (A) Illustration of known negative genetic interactions within the interaction network of nucleoporins of the inner ring and the linker nucleoporins Nup53 and Nup59. Black arrows mark negative genetic interactions upon deletion of corresponding genes. Red arrows mark negative genetic interactions resulting in synthetic lethality (s.l.) of corresponding double deletion strains. (B) Cells from ScNdc1 shuffle strains deleted in NUP59 and additionally expressing ScNdc1 variants encoded by LEU2 marker-containing pRS315 plasmids, as indicated, were spread onto FOA-Leu plates forcing plasmid loss of pRS316-ScNDC1. Growth on SDC-Ura-Leu served as control. (C) Growth of ndc1Δ, ndc1Δ nup53Δ, and ndc1Δ nup59Δ cells expressing WT ScNdc1 or ScNdc1ΔAH from pRS315-based plasmids was monitored on leucine-lacking plates containing different amounts of the membrane-fluidizing drug benzyl alcohol (BA). (D) Electron micrographs of ndc1Δ nup59Δ cells expressing ScNdc1ΔAH from a pRS315-based plasmid showing cytoplasmic nuclear membrane protrusions (I), which partially fuse back to the NE enclosing parts of the cytoplasm (II). Bars, 500 nm. (E) Electron micrograph of an ndc1Δ cell expressing ScNdc1ΔAH from a pRS315-based plasmid. Bar, 500 nm. (F) Graphical illustration of the observed interaction between the amphipathic motif of ScNdc1 and ScNup59.
Based on this genetic data, we expressed Ndc1ΔAH as well as a related construct, Ndc1 (AS)9, carrying instead of the AH a tandem alanine-serine linker sequence, in the nup59Δ ndc1Δ double disruption strain (Fig. 8 B). Strikingly, corresponding cells displayed a slow-growth phenotype compared to NUP59 cells expressing Ndc1 lacking the amphipathic motif (Fig. 8 B).
To further investigate these opposite effects exerted by Ndc1-AH in the single ndc1Δ versus ndc1Δ nup59Δ double disruption strains, we incubated cells with the membrane-fluidizing drug benzyl alcohol (BA) during a dot spot growth analysis (Fig. 8 C). This revealed that the already strong growth inhibition of the nup59Δ ndc1Δ cells expressing Ndc1ΔAH was converted into a complete growth inhibition by addition of 0.2% BA (Fig. 8 C). However, ndc1Δ or ndc1Δ nup53Δ cells complemented by Ndc1ΔAH did not show such a BA-induced growth inhibition (Fig. 8 C).
Ultrastructural analysis of cells expressing Ndc1ΔAH but lacking Nup59 revealed long nuclear membrane protrusions reaching deeply into the cytoplasm (Fig. 8 D), similar to what has been already observed in the case of ndc1Δ nup53Δ (Fig. 6 E [II]), ndc1Δ nup53Δ nup59Δ (Fig. S5 C), or ndc1Δ pom34Δ cells (Fig. S5 D). In contrast, these membrane abnormalities were not observed in ndc1Δ cells complemented by Ndc1ΔAH (Fig. 8 E).
Together, this data shows that the amphipathic motifs in Ndc1 and Nup59 exhibit a specific genetic interaction (Fig. 8 F) by which this linker Nup can perform a unique role at the NE not related to the Ndc1-AH function.
Discussion
In this study, we describe new functional motifs of the nuclear membrane protein Ndc1, which enlighten its complex physical and genetic interactions within the NE-associated nucleoporin network. The N-terminal part of Ndc1 mediates direct contact to the Y-shaped Nup84 complex, whereas a short amphipathic helix that functionally interacts with related motifs in the inner pore ring linker Nups Nup53 and Nup59 directly binds the pore membrane.
Conserved ALPS domains present in the β-propeller domains of both Nup120/Nup160 and Nup133 (Drin et al., 2007; Kim et al., 2014; Kosinski et al., 2016; von Appen et al., 2015) presumably contribute to the targeting the Nup84 complex to the curved nuclear pore membrane. Using in vitro binding assays, we found a direct interaction between the C-terminal α-helical domains of both CtNup120 and CtNup133 and the N-terminus of Ndc1, pointing to an additional mechanism of how the Nup84 complex is recruited to the pore membrane. Interestingly, in C. elegans embryos Nup160 as a Y-complex component is partially mislocalized in Ndc1-depleted cells (Mauro et al., 2022). It is thus conceivable that the Y-complex in addition to its ALPS motifs requires direct interactions with transmembrane nucleoporins such as Ndc1 for NPC localization. Notably, in vertebrates the Y-complex can bind the integral pore membrane protein POM121 (Mitchell et al., 2010). Here, it remains open whether vertebrate NDC1 could in addition to POM121 serve as a recruitment point for the Y-complex.
Membrane interaction motifs have been identified in several nucleoporins with crucial roles in NPC assembly and function (Hamed and Antonin, 2021). Here, we identify a membrane binding motif in a transmembrane nucleoporin. This motif is localized within the C-terminal half of Ndc1 and preferentially binds to highly curved membranes, and hence behaves like a classical ALPS (Vanni et al., 2013), despite possessing also charged lysine and arginine residues in its polar face (Fig. 4 B). This ALPS motif might be involved in targeting Ndc1 to the curved pore membrane and/or contribute to the curved shape of the pore membrane. In vertebrates, the N-terminus of Ndc1 is essential but not sufficient for NPC assembly (Eisenhardt et al., 2014). This hinted toward an essential function of the C-terminus of Ndc1 during NPC biogenesis, probably involving the membrane binding role described here. In the case of yeast Ndc1, which has a dual role in NPC and SPB insertion, it is assumed that it acts as a mobile and dynamic membrane protein, which can be recruited to assembly sites of both NPCs and SPBs.
Surprisingly, the essential yeast Ndc1 is dispensable for cell growth if Nup53 is absent (Fig. 6 A), and this suppression phenotype depends on the C-terminal amphipathic helix of Nup53 (Fig. 6 B). In in vitro NPC reconstitution assays the N-terminal transmembrane regions of vertebrates Ndc1 are only required if Nup53 has a functional C-terminal amphipathic helix (Eisenhardt et al., 2014) pointing to a conserved function of the Nup53-Ndc1 interaction. Ndc1 is not essential in Aspergillus nidulans, but this fungus also lacks obvious Nup53 orthologs (Liu et al., 2009; Osmani et al., 2006). The phylum Amoebozoa lacks both Ndc1 and Nup53 homologs or possesses Nup53 homologs with only poor sequence homology at the C-terminus (Neumann et al., 2010). This and our results are in line with the idea that the N-terminus of Ndc1 controls or modulates the membrane-bending activity of Nup53 (Eisenhardt et al., 2014).
