Abstract
Iota-toxin is produced by Clostridium perfringens type E strains and consists of two independent components, the enzymatic and binding components, referred to as Ia and Ib, respectively. A recombinant C. perfringens strain, strain 667/pMRP147, produced processed Ia and partially processed Ib, while a recombinant C. perfringens type A strain, strain TS133/pMRP147, in which the VirR-VirS two-component system is inactivated, produced only precursor forms of Ia and Ib. This suggests that iota-toxin is processed by a VirR-VirS-responsive protease, although not completely in the recombinant type A strain. The precursor forms of Ia and Ib were purified from cultures of the latter strain, and their proteolytic activation was examined. Treatment with proteases cleaved off small peptides (9 to 13 amino acid residues) and a 20-kDa peptide from the N termini of the Ia and Ib precursors, respectively, leading to their active forms. They were activated efficiently by α-chymotrypsin, pepsin, proteinase K, subtilisin, and thermolysin but only weakly by trypsin, as demonstrated by the cell-rounding assay. λ-Protease from the C. perfringens type E strain, which was found to be a zinc-dependent protease related to thermolysin, activated iota-toxin as efficiently as did α-chymotrypsin. These results suggest that λ-protease is most responsible for the activation of iota-toxin in type E strains.
Clostridium perfringens is a ubiquitous pathogen which causes food poisoning and gas gangrene in humans and digestive diseases in other animals. This organism is divided into five toxin types on the basis of the production of four major lethal toxins, alpha-, beta-, epsilon-, and iota-toxins (8). C. perfringens type E, which produces iota-toxin, has been implicated in the enterotoxemia of calves and lambs (22).
Iota-toxin is a member of the actin ADP-ribosylating iota-toxin family, which includes immunologically related clostridial toxins such as Clostridium spiroforme toxin (7) and Clostridium difficile ADP-ribosyltransferase (CDT) (17). They are binary toxins consisting of two independent polypeptides, enzymatic and binding components. The binding component of iota-toxin (Ib; Mr 80,000) is involved in the binding and internalization of the enzymatic component (Ia; Mr 47,500) (4). Ia catalyzes the ADP-ribosylation of monomeric G-actin of the muscle and nonmuscle type at Arg-177, leading to disorganization of the actin filaments (1, 24). The components of the iota-toxin family are interchangeable: Ib can internalize the enzymatic components of C. spiroforme toxin and CDT. In contrast, Clostridium botulinum C2 toxin, which is structurally and functionally related but immunologically unrelated to iota-toxin, does not translocate the enzymatic component of the iota-toxin family (4, 19).
It has been shown that iota-toxin is produced as inactive precursor (23). The proteolytic activation of an Ib precursor is accompanied by removal of a N-terminal 20-kDa peptide, and it has been generally considered that only Ib requires the proteolytic activation to exhibit biological activity. However, the mode of iota-toxin activation, especially an in vivo mechanism, has not been fully explained. In an attempt to better understand the activation mechanism, we determined the proteolytic cleavage sites for the Ia and Ib precursor forms by using various proteases such as trypsin, α-chymotrypsin, and proteases from C. perfringens. In addition, we compared the activating efficiency among these proteases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. perfringens strains NCIB 10748 (type E) (16), 667 (9), and TS133 (21) were used in this study. Strain 667 is a spontaneous lecithinase-negative strain as tested by culture on egg yolk agar and produces no toxin lethal for mice (9). Strain TS133, a gift from T. Shimizu, is a VirR-negative mutant of S13 (21). All the C. perfringens strains were grown in broth containing 30 g of Trypticase, 20 g of yeast extract, and 0.5 g of cysteine-HCl per liter (pH 7.2) under anaerobic conditions. Escherichia coli TG1 was used as the host for the construction of recombinant plasmids.
The pMRP108 insert (16) containing the iap and ibp genes under the control of their own promoter regions was transferred into the E. coli-C. perfringens shuttle vector, pJIR750 (2), yielding pMRP147. pMRP384 is a derivative of pMRP147 in which an AvaII DNA fragment within the iap gene has been deleted and which hence produces only Ib.
Electroporation of plasmid DNA into C. perfringens was performed by the method of Scott and Rood (20).
Purification of Ia and Ib.
