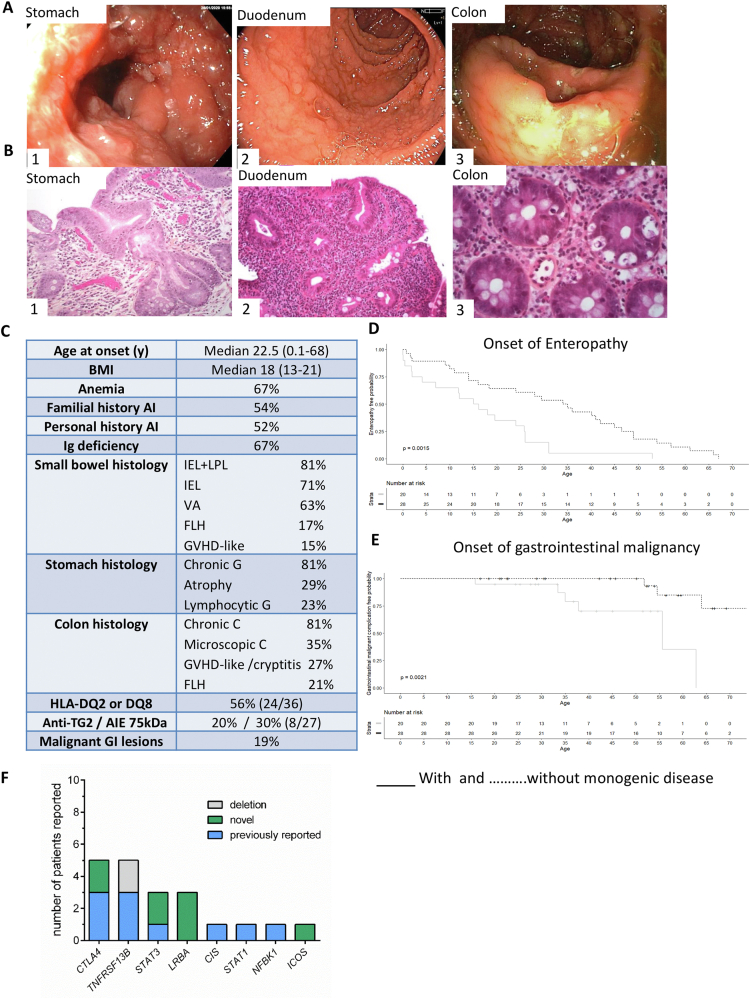
Autoimmune enteropathy (AIE) is a severe form of enteropathy characterized by chronic diarrhea refractory to any exclusion diet and associated with autoimmunity.1,2 Despite this clear definition, diagnosis is challenging. The presence of antienterocyte antibodies against the AIE-75kDa antigen3 is a strong but inconstant biomarker. In a recent cohort of 40 AIE patients, anti-enterocyte antibodies were reported in only 14% (4/28) of the cases, likely caused by the high frequency of patients with primary hypogammaglobulinemia.2 Moreover patients may display celiac anti-transglutaminase antibodies.4 The common histopathologic presentation of AIE includes intestinal villous atrophy with variable lymphocytic infiltration and various features of follicular lymphoid hyperplasia, cryptitis, graft-versus-host disease-like lesions, and loss of Paneth and goblet cells.2,4 This mixed histopathologic pattern combined with inefficacy of gluten-free diet allow differentiating AIE from celiac disease.1,2,4 Treatment remains challenging because AIE patients are only variably improved by steroids and immunomodulators.4 Therefore, identification of the underlying molecular mechanism is crucial to treat unresponsive patients. Genetic studies have revealed that several monogenic inborn errors of immunity, notably NFKB1 (nuclear factor kappa B subunit 1), CTLA4 (cytotoxic T-lymphocyte-associated protein 4) haploinsufficiency, and LRBA (LPS responsive beige-like anchor protein) deficiency may manifest by AIE only at adulthood (reviewed in5). The latter finding prompted us to screen a cohort of 48 adult patients with AIE by next-generation sequencing (Supplementary Methods). Representative endoscopic findings and histologic features are shown in Figure 1A and B. Clinical features are summarized in Figure 1C. Malnutrition (body mass index <19), anemia, and primary immunoglobulin deficiency were present in 67% of cases. Small bowel inflammation consisted mainly in villous atrophy in 30/48 (63%), and intraepithelial and/or lamina propria lymphocytosis in 39/48 patients (81%). Intestinal lymphocytes had a normal phenotype and TCRγ rearrangements showed a polyclonal profile in intestinal biopsies except in 2 patients who developed intestinal CD4+ T-cell lymphoproliferations. Chronic gastritis and colitis were present in 39/48 patients (81%). Eight patients with primary hypogammaglobulinemia and abnormal hepatic tests and/or liver imaging had biopsy-proven nodular regenerative hyperplasia. Main peripheral blood abnormalities were lymphopenia (34/48), particularly B lymphopenia, in keeping with the high frequency of primary hypogammaglobulinemia.