Overexpression of Ndc1 as well as its N-terminal domain, which includes all six transmembrane regions, induced membrane proliferation in the cytoplasm with NE-like double membrane structures with openings reminiscent of pores. This phenotype is different from the karmellae, membrane stacks associated with the outer nuclear membrane but devoid of pores and observed upon overexpression of membrane proteins like Hmg1 (Wright et al., 1988). It is also different from the observed phenotypes upon depletion of Spo7, Nem1, or Smp2, which typically show irregularly shaped nuclei consisting of two or more interconnected lobes (Santos-Rosa et al., 2005; Siniossoglou et al., 1998). Indeed, in this “flare” phenotype, observed in spo7Δ cells but also upon mitotic delay or blocking of the secretory pathway, the nuclear growth area typically co-localizes with the nucleolus (Campbell et al., 2006; Walters et al., 2014; Witkin et al., 2012), which we do not observe. In contrast, the NE expansions observed in the surviving NDC1-deletion strains (ndc1Δ nup53Δ, ndc1Δ nup53Δ nup59Δ, ndc1Δ pom34Δ) have a typical “flare” appearance in mitosis. This, and the larger cell size, is consistent with a cell cycle delay expected as Ndc1 has also an important function in nascent SPB integration (Chial et al., 1999; Lau et al., 2004; Madrid et al., 2006; Winey et al., 1993).
The pore-like structures observed in the extranuclear membrane proliferations upon Ndc1 overexpression are smaller in diameter as compared to NPCs (roughly 45 versus 67 nm). As these structures do not show the typical NPC’s electron density, they likely lack most nucleoporins, consistent with the absence of a GFP-Nup49 signal on these membrane proliferations. This supports the view that nucleoporins and especially the membrane coat can widen the pore diameter as recent molecular dynamics simulations suggest (Mosalaganti et al., 2022). This is also in line with observations on the mitotic NPC assembly process in human HeLa cells where membrane holes in the reforming nuclear envelope expand upon the assembly of NPC structures (Otsuka et al., 2018).
The observed dramatic changes in the NE morphology observed for NDC1-deleted cells that on top lack Nup53, Pom34, or both Nup53 and Nup59 are distinct from membrane changes observed upon Ndc1 overexpression but have a typical “flare” appearance in mitosis. This, and the larger cell size, is consistent with a cell cycle delay expected as Ndc1 has also an important function in nascent SPB integration (Chial et al., 1999; Lau et al., 2004; Madrid et al., 2006; Winey et al., 1993). These NE alterations might however also hint toward alterations in NE membrane homeostasis tightly linked to NPC biogenesis. The Apq12-Brr6-Brl1 complex, which is conserved in organisms undergoing closed mitosis (Tamm et al., 2011), transiently interacts with early NPC intermediates (Lone et al., 2015; Zhang et al., 2021; Zhang et al., 2018). The mechanistic role of this complex is not clear but a direct or indirect role in promoting phosphatidic acid accumulation at the curved membranes was suggested (Zhang et al., 2021). This, in turn, might stabilize or even induce membrane curvature via the conical-shaped phosphatidic acid (Zhukovsky et al., 2019). As Ndc1 interacts with Brl1 (Zhang et al., 2018), it is conceivable that in the absence of Ndc1, the Apq12-Brr6-Brl1 complex is not properly recruited to NPC assembly sites.
The three viable NDC1-deletion strains (ndc1Δ nup53Δ, ndc1Δ nup53Δ nup59Δ, and ndc1Δ pom34Δ) are sensitive toward the membrane-fluidizing drug benzyl alcohol (Fig. 7 A) suggesting defects in NE lipid homeostasis. Overexpression of long-chain fatty acid synthetase Faa3 rescued the BA sensitivity of ndc1Δ nup53Δ cells, similar to that observed for the pom152Δ mps2Δ strain (Friederichs et al., 2011). Presumably, increasing de novo lipid synthesis of long-chain fatty acid-containing membrane lipids stabilizes the curvature of the pore membrane and by this counteracts the increased membrane fluidity of cells caused by disturbances of the lipid homeostasis system.
In contrast to NUP53, the deletion of NUP59 did not suppress the lethal ndc1Δ disruption. We initially speculated that the differences in the amino acid composition of their C-terminal amphipathic helices are responsible for this discrepancy. However, expressing the chimeric protein Nup59-AHNup53 consisting of Nup59 and the amphipathic motif of Nup53 still allowed the suppressive growth of ndc1Δ cells. Thus, both orthologs possess distinct functions, which is consistent with previous results (Marelli et al., 1998; Miao et al., 2006; Onischenko et al., 2009). This is further supported by our genetic analyses of the amphipathic helix in the C-terminus of Ndc1 and Nup59 (Fig. 8 B). Cells deleted in NUP59 expressing Ndc1ΔAH display a severe growth defect and sensitivity toward BA. Nup59 is involved in the proper localization of Ndc1 to the NE overlapping in its function with the Pom152-Pom34 subcomplex (Onischenko et al., 2009). Presumably, the Ndc1-AH is involved in the correct positioning of Ndc1 at the NPCs for subsequent integration into the nucleoporin network. Ndc1ΔAH might not be targeted properly to the NE if Nup59 is absent and therefore corresponding cells behave similarly to those lacking Ndc1.
In conclusion, our work shows that Ndc1 has different features functional in NPC assembly. The N-terminal part of the protein, which contains its six transmembrane regions, provides a so far unrecognized targeting mechanism of the Nup84 complex to the NE, possibly enhancing the efficiency of its ALPS domain-mediated targeting to the curved pore membrane. Future studies on this question may help to clarify whether this interaction is conserved in vertebrates, where POM121 binds to the Y-complex. The C-terminal part of Ndc1 contains a previously unknown conserved amphipathic motif, which if overexpressed induces massive membrane alteration and changes in lipid homeostasis. Interestingly, this amphipathic motif genetically interacts with an amphipathic motif of Nup53, a non-essential binding partner of Ndc1. These membrane-binding and -bending motifs, if properly positioned via the direct Ndc1-Nup53 interaction, could cooperate in the integration of NPC into the two membranes of the nuclear envelope and, if necessary, counteract their membrane-deforming activities. During NPC formation, different membrane intermediates have to be passed through, in which the lipid bilayer is exposed to different bending stresses: It progresses from a rather flat membrane to a dimple-like deformation to the saddle-like membrane structure of the pore (Otsuka and Ellenberg, 2018; Weberruss and Antonin, 2016). Where in this process the critical balancing interplay between these motifs occurs remains open. We speculate that in the absence of both Ndc1 and Nup53, biophysical adaptations in nuclear membrane properties allow for (probably less efficient) NPC and SPB integration into the NE, a mode that is more sensitive to changes in membrane fluidity (Fig. 7 A). How loss of Pom34 could suppress the NDC1 deletion phenotype remains open given that no membrane bending activity has to our knowledge so far assigned to Pom34. Similarly, the structural arrangements of NPCs formed without Ndc1 and Nup53 or Pom34 remain open and especially the question of whether other nucleoporins occupy the vacant space.