Ia was purified from strain 667 or TS133 harboring pMRP147. Culture supernatants were prepared from the overnight cultures by centrifugation at 5,000 × g for 10 min. Proteins in the supernatant were precipitated with ammonium sulfate (70% saturation). The precipitate was dissolved in T buffer (10 mM Tris-HCl [pH 7.5]) and then dialyzed against T buffer. The dialysate was loaded onto a DEAE-Sephacel column (1.5 by 10 cm; Pharmacia, Orsay, France) equilibrated with T buffer. Proteins were eluted with a linear gradient of 0 to 0.1 M NaCl in T buffer. Fractions containing Ia, as tested by measuring the cytopathic activity in the presence of Ib, were combined and concentrated by precipitation with ammonium sulfate (70% saturation). Proteins were separated by gel filtration on a Superdex 200 column (2.6 by 60 cm; Pharmacia) equilibrated with T buffer. The Ia-containing fraction was stored at −80°C.
Ib was purified from cultures of strain 667 or TS133 harboring pMRP384. The supernatants from overnight cultures were subjected to precipitation with ammonium sulfate (70% saturation), dialyzed against T buffer, and dissolved in the same buffer. The proteins were loaded onto a DEAE-Sephacel column equilibrated with T buffer, and the column was washed with 0.1 M NaCl in T buffer and eluted with 0.2 M NaCl in T buffer. The eluate was dialyzed against 10 mM sodium citrate (pH 5) and loaded onto a DEAE-Sephacel column equilibrated with the same citrate buffer. Proteins were eluted with a liner gradient from 0 to 0.1 M in the citrate buffer. The Ib-containing fractions, as tested by measuring the cytopathic activity in the presence of Ia, were combined, concentrated by ammonium sulfate precipitation, and separated by gel filtration, as described above.
Proteolytic activation and amino acid sequencing of iota-toxin.
The iota-toxin components were activated in 10 mM Tris-HCl (pH 8) with 200 μg of proteases per ml unless otherwise stated. The proteases used in this study were trypsin-DPC (Serva, Heidelberg, Germany), proteinase K (Boehringer, Mannheim, Germany), α-chymotrypsin, papain, pepsin, subtilisin, thermolysin, thrombin, V8 protease (Sigma, L'Isle d'Abeau, France), and C. perfringens λ-protease, which was purified as described previously (10). After incubation at room temperature for 20 min, proteolysis was stopped by the addition of soybean trypsin inhibitor (Sigma) and Pefabloc (Boehringer) at a final concentration of 400 μg/ml each.
Proteins separated on a 0.1% sodium dodecyl sulfate (SDS)–10% polyacrylamide gel were transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore, St. Quentin, France). After amido black staining and destaining, the protein bands were cut out and subjected to Edman degradation on an Applied Biosystems 473A protein sequencer (Applied Biosystems, Norwalk, Conn.) for determination of the N-terminal amino acid sequence.
Cell-rounding assay.
Vero (African green monkey kidney) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. The cells were plated into a 96-well Falcon tissue culture plate (Becton Dickinson Labware, Oxnard, Calif.) and grown for 24 h to form a monolayer. Samples (100 μl) were serially diluted twofold with Dulbecco's modified Eagle's medium and then added to the monolayers. Changes in cell morphology were microscopically observed at the indicated times. Cytopathic units were expressed as the reciprocal of the highest dilution that caused rounding of 50% of the cells.
Ia mutants.
The recombinant plasmid pMRP189 containing the iap gene under the control of its own promoter was used for mutagenesis as previously described (15). Substitutions of Ile-10 to Ser and Ile-10 to Gly were performed using the oligonucleotides P666 (5′-GCAAGCAATTATGGTACAGATAGAGC-3′) and P664 (5′-GCAAGCAATTATTCTACAGATAGAGC-3′), respectively. The resulting plasmids, pMRP480 and pMRP481, respectively, were subjected to DNA sequencing to confirm the presence of the desired mutations and were transformed into E. coli strain BL21 (DE5). Ia protein mutants were purified from the bacterial sonicates by immunoaffinity chromatography using rabbit polyclonal antibodies against Ia (16) immobilized on a cyanogen bromide-activated Sepharose 4B (Pharmacia) column.
RESULTS
Production of processed and unprocessed iota-toxin components.
