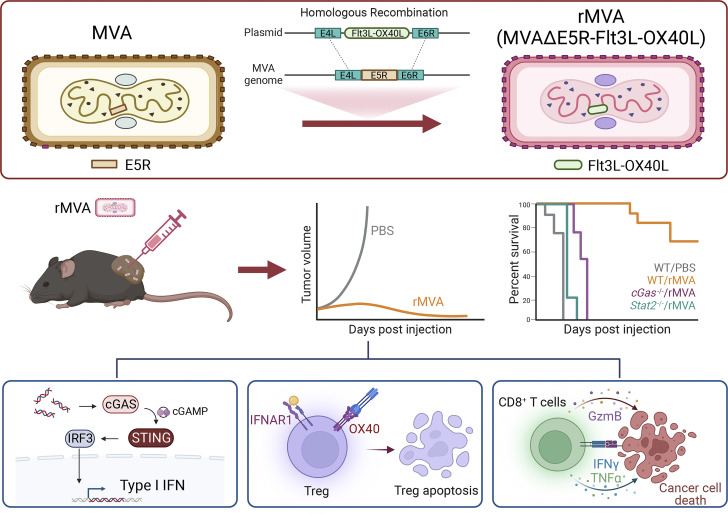
This study shows that intratumoral injection of engineered non-replicative recombinant modified vaccinia virus Ankara depletes and reprograms tumor-infiltrating regulatory T cells via OX40L–OX40 interaction and IFNAR signaling to enhance antitumor immunity.
Abstract
Effective depletion of immune suppressive regulatory T cells (Tregs) in the tumor microenvironment without triggering systemic autoimmunity is an important strategy for cancer immunotherapy. Modified vaccinia virus Ankara (MVA) is a highly attenuated, non-replicative vaccinia virus with a long history of human use. Here, we report rational engineering of an immune-activating recombinant MVA (rMVA, MVA∆E5R-Flt3L-OX40L) with deletion of the vaccinia E5R gene (encoding an inhibitor of the DNA sensor cyclic GMP-AMP synthase, cGAS) and expression of two membrane-anchored transgenes, Flt3L and OX40L. Intratumoral (IT) delivery of rMVA (MVA∆E5R-Flt3L-OX40L) generates potent antitumor immunity, dependent on CD8+ T cells, the cGAS/STING-mediated cytosolic DNA-sensing pathway, and type I IFN signaling. Remarkably, IT rMVA (MVA∆E5R-Flt3L-OX40L) depletes OX40hi regulatory T cells via OX40L/OX40 interaction and IFNAR signaling. Single-cell RNA-seq analyses of tumors treated with rMVA showed the depletion of OX40hiCCR8hi Tregs and expansion of IFN-responsive Tregs. Taken together, our study provides a proof-of-concept for depleting and reprogramming intratumoral Tregs via an immune-activating rMVA.
Graphical Abstract
Introduction
Immune checkpoint blockade (ICB) therapy utilizing antibodies targeting T cell inhibitory mechanisms has revolutionized how solid tumors are treated (Ribas and Wolchok, 2018; Wei et al., 2018; Zou et al., 2016). However, the majority of patients without pre-existing antitumor T cell responses do not respond to ICB therapy, and one-third of the initial responders develop acquired resistance to this line of therapy likely due to cancer immunoediting (Ribas and Wolchok, 2018; Schreiber et al., 2011; Zaretsky et al., 2016). Therefore, innovative approaches to rendering tumors sensitive to ICB therapy are urgently needed.
Viral-based cancer immunotherapy is a versatile and effective approach to alter tumor immunosuppressive microenvironment through multiple mechanisms, including the induction of innate immunity, immunogenic cell death in the infected immune and tumor cells, and activation of tumor-infiltrating dendritic cells (DCs) and antitumor CD8 and CD4 T cells, as well as depletion of immunosuppressive cells (Bommareddy et al., 2018; Davola and Mossman, 2019; Lemos de Matos et al., 2020; Russell et al., 2012; Workenhe and Mossman, 2014). As a result, intratumoral (IT) delivery of immunogenic viruses turns “cold” tumors into “hot” tumors, which renders them sensitive to other immunotherapeutic modalities including ICB (Chesney et al., 2018; Dai et al., 2017; Ribas et al., 2018; Wang et al., 2021; Zamarin et al., 2014).
Foxp3-expressing CD4+ regulatory T (Treg) cells are one of the immunosuppressive cells abundant in tumors and they promote tumor growth and metastasis (Plitas et al., 2016; Plitas and Rudensky, 2020; Sakaguchi et al., 2020). Systemic depletion of Tregs enhances antitumor immunity (Bos et al., 2013; Klages et al., 2010; Shimizu et al., 1999) and also provokes autoimmunity (Kim et al., 2007; Shimizu et al., 1999). How to selectively deplete immunosuppressive tumor-infiltrating Tregs without affecting peripheral Treg cells remains challenging. In this study, we devised a strategy to engineer recombinant modified vaccinia virus Ankara (rMVA) to deplete immune-suppressive Treg cells when delivered IT.
MVA is a highly attenuated vaccinia virus that belongs to the poxvirus family and has been used extensively as a vaccine vector (Gilbert, 2013; Liu et al., 2021; Volz and Sutter, 2017). MVA infection of dendritic cells induces type I IFN via the cytosolic DNA-sensing pathway mediated by the DNA sensor cyclic GMP–AMP synthase (cGAS) and downstream signaling molecules such as stimulator of IFN genes (STING; Dai et al., 2014). However, MVA encodes multiple inhibitors of the cytosolic nucleic acid-sensing pathways. In a previous study, we showed that IT heat-inactivated MVA (heat-iMVA) generates stronger antitumor immunity than IT live MVA, which requires CD8+ T cells, Batf3-dependent CD103+/CD8α cross-presenting DCs, and STING-mediated cytosolic DNA-sensing pathway (Dai et al., 2017). To improve MVA-based cancer immunotherapy, we performed a screen of vaccinia early genes for inhibiting the cGAS/STING pathway and identified vaccinia E5 as a major inhibitor of cGAS (Yang et al., 2021). MVA∆E5R, in which the E5R gene was deleted from the MVA genome, induces much higher levels of type I IFN compared with MVA (Yang et al., 2021). In addition, we engineered the virus to express two membrane-anchored transgenes, FMS-like tyrosine kinase 3 ligand (Flt3L) and OX40L. Flt3L is a growth factor for CD103+ DCs and plasmacytoid DCs (Liu and Nussenzweig, 2010). OX40L is a costimulatory ligand for OX40 (CD134), a member of the TNF receptor superfamily expressed on activated CD4 and CD8 T cells as well as Treg cells (Croft, 2009). It has been shown that OX40L blocks the generation of IL-10–producing Tregs in vitro in a human DC and CD4+ T cell coculture system (Ito et al., 2006). In addition, OX40L on DCs plays a crucial role in T cell priming and activation (Chen et al., 1999; Murata et al., 2000).
Our results provide evidence that IT rMVA (MVA∆E5R-hFlt3L-mOX40L) induces strong antitumor effects in immune-competent murine tumor models via the cGAS/STING-mediated DNA-sensing mechanism and type I IFN signaling. Depletion of CD8+ T cells renders tumors resistant to rMVA therapy. IT rMVA (MVA∆E5R-hFlt3L-mOX40L) dramatically reduces immunosuppressive OX40hi Tregs in the injected tumors via OX40L/OX40 interaction and IFNAR signaling. Ex vivo infection of human tumors with rhMVA (MVA∆E5R-hFlt3L-hOX40L) also reduces Tregs and activates CD8+ T cells. Collectively, our study strongly supports that rational engineering of MVA is an innovative strategy to enhance antitumor immunity by depleting IT OX40hi Tregs.
Results
Rational design of immune-activating rMVA (MVA∆E5R-hFlt3L-mOX40L)
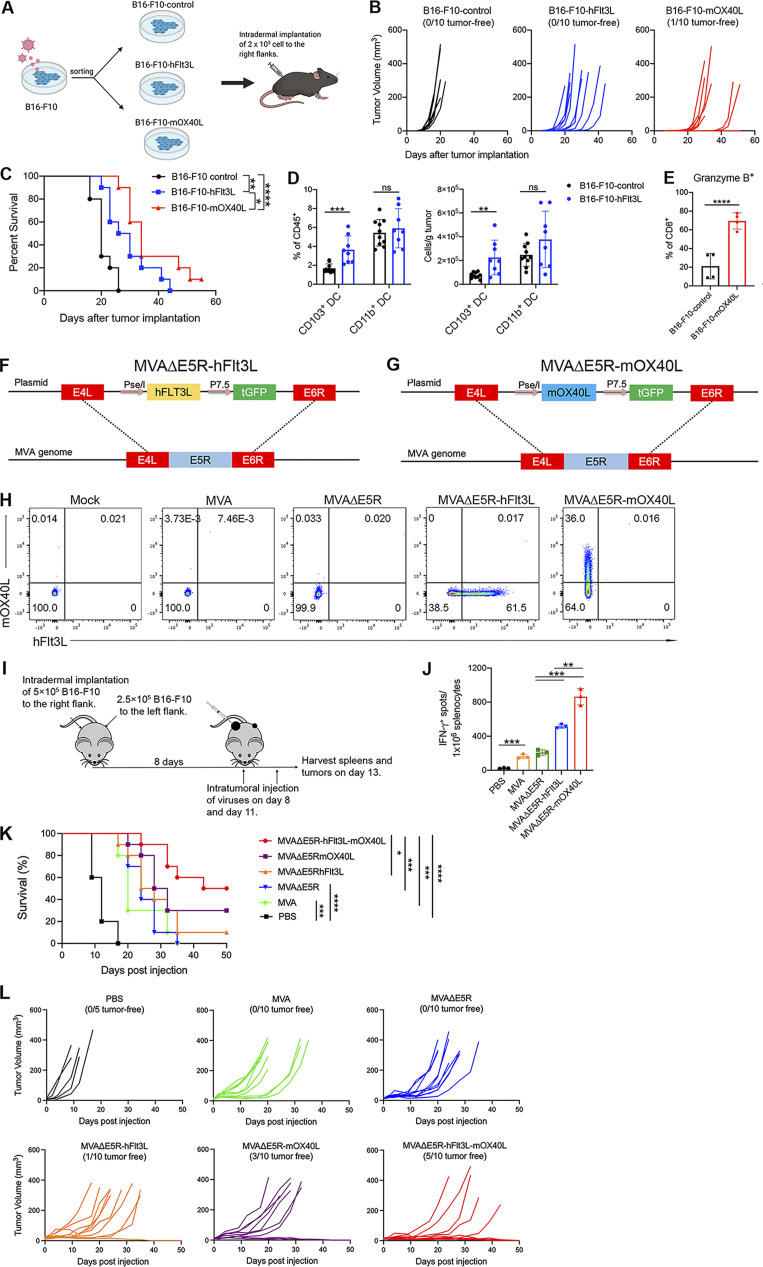
Our previous work demonstrated that Batf3-dependent CD103+ DCs are required for antitumor immunity induced by IT delivery of heat-iMVA (Dai et al., 2017). To investigate whether human Flt3L (hFlt3L) expression on tumor cells affects tumor growth and tumor-infiltrating myeloid cell populations, we constructed a murine melanoma B16-F10 stable cell line that expresses membrane-bound hFlt3L, and subsequently implanted either B16-F10-hFlt3L or the parental B16-F10 cells into WT C57BL/6J mice (Fig. S1 A). We observed that expressing hFlt3L on tumor cells delayed B16-F10 tumor growth and prolonged the survival of tumor-bearing mice (Fig. S1, B and C). The percentages of CD103+ DCs out of CD45+ cells and the absolute numbers of CD103+ DCs per gram of B16-F10-hFlt3L were increased in B16-F10-hFlt3L tumors compared with B16-F10 control tumors, whereas CD11b+ DCs were at similar levels in both tumors (Fig. S1 D). These results indicate that hFlt3L expression on tumor cell surfaces facilitates the development and proliferation of CD103+ DCs in the tumor microenvironment.
Figure S1.
Incremental engineering of MVA with deletion of E5R gene and expression of hFlt3L or mOX40L improves antitumor effects. (A) B16-F10 were transduced with retrovirus to generate hFlt3L- or mOX40L-expressing stable cell lines. C57BL/6J mice were intradermally implanted with 2 × 105 B16-F10-hFlt3L, B16-F10-mOX40L, or B16-F10 control cells. (B) Tumor growth curve (n = 10). (C) Kaplan–Meier survival curve (n = 10; *P < 0.05, **P < 0.01, ****P < 0.0001, Mantel-Cox test). (D) Percentages and absolute number of CD103+ DCs and CD11b+ DCs in B16-F10-hFlt3L or B16-F10-control tumors. Data are means ± SD (n = 8 or 10; **P < 0.01, ***P < 0.001, t test). A representative experiment is shown, repeated once. (E) Percentages Granzyme B+ CD8+ and Granzyme B+ CD4+ in B16-F10-hFlt3L or B16-F10-control tumors. Data are means ± SD (n = 4; ****P < 0.0001, t test). A representative experiment is shown, repeated once. (F and G) Schematic diagrams for the generation of MVAΔΕ5R-hFlt3L (F) or MVAΔΕ5R-mOX40L (G) through homologous recombination. (H) Representative flow cytometry plots of expression of hFlt3L or mOX40L by MVA, MVAΔΕ5R, MVAΔΕ5R-hFlt3L, MVAΔΕ5R-mOX40L, or mock-infected BHK21 cells. (I) Schematic diagram of IT MVA, MVAΔE5R, MVAΔΕ5R-hFlt3L, MVAΔΕ5R-mOX40L, or PBS in a bilateral B16-F10 melanoma implantation model. (J) IFN-γ+ splenocytes from MVA, MVAΔE5R, MVAΔΕ5R-hFlt3L, MVAΔΕ5R-mOX40L, or PBS-treated mice. Data are means ± SD (n = 3; *P < 0.05, ***P < 0.001, t test). (K) Kaplan–Meier survival curve of mice treated with MVA, MVAΔΕ5R, MVA∆E5R-hFlt3L, MVA∆E5R-mOX40L, MVA∆E5R-hFlt3L-mOX40L, or PBS in a unilateral B16-F10 implantation model (n = 10 in each virus group and n = 5 in PBS group; *P < 0.05, ***P < 0.001, ****P < 0.0001, Mantel-Cox test). (L) B16-F10 tumor volumes over time in C57BL/6J mice treated with MVA, MVAΔΕ5R, MVA∆E5R-hFlt3L, MVA∆E5R-mOX40L, MVA∆E5R-hFlt3L-mOX40L, or PBS.
OX40L is a costimulatory molecule that interacts with its receptor OX40 expressed on T cells (Croft et al., 2009). OX40L on activated DCs plays an important role in the generation of antigen-specific T cell responses (Murata et al., 2000). We constructed a B16-F10-mOX40L cell line that constitutively expresses murine OX40L (mOX40L) on its surface (Fig. S1 A). B16-F10-mOX40L tumors grew slower than the parental B16-F10 tumors after implantation (Fig. S1, B and C), with higher percentages of tumor-infiltrating Granzyme B+ CD8+ T cells compared with the parental B16-F10 cells (Fig. S1 E). The median survival of mice implanted with B16-F10-hFlt3L or B16-F10-mOX40L were 28 and 34 d, 8 or 14 d longer, respectively, than those implanted with the control B16-F10 (Fig. S1 C).
We recently discovered the vaccinia E5R gene encodes a major inhibitor of cGAS (Yang et al., 2021). Deleting the E5R gene from MVA resulted in much stronger induction of type I IFN from murine bone marrow–derived dendritic cells (BMDCs) compared with MVA (Yang et al., 2021). We designed the following rMVA viruses to evaluate the utility of hFlt3L and mOX40L individually expressed by MVA∆E5R (Fig. S1, F and G). The respective transgene expression was validated in MVA∆E5R-hFlt3L or MVA∆E5R-mOX40L infected BHK21 cells (Fig. S1 H). IT delivery of the two viruses resulted in higher numbers of IFN-γ+ T cells in the spleens compared with MVA or MVA∆E5R, as determined by ELISpot analysis (Fig. S1, I and J). These results suggest that expressing hFlt3L or mOX40L by rMVA improves antitumor efficacy.
