Abstract
Pancreatic cancer is one of the most lethal cancers worldwide, most notably in Europe and North America. Great strides have been made in combining the most effective conventional therapies to improve survival at least in the short and medium term. The start of treatment can only be made once a diagnosis is made, which at this point, the tumor volume is already very high in the primary cancer and systemically. If caught at the earliest opportunity (in circa 20% patients) surgical resection of the primary followed by combination chemotherapy can achieve 5-year overall survival rates of 30%–50%. A delay in detection of even a few months after symptom onset will result in the tumor having only borderline resectabilty (in 20%–30% of patients), in which case the best survival is achieved by using short-course chemotherapy before tumor resection as well as adjuvant chemotherapy. Once metastases become visible (in 40%–60% of patients), cure is not possible, palliative cytotoxics only being able to prolong life by few months. Even in apparently successful therapy in resected and borderline resectable patients, the recurrence rate is very high. Considerable efforts to understand the nature of pancreatic cancer through large-scale genomics, transcriptomics, and digital profiling, combined with functional preclinical models, using genetically engineered mouse models and patient derived organoids, have identified the critical role of the tumor microenvironment in determining the nature of chemo- and immuno-resistance. This functional understanding has powered fresh and exciting approaches for the treatment of this cancer.
Keywords: molecular subtypes, clonal evolution, plasticity, CYPY3A, targeted therapies, immunotherapy, persister
Graphical Abstract
Graphical Abstract.
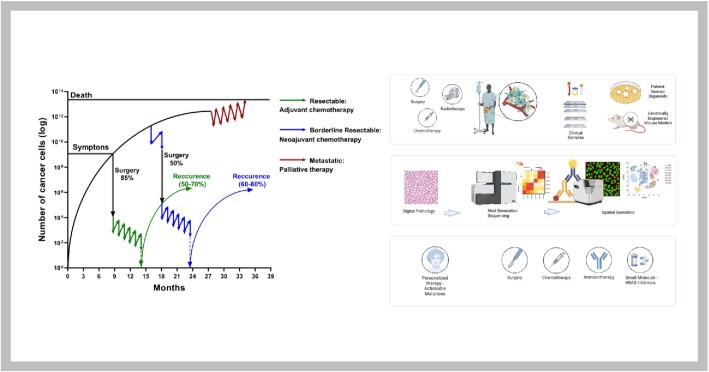
The current and emerging status of pancreatic cancer therapy. The left-hand panel shows a schematic representation of the estimated response rates to therapies based on tumor cell mass (the primary tumor and metastasis combined) and time assuming Gomp-Ex tumor cell growth rates. (A mathematical sigmoid function which describes growth as being slowest at the start and end of a given time period was proposed by Benjamin Gompertz in 1825 [Philosophical Transactions of the Royal Society of London]. This was modified for tumor cell growth by TE Wheldon in 1988 [Mathematical Models in Cancer Research], in which the cellular population expands exponentially [Gomp-Ex] on the assumption that initially there is no competition for resources. More complex models are needed to describe metastatic seeding and growth.) The start of treatment can only be made once a diagnosis is made, which at this point the tumor volume is already high in the primary cancer and in micrometastases around the body. If caught at the earliest opportunity (in circa 20% patients) surgical resection of the primary followed by combination chemotherapy can achieve 5-year overall survival rates of 30%–50%. A delay in detection of even a few months after symptom onset will result in the tumor having only borderline resectabilty (in 20%–30% of patients), in which case the best survival is achieved by using short-course chemotherapy before tumor resection as well as adjuvant chemotherapy. Once metastases become visible (in 40%–60% of patients), cure is not possible, palliative cytotoxics only being able to prolong life by a few months. The right-hand panel demonstrates the multiple approaches being undertaken to deepen the understanding of the nature of pancreatic cancer and its responses to different therapies. There is much to be gained from functional preclinical models (genetically engineered mouse models and patient-derived organoids), but the emphasis has shifted to deeper investigations of patient tumors within the context of clever multidisciplinary prospective clinical studies.
Introduction
Pancreatic cancer—pancreatic ductal adenocarcinoma (PDAC) is a highly lethal cancer with a mortality ranking in 2020 of seven worldwide, six in Northern Europe and four in the United States.1,2 From the outset, it is an aggressive tumor becoming invasive and metastatic within 11–13 months from detection and highly resistant to all forms of treatment.3,4 The mechanisms underlying this biological phenotype are not sufficiently understood but are being intensively investigated from many angles by different specialist teams around the world. In 2008, in depth sequencing of 24 PDAC tumors revealed an average of 63 genetic alterations, the majority of which are point mutations.5 These alterations defined a core set of 12 cellular signaling pathways and processes that were each genetically altered in 67 to 100% of the tumors. These 12 pathways (with the fraction of tumors with genetic alteration of at least one of the genes) comprised KRAS signaling (100%), DNA damage control (83%), regulation of G1/S phase transition (100%), TGFβ signaling (100%), apoptosis (100%), Hedgehog signaling (100%), homophilic cell adhesion (79%), integrin signaling, c-Jun N-terminal kinase signaling (96%), regulation of invasion (92%), small GTPase-dependent signaling (79%), and Wnt/Notch signaling (100%).5 To these, we can now also add genetic alterations in histone modulation (25%), SWI/SNF ATP-dependent chromatin remodeling complexes (20%), RNA processing (15%), and the ROBO/SLIT pathway (5%) (Table 1).6–9 The most common single gene alterations occur in KRAS (90%), TP53 (70%), CDKN2A (60%), and SMAD4 (40%) whilst TGFBR2, ARID1A (SWI/SNF subunit), KDM6A (histone demethylase), MLL3 (histone H3K4 methyltransferase), RBM10 (RNA-binding motif-10 regulating alternative splicing), BCORL1 (transcriptional corepressor), and ROBO2 (roundabout guidance receptor 2, limiting stromal T-cell infiltration) occur in 5%–10% of tumors.10,11
Table 1.
Pathogenic gene variants occurring in 5% or more pancreatic cancers
| Cellular Signaling Pathways and Processes | Pathogenic Gene Variants |
|---|---|
| KRAS signaling | KRAS, MAP2K4, RASGRP3 |
| DNA damage control | TP53, ERCC4, ERCC6, EP300, RANBP2, BRCA1/2, PALB2, ATM, ATR, MLH1, MSH2, MSH6, RPA1, STK11, FANCA, FANCC, ATF2 |
| Regulation of G1/S phase transition | CDKN2A, CHD1, APC2, FBXW7, |
| TGFβ signaling | SMAD4, SMAD3, TGFBR1, TGFBR2, BMPR2, ACVR1B, ACVR2A |
| Apoptosis | CASP10, VCP, CAD, HIP1 |
| Hedgehog signaling | TBX5, SOX3, LRP2, GLI1, GLI3, BMPR2, CREBBP |
| Homophilic cell adhesion | CDH1, FAT, PCDH15, PCDHB16, PCDHGA1 |
| Integrin signaling | ITGA4, LAMA1, LAMA4, LAMA5 FN1, ILK, |
| c-Jun N-terminal kinase signaling | MAP4K3, TNF, ATF2, NFATC3 |
| Regulation of invasion | ADAM11, DPP6, MEP1A, PCSK6, APG4A |
| Small GTPase-dependent signaling | AGHGEF7, ARHGEF9, CDC42BPA |
| Wnt/Notch signaling | JAG1, BCORL1, NF2, FBXW7, RNF43, MARK2, TLE4, MYC, PPP2R3A, WNT9A, MAP2, TSC2, GATA6. |
| Histone modulation | KDM6A, MLL2, MLL3, SETD2 |
| SWI/SNF ATP-dependent chromatin remodeling | ARID1A, ASD1B, PBRM1, SMARCA4 |
| RNA processing | RNMI0, SF3B1, U2AF1 |
| ROBO/SLIT pathway | ROBO1, ROBO2, SLIT2, MYCBP2 |
Genes underlined are most altered in pancreatic cancers.
The average number of genetic alterations in PDAC tumors is insufficient to explain the extremely poor prognosis and response to therapy, since other tumors with a much better prognosis have a larger number of average mutations notably breast and colorectal cancers whilst the genetic spectra of colorectal, brain, and pancreatic tumors are similar.5
To meet the challenge of a better understanding of how to effectively treat pancreatic cancer, there has been a huge investment in genomic and transcriptomic data acquisition.12–14 So far this has led to only marginal therapeutic advances, since the clinical context of this data collection has often been lacking.11,14 This paucity of progress was highlighted by Paul Nurse referring to Sydney Brenner, on receiving his Nobel Prize that we are “drowning in a sea of data and starving for knowledge.” Ole Petersen has again referred to this dilemma urging that to “re-establish a focus on what really matters, namely, to gain useable knowledge from data, it would seem a good idea to rethink our working and assessment culture and start by placing much more emphasis on theory and model building as well as context.”15 A multidisciplinary approach is required to build validatable models that must combine both empirical and reductionist approaches.16
Opportunities and Limitations of Current Therapies
Cytotoxic Therapies—The Empirical Drive
The treatment of pancreatic cancer has evolved into different therapies based upon the cancer stage of disease. Traditional staging relies on pathological staging using the tumor, lymph node and metastasis (TNM) system, typified by the Union for International Cancer Control (UICC) and the American Joint Commission on Cancer (AJCC) classifications.17,18 Given the central role of surgical resection in the overall management of pancreatic cancer, an empirical system has emerged comprising five stages: resectable, borderline resectable, locally unresectable, oligometastatic disease, and large volume metastatic disease.16,19,20
There have been several 1000 clinical trials conducted in pancreatic cancer over the past 50 years. Most of these studies have been phase I/II studies assessing the potential survival benefit (with at least manageable toxicity) of newer agents and other treatment modalities such as chemoradiation, but unfortunately most have met with failure. Today, there are over 3000 PDAC trials registered at clinicaltrials.gov.in with 1099 studies currently in phase I, 271 in phase I/II, 1441 in phase II, and 306 studies in phase III, including 254 neoadjuvant trials of which 23 are in phase III. The EU Clinical Trials Register presently records 487 PDAC trials, phase III in 87, of which 10 are neoadjuvant trials. In unravelling this huge amount of information, it is important to focus on well-conducted phase III randomized trials with appropriate control arms. The mainstay of systemic treatment is combination chemotherapy, and it is perhaps surprising how relatively few agents and combinations show sufficient efficacy to meet with regulatory approval or become standard-of-care4,21–29 (Table 2). In metastatic disease compared to gemcitabine monotherapy with a median overall rate of 5.7 months,22 combination treatments are a distinct advantage with median overall survival rates of 7.1 months for gemcitabine + capecitabine (GEM-CAP),30 8.7 months for gemcitabine + nab-paclitaxel (GnP),25 11.1 months for folinic acid + 5-fluorouracil (5FU) + irinotecan + oxaliplatin (FOLFIRINOX);24 and the combination of liposomal irinotecan + 5FU/leucovorin and oxaliplatin (NALIRIFOX) is also superior to GnP.29
Table 2.