Materials and methods
Media, yeast strains, plasmids, and genetic methods
Media preparation and microbiological techniques were performed according to standard protocols (Ausubel et al., 1992; Guthrie and Fink, 1991). S. cerevisiae strains used in this study are listed in Table S1. Yeast deletion strains were created using a homologous recombination-based approach and verified by colony PCR (Gueldener et al., 2002; Janke et al., 2004). A WT yeast strain endogenously expressing DsRed-HDEL was created by integrating the linearized plasmid pKW1803 (Madrid et al., 2006) into the trp1-1 locus. Cloning and plasmid propagation were performed according to standard procedures using E. coli DH5α. Site-directed mutagenesis was performed using a PCR-based method (Pfirrmann et al., 2013). Plasmids used in this study are listed in Table S2. Yeast and E. coli expression plasmids for proteins from C. thermophilum were generated using gene amplification from a cDNA library (Amlacher et al., 2011). Yeast expression plasmids based on YEplac112 additionally possess a LEU2 marker under the control of the truncated leu2d promoter increasing plasmid copy number if corresponding cells grow on media lacking leucine (Erhart and Hollenberg, 1983). Proteins expressed from the YEplac112-based plasmids are N-terminally tagged with two IgG binding units of Protein A from Staphylococcus aureus (Rigaut et al., 1999) followed by a TEV protease recognition sequence (ProtA-TEV). All plasmids generated were verified by sequencing.
Protein expression and purification
Full-length soluble nucleoporins from C. thermophilum carrying an N-terminal ProtA-TEV tag were expressed in S. cerevisiae under the control of the GAL1-10 promoter and affinity-purified using IgG beads. Cells were grown overnight in a raffinose medium (SRC-Leu) to an OD600 of 2 before dilution with galactose medium (2 × YPG). After 6 h of protein expression, cells were harvested, resuspended in lysis buffer (20 mM HEPES, pH 7.6, 150 mM NaCl, 50 mM KAc, 2 mM Mg(Ac)2, 5% (w/v) glycerol, 0.1% (w/v) NP-40 and protease inhibitor mix [Sigma-Aldrich]), and lysed mechanically using glass beads. After centrifugation at 39,000 × g at 4°C for 20 min, the supernatant was incubated with 0.5 ml of pre-equilibrated IgG-Sepharose slurry (GE Healthcare) at 4°C for 1 h. After three washing steps, bound proteins were subjected to TEV protease cleavage for 1 h at 16°C (TEV protease expressed and purified in-house). Eluted proteins were collected and further used for in vitro binding assays. Cells expressing the membrane nucleoporins CtNdc1 and CtPom152 were expressed as described earlier but resuspended in a differently composed lysis buffer (20 mM Tris, pH 7.6, 150 mM NaCl, 50 mM KAc, 2 mM Mg(Ac)2, 5% (w/v) glycerol, 2% (w/v) Triton X-100, 2 mM DTT and protease inhibitor mix) before immobilization on IgG beads for subsequent in vitro binding assays.
CtNdc1, CtNdc1 truncations, and SCL1/BC08 (Lorenz et al., 2015) for reconstitution in GUVs were expressed in E. coli BL21 codon plus (DE3) cells (EMD Millipore) from a modified pET28a plasmid containing an N-terminal MISTIC (membrane-integrating sequence for translation of integral membrane protein constructs) fragment (13 kD) from Bacillus subtilis, allowing high yield expression of membrane proteins in E. coli (Roosild et al., 2005) followed by a thrombin protease cleavage site. Both proteins were additionally expressed as C-terminal 6 × His fusion proteins enabling Ni-based affinity purification (Alves et al., 2017). Purification was done in 20 mM TRIS, pH 7.4, 500 mM NaCl, 8 mM Imidazole, 1% (w/v) cetyltrimethylammonium bromide (CTAB) on Ni-NTA magnetic beads (EMD). After washing proteins were eluted with 20 mM TRIS, pH 7.4, 500 mM NaCl, 400 mM Imidazole, 1% (w/v) CTAB followed by dialysis against PBS containing 1% (w/v) CTAB and 1 mM EDTA. MISTIC was finally cleaved off using thrombin protease (GE Healthcare).
Truncated versions of CtNup120 and CtNup133 used for in vitro binding assays were expressed and affinity-purified from E. coli BL21 codon plus (DE3) cells. Corresponding proteins were expressed from modified pPROEX-1 (Invitrogen)-based plasmids as N-terminal 6 × His-TEV fusion proteins. Cells were grown to an OD600 of 0.5 before IPTG-mediated induction (1 mM) of expression at 30°C for 3 h. Cells were harvested and resuspended in lysis buffer (20 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM DTT, and protease inhibitor mix). Lysis was performed using a high-pressure cavitation homogenizer (Microfluidizer 110 L, Microfluidics) followed by centrifugation at 39,000 × g at 4°C for 20 min. Precleared lysates were incubated with Ni-NTA beads (Macherey-Nagel) for 1 h at 4°C. After extensive washing in buffer containing 10 mM imidazole pH 8.0 bound proteins were eluted in buffer containing 200 mM imidazole.
Fluorescence microscopy of yeast cells
All Fluorescence microscopy was done using yeast cells grown overnight in selective media (plus 2 × adenine) to mid-exponential growth phase. 1 ml of the cell suspension was harvested and resuspended in 800 µl buffered glucose (100 mM HEPES, pH 7.7, 2% [w/v] glucose). The chromatin stain Hoechst 33258 (Sigma-Aldrich) was added (10 µg/ml) and the cells were afterward incubated in the dark for 3 min. Cells were fixed with formaldehyde (3% [w/v]) for 5 min, washed two times, and resuspended in 25 µl buffered glucose. Fluorescence microscopy was performed at room temperature using a fluorescence microscope (Imager Z1; Carl Zeiss) equipped with a 100 ×, NA 1.4 Plan-Apochromat oil immersion objective lens (Carl Zeiss). Imaging was done using channels for GFP, DsRed, mRFP, and Hoechst 33258 with different exposure times dependent on the fluorescence intensities of the used fluorophores. Pictures were acquired with a CCD camera (AxioCamM Rm; Carl Zeiss) and the AxioVision SE64 Rel. 4.9.1 imaging software (Carl Zeiss).