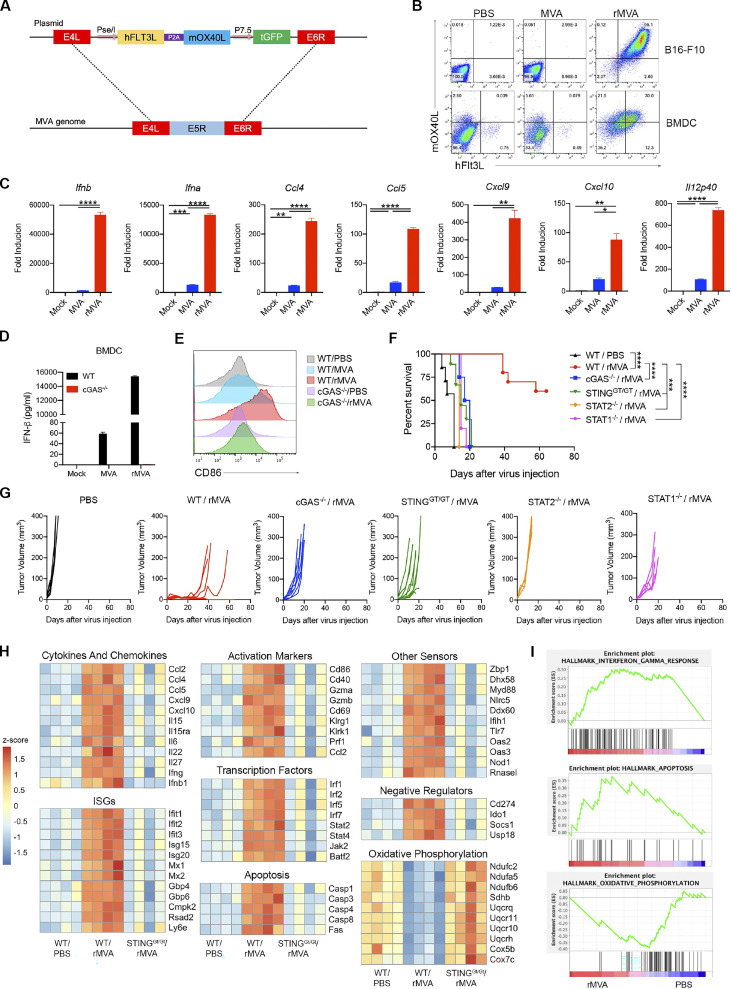
Based on these results, we designed an rMVA (MVA∆E5R-hFlt3L-mOX40L) by inserting two transgenes, membrane-bound hFlt3L and mOX40L, into the E5R locus (Fig. 1 A). hFlt3L and mOX40L are linked by a P2A self-cleaving sequence and their expression is driven by the vaccinia synthetic early/late promoter. Both transgenes were expressed efficiently on the surface of infected B16-F10 murine melanoma cells and murine BMDCs at 24 h after infection (Fig. 1 B). rMVA infection of BMDCs induced the expression of Ifnb, Ifna4, Ccl4, Ccl5, Cxcl9, Cxcl10, and Il12p40 genes. Infection of BMDCs with rMVA induced cGAS-dependent Ifnb gene expression and IFN-β protein secretion at higher levels compared with MVA (Fig. 1, C and D). rMVA infection also induces DC maturation, as manifested by CD86 upregulation, determined by FACS, in a cGAS-dependent manner (Fig. 1 E).
Figure 1.
IT injection of rMVA elicits strong antitumor immunity. (A) Schematic diagram for the generation of rMVA through homologous recombination. (B) Representative flow cytometry plots of expression of hFlt3L or mOX40L in rMVA-infected B16-F10 cells and BMDCs. (C) Relative mRNA expression levels of Ifnb, Ifna, Ccl4, Ccl5, Cxcl9, Cxcl10, and Il12p40 in BMDCs infected with MVA or rMVA. Data are means ± SD (n = 3; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, t test). A representative experiment is shown, repeated twice. (D) Concentrations of secreted IFN-β in the medium of WT or cGas−/− BMDCs infected with MVA or rMVA. Data are means ± SD. (E) Mean fluorescence intensity of CD86 expressed by WT or cGas−/− BMDCs infected with MVA or rMVA. (F) Kaplan–Meier survival curve of mice treated with rMVA or PBS in a unilateral B16-F10 implantation model (n = 5 ∼ 10; ***P < 0.001, ****P < 0.0001, Mantel-Cox test). A representative experiment is shown, repeated once. (G) Tumor growth curve of mice treated with rMVA or PBS in a unilateral B16-F10 implantation model. (H) Heatmap of gene expression from RNA-seq analysis of RNAs isolated from tumors implanted on WT or Stinggt/gt mice treated with or without IT delivery of rMVA. (I) Gene set enrichment analysis of the expression of genes involved IFN-γ response, apoptosis, and oxidative phosphorylation in tumors treated with rMVA vs. PBS control.
IT injection of rMVA elicits strong antitumor immune responses that are dependent on cGAS/STING-mediated DNA sensing and STAT1/STAT2-mediated IFNAR-signaling pathway
To test whether cGAS/STING and STAT1/STAT2 are important for IT rMVA-induced antitumor immunity, cGas−/−, Stinggt/gt (lacking function STING; Sauer et al., 2011), Stat1−/−, Stat2−/−, or age-matched WT C57BL/6J mice were implanted with B16-F10 melanoma intradermally. When the tumors were established, they were injected with rMVA twice weekly. Whereas IT rMVA resulted in tumor eradication or delayed tumor growth in WT mice, it failed to induce antitumor effects in Stat1−/− and Stat2−/− mice (Fig. 1, F and G). IT rMVA treatment of cGas−/− or StingGt/Gt mice extended median survival from 11 d in PBS control group to 18.5 d (P = 0.0002). However, all of the cGas−/− or StingGt/Gt mice died from tumor progression (Fig. 1, F and G). These results demonstrated that activation of the cGAS/STING-mediated cytosolic DNA-sensing pathway, as well as the IFNAR/STAT1/STAT2 signaling, by IT rMVA, is critical for the generation of antitumor immunity.
Incremental engineering of MVA improves antitumor efficacy
We used a unilateral B16-F10 murine melanoma model to compare the antitumor efficacy elicited by IT injection of either MVA, MVA∆E5R, MVA∆E5R-hFlt3L, MVA∆E5R-mOX40L, or rMVA (MVA∆E5R-hFlt3L-mOX40L). IT MVA∆E5R-hFlt3L-mOX40L significantly delayed tumor growth and resulted in 50% cure rate and extension of median survival from 12 d in the PBS mock-treatment control group to 46.5 d. IT MVA∆E5R-mOX40L resulted in 30% cure rate and extension of median survival to 30 d, and IT MVA∆E5R-hFlt3L resulted in 10% cure rate and extension of median survival to 26 d (Fig. S1, K and L). By contrast, IT MVA and MVA∆E5R did not result in cure but extended median survival to 20 and 24 d, respectively (Fig. S1, K and L).
IT injection of rMVA (MVA∆E5R-hFlt3L-mOX40L) is more effective than intravenous (IV) delivery in eradicating tumors in an immune-competent mouse model
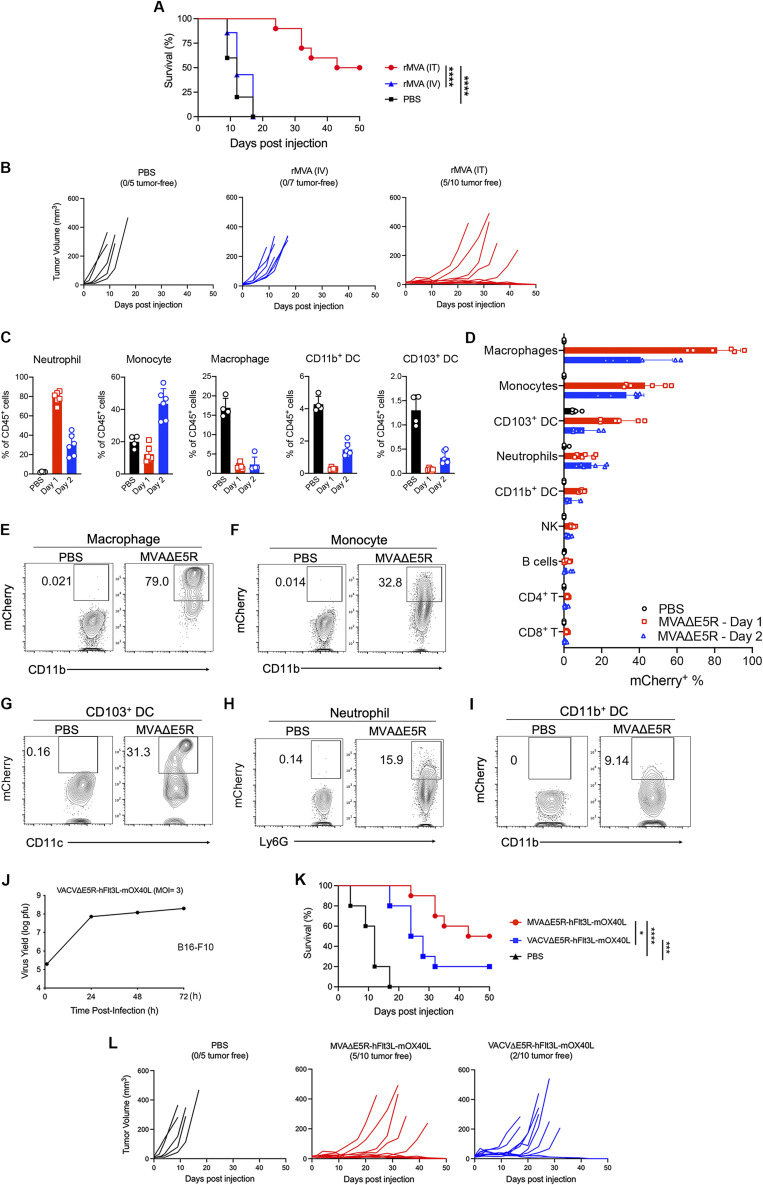
Systemic delivery of oncolytic vaccinia can potentially access tumors in distant sites. It has the advantage of easier delivery than IT administration, especially for internal tumors that are difficult to be injected. Here, we compared the antitumor efficacy of rMVA delivered twice weekly through IT vs. IV routes. IV delivery of the virus had no therapeutic benefits, whereas IT delivery of the virus achieved tumor control and 50% cure rate (Fig. S2, A and B). These results support the use of IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) as the preferred route.
Figure S2.
IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) is more efficacious in eradicating tumors than IV delivery of the virus. (A) Kaplan–Meier survival curve of mice treated with IT vs. IV delivery of rMVA in a unilateral B16-F10 implantation model (n = 5–10; ****P < 0.0001, Mantel-Cox test). (B) B16-F10 tumor volumes over time in C57BL/6J mice treated with IT vs. IV delivery of rMVA. PBS mock-treatment control was included. (C–I) Influx of myeloid cells into MVAE5R-treated tumors and induction of IFN-β and other inflammatory cytokine production in a cGAS/STING-dependent manner. (C) Percentages of neutrophils, monocytes, macrophages, CD103+ DCs, and CD11b+ DCs in the MVA∆E5R-treated tumors. Mice were intradermally implanted with B16-F10 cells. 7 d after implantation, tumors were injected with MVAΔE5R-mCherry or PBS as control and harvested 1 or 2 d after injection for myeloid cell analysis. Data are means ± SD (n = 4 ∼ 6). (D) Percentages of mCherry+ immune cells. Data are means ± SD (n = 4–6). NK, natural killer. (E–I) Representative flow cytometry plots of mCherry+ immune cells. (J–L) IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) is more effective than VACV∆E5R-hFlt3L-mOX40L in restricting tumor growth. (J) VACV∆E5R-hFlt3L-mOX40L replication curve in B16-F10 cells. Cells were infected at an MOI of 3. (K) Kaplan–Meier survival curve of mice treated with IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) vs. VACV∆E5R-hFlt3L-mOX40L in a unilateral B16-F10 implantation model (n = 10 in each virus group and n = 5 in PBS group; *P < 0.05, ***P < 0.001, ****P < 0.0001, Mantel-Cox test). (L) B16-F10 tumor volumes over time in C57BL/6J mice treated with IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) vs. VACV∆E5R-hFlt3L-mOX40L. PBS mock-treatment control was included.
IT rMVA (MVA∆E5R-hFlt3L-mOX40L) results in myeloid cell influx into the injected tumors and induces IFN-β and other inflammatory cytokine production in a cGAS/STING-dependent manner
To elucidate mechanisms of action of rMVA, we first investigated myeloid cell dynamics and determined which cell types are infected after IT viral therapy. To do that, we used the murine B16-F10 tumor implantation model and injected the tumors with MVA∆E5R expressing mCherry. At 1 or 2 d after injection, we harvested the tumors and analyzed tumor-infiltrating immune cells. IT MVA∆E5R-mCherry injection led to an influx of neutrophils 1 d after injection, which subsided after the second day when monocytes started to increase in the tumor microenvironment (Fig. S2 C). Among the myeloid cell populations, macrophages were most heavily infected, as determined by mCherry expression in the infected cells, followed by monocytes, neutrophils, CD103+, and CD11b+ DCs (Fig. S2, D–I). T, B, or natural killer cells, however, were largely not infected by IT-injected MVA∆E5R (Fig. S2 D).
To investigate the innate immune responses of tumors induced by IT rMVA and the role of the cytosolic DNA-sensing pathway in this process, we isolated tumors from WT or Stinggt/gt mice 1 d after treatment with rMVA or PBS (mock control) and subjected them to bulk RNA sequencing (RNA-seq) analyses. We observed striking upregulation of genes involved in immune activation, apoptosis, and downregulation of genes involved in oxidative phosphorylation (Fig. 1, H and I). We separated the immune activation genes into several categories, including cytokines and chemokines, interferon-stimulated genes (ISGs), activation markers, transcription factors, and other sensors. We found that rMVA treatment upregulated the expression of Ifnb1, Ifng, Il-15, Il15ra, Ccl2, Ccl4, Ccl5, Cxcl9, and Cxcl10 in a STING-dependent manner (Fig. 1, H and I). We also observed upregulation of DC activation markers, including CD86 and CD40, and T cell activation markers, including Gzma, Gzmb, Pfr1, and CD69, as dependent on STING (Fig. 1, H and I). Caspase 1, Caspase 3, Caspase 4, Caspase 8, and Fas gene expression was also induced by IT rMVA in WT mice but not in STING-deficient mice (Fig. 1, H and I). The expression of genes involved in oxidative phosphorylation was downregulated after IT rMVA injection in WT mice but not in STING-deficient mice (Fig. 1, H and I). Taken together, our results show that IT rMVA leads to infection and recruitment of myeloid cell populations and activation of innate immune responses in those cells via the cGAS/STING pathway.