FDA approved cytotoxic drugs and targeted agents for treating patients with PDAC
| Reference | Cytotoxics | Stage | FDA Approval |
|---|---|---|---|
| Burris et al.22 | Gemcitabine | Metastatic: first line | 1996 |
| Neoptolemos et al.31,32 | 5-fluoruracil/folinic acid | Adjuvant—post-surgery | NA:α 2004 |
| Moore et al.23 | Gemcitabine + *erlotinib | Metastatic: first line | 2005 |
| Conroy et al.24 | FOLFIRINOX | Metastatic: first line | 2010 |
| Von Hoff et al.25 | Gemcitabine + nab-paclitaxel | Metastatic: first line | 2013 |
| Wang-Gillam et al.26 | 5FU + nal-irinotecan | Metastatic: second line, post-gemcitabine | 2015 |
| Neoptolemos et al.27 | Gemcitabine + capecitabine | Adjuvant—post-surgery | 2016 |
| Conroy et al.28 | Modified FOLFIRINOX | Adjuvant—post-surgery | 2018 |
| Wainberg et al.29 | 5FU + nal-irinotecan + oxaliplatin | Locally advanced or metastatic: first line | 2022 |
| Reference | Targeted Agents | Pathogenic Gene Target, Prevalence | FDA Approval |
| Le et al.51 | Pembrolizumab | **Lymphocyte programmed cell death protein 1 (PD-1) receptor, tumors with microsatellite instability (MSI-Hi) or deficient mismatch repair (dMMR), 1%–3% | 2017 |
| Marabelle et al.52 | Pembrolizumab | ¶ PD-1 receptor, high tumor mutation burden (TMB ≥10 mutations/megabase), <1% | 2020 |
| Drilon et al.53 | Larotrectinib | † Inhibitor of tropomyosin receptor kinases (TRKs), tumor NTRK1/2/3 fusions, <1% | 2018 |
| Doebele et al.54 | Entrectinib | † TRK inhibitor, tumor NTRK1/2/3 fusions, <1% | 2019 |
| Golan et al.55 | Olaparib | PARP inhibitor, germline BRCA1/2, maintenance, 1%–5% | 2019 |
| FDA56 | Dostarlimab-gxly | ‡ PD-1 receptor, deficient mismatch repair (dMMR) tumors, 1%–2% | 2021 |
| Salama et al.57 | Dabrafenib + trametinib | ƒ BRAF/CRAF + MEK1/MEK2 inhibitors, BRAFv600E tumors, <1% | 2022 |
| Schram et al.65 | Zenocutuzumab | **Neuregulin ligand (NRG1) binding to HER2: HER3 heterodimers, tumor NRG1 fusions, <1% | 2022 |
| Reference | Emerging Targeted Agents | Pathogenic Gene Target, Prevalence | |
| Reiss et al.58 | Rucaparib | # PARP inhibitor. Germline or somatic BRCA1/2, or PALB2, 1%–5% | |
| Singhi et al.59 | Critzotinib | ALK, 0.16% all cases, 1.6% in patients < 50-years old | |
| Velthaus et al.60 | Lorlatinib | ROS1, 1% | |
| Strikler et al.61 | Sotorasib | Binds to KRASG12C—GDP in inactive state, KRASp.G12C tumors, 1%–2% | |
| Bekaii-Sabb et al.64 | Adagrasib | Binds to KRASG12C—GDP in inactive state, KRASp.G12C tumors, 1%–2% | |
| Heining et al.62 | Afatinib | Pan-EGFR tyrosine kinase inhibitor in KRAS wild-type tumors with NRG1 gene fusions, <1% | |
NA = not applicable as 5 fluoruracil/folinic acid were generic agents not requiring FD approval, but adjuvant chemotherapy first established as proof of principle.
Erlotinib is a small molecule tyrosine kinase inhibitor against EGFR, but the survival difference in combination with gemcitabine is a matter of only a few weeks but with considerable toxicity, so hardly used.
This applies to agnostic solid tumors (with defined mutations but of unknown tumor type) that have progressed following prior treatment and who have no satisfactory alternative treatment options.
High TMB (agnostic) as determined by an FDA-approved test, that have progressed following prior treatment and who have no satisfactory alternative treatment options.
Either metastatic or where surgical resection is likely to result in severe morbidity (agnostic), and who have no satisfactory alternative treatments, or whose cancer has progressed following treatment.
Recurrent or advanced solid tumors (agnostic), as determined by an FDA-approved dMMR test, that have progressed on or following prior treatment and who have no satisfactory alternative treatment options.
Unresectable or metastatic solid tumors (agnostic), who have progressed following prior treatment and have no satisfactory alternative treatment options.
Advanced pancreatic cancer who had received >16 weeks of platinum-based chemotherapy without evidence of resistance.
The best results are achieved in patients with resectable tumors followed by (adjuvant) chemotherapy.16,27,28,31–36 In the ESPAC-4 study with broad inclusion criteria, the addition of capecitabine to gemcitabine increased 5-year overall survival from 16.3% to 28.8%,27 whilst FOLFIRINOX in the PRODIGE24 study improved the 5-year overall survival to 43.2% compared to 31.4% with gemcitabine monotherapy, although with more selective criteria in patients <79 years.28,37
In patients with borderline resectable disease, the administration of chemotherapy prior to resection (neoadjuvant therapy) in addition to adjuvant therapy achieves superior survival compared to adjuvant therapy alone.38,39 In radiologically unresectable disease, a long course of combination chemotherapy can result in resectabilty rates of up to 60% in selected patients with a substantial improvement in median and 5-year survival rates compared to patients in whom the tumor cannot be removed.40,41 Better survival rates are also being reported in selected patients with oligometastatic disease to the lung and liver.16,42,43 The role of chemoradiation for survival improvement in pancreatic cancer is controversial, as proof of concept high-quality evidence is lacking, and indeed randomized controlled trials show negative outcomes in both the resectable and borderline locally advanced settings.31,32,39,44–51
Targeted Therapies—The Reductionist Drive
The evolution of cytotoxic therapies for the different stages of pancreatic cancer has largely been based on empirical approaches. A reductionist approach aiming to develop treatments based on targeting key pathogenic gene alterations has met with only limited success52–66 (Table 2). The prevalence of most of the pathogenic targets range from 0.1% to 5%, and the survival advantage of these agents is a matter of only a few months. The Know Your Tumor Registry is the largest pancreatic cancer targeted therapy programme.5 Of the 1856 patients referred, 282 (15.2%) patients had actionable mutations, but only 46 (2.5%) had matched therapy.67 Survival since diagnosis was possible in 677 patients with a median overall survival of 1.3 years in the 488 patients with no actionable alteration, 1.5 years in 143 patients with actionable mutations who received unmatched therapy, and 2.6 years in the 46 patients who had matched therapy.67 The targeted agent, olaparib, has now been licensed for patients with pancreatic cancer. In patients with germline mutations in the BRCA genes, olaparib can improve progression free survival from 3.8 to 7.4 months in patients that have not progressed after at least 4 months of platinum-based first-line chemotherapy.56 The fraction of PDAC patients with druggable driver alterations will considerably increase if the novel KRAS inhibitors that target the G12D mutation present in about 40% of PDAC patients68 are successful in clinical trials.
Mechanistic Insights into Therapy Response
PDAC Subtypes and Response to Therapy
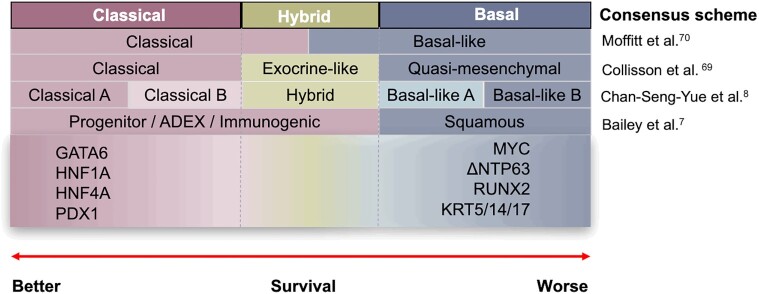
Transcriptomic profiling of PDAC has emerged as an alternative approach to better predict prognosis and response to therapy (Figure 1).7,8,69–71 Several large-scale transcriptomic studies, performed on predominantly resected nontreated samples, have identified two broad consensus subtypes (Moffitt subtypes—reviewed elsewhere11) of PDAC, namely:
Figure 1.
Transcriptomic subtype classifications and surrogate markers of PDAC. Classical tumors are associated with a better patient survival with surrogate markers: GATA6, HNF1A/4A, and PDX. Basal-like tumors have poorer outcomes with surrogate markers: MYC, ΔNTP63, RUNX2, and KRT5/14/17. Hybrid tumors show a mixed character of classical and basal with high plasticity after drug treatment.
Classical—well-differentiated tumors that express key pancreatic-specific transcription factors (TFs) GATA6, HNF1A, and PDX1, and are associated with better outcomes; and
Basal-like—less differentiated than Classical tumors with mesenchymal characteristics, including upregulated expression of ΔNP63 and TGFβ-signaling and associated with poorest patient outcomes.