Transmission electron microscopy
Yeast cells were grown overnight in liquid media to mid-exponential growth phase and resuspended in fixation buffer (0.2 M PIPES, pH 6.8, 0.2 M sorbitol, 2 mM MgCl2, 2 mM CaCl2, 2% [w/v] formaldehyde and 2% [w/v] glutaraldehyde) for 10 min at RT and 50 min at 4°C. After washing in 0.1 M phosphate-citrate buffer pH 5.8, the cells were resuspended in the same buffer containing 0.25 mg/ml Zymolyase 20T (Seikagaku Corp.) and incubated for 2 h at 30°C. The cells were washed three times in 0.1 M sodium acetate buffer, pH 6.1, and post-fixed with 2% (w/v) osmium tetroxide/1.5% (w/v) K4[Fe(CN)6] in 0.1 M sodium phosphate buffer, pH 7.4. After contrasting en bloc with 0.5% (w/v) uranyl acetate overnight and dehydrating using a graded dilution series of ethanol, the cells were embedded into glycid ether 100-based resin. Ultrathin sections were prepared with a Reichert ultracut S ultramicrotome (Leica), collected on grids, and contrasted with both uranyl acetate and lead citrate. Specimens were visualized using an electron microscope (EM 10 CR; Carl Zeiss) equipped with a 1 K CCD camera (Tröndle Restlichtverstärkersysteme, Moorenweis) at an acceleration voltage of 60 KV. Imaging was done by the software ImageSP (SYSPROG).
Lipid analysis
Yeast cells were grown overnight in corresponding media, harvested in a mid-exponential growth phase, and resuspended in 50 mM HEPES, pH 7.4, containing 0.8 mg/ml Zymolyase 100T (Carl Roth). Cells were mechanically lysed using glass beads. Lipidomics analyses were performed by a shotgun approach (Papagiannidis et al., 2021). Lipid extracts were prepared in presence of internal lipid standards via acidic Bligh-Dyer lipid extraction (Bligh and Dyer, 1959). The used master mix of lipid internal standards contained 50 pmol d7-PC mix (15:0/18:1-d7, Avanti Polar Lipids), 25 pmol PI (17:0/20:4, Avanti Polar Lipids), 50 pmol PE and 10 pmol PS (14:1/14:1, 20:1/20:1, 22:1/22:1, semi-synthesized [Özbalci et al., 2013]), 15 pmol PA (PA 17:0/20:4, Avanti Polar Lipids), 10 pmol PG (14:1/14:1, 20:1/20:1, 22:1/22:1), semi-synthesized [Özbalci et al., 2013]), 40 pmol DAG (17:0/17:0, Larodan), 40 pmol TAG (D7-TAG-Mix, LM-6000/D5-TAG 17:0,17:1,17:1, Avanti Polar Lipids), 10 pmol t-Cer (18:0, Avanti Polar Lipids) and 50 pmol ergosteryl ester (15:0 and 19:0).
The lipids in the organic phase were dried under a gentle stream of nitrogen at 37°C. Lipids were resuspended in 10 mM methanolic ammonium acetate and transferred to 96-well plates (Eppendorf Twintec 96). Mass spectrometry was performed on a Sciex QTRAP 6500+ mass spectrometer, equipped with chip-based (HD-D ESI Chip; Advion Biosciences) nano-electrospray infusion, and ionization (TriVersa NanoMate; Advion Biosciences; Özbalci et al., 2013). The following precursor ion (PREC) or neutral loss (NL) scanning modes were used: +PREC184 (PC), +PREC282 (t-Cer), +NL141 (PE), +NL185 (PS), +NL277 (PI), +NL189 (PG), +NL115 (PA), +NL77 (ergosterol), +PREC379 (ergosteryl ester). Mass spectrometric analysis of ergosterol was performed by applying one-step chemical derivatization to ergosterol acetate in the presence of 300 pmol (first and second replicate) or 100 pmol (third and fourth replicate) of the internal standard (22E)-Stigmasta-5,7,22-trien-3-beta-ol (R202967; Sigma-Aldrich) using 100 μl acetic anhydride/chloroform (1:12 v/v) overnight under argon atmosphere according to (Ejsing et al., 2009). Data evaluation was done using LipidView (Sciex) and ShinyLipids (in-house developed software).
Reconstitution of proteins into GUVs
Before reconstitution of the detergent-solubilized membrane proteins CtNdc1 and SCL1/BC08 into GUVs, the purified proteins were labeled using a succinimidyl ester of Alexa Fluor 488 dissolved in 200 mM NaHCO3, pH 8.4, containing 1% (w/v) CTAB. Detergent removal and subsequent formation of proteoliposomes were performed via gel filtration after incubation of the detergent-solubilized and labeled proteins with a lipid mixture mimicking the composition of the NE (Eisenhardt et al., 2014; Lorenz et al., 2015). For this, the protein-lipid mixture was loaded on a Sephadex G50 fine-filled Econo chromatography column equilibrated in sucrose buffer (10 mM HEPES, pH 7.5, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2), pelleted and resuspended in 20 mM HEPES, pH 7.4, 100 mM KCl and 1 mM DTT. GUVs containing either CtNdc1 or SCL1/BC08 were reconstituted via electroformation (Lorenz et al., 2015). For this, 5 μl of resuspended proteoliposomes were dried onto two 5 × 5 mm platinum gauzes (ALS) under vacuum for at least 1 h at room temperature. The gauzes were placed in parallel (5 mm distance) into a cuvette (UVette, Eppendorf) and submerged in 259 mM sucrose solution, and for 140 min an alternating current electric field with 10 Hz, 2.2 V was applied, followed by 20 min at 2 Hz at 42°C.
Successful reconstitution of CtNdc1 and SCL1/BC08 into GUVs was immediately monitored in 8-well glass chambers (Chambered #1.0 Borosilicate Coverglass System, LabTek) at room temperature on an inverted Olympus Fluoview 1,000 confocal laser scanning system with the FV10-ASW software using an UPlanSApo 60×/1.35 oil objective by light emission between 500 and 545 nm after argon laser-mediated excitation at 488 nm. Recruitment of Alexa Fluor 546-labeled nucleoporins to either CtNdc1-GUVs or SCL1/BC08-GUVs was monitored by the collection of emitted light between 570 and 625 nm after excitation at 546 nm. The pinhole was set to one airy unit. For Fig. S1 G, an inverted Zeiss LSM710 laser scanning confocal microscope, a Plan-Apochromatic 63×/1.4 NA Oil immersion objective lens, 488 and 561 nm excitation laser, and Zen black 2.3 SP1 software was used.