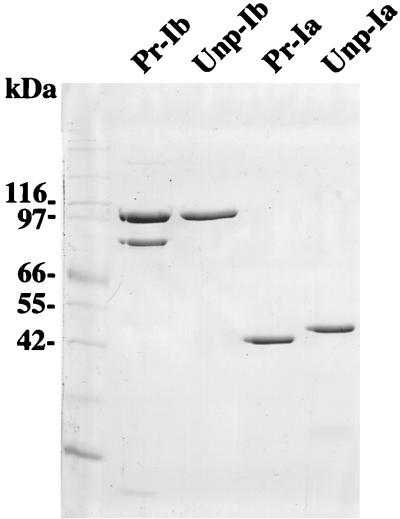
Recombinant C. perfringens strains were used to prepare iota-toxin components, since the production of iota-toxin in strain 667/pMRP147 was approximately 30-fold greater than in the wild-type strain NCIB 10748 as tested by measuring the cytopathic activity (data not shown). Ia purified from cultures of strain 667 harboring pMRP147 is referred to as processed Ia (Pr-Ia), and Ib purified from 667/pMRP384 is referred to as processed Ib (Pr-Ib), although the processing was partial (see below). Pr-Ia migrated as a single polypeptide with an apparent molecular mass of approximately 48 kDa on SDS-polyacrylamide gels, whereas Pr-Ib showed two bands at 80 and 100 kDa (Fig. 1).
FIG. 1.
SDS-PAGE of purified Pr-Ia, Unp-Ia, Pr-Ib, and Unp-Ib. A 1-μg portion of each component was electrophoresed on a 0.1% SDS–10% polyacrylamide gel.
The two-component system VirR-VirS regulates genes encoding various toxins and enzymes of C. perfringens such as alpha-toxin, theta-toxin, and collagenase (3, 21). The VirR-negative strain TS133 (21) harboring either pMRP147 or pMRP384 produced approximately the same amounts of iota-toxin components as did the 667 recombinant strains. However, Ia purified from TS133/pMRP147 had a slightly higher molecular weight than did Pr-Ia (Fig. 1), and hence it was termed unprocessed Ia (Unp-Ia). Ib purified from TS133/pMRP384, referred to as unprocessed Ib (Unp-Ib), showed only one band, corresponding to a 100-kDa polypeptide (Fig. 1). These results indicate that VirR was not involved in the control of iota-toxin production but could regulate the synthesis of a protease processing the iota-toxin components.
Activation of Ib.
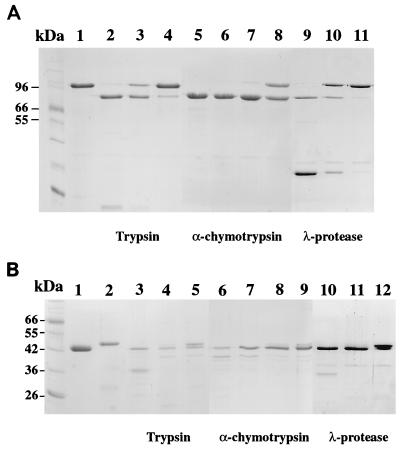
The maturation of Ib by proteases was examined using Unp-Ib, consisting only of high-molecular-weight polypeptide, which appears to be a precursor form (Fig. 1). α-Chymotrypsin and λ-protease but not trypsin efficiently processed Unp-Ib: the concentrations of α-chymotrypsin, λ-protease, and trypsin required for complete processing of the Unp-Ib were 0.5, 5, and 50 μg/ml, respectively, as demonstrated by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 2A). The N-terminal amino acid sequences of the Unp-Ib and Unp-Ib processed by these enzymes were determined (Fig. 3). Comparison of the N-terminal amino acid sequence of Unp-Ib with that deduced from the nucleotide sequence indicates that the N-terminal 28 amino acids correspond to a signal peptide. The N terminus of Pr-Ib determined for the lower-molecular-weight band of Pr-Ib begins with FFSA, indicating that Unp-Ib was cleaved at the C-terminal side of R, a trypsin-sensitive site. Unp-Ib processed by trypsin and α-chymotrypsin possessed the same N-terminal sequence, SAA, indicating that it was cleaved at an α-chymotrypsin cleavage site, F-S. This suggests that the activation of Unp-Ib by trypsin was due to residual activity of α-chymotrypsin in the trypsin preparation. λ-Protease cleaved Unp-Ib at a different site, generating Ib which was shorter by 1 amino acid residue than was Ib processed by α-chymotrypsin.