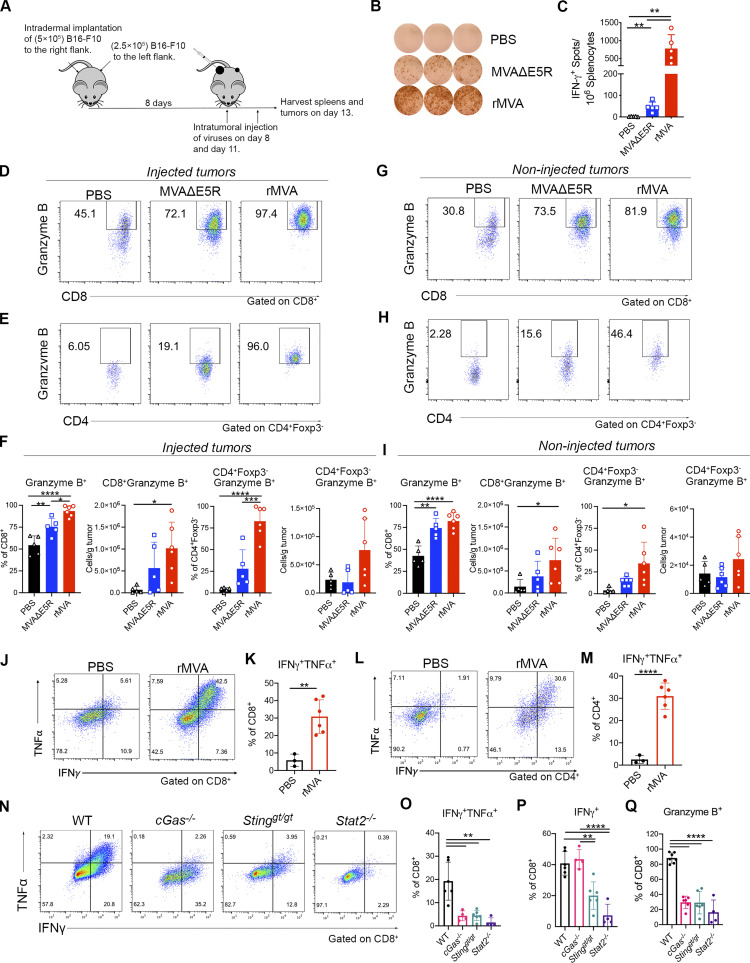
IT rMVA (MVA∆E5R-hFlt3L-mOX40L) generates stronger systemic and local antitumor immune responses compared with MVA∆E5R in a bilateral B16-F10 murine melanoma implantation model
To determine the immunological mechanism of rMVA-induced antitumor immune responses, we used a bilateral murine B16-F10 tumor implantation model. B16-F10 cells were intradermally implanted into both flanks of C57BL/6J mice. After tumors were established, we injected MVAΔE5R, rMVA, or PBS as a control to the right-side tumors twice, 3 d apart. Spleens and both tumors were harvested 2 d after the second injection (Fig. 2 A). IT rMVA generated the highest numbers of tumor-specific IFN-γ+ T cells in the spleens compared with those treated with MVA∆E5R or with PBS as determined by ELISpot assay (Fig. 2, B and C). In the injected tumors, IT rMVA resulted in stronger T cell activation with higher percentages and absolute numbers of granzyme B+ CD8+ and granzyme B+ Foxp3− CD4+ cells compared with MVA∆E5R or PBS control groups (Fig. 2, D and F). In the non-injected tumors, IT rMVA also induced more granzyme B+ CD8+ and granzyme B+ Foxp3− CD4+ T cells (Fig. 2, G–I), demonstrating that IT rMVA enhances T cell activation both locally and systemically. IT rMVA also induced IFN-γ+ TNF-α+ CD8+ and CD4+ T cells in the injected tumors, indicating enhanced T cell effector function (Fig. 2, J–M). Taken together, these results demonstrate that IT rMVA results in the activation of both CD8+ and CD4+ T cells in the injected and non-injected tumors and the generation of systemic antitumor immunity.
Figure 2.
IT rMVA generates strong systemic and local anti-tumor immune responses dependent on cGAS/STING/STAT2 pathways. (A) Schematic diagram of IT rMVA or MVAΔE5R for ELISpot assay and TIL analysis in a murine B16-F10 melanoma implantation model. (B) Representative images of IFN-γ+ spots from ELISpot assay. The experiment was repeated twice. (C) Statistical analysis of IFN-γ+ splenocytes from MVAΔE5R-, rMVA-, or PBS-treated mice. Data are means ± SD (n = 5 or 6; **P < 0.01, t test). (D and E) Representative flow cytometry plots of Granzyme B+ CD8+ (D) and Granzyme B+ CD4+ Foxp3− cells (E) in the injected tumors. (F) Percentages and absolute number of Granzyme B+ CD8+ and Granzyme B+ CD4+ Foxp3− cells in the injected tumors. Data are means ± SD (n = 5 or 6; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, t test). (G and H) Representative flow cytometry plots of Granzyme B+ CD8+ (G) and Granzyme B+ CD4+ Foxp3− cells (H) in the non-injected tumors. (I) Percentages and absolute number of Granzyme B+ CD8+ and Granzyme B+ CD4+ Foxp3− cells in the non-injected tumors. Data are means ± SD (n = 5 or 6; *P < 0.05, **P < 0.01, ****P < 0.0001, t test). (J–M) Representative flow cytometry plots and statistical analysis of IFNγ+TNFα+ CD8+ (J and K) and IFNγ+TNFα+ CD4+ cells (L and M) in the injected tumors. Data are means ± SD (n = 3 or 6; **P < 0.01, ****P < 0.0001, t test). The above experiment was repeated twice. (N) Representative flow cytometry plots of IFNγ+TNFα+ CD8+ T cells in the injected tumors harvested from WT, cGas−/−, Stinggt/gt, and Stat2−/− mice. (O) Percentages of IFNγ+TNFα+ CD8+ T cells in the injected tumors from WT, cGAS−/−, Stinggt/gt, and Stat2−/− mice. Data are means ± SD (n = 4 ∼ 6; **P < 0.01, t test). (P) Percentages of IFN-γ+ CD8+ T cells in the injected tumors from WT, cGAS−/−, Stinggt/gt and Stat2−/− mice. Data are means ± SD (n = 4 ∼ 6; **P < 0.01, ****P < 0.0001, t test). (Q) Percentages of Granzyme B+ CD8+ T cells in the injected tumors from WT, cGas−/−, Stinggt/gt, and Stat2−/− mice. Data are means ± SD (n = 4 ∼ 6; ****P < 0.0001, t test). A representative experiment is shown, repeated once.
rMVA (MVA∆E5R-hFlt3L-mOX40L)-induced antitumor immune responses are dependent on the cGAS/STING-mediated cytosolic DNA-sensing and STAT2-mediated type I IFN signaling pathways
We observed that the B16-F10–bearing cGAS, STING, or STAT2-deficient mice responded poorly to rMVA treatment (Fig. 1, F and G), which led us to hypothesize that the cGAS/STING-mediated cytosolic DNA-sensing and STAT2-dependent IFN signaling pathways are important for the generation of antitumor CD8+ T cell responses. To test that hypothesis, cGAS, STING, or STAT2-deficient and age-matched C57BL/6J control mice were intradermally implanted with B16-F10 cells into their right flanks. IT rMVA generated polyfunctional IFN-γ+ TNF-α+CD8+ and granzyme B+ CD8+ T cells in the injected tumors only in WT mice, whereas only IFN-γ+ CD8+ T cells were induced by IT rMVA in cGas−/− and Stinggt/gt mice (Fig. 2, N–Q). IT rMVA failed to induce either IFN-γ+, Granzyme B+ CD8+ cells, or IFN-γ+ TNF-α+ T cells in Stat2−/− mice (Fig. 2, N–Q). These results indicate that both the cGAS/STING and STAT2-mediated signaling pathways are crucial for rMVA-induced T cell activation.
CD8+ T cells are required for the antitumor effects induced by IT rMVA (MVA∆E5R-hFlt3L-mOX40L)
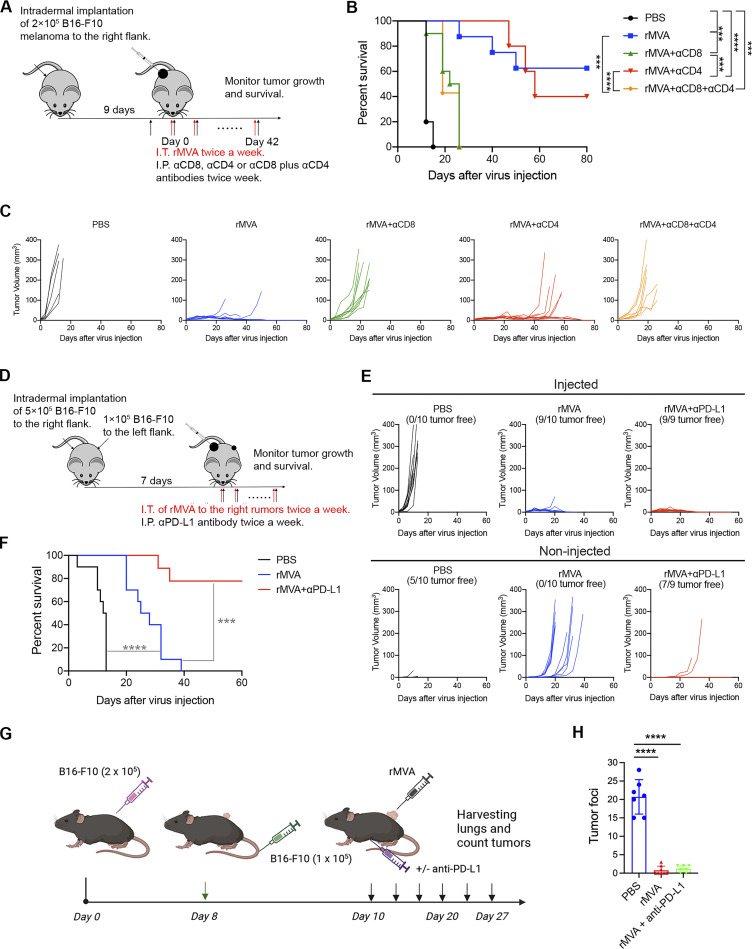
To determine which cell populations are essential for tumor eradication by IT rMVA, we performed an antibody depletion experiment using anti-CD8 and/or anti-CD4 antibodies during rMVA treatment. The depleting antibodies were first given intraperitoneally (IP) 2 d before IT rMVA and then were given at the same time when mice were treated with IT rMVA (Fig. 3 A). Depletion of CD8+ cells abrogated the therapeutic effect of rMVA (Fig. 3, A and C). Although CD4+ T cell depletion did not reduce the initial response to rMVA treatment, the antitumor response did not persist in the CD4+ T cell–depleted mice after IT rMVA treatment ended on day 42. 60% of mice died due to the recurrence of tumors. Mice with both CD4+ and CD8+ T cell–depleted behaved similarly to those with only CD8+ T cell depletion (Fig. 3, B and C). These results indicated that CD8+ T cells are required for tumor eradication in IT rMVA therapy, while CD4+ T cells are important for facilitating the generation of antitumor memory responses.
Figure 3.
CD8 T cells are required for rMVA-induced antitumor effects. (A) Schematic diagram of IT rMVA in C57BL/6J mice in the presence or absence of depleting antibodies for CD8 and/or CD4 in a unilateral B16-F10 melanoma implantation model. (B) Kaplan–Meier survival curve of mice treated with IT rMVA in the presence or absence of depleting antibodies for CD8 and/or CD4 (n = 5 ∼ 10; ***P < 0.001, ****P < 0.0001, Mantel-Cox test). A representative experiment is shown, repeated once. (C) Tumor volumes over days in mice under various treatment conditions. (D) Schematic diagram of IT rMVA in combination with IP αPD-L1 antibody in a bilateral B16-F10 melanoma implantation model. (E) Injected and non-injected tumor volumes over days in mice under various treatment conditions. (F) Kaplan–Meier survival curve of mice treated with IT rMVA alone or in combination with IP anti–PD-L1 antibody (n = 9 or 10; ***P < 0.001, ****P < 0.0001, Mantel-Cox test). A representative experiment is shown, repeated once. (G) Schematic diagram of testing whether IT rMVA results in restricting B16-F10 melanoma lung metastasis in C57BL/6J mice. (H) B16-F10 tumor foci on the surface of lungs of mice treated with PBS, rMVA, or the combination of IT rMVA and systemic delivery of anti–PD-L1 antibody (n = 7 or 8; ****P < 0.0001, t test).
Combination of IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) with systemic administration of anti–PD-L1 antibody provides systemic antitumor therapeutic effects
B16-F10 tumors respond poorly to ICB therapy. To test whether the combination of systemic delivery of ICB antibody and IT rMVA therapy can overcome the resistance to ICB therapy, we used a bilateral B16-F10 implantation model and compared the antitumor efficacy of IT rMVA alone vs. IT rMVA plus IP delivery of anti–PD-L1 antibody (Fig. 3 D). In the bilateral tumor implantation model, B16-F10 cells were implanted intradermally into the right and left flanks of C57BL/BJ mice (5 × 105 to the right flank and 1 × 105 to the left flank). At 7 d after implantation, the tumors at the right flank were injected with 4 × 107 PFU of rMVA or PBS. 250 µg αPD-L1 antibody was injected IP twice weekly in the combination group. Five out of 10 tumors did not grow out in the left flanks in the PBS mock-treated group because the PBS-injected tumors grew rapidly and the mice had to be euthanized early. IT rMVA alone eradicated nine out of 10 injected tumors and delayed the growth of non-injected tumors. However, 90% of mice died eventually due to the growth of non-injected tumors (Fig. 3, E and F). By contrast, the combination of IP anti–PD-L1 antibody and IT rMVA significantly improved the antitumor therapeutic efficacy. 80% of mice in the combination group rejected non-injected tumors and survived (Fig. 3, E and F). We have previously shown that IP delivery of anti–PD-L1 alone had no therapeutic benefits in the B16-F10 model (Dai et al., 2017). These results demonstrated that the combination of systemic delivery of anti–PD-L1 antibody and IT rMVA generated synergistic systemic antitumor therapeutic effects, leading to the eradication of both injected and non-injected tumors.
To test whether IT rMVA (MVA∆E5R-hFlt3L-mOX40L) generates systemic antitumor effects that restrict tumor metastasis, we performed the following experiment in which C57BL/6J mice were implanted intradermally on the right flank with 2 × 105 B16-F10 cells. On day 8 after implantation, mice were implanted IV with 1 × 105 B16-F10 cells to establish lung metastasis. rMVA (4 × 107 PFU) were injected into the right flank tumors twice weekly with or without systemic delivery of anti–PD-L1 antibody. PBS mock injection control was included. Lungs were harvested from PBS mock-treated tumor-bearing mice on day 20 after tumor implantation and on day 27 from mice treated with rMVA with or without anti–PD-L1 antibody. Tumor foci on the lung surfaces were counted under a dissecting microscope (Fig. 3 G). We observed that IT rMVA treatment of B16-F10 on the right flank resulted in a more than 18-fold reduction of tumor foci on the lung surfaces in the presence or absence of anti–PD-L1 antibody (Fig. 3 H).
rMVA (MVA∆E5R-hFlt3L-mOX40L) is more effective than oncolytic vaccinia VACV∆E5R-hFlt3L-mOX40L in delaying or eradicating tumors
In our previously published work (Wang et al., 2021), we compared the antitumor effects of live oncolytic vaccinia vs. its heat-inactivated version (by incubating the virus at 55°C for 1 h) and found that IT injection of heat-inactivated oncolytic viruses (OVs) generated more effective antitumor effects than live OVs, indicating that viral replication and oncolysis is not critical for viral-induced antitumor effects. Here, we engineered a replication-competent vaccinia virus, VACV∆E5R-hFlt3L-mOX40L, by inserting the same expression cassette encoding hFlt3L and mOX40L as that in rMVA into the E5R locus of WT vaccinia virus. VACV∆E5R-hFlt3L-mOX40L is replication-competent in B16-F10 cells when the cells were infected in vitro at a multiplicity of infection (MOI) of 3 (Fig. S2 J). IT injection of VACV∆E5R-hFlt3L-mOX40L resulted in the extension of median survival from 12 d in the PBS control group to 26 d, shorter than the 46.5 d median survival in the MVA∆E5R-hFlt3L-mOX40L group (Fig. S2, K and L). The cure rate in the VACV∆E5R-hFlt3L-mOX40L group was 20%, lower than 50% cure rate in the MVA∆E5R-hFlt3L-mOX40L group (Fig. S2, K and L). These results indicate that IT rMVA (MVA∆E5R-hFlt3L-mOX40L) is more efficacious than oncolytic vaccinia expressing the same transgenes.