Figure 1.
(A) Representative endoscopic aspects in patients with CTLA4 variants and AIE. (1) Ulcerative gastritis with pseudobstruction in patient P9. (2) Duodenal villous atrophy in P1. (3) Colitis with ulcerations in P9. (B) Representative histologic lesions in patients with AIE. (1) Chronic fundic atrophic gastritis with intestinal metaplasia and focal low-grade dysplasia (×200) in P9 carrying a CTLA4 variant. (2) Chronic duodenitis in P1 with CTLA4 mutation with severe villous blunting and increased IEL numbers (×200). (3) Colitis with GVHD-like lesions and cryptic apoptotic bodies (×400) in a patient without identified pathogenic variant (description of numbered patients is provided in Supplementary Table 1). (C) Clinical features of the 48 patients with AIE. (D) Kaplan-Meier enteropathy-free survival curve of patients with (solid line) or without molecular diagnosis (dashed line). (E) Kaplan-Meier gastrointestinal malignant-free survival curve of patients with (solid line) or without molecular diagnosis (dashed line). (F) Number of patients for each individual molecular diagnosis. anti-TG2, antitransglutaminase of type 2; AI, autoimmune; AIE-75kDa, autoimmune enteropathy-related 75-kilodalton antibody; BMI, body mass index; C, colitis; IEL, intestinal intraepithelial hyperlymphocytosis; FLH, follicular lymphoid hyperplasia; G, gastritis; GI, gastrointestinal; GVHD, graft-versus-host disease; LPL, lamina propria hyperlymphocytosis; VA, villous atrophy.
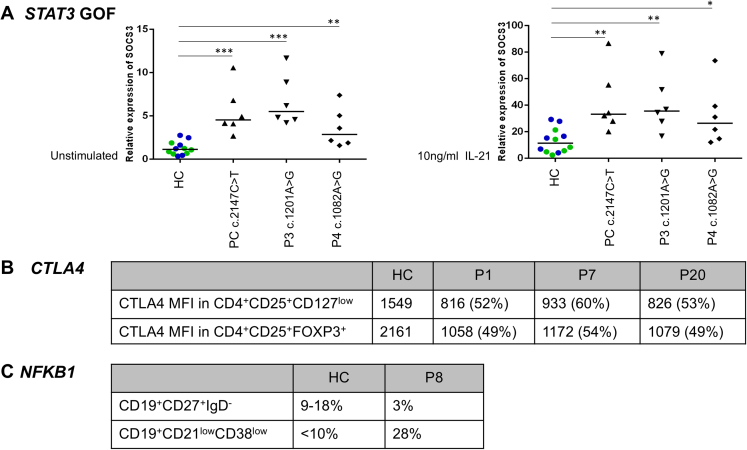
Pathogenic variants were identified in 20/48 patients (41.6%) (Supplementary Table 1). In 9 cases, the pathogenic variants were previously reported as disease causing. Five other cases displayed nonsense variants that were considered pathogenic according to American College of Medical Genetics and Genomics guidelines.6 Functional validation was performed for missense variants that had not been previously described in the literature (Supplementary Figure 1).
Supplementary Figure 1.
Functional validation of novel mutations. (A): RT-PCR of STAT3-target SOCS3 at baseline and in IL-21 activated-EBV cell lines of P3 and P4. Results are shown as relative expression normalized to RPLPO housekeeping gene and represent the mean of 6 independent experiments. ∗P < .05, ∗∗P < .01, ∗∗∗P < ,005 Mann-Whitney test; HC: results pooled from 2 healthy controls (HC1 in green, HC2 in blue), PC: positive control; (B): Decreased CTLA-4 mean fluorescence intensity (MFI) on circulating T-cells in P1, P7 and P20 as compared to healthy control (HC) by flow cytometry; (C): B-cell immunophenotyping in P8, showing decreased frequency of CD27+ and excess of CD21lowCD38low B-cells.