The Moffitt subtypes are prognostic for survival in patients with resected PDAC but fail to prognosticate in metastatic disease.8,70 These subtypes may also guide therapy choice. Basal-like tumors exhibit lower response rates to chemotherapy in locally advanced or metastatic PDAC.69,70 The COMPASS study (NCT02750657) retrospectively assessed the utility of the Classical and Basal-like subtypes for predicting survival and response to first-line mFOLFIRINOX and gemcitabine nab-paclitaxel in advanced PDAC.71,72 Data for the intent-to-treat population showed that overall survival was almost 4 months longer for patients having a Classical RNA expression signature compared with those having a Basal-like signature. Overall survival was more than 2 months longer for those having high versus low expression of GATA6, a surrogate biomarker for the Classical subtype. Among the subset of patients given mFOLFIRINOX, patients having the Classical signature had significantly better outcomes than those having the Basal-like signature, suggesting that GATA6-low Basal-like tumors may be more resistant to mFOLFIRINOX. Based on these findings, randomized clinical trials are currently underway to evaluate expression subtyping for therapy selection in PDAC including PASS-01-Pancreatic Adenocarcinoma Signature Stratification for Treatment (NCT04469556) PANCREAS-PurIST Classification-Guided Adaptive Neoadjuvant Chemotherapy by RNA Expression Profiling of EUS Aspiration Samples (NCT04683315), and the ESPAC6 Adjuvant Trial in Patients with Resected PDAC Randomized to Allocation of Oxaliplatin- or Gemcitabine-based Chemotherapy by Standard Clinical Criteria or by a Transcriptomic Treatment Specific Stratification Signature (NCT05314998).
Despite the apparent clinical utility of the Moffitt two subtype classification scheme, recent evidence suggests that the Moffitt subtypes fail to identify overlapping but clinically distinct neoplastic subtypes in locally advanced and metastatic PDAC. Chan-Seng-Yue et al. performed de novo classification of PDAC using transcriptomic data (excluding stromal input) from 206 patients with primary resectable (stage I and II) and 111 patients with advanced (stage III and IV) PDAC and identified five neoplastic subtypes referred to as Basal-like A, Basal-like B, Hybrid (also referred to as Intermediate Co-expressors), Classical A, and Classical B.8 This classification scheme splits the Classical and Basal-like subtypes into two subcategories and identifies tumors with Hybrid gene expression profiles that share common transcripts between the Classical and Basal-like subtypes. Based on this dataset, the Classical-like subtype was found to be more prevalent in patients with resectable PDAC whereas the Basal-like subtype was more prevalent in patients with advanced disease. Basal-like A and Basal-like B subtypes were also found to approximately distinguish metastatic disease from localized disease, with the Basal-like A subtype exhibiting greater resistance to chemotherapy.
Single-cell RNA sequencing of primary tumors and metastases has demonstrated that Classical, Hybrid, and Basal-like expression signatures segregate into distinct cell populations within the same tumor and that these subtypes represent a continuum of transcriptional states.8,73 Williams et al. recently performed a spatially resolved single-cell assessment of Classical, Basal, and Hybrid phenotypes in resectable and metastatic patient samples using a quantitative multimarker protein panel (Classical protein biomarkers: CLDN18.2, TFF1, GATA6; Basal-like protein biomarkers: KRT17, KRT5, S100A2).74 This analysis found that primary and metastatic samples exhibit considerable intratumoral subtype heterogeneity with very few tumors existing as purely Basal or purely Classical. While most tumors exhibited mixed Classica/Basal-like phenotypes, metastatic lesions exhibited a higher relative fraction of Basal-like to Classical cells when compared to primary tumors. Interestingly, 90% of primary tumors contained Hybrid cells with greater enrichment in metastatic biopsies. Cell–cell neighbor analysis of individual glands found that Classical, Hybrid, and Basal-like cells often co-exist in ordered chains further supporting the notion that these subtypes represent a continuum of cell states. Stratification of patient samples based on Classical, Hybrid, and Basal-like protein expression found that subtype fractional abundance was associated with survival outcomes. Patient tumors exhibiting a higher fraction of pure Classical cells were associated with better outcomes whereas patient tumors having mixed Classical and Basal cell fractions were associated with poorer outcomes.
Collectively, these findings point to increased subtype heterogeneity during disease progression with the enrichment of Basal-like and Hybrid cell populations in advanced disease. These studies also suggest that subtype plasticity may underpin disease progression and/or response to therapy with Hybrid cells acting as important transitional cell types in both resectable and metastatic PDAC. Recent ex-vivo analyses provide strong support for the role of subtype plasticity in therapy resistance.73,75 Shalek and colleagues have demonstrated that Classical subtype Patient Derived Organoids (PDOs) treated with growth factors such as TGFβ can transition towards a Basal-like transcriptional subtype via a Hybrid or intermediate co-expressor state.73 Importantly, TGFβ induced basal-like states were found to exhibit increased resistance to standard chemotherapies supporting earlier clinical findings. Subtype plasticity has also been observed in pancreatic cell lines treated with mFOLFIRINOX, wherein Basal-like subtypes were enriched following therapy.75
The reduction of PDAC tumor heterogeneity to a continuum of Classical to Basal-like transcriptional states, however, fails to recognize additional neoplastic lineages that contribute to disease progression and therapy resistance. Comparative analysis of chemo-naïve and post-treatment (chemoradiotherapy) patient tumors using single nuclei and spatial transcriptomics identified seven neoplastic lineage programmes that contribute to patient outcomes, namely Classical, Squamoid, Basaloid, Mesenchymal, Acinar-like, Neuroendocrine-like, and Neural-like progenitor.76 This refined cellular taxonomy partitioned the Basal-like subtype into discrete Squamoid, Basaloid, and Mesenchymal lineage programmes and identified additional Acinar-like, Neuroendocrine-like, and Neural-like progenitor cell lineages. Intriguingly, cells exhibiting the Neural-like progenitor cell lineage were significantly enriched in post-treatment samples. These cells expressed several genes involved in drug efflux, negative regulation of cell death and chemoresistance (eg, ABCB1, BCL2, PDGFD, and SPP1). Moreover, neural migration and axonal guidance genes (eg, SEMA3E, RELN and SEMA5A) were found to be expressed in Neural-like progenitors implicating these cells in tumor-neural crosstalk and possibly perinuclear invasion, which is associated with poorer patient outcomes.
Despite the stated simplicity of these findings, the genetic and nongenetic mechanisms controlling neoplastic lineage determination are complex and not well understood. Mutations in epigenetic modulators such as ARID1A and KDM6A are enriched in Basal-like tumors and are associated with metastatic progression.7,77–79 Moreover, biallelic loss of SMAD4 with GATA6 amplification and/or CDKN2A loss with mutant KRAS allele amplifications are commonly associated with Classical and Basal-like subtypes, respectively, although not exclusively.8 To add to this complexity, a wealth of evidence now suggests that crosstalk between neoplastic and stromal cells—cancer associated fibroblasts (CAFs) and immune cell subsets—may shape subtype identity with distinct communities of neoplastic and stromal cells (ecotypes) co-evolving within the same patient tumors.8,74,80–82 Precisely how these ecotypes evolve during cancer progression and in response to therapy are important questions for future research.
Spatially Confined Subtumor Microenvironments (ecotypes)
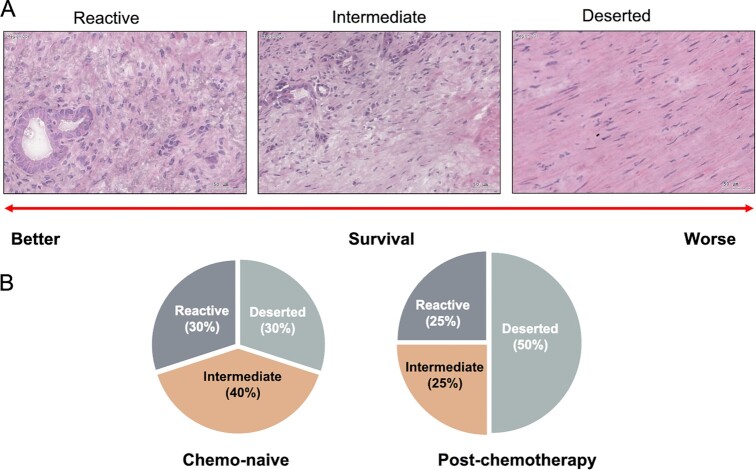
Recent evidence suggests that neoplastic and stromal cells self-organize into distinct spatially confined subtumor microenvironments (sub-TMEs).83 Based on histology, Grünwald and colleagues identified three recurrent sub-TMEs in PDAC: (i) “Deserted” containing myxoid stroma and keloid-like hyalinized collagen bundles, with thin, spindle-shaped fibroblasts; (ii) “Reactive” containing fibroblasts with enlarged nuclei and rounded morphology, inflammatory infiltrates, and few acellular components; and (iii) “Intermediate” containing intermediate levels of the Deserted and Reactive histotypes (Figure 2A). These sub-TMEs were found to co-exist within individual patient tumor samples with increased intratumoral sub-TME heterogeneity (ie, increased co-occurrence of different sub-TMEs within a single patient tumor sample) associated with advanced-stage PDAC and poor patient outcomes.
Figure 2.
TME subtypes of PDAC tumors. (A) Representative images of Deserted, Intermediate, and Reactive TME subtypes. (B) TME subtype switching from a relatively balanced Deserted/Intermediate/Reactive distribution pattern to a predominant Deserted state after patients have received chemotherapy.