IT rMVA (MVA∆E5R-hFlt3L-mOX40L) depletes OX40hi Tregs in the injected tumors
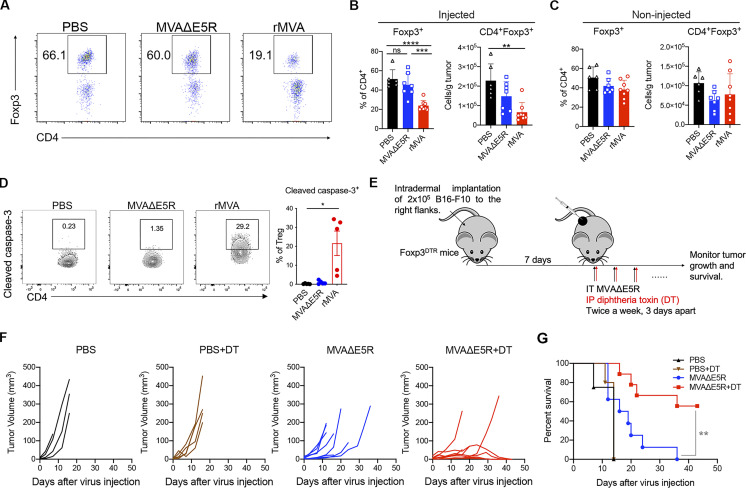
In addition to enhanced CD8+ and CD4+ T cell activation, we also observed that IT rMVA treatment resulted in a significant reduction of Tregs in the injected tumors (Fig. 4, A and B). The mean percentages of Tregs (Foxp3+CD4+) out of CD4+ T cells were 23, 45, and 51% in rMVA-, MVA∆E5R-, and PBS-treated tumors, respectively (Fig. 4, A and B). The absolute numbers of Tregs in rMVA-injected tumors were significantly reduced compared with the PBS-treated group (Fig. 4, A and B). In the non-injected tumors, however, we did not observe a reduction in the percentages of Tregs out of CD4+ T cells after rMVA treatment (Fig. 4 C). The percentages of cleaved caspase-3+ cells out of tumor-infiltrating Tregs from rMVA-treated tumors were much higher compared with those from PBS- or MVA∆E5R-treated tumors (Fig. 4 D). These results support that rMVA treatment triggers apoptosis in tumor-infiltrating Tregs.
Figure 4.
IT rMVA depletes OX40hi Tregs in the injected tumors to promote anti-tumor therapy. (A) Representative flow cytometry plots of Foxp3+CD4+ cells in the injected tumors. Mice were treated as described in Fig. 2 A. (B and C) Percentages and absolute number of Foxp3+CD4+ cells in the injected (B) and non-injected (C) tumors. Data are means ± SD (n = 6–8; **P < 0.01, ***P < 0.001, ****P < 0.0001, t test). (D–F) Mice were intradermally implanted with B16-F10 cells. Tumors were injected with rMVA, MVA∆E5R, or PBS as control after 7 d after implantation and harvested 2 d after injection. (D) Left: Representative flow cytometry plots of cleaved caspase-3+ Tregs in the injected tumors. The experiment was repeated twice. Right: Percentages of cleaved caspase-3 in tumor-infiltrating Tregs by flow cytometry. Data are means ± SD (n = 3–5; *P < 0.05, t test). (E) Schematic diagram of IT MVA∆E5R in the presence or absence of DT in a unilateral B16-F10 melanoma implantation model in Foxp3DTR mice. Three doses of DT (200 ng each per mouse) were administered to mice at −2, −1, and +1 d relative to the first MVA∆E5R injection at day 0. (F) Tumor volumes over time in mice treated with MVA∆E5R, MVA∆E5R + DT, DT alone, or PBS. (G) Kaplan–Meier survival curves of mice treated with MVA∆E5R, MVA∆E5R + DT, DT alone, or PBS (n = 5–10; **P < 0.01, Mantel-Cox test).
To determine whether Tregs play a negative role in rMVA-based virotherapy, we implanted B16-F10 cells intradermally into the right flanks of Foxp3-DTR mice. After tumors were established, we treated them with IT MVA∆E5R or PBS with or without diphtheria toxin (DT; Fig. 4 E). We chose MVA∆E5R instead of rMVA (MVA∆E5R-hFlt3L-mOX40L) in this experiment because IT MVA∆E5R does not reduce IT Treg whereas rMVA does. Three doses of DT (200 ng each per mouse) were administered to tumor-bearing mice at −2, −1, and +1 d relative to IT MVA∆E5R or PBS injection at day 0. MVA∆E5R or PBS was given twice weekly throughout the experiment (Fig. 4 E). Although IP administration of three doses of DT alone without virus treatment did not affect tumor growth or survival, the combination of DT and IT MVA∆E5R injection significantly improved therapeutic efficacy compared with IT MVA∆E5R alone (Fig. 4, F and G). These results suggest that Tregs play an inhibitory role in viral-based immunotherapy, and IT depletion of Tregs by rMVA (MVA∆E5R-hFlt3L-mOX40L) might be an important mechanism for potentiating antitumor immunity.
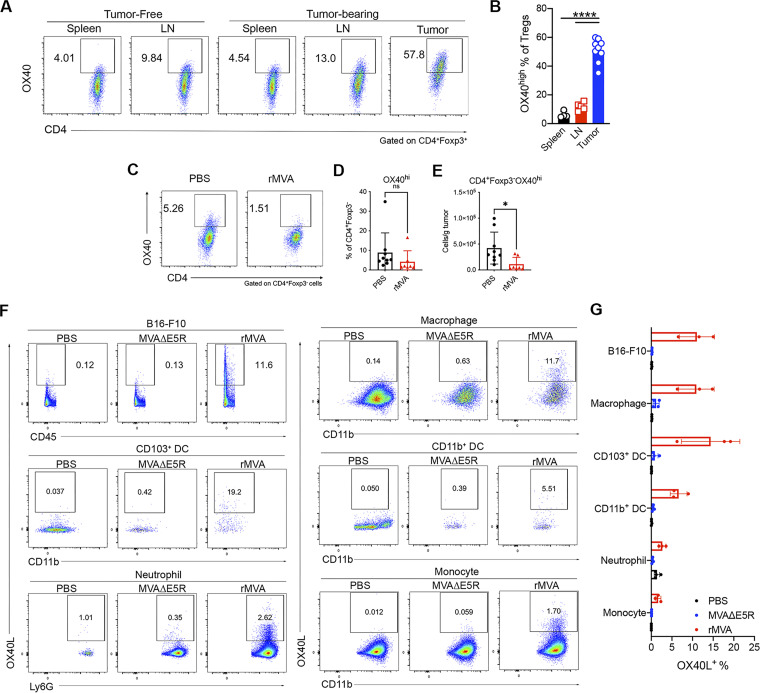
IT rMVA (MVA∆E5R-hFlt3L-mOX40L) preferentially depletes OX40hi Tregs via OX40L–OX40 interaction and IFNAR signaling
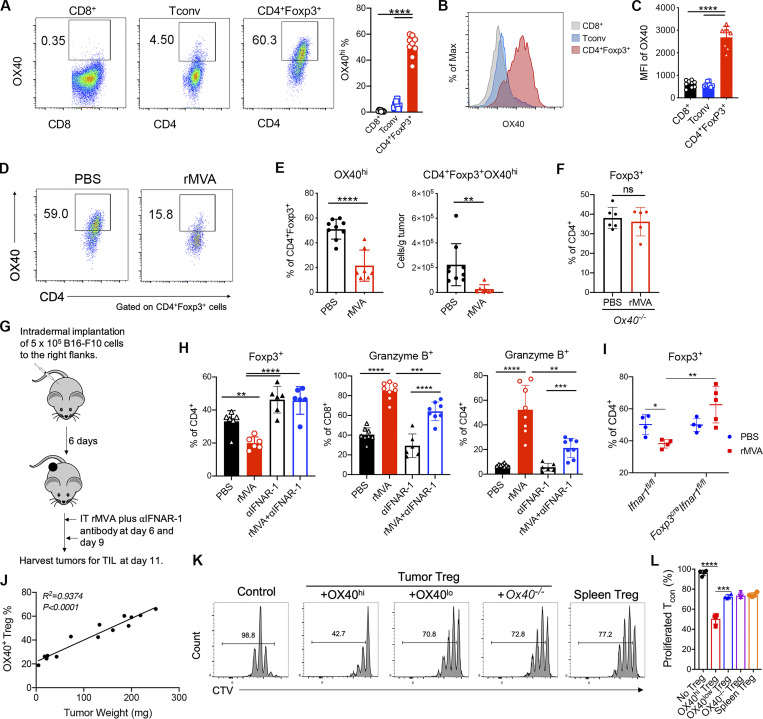
We hypothesized that OX40L expressed by rMVA-infected myeloid and tumor cells might be important in mediating the reduction of OX40hi Tregs. We first compared the surface expression of OX40 in various T cell populations within the tumor microenvironment. The mean percentages of OX40hi Tregs among CD4+ Tregs were 51% compared with 5.6% of OX40hi CD4+Foxp3− conventional T (Tconv) cells and 1% of OX40hi CD8+ T cells (Fig. 5 A). The mean fluorescence intensity of OX40 was higher in CD4+ Tregs than those in CD4+Foxp3− Tconv and CD8+ T cells (Fig. 5, B and C). OX40 expression levels in Tregs from spleens or lymph nodes were much lower than those from tumors (Fig. S3, A and B). IT rMVA treatment preferentially reduced the percentages of OX40hi Tregs out of total Tregs and the absolute numbers of OX40hi Tregs per gram of tumors in the injected tumors (Fig. 5, D and E). In OX40−/− mice, however, IT rMVA did not result in Treg reduction in the injected tumors (Fig. 5 F). In addition, IT rMVA also reduced the absolute numbers of OX40hi CD4+Foxp3− Tconv cells in the injected tumors (Fig. S3, C–E). Finally, we evaluated OX40L expression in both tumor cells and myeloid cells 2 d after IT rMVA. OX40L was detected on B16-F10, tumor-infiltrating macrophages, CD103+ DCs, CD11b+ DCs, neutrophils, and monocytes (Fig. S3, F and G). These results indicate that IT rMVA results in the OX40L expression in a variety of cell types including tumor and tumor-infiltrating myeloid cells and reduces tumor-infiltrating OX40hi Tregs likely via OX40–OX40L interaction.
Figure 5.
IT rMVA preferentially depletes OX40hi Tregs in the injected tumors in a type I IFN signaling dependent manner. (A) Representative flow cytometry plots of OX40 expression on tumor infiltrating CD8+, Tconv (CD4+Foxp3−), and CD4+Foxp3+ T cells in tumors 15 d after implantation. Mice were treated as described in Fig. 2 A. (B and C) Representative flow cytometry plots and statistical analysis of mean fluorescence intensity (MFI) of OX40 on tumor-infiltrating CD8+, Tconv (CD4+Foxp3−), and CD4+Foxp3+ T cells. Data are means ± SD in C (n = 6 ∼ 8; ****P < 0.0001, t test). (D) Representative flow cytometry plots of OX40hi CD4+Foxp3+ in the injected tumors. Mice were treated as described in Fig.2 A. The experiment was repeated twice. (E) Percentages and absolute number of OX40hi CD4+Foxp3+ T cells in the injected tumors. Data are means ± SD (n = 7 or 9; **P < 0.01, ****P < 0.0001, t test). (F) Percentages of CD4+Foxp3+ T cells in the injected tumors from WT and Ox40−/− mice. Mice were treated as described in Fig. 3 A. Data are means ± SD (n = 5 or 6; t test). A representative experiment is shown, repeated once. (G) Schematic diagram of IT rMVA in the presence or absence of IT αIFNAR-1 antibody in a unilateral B16-F10 melanoma implantation model. (H) Percentages of CD4+Foxp3+, CD8+Granzyme B+, and CD4+Granzyme B+ T cells in the injected tumors. Data are means ± SD (n = 6; **P < 0.01, ***P < 0.001, ****P < 0.0001, t test). A representative experiment is shown, repeated once. (I) Percentages of CD4+Foxp3+ T cells in the injected tumors in Ifnar1fl/fl and Foxp3creIfnar1fl/fl mice treated with IT rMVA or PBS. Data are means ± SD (n = 4–5; *P < 0.05, **P < 0.01, t test). A representative experiment is shown, repeated once. (J) Correlation of the percentages of OX40hi Tregs in the tumors with tumor weight. Mice were implanted with B16-F10 tumors intradermally. Tumors with different sizes were analyzed for OX40 expression on tumor-infiltrating Tregs. (K and L) Representative flow cytometry plots (K) and percentage of Tcon (conventional CD4+ T cells) proliferation (L) as measured by CellTrace Violet (CTV) dye dilution co-cultured with tumor OX40hi, OX40low, Ox40−/− Tregs, or spleen Tregs. Data are means ± SD in L (***P < 0.001, ****P < 0.0001, t test). A representative experiment is shown, repeated once.
Figure S3.
OX40 expression on T cells in lymphoid organs and in tumors and OX40L expression in tumors and tumor-infiltrating cells after IT rMVA. (A) Representative flow cytometry plots of OX40 expression on CD4+Foxp3+ T cells in the spleens, lymph nodes, or tumors from naive or B16-F10 tumor-bearing mice. The experiment was repeated twice. (B) Percentages of OX40hi Tregs in the spleens, lymph nodes, and tumors in the tumor-bearing mice (n = 7 or 9; ****P < 0.0001, t test). (C) Representative flow cytometry plots of OX40 expression on CD4+Foxp3− T cells in the PBS- or rMVA-injected tumors. (D and E) Percentages (D) and absolute numbers (E) of OX40hi CD4+Foxp3− Tconv cells in the injected tumors. Data are means ± SD (n = 7 or 9; *P < 0.05, t test). (F) Representative flow cytometry plots of OX40L expression on B16-F10 tumor cells or myeloid cells in the tumors injected with MVAΔE5R, rMVA, or PBS as control. The experiment was repeated once. (G) Percentages of OX40L+ B16-F10 cells or myeloid cells in the tumors. Data are means ± SD (n = 3–5).
To evaluate whether the IFNAR signaling pathway is involved in rMVA-mediated Treg reduction, we coadministered anti–IFNAR-1 antibody with rMVA into implanted B16-F10 melanoma twice, 3 d apart. Tumors were harvested 2 d after second injection (Fig. 5 G). Whereas IT rMVA decreased the percentages of OX40hi Treg out of Tregs as well as the percentages of Tregs out of CD4+ T cells, coadministration of anti–IFNAR-1 antibody with rMVA reversed the reduction (Fig. 5 H). In addition, coadministration of anti–IFNAR-1 with rMVA resulted in lower percentages of Granzyme B+ CD8+ and CD4+ T cells than IT rMVA alone (Fig. 5 H), consistent with the role of type I IFN in promoting CD8+ and CD4+ T cell activation.
To test whether IFNAR signaling on regulatory T cells plays a role in rMVA-mediated Treg depletion, we generated Foxp3CreIfnar1fl/fl mice. B16-F10 melanoma cells were implanted intradermally into the right flanks of age-matched Foxp3CreIfnar1fl/fl and Ifnar1fl/fl control mice. IT rMVA reduced Tregs in the Ifnar1fl/fl mice compared with PBS-treated controls (Fig. 5 I). However, IT rMVA failed to reduce Tregs in the Foxp3CreIfnar1fl/fl mice (Fig. 5 I). Taken together, our results provide strong evidence that IT rMVA results in the depletion of OX40hi Tregs in the injected tumors via OX40L–OX40 interaction, and this process is facilitated by IFNAR signaling on Tregs, suggesting that type I IFN induced by IT rMVA in the tumor microenvironment triggers Treg depletion.
Tumor-infiltrating OX40hi Tregs are more suppressive than OX40lo Tregs
We observed that in the murine B16-F10 melanoma model, the percentage of OX40hi Tregs in total Tregs positively correlated with tumor mass (Fig. 5 J), suggesting that OX40hi Tregs may represent an immunosuppressive cell population during tumor progression. To determine the functional differences between OX40hi, OX40lo, and OX40−/− Tregs, we intradermally implanted B16-F10 cells into Foxp3gfp and OX40−/−Foxp3gfp mice in a C57BL/6J background and FACS-sorted OX40hi and OX40lo IT Tregs from Foxp3gfp mice and OX40−/− Tregs from OX40−/− Foxp3gfp mice and compared their suppression function in vitro. Flow cytometry analysis showed that OX40hi Tregs isolated from tumors suppressed Tconv proliferation more strongly compared with tumor-infiltrating OX40lo Tregs or OX40−/− Tregs in vitro (Fig. 5, K and L). We also isolated splenic Tregs from tumor-bearing Foxp3gfp mice and found that the suppressive activities of these cells were similar to those of OX40lo Tregs or OX40−/− Tregs isolated from tumors (Fig. 5, K and L).