FIG. 2.
Proteolytic activation of Ia and Ib by various proteases. (A) SDS-PAGE of Ib after treatment with proteases. Lanes: 1, Unp-Ib without treatment; 2, 3, and 4, Unp-Ib treated with 50, 5, and 0.5 μg of trypsin per ml, respectively; 5, 6, 7, and 8, Unp-Ib treated with 5, 0.5, 0.05, and 0.005 μg of α-chymotrypsin per ml, respectively; lanes 9, 10, and 11, Unp-Ib treated with 5, 0.5, and 0.05 μg of λ-protease per ml, respectively. (B) SDS-PAGE of Ia after treatment with proteases. Lanes: 1, Pr-Ia without treatment; 2, Unp-Ia without treatment; 3, 4, and 5, Unp-Ia treated with 100, 50, and 5 μg of trypsin per ml, respectively; 6, 7, 8, and 9, Unp-Ia treated with 5, 0.5, 0.05, and 0.005 μg of α-chymotrypsin per ml, respectively; lanes 10, 11, and 12, Unp-Ia treated with 5, 0.5, and 0.05 μg of λ-protease per ml, respectively. A 1-μg portion of the toxin components was loaded on each lane.
FIG. 3.
The N-terminal sequences of unprocessed forms of Ia and Ib and the forms activated by proteolytic cleavage. The Ia and Ib components with and without protease treatment were separated by SDS-PAGE, transferred to an Immobilon membrane, and sequenced on an amino acid sequencer as described in Materials and Methods. Two N-terminal sequences were identified in Unp-Ia treated with trypsin. The peptide signal sequences are underlined.
Activation of Ia.
Unp-Ia purified from TS133/pMRP147 was larger than Pr-Ia from 667/pMRP147 (Fig. 1). The former seemed to be a precursor form, which requires proteolytic cleavage to be activated. Therefore, we examined the effects of the proteases on the processing of Unp-Ia. α-Chymotrypsin and λ-protease processed Unp-Ia more efficiently than did trypsin: the concentrations of α-chymotrypsin, λ-protease, and trypsin required for complete Unp-Ia processing were 0.05, 0.5, and 50 μg/ml, respectively (Fig. 2B). The N-terminal amino acid sequences of Pr-Ia, Unp-Ia, and Unp-Ia treated with the proteases were determined (Fig. 3). N-terminal sequencing of Unp-Ia showed that it starts at Q29, indicating that Unp-Ia was generated by cleaving off a possible signal peptide of 28 amino acids, similar to Unp-Ib. The processing of Unp-Ia to Pr-Ia was accompanied by removal of an additional 13 amino acid residues, resulting from cutting at R-A, a trypsin cleavage site. Two N-terminal sequences were obtained with Unp-Ia treated with trypsin. This indicates that trypsin cut two sites, one being the same R-A site as that described above and the other being the same Y-I site as that cut by α-chymotrypsin or λ-protease. The cleavage at the latter site could be due to the residual activity of α-chymotrypsin in the trypsin preparation.
Cytopathic activity.
All the Ia and Ib forms were used for the cell-rounding assay without further treatment or after treatment with the proteases (Fig. 4). For Unp-Ib plus Pr-Ia, no significant cytopathic activity was observed. In contrast, the combination of Pr-Ib and Unp-Ia exhibited a gradual increase in cytopathic activity during incubation with Vero cells, but the cytopathic activity was fourfold lower than that of Pr-Ib plus Pr-Ia (Fig. 4). These results indicate that the processed forms of both Ia and Ib are required for the full cytopathic activity of iota-toxin and also suggest that Unp-Ia possessed a low cytopathic activity or was converted gradually to the active form by a protease(s) associated with Vero cells.
FIG. 4.
Cytopathic activity of processed and unprocessed iota-toxin components. Vero cells were incubated with equimolar concentrations (10−7 M and serial dilutions) of Pr-Ib and Pr-Ia (■), Pr-Ib and Unp-Ia (×), and Unp-Ib and Pr-Ia (▴). The Vero cell rounding was recorded after 21 h of incubation. Means and standard deviations are shown (n = 4).