Deep molecular profiling of both Deserted and Reactive sub-TMEs revealed that while Deserted sub-TMEs were immune-poor (with sporadic T-cell/NK/B-cell signals), Reactive sub-TMEs were immune-rich comprising significant numbers of immune and stromal cell subsets including T cells, macrophages, endothelial cells, and CAFs. Higher levels of the immunosuppressive factors IDO-1 and PD-L1 and immunosuppressive cell types including FOXP3-expressing regulatory T cells (Tregs), CD11b, and CD15-expressing Myeloid-Derived Suppressor Cells (MDSCs) and CD206-expressing M2 Tumor-Associated Macrophages (TAMs) were also a feature of Reactive sub-TMEs. Moreover, Reactive sub-TMEs were enriched for diverse CAF phenotypes capable of expressing a host of proinflammatory factors including IL-1β, IL-6, TNF-a, and TGFβ. An assessment of PDAC subtype, revealed that Reactive sub-TMEs were associated with Basal-like (KRT5High/GATA6Low) phenotypes whereas Deserted sub-TMEs were associated with Classical-like (KRT5Low/GATA6High) phenotypes. Importantly, patient tumors exhibiting dominant Reactive Basal-like sub-TMEs were associated with advanced-stage PDAC and shortened disease-free survival. Encapsulating fibrosis following neoadjuvant therapy (chemotherapy and/or chemoradiotherapy) is associated with better patient outcomes in PDAC. In this study, neoadjuvant treated PDAC was associated with a higher frequency of Deserted-dominant sub-TMEs when compared to stage-matched chemo-naïve cases (Figure 2B). These sub-TMEs exhibited lower stromal immunoreactivity suggesting that chemotherapy may promote poorly vascularized, the extracellular matrix (ECM)-rich deserted sub-TMEs at the expense of aggressive Reactive sub-TME phenotypes.
Evidence derived from murine models of PDAC, demonstrate that crosstalk between neoplastic, stromal cells, and the ECM shape both neoplastic identity and localized subtumor microenvironments.80–86 Modulation of these key cell–cell interactions or signaling pathways can modify both neoplastic state and/or localized tumor microenvironments and expose therapeutic vulnerabilities. Developing an in-depth knowledge of these tumor–stromal interactions is therefore important to unlock new and more effective therapies for PDAC (Figure 3).11
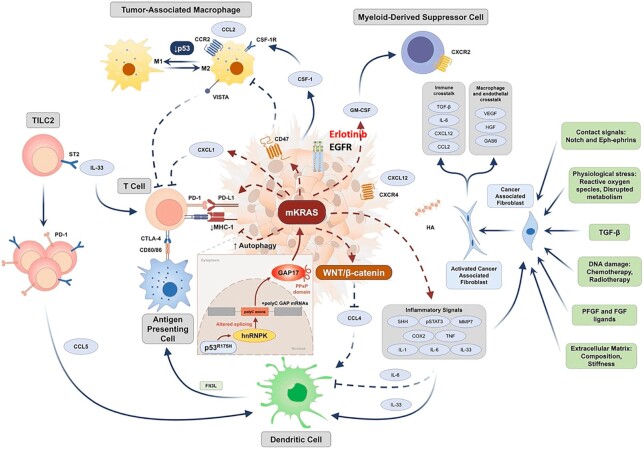
Figure 3.
The PDAC TME showing tumor cell and stromal cell interactions11. CCL2/4/5, CC-chemokine ligand 2/4/5; CCR, CC-chemokine receptor; COX2, cyclooxygenase 2; CSF-1, colony stimulating factor 1; CSF-1R, colony stimulating factor 1 receptor; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; CXCL1/12, CXC-chemokine ligand 1/12; CXCR4, CXC-chemokine receptor type 4; DC, conventional type 1 dendritic cell; FGF, fibroblast growth factor; Flt3L, Fms related receptor tyrosine kinase 3 ligand; GAS6, growth arrest-specific protein 6; GM-CSF, granulocyte-macrophage colony-stimulating factor; hnRNPK, heterogeneous nuclear ribonucleoprotein K; HA, hyaluronic acid; HGF, hepatocyte growth factor; IL-1/-6/-33, interleukin-1/-6/-33; MHC-1, major histocompatibility complex 1; PDGF, platelet-derived growth factor; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; SHH, sonic hedgehog; ST2, suppression of tumorigenicity 2; STAT3, signal transducer and activator of transcription 3; TAM, tumor-associated macrophage; TGF-β, transforming growth factor-β; VEGF, vascular endothelial growth factor; and VISTA, V-domain Ig suppressor of T-cell activation.
PDAC tumors are characterized by dense stroma, which consist mainly of host stromal cells including CAFs and an ECM.87 The ECM is a major component of the TME that exerts mechanical and biochemical properties on tumor cells. The ECM is thought to support tumor growth by supplying nutrients, activating mechano-signaling pathways that augment oncogenic signaling pathways, and by reducing blood vessel density.88 Fibrilla collagens such as collagen-I make up most of the ECM and can be produced by both tumor-resident CAFs and pancreatic cancer cells.89 Recent evidence has revealed that cleavage of collagen-I by matrix-metalloproteinases can activate discoidin domain receptor 1 (DDR1)–NF-κB–p62–NRF2 signaling resulting in mitochondrial biogenesis, micropinocytosis, tumor growth, and metastasis.86 Patient tumors enriched for intact collagen-I were associated with improved median survival compared to patient tumor enriched for cleaved collagen-I. An alternate study defined an oncogenic role for tumor-cell-produced collagen-I homodimers in PDAC.84 Collagen-I fibers typically assemble as heterodimers (a1, a2, a1–products of the COL1A1 and COL1A2 genes); however, due to methylation of the COL1A2 gene promoter collagen-I assembles predominantly as a homotrimer (a1, a1, a1) in PDAC cells. Targeted deletion of COL1A1 in murine models of PDAC demonstrated that loss of COL1A1 significantly perturbed PDAC development and resulted in increased overall survival. Importantly, loss of COL1A1 in pancreatic cancer cells was associated with the reduced expression of the MDSCs attractant C-X-C chemokine (CXC) l5 and an increase in the expression of the T-cell attractant CXCl16. This resulted in the reduced infiltration of CD11b/GR1 MDSCs and the increased infiltration of CD4 and CD8 T cells. Treatment with an immune checkpoint inhibitor (ICI) targeting PD-1 increased overall survival suggesting that selective immunotherapies may have utility in the context of collagen-I homotrimer loss.
Expression of inflammatory cytokines such as TGFβ and TNFα have long been known to accelerate PDAC progression.11 Recent evidence has revealed that TNFα-mediated cross-talk between macrophages and pancreatic cancer cells is important for maintaining Basal-like subtype identity.85 Treatment of pancreatic cancer cells with TNFα was sufficient to shift Classical-like cells toward a Basal-like state. Importantly, transcription factor networks driven by master regulators BRD4 and cJUN, were found to transduce TNFα signals and to activate Basal-like gene expression programmes. Pharmacological inhibition of the BRD4/cJUN transcriptional network was sufficient to revert Basal-like cells toward a Classical state. Highlighting the remarkable interdependence of different cells within the TME, activation of BRD4/cJUN transcriptional networks by macrophage secreted TNFα induced the expression of CCL2 in neoplastic cells, a key chemokine linked to macrophage recruitment. Additional studies have shown that ablation of myeloid cells using inhibitors for colony-stimulating factor 1 receptor (CSF-1R) targeting tumor-associated macrophages or CXC chemokine receptor (CXCR) 2 targeting neutrophils in murine models can also shift Basal-like tumors toward a Classical-like state.81,82 Importantly, these studies show that changes in neoplastic state are accompanied by increases in T-cell infiltrates and greater susceptibility to ICIs.
Halbrook et al. have demonstrated that intratumoral metabolic crosstalk between M2 macrophages and pancreatic cancer cells can promote pyrimidine-based chemotherapy resistance in PDAC.90 M2 macrophages release a spectrum of pyrimidines including deoxycytidine (a molecular analogue of gemcitabine), which both blocks the uptake of gemcitabine by equilibrative nucleoside (ENT) and concentrative nucleoside (CNT) transporters and incorporation of gemcitabine through molecular competition at deoxycytidine kinase (DCK). Patients with a low M2 macrophage burden exhibited improved responses to gemcitabine. Moreover, targeting macrophages using small molecule inhibitors (such as the CSF-1R inhibitor) increased the efficacy of gemcitabine in murine models of PDAC.
Mazzone and colleagues have demonstrated that PDAC cells express SLC4A4 a transporter that actively imports bicarbonate.91 The accumulation of bicarbonate in PDAC cells increases intracellular pH, which in turn buffers against the accumulation of H+ ions that are generated by glycolysis. This buffering allows glycolysis to proceed at a faster rate generating increased levels of H+/lactate, which PDAC cells readily export into the interstitial space. Localized accumulation of lactate and increases in TME acidity establish a protumorigenic immune microenvironment. It has long been established that the glycolytic capacity is an important driver of metastatic progression and escape from immune surveillance. Increases in lactate and TME acidity are thought to modulate tumor resident immune cells in several important ways. First, CD8 cells become dysfunctional in the presence of lactate. Second, Treg cells which comprise the lactate importer SLC16A1 actively take up and metabolize lactate to increase proliferation and suppress T-cells. Thirdly, lactate induces arginase-positive macrophage phenotypes that suppress antitumor immune responses. Additionally, immune suppressive cell surface receptors such as VISTA that are expressed on immunosuppressive macrophages require low pH to engage their cognate T-cell receptor p-Selectin through a mechanism that involves protonation of receptor histidine residues. Targeted deletion of the SLC4A4 gene or pharmacological inhibition of murine pancreatic cancer cells using the small molecule DIDS decreased extracellular H+/lactate levels and inhibited glycolysis. SLC4A4 targeting in orthotopic murine models of PDAC reinvigorated CD8+ T cells and reduced immunosuppressive Treg and macrophage infiltrates resulting in reduced tumor growth and metastases. Importantly, depletion of SLC4A4 was able to overcome immunotherapy resistance to immune checkpoint inhibitors and prolong survival.
Previous efforts to broadly target the TME in PDAC have been unsuccessful. Ablation of αSMApos myofibroblastic CAFs or ablation of sonic hedgehog paracrine signaling have accelerated metastasis and reduced survival.92 Combination therapies targeting both neoplastic and stromal cell populations may represent more effective therapeutic opportunities.
