Despite previous circulation of the highly transmissible and antibody evasive BA.2.75, BQ.1, XBB.1 and XBB.1.5 lineages, the share of infections caused by the SARS-CoV-2 lineage XBB.1.16 has gradually increased in India in early 2023, resulting in XBB.1.16 being the dominating SARS-CoV-2 lineage in India today. Since a similar trend may also take place in other countries and information on the biological properties of the XBB.1.16 lineage is scarce, we conducted a rapid assessment of the SARS-CoV-2 XBB.1.16 lineage with respect to its ability to enter cells and evade neutralisation by antibodies.
The newly emerged SARS-CoV-2 XBB.1.16 lineage (also dubbed as Arcturus), which harbours a unique combination of spike (S) protein mutations (Fig. 1a), was first described in India in January 2023 and rapidly became the dominating lineage (https://cov-spectrum.org/ [1]) (Fig. 1b). Here we performed a rapid assessment of the SARS-CoV-2 XBB.1.16 lineage regarding its ability to enter cells and evade neutralisation by antibodies using S protein-carrying pseudovirus particles (pp), which constitute a suitable model to study host cell entry of SARS-CoV-2 and its neutralisation [2]. For comparison, particles bearing the S protein of the ancestral B.1 lineage (B.1pp) or S proteins of Omicron sublineages BA.5 (BA.5pp), CH.1.1 (CH.1.1pp), XBB.1 (XBB.1pp), or XBB.1.5 (XBB.1.5pp) were used.
Fig. 1.
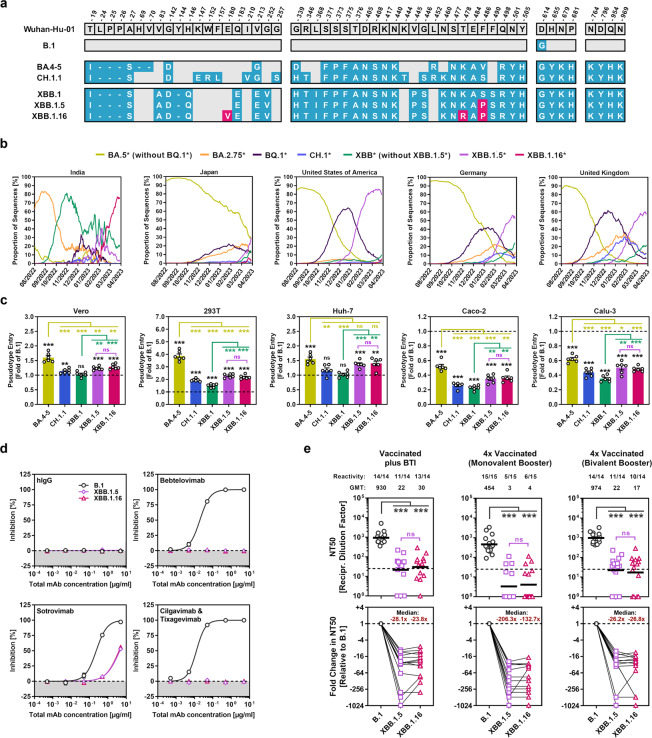
Host cell entry and neutralisation sensitivity of the SARS-CoV-2 XBB.1.16 lineage. a Mutations in the S proteins of SARS-CoV-2 lineages B.1, BA.4-5 (identical on protein level), CH.1.1, XBB.1, XBB.1.5 and XBB.1.16 compared to the S protein of the Wuhan-Hu-01 isolate. Unique mutations in the S proteins of XBB.1.5 and XBB.1.16 (compared to XBB.1) are highlighted in pink. b Relative frequency of SARS-CoV-2 lineages BA.5* (without BQ.1*), BA.2.75*, BQ.1*, CH.1*, XBB.1* (without XBB.1.5* and XBB.1.16), XBB.1.5*, and XBB.1.16* in selected countries. (Graphs are based on data obtained from https://cov-spectrum.org/). c Cell line tropism and entry efficiency of the SARS-CoV-2 XBB.1.16 lineage. Pseudovirus particles bearing the indicated S proteins were inoculated onto Vero (African green monkey, kidney), 293 T (human, kidney), Huh-7 (human, liver), Caco-2 (human, colon), and Calu-3 (human, lung) cells. Cell entry was analysed at 16–18 h postinoculation by measuring luciferase activity in cell lysates. Presented are the normalised mean data from six biological replicates (performed with four technical replicates) in which cell entry was normalised against particles carrying B.1 S protein (set as 1). Error bars represent the standard error of the mean (SEM). Statistical significance was assessed by two-tailed Student’s t-test with Welch correction (not significant [ns], p > 0.05; **p ≤ 0.01; ***p ≤ 0.001). Please see also Fig. S1. d Sensitivity of the SARS-CoV-2 XBB.1.16 lineage to neutralisation by monoclonal antibodies (mAb). Pseudotype particles harbouring the S protein of SARS-CoV-2 lineages B.1, XBB.1.5 or XBB.1.16 were preincubated with individual mAb or mAb cocktails for 30 min at room temperature, before being inoculated onto Vero cells (hIgG represents an unrelated control antibody). Pseudovirus entry was analysed at 16–18 h postinoculation and normalised against samples without mAb (= 0% inhibition). Data represent the mean of three biological replicates (performed with four technical replicates). Error bars indicate the SEM. e Sensitivity of the SARS-CoV-2 XBB.1.16 lineage to neutralisation by antibodies induced by vaccination or vaccination plus breakthrough infection. Pseudotype particles harbouring the S protein of SARS-CoV-2 