The result in Fig. 5 indicates that the proteases, especially λ-protease and α-chymotrypsin, efficiently activated Unp-Ib and Unp-Ia. Trypsin was inefficient in activating Unp-Ib and induced only a low level of activation of Unp-Ia, as tested by measuring the cytopathic activity on Vero cells. α-Chymotrypsin and λ-protease were equally potent in activating both Unp-Ib and Unp-Ia. It should be noted that the combination of Pr-Ib and Pr-Ia was activated further (twofold) by treatment with either α-chymotrypsin or λ-protease (data not shown). The Pr-Ib preparation showed two bands (Fig. 1), probably corresponding to a partial processing of Ib by a trypsin-like protease of 667/pMRP147, and treatment with α-chymotrypsin or λ-protease induced a complete activation of Pr-Ib. In addition, since Pr-Ia is shorter by 4 amino acids than Unp-Ia treated with the proteases and Pr-Ib is longer by 2 or 3 amino acids than Unp-Ib treated with the proteases (Fig. 3), such an additional activation may be due to removal of the extra amino acids from the Pr-Ib.
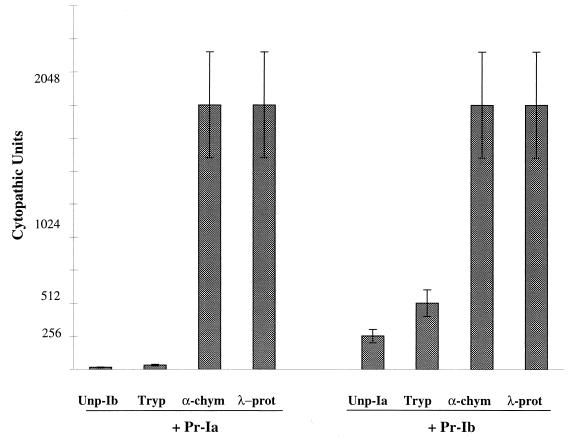
FIG. 5.
Effects of the protease treatment of unprocessed iota-toxin components on the cytopathic activity. Unp-Ib (10−7 M) was treated with protease for 20 min at room temperature, blocked with soybean trypsin inhibitor and Pefablock, and then combined with Pr-Ia (10−7 M). Unp-Ia was treated under the same conditions and then combined with Pr-Ib. The Vero cell rounding was recorded after 21 h of incubation. Means and standard deviations are shown (n = 4).
Proteases which activate iota-toxin components.
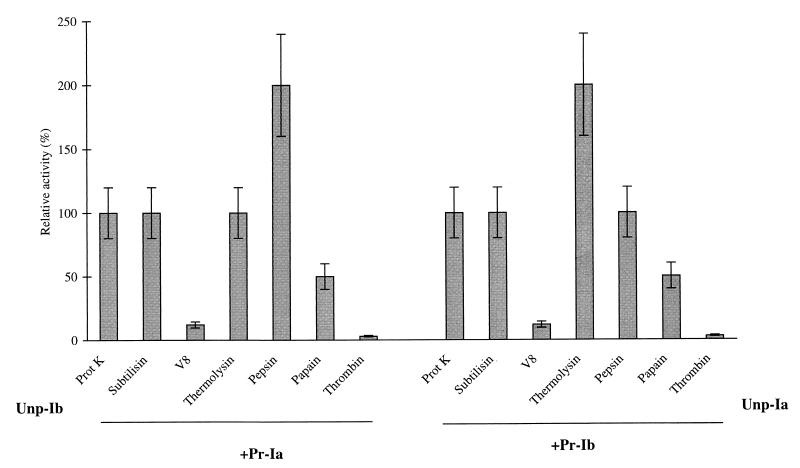
Various proteases were tested for their ability to activate Unp-Ia and Unp-Ib by using the cytopathic-effect assay on Vero cells. As shown in Fig. 6, the serine proteases (proteinase K and subtilisin), metalloprotease (thermolysin), and arginine proteases (pepsin) were at least as potent as α-chymotrypsin in activating Unp-Ia and Unp-Ib, whereas the cysteine protease (papain) was only half as effective. Thermolysin and pepsin induced a twofold increase in the activation of Unp-Ia and Unp-Ib, respectively, compared to that obtained with α-chymotrypsin. In contrast, V8 protease and thrombin were ineffective or only weakly effective in activating the iota-toxin components.