Before diagnosis, gluten-free diet (20/30; 67%) and, in patients with primary hypogammaglobulinemia, immunoglobulin replacement therapy (19/32; 59%) were ineffective in all cases. Steroids (34 patients; 71%), frequently budesonide (27/48), induced partial clinical remission and partial mucosal recovery in all except 2. Thirteen patients (27%) received immunomodulators and 26 (54%) at least 1 biotherapy. Anti–tumor necrosis factor-α agents (19 patients; 40%) induced partial and/or transient clinical remission without any histologic effect in all except 2. In 8 patients, molecular diagnosis enabled targeted therapy. Abatacept, a soluble fusion protein analogue to CTLA-4, induced durable (n = 3) or transient (n = 3) remission with relapse occurring after 2–3 years. In 1 patient with STAT3 gain of function (GOF) variant, JAK inhibitor ruxolitinib induced complete clinical and histologic remission still persistent after 4 years.7 In contrast, ruxolitinib had no effect in 1 patient carrying a STAT1 GOF variant.
Patients with identified pathogenic gene variants had earlier onset of enteropathy (P = .0015) (Figure 1D). Malignant gastrointestinal disorders were observed in 9 patients and developed more rapidly in those with identified genetic variants (P = .0021) (Figure 1D). Nevertheless, identification of a pathogenic variant did not significantly affect global survival. Four patients with (CTLA4, n = 2) or without (n = 2) identified pathogenic variants developed gastric dysplasia (n = 2) or gastric adenocarcinoma (n = 2). All 4 had atrophic gastritis, Helicobacter pylori being only detected in 1 patient before onset of gastric adenocarcinoma. Endoscopic mucosal resection was performed in all cases of gastric dysplasia, and total gastrectomy with radiotherapy in 1 patient with gastric cancer. Duodenal adenoma developed in 1 patient with STAT3 GOF variant and villous atrophy and was endoscopically resected. Colonic dysplastic adenomas occurred in 3 patients (CTLA4, n = 1; NFKB1, n = 1; no diagnosis, n = 1), all treated by endoscopic resection. Among them, high-grade dysplasia was observed in the mutated NFKB1 patient. Low-grade intestinal CD4+ T-cell lymphoproliferation was observed in 2 patients: 1 patient without identified variant but treated by azathioprine still displayed conspicuous CD4+ T-cell intraepithelial infiltration without detectable clonal TCRγ rearrangement 2 years after azathioprine discontinuation. The second patient with TNFRSF13B variant progressed toward fatal high-grade CD4+ T-cell lymphoma. Extraintestinal tumors were diagnosed in 2 other patients without identified pathogenic variants: 1 thymoma (4 years after first detection of hypogammaglobulinemia, a posteriori leading to diagnosis of Good syndrome) and 1 papillary thyroid cancer treated by surgery. Six patients died and 5 patients were lost to follow-up.
In summary, 40% of our adult AIE patients carried pathogenic variants providing clues for diagnosis, treatment, and follow-up. Diagnosis of AIE can be difficult and notably mistaken for refractory celiac disease. Because 30%–50% of adult AIE can display anti-transglutaminase antibodies4,8 identification of genetic variant allows diagnosis of AIE. Tailor-made therapeutics were available for more than half of the patients who received a molecular diagnosis (patients with variants in CTLA4, LRBA, STAT3, and STAT1; 12/20; 60%). Herein, 8 patients received such therapy. All patients responded except 1 patient with STAT1 GOF and very severe disease. Along this line, a recent study reported significant improvement in 14/17 patients with STAT1 or STAT3 GOF variants treated with JAK-inhibitors.9 Finally, this study highlights the risk of malignant transformation associated with AIE and their earlier onset in patients with identified variants, in keeping with previous data demonstrating increased risk of gastric neoplasia in CTLA4-mutated patients.10 Overall these observations stress the need for early genetic diagnosis and regular endoscopic follow-up of these patients.