lineage B.1, XBB.1.5, or XBB.1.16 were preincubated with plasma from (i) three- or four-times vaccinated individuals with breakthrough infection (BTI) between October 2022 and March 2023 in Germany (n = 14), (ii) four-times vaccinated individuals that received the monovalent BNT162b2/Comirnaty vaccine booster (n = 15), or (iii) four-times vaccinated individuals that received the bivalent BNT162b2/Comirnaty Original/Omicron BA.4-5 vaccine booster (n = 14). After an incubation period of 30 min at room temperature, samples were inoculated onto Vero cells. Pseudovirus entry was analysed at 16–18 h postinoculation, normalised against samples without plasma (0% inhibition), and the neutralising titre 50 (NT50, indicating the plasma dilution responsible for half-maximal inhibition) was calculated. Top panels: Presented are the geometric mean NT50 values (geometric mean titres, GMT) from a single biological replicate (conducted with four technical replicates). Numbers above the graphs represent reactivity rates (= proportion of plasma samples with detectable neutralising activity) and GMT. Statistical significance was assessed by Wilcoxon matched-pairs signed rank test (ns, p > 0.05; ***p ≤ 0.001). Please also see Table S1 and Figs. S2–S4. Bottom panels: Median fold change in neutralisation relative to B.1pp (set as 1). Individual plasma are connected by lines
In line with expectations, BA.5pp entered Vero (African green monkey, kidney), 293 T (human, kidney) and Huh-7 (human, liver) cells with higher efficiency compared to B.1pp (1.6–3.9x higher, respectively), while entry into Caco-2 (human, colon) and Calu-3 (human, lung) cells was less efficient (1.6–1.9x reduced, compared to B.1pp) [3] (Fig. 1c). CH.1.1pp and XBB.1pp displayed comparable to slightly higher entry efficiency for Vero, 293 T and Huh-7 cells compared to B.1pp (1.1x–1.9x higher, compared to B.1pp), while Caco-2 and Calu-3 cell entry was even less efficient as compared to BA.5pp (2.3–3.8x reduced, compared to B.1pp) (Fig. 1c). In accordance with literature, cell entry of XBB.1.5pp was generally increased compared to XBB.1pp (1.2–1.5x higher, compared to XBB.1pp) [4] and the same observation was made for XBB.1.16pp (1.3–1.6x higher, compared to XBB.1pp) (Fig. 1c).
As the mutations of XBB.1.16 might increase neutralisation evasion, we next investigated neutralisation sensitivity of XBB.1.16pp to clinically used monoclonal antibodies (mAbs) as well as antibodies elicited upon vaccination, or vaccination plus breakthrough infection (BTI). With respect to mAb neutralisation, XBB.1.16pp displayed partial resistance against Sotrovimab (effective concentration 50: 0.2 µg/ml [B.1] vs. 4.1 µg/ml [XBB.1.16]), and full resistance against Bebtelovimab, and a cocktail of Cilgavimab and Tixagevimab (Evusheld) (Fig. 1d), all of which is similar to what has been reported for XBB.1pp [5] and XBB.1.5pp (Fig. 1d) [4]. Finally, we analysed sensitivity of XBB.1.16pp to neutralisation by vaccination- or vaccination plus BTI-induced antibodies. For the latter, we selected plasma from vaccinated individuals that became infected between October 2022 and March 2023 in Germany, a period in which sublineages of BA.5, BQ.1, BA.2.75, CH.1, and XBB.1.5 were abundant (Fig. 1b), since we did not have access to samples from individuals with proven XBB.1.5 infection. All plasma showed high neutralising activity against B.1pp, while the same plasma displayed strongly reduced neutralising activities against XBB.1.16pp and XBB.1.5pp (23.8–90.0x reduced and 26.2–132.7x reduced compared to B.1pp, respectively) (Fig. 1e). No differences in neutralisation sensitivity were observed between XBB.1.16pp and XBB.1.5pp (Fig. 1e).