Liposome flotation assays
Liposome generation and flotation were performed as described in Vollmer et al. (2015). In short, E. coli polar lipids (Avanti Polar Lipids) dissolved in chloroform and supplemented with 0.2 mol % octadecyl rhodamin B chloride (R18; Thermo Fisher Scientific) were vacuum dried on a rotary evaporator, dissolved as liposomes in PBS by freeze/thawing cycles and extruded by passages through Nuclepore track-etched membranes (Whatman) with defined pore sizes using an Avanti Mini-Extruder to generate small unilamellar liposomes of defined sizes. For liposome flotations, proteins purified from either pGEX-4T3- or pET28a-based plasmids (6 μM) were mixed 1:1 with liposomes (5 mg/ml) and floated for 2 h at 55,000 rpm in a TLS-55 rotor (Beckman) at 25°C through a sucrose gradient. Binding efficiency was determined by Western blot analysis using an ImageQuant LAS-4000 system (Fuji) and the AIDA software, comparing band intensities of start materials with floated liposome fractions.
Miscellaneous
SDS-PAGE and Western blot analysis were performed using standard protocols (Laemmli, 1970; Towbin et al., 1979). PageRuler Unstained Protein ladder (Thermo Fisher Scientific), Unstained Protein Standard, Broad Range (10–200 kD; NEB), peqGOLD Protein-Marker I Unstained (VWR), and PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) were used as protein markers. Immunodetection of ProtA-tagged proteins was performed using Peroxidase Anti-Peroxidase Soluble Complex antibody (Sigma-Aldrich) in a 1:3,000 dilution. Brilliant Blue G-Colloidal Concentrate Electrophoresis Reagent (Sigma-Aldrich) served as Coomassie stain.
Online supplemental material
Fig. S1 shows supporting evidence for the Nup133/Nup120 Ndc1 interaction and controls. Fig. S2 shows that overexpression of the Ndc1 C-terminus impairs cell growth. Fig. S3 shows that Ndc1 C-terminus contains an amphipathic membrane binding motif. Fig. S4 shows that Nup53 or Nup59 overexpression-induced growth defects depend on their amphipathic helices. Fig. S5 shows that NDC1 deletion causes a mitotic delay of corresponding cells with “flare-like” NE expansions and alterations of nuclear shape. Table S1 lists yeast strains used in this study. Table S2 lists the plasmids used.
Supplementary Material
lists yeast strains used in this study.
lists plasmids used in this study.
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Acknowledgments
We are grateful to Hilmar Bading, Department of Neurobiology and Interdisciplinary Center for Neurosciences, University of Heidelberg, for providing the opportunity to carry out the electron microscopy work in his laboratory. We acknowledge the Confocal Microscopy Facility, core facilities of the Interdisciplinary Center for Clinical Research (IZKF), Aachen, within the Faculty of Medicine at RWTH Aachen University, and are thankful to Sabrina Ernst for her help.
E. Hurt and W. Antonin are recipients of grants from the Deutsche Forschungsgemeinschaft (Hu363/13-1, Hu363/13-2, Hu363/13-3; AN377/7-1). B. Brügger gratefully acknowledges the data storage service SDS@hd supported by the Ministry of Science.
Author contributions: I. Amm, W. Antonin, and E. Hurt designed the study and analyzed the data. All experiments were performed by I. Amm. and M. Kallas except TEM, which was carried out by A. Hellwig, the liposome flotation assays carried out by M. Tatarek-Nossol, the GUV binding assays carried out by M. Weberruss and M. Tatarek-Nossol, the in vitro Ndc1 binding assays carried out by J. Schwarz, and the lipidomics carried out by C. Lüchtenborg and B. Brügger. The manuscript was written by I. Amm, W. Antonin, and E. Hurt. All authors discussed the results and commented on the manuscript.
Data availability
Western blot data underlying all figures and supplementary figures are available in the source data file. Lipidomic data associated with this study are available from the Dryad digital repository: DOI: 10.5061/dryad.s7h44j1bw.
References
- Akey, C.W., Singh D., Ouch C., Echeverria I., Nudelman I., Varberg J.M., Yu Z., Fang F., Shi Y., Wang J., et al. 2022. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell. 185:361–378.e25. 10.1016/j.cell.2021.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti, M., Zimmerli C.E., Rantos V., Wilfling F., Ronchi P., Fung H.K.H., Lee C.W., Hagen W., Turoňová B., Karius K., et al. 2020. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature. 586:796–800. 10.1038/s41586-020-2670-5 [DOI] [PubMed] [Google Scholar]
- Alves, N.S., Astrinidis S.A., Eisenhardt N., Sieverding C., Redolfi J., Lorenz M., Weberruss M., Moreno-Andrés D., and Antonin W.. 2017. MISTIC-fusion proteins as antigens for high quality membrane protein antibodies. Sci. Rep. 7:41519. 10.1038/srep41519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlacher, S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., and Hurt E.. 2011. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 146:277–289. 10.1016/j.cell.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Antonin, W., Ellenberg J., and Dultz E.. 2008. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett. 582:2004–2016. 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Araki, Y., Lau C.K., Maekawa H., Jaspersen S.L., Giddings T.H. Jr, Schiebel E., and Winey M.. 2006. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol. Biol. Cell. 17:1959–1970. 10.1091/mbc.e05-07-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Kingston R.M., Seidman F.G., Struhl K., Moore D.D., Brent R., and Smith F.A.. 1992. Current Protocols in Molecular Biology. Greene Publishing and Wiley Interscience, New York [Google Scholar]
- Bernales, S., McDonald K.L., and Walter P.. 2006. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4:e423. 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Campbell, J.L., Lorenz A., Witkin K.L., Hays T., Loidl J., and Cohen-Fix O.. 2006. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol. Biol. Cell. 17:1768–1778. 10.1091/mbc.e05-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Smoyer C.J., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2014. The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J. Cell Biol. 204:523–539. 10.1083/jcb.201307043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial, H.J., Giddings T.H. Jr, Siewert E.A., Hoyt M.A., and Winey M.. 1999. Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl. Acad. Sci. USA. 96:10200–10205. 10.1073/pnas.96.18.10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial, H.J., Rout M.P., Giddings T.H. Jr, and Winey M.. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800. 10.1083/jcb.143.7.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, N.B., Murphy D.D., Grider T., Rueter S., Brasaemle D., and Nussbaum R.L.. 2002. Lipid droplet binding and oligomerization properties of the Parkinson’s disease protein alpha-synuclein. J. Biol. Chem. 277:6344–6352. 10.1074/jbc.M108414200 [DOI] [PubMed] [Google Scholar]
- D’Angelo, M.A., Anderson D.J., Richard E., and Hetzer M.W.. 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 312:440–443. 10.1126/science.1124196 [DOI] [PubMed] [Google Scholar]