FIG. 6.
Activation of unprocessed iota-toxin components by various proteases. The protease treatment and cytopathic assays were carried out as reported in the legend of Fig. 5. Iota-toxin components (10−7 M) were incubated with proteinase K (1 mg/ml), papain (1 mg/ml), thermolysin (1 mg/ml), subtilisin (1 mg/ml), V8 protease (10 U/ml), or thrombin (10 U/ml), in 50 mM Tris (pH 8) or with pepsin (1 mg/ml) in 10 mM acetate buffer (pH 4). The cytopathic activity on Vero cells was recorded after 21 h of incubation. The results are expressed as the relative activation compared to that obtained with α-chymotrypsin (1 mg/ml) (100% indicates the same level of activation as that obtained with α-chymotrypsin). Means and standard deviations are shown (n = 2).
λ-Protease cleavage site on Ia.
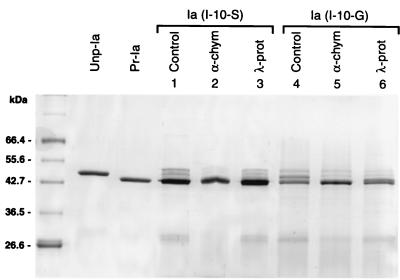
λ-Protease cleaves ɛ-prototoxin of C. perfringens D at two sites, Glu-Met and Tyr-Val (13). Based on such substrate preference and the sequence similarity between λ-protease and thermolysin, it has been suggested that λ-protease recognizes the N-terminal sides of amino acid residues displaying a bulky and hydrophobic side chain similarly to thermolysin. This was supported by determination of the λ-protease cleavage sites on Ia (Tyr-Ile) and Ib (Ser-Ala). To further strengthen this possibility, Ile-10 was replaced by Ser, a hydrophilic residue, or Gly, a hydrophobic one with the shortest side chain, both of which are insensitive to thermolysin. The mutant proteins were partially processed in E. coli. As shown in Fig. 7, λ-protease did not induce further processing of the mutant Ile-10–Ser, as expected. However, the mutant Ile-10–Gly was completely activated by λ-protease. This may be due to a broader substrate range for λ-protease than for thermolysin (5). In contrast, α-chymotrypsin, which cut Ia at the same site as λ-protease did (Tyr-Ile), recognizing the aromatic residues, cleaved both mutants and produced their fully processed forms.
FIG. 7.
Susceptibility of Ia mutants I-10–S and I-10–G to λ-protease and α-chymotrypsin. Ia mutants produced in E. coli BL21 (2 μg) were incubated without or with λ-protease (0.5 μg/ml) or α-chymotrypsin (0.5 μg/ml) for 20 min at room temperature, boiled for 2 min, and loaded on an SDS-polyacrylamide gel. Unp-Ia and Pr-Ia prepared from C. perfringens are shown as controls.
Since thermolysin is a metalloprotease, we have checked whether λ-protease cut iota-toxin components in a zinc-dependent manner. 1,10-Phenanthroline (1 mM), which is a zinc chelator, completely blocked the activation of Unp-Ia and Unp-Ib by λ-protease (100 μg/ml), as tested by measuring the cytopathic activity (data not shown). This confirms the previous observations (5) that λ-protease is a metalloprotease related to thermolysin but with a slightly different substrate specificity.
DISCUSSION
Iota-toxin is produced as an inactive protoxin, which requires proteolytic activation to exhibit its biological effects (23). Contrary to the general belief that it is only Ib from which a 20-kDa N-terminal peptide is liberated upon activation by proteolytic cleavage, our results show that this is also the case for Ia. Although the signal peptide of Ia was predicted to have 41 N-terminal amino acids (16), N-terminal sequencing of Unp-Ia showed that it has 28 amino acids. Two different mature forms of Ia are generated by proteolytic activation, depending on the enzymes: α-chymotrypsin and λ-protease removed 9 amino acids and trypsin and the protease from strain 667 removed 13 amino acids from the N terminus of the Ia propeptide. Both mature forms exhibited the same cytopathic activity. Pr-Ia and Unp-Ia treated with α-chymotrypsin or λ-protease showed the same level of cell rounding when they were combined with Unp-Ib treated with α-chymotrypsin (Fig. 5 and data not shown). Therefore, both types of mature forms seem to possess the same cytopathic activity.