Targeting the Immune Landscape of PDAC
Immune checkpoint inhibition (ICI) has revolutionized cancer care, with up to 70 distinct USA Food and Drug Administration label indications across more than 18 cancer types.93 Although PDAC patients exhibiting microsatellite instability (MSI) (<1%) may benefit from ICI, single-agent, and combinations of PD-1, PD-L1, or CTLA-4 inhibitors are ineffective in patients with advanced PDAC, objective response rates being <5%.94–96 Chemotherapy combined with ICIs, however, have been demonstrated to improve response rates in metastatic PDAC.97,98 The randomized phase 2 PRINCE trial which tested nivolumab (anti-PD-1) in combination with gemcitabine and nab-paclitaxel (GnP) resulted in significantly higher 1-year overall survival (58%) as compared to a historical 1-year survival of 35% (P = .006).98 The median progression free survival in the nivolumab plus GnP arm was 6.4 months (95% CI, 5.2–8.8), and the tumor objective response rate was 50% (95% CI, 32%–68%). Survival after nivolumab plus GnP correlated with a less repressive tumor microenvironment and higher numbers of activated, antigen-experienced T-cells at baseline. These findings point to molecularly identifiable subsets of patients that may obtain enduring clinical benefit from chemoimmunotherapy in metastatic PDAC.
Profiling of PDAC tumors indicates that as many as 20%–30% of patients exhibit moderate T-cell content, and that tumor immunogenic neo-epitopes and T-cell immunity can correlate with overall survival.99–101 Despite the prevalence of T-cells in at least a third of patients, tumor specific T-cell responses are largely suppressed by the presence of myeloid cells in the tumor microenvironment.102 The accumulation of tumor-modified myeloid cells derived from monocytes and neutrophils, termed MDSCs, and TAMs are associated with resistance to both chemotherapy and ICIs.93 Preclinical efficacy studies in murine models of PDAC have demonstrated that myeloid modulators can enhance responses to both chemotherapy or immunotherapy by increasing CD8+ T-cell infiltration.81,82 The most extensively studied first-generation myeloid modulators include drugs targeting CSF-1R, C-C chemokine receptor type 2 (CCR2), and CXCR2. Clinical trials of these drugs alone or in combination with chemotherapy are ongoing in PDAC although early indications suggest that single arm Phase I and II trials in unselected populations show poor efficacy and are challenged by treatment toxicity.
Therapeutic cancer vaccines have historically been unsuccessful in PDAC103; however, the recent success of a vaccine targeting mutant KRAS has reignited interest in this approach.104 KRAS mutants are found in 90% of pancreatic ductal adenocarcinomas with the G12D single acid mutation occurring in 41% of patients.6,7,105 Mutant KRAS epitopes are presented on multiple HLA alleles and can be recognized by CD8 + T-cell Receptors (TCRs) suggesting that engineered T-cell vaccines targeting mutant KRAS may elicit robust antitumor responses.106 In the recent landmark study, a patient with progressive metastatic pancreatic cancer was treated with a single infusion of autologous T-cells that had been genetically engineered to clonally express two allogeneic HLA-C*08:02–restricted TCRs targeting mutant KRAS G12D.104 Remarkably, this treatment induced robust and durable (>6 months) regression of visceral metastases with the engineered T-cells constituting more than 2% of all circulating peripheral-blood T-cells 6 months after cell transfer.104 The benefit of this approach, however, is limited to the small number of patients who have the HLA-C*08:02 allele, which is present in approximately 8% of white people and approximately 11% of black people. The development of better computational tools for identifying robust candidate epitopes and improved methods of vaccine delivery may expand the utility of this approach in larger groups in patients.
Cytotoxic or metabolic stress induced by chemotherapy and/or radiation can stimulate nucleic acid sensing pathways in both tumor cells and infiltrating immune cells to trigger type-I IFN signaling (INF-α and -β).107 Type-I IFN is required for robust immune surveillance by directly or indirectly stimulating T-cells, natural killer (NK) cells, or macrophages in the TME. Chemotherapy in combination with immune checkpoint or myeloid inhibitors has shown promise in triggering antitumor immune responses in PDAC, however these responses have not produced durable outcomes.93,103,107 Tumor mutational status or changes in protein expression may underpin resistance to immunotherapy. Loss of the tumor suppressors STK11, CDKN2A, PTEN, and STING reduction in β2-microglobin are all associated with poor responses to chemotherapy and/or ICIs.107 Homozygous deletion of chromosome 9p21.3 has been identified as a candidate biomarker of immunotherapy resistance in several cancers.108 Tumors with deletion of 9p21.3 exhibit increased resistance to immune checkpoint inhibitors and altered immune infiltrates. The 9p21.3 locus encompasses the suppressor genes CDKN2A and CDKN2B and a cluster of type-I IFN genes. (Type-I IFN-I comprises IFNα, IFNβ, IFNδ, IFNϵ, IFNκ, IFNω, and IFNτ genes; type-II IFN comprises the IFNγ gene). Tumors with deletion of 9p21.3 exhibit either loss of CDKN2A alone or the co-deletion of the IFN gene cluster. Type-I IFN responses are critical for adaptive immune responses suggesting that co-deletion of the 9p21.3 IFN gene cluster may underpin immune checkpoint therapy resistance.
To understand the impact of 9p21.3 homozygous loss on pancreatic cancer Scott Lowe and colleagues developed MACHETE (molecular alteration of chromosomes with engineered random repeats), a CRISPR-based approach that enables the targeted deletion of large contiguous genomic regions.108 Applying MACHETE to a syngeneic mouse model of pancreatic cancer, the Lowe group demonstrated that co-deletion of CDKN2A/CDKN2B and IFN genes combined to both activated cell proliferation and disrupt type-I IFN signaling within the TME. Disruption of type-I IFN signaling was associated with the accumulation of exhausted CD8+ T-cells that express markers of terminal differentiation leading to immune evasion, metastasis, and resistance to immune checkpoint blockade.
These findings have important clinical implications for the stratification of patients receiving immunotherapy. CDKN2A copy number loss is found in around 60% of PDAC patient; however, the loss of the type-I IFN gene cluster is less well characterized. KRAS copy number gains are also associated with CDKN2A loss and basal-like transcriptional states in advanced PDAC. Many targeted trial platforms test for CDKN2A/B deletion but fail to assess loss of IFN genes within the same locus. Accordingly, future immunotherapy clinical trials in PDAC should incorporate type-I IFN status into their trial design. Preclinical development of new immunotherapies for PDAC typically utilize genetically engineered mouse models driven by oncogenic KRAS mutations and TP53 loss due to their complex TMEs.81,82 These models, however, do not include deletions of the 9p21.3 locus. Accordingly, co-deletion of the 9p21.3 locus should be included in preclinical immunotherapy studies to develop more focused clinical trials.
PDAC Evolution, Drug Resistant Persisters, and Therapy
PDAC evolution is widely considered to involve the stepwise transformation of ductal epithelial from low-grade then high-grade dysplastic pancreatic intraepithelial neoplasia (PanINs) to invasive adenocarcinoma.14 Genomic profiling of these distinct developmental stages has identified a continuum of accumulating genomic alterations. Low-grade PanINs are almost universally associated with oncogenic mutations in KRAS while increased PanIN dysplasia is associated with frequent alterations in TP53, CDKN2A, and SMAD4. Greater than 40% of intermediate and invasive adenocarcinomas are associated with complex genomic changes including increases in polyploidization and chromothripsis (a phenomenon in which a chromosome or a portion of a chromosome undergoes DNA breakages and rearrangements leading to the loss, duplication, inversion, and translocation of large segments of DNA), which contribute to significant intratumorally heterogeneity.14
A recent landmark study by Scott Lowe and colleagues has demonstrated that loss of TP53 (altered in 70% of PDAC tumors) enables a deterministic pattern of genome evolution in PDAC.109 Using a mouse model of pancreatic cancer that reports sporadic loss of TP53 heterozygosity, the Lowe group traced the genomic trajectories of single cells from early dysplasia to frank PDAC. This analysis demonstrated that TP53 loss of heterozygosity initiates sequential phases of genome evolution including: (i) initial deletion of chromosomes 4 and 11 (encompassing CDKN2A and TP53 loci, respectively) as well as gain in chromosomes 5 (encompassing genes involved in PDAC proliferation or progression and TGFβ-signaling) and 6 (encompassing mutant KRAS); (ii) genome doubling; and (iii) the emergence of gains and amplifications in oncogenes, such as MYC. Importantly, evolutionary patterns observed in mice were reflected in human PDAC. Analysis of genomic data obtained from patient tumors harboring TP53-mutations demonstrated that deletion events in diploid genomes were associated with the accumulated loss of tumor suppressors on chromosomes 9p, 17p, and 18q. In contrast, diploid or polyploid genomes harboring biallelic TP53-mutations were associated with heterogeneous gains in KRAS, MYC, and GATA6—oncogenic events that drive metastasis and/or influence PDAC subtypes. Perhaps predictably, patients with polyploid genomes and biallelic TP53-mutations invariably presented with metastatic disease and had the poorest patient outcomes. Intriguingly, the observed gains and amplifications of oncogenes such as KRAS, MYC, and GATA6 in advanced PDAC mirror the emergence of Basal-like and Reactive sub-TMEs during PDAC progression strongly suggesting that these events are linked.
The ordered and deterministic evolution of PDAC toward increased genomic heterogeneity provides evidence that subclonal heterogeneity likely underpins poor responses to therapy in PDAC. In this context, the co-evolution of different subclones within the same tumor and at different stages along a deterministic path may explain why dissociated responses to therapy are common in PDAC. While these data point to a classical model of sequential genomic evolution in therapy resistance, accumulating evidence suggests that nongenetic priming or adaption contribute substantially to therapy resistance.110 New evidence from our laboratory suggests that drug tolerant cells or “persisters” can emerge from a pre-existing subpopulation of neoplastic cells following neoadjuvant chemotherapy (Figure 4).111 These persister-like cells adapt to chemotherapy by upregulating CYP3A5 and other co-expressed drug-metabolizing genes, which metabolize the prodrug irinotecan a constituent of FOLFIRINOX into nonactive forms (Figure 5). While persister cells are commonly associated with bacterial infections, there is growing evidence that analogous cell populations (alternatively referred to as “cancer stem cells” or “tumor-initiating cells”) may exist in tumors. Like bacterial persister cells, cancer persister cells may enter a dormant or quiescent state, making them resistant to chemotherapy and/or chemoradiotherapy. Chemotherapy may therefore kill most cancer cells but fail to eradicate a small population of persister cells that survive and eventually repopulate the tumor. In this model, the persister cell state resembles minimal residual disease from which relapse can occur if treatment is discontinued. The identification and characterization of persister cells may therefore lead to new strategies for combating drug-resistant cancers.