- Doucet, C.M., Esmery N., de Saint-Jean M., and Antonny B.. 2015. Membrane curvature sensing by amphipathic helices is modulated by the surrounding protein backbone. PLoS One. 10:e0137965. 10.1371/journal.pone.0137965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet, C.M., and Hetzer M.W.. 2010. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 119:469–477. 10.1007/s00412-010-0289-2 [DOI] [PubMed] [Google Scholar]
- Doucet, C.M., Talamas J.A., and Hetzer M.W.. 2010. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 141:1030–1041. 10.1016/j.cell.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin, G., and Antonny B.. 2010. Amphipathic helices and membrane curvature. FEBS Lett. 584:1840–1847. 10.1016/j.febslet.2009.10.022 [DOI] [PubMed] [Google Scholar]
- Drin, G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., and Antonny B.. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146. 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Dultz, E., and Ellenberg J.. 2010. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 191:15–22. 10.1083/jcb.201007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt, N., Redolfi J., and Antonin W.. 2014. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci. 127:908–921. 10.1242/jcs.141739 [DOI] [PubMed] [Google Scholar]
- Ejsing, C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., and Shevchenko A.. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106:2136–2141. 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguden, B. 2022. Benzyl alcohol increases diffusion limit of nuclear membrane in Saccharomyces cerevisiae cells. Eur. J. Biol. 81:26–30. 10.26650/EurJBiol.2022.1058174 [DOI] [Google Scholar]
- Erhart, E., and Hollenberg C.P.. 1983. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J. Bacteriol. 156:625–635. 10.1128/jb.156.2.625-635.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, S., Haque A., Imberdis T., Baru V., Barrasa M.I., Nuber S., Termine D., Ramalingam N., Ho G.P.H., Noble T., et al. 2019. Lipidomic analysis of α-synuclein neurotoxicity identifies stearoyl CoA desaturase as a target for Parkinson treatment. Mol. Cell. 73:1001–1014.e8. 10.1016/j.molcel.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, S., Richter R.P., and Görlich D.. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 314:815–817. 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- Friederichs, J.M., Ghosh S., Smoyer C.J., McCroskey S., Miller B.D., Weaver K.J., Delventhal K.M., Unruh J., Slaughter B.D., and Jaspersen S.L.. 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7:e1002365. 10.1371/journal.pgen.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24:2101–2102. 10.1093/bioinformatics/btn392 [DOI] [PubMed] [Google Scholar]
- Girard, V., Jollivet F., Knittelfelder O., Celle M., Arsac J.N., Chatelain G., Van den Brink D.M., Baron T., Shevchenko A., Kühnlein R.P., et al. 2021. Abnormal accumulation of lipid droplets in neurons induces the conversion of alpha-Synuclein to proteolytic resistant forms in a Drosophila model of Parkinson’s disease. PLoS Genet. 17:e1009921. 10.1371/journal.pgen.1009921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener, U., Heinisch J., Koehler G.J., Voss D., and Hegemann J.H.. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink G.R.. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1–863. [PubMed] [Google Scholar]
- Hamed, M., and Antonin W.. 2021. Dunking into the lipid bilayer: How direct membrane binding of nucleoporins can contribute to nuclear pore complex structure and assembly. Cells. 10:3601. 10.3390/cells10123601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz, B., Andres-Pons A., Kastritis P., and Beck M.. 2019. Structure and assembly of the nuclear pore complex. Annu. Rev. Biophys. 48:515–536. 10.1146/annurev-biophys-052118-115308 [DOI] [PubMed] [Google Scholar]
- Hellmuth, K., Lau D.M., Bischoff F.R., Künzler M., Hurt E., and Simos G.. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374–6386. 10.1128/MCB.18.11.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henríquez, R., Blobel G., and Aris J.P.. 1990. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J. Biol. Chem. 265:2209–2215. 10.1016/S0021-9258(19)39963-6 [DOI] [PubMed] [Google Scholar]
- Hodge, C.A., Choudhary V., Wolyniak M.J., Scarcelli J.J., Schneiter R., and Cole C.N.. 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123:141–151. 10.1242/jcs.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt, E., and Beck M.. 2015. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr. Opin. Cell Biol. 34:31–38. 10.1016/j.ceb.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Janke, C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jaspersen, S.L., and Ghosh S.. 2012. Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus. 3:226–236. 10.4161/nucl.20148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., Roberts B.L., Richardson W.D., and Smith A.E.. 1984. A short amino acid sequence able to specify nuclear location. Cell. 39:499–509. 10.1016/0092-8674(84)90457-4 [DOI] [PubMed] [Google Scholar]
- Kelley, K., Knockenhauer K.E., Kabachinski G., and Schwartz T.U.. 2015. Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 22:425–431. 10.1038/nsmb.2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J., Fernandez-Martinez J., Nudelman I., Shi Y., Zhang W., Raveh B., Herricks T., Slaughter B.D., Hogan J.A., Upla P., et al. 2018. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 555:475–482. 10.1038/nature26003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J., Fernandez-Martinez J., Sampathkumar P., Martel A., Matsui T., Tsuruta H., Weiss T.M., Shi Y., Markina-Inarrairaegui A., Bonanno J.B., et al. 2014. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol. Cell. Proteomics. 13:2911–2926. 10.1074/mcp.M114.040915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski, J., Mosalaganti S., von Appen A., Teimer R., DiGuilio A.L., Wan W., Bui K.H., Hagen W.J., Briggs J.A., Glavy J.S., et al. 2016. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 352:363–365. 10.1126/science.aaf0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, M.M., McMahon H.T., and Chernomordik L.V.. 2010. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 35:699–706. 10.1016/j.tibs.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson B., von Heijne G., and Sonnhammer E.L.. 2001. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kutay, U., Jühlen R., and Antonin W.. 2021. Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol. 31:1019–1033. 10.1016/j.tcb.2021.06.011 [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Larkin, M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lau, C.K., Delmar V.A., and Forbes D.J.. 2006. Topology of yeast Ndc1p: Predictions for the human NDC1/NET3 homologue. Anat. Rec. A. Discov. Mol. Cell. Evol. Biol. 288:681–694. 10.1002/ar.a.20335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.K., Giddings T.H. Jr, and Winey M.. 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell. 3:447–458. 10.1128/EC.3.2.447-458.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, O., Heath C.V., Amberg D.C., Dockendorff T.C., Copeland C.S., Snyder M., and Cole C.N.. 1995. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell. 6:401–417. 10.1091/mbc.6.4.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.H., and Hoelz A.. 2019. The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88:725–783. 