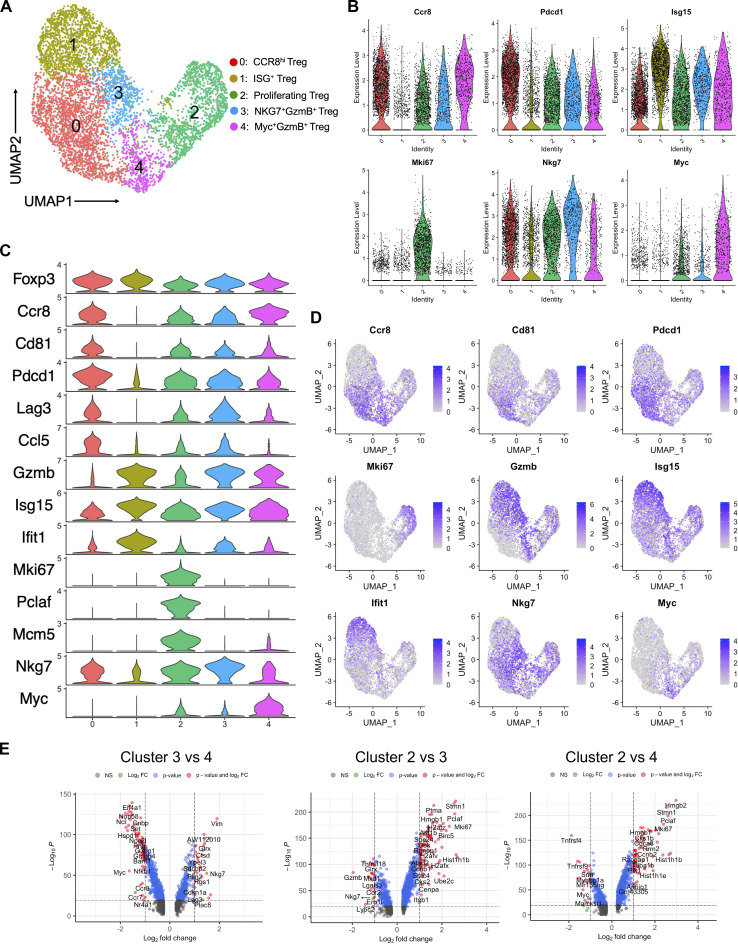
Single-cell RNA-seq (scRNA-seq) revealed reprogramming of Treg subpopulations after rMVA (MVA∆E5R-hFlt3L-mOX40L) treatment
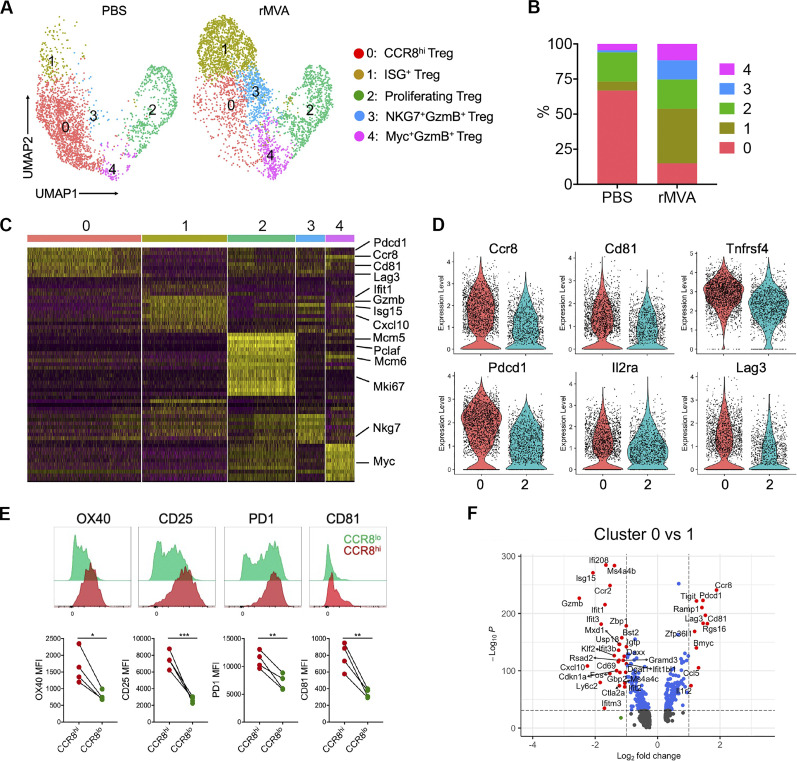
To evaluate how IT delivery of rMVA might alter the transcriptomes of tumor-infiltrating Treg populations, we performed scRNA-seq analyses of sorted Foxp3-GFP Tregs from tumors treated with IT rMVA or PBS twice, 3 d apart. Five different Treg populations (clusters 0–4) were identified in mouse B16-F10 tumors (Fig. 6, A–C; and Fig. S4 A). Two main Treg populations in the PBS mock-treated tumors, cluster 0 (CCR8hi Tregs) and cluster 2 (proliferating Tregs), comprised about 67 and 21% of Treg cells in PBS-treated tumors, respectively (Fig. 6, A–C). Upon IT rMVA treatment, cluster 0 (CCR8hi Tregs) was reduced from 67 to 15%, whereas the size of cluster 2 (proliferating Treg) remained stable. IT rMVA also resulted in increase of cluster 1 (ISG+ Tregs expressing Gzmb) from 6.5% in the PBS-treated tumors to 39% in rMVA-treated tumors, cluster 3 (NKG7+GzmB+ Tregs) from 1.5 to 13.7%, and cluster 4 (Myc+GzmB+ Tregs) from 4.5 to 11.7% (Fig. 6, A–C). Differential gene expression analysis revealed that Ccr8, Cd81, Tnfrsf4 (OX40), Il2ra, and T cell exhaustion markers such as Pdcd1 and Lag3 are expressed higher in cluster 0 compared with cluster 2 (Fig. 6, C and D; and Fig. S4, B–D). In addition, flow cytometry analyses confirmed higher OX40, CD25, PD-1, and CD81 expression in CCR8hi tumor-infiltrating Tregs compared with CCR8lo tumor-infiltrating Tregs (Fig. 6 E).
Figure 6.
scRNA-seq of Tregs in tumors treated with rMVA or PBS control. (A) UMAP visualization of single-cell transcriptomes of Treg cells isolated from tumors treated with rMVA or PBS control. Each dot corresponds to a single cell, color coded by cell cluster. (B) Proportion of each Treg subtype in all Treg cells from PBS- and rMVA-treated tumors. (C) Heatmap of genes differentially expressed according to cluster identity as in A. (D) Violin plots showing the expression levels of selected marker genes in clusters 0 and 2. (E) Representative FACS plot and statistical analysis of mean fluorescence intensity (MFI) showing the expressions of OX40, CD25, PD1, and CD81 on CCR8hi Tregs and CCR8lo IT Tregs. (F) Volcano plots showing genes differentially expressed between clusters 0 and 1.
Figure S4.
Single-cell transcriptomic analysis of Tregs in the tumors. (A) UMAP visualization of single-cell transcriptomes of Treg cells isolated from PBS- and rMVA-treated tumors. Each dot corresponds to a single cell and each color represents one cluster. (B) Violin plots showing the expression levels of selected marker genes in different Treg cell clusters. (C) Violin plots showing the expression levels of top enriched genes in different Treg cell clusters. (D) UMAP showing the expression of selected marker genes for each Treg cell cluster. (E) Volcano plots showing genes differentially expressed between clusters 3 vs. 4, clusters 2 vs. 3, and clusters 2 vs. 4.
Cluster 2 represents proliferating Treg cells with high expression of cell proliferation markers, including Mki67, Pclaf, Mcm5, and Mcm6 (Fig. 6 C and Fig. S4, B–E). Cluster 1 represents ISG+ Tregs, in which ISGs such as Isg15 and Ifit1 were upregulated (Fig. 6, C and F; and Fig. S4, B–D), which suggests that cluster 1 Treg cells are responsive to type I IFN induced by rMVA in the tumor immunosuppressive microenvironment. They also express lower levels of T cell exhaustion markers such as Pdcd1, Tigit, and Lag3 and higher levels of effector genes such as Gzmb and Cxcl10 compared with cluster 0 (Fig. 6 F and Fig. S4, B–D). In addition, cluster 1 Tregs express higher levels of CCR2 compared with cluster 0, suggesting that this population might be recruited to the tumors after IT rMVA (Fig. 6, F and G).
Figure S5.
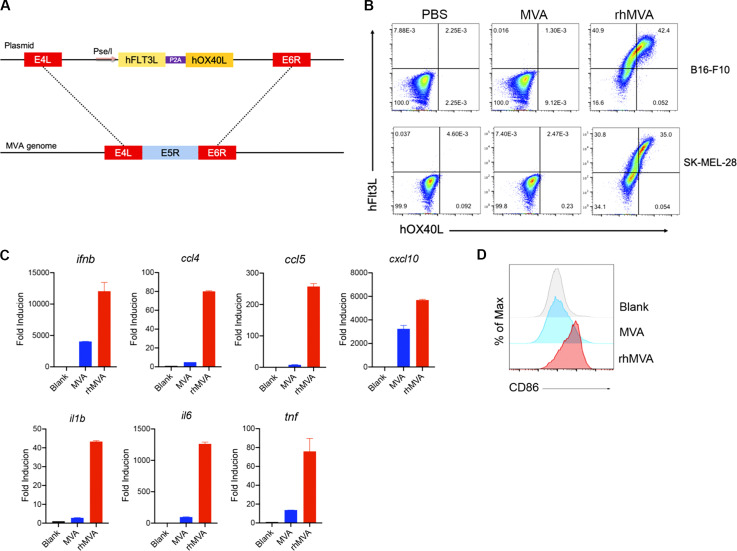
Clinical candidate rhMVA induces innate immunity and promotes maturation of human moDCs. (A) Schematic diagram for the generation of rhMVA through homologous recombination. (B) Representative flow cytometry plots of expression of hFlt3L or hOX40L by rMVA-infected B16-F10 cells and SK-MEL-28 cells. (C) Relative mRNA expression levels of ifnb, ccl4, ccl5, cxcl10, il1b, il6, and tnf in moDCs infected with MVA or rhMVA. A representative experiment is shown, repeated once. (D) Mean fluorescence intensity of CD86 expressed by human moDCs infected with MVA or rhMVA. A representative experiment is shown, repeated once.
Differential gene expression analysis between clusters 2 and 3 revealed that activation markers including Gzmb, Prf1, and Nkg7 as well as inflammatory chemokines such as Ccl4 and Ccl5 are expressed higher in cluster 3 compared with cluster 2 (Fig. S4, B–E). Cluster 4 Tregs express higher levels of Myc, Gzmb, Irf8, Tnfrsf4, and Tnfrsf9 compared with cluster 2 (Fig. S4, B–E). When compared with cluster 3, cluster 4 Tregs express higher levels of Myc and Myc-target genes, including Eif4a1, Eif5a, Fbl, Nop58, Nop56, Hnrnpab, G3bp1, C1qbp, and Cnbp. In addition, cluster 4 expresses higher levels of Irf8, Ccr7, and Ccr8 compared with cluster 3 (Fig. S4, B–E).
IFNAR1 signaling on Tregs is important in mediating rMVA (MVA∆E5R-hFlt3L-mOX40L)-induced antitumor effects
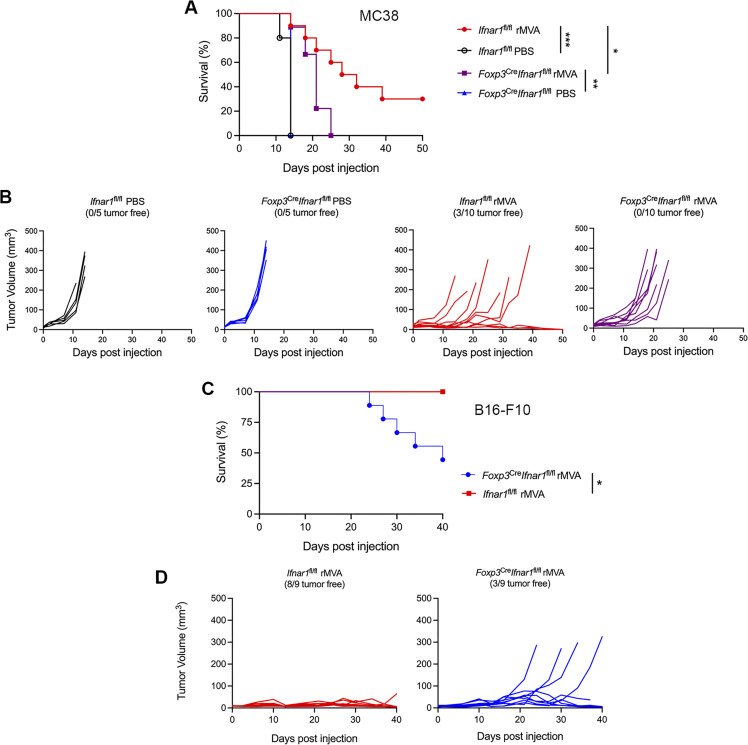
To test whether IFNAR signaling on Tregs plays a role in rMVA-induced antitumor effects, we intradermally implanted MC38 murine colon cancer cells into Ifnar1fl/fl and Foxp3CreIfnar1fl/fl mice. When the tumors were 3–4 mm in diameter, they were injected with rMVA or PBS twice weekly. We observed 30% cure in Ifnar1fl/fl mice and no cure in Foxp3creIfnar1fl/fl mice after rMVA treatment (Fig. 7, A and B). The median survival in Ifnar1fl/fl mice were extended from 14 d in PBS-treated mice to 30 d in rMVA-treated mice, whereas the median survival in Foxp3creIfnar1fl/fl mice was extended from 14 d in PBS-treated mice to 21 d (Fig. 7, A and B). In a B16-F10 melanoma model, IT rMVA resulted in eradication of tumors in eight out of nine Ifnar1fl/fl mice, but only in three out nine of the Foxp3creIfnar1fl/f mice (Fig. 7, C and D). These results indicate that IFNAR1 signaling on Tregs plays an important role in mediating rMVA-induced antitumor effects, likely through induction of apoptosis in Tregs and reprogramming of Tregs.
Figure 7.
IFNAR1 on Tregs are important for rMVA-induced antitumor effects in two murine tumor models. (A) Kaplan–Meier survival curve of MC38-bearing Ifnar1fl/fl and Foxp3CreIfnar1fl/fl mice treated with IT rMVA or PBS (n = 5 or 10; *P < 0.05, **P < 0.01, ***P < 0.001, Mantel-Cox test). (B) MC38 tumor volumes over days in Ifnar1fl/fl and Foxp3CreIfnar1fl/fl mice treated with IT rMVA or PBS control. (C) Kaplan–Meier survival curve of B16-F10-bearing Ifnar1fl/fl and Foxp3CreIfnar1fl/fl mice treated with IT rMVA (n = 9; *P < 0.05, Mantel-Cox test). (D) B16-F10 tumor volumes over days in Ifnar1fl/fl and Foxp3CreIfnar1fl/fl mice treated with IT rMVA.
IT rMVA (MVA∆E5R-hFlt3L-mOX40L) is effective in generating antitumor T cell responses and controlling tumor growth in murine A20 B cell lymphoma and triple-negative breast cancer (TNBC) models
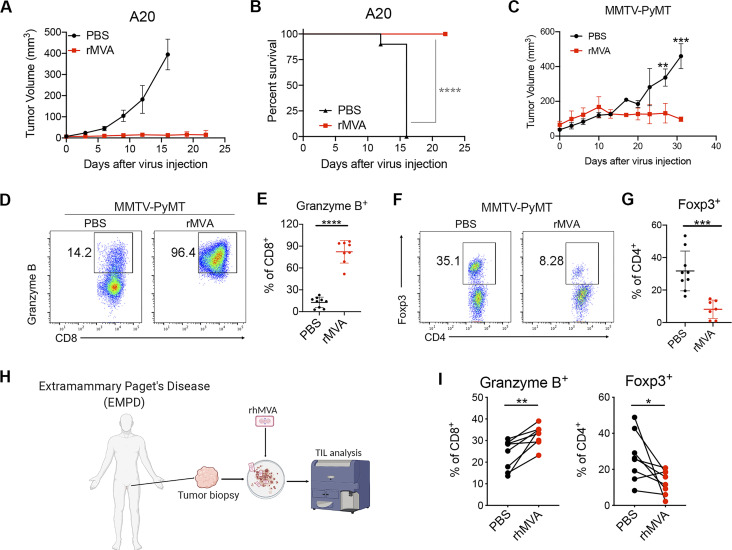
In addition to murine B16-F10 melanoma, we evaluated the therapeutic efficacy of IT rMVA in other murine tumor models. IT rMVA efficiently eradicated A20 B cell lymphoma tumors and resulted in 100% survival (Fig. 8, A and B). MMTV-PyMT is a transgenic mouse strain that develops multiple tumors in the mammary fat pads spontaneously, commonly used as a triple-negative breast tumor model. In the MMTV-PyMT mice, IT injection of rMVA resulted in delayed tumor growth compared with the PBS control group (Fig. 8 C). Similar to what we observed in the B16-F10 murine melanoma model, IT rMVA activated CD8+ T cells in the mouse mammary gland tumors and reduced intertumoral Tregs (Fig. 8, D–G).