Acknowledgments
The authors thank Université Paris Cité, Imagine Institute, Bioinformatics Platform, Paris, France; Université Paris Cité, Imagine Institute, Translational Genomics-Sylvain Hanein, Paris, France; Université Paris Cité, APHP Hôpital Necker, Imagine Institute, Centre de Ressources Biologiques, Paris, France; and The Centre de Référence Déficits Immunitaires Héréditaires.
Contributors from the AutoImmune Enteropathy Working Group: Sherine Khater∗, Department of Gastroenterology, AP-HP, Centre- Université de Paris, Hôpital Européen Georges Pompidou, Paris, France; Anis Khiat, Université de Paris Cité, INSERM UMR 1163 and Imagine Institute, Laboratory of Intestinal Immunity, Paris, France; Sascha Cording, Université de Paris Cité, INSERM UMR 1163 and Imagine Institute, Laboratory of Intestinal Immunity, Paris, France; Marianna Parlato, Université de Paris Cité, INSERM UMR 1163 and Imagine Institute, Laboratory of Intestinal Immunity, Paris, France; Marie-Agnès Dragon-Durey, Department of Immunology, AP-HP, Centre-Université Paris Cité, Hôpital Européen Georges Pompidou, Paris, France; Frédéric Beuvon, Department of Pathology, AP-HP, Centre-Université Paris Cité, Hôpital Cochin, Paris, France; Nicole Brousse, Department of Pathology AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Benoît Terris, Department of Pathology, AP-HP, Centre-Université Paris Cité, Hôpital Cochin, Paris, France. Capucine Picard, Study Center of Primary Immunodeficiency, AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Mathieu Fusaro, Study Center of Primary Immunodeficiency, AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Frédéric Rieux-Laucat, Université de Paris, INSERM UMR1163 and Imagine Institute, Immunogenetics of Pediatric Autoimmune Diseases, Paris, France; Marie-Claude Stolzenberg, Université de Paris, INSERM UMR1163 and Imagine Institute, Immunogenetics of Pediatric Autoimmune Diseases, Paris, France; Anne-Sophie Jannot, Department of Clinical Investigation and Clinical Epidemiology, AP-HP-Centre-Université Paris Cité, Hôpital Européen Georges Pompidou, Paris, France; Alexis Mathian, Department of Internal Medicine, AP-HP, Sorbonne Université, Hôpital Pitié Salpétrière, Paris, France; Matthieu Allez, Department of Gastroenterology, AP-HP, Nord-Université Paris Cité, Hôpital Saint Louis, Paris, France; Marion Malphettes, Department of Clinical Immunology, AP-HP, Nord-Université Paris Cité, Hôpital Saint Louis, Paris, France; Claire Fieschi, Department of Clinical Immunology, AP-HP, Nord-Université Paris Cité, Hôpital Saint Louis, Paris, France; Alexandre Aubourg, Department of Gastroenterology, CHRU de Tours, Tours, France; Camille Zallot, Department of Gastroenterology, CHRU de Nancy, Hôpitaux de Brabois, Nancy, France; Xavier Roblin, Department of Gastroenterology, CHU de Saint Etienne, Saint Etienne, France; Vered Abitbol, Department of Gastroenterology, AP-HP. Centre- Université de Paris, Hôpital Cochin, Paris, France; Arthur Belle, Department of Gastroenterology, AP-HP, Centre- Université de Paris, Hôpital Cochin, Paris, France; Pauline Wils, Department of Gastroenterology, CHRU de Lille, Lille, France; Morgane Cheminant, Department of Haematology, AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Tamara Matysiak-Budnik, Department of Gastroenterology, CHRU de Nantes, Hôpital Hôtel Dieu, Nantes, France; Lucine Vuitton, Department of Gastroenterology, CHRU de Besançon, Hôpital Jean Minjoz, Besançon, France; Philippe Pouderoux, Department of Gastroenterology, CHRU de Nîmes, Hôpital universitaire Carémeau, Nîmes, France; Laurent Abramowitz, Department of Gastroenterology, AP-HP, Nord-Université Paris Cité, Hôpital Bichat, Paris, France; Martin Castelle, Department of Pediatric Immunology and Hematology, AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Felipe Suarez, Department of Haematology, AP-HP. Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Olivier Hermine, Department of Haematology, AP-HP, Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Frank Ruemmele, Université de Paris Cité, INSERM UMR 1163 and Imagine Institute, Laboratory of Intestinal Immunity and Department of Paediatric Gastroenterology, AP-HP-Centre-Université Paris Cité, Hôpital Necker-Enfants Malades, Paris, France; Luc Mouthon, Service de Médecine Interne, Centre de Référence Maladies Autoimmunes Systémiques Rares d'Ile de France, AP-HP-Centre-Université Paris Cité, Hôpital Cochin, Paris, France.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by Institut National de la Santé et de la Recherche Médicale (INSERM) and Université de Paris; by ERC-2013-AdG-339407-IMMUNOBIOTA, Fondation des Maladies Rares, Association François Aupetit, Société Nationale Française de GastroEntérologie; by Agence Nationale de la Recherche (ANR-14-CE14-0026-01; ANR- 18-CE17-0001); by Government grants managed by Agence Nationale de la Recherche as part of the “Investment for the Future” program (ANR-10-IAHU-01 and ANR-18-RHUS-0010); and by Princesse Grace Fondation.