The protein sequencing of Unp-Ib indicates that the signal peptide of Ib also comprises 28 amino acid residues, although it was predicted to consist of 39 amino acids (16). Both signal peptides from Ia and Ib end at the same cleavage site (A-Q), which is a common cleavage site for signal peptidase (25). In contrast to the short propeptide(s) of Ia, the propeptide of Ib is extremely long (20 kDa). Ib shows significant similarity (34% identity) to the protective antigen (PA) of Bacillus anthracis (16), whose crystal structure has been identified (18); and the segment R207 to A211, containing the cleavage sites of the Ib propeptide, matches a surface-exposed loop of PA between the propeptide and the mature form. Therefore, the segment may adopt such a conformation, increasing its accessibility to the proteases like the loop of PA (18). The proteases used cut the three different sites within the segment, leading to three different mature forms of Ib (Fig. 3). The mature Ib form produced by λ-protease treatment was as active as that produced by α-chymotrypsin, 64-fold more active than that produced by trypsin (Fig. 5), and 2-fold more active than Pr-Ib obtained in strain 667/pMRP147 (Fig. 4 and 5). This may be due to differences in the sensitivity to the protease or may be due to the hindrance to Ib activity imposed by two hydrophobic residues, FF, at the N terminus.
C2 toxin is structurally related to iota-toxin, and the toxins have 31 to 41% amino acid sequence identity (6, 11, 16). In contrast to Ib, the binding component, C2-II, is efficiently activated by trypsin, which also releases a 20-kDa N-terminal peptide (14). In addition, the enzymatic component, C2-I, seems not to require proteolytic activation, since the sequence corresponding to the signal peptide and the 13 amino acids of the Ia propeptide is missing in C2-I. Despite their relatedness, iota-toxin and C2 toxin show different modes of processing.
The iota-toxin components are secreted as inactive forms, which are matured extracellularly. Unlike anthrax toxins, which are cleaved by furin, a eukaryotic cell-associated protease (12), Ia and Ib do not contain a furin cleavage site. Ia was activated only partially and Ib was not activated during the incubation with Vero cells (Fig. 4). Since iota-toxin can be activated by various proteases such as α-chymotrypsin, pepsin, proteinase K, thermolysin, subtilisin, and λ-protease, C. perfringens proteases or digestive proteases from the host are responsible for in vivo activation of iota-toxin. PCR detection showed that type E strains such as NCIB 10748 but not strain 667 possess a λ-protease-encoding gene (data not shown). The N-terminal sequences of Pr-Ia and Pr-Ib show that the proteolytic activation takes place at trypsin cleavage sites (Fig. 3). Thus, strain 667 seems to produce a trypsin-like enzyme, which we have found to be tightly regulated by VirR-VirS. In contrast, C. perfringens collagenase is only partially controlled by this two-component system (21). In addition, λ-protease, which is related to thermolysin and which is also produced by C. perfringens type D, is responsible for the activation of epsilon-toxin (13). On the basis of these facts, along with the finding that λ-protease activates iota-toxin more efficiently than do trypsin and the trypsin-like C. perfringens protease, it is implied that λ-protease is the main activator of iota-toxin in C. perfringens type E and in the host. A possible contribution of the host proteases, including α-chymotrypsin, to the activation of iota-toxin components must be addressed in an animal model using recombinant strains producing iota-toxin but not any clostridial proteases.
ACKNOWLEDGMENTS
We are especially grateful to J. D'Alayer for protein sequencing.
This work was supported by a DRET contract (96-129) and by funding from Institut Pasteur.