10.1146/annurev-biochem-062917-011901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.H., Stuwe T., Schilbach S., Rundlet E.J., Perriches T., Mobbs G., Fan Y., Thierbach K., Huber F.M., Collins L.N., et al. 2016. Architecture of the symmetric core of the nuclear pore. Science. 352:aaf1015. 10.1126/science.aaf1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.L., De Souza C.P., Osmani A.H., and Osmani S.A.. 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell. 20:616–630. 10.1091/mbc.e08-06-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone, M.A., Atkinson A.E., Hodge C.A., Cottier S., Martínez-Montañés F., Maithel S., Mène-Saffrané L., Cole C.N., and Schneiter R.. 2015. Yeast integral membrane proteins Apq12, Brl1, and Brr6 form a complex important for regulation of membrane homeostasis and nuclear pore complex biogenesis. Eukaryot. Cell. 14:1217–1227. 10.1128/EC.00101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M., Vollmer B., Unsay J.D., Klupp B.G., García-Sáez A.J., Mettenleiter T.C., and Antonin W.. 2015. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J. Biol. Chem. 290:6962–6974. 10.1074/jbc.M114.627521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, C.P., Makhnevych T., Marelli M., Aitchison J.D., and Wozniak R.W.. 2002. Karyopherins in nuclear pore biogenesis: A role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267–278. 10.1083/jcb.200203079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann, M., Kunze R., Buerer A., Aebi U., and Hurt E.. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21:387–397. 10.1093/emboj/21.3.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, A.S., Mancuso J., Cande W.Z., and Weis K.. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371. 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans, B.J., Anantharaman V., Aravind L., and Koonin E.V.. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3:1612–1637. 10.4161/cc.3.12.1316 [DOI] [PubMed] [Google Scholar]
- Mansfeld, J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103. 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Marelli, M., Aitchison J.D., and Wozniak R.W.. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. 10.1083/jcb.143.7.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., Lusk C.P., Chan H., Aitchison J.D., and Wozniak R.W.. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. 10.1083/jcb.153.4.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, S., and McMahon H.T.. 2008. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9:543–556. 10.1038/nrm2417 [DOI] [PubMed] [Google Scholar]
- Mauro, M.S., Celma G., Zimyanin V., Magaj M.M., Gibson K.H., Redemann S., and Bahmanyar S.. 2022. Ndc1 drives nuclear pore complex assembly independent of membrane biogenesis to promote nuclear formation and growth. Elife. 11:e75513. 10.7554/eLife.75513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema, A.C., Poolman B., and Veenhoff L.M.. 2013. Quantitative analysis of membrane protein transport across the nuclear pore complex. Traffic. 14:487–501. 10.1111/tra.12048 [DOI] [PubMed] [Google Scholar]
- Miao, M., Ryan K.J., and Wente S.R.. 2006. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 172:1441–1457. 10.1534/genetics.105.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.M., Mansfeld J., Capitanio J., Kutay U., and Wozniak R.W.. 2010. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J. Cell Biol. 191:505–521. 10.1083/jcb.201007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, D., Frey S., Fischer T., Güttler T., and Görlich D.. 2009. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28:2541–2553. 10.1038/emboj.2009.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti, S., Obarska-Kosinska A., Siggel M., Taniguchi R., Turoňová B., Zimmerli C.E., Buczak K., Schmidt F.H., Margiotta E., Mackmull M.T., et al. 2022. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science. 376:eabm9506. 10.1126/science.abm9506 [DOI] [PubMed] [Google Scholar]
- Neumann, N., Lundin D., and Poole A.M.. 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 5:e13241. 10.1371/journal.pone.0013241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen, S.A., Turman D.L., and Schwartz T.U.. 2020. Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat. Commun. 11:6060. 10.1038/s41467-020-19885-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko, E., Stanton L.H., Madrid A.S., Kieselbach T., and Weis K.. 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185:475–491. 10.1083/jcb.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, A.H., Davies J., Liu H.L., Nile A., and Osmani S.A.. 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell. 17:4946–4961. 10.1091/mbc.e06-07-0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., and Ellenberg J.. 2018. Mechanisms of nuclear pore complex assembly—two different ways of building one molecular machine. FEBS Lett. 592:475–488. 10.1002/1873-3468.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., Steyer A.M., Schorb M., Hériché J.K., Hossain M.J., Sethi S., Kueblbeck M., Schwab Y., Beck M., and Ellenberg J.. 2018. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol. 25:21–28. 10.1038/s41594-017-0001-9 [DOI] [PubMed] [Google Scholar]
- Outeiro, T.F., and Lindquist S.. 2003. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 302:1772–1775. 10.1126/science.1090439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özbalci, C., Sachsenheimer T., and Brügger B.. 2013. Quantitative analysis of cellular lipids by nano-electrospray ionization mass spectrometry. Methods Mol. Biol. 1033:3–20. 10.1007/978-1-62703-487-6_1 [DOI] [PubMed] [Google Scholar]
- Paci, G., Caria J., and Lemke E.A.. 2021. Cargo transport through the nuclear pore complex at a glance. J. Cell Sci. 134:jcs247874. 10.1242/jcs.247874 [DOI] [PubMed] [Google Scholar]
- Papagiannidis, D., Bircham P.W., Lüchtenborg C., Pajonk O., Ruffini G., Brügger B., and Schuck S.. 2021. Ice2 promotes ER membrane biogenesis in yeast by inhibiting the conserved lipin phosphatase complex. EMBO J. 40:e107958. 10.15252/embj.2021107958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S.S., and Rexach M.F.. 2008. Discovering novel interactions at the nuclear pore complex using bead halo: A rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics. 7:121–131. 10.1074/mcp.M700407-MCP200 [DOI] [PubMed] [Google Scholar]