Note: To access the supplementary material accompanying this article, visit the online version of Clinical Gastroenterology and Hepatology at www.cghjournal.org, and at https://doi.org/10.1016/j.cgh.2022.07.030.
Contributor Information
Nadine Cerf-Bensussan, Email: nadine.cerf-bensussan@inserm.fr.
Georgia Malamut, Email: georgia.malamut@aphp.fr.
Supplementary Methods
Study Design and Primary Goal
Forty-eight adult patients with AIE were consecutively recruited for genetic analysis through multidisciplinary consultation meetings in 9 adult French university hospitals (AP-HP-Centre-Université Paris Cité [n = 32], AP-HP-Nord Université Paris Cité [n = 8], AP-HP-Université Paris Sorbonne [n = 2], CHRU de Besançon [n = 1], CHRU de Nantes [n = 1], CHU de St Etienne [n = 1], CHRU de Tours [n = 1], CHRU de Nîmes [n = 1], and CHRU de Lille [n = 1]).
Inclusion Criteria
All patients had chronic diarrhea and histopathologic features of AIE refractory to any diet.1 Enteropathy induced by sartans, checkpoint inhibitors, and refractory celiac disease were excluded. Clinical and biologic data were assessed as described.2
Outcome
Date at onset of enteropathy and of gastrointestinal malignant complications were compared between patients with and without identified pathogenic variants. Response to treatment was assessed retrospectively and prospectively. Clinical remission was defined by 50% reduction in stool frequency and recovery of at least 50% of body weight loss. Histologic response was defined by total or partial mucosal healing. Follow-up extended from diagnosis of enteropathy to December 31, 2020.
Genetic Analysis
Genetic analysis was conducted from January 2015 to December 2020. Some patients were also investigated by other research laboratories (P9 and P19,3 P14,4 and P185). We published 1 case (P36,7).
Targeted Next-Generation Sequencing and Whole Exome sequencing
Next-generation sequencing was performed on genomic DNA extracted from peripheral blood mononuclear cells, using custom-made targeted panels (TNGS) dedicated to intestinal disorders6 or using whole exome sequencing in trios (P3, P4, P5).7 Panel for TNGS described in6 has been updated with genes described in Charbit-Henrion et al8 and in Uhlig et al.9 Genomic DNA libraries were captured by hybridization using either Agilent Sure Select All Exon V5 or V6 (Agilent, Les Ulis, France) for whole exome sequencing or biotinylated complementary 120-pb RNA baits designed with SureSelect SureDesign software to cover all exons of the selected genes for TNGS. Data analysis was performed using the in-house platform POLYWEB created by the Bioinformatics core facilities of Université Paris Cité and Institut Imagine to prioritize rare and potentially damaging variants according to the American College of Medical Genetics guidelines,10 OMIM identity, allele frequency in control populations (gnomAD), conservation (UCSC genome browser, 100 vertebrates), protein function and expression, and phenotypic assessment. Consequences of variants on protein function were predicted using Combined Annotation-Dependent Depletion score (https://cadd.gs.washington.edu) and Splice AI (https://spliceailookup.broadinstitute.org). All variants were confirmed by Sanger sequencing and familial segregation was analyzed when DNA from family members was available.