Figure 8.
IT rMVA elicits strong antitumor immunity in multiple murine tumor models. (A) Tumor volumes over days in BALB/c mice treated with IT rMVA or PBS control in an A20 B cell lymphoma implantation model. (B) Kaplan–Meier survival curve of mice treated with IT rMVA or PBS (n = 10; ****P < 0.0001, Mantel-Cox test). A representative experiment is shown, repeated once. (C) Tumor volumes over days in the MMTV-PyMT breast tumor model. Data are means ± SD (n = 5; **P <0.01, ***P < 0.001, t test). (D) Representative flow cytometry plots of Granzyme B+CD8+ T cells in the IT rMVA- or PBS-treated tumors from MMTV-PyMT mice. A representative experiment is shown, repeated twice. (E) Percentages of Granzyme B+ CD8+ T cells in the rMVA- or PBS-injected tumors. Data are means ± SD (n = 8; ****P < 0.0001, t test). (F) Representative flow cytometry plots of Foxp3+CD4+ T cells in the rMVA- or PBS-injected tumors. (G) Percentages of Foxp3+CD4+ T cells in the rMVA- or PBS-injected tumors. Data are means ± SD (n = 8; ***P < 0.001, t test). (H) Schematic diagram of ex vivo infection of human Extramammary Paget’s disease tumors with rhMVA (MVA∆E5R-hFlt3L-hOX40L). (I) Percentages of Granzyme B+CD8+ T cells and Foxp3+CD4+ T cells in the rhMVA- or PBS-treated tumor tissues. Data are means ± SD (n = 7; *P < 0.05, **P < 0.01, t test).
Clinical candidate rhMVA (MVA∆E5R-hFlt3L-hOX40L) induces innate immunity and promotes maturation of human monocyte-derived DCs (moDCs)
For clinical applications, we generated an rhMVA-expressing hFlt3L and human OX40L (hOX40L) with the deletion of E5R gene (Fig. S5 A). hFlt3l and hOX40L are membrane-bound ligands that were expressed on the surface of murine B16-F10 cells and human melanoma cell line, SK-MEL-28, after infection with rhMVA in vitro (Fig. S5 B). rhMVA induced higher levels of ifnb gene expression as well as ccl4, ccl5, cxcl10, il1b, il6, and tnf in moDCs compared with MVA (Fig. S5 C). rhMVA infection of human moDCs induced the expression of CD86 on the cell surface, which is indicative of DC maturation (Fig. S5 D).
To test whether ex vivo infection of human tumor samples with rhMVA could induce phenotypic changes of tumor-infiltrating lymphocytes (TILs), we obtained skin biopsy samples from patients with Extramammary Paget’s disease, infected the processed tissues with rhMVA, and analyzed TILs 24 h later (Fig. 8 H). rhMVA-infected samples exhibited upregulation of granzyme B on CD8+ T cells and reduction of Tregs compared with the paired control samples (Fig. 8 I). These results are consistent with what we observed in murine tumors treated with rMVA in vivo, supporting rhMVA as a potential clinical candidate for the treatment of human cancers.
Discussion
Preclinical and clinical studies have shown that viral-based immunotherapeutics can alter immunosuppressive tumor microenvironment to enhance antitumor effects and overcome resistance to immune checkpoint blockade antibody therapy. In this study, we engineered an rMVA, rMVA (MVA∆E5R-hFlt3L-mOX40L), with deletion of the vaccinia E5R gene, which encodes an inhibitor of cGAS, and with the expression of two membrane-bound transgenes, hFlt3L and mOX40L. Here, we show that rMVA activates innate immunity via the cGAS/STING pathway and the IFNAR positive feedback loop and reduces OX40 hi regulatory T cells via OX40L–OX40 interaction under the influence of type I IFN. As a result, IT rMVA dramatically enhances antitumor immunity, which synergizes with systemic delivery of ICB.
The rationale for the choice of genetic modification of the MVA (inserting Flt3L and OX40L and deleting E5R) includes the following: (i) Flt3L encodes a growth factor of CD103+ DCs and plasmacytoid DCs, and OX40L encodes a costimulatory molecule; and (ii) in this study, we found that OX40L expression by virally infected cells in the tumor microenvironment results in depleting OX40hi immune-suppressive Tregs.
Other transgenes that have been tested using vaccinia-based vectors include the super-agonist IL-15 (a fusion protein of IL-15 and IL-15Rα; Kowalsky et al., 2018), membrane-anchored IL-2 and IL-12 (Ge et al., 2020; Liu et al., 2020), IL-7/IL-12 co-expression (Nakao et al., 2020), and anti–CTLA-4 antibody (Semmrich et al., 2022). In addition to inserting transgenes into the viral vector, viral genes can be deleted to enhance the immune responses. For example, B18R encodes a soluble type I IFN receptor, which contributes to the virulence of vaccinia virus (Symons et al., 1995). Oncolytic vaccinia virus deleting B18R and TK encoding the thymidine kinase gene and expressing IFN-β (TK−/B18R−/IFN-β+) showed enhanced tumor selectivity and antitumor efficacy compared with (TK−/B18R) or WT vaccinia when delivered IV (Kirn et al., 2007). B18R (WR200) is fragmented and non-functional in MVA (Antoine et al., 1998). We chose to delete the E5R gene because E5R is a dominant inhibitor of cGAS and deleting the E5R gene from the MVA genome strongly induced type I IFN induction in myeloid cells (Yang et al., 2021).
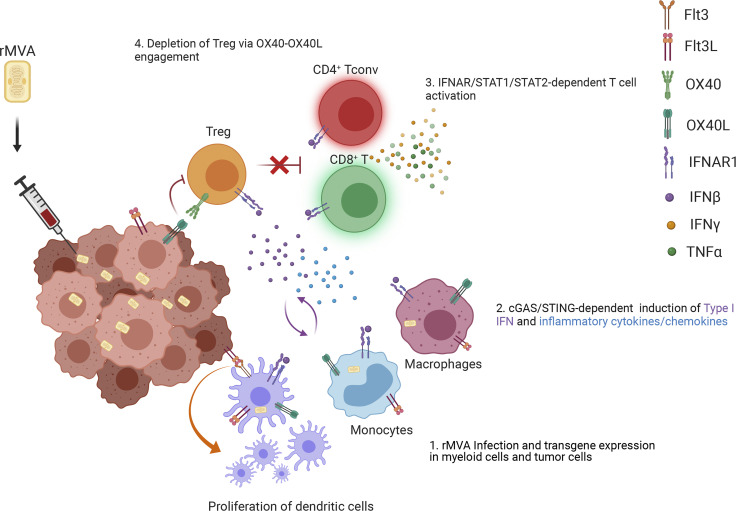
Here, we propose the following working model for the mechanisms of action of rMVA (Fig. 9). After IT injection with rMVA, fractions of tumor and tumor-infiltrating myeloid cells are infected by the virus, which leads to the expression of hFlt3L and mOX40L transgenes on the cell surface, as well as the induction of type I IFN and proinflammatory cytokines and chemokines via the cGAS/STING-dependent cytosolic DNA-sensing pathway from the resident and recruited tumor-infiltrating myeloid cells. Type I IFN plays an important role in activating DCs and tumor-infiltrating CD8+ and CD4+ T cells. In addition, OX40L expression on infected tumor and myeloid cells leads to the depletion of OX40hi Tregs via OX40L–OX40 interaction and IFNAR signaling, which promotes the antitumor activities of CD8+ and CD4+ T cells. Therefore, rMVA engages both innate and adaptive immunity to generate local and systemic antitumor effects, which are amplified in the presence of immune checkpoint blockade antibodies.
Figure 9.
Working model. IT injection of rMVA results in the infection of tumor-infiltrating myeloid cells, including macrophages, monocytes, and DCs, as well as tumor cells. This leads to the activation of cGAS/STING-mediated cytosolic DNA-sensing pathway and the production of type I IFN and cytokines and chemokines that are important for CD8+ and CD4+ T cell proliferation and activation (as indicated by Granzyme B, TNF, and IFN-γ expression). Flt3L expression of the tumor microenvironment facilitates the proliferation of CD103+ DCs in the tumors. OX40L expression by myeloid cell populations and tumor cells results in the depletion of OX40hi Tregs infiltrating the tumors via OX40L–OX40 ligation, which is promoted by type I IFN. This leads to the blunting of their inhibition on tumor-specific effector CD4+ and CD8+ T cells. Taken together, IT delivery of rMVA results in the alteration of tumor immunosuppressive microenvironment through activation of innate immunity and boosting of antitumor T cells by depletion of OX40hi regulatory T cells.
Our study demonstrates that IT rMVA reduces OX40hi Tregs in injected tumors via the OX40L–OX40 interaction, and this process is promoted by type I IFN in the tumor microenvironment. We found that OX40 is preferentially expressed by IT Tregs and its expression correlates with tumor weight in murine melanoma models. OX40hi Tregs isolated from tumors are more immunosuppressive compared with OX40low Tregs. Therefore, targeting IT OX40hi Tregs by rMVA expressing OX40L is a logical approach to deplete this cell population within the tumors but not in the periphery, thereby improving the efficacy of immunotherapy without unwanted autoimmunity.
Our scRNA-seq analyses of IT Tregs revealed two main populations, cluster 0 (CCR8hi Tregs) and cluster 2 (proliferating Tregs) in PBS-treated tumors, and that IT rMVA alters the Treg subpopulations by depleting cluster 0 (CCR8hi Tregs) and expanding cluster 1 (ISG+ Tregs) and cluster 3 (NKG7+GzmB+ Tregs). Cluster 0 (CCR8hi Tregs) has higher expression of OX40, CD25, and PD1 compared with cluster 2. Therefore, scRNA-seq data confirmed our earlier findings that IT rMVA depletes OX40hi Tregs in the injected tumors.
CCR8+ Tregs have been reported to play important roles in immune suppression in mice and humans (Barsheshet et al., 2017; Coghill et al., 2013; Plitas et al., 2016). Targeting CCR8+ Tregs using an anti-CCR8 antibody showed therapeutic benefits for cancer treatment and cancer vaccines in preclinical models (Villarreal et al., 2018). Similarly, OX40 modulating agents have been explored for enhancing antitumor effects through targeting Tregs. For example, the combination of anti-OX40 agonist antibody with cyclophosphamide triggers activation and apoptosis of IT Tregs while causing Treg expansion in the tumor-draining lymph nodes and spleens (Hirschhorn-Cymerman et al., 2009). This combination also promotes tumor-killing activities of antigen-specific adoptively transferred CD4+ T cells (Hirschhorn-Cymerman et al., 2012). In addition, IT delivery of low doses of anti–CTLA-4 and anti-OX40 antibodies together with TLR9 agonist CpG leads to Treg depletion at the injected site but not in the non-injected site and generates systemic antitumor immunity (Marabelle et al., 2013).
Compared with anti-OX40 agonist antibody approach, our engineered virus expressing OX40L is more specific in targeting OX40hi Tregs within the tumor microenvironment. Type I IFN–inducing ability of the virus also promotes Treg depletion. The combination of IT delivery of rMVA expressing mOX40L with systemic delivery of anti–PD-L1 antibody generates synergistic antitumor effects. This contrasts with two reports showing that concurrent administration of anti-PD1 antibody leads to reduced efficacy of anti-OX40 antibody due to apoptosis of activated T cells (Messenheimer et al., 2017; Shrimali et al., 2017).
Our study has several limitations. For example, we used a murine transplantable B16-F10 melanoma model in most of the in vivo studies. Although the key findings are replicable in MMTV-PyMT, a transgenic mouse TNBC model, we did not perform scRNA-seq of Tregs in the MMTV-PyMT model with or without rMVA treatment. In addition, we were not able to specifically address the role of OX40 on Tregs in this study due to lack of mouse model in which OX40 gene is deleted only in Treg cells. Although our data suggest that IT rMVA treatment leads to depletion of OX40hi Tregs via OX40L–OX40 interaction and type I IFN signaling likely through induction of Treg apoptosis, the detailed molecular mechanism underlying the crosstalk of the two pathways has not been explored. Finally, it would be interesting to examine the effects of rMVA on other immune cell populations including CD8+ and CD4+ T cells and tumor-infiltrating myeloid cells using scRNA-seq approaches in future studies.
There have been some setbacks in the clinical application of OVs in recent years, which could be due to the following reasons: (i) most of the OVs in late-stage clinical trials were designed decades ago with relatively limited capacity of immune modulation; (ii) viral-induced oncolysis was viewed as a major mechanism of tumor control at the time when the early OVs were designed; and (iii) many preclinical studies for these OVs were done in immune-deficient xenograft models. In recent years, with a better understanding of tumor immune-suppressive microenvironment, new generations of OVs have been designed to modulate the tumor microenvironment and turn cold tumors into hot tumors. These OVs are still in the early stages of clinical trials. The advantages of non-replicative viral-based therapy include: (i) it provides more potent innate immune activation, especially higher levels of type I IFN induction; (ii) it is safer in immune-compromised patient populations; and (iii) it provides strong transgene expression despite of being non-replicative.
In summary, our study revealed that IT delivery of engineered rMVA can lead to alteration of immunosuppressive tumor microenvironment through the induction of innate immunity and selective depletion of tumor-infiltrating Tregs. As MVA has been widely used in humans during smallpox eradication campaign with excellent safety records, we expect that the immune-activating rMVA can be safely administered IT in patients with injectable solid tumors. A first-in-human phase I clinical trial to evaluate the safety and tolerability of escalating doses of rhMVA alone and in combination with systemic checkpoint inhibitors in solid tumors will be initiated in the near future.
Materials and methods
Study design
The primary objectives of our study were (i) to engineer an immune-activating rMVA to enhance systemic antitumor immunity through IT delivery of the virus either alone or in combination with systemic delivery of immune checkpoint blockade for solid tumors and (ii) to elucidate the mechanism of action of rMVA in reprogramming the immunosuppressive tumor microenvironment. We constructed a series of rMVA viruses and performed immunological analyses and survival studies to determine the antitumor immunity induced by the viruses. Based on those results, we selected the best performing rMVA with deletion of the E5R gene and expression of two transgenes, hFlt3L and mOX40L, for more detailed studies in mice. Both unilateral and bilateral tumor implantation models in immune-competent syngeneic mice were used to assess the antitumor efficacy and immune responses induced by IT delivery of rMVA. The antitumor effects were assessed using a murine B16-F10 melanoma and MMTV-PyMT transgenic TNBC models in C57BL/6J background and an A20 B cell lymphoma model in BALB/c mice. Furthermore, to assess the contribution of the cGAS/STING-mediated cytosolic DNA-sensing pathway and type I IFN signaling pathway in rMVA-induced antitumor effects, cGAS-, STING-, STAT1-, and STAT2-deficient mice were used in comparison with WT mice. Lastly, we focused on elucidating the mechanisms and biological significance of rMVA-mediated Treg depletion and reprogramming using scRNA-seq and genetically engineered mouse strains, including Ox40−/−, Foxp3gfp, Foxp3DTR, and Foxp3cre-IFNAR1fl/fl mice.