Altogether, our data indicate that XBB.1.16pp and XBB.1.5pp display similar characteristics regarding cell line tropism, host cell entry efficiency and neutralisation evasion. This suggests that the recent increase in XBB.1.16 frequency in India may be linked to the immunisation background of the Indian population (e.g., high vaccination rate with vectored-adenovirus vaccines, pronounced circulation of BA.2.75 and early XBB.1 sublineages in the second half of 2022; Fig. 1b) or to specific properties of the XBB.1.16 lineage that could not be resolved within this study. Future studies are required to address these possibilities. For instance, it needs to be investigated whether XBB.1.16 has an advantage over XBB.1.5 when it comes to infection of individuals who have a history of infection by XBB.1.5 (or other XBB.1 sublineages).
Supplementary information
Acknowledgements
We gratefully acknowledge the originating laboratories responsible for obtaining the specimens, as well as the submitting laboratories where the genome data were generated and shared via GISAID, on which this research is based. We thank all study participants for their support and Janine Topal, Luis Manthey and Noemí Calderón Hampel for technical and logistical help. We further thank Stephan Ludwig, Andrea Maisner, Thomas Pietschmann and Gert Zimmer for providing reagents. SP acknowledges funding by the EU project UNDINE (grant agreement number 101057100), the Ministry for Science and Culture of Lower Saxony (Niedersächsisches Ministerium für Wissenschaft und Kultur; 14-76103-184, COFONI Network, including projects 7FF22, 6FF22, 10FF22) and the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG; PO 716/11-1). H-MJ received funding from BMBF (01KI2043, NaFoUniMedCovid19-COVIM: 01KX2021), Bavarian State Ministry for Science and the Arts (Bayerisches Staatsministerium für Wissenschaft und Kunst) and DFG through the research training groups RTG1660 and TRR130, the Bayerische Forschungsstiftung (Project CORAd) and the Kastner Foundation. GMNB acknowledges funding by the German Centre for Infection Research (Deutsches Zentrum für Infektionsforschung, DZIF; grant no 80018019238). Further, GMNB and AD-J acknowledge funding by a European Regional Development Fund (Defeat Corona, ZW7-8515131). The funding sources had no role in the design and execution of the study, the writing of the manuscript and the decision to submit the manuscript for publication.
Author contributions
Conceptualisation: SP and MH; Methodology: IN, AK, PA, SP and MH; Investigation: IN, AK, MVS and MH; Formal analysis: IN, AK and MH; Recruitment and plasma collection: AC, AD-J and GMNB; Resources: AC, AD-J, SRS, H-MJ and GMNB; Funding acquisition: H-MJ, GMNB, AD-J and SP; Writing – original draft: PA and MH; Writing – review & editing: all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
SP and MH conducted contract research (testing of vaccine sera for neutralising activity against SARS-CoV-2) for Valneva unrelated to this work. GMNB served as advisor for Moderna and SP served as advisor for BioNTech, unrelated to this work. All other authors declare no competing interests.
Footnotes
These authors contributed equally: Inga Nehlmeier, Amy Kempf.
Contributor Information
Stefan Pöhlmann, Email: spoehlmann@dpz.eu.
Markus Hoffmann, Email: mhoffmann@dpz.eu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-023-01030-z.
References
- 1.Chen C, Nadeau S, Yared M, Voinov P, Xie N, Roemer C, et al. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2022;38:1735–7. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora P, Zhang L, Nehlmeier I, Kempf A, Cossmann A, Dopfer-Jablonka A, et al. The effect of cilgavimab and neutralisation by vaccine-induced antibodies in emerging SARS-CoV-2 BA.4 and BA.5 sublineages. Lancet Infect Dis. 2022;22:1665–6. doi: 10.1016/S1473-3099(22)00693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Arora P, Nehlmeier I, Kempf A, Cossmann A, Schulz SR, et al. Profound neutralization evasion and augmented host cell entry are hallmarks of the fast-spreading SARS-CoV-2 lineage XBB.1.5. Cell Mol Immunol. 2023;20:419–22. doi: 10.1038/s41423-023-00988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arora P, Cossmann A, Schulz SR, Ramos GM, Stankov MV, Jack HM, et al. Neutralisation sensitivity of the SARS-CoV-2 XBB.1 lineage. Lancet Infect Dis. 2023;23:147–8. doi: 10.1016/S1473-3099(22)00831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.