REFERENCES
- 1.Aktories K, Bärmann M, Ohishi I, Tsuyama S, Jakobs K H, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 2.Bannam T L, Rood J I. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid. 1993;229:233–235. doi: 10.1006/plas.1993.1025. [DOI] [PubMed] [Google Scholar]
- 3.Ba-Thein W, Lyristis M, Ohtani K, Nisbet I T, Hayashi H. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol. 1996;178:2514–2520. doi: 10.1128/jb.178.9.2514-2520.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Considine R V, Simpson L L. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon. 1991;29:913–936. doi: 10.1016/0041-0101(91)90076-4. [DOI] [PubMed] [Google Scholar]
- 5.Fu J, Matsushita O, Katayama S, Jin S, Matsushita C, Minami J, Okabe A. Purification, characterization, and primary structure of Clostridium perfringens lambda-toxin, a thermolysin-like metalloprotease. Infect Immun. 1996;64:230–237. doi: 10.1128/iai.64.1.230-237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii N, Kubota T, Shirakawa S, Kimura K, Ohishi I, Moriishi K, Isogai E, Isogai H. Characterization of component-I gene of botulinum C2 toxin and PCR detection of its gene in clostridial species. Biochem Biophys Res Commun. 1996;220:353–359. doi: 10.1006/bbrc.1996.0409. [DOI] [PubMed] [Google Scholar]
- 7.Gibert M, Perelle S, Daube G, Popoff M R. Clostridium spiroforme toxin genes are related to C. perfringens iota toxin genes but have a different genomic localization. Syst Appl Microbiol. 1997;20:337–347. [Google Scholar]
- 8.Hatheway C L. Toxigenic clostridia. Clin Microbiol Rev. 1990;3:66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdman L V, Cato E P, Moore W E C. Anaerobe laboratory manual. 4th ed. Blacksburg: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 10.Jin F, Matsushita O, Katayama S, Jin S, Matsushita C, Minami J, Okabe A. Purification, characterization, and primary structure of Clostridium perfringens lambda-toxin, a thermolysin-like metalloprotease. Infect Immun. 1996;64:230–237. doi: 10.1128/iai.64.1.230-237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura K, Kubota T, Ohishi I, Isogai E, Isogai H, Fujii N. The gene component-II of botulinum C2 toxin. Vet Microbiol. 1998;62:27–34. doi: 10.1016/s0378-1135(98)00195-3. [DOI] [PubMed] [Google Scholar]
- 12.Klimpel K R, Molloy S S, Thomas G, Leppla S H. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minami J, Katayama S, Matsushita O, Matsushita C, Okabe A. Lambda-toxin of Clostridium perfringens activates the precursor of epsilon-toxin by releasing its N- and C-terminal peptides. Microbiol Immunol. 1997;41:527–535. doi: 10.1111/j.1348-0421.1997.tb01888.x. [DOI] [PubMed] [Google Scholar]
- 14.Ohishi I. Activation of botulinum C2 toxin by trypsin. Infect Immun. 1987;55:1461–1465. doi: 10.1128/iai.55.6.1461-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perelle S, Domenighini M, Popoff M R. Evidence that Arg-295, Glu-378 and Glu-380 are active-site residues of the ADP-ribosyltransferase activity of iota toxin. FEBS Lett. 1996;395:191–194. doi: 10.1016/0014-5793(96)01035-6. [DOI] [PubMed] [Google Scholar]
- 16.Perelle S, Gibert M, Boquet P, Popoff M R. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli. Infect Immun. 1993;61:5147–5156. doi: 10.1128/iai.61.12.5147-5156.1993. . (Author's correction, 63:4967, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff M R. Production of a complete binary toxin (actin-ADP-ribosylating toxin) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petosa C, Collier J R, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–938. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 19.Popoff M R, Boquet P. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem Biophys Res Commun. 1988;152:1361–1368. doi: 10.1016/s0006-291x(88)80435-2. [DOI] [PubMed] [Google Scholar]
- 20.Scott P T, Rood J I. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene. 1989;82:327–333. doi: 10.1016/0378-1119(89)90059-0. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J Bacteriol. 1994;176:1616–1623. doi: 10.1128/jb.176.6.1616-1623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Songer J G. Clostridial enteric disease of domestic animals. Clin Microbiol Rev. 1996;9:216–234. doi: 10.1128/cmr.9.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiles B G, Wilkins T D. Clostridium perfringens iota toxin: synergism between two proteins. Toxicon. 1986;24:767–773. doi: 10.1016/0041-0101(86)90101-7. [DOI] [PubMed] [Google Scholar]
- 24.Vandekerckhove J, Schering B, Bärmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;255:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 25.VonHeijne G, Abrahmsen L. Species-specific protein secretion in signal peptide design. Implication for protein secretion in foreign hosts. FEBS Lett. 1989;244:439–446. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]