- Peeters, B.W.A., Piët A.C.A., and Fornerod M.. 2022. Generating membrane curvature at the nuclear pore: A lipid point of view. Cells. 11:469. 10.3390/cells11030469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton, L.F., Rout M.P., and Blobel G.. 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 92:1187–1191. 10.1073/pnas.92.4.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic, S., Samanta D., Perriches T., Bley C.J., Thierbach K., Brown B., Nie S., Mobbs G.W., Stevens T.A., Liu X., et al. 2022. Architecture of the linker-scaffold in the nuclear pore. Science. 376:eabm9798. 10.1126/science.abm9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfirrmann, T., Lokapally A., Andréasson C., Ljungdahl P., and Hollemann T.. 2013. SOMA: A single oligonucleotide mutagenesis and cloning approach. PLoS One. 8:e64870. 10.1371/journal.pone.0064870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popken, P., Ghavami A., Onck P.R., Poolman B., and Veenhoff L.M.. 2015. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell. 26:1386–1394. 10.1091/mbc.E14-07-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranke, I.M., Morello V., Bigay J., Gibson K., Verbavatz J.M., Antonny B., and Jackson C.L.. 2011. α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J. Cell Biol. 194:89–103. 10.1083/jcb.201011118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B.. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Roosild, T.P., Greenwald J., Vega M., Castronovo S., Riek R., and Choe S.. 2005. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 307:1317–1321. 10.1126/science.1106392 [DOI] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., and Chait B.T.. 2000. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941. 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli, J.J., Hodge C.A., and Cole C.N.. 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 178:799–812. 10.1083/jcb.200702120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck, S., Gallagher C.M., and Walter P.. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127:4078–4088. 10.1242/jcs.154716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, T.U. 2016. The structure inventory of the nuclear pore complex. J. Mol. Biol. 428:1986–2000. 10.1016/j.jmb.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464. 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., and Hurt E.C.. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84:265–275. 10.1016/S0092-8674(00)80981-2 [DOI] [PubMed] [Google Scholar]
- Stavru, F., Hülsmann B.B., Spang A., Hartmann E., Cordes V.C., and Görlich D.. 2006. NDC1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519. 10.1083/jcb.200601001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe, T., Correia A.R., Lin D.H., Paduch M., Lu V.T., Kossiakoff A.A., and Hoelz A.. 2015. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 347:1148–1152. 10.1126/science.aaa4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm, T., Grallert A., Grossman E.P., Alvarez-Tabares I., Stevens F.E., and Hagan I.M.. 2011. Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle. J. Cell Biol. 195:467–484. 10.1083/jcb.201106076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, L.J., and Wente S.R.. 2009. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell. 8:1814–1827. 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierbach, K., von Appen A., Thoms M., Beck M., Flemming D., and Hurt E.. 2013. Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure. 21:1672–1682. 10.1016/j.str.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin T., and Gordon J.. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA. 76:4350–4354. 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upla, P., Kim S.J., Sampathkumar P., Dutta K., Cahill S.M., Chemmama I.E., Williams R., Bonanno J.B., Rice W.J., Stokes D.L., et al. 2017. Molecular architecture of the major membrane ring component of the nuclear pore complex. Structure. 25:434–445. 10.1016/j.str.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni, S., Vamparys L., Gautier R., Drin G., Etchebest C., Fuchs P.F., and Antonny B.. 2013. Amphipathic lipid packing sensor motifs: Probing bilayer defects with hydrophobic residues. Biophys. J. 104:575–584. 10.1016/j.bpj.2012.11.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova, E.G., Kurchashova S.Y., Polyakov V.Y., and Sheval E.V.. 2011. Self-organization of cellular structures induced by the overexpression of nuclear envelope proteins: A correlative light and electron microscopy study. J. Electron Microsc. 60:57–71. 10.1093/jmicro/dfq067 [DOI] [PubMed] [Google Scholar]
- Vollmer, B., Lorenz M., Moreno-Andrés D., Bodenhöfer M., De Magistris P., Astrinidis S.A., Schooley A., Flötenmeyer M., Leptihn S., and Antonin W.. 2015. Nup153 recruits the nup107-160 complex to the inner nuclear membrane for interphasic nuclear pore complex assembly. Dev. Cell. 33:717–728. 10.1016/j.devcel.2015.04.027 [DOI] [PubMed] [Google Scholar]
- Vollmer, B., Schooley A., Sachdev R., Eisenhardt N., Schneider A.M., Sieverding C., Madlung J., Gerken U., Macek B., and Antonin W.. 2012. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31:4072–4084. 10.1038/emboj.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen, A., Kosinski J., Sparks L., Ori A., DiGuilio A.L., Vollmer B., Mackmull M.T., Banterle N., Parca L., Kastritis P., et al. 2015. In situ structural analysis of the human nuclear pore complex. Nature. 526:140–143. 10.1038/nature15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, A.D., May C.K., Dauster E.S., Cinquin B.P., Smith E.A., Robellet X., D’Amours D., Larabell C.A., and Cohen-Fix O.. 2014. The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Curr. Biol. 24:2861–2867. 10.1016/j.cub.2014.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A.M., Procter J.B., Martin D.M., Clamp M., and Barton G.J.. 2009. Jalview Version 2: A multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberruss, M., and Antonin W.. 2016. Perforating the nuclear boundary—how nuclear pore complexes assemble. J. Cell Sci. 129:4439–4447. 10.1242/jcs.194753 [DOI] [PubMed] [Google Scholar]
- West, R.R., Vaisberg E.V., Ding R., Nurse P., and McIntosh J.R.. 1998. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 9:2839–2855. 10.1091/mbc.9.10.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Hoyt M.A., Chan C., Goetsch L., Botstein D., and Byers B.. 1993. NDC1: A nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122:743–751. 10.1083/jcb.122.4.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin, K.L., Chong Y., Shao S., Webster M.T., Lahiri S., Walters A.D., Lee B., Koh J.L., Prinz W.A., Andrews B.J., and Cohen-Fix O.. 2012. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr. Biol. 22:1128–1133. 10.1016/j.cub.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R.W., Blobel G., and Rout M.P.. 1994. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 125:31–42. 10.1083/jcb.125.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R., Basson M., D’Ari L., and Rine J.. 1988. Increased amounts of HMG-CoA reductase induce “karmellae”: A proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107:101–114. 10.1083/jcb.107.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Khan A., Vitale J., Neuner A., Rink K., Lüchtenborg C., Brügger B., Söllner T.H., and Schiebel E.. 2021. A short perinuclear amphipathic α-helix in Apq12 promotes nuclear pore complex biogenesis. Open Biol. 11:210250. 10.1098/rsob.210250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Neuner A., Rüthnick D., Sachsenheimer T., Lüchtenborg C., Brügger B., and Schiebel E.. 2018. Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol. 217:877–894. 10.1083/jcb.201706024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovsky, M.A., Filograna A., Luini A., Corda D., and Valente C.. 2019. Phosphatidic acid in membrane rearrangements. FEBS Lett. 593:2428–2451. 10.1002/1873-3468.13563 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains used in this study.
lists plasmids used in this study.
is the source file for Fig. 1.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Data Availability Statement
Western blot data underlying all figures and supplementary figures are available in the source data file. Lipidomic data associated with this study are available from the Dryad digital repository: DOI: 10.5061/dryad.s7h44j1bw.