Animals were assigned to various experimental groups at random. For survival studies, sample sizes of 8–10 mice were used, and the experiments were performed at least twice. For experiments designed to evaluate the tumor immune cell infiltrates, three to five mice were used for each experiment and the experiments were performed at least two to three times.
To explore the translational potential of our study, we engineered the version suitable for human use, rhMVA (MVA∆E5R-hFlt3L-hOX40L), and performed infection of rhMVA in human monocyte-derived DCs and tumor samples ex vivo.
Cell lines
BHK-21 (baby hamster kidney cell, ATCC CCL-10) cells were cultured in Eagle’s minimal essential medium containing 10% FBS, 0.1 mM nonessential amino acids, penicillin, and streptomycin. The murine melanoma cell line B16-F10 was originally obtained from I. Fidler (MD Anderson Cancer Center). The A20 B cell lymphoma cell line was obtained from ATCC. Both B16-F10 and A20 were maintained in RPMI-1640 medium supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol, penicillin, and streptomycin.
B16 cell line expressing mOX40L or hFlt3L was created by transduction into B16 cells with vesicular stomatitis virus G protein-pseudotyped murine leukemia viruses containing pQCXIP-mOX40L or pQCXIP-hFlt3L. Cells were selected and maintained in growth media including 2 μg/ml puromycin for selection of stably transduced cells.
Viruses
The MVA virus was provided by G. Sutter (University of Munich, Munich, Germany). MVAΔE5R, MVAΔE5R-hFlt3L, MVAΔE5R-mOX40L, rMVA (MVA∆E5R-hFlt3L-mOX40L), and rhMVA (MVA∆E5R-hFlt3L-hOX40L) were generated by transfecting pUC57-based plasmids into BHK-21 cells that were infected with MVA at MOI 0.05. Recombinant viruses were purified after four to six rounds of plaque selection based on the fluorescence marker. Viruses were propagated in BHK-21 cells and purified through a 36% sucrose cushion. PCR and DNA sequencing were performed to verify the purity of the recombinant viruses. Viral titers were determined using BHK-21 cells.
Mice
Female C57BL/6J mice and BALB/cJ between 6 and 10 wk of age were purchased from the Jackson Laboratory (stock #000664 and stock #000651) and used for the preparation of BMDCs and for in vivo experiments. These mice were maintained in the animal facility at the Sloan Kettering Institute. All procedures were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of Sloan Kettering Cancer Institute. Stinggt/gt mice were generated in the laboratory of Dr. Russell Vance (University of California, Berkeley, Berkeley, CA, USA; Sauer et al., 2011). Foxp3gfp, Foxp3DTR, and Foxp3YFP-cre mice were generated in the laboratory of Dr. Alexander Y. Rudensky (Memorial Sloan Kettering Cancer Center; Fontenot et al., 2005; Kim et al., 2007; Rubtsov et al., 2008). MMTV-PyMT mice were provided by Ming Li (Memorial Sloan Kettering Cancer Center; Franklin et al., 2014). cGas−/− mice were generated in Herbert (Skip) Virgin’s laboratory (Washington University, St. Louis, MO, USA; Schoggins et al., 2014). Ifnar1fl/fl, Stat1−/−, Stat2−/−, and Ox40−/− were purchased from Jackson Laboratory. OX40−/−Foxp3gfp and Foxp3creIfnar1fl/fl mice were bred in our lab.
TIL isolation and flow cytometry
For TIL or myeloid cells analysis, tumors were minced prior to incubation with Liberase (1.67 Wünsch U/ml) and DNaseI (0.2 mg/ml) for 30 min at 37°C. Tumors were then homogenized by gentle MACS dissociator and filtered through a 70-μm nylon filter. Cell suspensions were washed and resuspended with complete RPMI. For cytokine production analysis, cells were restimulated with Cell Stimulation Cocktail (Thermo Fisher Scientific) and GolgiPlug (BD Biosciences) in complete RPMI for 6 h at 37°C. Cells were incubated with appropriate antibodies for surface labeling for 30 min at 4°C after staining dead cells with LIVE/DEAD Fixable Aqua Stain (Thermo Fisher Scientific). Cells were fixed and permeabilized using Foxp3 fixation and permeabilization kit (Thermo Fisher Scientific) for 1 h at 4°C and then stained for Granzyme B, Foxp3, IFNγ, and TNFα.
To analyze transgene expression, cells were infected with various viruses at an MOI of 10 or mock-infected. At 24 h after infection, cells were collected and the cell viability was determined by labeling with LIVE/DEAD Fixable Aqua Stain (Thermo Fisher Scientific) 15 min at 4°C. Cells were then sequentially stained with hFlt3L primary antibody, PE-conjugated goat-anti-mouse IgG antibody, and AF647-conjugated anti-mOX40L antibody at 4°C, 15 min for each step.
For DC maturation assay, cells were infected with virus at an MOI of 10 and collected at 16 h after infection. Then cells were stained with anti-CD86 antibody for surface labeling for 30 min at 4°C. LIVE/DEAD Fixable Aqua Stain (Thermo Fisher Scientific) was used to stain dead cells. Cells were analyzed using the BD LSRFortessa flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Treestar).
RNA isolation and real-time PCR
For the generation of BMDCs, the bone marrow cells (5 million cells in each 15 cm cell culture dish) were cultured in RPMI-1640 medium supplemented with 10% FBS in the presence of 30 ng/ml GM-CSF (BioLegend) for 10–12 d.
To generate human monocyte-derived DCs, peripheral blood mononuclear cells were prepared by centrifugation on a Ficoll gradient. Monocytes layer was collected and plated onto a tissue culture dish. After 1 h, non-adherent cells were washed off. The remaining cells were cultured for 5–7 d in RPMI-1640 supplemented with antibiotics (penicillin and streptomycin) and 10% FCS in the presence of 1,000 IU/ml GM-CSF (PeproTech) and 500 IU/ml IL-4 (PeproTech).
Cells were infected with various viruses at an MOI of 10 for 1 h or mock-infected. The inoculum was removed and the cells were washed with PBS twice and incubated with fresh medium. RNA was extracted from whole-cell lysates with RNeasy Plus Mini kit (Qiagen) and reverse-transcribed with cDNA synthesis kit (Thermo Fisher Scientific). Real-time PCR was performed in triplicate with SYBR Green PCR Master Mix (Life Technologies) and Applied Biosystems 7500 Real-time PCR Instrument (Life Technologies) using gene-specific primers. Relative expression was normalized to the levels of GAPDH. The primer sequences for quantitative real-time PCR are listed in Table S1.
Tumor challenge and treatment
For tumor immune cells analysis, B16-F10 cells were implanted intradermally into right and left flanks of the mice (5 × 105 to the right flank and 2.5 × 105 to the left flank). At 7–9 d after implantation, the tumors at the right flank were injected with 4 × 107 PFU of rMVA (MVA∆E5RhFlt3L-mOX40L), MVAΔE5R, or PBS twice, 2 or 3 d apart. Tumors, spleens, and/or tumor-draining lymph nodes were harvested 2 d after second injection. In some experiments, 50 μg of αIFNAR-1 antibody (MAR1-5A3, BioXcell) were injected into the tumors together with rMVA.
For survival experiments, 2 × 105 B16-F10 cells were implanted intradermally into the shaved skin on the right flank of WT C57BL/6J mice or age-matched cGas−/−, Stinggt/gt, Stat2−/−, and Stat1−/− mice. In some experiments, 2 × 105 A20 cells were implanted intradermally into the right flank of WT BALB/cJ mice. At 6–9 d after implantation, tumor sizes were measured and tumors that were 3 mm in diameter or larger were injected with 4 × 107 PFU of rMVA or PBS when the mice were under anesthesia. Viruses were injected twice weekly as specified in each experiment and tumor sizes were measured twice a week. Tumor volumes were calculated according to the following formula: l (length) × w (width) × h (height)/2. Mice were euthanized for signs of distress or when the diameter of the tumor reached 10 mm. For depletion of T cells, depletion antibodies for CD8+ and CD4+ cells (200 μg of clone 2.43 and GK1.5, BioXcell) were injected IP twice weekly starting 1 d before viral injection, and they were used until the animals either died, were euthanized, or were completely clear of tumors.
For depletion of Tregs, 2 × 105 B16-F10 cells were implanted intradermally into the shaved skin on the right flank of Foxp3DTR mice. Three doses of DT (200 ng each per mouse) were administered to tumor-bearing mice at −2, −1, and +1 d relative to IT MVA∆E5R or PBS injection at day 0. MVA∆E5R was injected IT twice weekly.
In the bilateral tumor implantation model, B16-F10 cells were implanted intradermally into right and left flanks of C57BL/6J mice (5 × 105 to the right flank and 1 × 105 to the left flank). At 7 d after implantation, the tumors at the right flank were injected with 4 × 107 PFU of rMVA (MVA∆E5R-hFlt3L-mOX40L) or PBS. 250 μg αPD-L1 antibody (10F.9G2; BioXcell) was injected IP twice weekly.
For the tumor rechallenge study, the survived mice (more than 40 d after initiation of IT virotherapy) were rechallenged with intradermal delivery of a lethal dose of B16-F10 (1 × 105 cells) at the contralateral side.
ELISpot assay
Spleens were mechanically disrupted by gentleMACS dissociator, and red blood cells were lysed by ACK lysing buffer. 1 × 106 splenocytes were cocultured with 2.5 × 105 irradiated B16-F10 in complete RPMI medium overnight. IFNγ+ splenocytes were detected by Mouse IFNγ ELISPOT kit (BD Biosciences)
In vitro Treg suppression assay
5 × 105 WT B16-F10 cells were implanted intradermally into the right and left flanks of Foxp3gfp and OX40−/−Foxp3gfp mice. Tumors and spleens were harvested when tumor sizes reached 5 mm in diameter or larger and processed into single-cell suspensions as described above. Cells were stained with anti-CD45.2 (AF700), CD4 (Pacific Blue), and OX40 (PE) antibodies for 30 min at 4°C. LIVE/DEAD Fixable Aqua Stain (Thermo Fisher Scientific) was used to stain dead cells. Treg cells were sorted into CD4+GFP+OX40hi and CD4+GFP+OX40lo populations. Naive CD4+ T cells were sorted from mouse spleen. Spleens from CD45.1 congenic mouse were harvested, chopped, and digested with Collagenase D (2.5 mg/ml) and DNaseI (50 μg/ml) for 30 min at 37°C. CD11c+ DCs were then isolated by CD11c MicroBeads (Miltenyi Biotec). CellTrace Violet–labeled 4 × 104 native CD4+ T cells were cocultured with 1 × 105 CD11c+ DCs. Purified CD4+GFP+OX40hi, CD4+GFP+OX40lo, or CD4+GFP+OX40−/− cells were seeded in indicated ratios and cultured in complete RPMI supplemented with 1 μg/ml anti-CD3 antibody for 3 d. Cell were then collected and stained with APC anti-CD4 antibody for 30 min at 4°C after staining dead cells with Zombie Nir. Data were acquired using the BD LSRFortessa flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Tree Star).
Bulk tumor RNA-seq
5 × 105 WT B16-F10 cells were implanted intradermally into right and left flanks of WT C57BL/6J or age-matched STINGGt/Gt mice. At 7 d after implantation, the tumors were injected with 4 × 107 PFU of rMVA or PBS. Tumors were harvested 1 d after injection and processed into a single-cell suspension as described previously. RNA was extracted from whole-cell lysates with RNeasy Plus Mini kit (Qiagen) for RNA-seq.
Library prep and sequencing: Following RNA isolation, total RNA integrity was checked using a 2100 Bioanalyzer (Agilent Technologies). RNA concentrations were measured using the NanoDrop system (Thermo Fisher Scientific, Inc). Preparation of RNA sample library and RNA-seq was performed by the Genomics Core Laboratory at Weill Cornell Medicine. Messenger RNA was prepared using TruSeq Stranded mRNA Sample Library Preparation kit (Illumina), according to the manufacturer’s instructions. The normalized cDNA libraries were pooled and sequenced on Illumina NovaSeq6000 sequencer with pair-end 50 cycles.
scRNA-seq and data analysis
Library preparation and sequencing
5 × 105 WT B16-F10 cells were implanted intradermally into right flank of Foxp3GFP mice. When tumor diameter reached 5 mm, the tumors were injected twice on two consecutive days with 4 × 107 PFU of rMVA in each injection. 2 d after second injection, mice were euthanized and tumors were harvested and processed into single-cell suspensions as described before. CD4+GFP+ populations from tumors were purified by FACS sorting. We performed single-cell 5′ gene expression profiling on the single-cell suspension using the Chromium Single Cell V(D)J Solution from 10× Genomics according to the manufacturer’s instructions. Cell-barcoded 5′ gene expression libraries were sequenced on an Illumina NovaSeq6000 system.
Data preprocessing and clustering
Fastq files were processed using Cell Ranger v3.0 (10X Genomics). Reads were aligned to the mouse genome mm10 from ENSEMBL GRCm38 and analyzed using the Seurat R package (v4.0.4; Stuart et al., 2019). All cells expressing <200 or >6,000 genes were removed as well as cells that contained >5% mitochondrial counts. Samples were merged and normalized. The default parameters of Seurat were used unless mentioned otherwise. Briefly, 2,000 variable genes were identified for the clustering of all cell types, and principal component analysis was applied to the dataset to reduce dimensionality after regressing for the number of unique molecular identifiers. The 20 most informative principal components were used for clustering and Uniform Manifold Approximation and Projection for dimension reduction (UMAP). To characterize each cluster, we applied both the FindMarkers procedure in Seurat, which identified markers using log fold changes (FC) of mean expression. To identify differentially expressed genes between two groups of clusters, FindMarkers functions in Seurat and Enhanced Volcano R package (v1.8.0) were used with a cutoff for log2FC as 1 and cutoff for P value as 10e−32.
Human tumor specimens
Fresh biopsy samples from patients with Extramammary Paget’s disease were obtained at the dermatology service in the Department of Medicine of Memorial Sloan Kettering Cancer Center. Written informed consent was obtained from patients enrolled in the protocol approved by Memorial Sloan Kettering Cancer Center Institutional Review Board. Studies were conducted in accordance with National Institutes of Health and institutional guidelines for human subject research. Tumor tissues were cut into small pieces using a pair of fine scissors. They were infected with rhMVA (MVA∆E5R-hFlt3L-hOX40L) or mock-infected. Cells were collected after 24 h and processed for FACS analyses.
Statistical analysis
Two-tailed unpaired Student’s t test was used for comparisons of two groups in the studies. Survival data were analyzed by log-rank (Mantel-Cox) test. The P values deemed significant are indicated in the figures as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. The numbers of animals included in the study are discussed in each figure legend.
Online supplemental material
Fig. S1 shows mice implanted with B16-F10 murine melanoma cell line overexpressing hFlt3L or mOX40L had longer survival compared with those implanted with the parental cell line. Incremental engineering of MVA by deleting E5R gene and expressing hFlt3L or mOX40L improves antitumor effects. Fig. S2 demonstrates that IT delivery of rMVA (MVA∆E5R-hFlt3L-mOX40L) is more efficacious in eradicating tumors than IV delivery of the virus. It also shows that IT rMVA resulted in the recruitment of neutrophils and monocytes into the injected tumors. IT rMVA (MVA∆E5R-hFlt3L-mOX40L) is more effective than oncolytic VACV∆E5R-hFlt3L-mOX40L in restricting tumor growth. Fig. S3 shows that IT Tregs expressed higher levels of OX40 compared with Tregs in lymphoid organs. It also shows OX40L expression in tumors and tumor-infiltrating immune cells after IT rMVA. Fig. S4 shows single-cell transcriptomic analysis of Tregs in the tumors treated with rMVA or PBS as a control, demonstrating that IT rMVA resulted in the emergence of ISG+ Tregs expressing GzmB and two additional GzmB+ Tregs. Fig. S5 shows the design of clinical candidate rhMVA (MVA∆E5R-hFlt3L-hOX40L) and characterization of transgene expression in infected human melanoma cells and immune-activating functions in human monocyte-derived DCs. Table S1 shows the primer sequences used for quantitative real-time PCR analyses for selected mouse and human genes.
Lead contact
Further information and requests for resources and reagents used in this study should be directed to and will be fulfilled by the lead contact and the corresponding author, Liang Deng (dengl@mskcc.org).