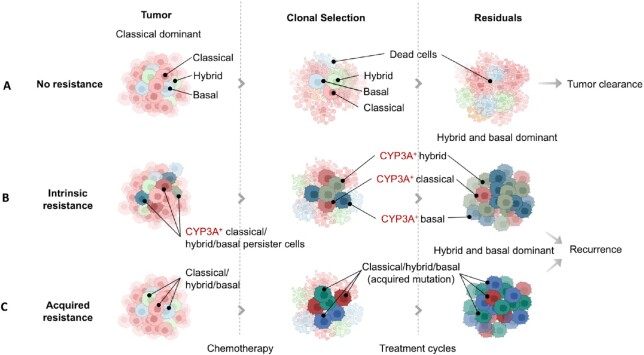
Figure 4.
Drug resistance in PDAC patient after cytotoxic chemotherapy. (A) Tumor cells without drug resistance can be completely eradicated after a full (6-month) course of effective chemotherapy. (B) Tumor cell plasticity during chemotherapy results in cells with intrinsic mechanisms of cytotoxic drug resistance (such as CYP3A) to persist and become enriched as the sensitive cell types are killed. (C) Chemotherapy may drive the clonal evolution of cell types to a more Basal-like subtype with new mutations resulting in acquired resistance. Within any one tumor there will be heterogeneity with differential proportions in sensitive and resistant cell types and sub-TMEs, determining the rate of clonal evolution and ultimately survival or death.
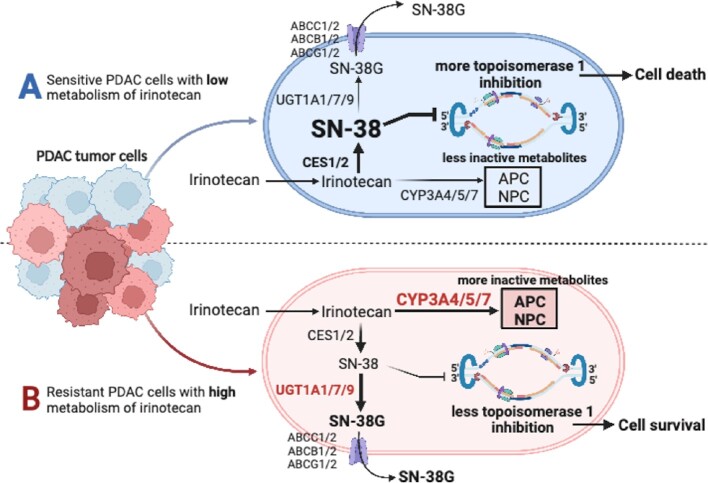
Figure 5.
(A) In sensitive PDAC cells following uptake, irinotecan (a component of the FOLFIRINOX and other regimens) is converted to the active metabolite SN-38 by carboxyl esterases, which then kills the cell by inhibition of topoisomerase I. (B) In resistant PDAC cells, CYP3A isoforms 4/5/7 interfere with the metabolism of irinotecan to SN-38 by direct and indirect mechanisms resulting in cell survival. CES: carboxylesterase; CYP: cytochrome P450 isoenzymes; UGT uridine diphosphate glucuronosyltransferase isoenzymes; ABC: ATP-binding cassette transporters, the multidrug resistance-associated protein-1 is encoded by ABCC1; APC: inactive metabolite,7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyl-oxy-camptothecin; and NPC: inactive metabolite 7-ethyl-10-[4-amino-1-piperidino] carbonyl-oxy-camptothecin).
Precision medicine is built on the premise that actionable genomic events can guide therapeutic decisions. The emerging picture in PDAC is that this premise is an enormous oversimplification and that complex genomic and nongenomic events underpin responses to therapy.
Evolving Therapeutic Opportunities
So, an important concept is to better understand how we can use chemotherapy and better understand the mechanisms of pancreatic cancer chemoresistance and chemosensitivity.16 To this extent, we require a deeper understanding of the development of the tumor mass (the TME), plasticity of molecular subtypes, clonal evolution, and metastasis (Figures 1-3).11 Different molecular subtypes are associated with differential clinical responses to chemotherapy-based regimens.8 Stromal mediated chemoresistance to gemcitabine can be induced by TAMs by upregulation of PDAC cell cytidine deaminase, or transfer of miR-365 to PDAC cells by TAM-derived exosomes, and in mtKRAS PDAC cells with a gain-of-function mtTP53 induced chemoresistance through CAFs via NFκB/TNFα signaling leading to secretion of perlecan (Figure 3).112–114 Chemotherapy can induce drug-tolerant persister cell phenotypes from distinct tumor cell lineages leading to tumor relapse.11,83,111,115 The acquired drug-tolerant persister cells do not in the main involve mutations that confer therapy resistance, suggesting that alternative mechanisms, including phenotypic plasticity driven by stromal interactions, may be involved.116 Both chemotherapy using FOLFIRINOX and chemoradiotherapy can induce a transformation to a predominant Basal-like subtype from a Classical-like subtype to associated with greater chemoresistance and reduced patient survival.75,76,111,117 Platinum therapy after adjuvant or first-line treatment results in recurrent disease associated an increased mutational burden notably neurofibromin (NF1), AKT1, PIK3CA, and serine/threonine kinase 11 (STK11), activating MAPK/ERK and PI3K-AKT signaling, and develops from a synchronous or metachronous primaries (monophyletic and polyphyletic origins).118 Pancreatic cancer has relatively few neo-antigenic targets, and is embedded in an immunosuppressive cold TME, which hinders intratumoral CD8+ T-cell infiltration and activation rendering single agent immunotherapies with immune checkpoint inhibitors mostly ineffective.119 Chemoradiotherapy may also impair immunity in PDAC resulting in an increased rate of metastases whilst organs targeted with radiotherapy may also promote metastases to the radiotherapy-directed site.120,121 Learning how to take advantage of modifying the deleterious effects of post-therapy cellular plasticity and persister cell enrichment, whilst reversing the cold immune TME is fundamental to harnessing clever clinical trials closely linked to multiple omics approaches in order to gain fundamental traction on treating pancreatic cancer. Strongly declared hypothesis driven research is required to deal with the increasing challenge of entropy of information.122
Acknowledgement
Professor Frank Bergmann (Departments of Pathology at the Universities of Heidelberg and Darmstadt) provided the histological figures in Figure 2.
Contributor Information
Peter Bailey, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany; School of Cancer Sciences, University of Glasgow, Glasgow, G61 1QH, UK.
Xu Zhou, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
Jingyu An, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
Teresa Peccerella, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
Kai Hu, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
Christoph Springfeld, Department of Medical Oncology, National Center for Tumor Disease (NCT), Heidelberg University Hospital, Heidelberg, Germany.
Markus Büchler, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
John P Neoptolemos, Department of General, Visceral and Transplantation Surgery, Heidelberg University Hospital, Heidelberg 69120, Germany; Section Surgical Research, University Clinic Heidelberg, Heidelberg 69120, Germany.
Funding
P.B. is funded by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement Nº 861196 (PRECODE). C.S. has grants from the Stiftung Deutsche Krebshilfe and the Bundesministerium für Bildung und Forschung (BMBF). J.P.N. has grants from the Stiftung Deutsche Krebshilfe, Heidelberger Stiftung Chirurgie, Dietmar Hopp Stiftung, and the BMBF. X.Z., J.A., and K.H. have grants from the China Scholarship Council.
Conflict of Interest Statement
C.S. declares serving on advisory boards for AstraZeneca, Bayer, Eisai, lncyte, MSD, Roche, and Servier. J.P.N. holds the position of Editorial Board Member for Function and is blinded from reviewing or making decisions for the manuscript. The remaining authors have no conflict of interest.
Data Availability Statement
The data underlying this article are available in the article.
References
- 1. Global Cancer Observatory . International Agency for Research on Cancer. Cancer today. Accessed 12 February 2023. https://gco.iarc.fr/today/home.