Functional Validation
Functional validation was performed in patients carrying missense variants not previously reported. In P4 with STAT3 GOF mutation, transcription of Suppressor of Cytokine Signaling 3 (SOCS3) was studied in EBV transformed B cell lines as described.6,7 In P9, B-cell subsets were assessed by flow cytometry.3 In P18 and P19, LRBA expression was assessed by flow cytometry and Western blot.5,11 In P1, P7, and P20, CTLA4 expression was assessed by flow cytometry.3,11
Statistical Analysis
Frequencies were compared using the Fisher exact test. Global survival, enteropathy-free survival, and gastrointestinal malignant/premalignant-free survival were estimated using Kaplan-Meier estimates and survival differences using the log-rank test. For each analysis, comparison was performed between patients with and without identified pathogenic variant. Statistical analyses used the R software version 4.0.2 (Vienna, Austria). P < .05 was considered statistically significant.
Supplementary Appendix
References
- 1.Schiepatti A., et al. Gut. 2022 Jun 8 gutjnl-2021-326645. [Google Scholar]
- 2.Villanacci V., et al. Clin Immunol. 2019;207:10–17. doi: 10.1016/j.clim.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi I., et al. Gastroenterology. 1999;117:823–830. doi: 10.1016/s0016-5085(99)70340-9. [DOI] [PubMed] [Google Scholar]
- 4.Akram S., et al. Clin Gastroenterol Hepatol. 2007;5:1282–1290. doi: 10.1016/j.cgh.2007.05.013. quiz 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbit-Henrion F., et al. Mucosal Immunol. 2021;14:1017–1037. doi: 10.1038/s41385-021-00398-3. [DOI] [PubMed] [Google Scholar]
- 6.Richards S., et al. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parlato M., et al. Gastroenterology. 2019;156:1206–1210. doi: 10.1053/j.gastro.2018.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scialom S., et al. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125024. e0125024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes L.R., et al. J Allergy Clin Immunol. 2018;142:1665–1669. doi: 10.1016/j.jaci.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwab C., et al. J Allergy Clin Immunol. 2018;142:1932–1946. doi: 10.1016/j.jaci.2018.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Villanacci V., et al. Clin Immunol. 2019;207:10–17. doi: 10.1016/j.clim.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Scialom S., et al. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fusaro M., et al. J Allergy Clin Immunol. 2021;147 doi: 10.1016/j.jaci.2020.05.052. 737–737. [DOI] [PubMed] [Google Scholar]
- 4.Dragon-Durey M.A., et al. J Immunol. 2001;166:7612–7616. doi: 10.4049/jimmunol.166.12.7612. [DOI] [PubMed] [Google Scholar]
- 5.Besnard C., et al. Clin Immunol. 2018;188:52–57. doi: 10.1016/j.clim.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Charbit-Henrion F., et al. J Crohns Colitis. 2018;12:1104–1112. doi: 10.1093/ecco-jcc/jjy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parlato M., et al. Gastroenterology. 2019;156:1206–1210. doi: 10.1053/j.gastro.2018.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charbit-Henrion F., et al. Mucosal Immunol. 2021;14:1017–1037. doi: 10.1038/s41385-021-00398-3. [DOI] [PubMed] [Google Scholar]
- 9.Uhlig H.H., et al. J Pediatr Gastroenterol Nutr. 2021;72:456–473. doi: 10.1097/MPG.0000000000003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S., et al. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazerolles F., et al. Front Immunol. 2018;9:718. doi: 10.3389/fimmu.2018.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Schubert D., et al. Nat Med. 2014;20:1410–1416. doi: 10.1038/nm.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuijnenburg P., et al. J Allergy Clin Immunol. 2018;142:1285–1296. doi: 10.1016/j.jaci.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fusaro M., et al. J Allergy Clin Immunol. 2021;147:734–737. doi: 10.1016/j.jaci.2020.05.046. [DOI] [PubMed] [Google Scholar]
- 4.Arts P., et al. Genome Med. 2019;11:38. doi: 10.1186/s13073-019-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J.J., et al. J Allergy Clin Immunol. 2010;126:1234–1241. doi: 10.1016/j.jaci.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragon-Durey M.A., et al. J Immunol. 2001;166:7612–7616. doi: 10.4049/jimmunol.166.12.7612. [DOI] [PubMed] [Google Scholar]
- 7.Uzel G., et al. J Allergy Clin Immunol. 2013;131:1611–1623. doi: 10.1016/j.jaci.2012.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.