Materials availability
Materials generated in our laboratory are available upon request.
Supplementary Material
shows the primer sequences used for quantitative real-time PCR analyses for selected mouse and human genes.
Acknowledgments
We thank the Flow Cytometry Core Facility and Molecular Cytology Core Facility at the Sloan Kettering Institute and Genomic core at Weill Cornell Medical College. We thank Ming O. Li (Sloan Kettering Institute) for review of the manuscript.
This work was supported by National Institutes of Health grants K-08 AI073736 (L. Deng), R56AI095692 (L. Deng), R03 AR068118 (L. Deng), R01 CA56821 (J.D. Wolchok), and R01 AI091707 (C.M. Rice); the Charles H. Revson Senior Fellowship in Biomedical Science (J.M. Luna); Black Family Metastasis Center Fellowship (J.M. Luna); Society of Memorial Sloan Kettering research grant (L. Deng); Memorial Sloan Kettering Technology Development Fund (L. Deng); Parker Institute for Cancer Immunotherapy Career Development Award (L. Deng); Cycle for Survival grant (L. Deng); and sponsored research agreement from IMVAQ Therapeutics (L. Deng). This work was supported in part by Swim Across America (J.D. Wolchok, T. Merghoub) and Ludwig Institute for Cancer Research (J.D. Wolchok, T. Merghoub). This research was also funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Author contributions: N. Yang, Y. Wang, and L. Deng were involved in all aspects of this study, including conceiving the project, designing and performing experiments, data analyses and interpretation, and manuscript writing. S. Liu and S.B. Tariq assisted some mouse experiments and human tumor ex vivo infection experiments and analyzed the data. G. Mazo, J. Wang, and W. Yan assisted with construct designs and viral engineering. A. Tan and T. Zhang analyzed the bulk RNA-seq data from tumors. A. Rossi provided human tumor samples. J. Choi contributed to resources. J.D. Wolchok, T. Merghoub, C.M. Rice, J. Luna, and J.Z. Xiang assisted in experimental design, data interpretation, and manuscript preparation. All authors are involved in manuscript preparation. L. Deng provided overall supervision of the study.
Data availability
The RNA-seq data reported in this study have been deposited in the Gene Expression Omnibus database under the accession numbers: GSE188496 and GSE192563.
References
- Antoine, G., Scheiflinger F., Dorner F., and Falkner F.G.. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology. 244:365–396. 10.1006/viro.1998.9123 [DOI] [PubMed] [Google Scholar]
- Barsheshet, Y., Wildbaum G., Levy E., Vitenshtein A., Akinseye C., Griggs J., Lira S.A., and Karin N.. 2017. CCR8+FOXp3+ Treg cells as master drivers of immune regulation. Proc. Natl. Acad. Sci. USA. 114:6086–6091. 10.1073/pnas.1621280114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy, P.K., Shettigar M., and Kaufman H.L.. 2018. Integrating oncolytic viruses in combination cancer immunotherapy. Nat. Rev. Immunol. 18:498–513. 10.1038/s41577-018-0014-6 [DOI] [PubMed] [Google Scholar]
- Bos, P.D., Plitas G., Rudra D., Lee S.Y., and Rudensky A.Y.. 2013. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J. Exp. Med. 210:2435–2466. 10.1084/jem.20130762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A.I., McAdam A.J., Buhlmann J.E., Scott S., Lupher M.L. Jr, Greenfield E.A., Baum P.R., Fanslow W.C., Calderhead D.M., Freeman G.J., and Sharpe A.H.. 1999. Ox40-ligand has a critical costimulatory role in dendritic cell:T cell interactions. Immunity. 11:689–698. 10.1016/S1074-7613(00)80143-0 [DOI] [PubMed] [Google Scholar]
- Chesney, J., Puzanov I., Collichio F., Singh P., Milhem M.M., Glaspy J., Hamid O., Ross M., Friedlander P., Garbe C., et al. 2018. Randomized, open-label phase II study evaluating the efficacy and safety of talimogene laherparepvec in combination with ipilimumab versus ipilimumab alone in patients with advanced, unresectable melanoma. J. Clin. Oncol. 36:1658–1667. 10.1200/JCO.2017.73.7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill, J.M., Fowler K.A., West M.L., Fulton L.M., van Deventer H., McKinnon K.P., Vincent B.G., Lin K., Panoskaltsis-Mortari A., Cook D.N., et al. 2013. CC chemokine receptor 8 potentiates donor Treg survival and is critical for the prevention of murine graft-versus-host disease. Blood. 122:825–836. 10.1182/blood-2012-06-435735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, M. 2009. The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol. 9:271–285. 10.1038/nri2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft, M., So T., Duan W., and Soroosh P.. 2009. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol. Rev. 229:173–191. 10.1111/j.1600-065X.2009.00766.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, P., Wang W., Cao H., Avogadri F., Dai L., Drexler I., Joyce J.A., Li X.D., Chen Z., Merghoub T., et al. 2014. Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog. 10:e1003989. 10.1371/journal.ppat.1003989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, P., Wang W., Yang N., Serna-Tamayo C., Ricca J.M., Zamarin D., Shuman S., Merghoub T., Wolchok J.D., and Deng L.. 2017. Intratumoral delivery of inactivated modified vaccinia virus Ankara (iMVA) induces systemic antitumor immunity via STING and Batf3-dependent dendritic cells. Sci. Immunol. 2:eaal1713. 10.1126/sciimmunol.aal1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davola, M.E., and Mossman K.L.. 2019. Oncolytic viruses: How “lytic” must they be for therapeutic efficacy?. OncoImmunology. 8:e1581528. 10.1080/2162402X.2019.1596006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot, J.D., Rasmussen J.P., Williams L.M., Dooley J.L., Farr A.G., and Rudensky A.Y.. 2005. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 22:329–341. 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Franklin, R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., and Li M.O.. 2014. The cellular and molecular origin of tumor-associated macrophages. Science. 344:921–925. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, Y., Wang H., Ren J., Liu W., Chen L., Chen H., Ye J., Dai E., Ma C., Ju S., et al. 2020. Oncolytic vaccinia virus delivering tethered IL-12 enhances antitumor effects with improved safety. J. Immunother. Cancer. 8:e000710. 10.1136/jitc-2020-000710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, S.C. 2013. Clinical development of modified vaccinia virus Ankara vaccines. Vaccine. 31:4241–4246. 10.1016/j.vaccine.2013.03.020 [DOI] [PubMed] [Google Scholar]
- Hirschhorn-Cymerman, D., Budhu S., Kitano S., Liu C., Zhao F., Zhong H., Lesokhin A.M., Avogadri-Connors F., Yuan J., Li Y., et al. 2012. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J. Exp. Med. 209:2113–2126. 10.1084/jem.20120532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn-Cymerman, D., Rizzuto G.A., Merghoub T., Cohen A.D., Avogadri F., Lesokhin A.M., Weinberg A.D., Wolchok J.D., and Houghton A.N.. 2009. OX40 engagement and chemotherapy combination provides potent antitumor immunity with concomitant regulatory T cell apoptosis. J. Exp. Med. 206:1103–1116. 10.1084/jem.20082205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Wang Y.H., Duramad O., Hanabuchi S., Perng O.A., Gilliet M., Qin F.X., and Liu Y.J.. 2006. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc. Natl. Acad. Sci. USA. 103:13138–13143. 10.1073/pnas.0603107103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Kirn, D.H., Wang Y., Le Boeuf F., Bell J., and Thorne S.H.. 2007. Targeting of interferon-beta to produce a specific, multi-mechanistic oncolytic vaccinia virus. PLoS Med. 4:e353. 10.1371/journal.pmed.0040353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages, K., Mayer C.T., Lahl K., Loddenkemper C., Teng M.W., Ngiow S.F., Smyth M.J., Hamann A., Huehn J., and Sparwasser T.. 2010. Selective depletion of Foxp3+ regulatory T cells improves effective therapeutic vaccination against established melanoma. Cancer Res. 70:7788–7799. 10.1158/0008-5472.CAN-10-1736 [DOI] [PubMed] [Google Scholar]
- Kowalsky, S.J., Liu Z., Feist M., Berkey S.E., Ma C., Ravindranathan R., Dai E., Roy E.J., Guo Z.S., and Bartlett D.L.. 2018. Superagonist IL-15-armed oncolytic virus elicits potent antitumor immunity and therapy that are enhanced with PD-1 blockade. Mol. Ther. 26:2476–2486. 10.1016/j.ymthe.2018.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos de Matos, A., Franco L.S., and McFadden G.. 2020. Oncolytic viruses and the immune system: The dynamic duo. Mol. Ther. Methods Clin. Dev. 17:349–358. 10.1016/j.omtm.2020.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., and Nussenzweig M.C.. 2010. Origin and development of dendritic cells. Immunol. Rev. 234:45–54. 10.1111/j.0105-2896.2009.00879.x [DOI] [PubMed] [Google Scholar]
- Liu, R., Americo J.L., Cotter C.A., Earl P.L., Erez N., Peng C., and Moss B.. 2021. One or two injections of MVA-vectored vaccine shields hACE2 transgenic mice from SARS-CoV-2 upper and lower respiratory tract infection. Proc. Natl. Acad. Sci. USA. 118:e2026785118. 10.1073/pnas.2026785118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Dai E., Liu Z., Ma C., Guo Z.S., and Bartlett D.L.. 2020.In situ therapeutic cancer vaccination with an oncolytic virus expressing membrane-tethered IL-2. Mol. Ther. Oncolytics. 17:350–360. 10.1016/j.omto.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabelle, A., Kohrt H., Sagiv-Barfi I., Ajami B., Axtell R.C., Zhou G., Rajapaksa R., Green M.R., Torchia J., Brody J., et al. 2013. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J. Clin. Invest. 123:2447–2463. 10.1172/JCI64859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenheimer, D.J., Jensen S.M., Afentoulis M.E., Wegmann K.W., Feng Z., Friedman D.J., Gough M.J., Urba W.J., and Fox B.A.. 2017. Timing of PD-1 blockade is critical to effective combination immunotherapy with anti-OX40. Clin. Cancer Res. 23:6165–6177. 10.1158/1078-0432.CCR-16-2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, K., Ishii N., Takano H., Miura S., Ndhlovu L.C., Nose M., Noda T., and Sugamura K.. 2000. Impairment of antigen-presenting cell function in mice lacking expression of OX40 ligand. J. Exp. Med. 191:365–374. 10.1084/jem.191.2.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao, S., Arai Y., Tasaki M., Yamashita M., Murakami R., Kawase T., Amino N., Nakatake M., Kurosaki H., Mori M., et al. 2020. Intratumoral expression of IL-7 and IL-12 using an oncolytic virus increases systemic sensitivity to immune checkpoint blockade. Sci. Transl. Med. 12:eaax7992. 10.1126/scitranslmed.aax7992 [DOI] [PubMed] [Google Scholar]
- Plitas, G., Konopacki C., Wu K., Bos P.D., Morrow M., Putintseva E.V., Chudakov D.M., and Rudensky A.Y.. 2016. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 45:1122–1134. 10.1016/j.immuni.2016.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitas, G., and Rudensky A.Y.. 2020. Regulatory T cells in cancer. Annu. Rev. Cancer Biol. 4:459–477. 10.1146/annurev-cancerbio-030419-033428 [DOI] [Google Scholar]
- Ribas, A., Dummer R., Puzanov I., VanderWalde A., Andtbacka R.H.I., Michielin O., Olszanski A.J., Malvehy J., Cebon J., Fernandez E., et al. 2018. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell. 174:1031–1032. 10.1016/j.cell.2018.07.035 [DOI] [PubMed] [Google Scholar]
- Ribas, A., and Wolchok J.D.. 2018. Cancer immunotherapy using checkpoint blockade. Science. 359:1350–1355. 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov, Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr, et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28:546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Russell, S.J., Peng K.W., and Bell J.C.. 2012. Oncolytic virotherapy. Nat. Biotechnol. 30:658–670. 10.1038/nbt.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, S., Mikami N., Wing J.B., Tanaka A., Ichiyama K., and Ohkura N.. 2020. Regulatory T cells and human disease. Annu. Rev. Immunol. 38:541–566. 10.1146/annurev-immunol-042718-041717 [DOI] [PubMed] [Google Scholar]
- Sauer, J.D., Sotelo-Troha K., von Moltke J., Monroe K.M., Rae C.S., Brubaker S.W., Hyodo M., Hayakawa Y., Woodward J.J., Portnoy D.A., and Vance R.E.. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79:688–694. 10.1128/IAI.00999-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins, J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., et al. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 505:691–695. 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, R.D., Old L.J., and Smyth M.J.. 2011. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science. 331:1565–1570. 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- Semmrich, M., Marchand J.B., Fend L., Rehn M., Remy C., Holmkvist P., Silvestre N., Svensson C., Kleinpeter P., Deforges J., et al. 2022. Vectorized Treg-depleting αCTLA-4 elicits antigen cross-presentation and CD8+ T cell immunity to reject ‘cold’ tumors. J. Immunother. Cancer. 10:10. 10.1136/jitc-2021-003488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, J., Yamazaki S., and Sakassguchi S.. 1999. Induction of tumor immunity by removing CD25+CD4+ T cells: A common basis between tumor immunity and autoimmunity. J. Immunol. 163:5211–5218. 10.4049/jimmunol.163.10.5211 [DOI] [PubMed] [Google Scholar]
- Shrimali, R.K., Ahmad S., Verma V., Zeng P., Ananth S., Gaur P., Gittelman R.M., Yusko E., Sanders C., Robins H., et al. 2017. Concurrent PD-1 blockade negates the effects of OX40 agonist antibody in combination immunotherapy through inducing T-cell apoptosis. Cancer Immunol. Res. 5:755–766. 10.1158/2326-6066.CIR-17-0292 [DOI] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., W.M. Mauck, 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902 e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons, J.A., Alcamí A., and Smith G.L.. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 81:551–560. 10.1016/0092-8674(95)90076-4 [DOI] [PubMed] [Google Scholar]
- Villarreal, D.O., L’Huillier A., Armington S., Mottershead C., Filippova E.V., Coder B.D., Petit R.G., and Princiotta M.F.. 2018. Targeting CCR8 induces protective antitumor immunity and enhances vaccine-induced responses in colon cancer. Cancer Res. 78:5340–5348. 10.1158/0008-5472.CAN-18-1119 [DOI] [PubMed] [Google Scholar]
- Volz, A., and Sutter G.. 2017. Modified vaccinia virus Ankara: History, value in basic research, and current perspectives for vaccine development. Adv. Virus Res. 97:187–243. 10.1016/bs.aivir.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Liu S., Dai P., Yang N., Wang Y., Giese R.A., Merghoub T., Wolchok J., and Deng L.. 2021. Elucidating mechanisms of antitumor immunity mediated by live oncolytic vaccinia and heat-inactivated vaccinia. J. Immunother. Cancer. 9:e002569. 10.1136/jitc-2021-002569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, S.C., Duffy C.R., and Allison J.P.. 2018. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8:1069–1086. 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- Workenhe, S.T., and Mossman K.L.. 2014. Oncolytic virotherapy and immunogenic cancer cell death: Sharpening the sword for improved cancer treatment strategies. Mol. Ther. 22:251–256. 10.1038/mt.2013.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., Wang Y., Dai P., Li T., Zierhut C., Tan A., Zhang T., Pan H., Li Z., Ordureau A., et al. 2021. Vaccinia E5 is a major inhibitor of the DNA sensor cGAS. bioRxiv. 10.1101/2021.10.25.465197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin, D., Holmgaard R.B., Subudhi S.K., Park J.S., Mansour M., Palese P., Merghoub T., Wolchok J.D., and Allison J.P.. 2014. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci. Transl. Med. 6:226ra32. 10.1126/scitranslmed.3008095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky, J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. 2016. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375:819–829. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, W., Wolchok J.D., and Chen L.. 2016. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8:328rv4. 10.1126/scitranslmed.aad7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the primer sequences used for quantitative real-time PCR analyses for selected mouse and human genes.
Data Availability Statement
The RNA-seq data reported in this study have been deposited in the Gene Expression Omnibus database under the accession numbers: GSE188496 and GSE192563.