- 2. Cronin KA, Scott S, Firth AUet al. Annual report to the nation on the status of cancer, part 1: national cancer statistics. Cancer. 2022;128(24):4251–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu J, Blackford AL, Dal Molin M, Wolfgang CL, Goggins M. Time to progression of pancreatic ductal adenocarcinoma from low-to-high tumour stages. Gut. 2015;64(11):1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15(6):333–348. [DOI] [PubMed] [Google Scholar]
- 5. Jones S, Zhang X, Parsons DWet al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waddell N, Pajic M, Patch AMet al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bailey P, Chang DK, Nones Ket al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. [DOI] [PubMed] [Google Scholar]
- 8. Chan-Seng-Yue M, Kim JC, Wilson GWet al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52(2):231–240. [DOI] [PubMed] [Google Scholar]
- 9. Escobar-Hoyos LF, Penson A, Kannan Ret al. Altered RNA splicing by mutant p53 activates oncogenic RAS signaling in pancreatic cancer. Cancer Cell. 2020;38(2):198–211.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pelosi E, Castelli G, Testa U. Pancreatic cancer: molecular characterization, clonal evolution and cancer stem cells. Biomedicines. 2017;5(4):65. doi:10.3390/biomedicines5040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Z, Hu K, Bailey Pet al. Clinical impact of molecular subtyping of pancreatic cancer. Front Cell Dev Biol. 2021;9:743908.doi:10.3389/fcell.2021.743908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alexandrov LB, Nik-Zainal S, Wedge DCet al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium . Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22(3):131–142. [DOI] [PubMed] [Google Scholar]
- 15. Petersen OH. Are scientists sufficiently ambitious?. Function (Oxf). 2022;3(5):zqac048. doi:10.1093/function/zqac048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Springfeld C, Ferrone CR, Katz AWet al. Neoadjuvant therapy for pancreatic cancer. Nat Rev Clin Oncol. Published online 2023: In press. doi:10.1038/s41571-023-0746-1. [DOI] [PubMed] [Google Scholar]
- 17. Brierly JD, Gospodarowicz MK, Wittekind C. Union for International Cancer Control TNM Classification of Malignant Tumours. 8th ed. Wiley; New York, 2016. [Google Scholar]
- 18. Amin MB, American Joint Committee on Cancer, American Cancer Society . AJCC Cancer Staging Manual. 8th ed. American Joint Committee on Cancer, Springer; New York, 2017. [Google Scholar]
- 19. Ducreux M, Cuhna AS, Caramella Cet al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(5):v56–v68.. doi:10.1093/annonc/mdv295. [DOI] [PubMed] [Google Scholar]
- 20. Tempero MA, Malafa MP, Al-Hawary Met al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19(4):439–457. [DOI] [PubMed] [Google Scholar]
- 21. Springfeld C, Jäger D, Büchler MWet al. Chemotherapy for pancreatic cancer. La Presse Médicale. 2019;48(3):e159–e174. [DOI] [PubMed] [Google Scholar]
- 22. Burris HA, Moore MJ, Andersen Jet al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6):2403–2413. [DOI] [PubMed] [Google Scholar]
- 23. Moore MJ, Goldstein D, Hamm Jet al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966. [DOI] [PubMed] [Google Scholar]
- 24. Conroy T, Desseigne F, Ychou Met al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. [DOI] [PubMed] [Google Scholar]
- 25. Von Hoff DD, Ervin T, Arena FPet al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang-Gillam A, Li CP, Bodoky Get al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet North Am Ed. 2016;387(10018):545–557. [DOI] [PubMed] [Google Scholar]
- 27. Neoptolemos JP, Palmer DH, Ghaneh Pet al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet North Am Ed. 2017;389(10073):1011–1024. [DOI] [PubMed] [Google Scholar]
- 28. Conroy T, Hammel P, Hebbar Met al. FOLFIRINOX or Gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379(25):2395–2406. [DOI] [PubMed] [Google Scholar]
- 29. Wainberg ZA, Melisi D, Macarulla Tet al. NAPOLI-3: a randomized, open-label phase 3 study of liposomal irinotecan + 5-fluorouracil/leucovorin + oxaliplatin (NALIRIFOX) versus nab-paclitaxel + gemcitabine in treatment-naïve patients with metastatic pancreatic ductal adenocarcinoma (mPDAC). J Clin Oncol. 2023;41(4):LBA661–LBA661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cunningham D, Chau I, Stocken DDet al. Phase III randomized comparison of gemcitabine versus gemcitabine plus capecitabine in patients with advanced pancreatic cancer. J Clin Oncol. 2009;27(33):5513–5518. [DOI] [PubMed] [Google Scholar]
- 31. Neoptolemos JP, Dunn JA, Stocken DDet al. Adjuvant chemoradiotherapy and chemotherapy in resectable pancreatic cancer: a randomised controlled trial. Lancet North Am Ed. 2001;358(9293):1576–1585. [DOI] [PubMed] [Google Scholar]
- 32. Neoptolemos JP, Stocken DD, Friess Het al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350(12):1200–1210. [DOI] [PubMed] [Google Scholar]
- 33. Oettle H, Post S, Neuhaus Pet al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297(3):267–277. [DOI] [PubMed] [Google Scholar]
- 34. Neoptolemos JP, Stocken DD, Bassi Cet al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010;304(10):1073–1081. [DOI] [PubMed] [Google Scholar]
- 35. Uesaka K, Boku N, Fukutomi Aet al. Adjuvant chemotherapy of S-1 versus gemcitabine for resected pancreatic cancer: a phase 3, open-label, randomised, non-inferiority trial (JASPAC 01). Lancet North Am Ed. 2016;388(10041):248–257. [DOI] [PubMed] [Google Scholar]
- 36. Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16(1):11–26. [DOI] [PubMed] [Google Scholar]
- 37. Conroy T, Castan F, Lopez Aet al. Five-year outcomes of FOLFIRINOX vs Gemcitabine as adjuvant therapy for pancreatic cancer: a randomized clinical trial. JAMA Oncol. 2022;8(11):1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Versteijne E, van Dam JL, Suker Met al. Neoadjuvant chemoradiotherapy versus upfront surgery for resectable and borderline resectable pancreatic cancer: long-term results of the Dutch randomized PREOPANC trial. JCO. 2022;40(11):1220–1230. [DOI] [PubMed] [Google Scholar]
- 39. Ghaneh P, Palmer D, Cicconi Set al. Immediate surgery compared with short-course neoadjuvant gemcitabine plus capecitabine, FOLFIRINOX, or chemoradiotherapy in patients with borderline resectable pancreatic cancer (ESPAC5): a four-arm, multicentre, randomised, phase 2 trial. Lancet Gastroenterol Hepatol. 2023;8(2):157–168. [DOI] [PubMed] [Google Scholar]
- 40. Hackert T, Sachsenmaier M, Hinz Uet al. Locally advanced pancreatic cancer: neoadjuvant therapy with Folfirinox results in resectability in 60% of the patients. Ann Surg. 2016;264(3):457–463. [DOI] [PubMed] [Google Scholar]
- 41. Kunzmann V, Siveke JT, Algul Het al. Nab-paclitaxel plus gemcitabine versus nab-paclitaxel plus gemcitabine followed by FOLFIRINOX induction chemotherapy in locally advanced pancreatic cancer (NEOLAP-AIO-PAK-0113): a multicentre, randomised, phase 2 trial. Lancet Gastroenterol Hepatol. 2021;6(2):128–138. [DOI] [PubMed] [Google Scholar]
- 42. Hank T, Klaiber U, Hinz Uet al. Oncological outcome of conversion surgery after preoperative chemotherapy for metastatic pancreatic cancer. Ann Surg. Publish Ahead of Print. Published online 27 June2022. doi:10.1097/SLA.0000000000005481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Homma Y, Endo I, Matsuyama Ret al. Outcomes of lung metastasis from pancreatic cancer: a nationwide multicenter analysis. J Hepato Biliary Pancreat. 2022;29(5):552–561. [DOI] [PubMed] [Google Scholar]
- 44. Sultana A, Tudur Smith C, Cunningham Det al. Systematic review, including meta-analyses, on the management of locally advanced pancreatic cancer using radiation/combined modality therapy. Br J Cancer. 2007;96(8):1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chauffert B, Mornex F, Bonnetain Fet al. Phase III trial comparing intensive induction chemoradiotherapy (60 Gy, infusional 5-FU and intermittent cisplatin) followed by maintenance gemcitabine with gemcitabine alone for locally advanced unresectable pancreatic cancer. Definitive results of the 2000-01 FFCD/SFRO study. Ann Oncol. 2008;19(9):1592–1599. [DOI] [PubMed] [Google Scholar]
- 46. Barhoumi M, Mornex F, Bonnetain Fet al. Locally advanced unresectable pancreatic cancer: induction chemoradiotherapy followed by maintenance gemcitabine versus gemcitabine alone: definitive results of the 2000-2001 FFCD/SFRO phase III trial. Cancer/Radiothérapie. 2011;15(3):182–191. [DOI] [PubMed] [Google Scholar]
- 47. Liao WC, Chien KL, Lin YLet al. Adjuvant treatments for resected pancreatic adenocarcinoma: a systematic review and network meta-analysis. Lancet Oncol. 2013;14(11):1095–1103. [DOI] [PubMed] [Google Scholar]
- 48. Neoptolemos JP, Cox TF. Bayesian analysis unravels pancreas-cancer adjuvant therapy. Lancet Oncol. 2013;14(11):1034–1035. [DOI] [PubMed] [Google Scholar]
- 49. Katz MHG, Shi Q, Meyers Jet al. Efficacy of preoperative mFOLFIRINOX vs mFOLFIRINOX Plus hypofractionated radiotherapy for borderline resectable adenocarcinoma of the pancreas: the A021501 phase 2 randomized clinical trial. JAMA Oncol. 2022;8(9):1263, doi:10.1001/jamaoncol.2022.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hammel P, Huguet F, van Laethem JLet al. Effect of chemoradiotherapy vs chemotherapy on survival in patients with locally advanced pancreatic cancer controlled after 4 months of Gemcitabine with or without Erlotinib: the LAP07 randomized clinical trial. JAMA. 2016;315(17):1844–1853. [DOI] [PubMed] [Google Scholar]
- 51. Springfeld C, Neoptolemos JP. The role of neoadjuvant therapy for resectable pancreatic cancer remains uncertain. Nat Rev Clin Oncol. 2022;19(5):285–286. [DOI] [PubMed] [Google Scholar]
- 52. Le DT, Durham JN, Smith KNet al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marabelle A, Fakih M, Lopez Jet al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353–1365. [DOI] [PubMed] [Google Scholar]
- 54. Drilon A, Laetsch TW, Kummar Set al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doebele RC, Drilon A, Paz-Ares Let al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21(2):271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Golan T, Hammel P, Reni Met al. Maintenance Olaparib for Germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. FDA grants accelerated approval to dostarlimab-gxly for dMMR advanced solid tumors. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-regular-approval-dostarlimab-gxly-dmmr-endometrial-cancer.
- 58. Salama AKS, Li S, Macrae ERet al. Dabrafenib and Trametinib in patients with tumors with BRAF(V600E) mutations: results of the NCI-MATCH trial subprotocol H. J Clin Oncol. 2020;38(33):3895–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reiss KA, Mick R, O'Hara MHet al. Phase II study of maintenance Rucaparib in patients with platinum-sensitive advanced pancreatic cancer and a pathogenic germline or somatic variant in BRCA1, BRCA2, or PALB2. J Clin Oncol. 2021;39(22):2497–2505. [DOI] [PubMed] [Google Scholar]
- 60. Singhi AD, Ali SM, Lacy Jet al. Identification of targetable ALK rearrangements in pancreatic ductal adenocarcinoma. J Natl Compr Canc Netw. 2017;15(5):555–562. [DOI] [PubMed] [Google Scholar]
- 61. Velthaus JL, Iglauer P, Simon Ret al. Lorlatinib induces durable disease stabilization in a pancreatic cancer patient with a ROS1 p.L1950F mutation: case report. Oncol Res Treat. 2021;44(9):495–502. [DOI] [PubMed] [Google Scholar]
- 62. Strickler JH, Satake H, George TJet al. Sotorasib in KRAS p.G12C-mutated advanced pancreatic cancer. N Engl J Med. 2023;388(1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heining C, Horak P, Uhrig Set al. NRG1 Fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 2018;8(9):1087–1095. [DOI] [PubMed] [Google Scholar]
- 64. Lee MS, Pant S. Personalizing medicine with germline and somatic sequencing in advanced pancreatic cancer: current treatments and novel opportunities. Am Soc Clin Oncol Educ Book. 2021;41:1–13. [DOI] [PubMed] [Google Scholar]
- 65. Bekaii-Saab TS, Spira AI, Yaeger Ret al. KRYSTAL-1: updated activity and safety of adagrasib (MRTX849) in patients (Pts) with unresectable or metastatic pancreatic cancer (PDAC) and other gastrointestinal (GI) tumors harboring a KRASG12C mutation. J Clin Onco. 2022;40(4):519–519. [Google Scholar]
- 66. Jones MR, Williamson LM, Topham JTet al. NRG1 Gene fusions are recurrent, clinically actionable Gene rearrangements in KRAS wild-type pancreatic ductal adenocarcinoma. Clin Cancer Res. 2019;25(15):4674–4681. [DOI] [PubMed] [Google Scholar]
- 67. Pishvaian MJ, Blais EM, Brody JRet al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the know your tumor registry trial. Lancet Oncol. 2020;21(4):508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018;8(9):a031435. doi:10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Collisson EA, Sadanandam A, Olson Pet al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Moffitt RA, Marayati R, Flate ELet al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10):1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. O'Kane GM, Grünwald BT, Jang GHet al. GATA6 Expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin Cancer Res. 2020;26(18):4901–4910. [DOI] [PubMed] [Google Scholar]
- 72. O'Kane GM, Fischer S, Denroche Ret al. Integrative molecular profiling and response to chemotherapy on the COMPASS trial. J Clin Onco. 2019;37(4):188–188. [Google Scholar]
- 73. Raghavan S, Winter PS, Navia AWet al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184(25):6119–6137.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Williams HL, Dias Costa A, Zhang Jet al. Spatially resolved single-cell assessment of pancreatic cancer expression subtypes reveals co-expressor phenotypes and extensive intratumoral heterogeneity. Cancer Res. 2023;83(3):441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Porter RL, Magnus NKC, Thapar Vet al. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2019;116(52):26835–26845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hwang WL, Jagadeesh KA, Guo JAet al. Single-nucleus and spatial transcriptomics of archival pancreatic cancer reveals multi-compartment reprogramming after neoadjuvant treatment. bioRxiv 2020.08.25.267336. Published online 25 August 2020. doi:10.1101/2020.08.25.267336. [Google Scholar]
- 77. Hayashi A, Fan J, Chen Ret al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. 2020;1(1):59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kalisz M, Bernardo E, Beucher Aet al. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J. 2020;39(9):e102808. doi:10.15252/embj.2019102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33(3):512–526.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Steele NG, Carpenter ES, Kemp SBet al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat Cancer. 2020;1(11):1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Candido JB, Morton JP, Bailey Pet al. CSF1R+ Macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 2018;23(5):1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Steele CW, Karim SA, Leach JDGet al. CXCR2 Inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell. 2016;29(6):832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Grunwald BT, Devisme A, Andrieux Get al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell. 2021;184(22):5577–5592.e18. [DOI] [PubMed] [Google Scholar]
- 84. Chen Y, Yang S, Tavormina Jet al. Oncogenic collagen I homotrimers from cancer cells bind to α3β1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell. 2022;40(8):818–834.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tu M, Klein L, Espinet Eet al. TNF-α-producing macrophages determine subtype identity and prognosis via AP1 enhancer reprogramming in pancreatic cancer. Nat Cancer. 2021;2(11):1185–1203. [DOI] [PubMed] [Google Scholar]
- 86. Su H, Yang F, Fu Ret al. Collagenolysis-dependent DDR1 signalling dictates pancreatic cancer outcome. Nature. 2022;610(7931):366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21(4):217–238. [DOI] [PubMed] [Google Scholar]
- 88. Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer—clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17(9):527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tian C, Clauser KR, Öhlund Det al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci USA. 2019;116(39):19609–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Halbrook CJ, Pontious C, Kovalenko Iet al. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 2019;29(6):1390–1399.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cappellesso F, Orban MP, Shirgaonkar Net al. Targeting the bicarbonate transporter SLC4A4 overcomes immunosuppression and immunotherapy resistance in pancreatic cancer. Nat Cancer. 2022;3(12):1464–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Helms E, Onate MK, Sherman MH. Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discov. 2020;10(5):648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sharma P, Siddiqui BA, Anandhan Set al. The next decade of Immune checkpoint therapy. Cancer Discov. 2021;11(4):838–857. [DOI] [PubMed] [Google Scholar]
- 94. Royal RE, Levy C, Turner Ket al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33(8):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Patnaik A, Kang SP, Rasco Det al. Phase I study of Pembrolizumab (MK-3475; Anti-PD-1 Monoclonal Antibody) in patients with advanced solid tumors. Clin Cancer Res. 2015;21(19):4286–4293. [DOI] [PubMed] [Google Scholar]
- 96. Brahmer JR, Tykodi SS, Chow LQMet al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Byrne KT, Vonderheide RH. CD40 Stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Rep. 2016;15(12):2719–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Padrón LJ, Maurer DM, O'Hara MHet al. Sotigalimab and/or nivolumab with chemotherapy in first-line metastatic pancreatic cancer: clinical and immunologic analyses from the randomized phase 2 PRINCE trial. Nat Med. 2022;28(6):1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Balachandran VP, Łuksza M, Zhao JNet al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Balli D, Rech AJ, Stanger BZ, Vonderheide RH. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res. 2017;23(12):3129–3138. [DOI] [PubMed] [Google Scholar]
- 101. Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell localization, activation, and clonal expansion in human pancreatic ductal adenocarcinoma. Cancer Immunol Res. 2017;5(11):978–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bockorny B, Grossman JE, Hidalgo M. Facts and hopes in immunotherapy of pancreatic cancer. Clin Cancer Res. 2022;28(21):4606–4617., Published online 1 July 2022. doi:10.1158/1078-0432.CCR-21-3452. [DOI] [PubMed] [Google Scholar]
- 103. Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat Cancer. 2022;3(3):272–286. [DOI] [PubMed] [Google Scholar]
- 104. Leidner R, Sanjuan Silva N, Huang Het al. Neoantigen T-cell receptor gene therapy in pancreatic cancer. N Engl J Med. 2022;386(22):2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Biankin AV, Waddell N, Kassahn KSet al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bailey P, Chang DK, Forget MAet al. Exploiting the neoantigen landscape for immunotherapy of pancreatic ductal adenocarcinoma. Sci Rep. 2016;6(1):35848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cattolico C, Bailey P, Barry ST. Modulation of type I interferon responses to influence tumor-immune cross talk in PDAC. Front Cell Dev Biol. 2022;10:816517. doi:10.3389/fcell.2022.816517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Barriga FM, Tsanov KM, Ho YJet al. MACHETE identifies interferon-encompassing chromosome 9p21.3 deletions as mediators of immune evasion and metastasis. Nat Cancer. 2022;3(11):1367–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Baslan T, Morris JP, Zhao Zet al. Ordered and deterministic cancer genome evolution after p53 loss. Nature. 2022;608(7924):795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cabanos HF, Hata AN. Emerging insights into targeted therapy-tolerant persister cells in cancer. Cancers. 2021;13(11):2666. doi:10.3390/cancers13112666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhou X, Kurilov R, Neoptolemos JPet al. Abstract A010: persister cell phenotypes contribute to poor patient outcomes after neoadjuvant chemotherapy in PDAC. Cancer Res. 2022;82(22):A010. doi:10.1158/1538-7445.PANCA22-A010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Weizman N, Krelin Y, Shabtay-Orbach Aet al. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene. 2014;33(29):3812–3819. [DOI] [PubMed] [Google Scholar]
- 113. Binenbaum Y, Fridman E, Yaari Zet al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78(18):5287–5299. [DOI] [PubMed] [Google Scholar]
- 114. Vennin C, Melenec P, Rouet Ret al. CAF hierarchy driven by pancreatic cancer cell p53-status creates a pro-metastatic and chemoresistant environment via perlecan. Nat Commun. 2019;10(1):3637. doi:10.1038/s41467-019-10968-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shen S, Vagner S, Robert C. Persistent cancer cells: the deadly survivors. Cell. 2020;183(4):860–874. [DOI] [PubMed] [Google Scholar]
- 116. Sharma SV, Lee DY, Li Bet al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Aung KL, Fischer SE, Denroche REet al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin Cancer Res. 2018;24(6):1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sakamoto H, Attiyeh MA, Gerold JMet al. The evolutionary origins of recurrent pancreatic cancer. Cancer Discov. 2020;10(6):792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bear AS, Vonderheide RH, O'Hara MH. Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell. 2020;38(6):788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Seifert L, Werba G, Tiwari Set al. Radiation therapy induces macrophages to suppress T-cell responses against pancreatic tumors in mice. Gastroenterology. 2016;150(7):1659–1672.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nolan E, Bridgeman VL, Ombrato Let al. Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization. Nat Cancer. 2022;3(2):173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article.